Abstract
Ras1 is a small GTPase in the budding yeast Saccharomyces cerevisiae that regulates nutrient signaling. It has been shown that Ras1 undergoes phosphorylation, but the functional consequences and regulation of Ras1 phosphorylation remain unknown. Here we identify Ser-226 as an important residue for Ras1 phosphorylation, as mutating this residue to an alanine drastically diminishes the level of Ras1 phosphorylation. Notably, phosphorylated Ras1 accumulates as the cells approach the stationary phase of growth. Likewise, subjecting cells to nitrogen starvation also elevates the level of Ras1 phosphorylation. Interestingly, blocking Ras1 phosphorylation diminishes the level of autophagy and also renders the cells more sensitive to heat shock. Together, these data suggest a role of Ras1 phosphorylation in modulating nutrient signaling and stress response.
Keywords: G protein, phosphorylation, Ras protein, Saccharomyces cerevisiae, signal transduction
Introduction
A fundamental property of any living system is its ability to respond to environmental cues and mount an appropriate response to adapt to changes in nutrient availability and cellular stress. This property is especially important for single-cell organisms such as yeast. Indeed, the budding yeast Saccharomyces cerevisiae has been widely used as a model organism to study the molecular basis of cell signaling in response to environmental cues.
Like every living system, yeast requires both carbon and nitrogen sources for its growth and proliferation. In yeast, glucose is the preferred carbon source, and sophisticated signaling pathways exist in yeast to detect and respond to the availability of glucose (1, 2). At the core of these signaling pathways are Ras1 and Ras2, the only two members of the Ras GTPase family in yeast (3). Through a still not well characterized mechanism, the presence of glucose leads to activation of Cdc25, the exchange factor for both Ras1 and Ras2. The resulting GTP-bound Ras1 and Ras2 directly bind adenylate cyclase (Cyr1) and stimulate its production of cAMP, which consequently activates the cAMP-dependent protein kinase (PKA) (1, 4). The signaling pathway that senses and responds to nitrogen availability is mediated by the target of rapamycin (TOR), a highly conserved serine/threonine kinase that phosphorylates and thereby controls the activity of a variety of proteins that are important for protein translation, ribosome biogenesis, and cell growth (5). Given that both Ras/PKA and target of rapamycin regulate nutrient signaling, mechanisms must exist to coordinate these two pathways to allow cells to accurately align the growth and proliferation with the availability of nutrients (6).
The absence of nutrients triggers a variety of cellular responses that include switching off anabolic pathways and turning on catabolic pathways to maintain the cell viability. Autophagy is one such process that is triggered by nitrogen starvation (7, 8). In this process, a portion of cytoplasm is wrapped inside a double-membrane vesicle (autophagosome) and delivered to the lysosome (yeast vacuole) for degradation, resulting in building blocks such as amino acids that can be used in the biosynthetic pathway essential for cell viability. Under nutrient-replete conditions, autophagy occurs at a basal level, which is important for the maintenance of cellular homeostasis, including clearing damaged proteins and organelles (8). Under nutrient-depleted conditions, especially in the absence of nitrogen, autophagy is markedly induced to recycle cellular materials to generate usable nitrogen sources for essential processes. Interestingly, it appears that the extent of autophagy can be regulated by the activation level of Ras, a key component of the glucose signaling pathway in yeast (9). Specifically, it has been demonstrated that expressing a constitutively activated Ras2 (Ras2G12V) can substantially impair autophagy. Thus, regulating the activation level of Ras proteins could be a molecular mechanism to coordinate the cellular responses to both carbon and nitrogen availability.
Ras signaling in yeast is turned off mainly via the action of RasGAPs Ira1 and Ira2 (10). Disrupting these proteins can lead to constitutive activation of Ras and PKA (10). Interestingly, both Ras1 and Ras2 undergo phosphorylation (11, 12), suggesting that the activity of these proteins may also be regulated by phosphorylation. There are a few studies that characterized Ras2 phosphorylation, among which one recent work suggests that phosphorylation of Ras2 on the Ser-214 residue plays a role in negatively regulating Ras/PKA signaling (12, 13). Notably, phosphorylation of Ras1 remains largely unexplored other than the demonstrations that it does undergo phosphorylation and that phosphorylation primarily occurs on serine residues (11, 12). In this study, we have further characterized Ras1 phosphorylation. We find that phosphorylation of Ras1 is regulated by nutrient status and requires a critical serine residue. Interestingly, blocking phosphorylation of Ras1 diminishes the extent of basal macroautophagy and renders cells more sensitive to heat shock.
Results
Ras1 Phosphorylation Is Regulated by the Nutrient Status
It has been demonstrated very convincingly that Ras1 undergoes phosphorylation and that phosphorylation occurs primarily on serine residues (11, 12). However, the functional consequences and regulation of Ras1 phosphorylation have not been characterized. Given the role of Ras proteins in regulating nutrient signaling in yeast, we became interested in whether nutrient status and/or the growth stages could impact Ras1 phosphorylation. Thus, we elected to monitor the phosphorylation status of Ras1 in different growth stages.
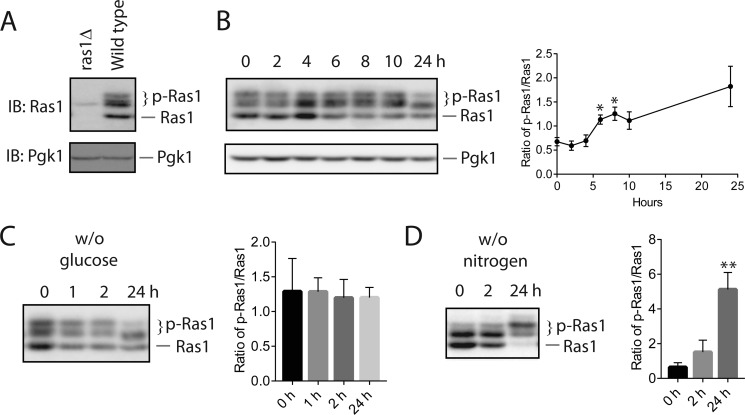
To detect Ras1, we used a monoclonal antibody that is directed against Ras (Abcam, ab79973). The specificity of the antibody is established by comparing the band patterns of wild-type versus ras1Δ cells. As shown in Fig. 1A, in the cell extract prepared from wild-type cells, the Ras antibody detected a series of bands with different mobility, whereas, in the extract prepared from ras1Δ cells, only a very faint band was detected, which presumably corresponds to Ras2. As documented in earlier work on Ras1 phosphorylation (11, 12), the bands with slower mobility represent phosphorylated forms of Ras1 (p-Ras1) because their mobility is sensitive to phosphatase treatment, whereas the band with the fastest mobility represents the non-phosphorylated form. After validation of the antibody, we monitored the level of phosphorylated Ras1 in cells that were in different growth stages. For this purpose, we let mid-log phase cells continue to grow in the same medium, removed an aliquot of cell culture at the indicated times, and monitored the level of both phosphorylated and non-phosphorylated Ras1. As shown in Fig. 1B, there is a dynamic change in the level of phosphorylated Ras1. Specifically, after the 4-h time point, there is a gradual shift of Ras1 from the non-phosphorylated form to the phosphorylated form.
FIGURE 1.
Nitrogen starvation elevates Ras1 phosphorylation. A, whole cell extracts from wild-type cells and ras1Δ cells were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies (ab79973, Abcam). Equal loading of each lane was confirmed via immunoblotting (IB) with anti-Pgk1. p-Ras1, phosphorylated Ras1; Ras1, unphosphorylated Ras1. B, wild-type cells (BY4741) were grown to mid-log phase in yeast extract peptone dextrose (YPD) medium (time 0) and continued to grow for the indicated times. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. Equal loading of each lane was confirmed via immunoblotting with anti-Pgk1. Quantification of immunoblots by densitometry from three independent experiments is shown in the right panel. The difference between each time point and time 0 was statistically analyzed (*, p < 0.050). C and D, wild-type cells (BY4741) were grown in YPD medium to mid-log phase and then switched to medium without glucose (w/o glucose, C) or without nitrogen (w/o nitrogen, D) for the indicated times. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. The data shown are representative of three independent experiments. Quantification of immunoblots by densitometry from three independent experiments is shown in the right panels. The difference between each time point and time 0 was statistically analyzed (**, p < 0.010).
Next, we examined whether phosphorylation of Ras1 is influenced by a lack of glucose or nitrogen. For this purpose, we shifted mid-log phase culture to medium either lacking glucose or lacking nitrogen and examined the level of phosphorylated Ras1. As shown in Fig. 1C, subjecting cells to glucose starvation did not lead to any obvious increase in Ras1 phosphorylation. Interestingly, subjecting cells to nitrogen starvation clearly causes a shift of Ras1 to the phosphorylated form, which is already apparent at the time point of 2 h. After prolonged nitrogen starvation, the majority of Ras1 exists as the phosphorylated form. These data indicate that the level of Ras1 phosphorylation is regulated by nutrient status. In particular, a lack of nitrogen triggers a marked elevation of Ras1 phosphorylation, raising the possibility that this modification may serve a role in regulating cellular responses to the availability of nitrogen.
The Residue Ser-226 Is Required for Ras1 Phosphorylation
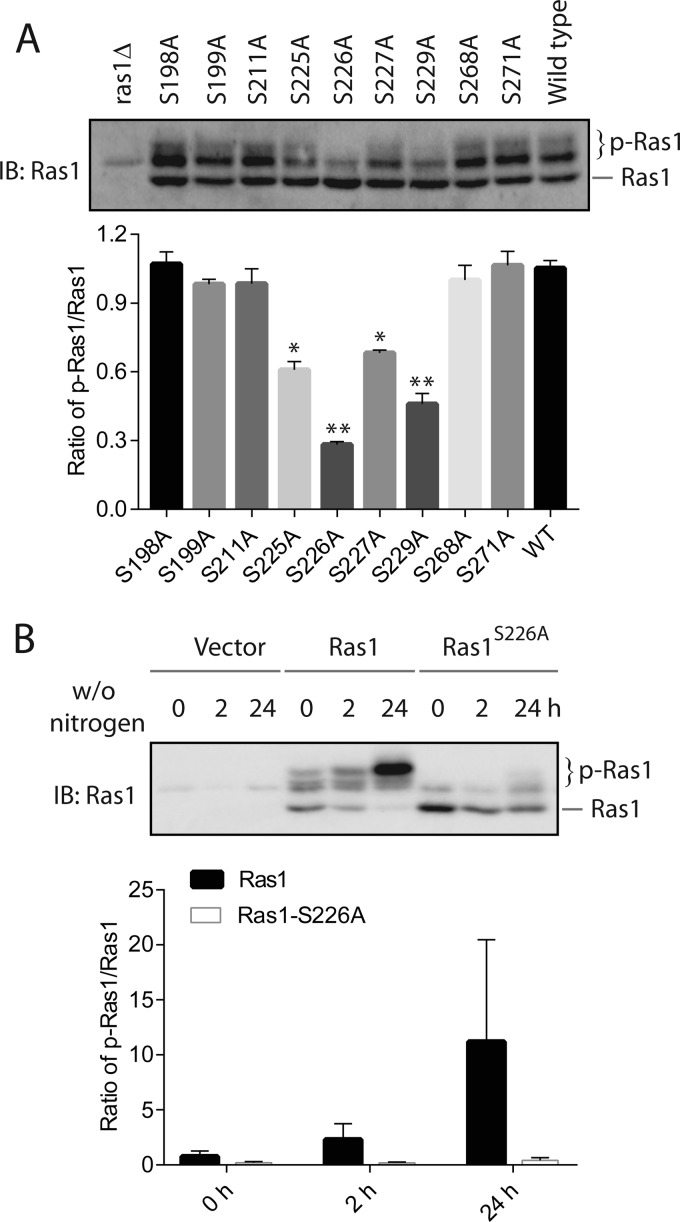
To understand the potential function of Ras1 phosphorylation, we sought to identify point mutants that block this modification. Previous work indicated that phosphorylation of Ras1 occurs primarily on serine residues (12). A search of a database generated via global phosphor-proteomics analysis (phosphoGRID) indicated that a few serine residues on Ras1 may undergo phosphorylation. Specifically, these residues are Ser-198, Ser-199, Ser-211, Ser-225, Ser-226, Ser-227, Ser-229, and Ser-268. To examine whether any of these residues might be critical for Ras1 phosphorylation, we individually mutated each of them to an alanine and examined whether the mutation alters the level of Ras1 phosphorylation. As shown in Fig. 2A, substituting the residues Ser-225, Ser-226, Ser-227, and Ser-229 to an alanine diminished the level of Ras1 phosphorylation to various extents, suggesting that the region between Ser-225 and Ser-229 is critical for Ras1 phosphorylation. Given that the largest diminishment of Ras1 phosphorylation occurs when Ser-226 is mutated to an alanine, we asked whether nitrogen starvation-induced Ras1 phosphorylation also requires this residue. As shown in Fig. 2B, the marked accumulation of Ras1 phosphorylation induced by nitrogen starvation is largely blocked in the Ras1S226A mutant. We conclude that the residue Ser-226 is required for both basal and nitrogen starvation-induced phosphorylation of Ras1.
FIGURE 2.
Ser-226 is required for both basal and nitrogen starvation-induced Ras1 phosphorylation. A, ras1Δ cells were transformed with either an empty vector or the pRS315 plasmids that express various Ras1 alleles under the control of the RAS1 promoter. Cells were grown in selective medium to mid-log phase, and whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. Quantification of immunoblots (IB) from two independent experiments by densitometry is shown in the bottom panel. The difference between each mutant and wild type was statistically analyzed (*, p < 0.050; **, p < 0.01)). B, ras1Δ cells were transformed with either an empty vector or the pRS315 plasmids that express either wild-type Ras1 or the Ras1S226A mutant under the control of the RAS1 promoter. Cells were grown in selective medium to mid-log phase and then switched to medium without nitrogen (w/o nitrogen) for the indicated times. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. The data shown are representative of three independent experiments. Quantification of immunoblots by densitometry from three independent experiments is shown in the bottom panel.
Glycogen Synthase Kinase Is Partially Required for Ras1 Phosphorylation
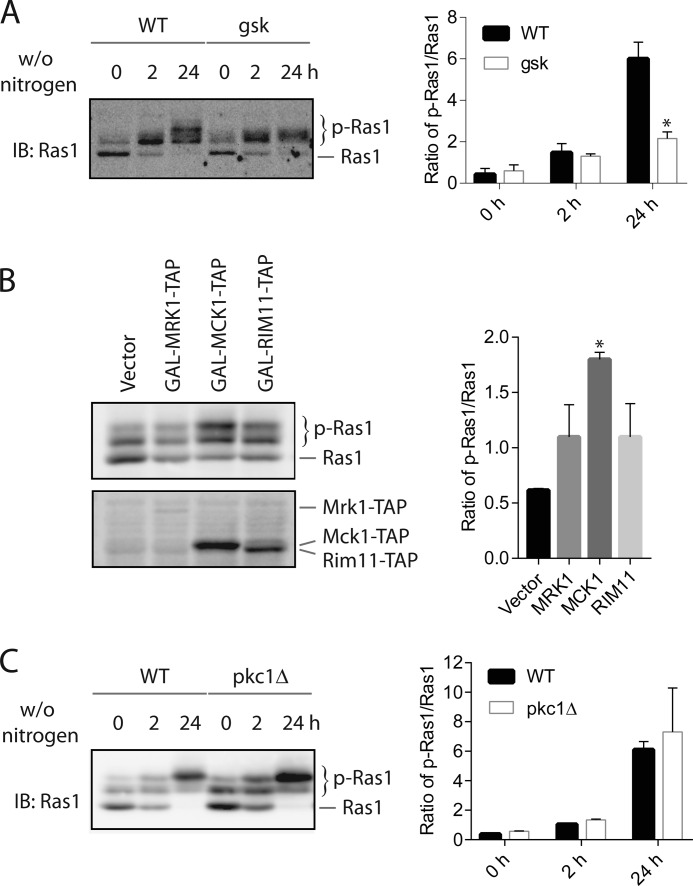
Next, we sought to identify the kinase that is required for Ras1 phosphorylation. One plausible candidate kinase is glycogen synthase kinase (GSK);2 it is known that this enzyme is active under the condition of nitrogen starvation (14). In addition, the region between Ser-225 and Ser-229 fits fairly well with the consensus sequence for glycogen synthase kinase (15). There are four isoforms of glycogen synthase kinase in yeast, and they are Mrk1, Mck1, Rim11, and Ygk3 (YOL128C). To determine whether any of these yeast glycogen synthase kinases is responsible for Ras1 phosphorylation, we compared the level of Ras1 phosphorylation in the wild type versus mutants that lack each individual glycogen synthase kinase with or without nitrogen starvation. We found that deleting any individual glycogen synthase kinase does not significantly affect basal or nitrogen starvation-induced Ras1 phosphorylation (data not shown). The lack of effect may be due to the redundancy of GSK isoforms, as demonstrated by other studies (14, 16). Accordingly, we analyzed Ras1 phosphorylation in a mutant that lacks three GSK isoforms (Mck1, Rim11, and Ygk3) (16). As shown in Fig. 3A, nitrogen starvation-induced Ras1 phosphorylation still occurs in the mutant at the early time point (2 h after nitrogen starvation), but the further phosphorylation induced by prolonged nitrogen starvation (24 h after nitrogen starvation) is partially blocked in the mutant. To provide additional evidence that GSK might be the key kinase that phosphorylates Ras1 here, we examined the effect of overexpressing GSK isoforms on the level of Ras1 phosphorylation. The prediction is that overexpression of GSK would lead to an increased level of Ras1 phosphorylation. As shown in Fig. 3B, overexpressing MRK1, MCK1, or RIM11 indeed resulted in an elevated level of Ras1 phosphorylation.
FIGURE 3.
Glycogen synthase kinase is partially required for Ras1 phosphorylation. A, the wild type and the mutant (gsk) lacking three yeast glycogen synthase kinases (Mck1, Rim11, and Ygk3) were grown in YPD medium to mid-log phase and then switched to medium without nitrogen (w/o nitrogen) for the indicated time. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. Quantification of immunoblots (IB) by densitometry from three independent experiments is shown in the right panel. The difference between the wild type and gsk mutant was statistically analyzed (*, p < 0.050). B, wild-type cells were transformed with either plasmids that express MRK1, MCK1, or RIM11 under the control of the GAL1 promoter or an empty vector and grown to mid-log phase in galactose-containing medium. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. Quantification of immunoblots by densitometry from three independent experiments is shown in the right panel. The difference between the vector and each overexpression construct was statistically analyzed (*, p < 0.050). C, wild-type and pkc1Δ cells were grown in YPD medium supplemented with 1 m sorbitol (to maintain the viability of the pkc1Δ cells). Cells were grown to mid-log phase and switched to medium without nitrogen for the indicated time. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras antibodies. Quantification of immunoblots by densitometry from two independent experiments is shown in the right panel.
Another candidate kinase for Ras1 phosphorylation is PKC, because nitrogen starvation is known to activate this kinase (17, 18) and K-Ras is known to be phosphorylated by PKC in mammals (19). Yeast has only a single protein kinase C (i.e. Pkc1), which is essential for the viability of yeast because of its important function in maintaining yeast cell wall integrity. However, the pkc1Δ mutant can be kept viable in medium supplemented with a high concentration of osmo-stabilizer such as 1 m sorbitol (17). To examine whether Pkc1 is required for Ras1 phosphorylation, we grew both the wild-type and pkc1Δ mutant in medium supplemented with 1 m sorbitol and examined the level of Ras1 phosphorylation. As shown in Fig. 3C, neither basal nor nitrogen starvation-induced phosphorylation of Ras1 are impaired in the pkc1Δ mutant, indicating that Pkc1 is not required for Ras1 phosphorylation.
Phosphorylation Modulates Ras1 Properties
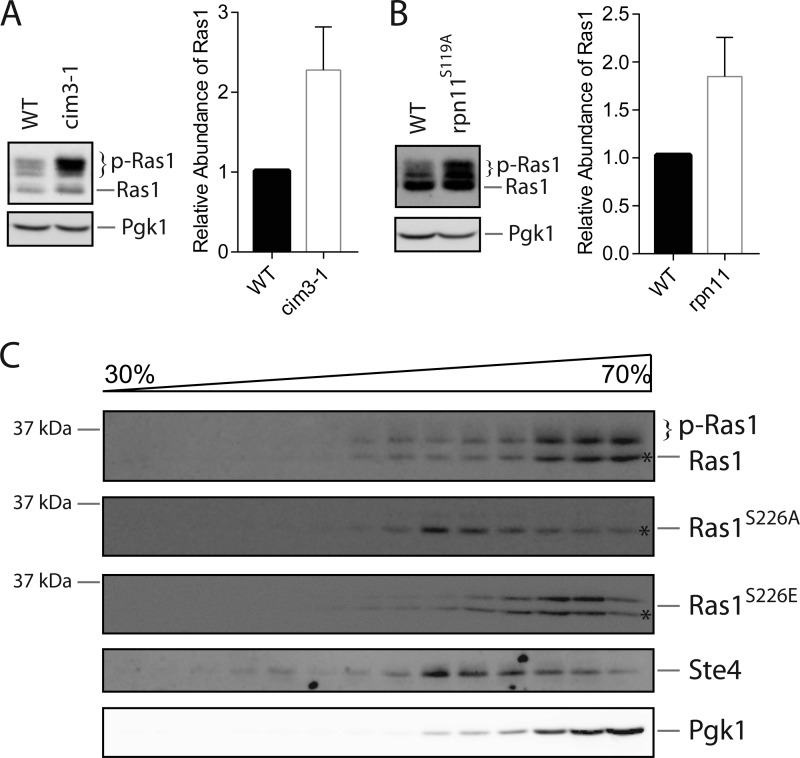
To investigate whether phosphorylation may impact the properties of Ras1, we first considered its potential role in regulating the abundance and/or stability of Ras1, as phosphorylation is a common mechanism that targets protein for ubiquitination and the subsequent degradation in the proteasome (20). In addition, as shown in Fig. 2A, the protein level of Ras1S226A is indeed higher than that of Ras1, albeit modestly. To investigate whether the abundance of Ras1 is regulated via the proteasome, we examined whether disrupting proteasome activity has any effect on Ras1 protein. For this purpose, we compared Ras1 in the wild-type versus a mutant (cim3-1) with disrupted proteasomal activity. As shown in Fig. 4A, similar levels of non-phosphorylated Ras1 are present in the wild-type and the cim3-1 mutant. Notably, the level of phosphorylated Ras1 is clearly higher in the cim3-1 mutant. There was no obvious accumulation of higher molecular weight species of Ras1 in the cim3-1 mutant that may represent poly-ubiquitinated Ras1. To ascertain the result, we also examined the level of Ras1 in a different proteasome-deficient mutant that contains a catalytically inactive Rpn11, a metalloprotease subunit of the 19S proteasome (21). Again, we observed a clear accumulation of phosphorylated Ras1 in the mutant (Fig. 4B). Together, these findings indicate that a functional proteasome has a role in preventing the abnormal accumulation of phosphorylated Ras1.
FIGURE 4.
Accumulating phosphorylated Ras1 in the proteasome-deficient mutants. A and B, a proteasome-deficient mutant (cim3-1 or rpn11S119A) and its isogeneic wild type were grown in YPD medium to mid-log phase. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras or anti-Pgk1 antibodies. Quantification of immunoblots by densitometry from three independent experiments is shown in the right panels. C, cells expressing either Ras1, Ras1S226A, or Ras1S226E under the control of the RAS1 promoter were grown to mid-log phase. Whole cell extracts were prepared, separated by sucrose gradient fractionation, resolved by 10% SDS-PAGE, and probed with anti-Ras, anti-Ste4, or anti-Pgk1 antibodies. The asterisks denote the position of Ras1 species with the same mobility on SDS-PAGE.
Next, we considered the possibility that phosphorylation may regulate the subcellular localization of Ras1. For this purpose, we compared the subcellular localization of the wild type and the Ras1 mutants using a well established sucrose density gradient fractionation method (22). As demonstrated previously by others (23), in addition to plasma membrane localization, a substantial fraction of Ras1 is present in both the cytosolic fraction and the intramembrane fraction (Fig. 4C). Interestingly, compared with wild-type Ras1, more Ras1S226A is present in the fractions corresponding to the plasma membrane, suggesting that phosphorylation promotes a relocalization of Ras1 from the plasma membrane to the cytosol. To further test this model, we also examined the localization of Ras1S226E, a Ras1 variant that potentially mimics phosphorylated Ras1 (at the Ser-226 site). As shown in Fig. 4C, the subcellular localization of Ras1S226E is similar to Ras1 and is slightly more cytosolic, which is consistent with the prediction of our model.
Phosphorylation of Ras1 Regulates the Extent of Basal Macroautophagy
Next, we investigated the physiological roles of Ras1 phosphorylation. In response to nitrogen starvation, cells undergo autophagy, a catabolic process that allows cells to degrade bulky materials in the lysosome (also called the vacuole in yeast) to recycle nutrients for the maintenance of viability. One process that is related to autophagy is the cytoplasm-to-vacuole targeting (Cvt) pathway, which is a biosynthetic pathway that serves to deliver vacuolar proteases such as aminopeptidase 1 (Ape1) to the vacuole (24). There are extensive similarities between the Cvt pathway and autophagy. In fact, these two pathways share a common set of proteins and machinery to deliver materials to the vacuole (25, 26). It has been demonstrated previously that activated Ras inhibits both macroautophagy and the Cvt pathway (9). Given that phosphorylation of Ras1 is enhanced by nitrogen starvation, we became interested in exploring whether phosphorylation of Ras1 may serve any role in regulating the Cvt pathway and/or the extent of macroautophagy.
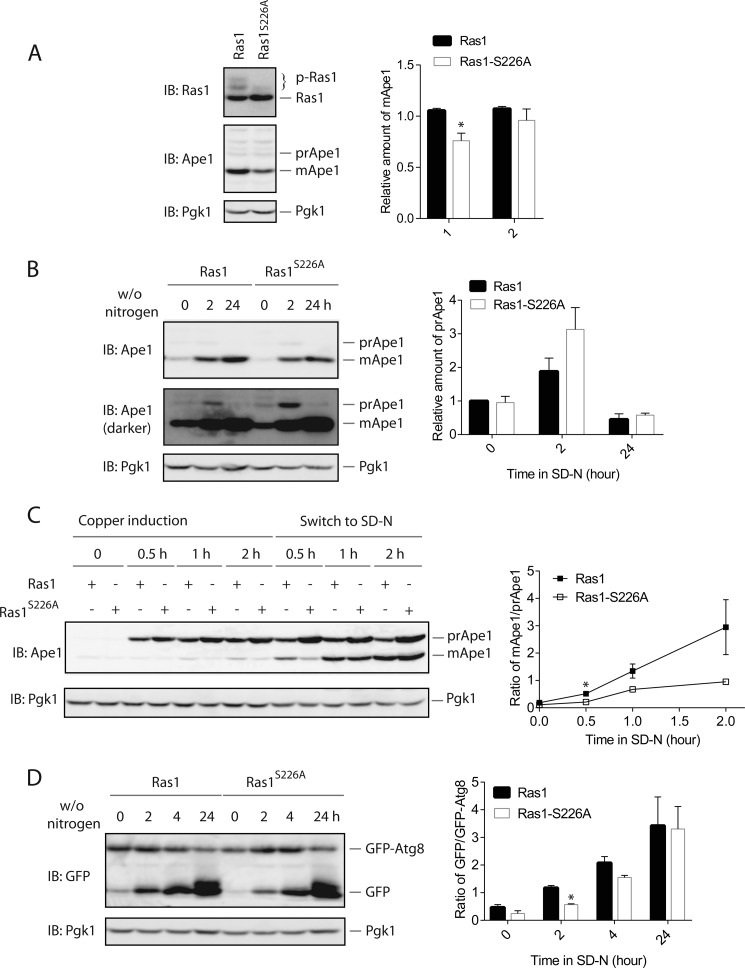
The Cvt pathway can be conveniently examined by monitoring the level of mature Ape1 (mApe1) (27). Ape1 is synthesized in the cytosol as an inactive precursor form (prApe1) that is delivered to the vacuole via the Cvt pathway. When in the vacuole, prApe1 is converted to its mature form (mApe1) after the removal of its N-terminal segment by a vacuolar protease. As shown in Fig. 5A, under the nutrient-replete condition, the level of mApe1 is less in the Ras1S226A mutant, indicating that the basal activation of the Cvt pathway is less in the Ras1S226A mutant. Under the nitrogen starvation condition, a marked accumulation of mApe1 occurred in both the wild type and the Ras1S226A mutant (Fig. 5B). Notably, there is a modest increase in the level of prApe1 in the Ras1S226A mutant (Fig. 5B), indicating that the delivery and/or processing of prApe1 induced by nitrogen starvation is less effective when the phosphorylation of Ras1 is blocked.
FIGURE 5.
Blocking Ras1 phosphorylation impairs the Cvt pathway and macroautophagy. A, cells expressing either Ras1 or Ras1S226A under the control of the RAS1 promoter were grown in selective medium to mid-log phase. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ras (top panel), anti-Ape1 (center panel), or anti-Pgk1 (bottom panel) antibodies. Quantification of immunoblots (IB) by densitometry from three independent experiments is shown in the right panel. Group 1 is a direct comparison of the mApe1 level between Ras1 and Ras1-S226A, whereas group 2 is a comparison of the mApe1 level between Ras1 and Ras1-S226A that is normalized to the expression level of Ras1 protein. It appears that the signaling difference can be partially accounted for by the difference in the expression of Ras1 protein. The difference between Ras1 and Ras1-S226A was statistically analyzed (*, p < 0.050). B, the same cells as shown in A were grown in selective medium to mid-log phase and switched to medium without nitrogen (w/o nitrogen) for the indicated times. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ape1 or anti-Pgk1 antibodies. Quantification of immunoblots by densitometry from three independent experiments is shown in the right panel. C, the same cells as shown in A and B were transformed with a plasmid that expresses Ape1 under the control of the CUP1 promoter. Cells were grown to early mid-log phase, treated with 10 μm Cu2+ to induce the expression of Ape1, and then shifted to medium without nitrogen (SD-N) for the indicated times. Whole cell extracts were resolved by 8% SDS-PAGE and probed with anti-Ape1 or anti-Pgk1 antibodies. Quantification of immunoblots by densitometry from three independent experiments is shown in the right panel. The difference between Ras1 and Ras1-S226A was statistically analyzed (*, p < 0.050). D, the same cells as shown in A and B were transformed with a plasmid that expresses GFP-Atg8 under the control of the ATG8 promoter. Cells were grown in selective medium to mid-log phase and switched to medium without nitrogen for the indicated times. Whole cell extracts were resolved by 10% SDS-PAGE and probed with anti-GFP or anti-Pgk1 antibodies. GFP-Atg8, full-length protein composed of GFP fused to the N-terminal of Atg8; GFP, free GFP originated from the degradation of GFP-Atg8 in the vacuole. Quantification of immunoblots by densitometry from two independent experiments is shown in the right panel. The difference between Ras1 and Ras1-S226A was statistically analyzed (*, p < 0.050).
It is possible that Ras1 phosphorylation might be required for the expression of Ape1, which could account for the lower basal mApe1 in the Ras1S226A mutant. To directly test whether Ras1 phosphorylation is required for optimal delivery and/or processing of Ape1, we expressed Ape1 under the control of an inducible CUP1 promoter and compared the relative level of prApe1 and mApe1 in the wild type versus the Ras1S226A mutant. As shown in Fig. 5C, addition of 100 μm Cu2+ (final concentration) rapidly induced the production of prApe1. After switching the cells to the nitrogen starvation condition, we could easily detect a conversion of prApe1 to mApe1 in both wild-type and the Ras1S226A cells (Fig. 5C). However, the extent of conversion from prApe1 to mApe1 is clearly less in the Ras1S226A mutant compared with the wild-type at every time point, which clearly indicates that optimal delivery and/or processing of Ape1 requires phosphorylation of Ras1.
The extent of macroautophagy can be examined via tracking the processing of GFP-Atg8, a fusion protein between a GFP and Atg8 (27, 28). During autophagy, GFP-Atg8 is delivered to the vacuole and degraded. Because of the higher stability of GFP, degradation of GFP-Atg8 leads to an easily detectable conversion of full-length GFP-Atg8 to free GFP. The ratio between the level of free GFP and the full-length GFP-Atg8 can be used to quantify the extent of macroautophagy. As shown in Fig. 5D, blocking Ras1 phosphorylation decreased the extent of macroautophagy, especially for both the basal level and the earlier time points of nitrogen starvation. Together, these data indicate that phosphorylation of Ras1 plays a role in fine-tuning the extent of basal macroautophagy as well as the early stage of macroautophagy induction.
Phosphorylation of Ras1 Affects Sensitivity to Heat Shock
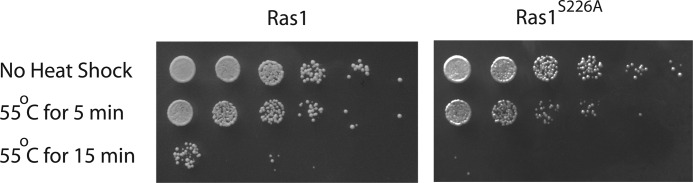
It has been shown previously that expressing a constitutively active Ras2 leads to hyperactivation of protein kinase A and, consequently, inhibition of autophagy (9). It is possible that the diminished autophagy displayed by the Ras1S226A mutant is due to a higher activation level of protein kinase A. It has been demonstrated previously that hyperactivation of protein kinase A often renders cells more sensitive to heat shock at 55 °C (6, 29). Accordingly, we compared the heat shock sensitivity of the wild type and Ras1S226A mutant. As shown in Fig. 6, the Ras1S226A mutant indeed is more sensitive to heat shock, which suggests a role of Ras1 phosphorylation in stress response.
FIGURE 6.
Blocking Ras1 phosphorylation renders cells more sensitive to heat shock. Cells expressing either Ras1 or Ras1S226A under the control of the RAS1 promoter were grown to stationary phase and then treated or not treated by heat shock at 55 °C for the indicated times. About 5 μl of serially diluted cells were placed on selective medium, grown for 3 days, and photographed. The data shown are representative of three independent experiments.
Discussion
Ras1 is a yeast homolog of the human Ras proteins. It has been demonstrated that Ras1 undergoes phosphorylation, but the functional consequences and regulation of this modification remain unclear. Our study demonstrates that phosphorylation of Ras1 is responsive to nutrient status. In particular, nitrogen starvation significantly elevates the level of Ras1 phosphorylation. We identify Ser-226 as a residue that is required for proper Ras1 phosphorylation. Consistent with a role of Ras1 phosphorylation in regulating nutrient signaling, we find that the Ras1S226A mutant has a reduced level of macroautophagy as well as a diminished activation of the Cvt pathway. In addition, we find that the Ras1S226A mutant is more sensitive to heat shock. Together, the findings reveal a role of phosphorylation of Ras1 in regulating stress response as well as in fine-tuning the extent of autophagy.
Autophagy has gained enormous interest in recent years, and it has been implicated in the regulation of numerous biological processes (30). Autophagy is a double-edged sword, and either too high or too low of a level can have a devastating effect on normal cellular physiology (31). Thus, not surprisingly, a multitude of mechanisms exist to fine-tune the level of autophagy. In some cases, the magnitude of regulation is modest (32), which may attest to the crucial importance of fine-tuning in autophagy. Our findings that Ras1 phosphorylation is promoted by nitrogen starvation and that blocking Ras1 phosphorylation diminishes the level of autophagy suggest that phosphorylation of Ras1 serves as a new mechanism to align nutrient status and the extent of autophagy. It is important to point out that the main defects we have observed with the Ras1S226A mutant are decreased basal macroautophagy as well as a delay in autophagy induction. Given that basal autophagy is important in maintaining proteostasis by clearing out damaged proteins, in the future, it would be interesting to examine whether phosphorylation of Ras1 may play a role in the regulation of proteostasis.
How does phosphorylation of Ras1 inhibit downstream signaling activity such as the extent of macroautophagy? One possibility is that phosphorylation down-regulates Ras1-mediated signaling by targeting the protein for degradation. Consistent with this model, we find that phosphorylated Ras1 is accumulated in proteasome-deficient mutants (both cim3-1 and rpn11S119A), suggesting that phosphorylation of Ras1 might be a precursor for its degradation in the proteasome. It would have been informative to directly compare the in vivo stability of phosphorylated versus non-phosphorylated Ras1, but this is technically challenging, as it is not easy to distinguish the effect of accelerated degradation from that of enhanced dephosphorylation of phosphorylated Ras1. In this regard, identification of a specific phosphatase that modulates Ras1 phosphorylation status might be helpful. It should be pointed out that the accumulation of phosphorylated Ras1 in proteasome-deficient mutants may be due to other indirect effects, especially considering the observations that the primarily accumulated species in proteasome-deficient mutants is phosphorylated Ras1 and that there is no evidence of accumulation of poly-ubiquitinated Ras1. One reasonable possibility is that kinase, such as glycogen synthase kinase, which is responsible for Ras1 phosphorylation, has a higher activity in proteasome-deficient mutants.
Interestingly, our sucrose gradient fractionation experiments indicate that phosphorylation could impact Ras1 activity via altering its subcellular localization. Compared with wild-type Ras1, the Ras1S226A mutant displays substantially more plasma membrane localization, which could potentially explain the observed enhanced activity of the mutant, as the known effector for Ras1 (i.e. adenylate cyclase) is primarily found in the plasma membrane (33, 34). It is worth noting that our localization experiments did not rely on either the epitope-tagged or overexpressed version of Ras1 protein, which may have unintended consequences for its subcellular localization. It is unclear whether the Ras1 protein found in the cytosolic fractions is functional and what makes the protein not membrane-associated. To answer these questions, in the future, it might be helpful to identify proteins that specifically interact with Ras1 in these fractions.
In summary, our data clearly demonstrate that nitrogen starvation induces Ras1 phosphorylation and that Ser-226 is critical for Ras1 phosphorylation. Blocking Ras1 phosphorylation decreases the extent of basal macroautophagy and renders cells more sensitive to heat shock. Together, these findings suggest a role of Ras1 phosphorylation in nutrient signaling and stress responses.
Experimental Procedures
Strains and Plasmids
Standard methods for the growth, maintenance, and transformation of yeast and bacteria and for the manipulation of DNA were used throughout. The S. cerevisiae strains used in this study are BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ); BY4741-derived mutants lacking MCK1, MRK1, RIM11, RAS1, or RAS2 (Research Genetics, Huntsville, AL); YPH499 (MATa ura3-52 lys2-801am ade2-101oc trp1-Δ 63 his3-Δ 200 leu2-Δ 1); YPH499-derived mutants lacking PKC1 (pkc1::LEU2, generously provided by Dr. J. Thorner, University of California, Berkeley, CA) (17); a triple gsk mutant (GSK3-mck1 rim11 yol128c, MATα his3 leu2 ura3 trip1 ade2 mck1::TRP1 rim11:HIS3 yol128c::LEU2, generously provided by Dr. Claudina Rodrigues-Pousada, the Instituto de Tecnologia Quimica e Biologica of the Universidade Nova de Lisboa (ITQB-UNL), Portugal) (16); MY316 (WT) and MY316-derived rpn11 mutant MY320 (rpn11-S119A) (generously provided by Dr. Michael Glickman) (21); and MHY753 (WT) and MHY753-derived cim3 mutant MHY754 (MHY753, cim3-1) (35).
The expression plasmids used in this study that have been described previously are 416-GFP-ATG8 (generously provided by Dr. D. Klionsky, University of Michigan) and BG1805-GAL-MCK1, BG1805-GAL-MRK1, and BG1805-GAL-RIM11 (purchased from Openbiosystems). The plasmid pRS315-RAS1, which expresses RAS1 under the control of its own promoter, was constructed via amplifying the RAS1 gene using PCR primers that anneal 600 bp upstream (5′-CGC GGA TCC ACT GAA ACA TCT CGA TAT AAA G-3′) or 300 bp downstream (5′ATA AGA ATG CGG CCG CAA CTC TTT TAC TAG A-3′) of the open reading frame. The PCR product was subcloned by digestion with BamHI and NotI and ligation to pRS315. Site-directed mutagenesis was used to generate various Ser-to-Ala mutations as indicated. The plasmid pRS315-RAS1 served as the template for the mutagenesis. All constructs were confirmed by sequencing.
The pYES-APE1 plasmid that expresses APE1 under the control of the CUP1 promoter was constructed by the following steps. The CUP1 promoter was amplified using a forward PCR primer (5′ CCC AAG CTT CCA GGA CCG ACA TTT GGG CGC 3′) and a reverse PCR primer (5′ CCC AAG CTT AGA TCG CAG TTT GTT TTT CTT AAT ATC 3′). The PCR product was subcloned by digestion with HindIII and ligation to pYES2.1/V5-His-TOPO, which has been engineered to have a HindIII site. The APE1 fragment was amplified using a forward PCR primer (5′ GGA AGA TCG ATG GAG GAA CAA CGTGAA ATA C 3′) and a reverse PCR primer (5′ T CCC CCC GGG TCA CAA CTC GCC GAA TTC ATC 3′). The PCR product was subcloned to the above vector, which has been engineered to have an EcoRI site.
Sucrose Gradient Fractionation
Sucrose gradient fractionation was conducted as described previously (22). Spheroplasts were prepared by incubation with 20 μg/ml of zymoylase in SK buffer (1.2 m sorbitol and 0.1 m KPO4 (pH 7.5)) for 45 min at 30 °C. All subsequent steps were carried out at 4 °C. The resulting spheroplasts were washed once with ice-cold SK buffer and resuspended in lysis buffer C (0.8 m sucrose, 20 mm triethanolamine hydrochloride (pH 7.2), 1 mm EDTA, 1 mm dithiothreitol, 0.2 mm phenylmethylsulfonyl fluoride, and 1 Roche protease inhibitor mixture tablet per 50 ml) and disrupted by 15 strokes in a motorized homogenizer. The lysate was cleared of unbroken cells with 15 min of 500 × g centrifugation. Powdered sucrose was added to each sample to make 70% total sucrose, followed by mixing on a stir plate at 4 °C for 1 h. They were then carefully overlaid with 60%, 50%, 40%, and 30% sucrose solutions and subjected to ultracentrifugation at 190,000 × g for 19 h. Fifteen equal portions (300 μl) were drawn from the top of each tube, mixed with 2× SDS sample buffer, boiled at 100 °C for 5 min, cooled, and resolved on 10% SDS-PAGE. Blots were probed with anti-Ras (Abcam, ab79973) for examining Ras1 localization. Membrane fraction markers were used to establish which fractions corresponded to which membranes (anti-Ste4 (Duane Jenness, University of Massachusetts) for the plasma membrane and anti-Pgk1 (Jeremy Thorner, University of California) for cytosolic fractions).
Phosphorylation and Autophagy Bioassays
Phosphorylation of Ras1 was monitored by immunoblotting of whole cell extracts using antibodies that recognize both phosphorylated and non-phosphorylated Ras1. Macroautophagy was monitored by following the processing of GFP-Atg8 with or without nitrogen starvation, as described previously (27). For all immunoblotting analyses, mid-log-phase cells were grown on the appropriate medium and then treated or not treated with nitrogen starvation as indicated. Proteins were extracted via trichloroacetic precipitation following procedures described previously (36). Whole cell extracts were resuspended in boiling SDS-PAGE sample buffer (62.5 mm Tris-HCl (pH 6.8), 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, and 0.0005% bromphenol blue) for 5 min. Following SDS-polyacrylamide gel electrophoresis and transfer to nitrocellulose, the membrane was probed with antibodies to Ras at 1:1000 (Abcam, ab79973), GFP at 1:5000 (Abcam, ab13970), Ape1 at 1:5000 (a generous gift from Dr. Daniel Klionsky, University of Michigan), and Pgk1 at 1:75,000 (a generous gift from Dr. Jeremy Thorner, University of California). Immunoreactive species were visualized by enhanced chemiluminescence detection (Pierce) of horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad), anti-Rat IgG (Abcam), or anti-chicken IgY (Abcam). Specificity of detection was established using ras1Δ cell extracts as negative controls. All experiments have been repeated at least three times.
Data Quantification and Statistical Analysis
Band intensities of Western blotting analyses were quantified using ImageJ software from the National Institutes of Health. Where indicated, the data were statistically analyzed by t test, with p <0.050 considered significant.
Author Contributions
X. J. identified Ser-226 as a critical residue for Ras1 phosphorylation, conducted experiments that compared the properties of WT and Ras1S226A mutants, and identified GSK as responsible for full phosphorylation of Ras1. S. Starke and P. D. examined the effect of overexpressing GSK on the abundance and stability of Ras1. Y. L. cloned RAS1 and examined Ras1 in the proteasome-deficient strains. S. Sethupathi made the CUP1-APE1 construct. G. K. made the FLAG-tagged GSK constructs. Y. W. designed the experiments, identified nitrogen starvation as a condition that induces Ras1 phosphorylation, and wrote the manuscript.
Acknowledgments
We thank Drs. Paul K. Herman, Claudina Rodrigues-Pousada, Daniel Klionsky, Henrik G. Dohlman, and Jeremy Thorner for generously providing strains, plasmids, and antibodies. We also thank Dr. Jonathan Fisher for editing the manuscript.
This work was supported by National Institutes of Health Grant 1R15GM106330-01 (to Y. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GSK
- glycogen synthase kinase
- Cvt
- cytoplasm-to-vacuole targeting.
References
- 1. Broach J. R. (2012) Nutritional control of growth and development in yeast. Genetics 192, 73–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rolland F., Winderickx J., and Thevelein J. M. (2002) Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2, 183–201 [DOI] [PubMed] [Google Scholar]
- 3. Tamanoi F. (2011) Ras signaling in yeast. Genes Cancer 2, 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santangelo G. M. (2006) Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loewith R., and Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramachandran V., and Herman P. K. (2011) Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics 187, 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reggiori F., and Klionsky D. J. (2013) Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parzych K. R., and Klionsky D. J. (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal. 20, 460–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Budovskaya Y. V., Stephan J. S., Reggiori F., Klionsky D. J., and Herman P. K. (2004) The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279, 20663–20671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., and Toh-e A. (1990) S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60, 803–807 [DOI] [PubMed] [Google Scholar]
- 11. Cobitz A. R., Yim E. H., Brown W. R., Perou C. M., and Tamanoi F. (1989) Phosphorylation of RAS1 and RAS2 proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 86, 858–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whistler J. L., and Rine J. (1997) Ras2 and Ras1 protein phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 272, 18790–18800 [DOI] [PubMed] [Google Scholar]
- 13. Xiaojia B., and Jian D. (2010) Serine214 of Ras2p plays a role in the feedback regulation of the Ras-cAMP pathway in the yeast Saccharomyces cerevisiae. FEBS Lett. 584, 2333–2338 [DOI] [PubMed] [Google Scholar]
- 14. Xiao Y., and Mitchell A. P. (2000) Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol. Cell Biol. 20, 5447–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fiol C. J., Mahrenholz A. M., Wang Y., Roeske R. W., and Roach P. J. (1987) Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262, 14042–14048 [PubMed] [Google Scholar]
- 16. Pereira J., Pimentel C., Amaral C., Menezes R. A., and Rodrigues-Pousada C. (2009) Yap4 PKA- and GSK3-dependent phosphorylation affects its stability but not its nuclear localization. Yeast 26, 641–653 [DOI] [PubMed] [Google Scholar]
- 17. Roelants F. M., Torrance P. D., and Thorner J. (2004) Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 150, 3289–3304 [DOI] [PubMed] [Google Scholar]
- 18. Torres J., Di Como C. J., Herrero E., and De La Torre-Ruiz M. A. (2002) Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 277, 43495–43504 [DOI] [PubMed] [Google Scholar]
- 19. Bivona T. G., Quatela S. E., Bodemann B. O., Ahearn I. M., Soskis M. J., Mor A., Miura J., Wiener H. H., Wright L., Saba S. G., Yim D., Fein A., Pérez de Castro I., Li C., Thompson C. B., Cox A. D., and Philips M. R. (2006) PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell 21, 481–493 [DOI] [PubMed] [Google Scholar]
- 20. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 21. Maytal-Kivity V., Reis N., Hofmann K., and Glickman M. H. (2002) MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song J., Hirschman J., Gunn K., and Dohlman H. G. (1996) Regulation of membrane and subunit interactions by N-myristoylation of a G protein α subunit in yeast. J. Biol. Chem. 271, 20273–20283 [DOI] [PubMed] [Google Scholar]
- 23. Belotti F., Tisi R., Paiardi C., Rigamonti M., Groppi S., and Martegani E. (2012) Localization of Ras signaling complex in budding yeast. Biochim. Biophys. Acta 1823, 1208–1216 [DOI] [PubMed] [Google Scholar]
- 24. Scott S. V., Hefner-Gravink A., Morano K. A., Noda T., Ohsumi Y., and Klionsky D. J. (1996) Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. U.S.A. 93, 12304–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Umekawa M., and Klionsky D. J. (2012) The cytoplasm-to-vacuole targeting pathway: a historical perspective. Int. J. Cell Biol. 2012, 142634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lynch-Day M. A., and Klionsky D. J. (2010) The Cvt pathway as a model for selective autophagy. FEBS Lett. 584, 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheong H., and Klionsky D. J. (2008) Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 451, 1–26 [DOI] [PubMed] [Google Scholar]
- 28. Guimaraes R. S., Delorme-Axford E., Klionsky D. J., and Reggiori F. (2015) Assays for the biochemical and ultrastructural measurement of selective and nonselective types of autophagy in the yeast Saccharomyces cerevisiae. Methods 75, 141–150 [DOI] [PubMed] [Google Scholar]
- 29. Li Y., and Wang Y. (2013) Ras/cAMP-dependent protein kinase signaling is negatively regulated by a deubiquitinating enzyme Ubp3 in yeast. J. Biol. Chem. 288, 11358–11365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klionsky D. J., and Codogno P. (2013) The mechanism and physiological function of macroautophagy. J. Innate Immun. 5, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shintani T., and Klionsky D. J. (2004) Autophagy in health and disease: a double-edged sword. Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gohla A., Klement K., Piekorz R. P., Pexa K., vom Dahl S., Spicher K., Dreval V., Häussinger D., Birnbaumer L., and Nürnberg B. (2007) An obligatory requirement for the heterotrimeric G protein Gi3 in the antiautophagic action of insulin in the liver. Proc. Natl. Acad. Sci. U.S.A. 104, 3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., and Wigler M. (1985) In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40, 27–36 [DOI] [PubMed] [Google Scholar]
- 34. Thevelein J. M. (1992) The RAS-adenylate cyclase pathway and cell cycle control in Saccharomyces cerevisiae. Antonie van Leeuwenhoek 62, 109–130 [DOI] [PubMed] [Google Scholar]
- 35. Ghislain M., Udvardy A., and Mann C. (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366, 358–362 [DOI] [PubMed] [Google Scholar]
- 36. Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., and Dohlman H. G. (2006) Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein α subunit at the endosome. Cell 126, 191–203 [DOI] [PubMed] [Google Scholar]