Abstract
The HIV-1 transactivator protein Tat is a critical regulator of HIV transcription primarily enabling efficient elongation of viral transcripts. Its interactions with RNA and various host factors are regulated by ordered, transient post-translational modifications. Here, we report a novel Tat modification, monomethylation at lysine 71 (K71). We found that Lys-71 monomethylation (K71me) is catalyzed by KMT7, a methyltransferase that also targets lysine 51 (K51) in Tat. Using mass spectrometry, in vitro enzymology, and modification-specific antibodies, we found that KMT7 monomethylates both Lys-71 and Lys-51 in Tat. K71me is important for full Tat transactivation, as KMT7 knockdown impaired the transcriptional activity of wild type (WT) Tat but not a Tat K71R mutant. These findings underscore the role of KMT7 as an important monomethyltransferase regulating HIV transcription through Tat.
Keywords: human immunodeficiency virus (HIV), post-translational modification (PTM), protein methylation, transcription regulation, viral transcription
Introduction
The HIV-1 epidemic remains a global health problem despite the growing availability of potent antiretroviral therapies. These therapies are not curative, as latent, transcriptionally silent virus can spontaneously reactivate from sanctuaries and rapidly rekindle viral infection after withdrawal of therapy (1–3). Therefore, the molecular mechanisms of the activation and suppression of HIV transcription are of great interest.
HIV encodes its own viral transactivator, Tat, which activates HIV transcription and facilitates its own production in a positive feedback loop. Tat is a small protein, typically found in a full-length form of ∼101 amino acids (aa)3 or as a splice variant (72 aa) encoded only by the first exon of the tat open reading frame. Both isoforms efficiently transactivate the HIV promoter in the 5′ long terminal repeat (LTR). Tat interacts with the positive transcriptional elongation factor b (P-TEFb), and viral RNA through several well-characterized domains that can be found in its one-exon form: a cysteine-rich domain (aa 22–37) and a highly conserved core domain (aa 41–48), both of which participate in binding of P-TEFb (4–6). The neighboring arginine-rich motif (ARM; aa 49–57) interacts with a specific stem-loop RNA structure called transactivation response element (TAR) located in the 5′ extremities of all viral transcripts (7–10). Specific binding of the Tat ARM to TAR requires the coordinated binding of P-TEFb to Tat, as the cyclin T1 subunit of P-TEFb binds both the Tat cysteine-rich and core domains and loop sequences of TAR (11). C-terminal to these well-characterized domains is a glutamine-rich motif (aa 59–72). When expressed as a peptide, this region adopts a conserved α-helical structure that is stabilized upon binding to TAR RNA and is implicated in T-cell apoptosis (12, 13).
Tat is regulated by a number of post-translational modifications including phosphorylation, acetylation, methylation, and polyubiquitylation (14). The Tat ARM is highly modified at lysine and arginine residues. These modifications, including acetylation of Lys-50/51 and methylation of Lys-51 and Arg-52/53, regulate TAR and P-TEFb binding positively (K51me) or negatively (K50ac/K51ac; R52me2/R53me2) (15–20). The role of post-translational modifications in the glutamine-rich motif is still unclear. Several phosphorylation sites in this domain (Ser-62, Thr-64, Ser-68) enhance transcriptional activity, but are not well conserved among viral isolates (21, 22). In contrast, lysine 71 is a highly conserved residue found in 74% of HIV-1 isolates across all clades reported in the HIV-1 sequence compendium (22). At this residue, polyubiquitylation at Lys-71 is required for full transactivation, but does not affect Tat stability (23).
We previously reported that Lys-51, within the Tat ARM, is monomethylated (K51me) by the methyltransferase KMT7 (also called SET7/9) (17). Monomethylation at Lys-51 enhanced TAR RNA binding of Tat, and increased trimolecular complex formation between Tat, TAR, and P-TEFb. Interestingly, KMT7 itself was found to bind TAR RNA in band-shift assays, indicating that it could be recruited to the HIV promoter before Tat was produced (17). In in vitro methylation assays of ARM peptides (aa 44–59), we observed that that KMT7 fails to methylate residues other than Lys-51 in Tat (17, 18). However, when we conducted the same in vitro assays using purified one-exon Tat proteins (Tat72), we observed additional methylation on Tat indicating that one or more additional methylation sites existed outside of the ARM region. In this study, we sought to further understand the role of KMT7 in the Tat transcription cycle by identifying and characterizing the additional modification(s). We hypothesized that understanding the full extent of Tat methylation would bring new insight into the regulation of Tat function and the role of KMT7 as an HIV-1 transcriptional cofactor.
Results
In Vitro Methylation of Tat72 Peptides Reveals a Novel KMT7 Methylation Site at Lys-71
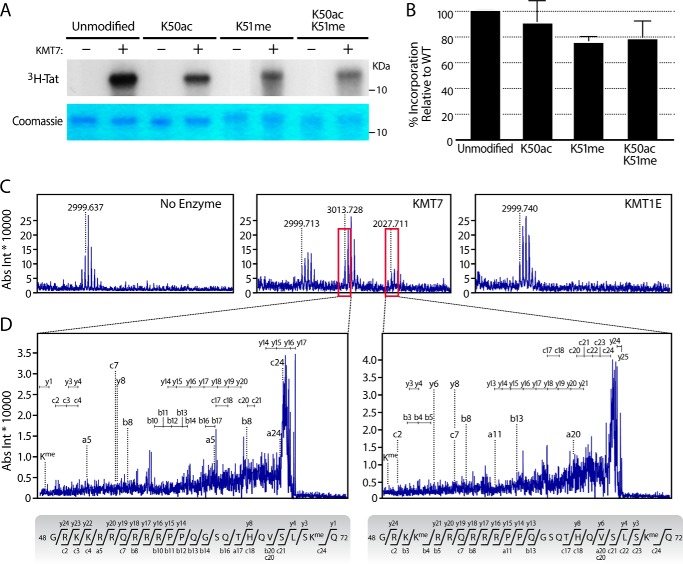
To determine whether Lys-51 is the only KMT7 methylation site in Tat, we performed in vitro methylation reactions with Tat peptides spanning aa 1–72 (Tat72). We observed substantial incorporation of [3H]S-adenosyl methionine ([3H]SAM) in Tat72 proteins carrying monomethylated Lys-51, indicating additional KMT7 methylation sites in Tat (Fig. 1, A and B). In Tat72 proteins with an acetyl group at Lys-50, [3H]SAM incorporation was also slightly decreased. This is consistent with previous observations that this modification reduces access of KMT7 to Lys-51 (18).
FIGURE 1.
HIV-1 Tat is monomethylated at Lys-71 by KMT7. In vitro methylation assays were done with 3 μg of synthesized Tat72 proteins that were unmodified, acetylated (K50ac), methylated (K51me), or both acetylated and methylated, with or without 1 μg of recombinant KMT7 in the presence of [3H]SAM. A, top, representative autoradiogram. This image is uncropped and from one gel; the difference in background is an imperfection of the film. Bottom, Coomassie Blue stain for Tat. Molecular weight markers are noted in kilodaltons (kDa) B, quantification of three autoradiograms with ImageJ (mean ± S.E.). C, in vitro methylation assays on unmodified Tat proteins with indicated enzymes were subjected to MS, shown are the zoomed regions containing Tat ions corresponding to aa 48–72. Boxed in red are peaks indicating modified Tat ions. D, MS/MS spectra of the ions boxed in 1C, and their corresponding peptide sequences. Ion annotations are found in supplemental Table S-1.
To specifically identify additional KMT7 methylation sites in Tat, we performed in vitro methylation reactions using non-radiolabeled SAM and subjected modified Tat proteins to MALDI-TOF MS/MS analysis developed to analyze Tat (18). This analysis revealed monomethylation at a single additional site, Lys-71 (Fig. 1, C and D, supplemental Table S-1) in two distinct peptides in reactions with KMT7, but not in reactions with a control enzyme KMT1E (also called SETDB1) or no enzyme (Fig. 1C). In one peptide, only Lys-71 was monomethylated (Fig. 1D, left); in the other, both Lys-51 and Lys-71 were monomethylated (Fig. 1D, right). Neither Lys-51 nor Lys-71 was dimethylated, underscoring the function of KMT7 as a monomethyltransferase in Tat.
KTM7 Modifies Tat at Lys-71 in Vitro
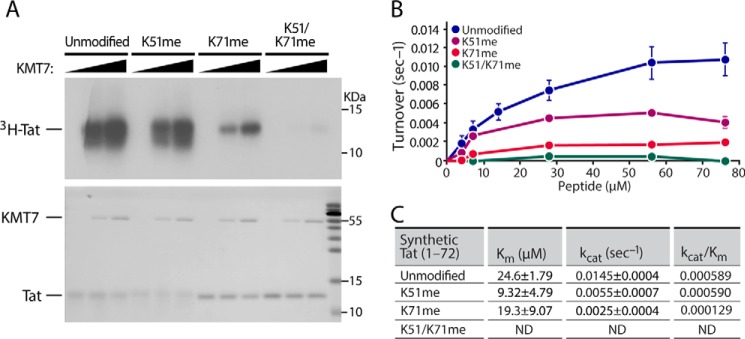
Next, we tested whether pre-modification of Lys-71 in Tat72 peptides affects in vitro methylation by KMT7. Premethylation of Lys-71 markedly decreased [3H]SAM incorporation while Lys-51 pre-methylation had a lesser effect. Finally, premethylation of both Lys-51 and Lys-71 abolished methylation of Tat, demonstrating that there are no additional targets for KMT7 in Tat (Fig. 2A). These findings support Lys-51 and Lys-71 as the sole sites for KMT7 monomethylation in Tat72.
FIGURE 2.
KMT7 preferentially monomethylates Tat Lys-71 in vitro. A, reactions were done as in Fig. 1A with 2 μg of synthesized Tat72 proteins that were unmodified, monomethylated at Lys-51 or Lys-71 or both, and incubated with 0, 1, or 2 μg of purified KMT7 in the presence of [3H]SAM. Top, representative autoradiogram. Bottom, Coomassie Blue stain for Tat and KMT7. B, kinetic assays were done with indicated concentrations of Tat peptides and 1 μg of recombinant KMT7. Assays were conducted in triplicate; error bars indicate S.E. C, activity calculated by fitting data to Michaelis-Menten equation with SigmaPlot 11 after correction for methylation in the control reactions with either the enzyme or the substrate removed. ND, not determined in doubly modified peptides.
To better quantify dynamics of KMT7-mediated methylation of Tat72 proteins, we used pre-modified Tat proteins in a modified kinetic radiometric assay (24). After testing for linearity with time and KMT7 enzyme concentrations, kinetics assays were performed with various concentrations of Tat72 proteins (Fig. 2B). Methyltransferase activity was hyperbolic, and all Tat proteins except the Tat K51/K71me doubly modified form followed Michaelis-Menten kinetics. The doubly modified form had very little methylation, consistent with our observations using autoradiography (Fig. 2A).
The catalytic turnover (kcat) and Michaelis constant (Km) of the Tat K51me proteins were 62% lower relative to the unmodified proteins. Interestingly, the kcat of the K71me proteins decreased by 83% but the Km decreased only 21%. Thus, the methylation efficiency (kcat/Km) on K51me protein was similar to that of the unmodified protein, but the K71me was 78% less than that on control proteins. Furthermore, almost no methylation was detected in the K51/K71me protein. Together, these data support the model that Lys-71 is preferred over Lys-51 as a target for KMT7 in Tat (Fig. 2, B and C) and there are no additional sites in Tat72 for KMT7 monomethylation.
MS of Tat Purified from HEK293T and Jurkat A2 Cells Reveals Monomethylation at Lys-71 exists in Vivo
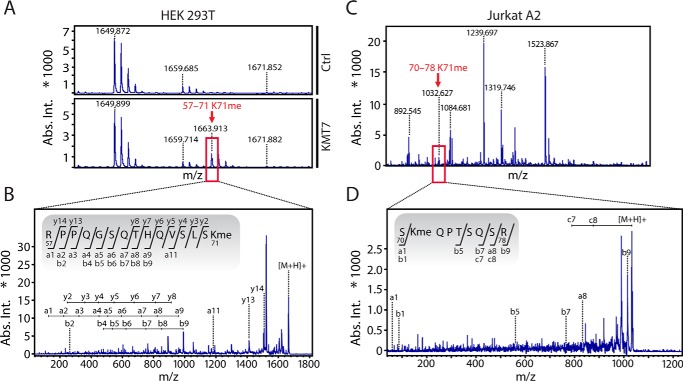
To examine Lys-71 methylation in vivo, we coexpressed Tat101 bearing a C-terminal FLAG tag together with either a KMT7 or empty vector in HEK293T cells for 24 h. We first purified Tat from lysates by FLAG immunoprecipitation, followed by SDS-PAGE separation. The separated Tat protein was subjected to MALDI-TOF MS/MS, which showed that Lys-71 was monomethylated only when KMT7 was overexpressed (Fig. 3, A and B; supplemental Table S-2). This suggests that the fraction of Tat that is naturally monomethylated at Lys-71 in HEK293T cells may be small compared with the unmodified form.
FIGURE 3.
In vivo detection of Tat K71me in HEK293T and Jurkat A2 cell lines. A, WT Tat101-FLAG and KMT7 were overexpressed in HEK293T cells and analyzed by MS after FLAG affinity purification. A zoomed region of the initial MS spectrum shows the K71me containing ion found in the presence of KMT7 overexpression, but not the control. The spectrum is representative of two independent experiments. B, MS/MS of the ion containing K71me indicated by a red box in 3A and sequence of the fragment. C, Tat101-FLAG was FLAG-affinity-purified from Jurkat A2 cells after induction with TNFα and analyzed by MS. Depicted is the full spectrum with a Tat K71me-carrying ion (aa 70–78). D, MS/MS of the ion containing K71me indicated by a red box in 3C and sequence of the fragment. Ion annotations are found in supplemental Table S-2.
Next, we analyzed Tat methylation in J-Lat A2 T cells, in which expression of FLAG-tagged Tat101 is controlled by its natural promoter and induced by stimuli such as tumor necrosis factor α (TNFα) (25). Tat expression was induced with TNFα, FLAG-affinity-purified, and analyzed by MS. We identified a peptide fragment of 1032.627 Da, corresponding to Tat aa 70–78 with a monomethyl group at Lys-71 (Fig. 3C). MS/MS confirmed the monomethylation at Lys-71 (Fig. 3D, supplemental Table S-2). Together these findings show that Tat Lys-71 is monomethylated in Jurkat T cells under conditions mimicking natural HIV infection and that this modification can be induced by KMT7 overexpression in 293T cells.
Generating Antibodies Specific for K71me
We confirmed these results using newly generated modification-specific polyclonal antibodies. We previously published methods to produce and characterize mono-, di-, and tri-methyl Tat-specific antibodies at Lys-51 (26). Following these methods, we focused on two chemically synthesized K71me 11-mer peptides (type 1 and type 2) (Fig. 4A). Peptide 1 (aa 63–72) ends with the first exon, and peptide 2 (aa 67–76) spans both Tat exons. Cysteine residues are added to the C-terminal end of the peptides for purification purposes as described (26). After antigen-purification of the modification-specific antibodies, we performed dot-blot analysis of various chemically modified Tat proteins (Fig. 4B). Type 1 antibodies reacted with all synthetic Tat72 proteins (WT, K51me, K71me, K50Ac, and K51/K71me), but type 2 antibodies detected only the K71me Tat72 proteins. Similar results were obtained by SDS-PAGE and Western blotting (Fig. 4C). The type 2 antibodies were highly specific for Tat72 proteins carrying a monomethyl group at K71, and the type 1 antibodies recognized all Tat proteins. Type 2 antibodies detected several cellular bands in whole cell lysates (data not shown). Therefore, for further analysis, we used type 2 TatK71me antibodies on FLAG-immunopurified samples.
FIGURE 4.
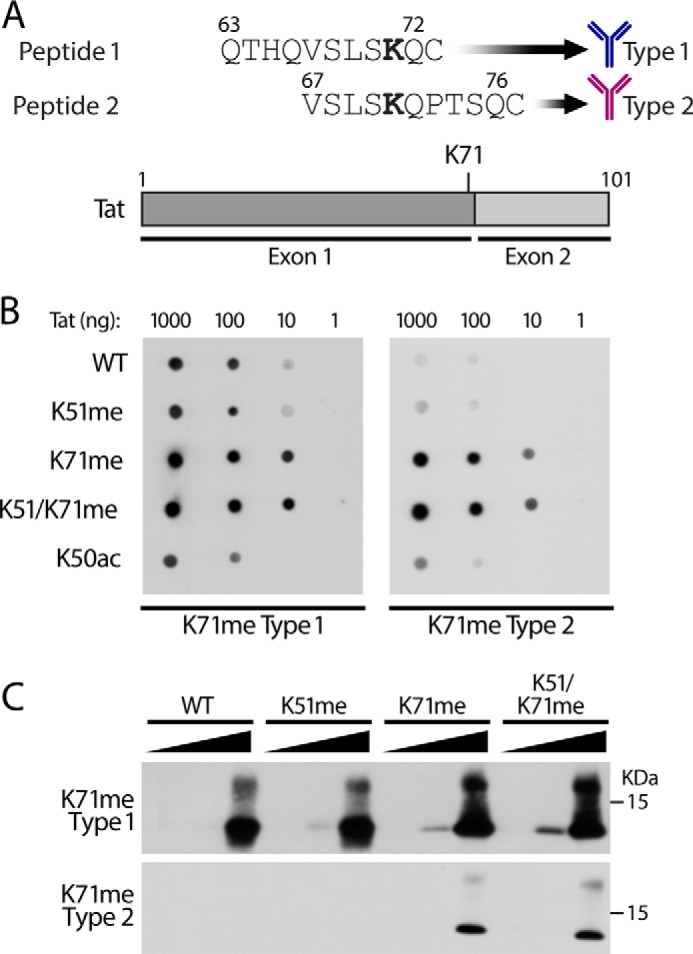
Generation of K71me-specific antibodies. A, schematic of the synthetic methylated peptides used as antigens to generate K71me Tat antibodies. B, dot blots with synthetically modified Tat72 proteins (K51me, K71me, K51/K71me, and K50ac) incubated with antigen-purified type 1 or type 2 K71me antibodies. C, Western blot analysis of indicated Tat72 proteins (1, 10, 100 ng each) with type1 antibodies (top) and type 2 antibodies (bottom).
Detecting Tat K71me by Western Blot Analysis in Vivo
To confirm the specificity of the type 2 antibodies in vivo, we overexpressed WT or FLAG-tagged Tat101 proteins carrying mutations at Lys-71, Lys-51, or Lys-50 in 293T cells. After FLAG immunoprecipitation, we detected K71me Tat only in WT, Lys-50 and Lys-51 mutants, but not in Lys-71 mutants (Fig. 5A). This finding indicates that endogenous KMT7 is sufficient to methylate Lys-71 in cells. Furthermore, we were able to increase TatK71me by coexpressing Tat101 with WT KMT7 relative to coexpression with a catalytically inactive KMT7 mutant (H297A). Importantly, no change in methylation was observed on the K71R mutant (Fig. 5B).
FIGURE 5.
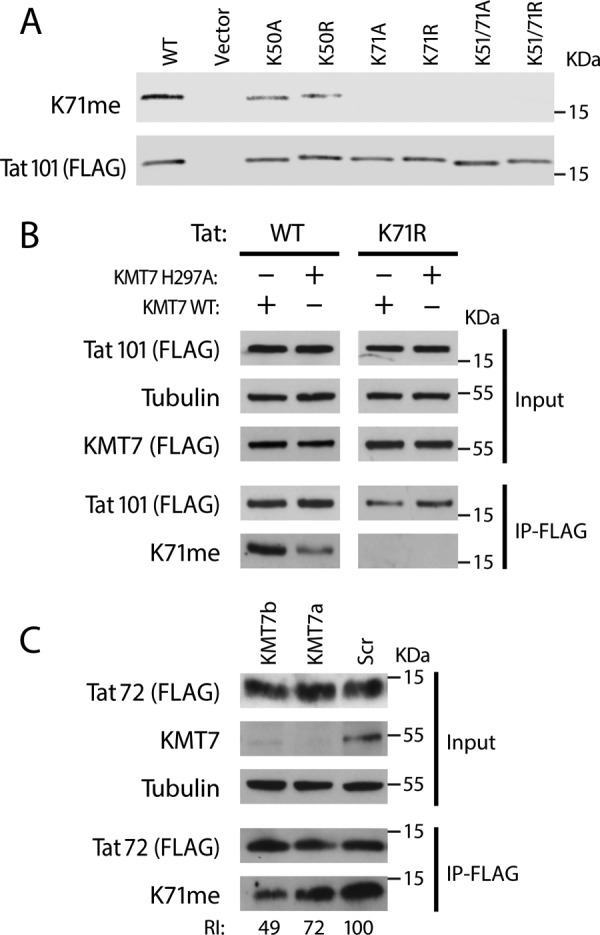
Detection of K71me Tat methylation in vivo. Lysates from 293T or Jurkat cells were FLAG-immunopurified and blotted with indicated antibodies including type 2 Tat K71me antibodies. A, Western blot analysis of indicated Tat101-FLAG mutants expressed in HEK293T cells. B, Western blot analysis of Wt or K71R Tat101-FLAG co-expressed with Wt or catalytically inactive KMT7 mutant in HEK293T cells. Tat Wt and K71R blots were cropped from the same gel and had the same exposure times. C, Western blot analysis of FLAG-purified Tat72-FLAG isolated from stable cells lines transduced with shRNAs against KMT7 (KMT7a or KMT7b) or a scrambled control. Western blots were performed with indicated antibodies. ImageJ was used for quantification.
Next, we determined whether the K71me antibodies are able to detect Tat72 in T cells. For this, we used an HIV-1 mini-genome to generate Jurkat cell lines that stably express Tat72, as described for the Jurkat A2 cells (27). We sorted for cells with high-level GFP expression, and transduced them with lentiviruses containing one of two shRNAs targeting KMT7 (a and b) or a scrambled shRNA (Scr). The lentiviral constructs also contained a puromycin resistance gene, which allowed us to select for shRNA-expressing cells. After selection, shRNA-expressing cells were lysed and subjected to FLAG immunoprecipitation, and Tat K71me levels were determined by Western blotting (Fig. 5C). Knockdown of KMT7 was robust; in accordance, Tat K71me levels were reduced by 51% in cells treated with the KMT7b shRNA and by 28% in cells treated with KMT7a shRNA (Fig. 5B, bottom). These results underscore the role of KMT7 as an important K71 methyltransferase for both forms of Tat in T cells. Since a substantial amount of Tat remained methylated at Lys-71 despite the knockdown, we suspect that a small residual pool of KMT7 may be sufficient to modify a proportion of Tat proteins, or perhaps another enzyme is capable of monomethylating Tat in cells.
K71me Does Not Affect Tat Half-life
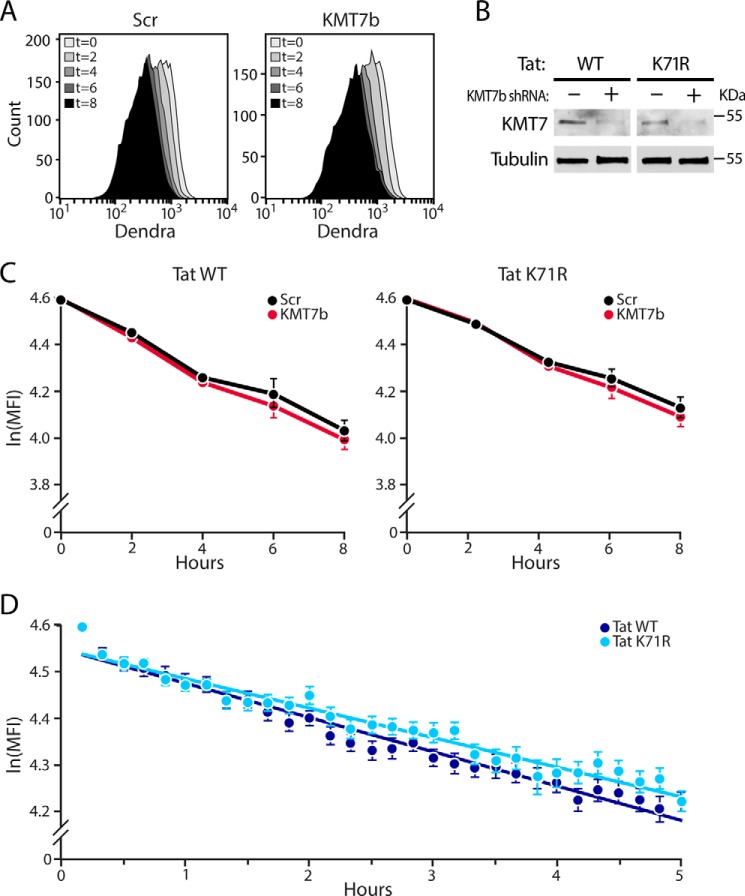
A number of substrates modified by KMT7 are altered in their stability by downstream changes in their polyubiquitylation (28). Because Lys-71 has previously been reported as a site for polyubiqitination (23), we examined whether Lys-71 methylation by KMT7 affects the stability of Tat in Jurkat cell lines expressing fluorescent Tat101-Dendra fusion proteins generated for this purpose (25). The Tat-Dendra system has been used to determine the Tat half-life in flow cytometry and microscopy studies (29, 30). To knock down KMT7, we transduced Tat-Dendra cells with the lentiviral vectors expressing scrambled or KMT7b shRNAs used for Fig. 5B. After puromycin selection, we monitored effects of KMT7 knockdown on Tat half-life only in successfully transduced cells. Knockdown of KMT7 was confirmed by Western blotting (Fig. 6B). Expression of WT or K71R Tat-Dendra proteins was induced with TNFα. After 16–20 h of TNFα treatment, cycloheximide was added to inhibit de novo production of Tat. Cells were fixed at various times, and Tat expression was determined by flow cytometry of Dendra (Fig. 6A). Tat stability did not differ in cells transduced with control and KMT7 shRNAs, excluding any prominent effect of K71 methylation on Tat stability (Fig. 6C). In cells expressing scrambled shRNA, the K71R Tat mutant had a slightly longer half-life than WT Tat (10.9 versus 9.0 h, p = 0.043). However, time-lapse single-cell microscopy of Tat-Dendra cells revealed no statistical difference in the half-lives of WT and K71R Tat in the absence of shRNAs (Fig. 6D). Together, these data affirm previous findings that Lys-71 modifications are not involved in Tat protein stability (29).
FIGURE 6.
Lys-71 mutation or KMT7 knockdown does not affect Tat stability. A, flow cytometry histograms showing mean Tat-Dendra fluorescence (MFI) at indicated times (0–8 h) in WT cells stably transduced with KMT7b or scrambled shRNAs and treated with cycloheximide. B, Western blot analysis confirms knockdown of KMT7 in Tat-Dendra J-Lat lysates after puromycin selection. C, measurements of Tat half-life by flow cytometry. MFIs were normalized to t = 0 after removing background, natural log transformed, and plotted over time as mean ln(MFI) ± S.E. of four independent experiments in cells transduced with lentiviruses containing KMT7b or scrambled shRNAs and expressing WT or K71R Tat. D, time-lapse single cell microscopy of WT or K71R Tat-Dendra J-Lats each point represents the MFI of 50 independent cells that were tracked for at least 2 h.
Lys-71 Methylation Enhances Tat Transactivation
Previously, we found that KMT7-mediated monomethylation of Tat Lys-51 enhances interactions between Tat and TAR RNA and P-TEFb and activates HIV gene expression (17). To assess the effect of Lys-71 monomethylation on Tat transactivation, we transfected plasmids expressing WT or mutant Tat101 (K51R, K71R, or K51/71R) into TZMBL cells, which express firefly luciferase from the integrated HIV LTR when functional Tat is expressed (31). Transactivation was ∼50% lower in the Tat K51R and Tat K71R mutants than in WT Tat at three different plasmid concentrations, confirming that these residues are important for Tat transactivation (17, 23). Transactivation was almost completely lost when both Lys-51 and Lys-71 were mutated, underscoring their combined importance in Tat transactivation. All Tat proteins were expressed at similar levels as confirmed by Western blot analysis (Fig. 7A).
FIGURE 7.
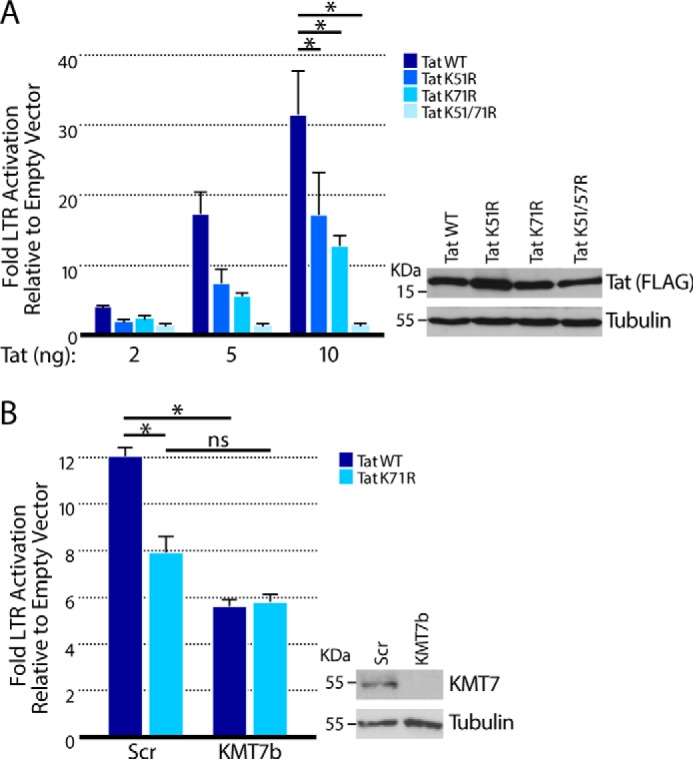
Tat K71me regulates Tat transactivation. A, TZMBL cells were transfected with increasing concentrations of plasmids (2, 5, and 10 ng) expressing WT or mutant Tat101. Luciferase values are shown from four independent experiments measured as fold expression over empty vector (mean ± S.E.). Western blot analysis was performed in cells transfected with 100 ng of Tat-FLAG plasmid and blotted with anti-FLAG antibodies. B, luciferase values in TZMBL cells transduced with KMT7b or scrambled shRNAs and then transfected with WT or TatK71R Tat-expressing plasmids (20 ng). KMT7 knockdown is visualized by Western blotting. Values are mean ± S.E. from four independent experiments. Significance was calculated with a one-sided t test. *, p < 0.05.
To test whether monomethylation at Lys-71 by KMT7 contributes to Tat transactivation, we knocked down KMT7 expression using KMT7b shRNAs in TZMBL cells before transfection with WT Tat or K71R mutant Tat101. Knockdown of KMT7 decreased the transcriptional activity of Tat WT but had no effect on K71R mutant Tat. Evidently, KMT7 activates Tat transactivation, at least in part, through Lys-71 monomethylation (Fig. 7B). Knockdown of KMT7 was confirmed by Western blotting (Fig. 7B). These findings indicate that Lys-71 monomethylation, rather than regulating Tat stability, plays a positive role in the transcriptional activity of Tat.
Discussion
HIV-1 Tat is a potent viral transactivator that undergoes extensive post-translational modifications. Here, we expand the role of KMT7 in Tat function by identifying Lys-71 as a second monomethylation site. We found that Lys-71 is monomethylated in both functional HIV-1 Tat101 and Tat72 splice variants, underscoring its importance throughout the HIV-1 life cycle. We identified KMT7 as a robust Tat Lys-71 monomethyltransferase in both in vitro and in vivo assays. Although KMT7 substrates often have perturbed stability, mutation of Lys-71 or knock down of KMT7 did not affect the stability of Tat. Instead, Lys-71 monomethylation is important for the transcriptional activity of Tat through a yet unresolved mechanism.
Notably, in vitro methylation and enzymology experiments indicate that the preferred methylation site of KMT7 is Lys-71 rather than Lys-51, previously identified as a target of KMT7. Although KMT7 has no known stringent site specificity, two consensus sequences in KMT7 targets have been described: (K,R)−2-(S,T,A)−1-Kme0-X+1 (where X is a polar residue) and a newer sequence (G,R,H,K,P,S,T)−3-(K>R)−2-(S>K,Y,A,R,T,P,N)−1-Kme0-(Q,N)+1-(A,Q,G,M, S,P,T,Y,V)+2 (32, 33). Comparing the sequences of K71me (S−3L−2S−1Kme0Q+1P+2) and K51me (G−3R−2K−1Kme0R+1R+2), Lys-71 is more closely aligned with the newer consensus sequence than Lys-51, possibly explaining why Lys-71 was the preferred target in our in vitro studies (32).
Our finding that KMT7 methylates Tat at two sites is consistent with reports that KMT7 often has multiple targets within individual substrates. For example, the RelA subunit of the NF-κB transcription factor has three KMT7 monomethylation sites (K37me, K314me, K315me) and the PCAF acetyltransferase has two (K78me, K89me) (34–36). Since the addition of a monomethyl group is a rather subtle modification (14 Da), it is likely that multiple monomethylation sites act in concert to mediate appropriate regulation of the substrate protein by KMT7.
Two main consequences of KMT7-mediated methylation have emerged: alteration of protein stability and the regulation of interactions between nucleic acids and proteins (32). The oncogene p53 and estrogen receptor α are both stabilized upon monomethylation by KMT7 (37, 38). In contrast, RelA of NF-κB and DNMT1 are both destabilized upon monomethlyation of one or more sites by KMT7 (35, 39). One review regarded KMT7 as a bona fide protein stability modifier, proposing the presence of a methyl/ubiquityl switch that can regulate the stability of substrates (28). However, polyubiquitination of Tat at Lys-71 has been linked to enhanced transcriptional activity, not degradation (23). Indeed, we found that KMT7 knockdown did not significantly affect the stability of WT or K71R Tat.
Early structural studies of Tat isolates suggested that the glutamine-rich motif (aa 59–72) has a degenerate α-helical structure, consistent across different Tat isolates and adopted in the presence of TAR RNA (12). This structure is adopted partially through conserved glutamine-RNA hydrogen bonds (Gln-60, Gln-63, Gln-66, Gln-72) and an electrostatic interaction between Lys-71 and the TAR phosphodiester backbone at nucleotides 31–35 (12, 13). Lys-71 monomethylation could thus enhance these interactions by stabilizing the electrostatic interaction between Lys-71 and the TAR loop (13, 17). Previously, we showed that methylation of Lys-51 by KMT7 activates appropriate Tat/TAR/P-TEFb binding (17). Lys-51 lies in the ARM of Tat, which binds TAR RNA in the bulge region. Therefore, methylation of Lys-51 and Lys-71 by KMT7 might coordinately enhance the binding of TAR RNA to multiple residues in Tat, potentially positioning the RNA properly with respect to Tat and P-TEFb. Future experiments are necessary to explore this model and elucidate the mechanism by which KMT7 activates Tat transactivation by monomethylating Lys-71.
Experimental Procedures
Materials
HEK293T, Jurkat, and TZMBL cells were obtained from American Type Culture Collection. Anti-FLAG M2 affinity gel (A2220) and anti-FLAG monoclonal rabbit antibodies (F7425) were from Sigma-Aldrich. Tubulin antibodies (ab15246) were from Abcam and KMT7 antibodies (Clone 5F2.3, 04-805) were from Millipore. KMT7 siRNAs (4392420) were from ThermoFisher. Tat72 proteins (unmodified, Tat K71me, Tat K51me, Tat K50Ac, and Tat K51/K71me) were synthesized by PSL Peptide Specialty Laboratories (Heidelberg, Germany). TNFα (ThermoFisher, PHC3011) was resuspended in water at 100 ng/μl. Cycloheximide (MP Biomedicals, 02100183) was resuspended in water at 10 mg/ml.
In Vitro Methylation
Reactions were carried out with 3 μg of synthesized Tat peptides and 0, 1, or 2 μg of purified KMT7 in a solution of 0.1 m Bicine, pH 8.2, 60 μm [3H]SAM. After incubating the mixtures for 18h at room temperature, the methylation reactions were run on 15% SDS-PAGE gels, which were stained with Coomassie Blue, and destained overnight. Methylation was detected by autoradiography (22-h exposure).
Mass Spectrometry
In vitro modified and in vivo purified Tat peptides were analyzed by MALDI-TOF tandem mass spectrometry (MS) as described (18).
Enzymology
Methyltransferase activity was measured with a modified radiometric assay (24). Kinetic assays were performed with various concentrations of synthetic Tat72 peptides (unmodified, Tat K71me, Tat K51me, and Tat K51/K71me). The 20-μl reactions contained Tat peptides, 0.1 m Bicine, pH 8.2, 60 μm [3H]SAM (3.4Ci/mmol), and 1 μg of purified full-length KMT7 (40). The reactions were incubated for 1 min at 37 °C. To terminate the reaction and ensure full precipitation of the substrates, 0.5 ml of 10% TCA and 5 μl of a bovine serum albumin (BSA) solution (10 g/100 ml of water) were added. After this addition the mixture was then vortexed, incubated on ice for 3 min, and centrifuged at 14,000 rpm for 3 min. To process the reaction, the protein pellets were washed with 150 μl of 0.1 m NaOH, re-precipitated in TCA, vortexed and centrifuged at 14,000 rpm for 3 min. The pelleted proteins were dissolved in 50 μl of formic acid, diluted by half with water, mixed with 1.25 ml of Bio-Safe II scintillant (Research Products International), and subjected to liquid scintillation. Activity was calculated after correcting for methylation in control reactions lacking either the enzyme or the substrate. The assays were performed in triplicate, and the data were plotted and fitted with SigmaPlot 11 (Systat Software) to the Michaelis-Menten equation.
Tat K71me Antibody Purification
Peptides for antibody synthesis (K71me1 Type 1, K71me1 Type 2; Fig. 4A) were synthesized by PSL Peptide Specialty Laboratories and injected into rabbits and antibodies were purified from serum with antigenic peptides as described (26). Purified antibodies were eluted under acidic conditions, resuspended in 1% BSA and 0.1% sodium azide, and stored at −80 °C.
Dot Blot Analysis of K71me Antibodies
Tat peptides were serially diluted in water and spotted on a 0.2 μm Hybond ECL membrane. Membranes were air-dried, and nonspecific binding was blocked with nonfat dry milk (5 g/100 ml in TBST consisting of 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20) for 1 h at room temperature. Membranes were incubated with K71me antibodies diluted in blocking buffer for 1 h, washed five times with TBST, then incubated with HRP-conjugated anti-rabbit IgG (Jackson ImmunoResearch) at a concentration of 16 ng/ml in blocking buffer, washed 3–5 times with TBST, and analyzed using standard ECL substrate.
Detection of K71me in Vivo
Tat mutants (K71A, K71R, K51/71A, K51/71R) were generated by site-directed mutagenesis of previously described Tat constructs (17). KMT7 expression vectors (WT and H297A) were also previously described (17). For experiments conducted in HEK293T cells, Tat constructs (2 μg) or Tat and KMT7 (2.5 and 6 μg, respectively) were transfected into cells using Lipofectamine 2000 and incubated for 24 h prior to lysis. For experiments conducted in Jurkat cells, WT and K71R Tat72-expressing cells were produced as described (27). After sorting, cells were transduced with lentiviruses containing control short hairpin RNAs (shRNAs) or KMT7 shRNAs (KMT7a: GCACTTTATGGGAAATTTA; KMT7b: GTAGCTGTGGGACCTAATA) and a puromycin resistance cassette. After 1 week of puromycin selection at 2 μg/ml, cells were lysed using IP-lysis buffer (150 mm NaCl, 50 mm Tris, pH 7.4, 1 mm EDTA, 0.5% v/v Nonidet P-40 substitute) and Tat-FLAG was immunoprecipitated from 500 μg of protein using anti-FLAG M2 affinity gel. Immunoprecipitated Tat was analyzed using either anti-FLAG antibodies (0.16 μg/ml) or K71me antibodies (1.75 ng/ml) diluted in blocking buffer. After washing, membranes were incubated with a HRP-conjugated anti-rabbit antibody (160 ng/ml). All K71me Western blot analysis performed at least twice.
Measuring the Half Life of Tat
WT LTR-Tat101-Dendra (LTD) constructs were subjected to site-directed mutagenesis to generate the K71R mutation in Tat. WT or K71R LTD constructs, a lentiviral construct (pCMV-ΔR8.91) and VSV-G pseudotyped envelope plasmid were co-transfected into HEK293T cells to produce lentivirus, as described (41). Jurkat T cells were infected with either WT or K71R LTD lentiviral vectors to generate polyclonal J-Lat populations as described (25). After resilencing of the LTR, LTD jurkats were infected with KMT7b shRNAs and selected with puromycin for at least 1 week. For protein stability experiments, Tat expression was induced with TNFα (10 ng/ml) for 16 h followed by treatment with cycloheximide (10 μg/ml). Cells were fixed at various times in 2% paraformaldehyde (Alfa Aesar), incubated at 4 °C for at least 1 h, and analyzed on a FACSCalibur DxP8 (Cytek). Flow cytometry data were analyzed by FlowJo X. Data were normalized to 100% at time 0 and natural log transformed to produce a linear half-life curve, from which the slope was calculated. Two-tailed Z-tests were performed on averaged slope values from at least four independent experiments. The equation t(1/2) = ln(2)/-slope was used to generate half-life values. Time-lapse microscopy experiments were performed as described (29).
Luciferase Assays
TZMBL cells (1 × 105) were transfected with a total of 100 ng of DNA containing 1, 2, 5, or 10 ng of Tat-expressing plasmids (WT, K51R, K71R, K51/71R) or empty vector using X-tremeGENE 9 (Roche Diagnostics) as recommended by the manufacturer. The cells were incubated for 48 h and lysed in 1× Promega Passive Lysis Buffer. Luciferase assays were processed with the Promega Dual-Luciferase Reporter Assay System and measured on a Monolight 2010 luminometer. Experiments were conducted with four independent biological replicates with technical duplicates and the statistical significance of differences was determined with one-sided t tests.
Author Contributions
I. A., H. R., D. B., L. D., N. S., K. H., S. P., and K. K. contributed to experiment design and data collection. I. A., H. R., and D. B. performed luciferase assays and Western blot analysis. L. D., S. P., and K. K. performed in vitro methylation and enzymology assays. N. S. and K. H. performed in vitro methylation and mass spectrometry. M. S. provided Tat proteins. K. A. and I. A. performed half-life experiments, computational, and statistical analysis of data. R. T. provided active KMT7. L. W., R. H., M. K., and M. O. supervised the experiments and helped with data interpretation. I. A. and M. O. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Ott, Weinberger, and Verdin laboratories for helpful discussions, reagents, and expertise. We thank John Carroll for graphics, Stephen Ordway for editorial support, and Veronica Fonseca for administrative assistance.
This work was supported by the University of California San Francisco, Gladstone Institute of Virology and Immunology Center for AIDS Research, a collaboration with JT Pharma, Gladstone Institutes, CA AIDS Research Program, and Grants R01AI083139, U19AI096113, T32IA7334-26, and P30AI027763 from the National Institutes of Health, and Grant ID F13-GI-316 from the California HIV/AIDS Research Program (CHRP). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S-1 and S-2.
- aa
- amino acid
- LTR
- long terminal repeat
- TAR
- transactivation response element
- ARM
- arginine-rich motif
- SAM
- S-adenosyl methionine
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
References
- 1. Dahabieh M. S., Battivelli E., and Verdin E. (2015) Understanding HIV latency: the road to an HIV cure. Annu. Rev. Med. 66, 407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mbonye U., and Karn J. (2014) Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology 454–455, 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruner K. M., Hosmane N. N., and Siliciano R. F. (2015) Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. 23, 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang X., Gold M. O., Tang D. N., Lewis D. E., Aguilar-Cordova E., Rice A. P., and Herrmann C. H. (1997) TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. U.S.A. 94, 12331–12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu Y., Pe'ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M. B., and Price D. H. (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garber M. E., Wei P., KewalRamani V. N., Mayall T. P., Herrmann C. H., Rice A. P., Littman D. R., and Jones K. A. (1998) The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12, 3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taube R., Fujinaga K., Irwin D., Wimmer J., Geyer M., and Peterlin B. M. (2000) Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1. J. Virol. 74, 892–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Selby M. J., Bain E. S., Luciw P. A., and Peterlin B. M. (1989) Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 3, 547–558 [DOI] [PubMed] [Google Scholar]
- 9. Ivanov D., Kwak Y. T., Nee E., Guo J., Garcia-Martínez L. F., and Gaynor R. B. (1999) Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J. Mol. Biol. 288, 41–56 [DOI] [PubMed] [Google Scholar]
- 10. Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., and Gaynor R. B. (1989) Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 8, 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei P., Garber M. E., Fang S. M., Fischer W. H., and Jones K. A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462 [DOI] [PubMed] [Google Scholar]
- 12. Campbell G. R., Pasquier E., Watkins J., Bourgarel-Rey V., Peyrot V., Esquieu D., Barbier P., de Mareuil J., Braguer D., Kaleebu P., Yirrell D. L., and Loret E. P. (2004) The glutamine-rich region of the HIV-1 Tat protein is involved in T-cell apoptosis. J. Biol. Chem. 279, 48197–48204 [DOI] [PubMed] [Google Scholar]
- 13. Loret E. P., Georgel P., Johnson W. C. Jr., and Ho P. S. (1992) Circular dichroism and molecular modeling yield a structure for the complex of human immunodeficiency virus type 1 trans-activation response RNA and the binding region of Tat, the trans-acting transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 89, 9734–9738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ott M., Geyer M., and Zhou Q. (2011) The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10, 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sivakumaran H., van der Horst A., Fulcher A. J., Apolloni A., Lin M. H., Jans D. A., and Harrich D. (2009) Arginine methylation increases the stability of human immunodeficiency virus type 1 Tat. J. Virol. 83, 11694–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie B., Invernizzi C. F., Richard S., and Wainberg M. A. (2007) Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J. Virol. 81, 4226–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pagans S., Kauder S. E., Kaehlcke K., Sakane N., Schroeder S., Dormeyer W., Trievel R. C., Verdin E., Schnolzer M., and Ott M. (2010) The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe 7, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakane N., Kwon H. S., Pagans S., Kaehlcke K., Mizusawa Y., Kamada M., Lassen K. G., Chan J., Greene W. C., Schnoelzer M., and Ott M. (2011) Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog., 10.1371/journal.ppat.1002184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ott M., Schnölzer M., Garnica J., Fischle W., Emiliani S., Rackwitz H. R., and Verdin E. (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol. 9, 1489–1492 [DOI] [PubMed] [Google Scholar]
- 20. Mujtaba S., He Y., Zeng L., Farooq A., Carlson J. E., Ott M., Verdin E., and Zhou M. M. (2002) Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell 9, 575–586 [DOI] [PubMed] [Google Scholar]
- 21. Endo-Munoz L., Warby T., Harrich D., and McMillan N. A. (2005) Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol. J. 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foley B., L. T., Apetrei C., Hahn B., Mizrachi I., Mullins J., Rambaut A., Wolinsky S., Korber B. (2015) HIV Sequence Compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory [Google Scholar]
- 23. Brès V., Kiernan R. E., Linares L. K., Chable-Bessia C., Plechakova O., Tréand C., Emiliani S., Peloponese J. M., Jeang K. T., Coux O., Scheffner M., and Benkirane M. (2003) A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 5, 754–761 [DOI] [PubMed] [Google Scholar]
- 24. Houtz R. L., Royer M., and Salvucci M. E. (1991) Partial purification and characterization of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit ϵN-methyltransferase. Plant Physiol. 97, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jordan A., Bisgrove D., and Verdin E. (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pagans S., Sakane N., Schnölzer M., and Ott M. (2011) Characterization of HIV Tat modifications using novel methyl-lysine-specific antibodies. Methods 53, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jordan A., Defechereux P., and Verdin E. (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20, 1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang X. D., Lamb A., and Chen L. F. (2009) Methylation, a new epigenetic mark for protein stability. Epigenetics 4, 429–433 [DOI] [PubMed] [Google Scholar]
- 29. Razooky B. S., Pai A., Aull K., Rouzine I. M., and Weinberger L. S. (2015) A hardwired HIV latency program. Cell 160, 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh A., Razooky B. S., Dar R. D., and Weinberger L. S. (2012) Dynamics of protein noise can distinguish between alternate sources of gene-expression variability. Mol. Syst. Biol. 8, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montefiori D. C. (2009) Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485, 395–405 [DOI] [PubMed] [Google Scholar]
- 32. Del Rizzo P. A., and Trievel R. C. (2011) Substrate and product specificities of SET domain methyltransferases. Epigenetics 6, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dhayalan A., Kudithipudi S., Rathert P., and Jeltsch A. (2011) Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem. Biol. 18, 111–120 [DOI] [PubMed] [Google Scholar]
- 34. Ea C. K., and Baltimore D. (2009) Regulation of NF-κB activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. U.S.A. 106, 18972–18977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang X. D., Huang B., Li M., Lamb A., Kelleher N. L., and Chen L. F. (2009) Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 28, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masatsugu T., and Yamamoto K. (2009) Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem. Biophys. Res. Commun. 381, 22–26 [DOI] [PubMed] [Google Scholar]
- 37. Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., and Reinberg D. (2004) Regulation of p53 activity through lysine methylation. Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 38. Subramanian K., Jia D., Kapoor-Vazirani P., Powell D. R., Collins R. E., Sharma D., Peng J., Cheng X., and Vertino P. M. (2008) Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Estève P. O., Chin H. G., Benner J., Feehery G. R., Samaranayake M., Horwitz G. A., Jacobsen S. E., and Pradhan S. (2009) Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 106, 5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Couture J. F., Collazo E., Hauk G., and Trievel R. C. (2006) Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 13, 140–146 [DOI] [PubMed] [Google Scholar]
- 41. Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., and Naldini L. (1998) A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.