Abstract
Rearranged during transfection (RET), a receptor tyrosine kinase that is activated by the glial cell line-derived neurotrophic factor family ligands (GFLs), plays a crucial role in the development and function of the nervous system and additionally is required for kidney development and spermatogenesis. RET encodes a transmembrane receptor that is 20 exons long and produces two known protein isoforms differing in C-terminal amino acid composition, referred to as RET9 and RET51. Studies of human pheochromocytomas identified two additional novel transcripts involving the skipping of exon 3 or exons 3, 4, and 5 and are referred to as RETΔE3 and RETΔE345, respectively. Here we report the presence of RetΔE3 and RetΔE345 in zebrafish, mice, and rats and show that these transcripts are dynamically expressed throughout development of the CNS, peripheral nervous system, and kidneys. We further explore the biochemical properties of these isoforms, demonstrating that, like full-length RET, RETΔE3 and RETΔE345 are trafficked to the cell surface, interact with all four GFRα co-receptors, and have the ability to heterodimerize with full-length RET. Signaling experiments indicate that RETΔE3 is phosphorylated in a similar manner to full-length RET. RETΔE345, in contrast, displays higher baseline autophosphorylation, specifically on the catalytic tyrosine, Tyr905, and also on one of the most important signaling residues, Tyr1062. These data provide the first evidence for a physiologic role of these isoforms in RET pathway function.
Keywords: alternative splicing, cell signaling, neurotrophic factor, receptor tyrosine kinase, signal transduction
Introduction
RET is a receptor tyrosine kinase that is critical for kidney morphogenesis, spermatogenesis, and development of the nervous system (1–4). RET is activated by a family of four growth factors known as the glial cell line-derived neurotrophic factor (GDNF)2 family ligands (GFLs), which includes GDNF, neurturin, artemin, and persephin (1). Each GFL binds to one of four cognate glycosylphosphatidylinositol-anchored co-receptors known as the GDNF family receptor αs (GFRαs) (2, 5). The GFL-GFRα complex binds to RET, inducing RET dimerization and subsequent autophosphorylation on multiple tyrosine residues within the intracellular tyrosine kinase domain. This enhances tyrosine kinase activity and initiates the association of adaptor proteins and enzymes that trigger multiple second messenger cascades (6).
The presence of two major Ret isoforms, RET9 and RET51, has been extensively described in the literature, and a third isoform, RET43, has also been observed in humans (7–9). Ret is 20 exons long, and Ret9 and Ret51 transcripts differ in alternative splicing of intron 19. Intron 19 in the Ret51 transcript is excised properly, whereas, in the Ret9 transcript, the intron is retained, changing the reading frame and inserting a premature stop codon into the amino acid sequence. This creates a unique nine-amino acid C-terminal sequence for RET9 and a unique 51-amino acid C-terminal sequence for RET51. Interestingly, these two different isoforms display marked differences in their degradation and function (7, 10, 11).
Three additional Ret transcripts have been reported in various tumor sources as well as adult human tissues (12). These novel transcripts are a product of exon skipping in the 5′ region of RET, which encodes for the extracellular domain of the protein. Skipping of exons 3 (RETΔE3) or exons 3, 4 and 5 (RETΔE345) gives rise to transcripts that encode for full-length Ret proteins but with deletions in the extracellular domain, specifically cadherin-like domain 1 (CLD1) or CLD1–3, respectively. These deletions are hypothesized to change the extracellular domain structure as well as binding to GFL-GFRα complexes and may impact the overall stability of the proteins. The skipping of exons 3 and 4 results in a frameshift that culminates in a premature stop. This protein encodes only a small portion of the RET extracellular domain with no transmembrane or intracellular domain. Because this transcript is likely to be subjected to nonsense-mediated decay, it is unlikely to be involved in RET signal transduction.
Here we show that RetΔE3 and RetΔE345 transcripts are conserved in vertebrates and that the mRNA and proteins of these splice variants are expressed throughout the nervous system in mice. We also show that these isoforms are trafficked to the cell surface and that both isoforms interact with all four GFRαs. Additionally, we find that RETΔE3 is phosphorylated to a similar level as full-length RET and is activated in a GDNF-dependent manner. However, RETΔE345 displays higher baseline autophosphorylation, specifically on the catalytic tyrosine Tyr905, and also on an additional signaling tyrosine, Tyr1062. Interestingly, RETΔE345 is not activated in a GDNF-dependent manner. Taken together, these isoforms may have unique and important unidentified roles in the development and maintenance of the nervous system and kidneys as well as in the pathophysiology of neuroendocrine gland diseases.
Results
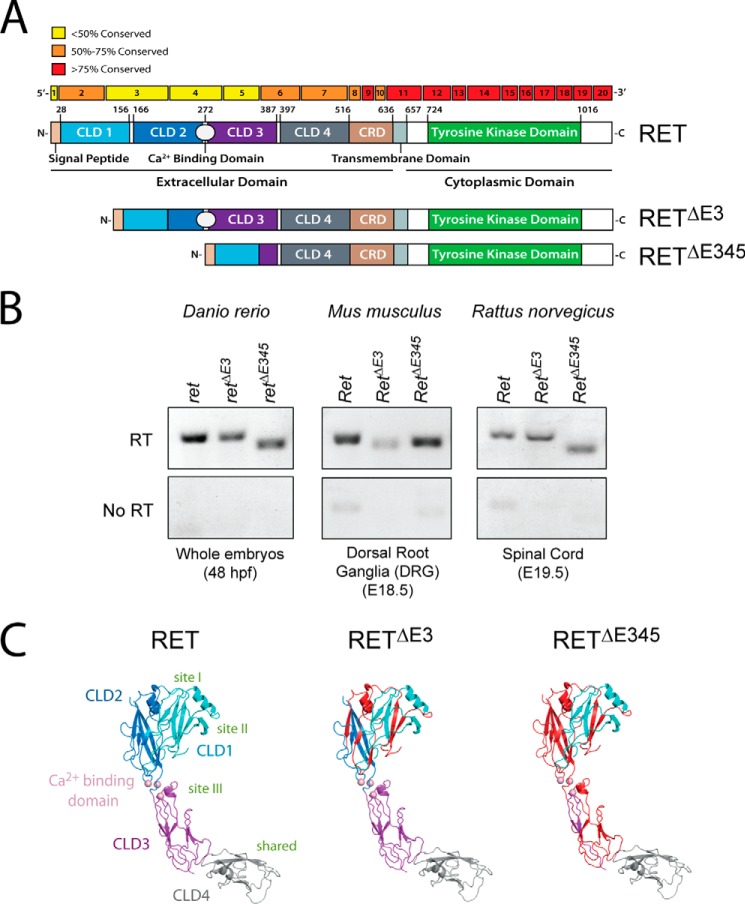
Exons 3, 4, and 5 of Ret Are Not Highly Conserved, but RetΔE3 and RetΔE345 Transcripts Are Observed in Several Organisms
It has been described previously that the intracellular domain of RET, which contains a tyrosine kinase domain, is more highly conserved than the extracellular domain (13). To better understand the level of conservation of amino acids encoded by each exon, we compared sequences from zebrafish (Danio rerio), mouse (Mus musculus), rat (Rattus norvegicus), and human (Homo sapiens). We found that the entire intracellular domain was more than 75% conserved at the amino acid level. This was not surprising because mutations in many of these highly conserved regions give rise to loss of function (e.g. Hirschsprung disease) or gain of function (e.g. multiple endocrine neoplasia type 2A (MEN2A) and 2B (MEN2B)) phenotypes (Fig. 1A). Interestingly, exons 3, 4, and 5 are less than 50% conserved between these species. Although this lack of conservation is suggestive that these regions of RET are not as functionally important, it could also be interpreted as allowing for amino acid flexibility between the different RET isoforms.
FIGURE 1.
Removal of exons 3 or 3, 4, and 5 of RET creates proteins with large deletions in the extracellular domain, and transcripts encoding these receptors are expressed in multiple organisms. A, comparison of the amino acid sequences of RET between zebrafish, mice, rats, and humans was performed, and the conservation within each exon is indicated by color. Interestingly, the amino acids encoded in exons 3, 4, and 5 are the least conserved. Additionally, the impact of exon skipping on the extracellular domain within the protein is shown. B, RetΔE3 and RetΔE345 transcripts are detectable in zebrafish (D. rerio), mice (M. musculus), and rats (R. norvegicus) by qPCR. Like human RETΔE3 and RETΔE345, alternative splicing of exon 3 or exons 3, 4, and 5 in these vertebrates are predicted to encode full-length, transmembrane RET proteins with deletions in the extracellular domain. C, deleted regions are shown in red, projected onto a structural model for RET cadherin domains 1–4 derived from PDB code 4UX8.
Our initial sequence analysis suggested that RetΔE3 and RetΔE345 transcripts could be expressed in additional vertebrates other than H. sapiens. These transcripts were originally identified in human kidney and substantia nigra fetal tissues (12). Sequence analysis demonstrated that exon skipping also encodes potential full-length transcripts in zebrafish, mice, and rats (Fig. 1A). To determine whether these novel transcripts were expressed in these organisms, species-specific primers were created to identify each of the Ret transcripts: full-length Ret, RetΔE3, and RetΔE345. RT-PCR analysis identified the presence of RetΔE3 and RetΔE345 from 48 hours post fertilization zebrafish embryos, E19.5 rat dorsal root ganglia, and E18.5 mouse spinal cord (Fig. 1B). Sequence analysis confirmed these amplicons to be RetΔE3 and RetΔE345 (data not shown). This is the first time these transcripts have been identified in vertebrates other than H. sapiens. Taken together, these data indicate that 5′ exon skipping is conserved and results in the expression of RetΔE3 and RetΔE345 transcripts.
Exon Skipping Creates Large Deletions in the Extracellular Domain of RET
To understand the impact deletions in the extracellular domain would have on RET, we analyzed the deletions in terms of a CLD1–4 model derived from electron microscopy and small angle x-ray scattering analyses (14). The main consequence of deletion of exon 3 is the removal of half of CLD1 (residues 113–150) and half of CLD2 (residues 154–208). Each domain adopts a β sandwich cadherin fold comprised of two β sheets separated by numerous buried hydrophobic residues. Elimination of exon 3 removes one sheet from CLD1 and several β strands from both sheets of CLD2 (Fig. 1C). Therefore, it is likely that both CLD domains are substantially perturbed leaving β strands without the cadherin fold structure and exposing some hydrophobic residues. Exon 3 removal also eliminates a highly constrained Cis-Pro loop (Cys137-Cys142) unique to higher vertebrate RET sequences. The portion of CLD1–2 removed coincides with regions stabilizing the clam shell arrangement of CLD1–2 (15). This would result in a more open and extended arrangement for the remaining parts of CLD1 and CLD2 with much greater flexibility between the domains. An odd number of cysteine residues (three in total) are deleted by removing exon 3, including the Cys157-Cys197 disulfide. Cys166 is also removed, leaving its partner Cys243 unpaired but buried. The two unpaired cysteines, Cys87 and Cys216, which constitute a known folding bottleneck in wild-type RET, are left untouched. We expect RETΔE3 to be a less stable transmembrane protein with a short half-life that retains at least some ligand-binding properties.
For RETΔE345, removal of exons 3, 4, and 5 effectively eliminates much of CLD1–3 but leaves CLD4-cysteine-rich domain (CRD) intact (Fig. 1C). This region by itself was shown to be insufficient to bind ligand but retains at least one important binding epitope (i.e. necessary for ligand binding but not sufficient) (14). Many receptor tyrosine kinases have extracellular domains that have autoinhibitory functions in the absence of ligand. Deletion of such regions can lead to autoactivation in a ligand-independent manner. By analogy with these other systems, removal of CLD1–3 could leave a truncated form of RET that would be constitutively activated in the absence of ligand and likely would be unresponsive to the GFLs.
RetΔE3 and RetΔE345 Are Co-expressed in Several Tissues with Ret throughout Development
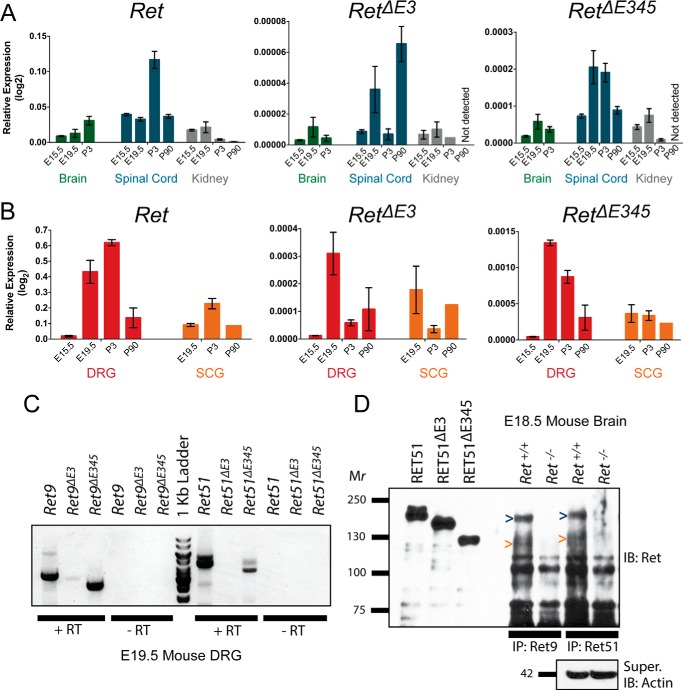
To determine where and when RetΔE3 and RetΔE345 transcripts are expressed and whether their expression levels change over time, we analyzed tissues in which full-length Ret is expressed at several developmental time points. Within the central nervous system, we analyzed the brain, as signaling via Ret is important for the survival of central noradrenergic neurons and dopaminergic neurons in the ventral midbrain, as well as the spinal cord, where GDNF/GFRα1/Ret signaling is critical for axon guidance into the hind limbs and survival of γ motor neurons (16–21). In the peripheral nervous system, we analyzed sensory neurons of the dorsal root ganglia (DRG), where neurturin/GFRα2/Ret signaling is important for the survival and maintenance of mechanoreceptors during development (22, 23). Additionally, shortly after birth, a subclass of nociceptors transition from being TrkA-positive to expressing GFRα1/Ret to support the development and survival of nonpeptidergic nociceptors (24–26). Sympathetic neurons of the superior cervical ganglia (SCG) were also analyzed, as it has been well established that artemin/GFRα3/Ret signaling is necessary for proper sympathetic chain migration and axon guidance during development (27–29). Last, the kidney was analyzed because GDNF/GFRα1/Ret signaling is crucial for ureteric bud branching and morphogenesis of the kidney (30–34). Using qPCR, we examined the relative expression of Ret, RetΔE3, and RetΔE345 in each of these tissues in mice and found that they were detected in all five tissues. The relative expression of Ret, RetΔE3, and RetΔE345 was lower in the brain, spinal cord, and kidney (Fig. 2A) compared with the DRG and SCG (Fig. 2B). Interestingly, in the spinal cord, DRG, and SCG, there was a significant increase of RetΔE3 expression at E19.5 that preceded a significant increase of Ret at P3. The expression of all three transcripts was highest in the DRG, particularly at E19.5 and P3, when the IB4+ subpopulation of nociceptors is emerging.
FIGURE 2.
RetΔE3 and RetΔE345 are dynamically expressed throughout development, and RET proteins with similar molecular weights as RETΔE345 are detect in vivo. A, expression of Ret, RetΔE3, and RetΔE345 in mouse brain (E15.5, E19.5, P3), spinal cord (E15.5, E19.5, P3, adult), and kidney (E15.5, E19.5, P3, adult) by qPCR is shown as relative expression compared with actin. B, expression of Ret, RetΔE3, and RetΔE345 in mouse sympathetic (E19.5, P3, adult) and DRG sensory neurons (E15.5, E19.5, P3, adult) by qPCR is graphed as in A. C, cDNA from E19.5 DRGs was synthesized from poly(A) RNA, and RT-PCR was performed to determine the presence of Ret transcripts with both 5′ and 3′ alternative splicing. Ret9, Ret9ΔE3, Ret9ΔE345, Ret51, and Ret51ΔE345 transcripts were identified. D, E18.5 mouse brains were lysed, and immunoprecipitations (IP) for RET9 or RET51 were performed. Immunoblotting for RET detected full-length RET9 and RET51, as expected. The presence of RET9 and RET51 proteins at the same molecular weight as RETΔE345 suggested the existence of RET9ΔE345 and RET51ΔE345 isoforms.
5′ and 3′ Alternative Splicing of Ret transcripts Are Not Mutually Exclusive
Alternative splicing of intron 19 in Ret allows for the translation of two isoforms of Ret, RET9 and RET51. We sought to determine whether alternative splicing could occur simultaneously, both 5′ and 3′ in Ret transcripts, allowing for increased diversity of encoded RET proteins. cDNA was isolated from E19.5 mouse DRGs, as we found this tissue to have the highest relative expression of Ret, RetΔE3, and RetΔE345 (Fig. 2B). Forward primers specific for each 5′ splicing event were paired with reverse primers that would detect either Ret9 or Ret51 alternative splicing. Full-length Ret9 and Ret51 transcripts were detected, as expected, at 3.33 kb and 4.38 kb, respectively. We observed RetΔE3 and RetΔE345 with intron 19 retention, thus encoding RetΔE3 and RetΔE345 transcripts with amplicons of 3.05 kb and 2.61 kb, respectively. However, we were only able to detect RetΔE345 with a properly excised intron 19 at an amplicon size of 3.66 kb, encoding a Ret51ΔE345 transcript. Although we were unable to detect a Ret51ΔE3 transcript (4.10 kb), this is likely due to the generally low expression levels of RetΔE3. Taken together, these data indicate that the Ret locus encodes for at least three previously unidentified transcripts and at least five total Ret isoforms: Ret9, Ret9ΔE3, Ret9ΔE345, Ret51, and Ret51ΔE345.
RET Proteins of the Same Molecular Weight as RETΔE345 Are Detected in Vivo
Although we can detect RetΔE3 and RetΔE345 mRNA transcripts, which are likely being processed for translation, we sought to detect the presence of these isoforms at the protein level. Plasmids encoding human RET51, RET51ΔE3, and RET51ΔE345 were transfected into NIH/3T3 cells, and lysates were run as size standards to aid in detecting Ret isoforms of the appropriate molecular weights from E18.5 mouse brain. The molecular weight of native RET51 protein is 120 kDa, but because of posttranslational modifications, specifically glycosylation of the extracellular domain, RET51 has a molecular weight of ∼180 kDa. The native molecular weights of RETΔE3 and RETΔE345 were predicted to be 109 kDa and 93 kDa, respectively. In transfected cells, we consistently observe the mature, processed proteins to have molecular weights of ∼150 kDa (RETΔE3) and ∼125 kDa (RETΔE345) (Fig. 2D).
Whole brains from Ret+/+ or Ret−/− E18.5 littermate mice were lysed and divided in two, and immunoprecipitations were performed to isolate either RET9 or RET51 proteins. Although the qPCR data suggested that Ret transcripts have an overall lower relative expression in the brain compared with PNS tissues, the brain was chosen for analysis because of the abundant amount of protein that could be isolated. Ret−/− mice were used as a control to demonstrate the specificity of RET immunoblotting and to show the potential presence of RET proteins at the same molecular weight as RETΔE3 and RETΔE345. Because we previously observed more highly expressed Ret9ΔE345 and Ret51ΔE345 transcripts compared with those of RetΔE3, we hypothesized that we would be most able to detect both RET9ΔE345 and RET51ΔE345 proteins. By immunoblotting for RET, we were able to detect full-length RET9 and RET51 proteins, as expected (Fig. 2D). Additionally, we observed RET9 and RET51 bands with similar molecular weights to RETΔE345, suggesting the presence of RET9ΔE345 and RET51ΔE345 proteins in mouse brain (Fig. 2D). Because the antibodies used for immunoprecipitations of RET9 and RET51 are to the C terminus, and the RET antibody for immunoblotting is to the common N-terminal region, this RET protein is unlikely to be a degradation product. However, we cannot exclude the possibility that this is an immature, non-glycosylated, full-length RET protein.
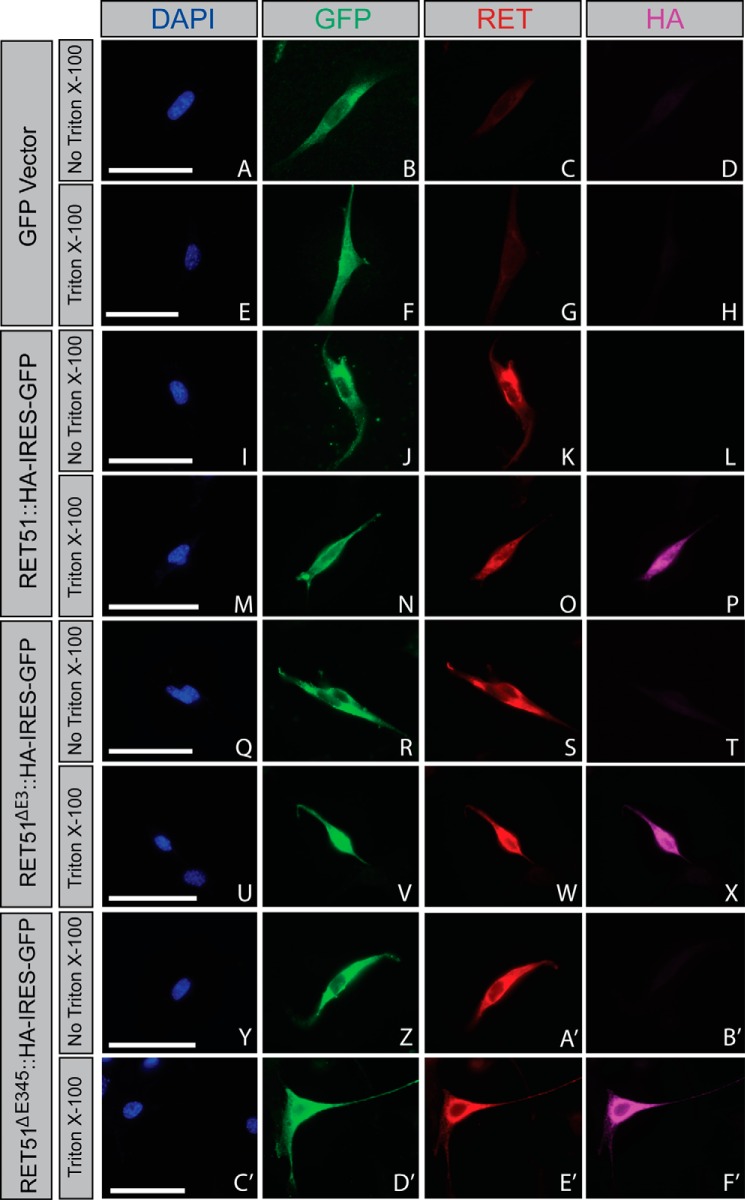
RETΔE3 and RETΔE345 Are Trafficked to the Cell Surface
To examine the biochemical properties of RETΔE3 and RETΔE345, we transitioned to an in vitro system for these experiments. Although RETΔE3 and RETΔE345 encode proteins that may be capable of signaling, this does not guarantee that they will be correctly trafficked to the cell surface for activation. To determine whether RETΔE3 and RETΔE345 are trafficked to the plasma membrane, NIH/3T3 cells were transfected with C-terminally epitope-tagged Ret constructs that also contain an IRES-GFP to serve as an indicator for transfected cells. Taking advantage of a RET antibody specific for the extracellular domain and using an HA antibody to label the C terminus of RET, we were able to selectively visualize RET located on the plasma membrane. Full-length RET was detected on the cell surface of transfected NIH/3T3 cells, as expected (Fig. 3K). Permeabilization revealed the C-terminal HA tag, confirming the integrity of the plasma membrane (Fig. 3P). Similar to full-length RET, both RETΔE3 and RETΔE345 were trafficked to the cell surface, as determined with the extracellular RET antibody in non-permeabilized cells (Fig. 3, S and A'). Transfection of GFP alone showed no labeling with the RET and HA antibodies, regardless of permeabilization, confirming the specificity of our immunostaining (Fig. 3, A–H). A cell surface biotinylation experiment was performed as a secondary measure to confirm these results, and we again observed trafficking of full-length RET, RETΔE3, and RETΔE345 to the cell surface (data not shown). Thus, RETΔE3 and RETΔE345 are trafficked properly to the cell surface, where they can participate in GFL-mediated activation.
FIGURE 3.
RETΔE3 and RETΔE345 are trafficked to the cell surface. NIH/3T3 cells were transfected with GFP (A–H), Ret51::HA (I–P), Ret51ΔE3::HA (Q–X), or Ret51ΔE345::HA (Y–F'). Constructs for the RET51 isoforms were bicistronic, allowing for expression of GFP as an indicator for positive transfection, which was encoded by an IRES-GFP following RET cDNA sequences. Cells were washed in 1× PBS and fixed briefly in 4% paraformaldehyde 24 h post-transfection. Cells were then blocked in immunofluorescence blocking solution with or without Triton X-100 to allow for cell membrane permeabilization. In the absence of Triton X-100, antibodies specific for the RET extracellular domain were able to bind to RET51::HA (K), RET51ΔE3::HA (S), and RET51ΔE345::HA (A'). However, using an antibody against the HA epitope that would detect the C terminus located intracellularly for RET51::HA (L), RET51ΔE3::HA (T), and RET51ΔE345::HA (B') showed no binding/fluorescence in the absence of Triton X-100. When immunofluorescence blocking solution with Triton X-100 was used, fluorescence was detected using the anti-HA antibody for RET51::HA (P), RET51ΔE3::HA (X), and RET51ΔE345::HA (F'). These data show that RET51ΔE3 and RET51ΔE345 are trafficked to the cell surface similarly as full-length RET51. Scale bars represent 20 μm.
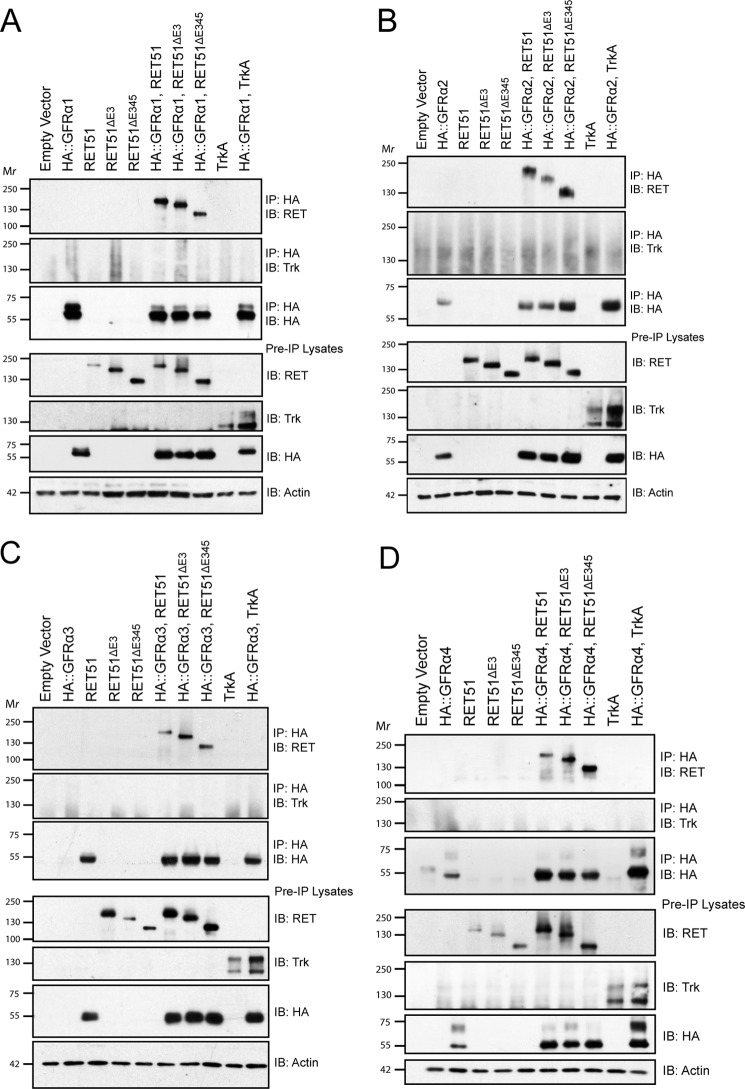
RETΔE3 and RETΔE345 Bind to GFRα1, GFRα2, GFRα3, and GFRα4
Previous structural analysis (Fig. 1C) suggested that interactions between RETΔE3 and the GFRαs may be unstable and that RETΔE345 would be unlikely to bind to the co-receptors because of deletions of important binding residues in CLD1 and CLD3. To confirm these results, we co-transfected HEK293T cells with RETΔE3 or RETΔE345 and HA epitope-tagged GFRα constructs (HA::GFRα1 (mouse), HA::GFRα2 (rat), HA::GFRα3 (mouse), and HA::GFRα4 (chicken)). Immunoprecipitations were performed using a stringent immunoprecipitation buffer (modified radioimmune precipitation assay buffer) to eliminate any weak, nonspecific interactions. Surprisingly, we found that both RETΔE3 and RETΔE345 bind to each of the four GFRαs (Fig. 4). Co-transfection of the GFRαs with TrkA, another receptor tyrosine kinase, was performed as a negative control to show selectivity of binding between the GFRαs and RET (Fig. 4). As expected, TrkA did not associate with any of the GFRαs. There were no apparent differences between the four different GFRαs in regard to the extent of their association with RETΔE3 and RETΔE345, suggesting that these splice variants could potentially be activated by all four GFLs (Fig. 4).
FIGURE 4.
RETΔE3 and RETΔE345 bind to GFRα1, GFRα2, GFRα3, and GFRα4. RET51, RET51ΔE3, and RET51ΔE345 were co-transfected with HA epitope-tagged GFRα1 (A), GFRα2 (B), GFRα3 (C), or GFRα4 (D). Immunoprecipitations (IP) were performed using an HA antibody to select for GFRαs. Immunoblotting (IB) for RET showed interaction of RET51ΔE3 and RET51ΔE345 with all four GFRα proteins. Co-transfection of HA::GFRα1, HA::GFRα2, HA::GFRα3, or HA::GFRα4 with TrkA was performed as a negative control, and, as expected, TrkA did not associate with any of the GFRαs.
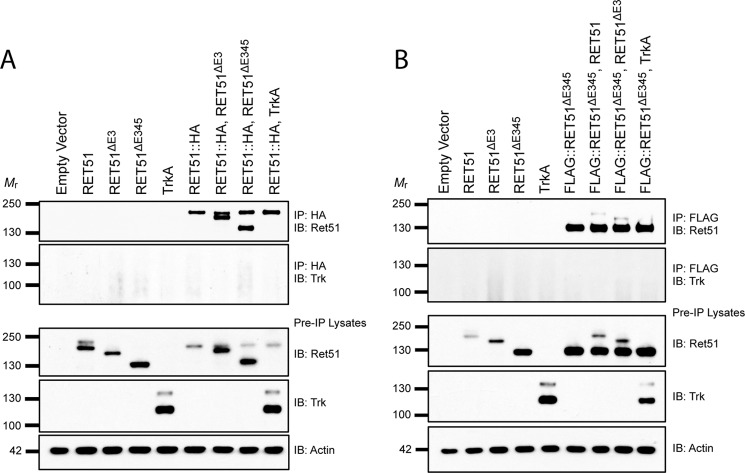
RETΔE3 and RETΔE345 Heterodimerize with Full-length RET
In addition to being potential signaling molecules on their own, we observed that these transcripts are normally expressed in tissues where full-length RET is also expressed (Fig. 2). Although these isoforms may be present in different cells within these tissues, these observations raise the question of whether RETΔE3 and RETΔE345 could heterodimerize with full-length RET and affect its function. To test this possibility, HEK293T cells were co-transfected with HA epitope-tagged RET51 and untagged RETΔE3 or RETΔE345. Immunoprecipitations were performed using an HA antibody to select for full-length RET51 and determine whether the different isoforms could interact with one another (Fig. 5A). In an additional experiment, FLAG-tagged RET51ΔE345 (FLAG:: RET51ΔE345) was co-transfected with RET51 and RET51ΔE3, and immunoprecipitations were performed using a FLAG antibody to select for RET51ΔE345 (Fig. 5B). We found that both RET51ΔE3 and RET51ΔE345 associated with full-length RET51 and that RET51ΔE3 and RET51ΔE345 could also bind to one another (Fig. 5). This is consistent with former experiments suggesting that overexpression of RET proteins allows for ligand-independent dimerization via the transmembrane domain (35).
FIGURE 5.
RETΔE3 and RETΔE345 can heterodimerize with full-length RET. A, HA epitope tagged RET51 (RET51::HA) was co-transfected with untagged RET51ΔE3 or RET51ΔE345 in HEK293T cells. Immunoprecipitations (IP) were performed with an HA antibody to select for RET51::HA. Immunoblotting (IB) for RET51 showed co-immunoprecipitation of RET51ΔE3 and RET51ΔE345 with full-length RET51. B, a similar experiment was performed using FLAG epitope tagged RET51ΔE345 (FLAG:: RET51ΔE345). Immunoprecipitations with an anti-FLAG antibody selected for FLAG:: RET51ΔE345. Immunoblotting for RET51 demonstrated the association of RET51 and RET51ΔE3 with FLAG:: RET51ΔE345. Co-transfections with TrkA were performed for both experiments as a negative control, and immunoblotting for TrkA showed no binding with either RET51::HA or FLAG::RET51ΔE345, as expected.
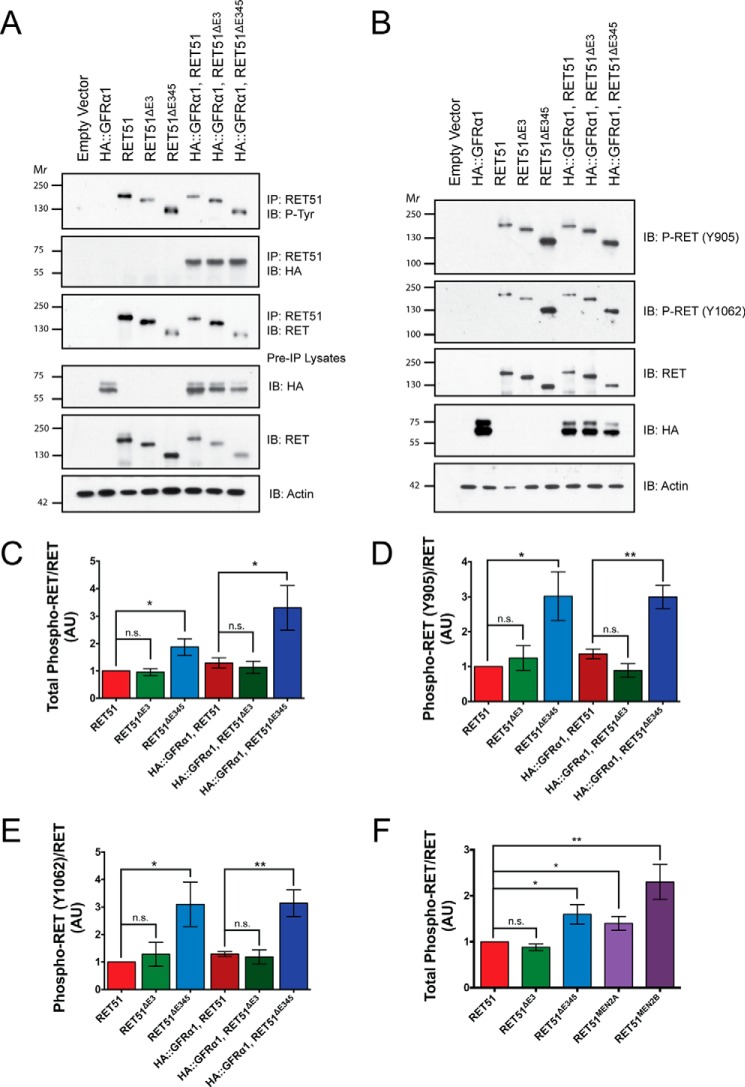
RETΔE345 Has Increased Ligand-independent Activation Compared with RET and RETΔE3
To determine levels of activation of RETΔE3 and RETΔE345, the basal phosphorylation of tyrosine residues of the RET isoforms were compared with full-length RET phosphorylation. Overexpression of the RET isoforms in vitro, either in the presence or absence of a GFRα, allows for ligand-independent dimerization and activation of RET. The level of activation can be assessed by determining the summated level of all of the phosphorylated tyrosines using a pan-phosphotyrosine antibody, or individual tyrosine residues can be analyzed using residue-specific RET phosphotyrosine antibodies. We evaluated the level of total phosphotyrosine (Tyr(P)) between the RET isoforms and individually evaluated phosphorylation at two tyrosine residues of RET, Tyr905 and Tyr1062. Tyrosine 905 in the RET kinase domain is an autocatalytic tyrosine that is conserved in many receptor tyrosine kinases and is a binding site for GRB7 and GRB10 (36–38). Additionally, mutation of tyrosine 905 to phenylalanine (Y905F) impairs the kinase activity of RET (39). Tyrosine 1062 is a binding site for SHC, Dok4/5, IRS-1, and FRS-2 when phosphorylated and is a binding site for Enigma in a phosphorylation-independent manner (36, 40–47). Interaction between Tyr1062 and these adaptor proteins leads to activation of the Ras/ERK and PI3K/AKT pathways (41, 42, 45, 46, 48).
To evaluate RET phosphorylation, NIH/3T3 cells were transfected with RET51, RET51ΔE3, or RET51ΔE345 constructs in the presence or absence of GFRα1, and the total levels of phosphorylated RET or the level of phosphorylation at Tyr905 and Tyr1062 were assessed. To analyze total Tyr(P) levels of RET, immunoprecipitations of RET51 were performed, followed by immunoblotting for Tyr(P) (Fig. 6A). An increase in the level of RETΔE345 phosphorylation compared with full-length RET was observed, but this was not statistically significant (p = 0.0837). We did detect, however, a significant increase in the Tyr(P) level of RETΔE345 co-expressed with GFRα1 compared with full-length RET co-expressed with GFRα1 (p = 0.0366). The phosphorylation of RETΔE3 co-expressed with GFRα1 or expressed alone was not significantly different from full-length Ret (p = 0.6231).
FIGURE 6.
RETΔE345 displays increased ligand-independent activation compared with full-length RET. A, RET51 was co-transfected with HA epitope-tagged GFRα1 in NIH/3T3 cells. Immunoprecipitations (IP) were performed with a RET51 antibody to select for the RET isoforms. Immunoblotting (IB) for phospho-tyrosine and RET was performed, and integrated density values were calculated for each. B, preimmunoprecipitation lysates from the previous experiment were subjected to SDS-PAGE, and Western blotting was performed to determine the levels of residue-specific tyrosine phosphorylation of RET. Immunoblotting was performed for phospho-RET (Tyr905), phospho-RET (Tyr1062), and RET, and integrated density values were calculated for each. Ratios of phospho-RET/RET (C), phospho-RET (Tyr905)/RET (D), and phospho-RET (Tyr1062)/RET (E) are reported, with values normalized to RET51. F, expression plasmids encoding oncogenic RET proteins, RET51MEN2A and RET51MEN2B, were transfected into NIH/3T3 cells, and integrated density values were calculated and analyzed for phospho-RET/RET for all RET isoforms. *, p < 0.05; **, p < 0.01; n.s., not significant.
To determine levels of Tyr(P)905 and Tyr(P)1062, pre-immunoprecipitation lysates were collected from samples in the previous experiments, and immunoblotting was performed using antibodies specific for these two phosphorylated tyrosine residues (Fig. 6B). Levels of Tyr(P)905 and Tyr(P)1062 were significantly elevated for RETΔE345 compared with RET (p = 0.0439 and p = 0.0411, respectively) and also for RETΔE345 co-expressed with GFRα1 compared with full-length RET co-expressed with GFRα1 (p = 0.0106 and p = 0.0096, respectively). However, levels of Tyr(P)905 and Tyr(P)1062 were unchanged for RETΔE3 compared with RET and also for RETΔE3 co-expressed with GFRα1 compared with full-length RET co-expressed with GFRα1. Taken together, these data suggest that RETΔE3 has a similar activation level as full-length RET, whereas RETΔE345 has an elevated activation level compared with RET in a ligand-independent manner. Additionally, the lower total phosphotyrosine level for RETΔE345 compared with Tyr(P)905 and Tyr(P)1062 suggests that one or more of the other tyrosine residues in RET is not phosphorylated as highly as Tyr905 and Tyr1062 compared with full-length RET.
Because we consistently observed an increased level of tyrosine phosphorylation of the RETΔE345 isoform compared with full-length RET, we wondered whether this level of receptor activation was similar to oncogenic RET mutations, particularly those found in MEN2. Human RET point mutations known to be causal for either MEN2A or MEN2B were introduced into the RET51 expression plasmid, specifically C634R (RET51MEN2A) and M918T (RET51MEN2B). NIH/3T3 cells were transfected with RET51, RET51ΔE3, RET51ΔE345, RET51MEN2A, or RET51MEN2B constructs. Immunoprecipitations for RET51 were performed to isolate RET proteins, followed by immunoblotting for Tyr(P) as described previously. We observed a significant increase in the level of phosphorylation of both RETMEN2A and RETMEN2B oncogenic proteins compared with full-length RET (p = 0.0282 and p = 0.0097, respectively) (Fig. 6F). The levels of phosphorylation of RETMEN2A and RETMEN2B compared with RETΔE345 showed no significant difference (p = 0.4898 and p = 0.1712, respectively). These data indicate that the RETΔE345 isoform is phosphorylated at similar levels as oncogenic RET mutations.
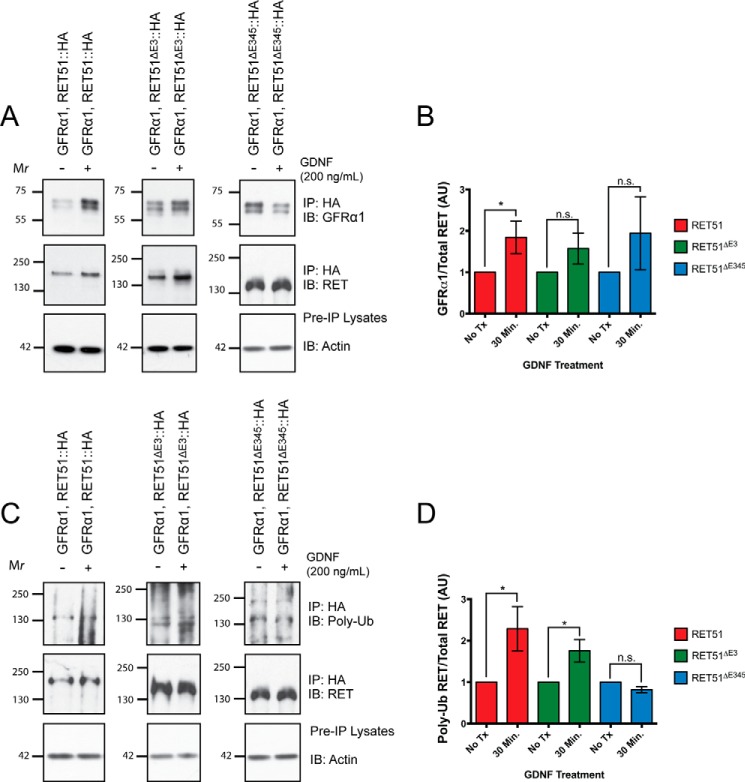
RETΔE3 Is Activated in a GDNF-dependent Manner but RETΔE345 Is Not
Although differences are observed in the level of ligand-independent activation for RETΔE3 and RETΔE345, we wondered whether these two receptors could be activated in a GDNF-dependent manner. To evaluate this, Neuro2A cells were co-transfected with HA epitope-tagged RET51, RET51ΔE3, or RET51ΔE345 and with untagged GFRα1. Twenty-four hours post-transfection, the culture medium was removed, and cells were stimulated with 200 ng/ml recombinant mouse GDNF for 30 min. Cells were detergent-extracted, and immunoprecipitations for HA were performed to isolate the exogenous RET isoforms. Immunoblotting for GFRα1 was performed to determine the level of GDNF-dependent association of GFRα1 to RET (Fig. 7A). For our positive control, where GFRα1 and RET51 were co-expressed, we observed a significant GDNF-dependent association of these proteins after 30 min of stimulation (p = 0.0373) (Fig. 7B). Under conditions where GFRα1 and RET51ΔE3 or RET51ΔE345 were co-expressed, we observed a trend of GDNF-dependent association of GFRα1 to both RET isoforms, but this ligand-dependent interaction was not statistically significant (p = 0.1750 and p = 0.3269, respectively).
FIGURE 7.
RETΔE3 is activated in a GDNF-dependent manner but RETΔE345 is not. A, HA epitope-tagged RET constructs were co-transfected with GFRα1 into Neuro2A cells. Twenty-four hours post-transfection, the medium was removed, and fresh medium was added to cells containing 200 ng/ml GDNF. Cells were stimulated for 30 min, and immunoprecipitations (IP) were performed from lysates with an HA antibody to selectively pull down the RET isoforms. Immunoblotting (IB) for GFRα1 and RET was performed, and integrated density values were calculated for each. B, quantification the of GFRα1/RET ratios. C, immunoblotting for poly-ubiquitin was performed, and integrated density values were calculated for poly-ubiquitinated RET and total RET. D, quantification of poly-ubiquitinated RET/RET ratios. n.s., not significant. AU is arbitrary units. No Tx indicates cells that were not stimulated with GDNF. Asterisks indicate p < 0.05.
To determine whether the RET isoforms were activated in a GDNF-dependent manner, immunoblotting for poly-ubiquitin was performed. We have shown previously that, upon ligand activation and autophosphorylation, RET is rapidly poly-ubiquitinated and targeted for degradation (49). Because ectopic expression of RET results in high levels of autophosphorylation in the absence of ligand, we were not able to use phosphotyrosine immunoblotting as a measure of RET activation upon GDNF stimulation. We observed a significant increase in poly-ubiquitinated RET51 (p = 0.0370) following GDNF stimulation, as expected, and also observed a significant increase in poly-ubiquitinated RET51ΔE3 (p = 0.0493), suggesting that this isoform is activated in a GDNF-dependent manner. Interestingly, we did not observe a difference in the level of poly-ubiquitinated RET51ΔE345 following GDNF stimulation (p = 0.0696). These data suggest that, like full-length RET, RETΔE3 is activated in a GDNF-dependent manner but RETΔE345 is not.
Discussion
Alternative splicing of precursor mRNA is one of many processes that mediates gene regulation in metazoans, and the frequency of alternative splicing increases with species complexity (50, 51). For example, of the ∼25,000 genes encoded by the human genome, ∼95% produce transcripts that are alternatively spliced (50, 51). By expanding the proteome through the synthesis of various protein isoforms, alternative splicing allows for increased protein diversity, with isoforms performing different biological functions.
Here we provide evidence for two alternative splicing events in Ret that, in combination with Ret9 or Ret51 alternative splicing, give rise to at least five Ret transcripts (Fig. 2C). The splicing events for RETΔE3 and RETΔE345 have only been described previously in human tissues (12). Because an additional Ret transcript, RET43, is only expressed in humans, we examined the expression of RETΔE3 and RETΔE345 in additional vertebrate tissues to determine whether these might also be human-specific transcripts (8, 52). To this end, we discovered that RETΔE3 and RETΔE345 are not human-specific RET transcripts but are also expressed in normal developing tissues of zebrafish, mice, and rats (Fig. 1C). Functionally important transcripts are transcriptionally conserved between species, arguing that RETΔE3 and RETΔE345 have physiologically important functions.
Analysis of an experimentally validated structural model for the RET cadherin-like domains 1–4 was made to evaluate the consequence of exon deletions of RET alternative splicing on the ligand binding and activation characteristics of RETΔE3 and RETΔE345 (Fig. 1B). Previous experiments indicate that the GDNF-GFRα1 complex binds directly to CLD4 and the cysteine-rich domain (53). This suggested that both RETΔE3 and RETΔE345 should be able to bind to the GDNF-GFRα1 complex, and indeed this interaction was observed not only for GFRα1 but also for GFRα2, GFRα3, and GFRα4 (Fig. 4). It should be noted that, in our experiments, the chicken GFRα4 construct contained all three cysteine-rich domains, unlike mammalian GFRα4, which lacks domain 1. Although it was originally thought that CLD1–3 of RET was involved in direct binding from results obtained through mutagenesis studies, a direct interaction between CLD1–3 and the GDNF-GFRα1 complex has not been observed (53, 54). Instead, it has been proposed that CLD1–3 contributes to the stability of the tertiary structure of the GDNF-GFRα1-RET complex by forming a secondary dimerization site to trap the GDNF-GFRα1 binary complex (14). These studies predict that RETΔE3, which forms a fused CLD1-CLD2 region, likely gives rise to a less stable tertiary GDNF-GFRα1-RET complex compared with full-length RET. This may result in a receptor variant with differential ligand binding specificities. FGF receptors are a subfamily of four receptor tyrosine kinases that are receptors for FGFs, of which there are 18 ligands (55). Alternative splicing of the FGF receptors in the region coding for the extracellular ligand-binding domain of the receptors allows for differential ligand binding. It is possible that the alternative splicing responsible for RETΔE3 similarly modifies GFL-GFRα affinity. Alternatively, the truncated extracellular domain of RETΔE3 may cause alterations in the conformational changes that occur upon GFL-GFRα binding that induces receptor autophosphorylation. This would cause different autophosphorylation kinetics in RETΔE3 compared with full-length RET and, therefore, differential activation of downstream second messenger cascades. Biochemical experiments using an in vitro system that allows for the monitoring of autophosphorylation upon direct ligand activation of RETΔE3 or full-length RET in isolation will be necessary to distinguish between these possibilities.
The large deletion in the extracellular domain of RETΔE345, including the Ca2+ binding domain, which is important for RET folding and ligand-dependent signaling, is predicted to have dramatic effects on RETΔE345 activation. Indeed, it is likely that RETΔE345 functions as a constitutively active form of RET, consistent with the mechanistic data shown in Fig. 6. Autophosphorylation of RETΔE345 is increased compared with full-length RET in the presence and absence of GFRα1 in vitro (Fig. 6B). Constitutively active receptor tyrosine kinases have not been found in normal tissues and usually arise from germline and somatic mutations, where they are most commonly found to cause neoplasia. Activating mutations in RET have been shown to be responsible for multiple endocrine neoplasia, type 2, including neoplasias such as medullary thyroid carcinoma and pheochromocytomas (56). Unlike other constitutively active forms of RET, RETΔE345 may be posttranscriptionally or translationally regulated in vivo in a manner that is not recapitulated in the in vitro system used here. In this way, the signaling of RETΔE345 may be under tight regulation for this isoform to perform specific functions in vivo.
To interrogate the functions of RETΔE3 and RETΔE345 in primary neurons and tissues, we attempted to generate isoform-specific antibodies. Likewise, we also tried to create siRNAs to selectively knock down RetΔE3 and RetΔE345 transcripts. Neither of these experimental approaches were successful at targeting RETΔE3 and RETΔE345 without also affecting full-length RET. Therefore, to determine the physiologic functions of RETΔE3 and RETΔE345, a transgenic approach in a vertebrate will be required to selectively remove these individual transcripts. This could be accomplished by deleting specific introns and fusing the flanking exons in-frame to inhibit exon skipping (e.g. remove intron 3 and fuse exons 2 and 3 together to inhibit alternative splicing for RetΔE3). How this approach would affect RET expression and other splicing events, however, is unknown. In addition, because of the invariably coincident expression of RetΔE3 and RetΔE345 in the same tissues as Ret, it is possible that full-length RET may compensate for the functions of RETΔE3 and RETΔE345. As an alternative approach, production of knockin animals in which only RetΔE3 or RetΔE3 is expressed would determine whether these isoforms are capable of supporting the normal development of the nervous system and kidneys.
Last, to gain a mechanistic understanding of the factors responsible for the alternative splicing of Ret, it would be interesting to explore RNA motifs encoding exon splicing enhancers within the locus that impact the formation of RetΔE3 and RetΔE345. Understanding the spliceosome machinery involved in this process may help us understand why the expression of these transcripts are elevated in tumors, such as in pheochromocytomas (12). Overall, it may be necessary to return to a disease model, such as neuroendocrine gland tumors, where these RET transcripts were originally identified and use the pathobiology to understand the signaling capacities of RETΔE3 and RETΔE345 to identify their functions in normal development.
Experimental Procedures
Molecular Modeling
This analysis was performed as described previously (14). Models of cadherin-like domains 1–4 were taken from PDB code 4UX8, defined by a combination of EM and small angle x-ray scattering analyses. The figure was made using PyMOL software (Schrödinger).
RNA Isolation, RT-PCR, and qPCR
Total RNA was isolated from zebrafish, mouse, or rat tissues using TRIzol reagent (Thermo Fisher Scientific) according to the instructions of the manufacturer. cDNA synthesis was performed from 1 μg of total RNA to generate poly(A) first-strand cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR kit (Life Technologies). For RT-PCR reactions with amplicons of less than 200 bp, GoTaq Green Master Mix (Promega) and species-specific oligonucleotide primers for Ret, RetΔ3, and RetΔ345 were used. RT-PCR reactions with amplicons greater than 200 bp were performed using the Phusion high-fidelity DNA polymerase kit (New England Biolabs) following the instructions of the manufacturer. qPCR was performed using FastStart Universal SYBR Green Master Mix (Roche) and run on a QuantStudio 6 Flex real-time PCR system (Thermo Fisher Scientific). Quantitative analyses were performed by calculating the ΔCt (Ct of transcript divided by Ct of the housekeeping gene actin) for each gene analyzed and transforming that value to log2. These values are reported as the relative expression.
Primer Sequences
The primers used for this study were as follows: ZF.RetFL.F, 5′-TCG TAG TTT ACG CAG CGG CTC A-3′; ZF.RetFL.R, 5′-TCG CGA TTT TCA GTG ATG TG-3′; ZF.Ret3.F, 5′-CAG TAG TTT ACG ATC TTC TGT ACC G-3′; ZF.Ret3.R, 5′-TTC AGT GTC AGT CCC GTT GA-3′; ZF.Ret345.F, 5′-CAG TAG TTT ACA GCT GAA ACT CAG TC-3′; ZF.Ret345.R, 5′-TGA CAT TGG AGA AGC GAA TG-3′; Mouse.RetFL.F, 5′-AGC ATC CGC AAT GGT GGT TT-3′; Mouse.RetFL.R, 5′-TGT TCT CCC TGA CTC GGA AG-3′; Mouse.Ret3.F, 5′-AGC ATC CGC AGG GAT AGT CT-3′; Mouse.Ret3.R, 5′-ACA CTG TCA CTG GGA AGG AC-3′; Mouse.Ret345.F, 5′-AGC ATC CGC AAG CTG ATT CT-3′; Mouse.Ret345.R, 5′-TTC ACT GGG AAG GAG TAG GC-3′; Rat.RetFL.F, 5′-AGC ATC CGC AAT GGC GGC TT-3′; Rat.RetFL.R, 5′-TAG CAT GCG GAA CTG GTA GA-3′; Rat.Ret3.F, 5′-AGC ATC CGC AGG GAC GGT CT-3′; Rat.Ret3.R, 5′-CCG CTT AAA CTC CAC CAC AG-3′; Rat.Ret345.F, 5′-AGC ATC CGC AAG CTG GTT CT-3′; Rat.Ret345.R, 5′-TAG GCC ATG GGT AGG TTC AG-3′; Mouse.Ret9.R, 5′-ATT TAC TGT CCA TTG CAA GGC-3′; and Mouse.Ret51.R, 5′-CCT ATC AGT GCT TTA AGT CTG-3′.
Mice
Wild-type C57BL/6J mice were purchased from The Jackson Laboratories (Bar Harbor, ME). Ret−/− mice have been described previously and were maintained in a mixed genetic background (57). For timed matings, noon of the day on which a vaginal plug was detected was considered E0.5. All housing and procedures performed on mice were approved by the University of Michigan Animal Care and Use Committee.
Detergent Extraction and Preclearing of Whole Tissues
Tissues were harvested, placed in a 2-ml tube with 250 μl of immunoprecipitation buffer lacking Nonidet P-40 (TBS (pH 7.4), 10% glycerol, 500 μm sodium vanadate, and protease inhibitors) along with a steel grinding ball (5 mm, 69989, Qiagen, Valencia, CA) and mechanically homogenized using a TissueLyzer II (Qiagen) for 2 min at a frequency of 20 Hz. Following this, 250 μl of 2% Nonidet P-40-containing immunoprecipitation buffer was added to homogenates and incubated for 1 h at 4 °C under gentle agitation. Homogenates were then centrifuged for 10 min at 16,000 × g and subjected to an initial preclearing step with protein A and protein G alone at 4 °C for 2 h under gentle agitation. A second preclearing step was performed with protein A, protein G, and a species-matched nonspecific control IgG for 2 h under gentle agitation.
Immunoprecipitations and Immunoblotting
Plates were placed on ice, gently washed twice with 1× PBS (pH 7.4), and lysed with modified radioimmune precipitation assay buffer (TBS (pH 7.4), 10% glycerol, 1% Triton X-100, 0.1% SDS, 500 μm sodium vanadate, and protease inhibitors). Proteins were immunoprecipitated using anti-RET9 (goat, Santa Cruz Biotechnology), anti-RET51 (goat, Santa Cruz Biotechnology), anti-HA (mouse, Millipore), or anti-FLAG (rabbit, Cell Signaling Technology) selective antibodies. Immunoprecipitates were subjected to SDS-PAGE, followed by electroblotting onto PVDF membranes (Immobilon P, Millipore). Immunoblotting was performed on blots using antibodies selective for RET (1:500, goat, R&D Systems), HA (1:5000, rabbit, Cell Signaling Technology), Trk (1:1000, rabbit, Santa Cruz Biotechnology), phospho-tyrosine (1:1000, mouse, Millipore), phospho-RET (Tyr905) (1:500, rabbit), phospho-RET (Tyr1062, 1:500, rabbit), GFRα1 (1:500, goat, R&D Systems), or poly-ubiquitin (1:500, mouse, Enzo Life Sciences). Phospho-RET (Tyr905) and phospho-RET (Tyr1062) have been described previously (58). Lysates collected prior to immunoprecipitations served as loading controls to assess protein expression levels and were also subjected to immunoblotting for actin (1:1000, goat, Santa Cruz Biotechnology). Each biochemical experiment was performed three to four times with similar results.
Mammalian Cell Culture and Transfections
NIH/3T3 (ATCC), HEK293T (ATCC), and Neuro2A cells (ATCC) were maintained at 37 °C with 5% CO2 in DMEM supplemented with FBS (10% by volume) and a penicillin/streptomycin/glutamine mixture. Cells were plated into 6-well tissue culture plates and allowed to proliferate until cells were 70% confluent. Transfections of cells were performed using Lipofectamine 2000 according to the instructions of the manufacturer (Invitrogen). For experiments testing the interaction of proteins, a total of 4 μg of plasmid DNA was added per well using a GFP plasmid as a control for transfecting equal amounts of DNA. For in vitro ligand-independent activation experiments, 2 μg of HA::GFRα1 and 1 μg of one of the RET constructs were transfected for a total of 3 μg of DNA per condition, with a GFP plasmid used as a control for transfecting equal amounts of DNA. RET51ΔE3 and RET51ΔE345 constructs were subcloned from RET51 constructs into pcDNA3.1 (Thermo Fisher Scientific). A Ret51 construct encoding mouse RET51 fused at the C terminus with an HA epitope tag (RET51::HA) was acquired from Dr. Ben Allen. This plasmid was subcloned to create similarly tagged Ret51ΔE3 and Ret51ΔE345 constructs, which were subcloned into pLentilox-IRES-GFP (University of Michigan Vector Core). For in vitro ligand-dependent activation experiments, 3 μg of HA epitope-tagged Ret51, Ret51ΔE3, or Ret51ΔE345 was co-transfected with 3 μg of untagged GFRα1. Twenty-four hours post-transfection, the medium was aspirated, and cells were treated with 200 ng/ml recombinant mouse GDNF (Peprotech) in fresh cell culture medium for 30 min. The HA::GFRα1 (mouse) plasmid was generously donated by Dr. Ben Allen. The HA::GFRα2 (rat), HA::GFRα3 (mouse), and HA::GFRα4 (chicken) plasmids were generously donated by Dr. Carlos Ibáñez. The TrkA (rat) plasmid was generously donated by Dr. Christin Carter-Su. Constructs encoding human RET51 harboring MEN2A (C634R) and MEN2B (M918T) point mutations in RET were created using the QuikChange II XL site-directed mutagenesis kit (Agilent).
Quantification of Immunoblots
Scanned images of x-ray films were imported into ImageJ (National Institutes of Health) and processed using the gel analysis tool. Integrated density values obtained from immunoblotting were reported as mean ± S.E. with arbitrary units on the vertical axis. Values were normalized to the level of RET51 phosphorylation in each experiment. Statistical analyses were performed using Student's t test for each experiment, and all biochemical experiments were performed at least three times with similar results.
Immunocytochemistry
Transfected cells that were plated on glass coverslips were fixed for 8 min in 4% paraformaldehyde. Cells were washed twice in 1× PBS and briefly blocked in immunofluorescence blocking solution (3% BSA and 1% normal donkey serum in 1× PBS (pH 7.4)) with or without 0.1% Triton X-100. Primary antibodies were diluted in blocking buffer (RET (1:50, goat, R&D Systems), HA (1:1000, mouse, Millipore)), and slides were incubated at 4 °C overnight. Cells were washed twice with 1× PBS and then incubated at room temperature for 1 h with secondary antibodies (donkey anti-mouse Alexa Fluor 633 (Thermo Fisher Scientific), donkey anti-goat 543 (Biotium)) diluted in blocking solution. Last, cells were washed three times in 1× PBS and mounted with DAPI Fluoromount-G (Southern Biotech). Cells were imaged using a Zeiss Axiovert 200 M epifluorescence microscope with a ×40 objective.
Author Contributions
N. A. G. conducted the experiments, analyzed the results, and wrote the paper. J. K. V. and S. S. N. performed experiments. N. Q. M. performed the structure-based predictions of the impact of exon deletions. B. A. P. conceived the idea for the project, contributed intellectual advice, and edited the manuscript.
Acknowledgments
We thank Dr. Benjamin Allen for reagents and critical scientific comments on the manuscript. Additionally, we thank Dr. Carlos Ibáñez, Dr. Christin Carter-Su, and Dr. Frank Costantini for reagents. We also thank the Pierchala laboratory for helpful scientific discussions.
This work was supported by National Institutes of Health Grant R01 NS089585 (to B. A. P.). Additional funding support came from the University of Michigan Cellular and Molecular Biology Predoctoral Fellowship T32-GM007315 (to N. A. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GDNF
- glial cell line-derived neurotrophic factor
- GFL
- glial cell line-derived neurotrophic factor family ligand
- GFR
- glial cell line-derived neurotrophic factor family receptor
- MEN
- multiple endocrine neoplasia
- E19.5
- embryonic day 19.5
- CLD
- cadherin-like domain
- DRG
- dorsal root ganglion/ganglia
- SCG
- superior cervical ganglion/ganglia
- qPCR
- quantitative PCR
- P3
- postnatal day 3
- RET
- rearranged during transfection
- IRES
- internal ribosome entry site.
References
- 1. Airaksinen M. S., and Saarma M. (2002) The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3, 383–394 [DOI] [PubMed] [Google Scholar]
- 2. Baloh R. H., Enomoto H., Johnson E. M. Jr., and Milbrandt J. (2000) The GDNF family ligands and receptors-implications for neural development. Curr. Opin. Neurobiol. 10, 103–110 [DOI] [PubMed] [Google Scholar]
- 3. Meng X., Lindahl M., Hyvönen M. E., Parvinen M., de Rooij D. G., Hess M. W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M., Pichel J. G., Westphal H., Saarma M., and Sariola H. (2000) Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493 [DOI] [PubMed] [Google Scholar]
- 4. Naughton C. K., Jain S., Strickland A. M., Gupta A., and Milbrandt J. (2006) Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol. Reprod. 74, 314–321 [DOI] [PubMed] [Google Scholar]
- 5. Airaksinen M. S., Titievsky A., and Saarma M. (1999) GDNF family neurotrophic factor signaling: four masters, one servant? Mol. Cell. Neurosci. 13, 313–325 [DOI] [PubMed] [Google Scholar]
- 6. Wells S. A. Jr., Santoro M. (2009) Targeting the RET pathway in thyroid cancer. Clin. Cancer Res. 15, 7119–7123 [DOI] [PubMed] [Google Scholar]
- 7. de Graaff E., Srinivas S., Kilkenny C., D'Agati V., Mankoo B. S., Costantini F., and Pachnis V. (2001) Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 15, 2433–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carter M. T., Yome J. L., Marcil M. N., Martin C. A., Vanhorne J. B., and Mulligan L. M. (2001) Conservation of RET proto-oncogene splicing variants and implications for RET isoform function. Cytogenet. Cell Genet. 95, 169–176 [DOI] [PubMed] [Google Scholar]
- 9. Tahira T., Ishizaka Y., Itoh F., Sugimura T., and Nagao M. (1990) Characterization of ret proto-oncogene mRNAs encoding two isoforms of the protein product in a human neuroblastoma cell line. Oncogene 5, 97–102 [PubMed] [Google Scholar]
- 10. Tsui-Pierchala B. A., Milbrandt J., and Johnson E. M. Jr. (2002) NGF utilizes c-Ret via a novel GFL-independent, inter-RTK signaling mechanism to maintain the trophic status of mature sympathetic neurons. Neuron 33, 261–273 [DOI] [PubMed] [Google Scholar]
- 11. Tsui C. C., and Pierchala B. A. (2010) The differential axonal degradation of Ret accounts for cell-type-specific function of glial cell line-derived neurotrophic factor as a retrograde survival factor. J. Neurosci. 30, 5149–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lorenzo M. J., Eng C., Mulligan L. M., Stonehouse T. J., Healey C. S., Ponder B. A., and Smith D. P. (1995) Multiple mRNA isoforms of the human RET proto-oncogene generated by alternative splicing. Oncogene 10, 1377–1383 [PubMed] [Google Scholar]
- 13. Anders J., Kjar S., and Ibáñez C. F. (2001) Molecular modeling of the extracellular domain of the RET receptor tyrosine kinase reveals multiple cadherin-like domains and a calcium-binding site. J. Biol. Chem. 276, 35808–35817 [DOI] [PubMed] [Google Scholar]
- 14. Goodman K. M., Kjær S., Beuron F., Knowles P. P., Nawrotek A., Burns E. M., Purkiss A. G., George R., Santoro M., Morris E. P., and McDonald N. Q. (2014) RET recognition of GDNF-GFRα1 ligand by a composite binding site promotes membrane-proximal self-association. Cell Rep. 8, 1894–1904 [DOI] [PubMed] [Google Scholar]
- 15. Kjaer S., Hanrahan S., Totty N., and McDonald N. Q. (2010) Mammal-restricted elements predispose human RET to folding impairment by HSCR mutations. Nat. Struct. Mol. Biol. 17, 726–731 [DOI] [PubMed] [Google Scholar]
- 16. Arenas E., Trupp M., Akerud P., and Ibáñez C. F. (1995) GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron 15, 1465–1473 [DOI] [PubMed] [Google Scholar]
- 17. Horger B. A., Nishimura M. C., Armanini M. P., Wang L. C., Poulsen K. T., Rosenblad C., Kirik D., Moffat B., Simmons L., Johnson E. Jr., Milbrandt J., Rosenthal A., Bjorklund A., Vandlen R. A., Hynes M. A., and Phillips H. S. (1998) Neurturin exerts potent actions on survival and function of midbrain dopaminergic neurons. J. Neurosci. 18, 4929–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonanomi D., Chivatakarn O., Bai G., Abdesselem H., Lettieri K., Marquardt T., Pierchala B. A., and Pfaff S. (2012) Ret is a multifunctional coreceptor that integrates diffusible- and contact- axon guidance signals. Cell 148, 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shneider N. A., Brown M. N., Smith C. A., Pickel J., and Alvarez F. J. (2009) γ Motor neurons express distinct genetic markers at birth and require muscle spindle-derived GDNF for postnatal survival. Neural Dev. 4, 10.1186/1749-8104-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gould T. W., Yonemura S., Oppenheim R. W., Ohmori S., and Enomoto H. (2008) The neurotrophic effects of glial cell line-derived neurotrophic factor on spinal motoneurons are restricted to fusimotor subtypes. J. Neurosci. 28, 2131–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kramer E. R., Knott L., Su F., Dessaud E., Krull C. E., Helmbacher F., and Klein R. (2006) Cooperation between GDNF/Ret and ephrinA/EphA4 signals for motor-axon pathway selection in the limb. Neuron 50, 35–47 [DOI] [PubMed] [Google Scholar]
- 22. Luo W., Enomoto H., Rice F. L., Milbrandt J., and Ginty D. D. (2009) Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron 64, 841–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bourane S., Garces A., Venteo S., Pattyn A., Hubert T., Fichard A., Puech S., Boukhaddaoui H., Baudet C., Takahashi S., Valmier J., and Carroll P. (2009) Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by Ret signaling. Neuron 64, 857–870 [DOI] [PubMed] [Google Scholar]
- 24. Molliver D. C., Wright D. E., Leitner M. L., Parsadanian A. S., Doster K., Wen D., Yan Q., and Snider W. D. (1997) IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron 19, 849–861 [DOI] [PubMed] [Google Scholar]
- 25. Luo W., Wickramasinghe S. R., Savitt J. M., Griffin J. W., Dawson T. M., and Ginty D. D. (2007) A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron 54, 739–754 [DOI] [PubMed] [Google Scholar]
- 26. Franck M. C., Stenqvist A., Li L., Hao J., Usoskin D., Xu X., Wiesenfeld-Hallin Z., and Ernfors P. (2011) Essential role of Ret for defining non-peptidergic nociceptor phenotypes and function in the adult mouse. Eur. J. Neurosci. 33, 1385–1400 [DOI] [PubMed] [Google Scholar]
- 27. Nishino J., Mochida K., Ohfuji Y., Shimazaki T., Meno C., Ohishi S., Matsuda Y., Fujii H., Saijoh Y., and Hamada H. (1999) GFR α3, a component of the artemin receptor, is required for migration and survival of the superior cervical ganglion. Neuron 23, 725–736 [DOI] [PubMed] [Google Scholar]
- 28. Enomoto H., Crawford P. A., Gorodinsky A., Heuckeroth R. O., Johnson E. M. Jr., and Milbrandt J. (2001) RET signaling is essential for migration, axonal growth and axon guidance of developing sympathetic neurons. Development 128, 3963–3974 [DOI] [PubMed] [Google Scholar]
- 29. Honma Y., Araki T., Gianino S., Bruce A., Heuckeroth R., Johnson E., and Milbrandt J. (2002) Artemin is a vascular-derived neurotrophic factor for developing sympathetic neurons. Neuron 35, 267–282 [DOI] [PubMed] [Google Scholar]
- 30. Moore M. W., Klein R. D., Fariñas I., Sauer H., Armanini M., Phillips H., Reichardt L. F., Ryan A. M., Carver-Moore K., and Rosenthal A. (1996) Renal and neuronal abnormalities in mice lacking GDNF. Nature 382, 76–79 [DOI] [PubMed] [Google Scholar]
- 31. Pichel J. G., Shen L., Sheng H. Z., Granholm A. C., Drago J., Grinberg A., Lee E. J., Huang S. P., Saarma M., Hoffer B. J., Sariola H., and Westphal H. (1996) Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382, 73–76 [DOI] [PubMed] [Google Scholar]
- 32. Sánchez M. P., Silos-Santiago I., Frisén J., He B., Lira S. A., and Barbacid M. (1996) Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382, 70–73 [DOI] [PubMed] [Google Scholar]
- 33. Cacalano G., Fariñas I., Wang L. C., Hagler K., Forgie A., Moore M., Armanini M., Phillips H., Ryan A. M., Reichardt L. F., Hynes M., Davies A., and Rosenthal A. (1998) GFRα1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron 21, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Enomoto H., Araki T., Jackman A., Heuckeroth R. O., Snider W. D., Johnson E. M. Jr., and Milbrandt J. (1998) GFR α1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron 21, 317–324 [DOI] [PubMed] [Google Scholar]
- 35. Kjaer S., Kurokawa K., Perrinjaquet M., Abrescia C., and Ibáñez C. F. (2006) Self-association of the transmembrane domain of RET underlies oncogenic activation by MEN2A mutations. Oncogene 25, 7086–7095 [DOI] [PubMed] [Google Scholar]
- 36. Durick K., Wu R.-Y., Gill G. N., and Taylor S. S. (1996) Mitogenic signaling by Ret/ptc2 requires association with Enigma via a LIM domain. J. Biol. Chem. 271, 12691–12694 [DOI] [PubMed] [Google Scholar]
- 37. Pandey A., Duan H., Di Fiore P. P., and Dixit V. M. (1995) The Ret receptor protein tyrosine kinase associates with the SH2-containing adapter protein Grb10. J. Biol. Chem. 270, 21461–21463 [DOI] [PubMed] [Google Scholar]
- 38. Pandey A., Liu X., Dixon J. E., Di Fiore P. P., and Dixit V. M. (1996) Direct association between the Ret receptor tyrosine kinase and the Src homology 2-containing adapter protein Grb7. J. Biol. Chem. 271, 10607–10610 [DOI] [PubMed] [Google Scholar]
- 39. Iwashita T., Asai N., Murakami H., Matsuyama M., and Takahashi M. (1996) Identification of tyrosine residues that are essential for transforming activity of the ret proto-oncogene with MEN2A or MEN2B mutation. Oncogene 12, 481–487 [PubMed] [Google Scholar]
- 40. Lorenzo M. J., Gish G. D., Houghton C., Stonehouse T. J., Pawson T., Ponder B. A., and Smith D. P. (1997) RET alternative splicing influences the interaction of activated RET with the SH2 and PTB domains of Shc, and the SH2 domain of Grb2. Oncogene 14, 763–771 [DOI] [PubMed] [Google Scholar]
- 41. Asai N., Murakami H., Iwashita T., and Takahashi M. (1996) A mutation at tyrosine 1062 in MEN2A-Ret and MEN2B-Ret impairs their transforming activity and association with shc adaptor proteins. J. Biol. Chem. 271, 17644–17649 [DOI] [PubMed] [Google Scholar]
- 42. Arighi E., Alberti L., Torriti F., Ghizzoni S., Rizzetti M. G., Pelicci G., Pasini B., Bongarzone I., Piutti C., Pierotti M. A., and Borrello M. G. (1997) Identification of Shc docking site on Ret tyrosine kinase. Oncogene 14, 773–782 [DOI] [PubMed] [Google Scholar]
- 43. Grimm J., Sachs M., Britsch S., Di Cesare S., Schwarz-Romond T., Alitalo K., and Birchmeier W. (2001) Novel p62dok family members, dok-4 and dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J. Cell Biol. 154, 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hayashi H., Ichihara M., Iwashita T., Murakami H., Shimono Y., Kawai K., Kurokawa K., Murakumo Y., Imai T., Funahashi H., Nakao A., and Takahashi M. (2000) Characterization of intracellular signals via tyrosine 1062 in RET activated by glial cell line-derived neurotrophic factor. Oncogene 19, 4469–4475 [DOI] [PubMed] [Google Scholar]
- 45. Kurokawa K., Iwashita T., Murakami H., Hayashi H., Kawai K., and Takahashi M. (2001) Identification of SNT/FRS2 docking site on RET receptor tyrosine kinase and its role for signal transduction. Oncogene 20, 1929–1938 [DOI] [PubMed] [Google Scholar]
- 46. Melillo R. M., Santoro M., Ong S. H., Billaud M., Fusco A., Hadari Y. R., Schlessinger J., and Lax I. (2001) Docking protein FRS2 links the protein tyrosine kinase RET and its oncogenic forms with the mitogen-activated protein kinase signaling cascade. Mol. Cell. Biol. 21, 4177–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Melillo R. M., Carlomagno F., De Vita G., Formisano P., Vecchio G., Fusco A., Billaud M., and Santoro M. (2001) The insulin receptor substrate (IRS)-1 recruits phosphatidylinositol 3-kinase to Ret: evidence for a competition between Shc and IRS-1 for the binding to Ret. Oncogene 20, 209–218 [DOI] [PubMed] [Google Scholar]
- 48. Besset V., Scott R. P., and Ibáñez C. F. (2000) Signaling complexes and protein-protein interactions involved in the activation of the Ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J. Biol. Chem. 275, 39159–39166 [DOI] [PubMed] [Google Scholar]
- 49. Pierchala B. A., Milbrandt J., and Johnson E. M. Jr. (2006) Glial cell line-derived neurotrophic factor-dependent recruitment of Ret into lipid rafts enhances signaling by partitioning Ret from proteasome-dependent degradation. J. Neurosci. 26, 2777–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pan Q., Shai O., Lee L. J., Frey B. J., and Blencowe B. J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 51. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., and Burge C. B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Myers S. M., Eng C., Ponder B. A., and Mulligan L. M. (1995) Characterization of RET proto-oncogene 3′ splicing variants and polyadenylation sites: a novel C-terminus for RET. Oncogene 11, 2039–2045 [PubMed] [Google Scholar]
- 53. Amoresano A., Incoronato M., Monti G., Pucci P., de Franciscis V., and Cerchia L. (2005) Direct interactions among Ret, GDNF, and GFRα1 molecules reveal new insights into the assembly of a functional three-protein complex. Cell Signal. 17, 717–727 [DOI] [PubMed] [Google Scholar]
- 54. Kjaer S., and Ibáñez C. F. (2003) Identification of a surface for binding to the GDNF-GFR α 1 complex in the first cadherin-like domain of RET. J. Biol. Chem. 278, 47898–47904 [DOI] [PubMed] [Google Scholar]
- 55. Turner N., and Grose R. (2010) Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 56. Mulligan L. (2014) RET revisited: expanding the oncogenic portfolio. Nat. Rev. Cancer 14, 173–186 [DOI] [PubMed] [Google Scholar]
- 57. Schuchardt A., D'Agati V., Larsson-Blomberg L., Costantini F., and Pachnis V. (1994) Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380–383 [DOI] [PubMed] [Google Scholar]
- 58. Tsui-Pierchala B. A., Ahrens R. C., Crowder R. J., Milbrandt J., and Johnson E. M. Jr. (2002) The long and short isoforms of Ret function as independent signaling complexes. J. Biol. Chem. 277, 34618–34625 [DOI] [PubMed] [Google Scholar]