Abstract
Phosphodiesterase 6 (PDE6) is the effector enzyme in the phototransduction cascade and is critical for the health of both rod and cone photoreceptors. Its dysfunction, caused by mutations in either the enzyme itself or AIPL1 (aryl hydrocarbon receptor-interacting protein-like 1), leads to retinal diseases culminating in blindness. Progress in research on PDE6 and AIPL1 has been severely hampered by failure to express functional PDE6 in a heterologous expression system. Here, we demonstrated that AIPL1 is an obligate chaperone of PDE6 and that it enables low yield functional folding of cone PDE6C in cultured cells. We further show that the AIPL1-mediated production of folded PDE6C is markedly elevated in the presence of the inhibitory Pγ-subunit of PDE6. As illustrated in this study, a simple and sensitive system in which AIPL1 and Pγ are co-expressed with PDE6 represents an effective tool for probing structure-function relationships of AIPL1 and reliably establishing the pathogenicity of its variants.
Keywords: chaperone, phosphodiesterases, photoreceptor, phototransduction, protein folding
Introduction
Cyclic nucleotide phosphodiesterases of the sixth family (PDE6)2 are the key effectors in the visual transduction cascade in rod and cone photoreceptors. In the dark, activity of the PDE6 catalytic dimers is restrained by two tightly bound inhibitory γ-subunits (Pγ). This allows cGMP to maintain depolarizing “dark” current through a cGMP-gated channel in the photoreceptor plasma membrane. Photoexcitation leads to G-protein-mediated activation of PDE6 followed by a drop in cytoplasmic cGMP, channel closure, and propagation of an electrical signal to downstream retinal neurons (1, 2). In addition to being essential to photoreceptor physiology, PDE6 is critical to the health and survival of rods and cones. Malfunctions caused by mutations in genes that encode either PDE6 or its putative chaperone, aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), lead to severe blinding retinal diseases. Mutations in the PDE6A, PDE6B, and PDE6G genes, which encode the catalytic PDE6AB subunits and Pγ of rod PDE6, respectively, are responsible for a significant proportion of cases of recessive retinitis pigmentosa (3–5) and can also lead to autosomal dominant congenital stationary night blindness (6). Mutations in their cone counterparts, PDE6C and PDE6H, cause autosomal recessive achromatopsia (7–10).
PDE6 deficiency also appears to underlie one of the most severe forms of Leber congenital amaurosis (LCA type 4), a condition caused by mutations in the AIPL1 gene (11, 12). LCA is an early onset inherited retinopathy and one of the main causes of blindness in children (13). The link between PDE6 and AIPL1 was discovered in studies of AIPL1 knock-out and knockdown mouse models, which revealed rapid severe retinal degeneration and a marked reduction in PDE6 protein levels and activity prior to the loss of photoreceptor cells (14, 15). Thus, the animal models recapitulated the hallmarks of LCA and suggested that AIPL1 is a potential chaperone of PDE6. This notion is consistent with the fact that AIPL1 contains an FK506-binding protein (FKBP) domain as well as a tetratricopeptide (TPR) domain, both of which are often found in proteins with chaperone activity (Fig. 1) (16, 17). Notably, AIPL1 (hence its name) shares domain organization and 50% sequence identity (71% similarity) with AIP (aryl hydrocarbon receptor-interacting protein) (11). AIP is expressed in various tissues where it acts as a co-chaperone with HSP90 in the maturation of aryl hydrocarbon receptor and other nuclear receptors (18). However, AIPL1 has never directly been shown to act as a chaperone for PDE6 or any other protein.
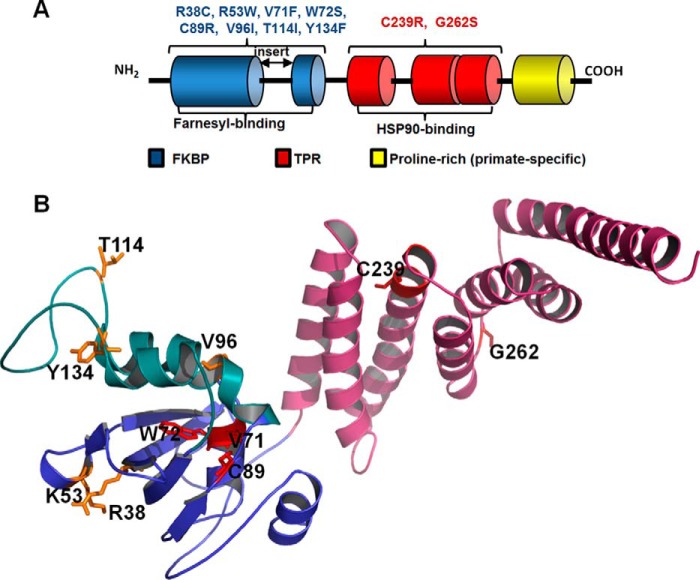
FIGURE 1.
Domains and LCA4 linked mutations of AIPL1. A, schematic representation of the domain structure of AIPL1. All vertebrate AIPL1 proteins consist of an FKBP domain (blue) and a TPR domain with three tetratricopeptide repeats (red). In addition, primate AIPL1 proteins contain a proline-rich region of unknown function (yellow). The “insert” region, which links the last two β-strands in the core FKBP domain, distinguishes the FKBP domains of AIPL1 and AIP from the classical FKBP domain of FKBP12. The FKBP domain of AIPL1 interacts with the prenyl modifications of PDE6, whereas the TPR domain binds HSP90. The diagram indicates which LCA4-linked mutations of AIPL1 were analyzed in this study. B, residues mutated in LCA patients and investigated in this study mapped to the solution structure of mouse AIPL1 (32). Features include the core FKBP domain (blue), the “insert” region (green), and the TPR domain (pink). Residues substituted in AIPL1 mutants that failed to chaperone PDE6 (V71F, W72S, C89R, and C239R) are shown as red sticks; residues substituted in mutant forms that are apparently benign are shown as yellow sticks.
For many years, progress on understanding the structure-function relationships of PDE6 and AIPL1, as well as the mechanisms whereby mutant forms of these proteins contribute to retinal diseases, has been impeded by the inability to express functional PDE6 in a heterologous system (15, 19–22). Here we report a pioneering yet remarkably simple PDE6 expression system in cultured HEK293T cells. Specifically, we used cone PDE6C, which functions as a homodimer. Our results demonstrate that AIPL1 is absolutely necessary for the expression of active PDE6. Moreover, the AIPL1-dependent production of functional enzyme is markedly augmented in the presence of Pγ, revealing a novel role for the latter in the maturation of PDE6.
Using heterologous expression of PDE6C as a unique readout of AIPL1 chaperone function, we demonstrate exclusive contributions of the FKBP and TPR domains to the folding of PDE6. Given that a high degree of polymorphism in the AIPL1 gene has made it difficult to reliably establish disease causation (23, 24), we also screened a panel of AIPL1 mutants linked to LCA, identifying those that failed to chaperone PDE6 and those that retain the ability to fold PDE6, with some behaving differently than suggested based on computational analysis. Thus, this novel system is an excellent tool for validating the pathogenicity of AIPL1 mutations.
Results
AIPL1 Is an Obligatory Chaperone of PDE6 and Is Assisted by Pγ
Our approach to developing an expression system for PDE6 was based on the hypothesis that the known failure of PDE6 to fold into a functional conformation outside of photoreceptors is due to a requirement for specific chaperone proteins. An obvious first candidate chaperone for PDE6 is AIPL1 (14, 15). To explore the requirements for expression of functional PDE6 in a heterologous system, we cloned sequences encoding the FLAG-tagged human PDE6C (FLAG-PDE6C), EGFP-tagged mouse cone Pγ (EGFP-Pγ), and HA-tagged mouse AIPL1 (AIPL1-HA) into the pcDNA3.1 vector. HEK293T cells were transfected with PDE6C, alone or in combination with Pγ, AIPL1, or both. 48 h post-transfection, the cells were examined by immunofluorescence microscopy, and lysates were prepared and analyzed for protein expression and PDE6 activity. When PDE6C was expressed alone, its signal was observed throughout the cell except in the nucleus (Fig. 2A). The intracellular localization of PDE6C did not change noticeably on co-expression with AIPL1, either alone (Fig. 2B) or in conjunction with Pγ (Fig. 2C). AIPL1 was present mainly in the cytoplasm, but also in the nucleus (excluding the nucleoli) (Fig. 2, B and C). EGFP-Pγ was distributed throughout the cytoplasm and the nucleus (Fig. 2C). Western blotting revealed that the expression of PDE6C was similar in all lysates (Fig. 2D). PDE activity assays indicated that in the PDE6C and PDE6C+Pγ lysates, cGMP hydrolysis did not exceed the very low levels in lysates from untransfected HEK293T cells (∼7 pmol cGMP hydrolyzed/mg protein/min) (Fig. 2E). To determine whether the inhibitory Pγ-subunit possibly masked PDE6C activity in the PDE6C+Pγ lysate, this lysate was subjected to limited treatment with trypsin. Trypsin selectively degrades Pγ, thus relieving full enzymatic activity of native PDE6 (25, 26). Trypsin treatment had no effect, indicating that no functional PDE6C was present in the PDE6C+Pγ lysates (Fig. 2E). However, in the PDE6C + AIPL1 lysates, the rate of cGMP hydrolysis was ∼20-fold higher than that in lysates from untransfected cells (Fig. 2E). These results suggest that AIPL1 is absolutely required for heterologous expression of active PDE6C. Notably, Pγ enhanced the AIPL1-assisted expression of functional PDE6C by ∼160-fold, as was evident from limited treatment of the PDE6C + AIPL1 + Pγ lysate with trypsin as compared with the PDE6C+AIPL1 lysate (Fig. 2E). Because Pγ does not affect the levels of PDE6C protein, our results suggest that it dramatically increases the proportion of correctly folded, functional PDE6 produced in the presence of AIPL1. The key characteristics of recombinant PDE6C in the PDE6C + AIPL1 + Pγ lysate (Km cGMP 15.6 ± 1.5 μm, Ki(Pγ) 94 ± 14 pm; mean ± S.E., n = 3) after trypsin treatment were comparable with those reported for native trypsin-activated PDE6C (27, 28). Given the effect of Pγ on the production of functional PDE6, the expression system was simplified for subsequent studies by subcloning FLAG-PDE6C and EGFP-Pγ into a single pcDNA3.1 vector with the two DNA sequences separated by an internal ribosome entry site (IRES).
FIGURE 2.
Expression of functional PDE6 requires AIPL1 and is markedly enhanced by Pγ. A–C, confocal immunofluorescence images of HEK293T cells transfected with PDE6C alone (red, anti-FLAG; blue, TO-PRO3 nuclear stain) (A), co-transfected with PDE6C (red, anti-FLAG) and AIPL1 (green, anti-HA) (B), or co-transfected with PDE6C (red, anti-PDE6C), AIPL1 (blue, anti-HA), and Pγ (green, EGFP fluorescence) (C). D, Western blot analysis of lysates of HEK293T cells transfected with PDE6C, PDE6C + Pγ, PDE6C + AIPL1, and PDE6C + AIPL1 + Pγ using anti-FLAG (PDE6C), anti-EGFP (Pγ), and anti-HA (AIPL1) antibodies. Lanes contain equal amounts of protein. E, cGMP hydrolysis in lysates of transfected HEK293T cells. UN, untransfected control; (t), limited treatment with trypsin to remove Pγ (mean ± S.E., n ≥ 3).
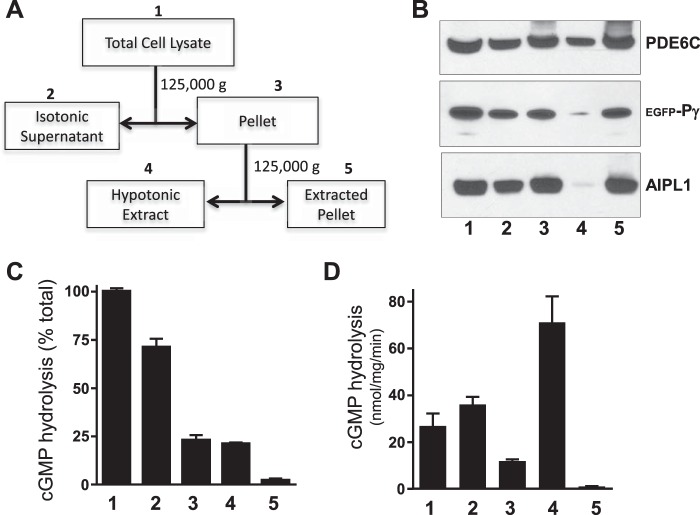
PDE6 is anchored to the membrane by C-terminal isoprenylation of its catalytic subunits (29), and it can be dissociated from photoreceptor membranes using hypotonic buffer (30). However, whereas rod PDE6 is predominantly found at the membrane, the majority of cone PDE6C is found in soluble retinal fractions (27). Under our experimental conditions, ∼25% of PDE6C activity was associated with the membranes of HEK293T cells in isotonic buffer (Fig. 3, A and C), but most was released into hypotonic buffer (Fig. 3, A and C). Specific PDE6C activity was highest in the hypotonic extract (Fig. 3D, fraction 4). Using immunoblotting with PDE6com antibodies (31), we matched the PDE6C protein level in fraction 4 with the standard preparation of bovine trypsin-activated PDE6AB and measured the respective PDE6 activities. Hence, we estimated that ∼50–60% of PDE6C in fraction 4 is functional (not shown). Inactive/misfolded PDE6C protein was trapped along with AIPL1 and Pγ in the hypotonically extracted membranes (Fig. 3B). Thus, the membrane binding properties of functional recombinant PDE6C in HEK293T cells are similar to those of the native enzyme.
FIGURE 3.
Distribution of folded PDE6C between membrane and soluble fractions of transfected HEK293T cells. A, scheme for fractionation of lysates from HEK293T cells co-transfected with PDE6C, AIPL1 and Pγ. Details are provided under “Experimental Procedures.” B, Western blot analysis of PDE6C, AIPL1, and Pγ in the fractions obtained as outlined in A. C and D, cGMP hydrolysis in each fraction following limited trypsin treatment to remove Pγ expressed as a percentage of that in total HEK293 cell lysates (C) or activity normalized to protein content in each fraction (D) (mean ± S.E., n = 3).
The FKBP and TPR Domains of AIPL1 Uniquely Contribute to the Folding of PDE6
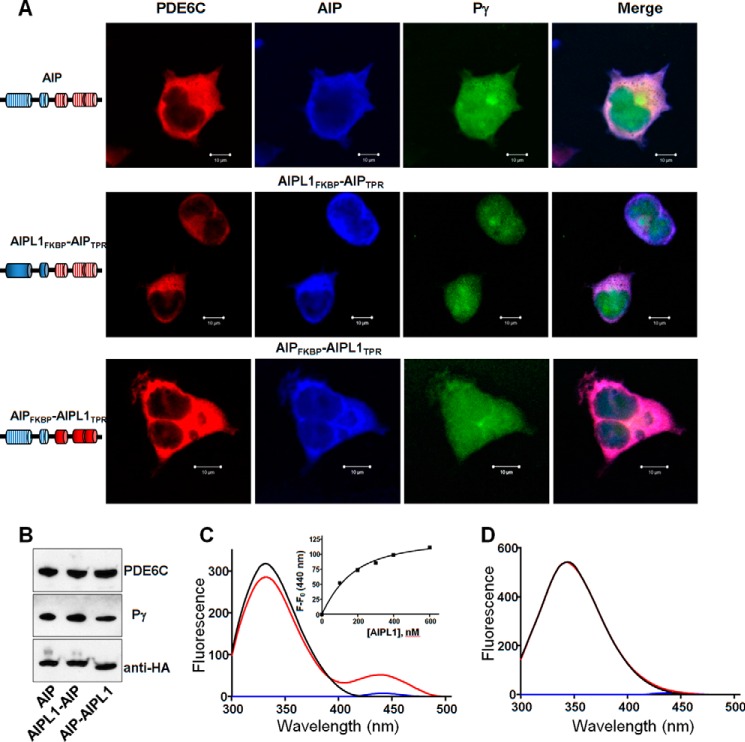
Considering the high degree of sequence conservation between AIPL1 and AIP, we investigated the possibility that AIP can aid in the expression of functional PDE6 in the heterologous system. Despite robust expression of AIP in transfected HEK293T cells (Fig. 4, A and B), no PDE6 activity was detected in its presence, regardless of whether or not Pγ was present in the cells (not shown). The inability of AIP to chaperone PDE6 makes it an excellent tool for probing the roles of the AIPL1 FKBP and TPR domains. Specifically, we constructed and tested two HA-tagged chimeras, one containing AIPL1FKBP and AIPTPR, and the other containing AIPFKBP and AIPL1TPR (Fig. 4, A and B). Both chimeras completely failed to chaperone PDE6C in transfected HEK293T cells (not shown), suggesting that each domain of AIPL1 plays a unique role in chaperoning PDE6.
FIGURE 4.
The AIPL1 FKBP and TPR domains uniquely contribute to the chaperoning of PDE6C. A, confocal immunofluorescence images of HEK293T cells co-transfected with PDE6C (red, anti-FLAG), Pγ (green, EGFP fluorescence) and either AIP or an AIPL1-AIP chimera (blue, anti-HA). B, Western blotting analysis of lysates from HEK293T cells co-transfected with PDE6C, Pγ, AIP, or one of two AIPL1-AIP chimeras. C, binding of AIPL1 to the farnesylated C terminus of PDE6. Emission spectra (λex 280 nm) for AMCA-CtPDE6A-farnesyl (100 nm, blue line), AIPL1 (100 nm, black line), and AIPL1 mixed with AMCA-CtPDE6A-farnesyl (red line; corrected for the probe alone). Inset, the FRET data were fit with an equation for binding with ligand depletion. Kd = 84 ± 12 nm (mean ± S.E., n = 3). D, AIP lacks binding to the farnesylated C terminus of PDE6. Emission spectra (λex 280 nm) for AMCA-CtPDE6A-farnesyl (100 nm, blue line), AIP (300 nm, black line), or AIP mixed with AMCA-CtPDE6A-farnesyl (red line) show no appreciable FRET signal.
Previously, we demonstrated that AIPL1 via its FKBP domain interacts with the farnesyl moiety of PDE6 (32). We therefore investigated whether AIP is also capable of similar interaction. Specifically we used a C-terminal peptide of PDE6A labeled with AMCA in a Trp FRET assay, which shows strong binding of the probe to AIPL1 (Fig. 4C). In contrast, AIP did not appreciably bind the prenylated peptide (Fig. 4D). Thus, the unique role of AIPL1FKBP appears to involve its interaction with the prenylated C terminus of PDE6.
The Spectrum of Chaperone Activities of AIPL1 Mutants Linked to LCA Reveals Pathogenic and Apparently Benign Variants
Most of the missense mutations in AIPL1 linked to LCA lie in the FKBP and TPR domains (Fig. 1) (11, 23, 33–36). We have examined a panel of the FKBP mutants (R38C, K53W (R53W in human AIPL1), V71F, W72S, C89R, V96I, T114I, and Y134F) and TPR mutants (C239R and G262S) for their ability to chaperone PDE6C, both in the absence and in the presence of Pγ. In transfected HEK293T cells, most of the AIPL1 mutants were expressed at levels comparable with WT AIPL1 (Fig. 5A). cGMP hydrolysis in lysates from HEK293T cells co-transfected with PDE6C and the V71F, W72S, C89R, or C239R mutant form of AIPL1 was not significantly different from the background level, and Pγ did not rescue the lack of chaperone activity of these mutants (Fig. 5, B and C). In contrast, when the R38C, V96I, T114I, Y134F, or G262S mutants were used in such an experiment, activity did not differ significantly from levels measured with WT AIPL1, and they were greatly enhanced in the presence of Pγ (Fig. 5, B and C). A modest 2-fold reduction with respect to chaperone activity of the WT protein that was observed for K53W may have been due to its lower expression level (Fig. 5).
FIGURE 5.
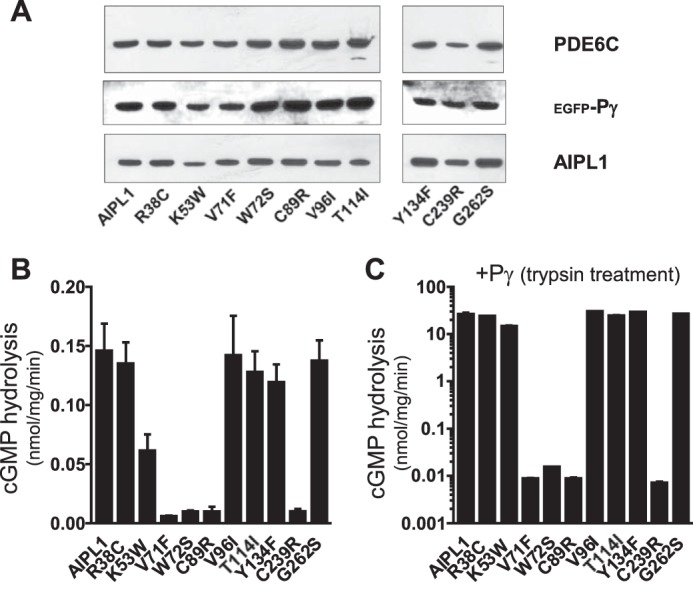
Chaperone activity of AIPL1 mutants linked to LCA. A, Western blot analyses of lysates from HEK293T cells co-transfected with PDE6C, Pγ, and various AIPL1 mutants. Lanes contain equal amounts of protein. B, cGMP hydrolysis in lysates of HEK293T cells co-transfected with PDE6C and AIPL1 mutants. C, cGMP hydrolysis in HEK293T cell lysates co-transfected with PDE6C, AIPL1 mutants, and Pγ. The samples were treated with trypsin to remove Pγ (mean ± S.E., n ≥ 3).
We next investigated the subcellular localization of the AIPL1 mutants. For this analysis, we used COS7 cells rather than HEK293T cells because their adherence and morphology are better suited for detecting potential differences in protein localization. First, we verified that AIPL1 and Pγ chaperone PDE6C in COS7 cells. The rate of cGMP hydrolysis in lysates from untransfected COS7 cells was 23 ± 2 pmol cGMP hydrolyzed/mg/min, whereas in the PDE6C + AIPL1 and PDE6C + AIPL1 + Pγ lysates, it was 47 ± 5 and 205 ± 25 pmol cGMP hydrolyzed/mg/min, respectively. The AIPL1 mutant W72S, lacking the PDE6C chaperone activity in HE293T cells, also failed to chaperone PDE6C in COS7 cells. The effects of AIPL1 and Pγ on expression of functional PDE6C in COS7 cells were not as robust as they were in HEK293T cells. Immunofluorescence microscopy showed WT-like cytoplasmic and nuclear localization for all but two AIPL1 mutants, W72S and C89R (Fig. 6, A and B), with massive nuclear and perinuclear inclusion bodies forming in the cells (Fig. 6B). We estimated that these inclusion bodies were present in 68 and 87% of the cells expressing W72S and C89R, respectively. Furthermore, bacterially expressed and purified AIPL1 mutants lacking the PDE6 chaperone activity, V71F, W72S, C89R, and C239R, had a tendency to aggregate at concentrations of >200 μm (not shown).
FIGURE 6.
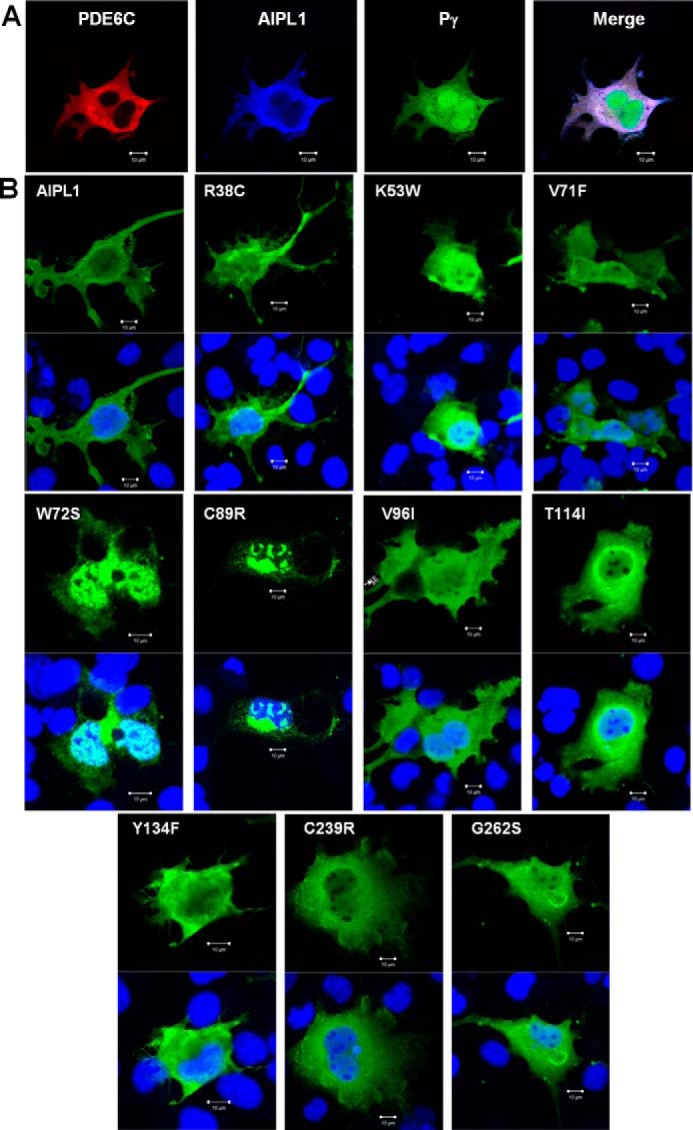
Intracellular localization of AIPL1 mutants in COS7 cells. A, immunofluorescence images of COS7 cells co-transfected with PDE6C (red, anti-PDE6C), AIPL1 (blue, anti-HA), and Pγ (green, EGFP fluorescence). B, immunofluorescence images of COS7 cells transfected with AIPL1 mutants (AIPL1, green; TO-PRO3 nuclear stain, blue). Cells transfected with the AIPL1 mutant proteins W72S and C89R show formation of nuclear and perinuclear inclusion bodies.
Discussion
Eleven families of PDEs have been identified in mammalian tissues based on sequence, substrate selectivity, and regulation (37). PDE6 is an enzyme with remarkable features that make it an exceptionally well suited effector in the phototransduction cascade. First, PDE6 is able to hydrolyze cGMP at a diffusion-limited rate that is 100–1000-fold higher than the catalytic activity of any other PDE. This unparalleled enzymatic efficiency of PDE6 is critical for the amplification of phototransduction and for the ability of rods to respond to single photons (2). The second unique attribute of PDE6 is its tight association with Pγ-subunits, which inhibit its catalytic activity in the dark and impart the ability to be activated by transducin in response to light (38, 39). Our findings demonstrate a third distinction of PDE6: the requirement of a specialized photoreceptor-specific chaperone, AIPL1, for proper folding. This requirement was previously postulated for two reasons: the relative ease of heterologous expression of various PDEs other than PDE6 and the destabilization of PDE6 in mice lacking AIPL1 (14, 15, 40). Our data demonstrate that AIPL1 is an obligatory chaperone for PDE6, whereas Pγ acts as potent co-chaperone that markedly enhances production of the folded enzyme provided that AIPL1 is present. Our results extend the findings from AIPL1 knock-out mice and also provide a simple explanation for the apparently paradoxical phenotype of a mouse with disrupted Pde6g gene (41). Removal of the inhibitory Pγ subunit from holoPDE6 is expected to activate the enzyme. Instead, deletion of Pγ in mouse rods led to dramatically reduced PDE6 activity and retinal degeneration, although initial levels of PDE6AB protein appeared normal (41). Our discovery of the profound effect of Pγ on PDE6 expression in cultured cells suggests that the levels of functional PDE6 in Pde6g−/− rods are at least 2 orders of magnitude lower than in WT rods. Such a reduction in functional PDE6 is certain to cause an elevation of cGMP and photoreceptor death (42–44).
Why does PDE6 require a specialized chaperone? This could have to do with the details of its conformation. Although PDE6 shares its overall domain organization with several PDEs, e.g. PDE2 and PDE5 likewise contain two N-terminal regulatory GAF domains (domains named for their presence in cGMP-regulated PDEs, adenylyl cyclases, and the Escherichia coli protein Fh1A) and a C-terminal catalytic domain, (37), the orientation of catalytic domains in holoPDE6 differs from that in the apo PDE2 dimer. Catalytic domains in PDE2 adopt a “closed” conformation, with the catalytic pockets occluded at the dimer interface (45). Thus, the activation of PDE2 by noncatalytic cGMP-binding to the GAF domain is thought to involve a transition to the “open” conformation with separation of the catalytic domains (45). In holoPDE6, in contrast, a closed conformation is not possible because it would be incompatible with binding of Pγ; the catalytic domains are persistently in the open conformation, and the catalytic pockets are occluded by the Pγ subunits (46–48). We hypothesize that AIPL1 is required for PDE6 to fold into the open conformation and that Pγ contributes to its chaperone activity by stabilizing this conformation in nascent PDE6. Our recent finding that AIPL1 interacts directly with Pγ suggests that Pγ may also act as an adaptor, increasing the affinity of AIPL1 for PDE6 during maturation of the enzyme (49). This mechanism would be akin to that whereby Pγ modulates activity of the RGS9 GAP (GTPase accelerating protein) complex. Although Pγ itself is not a GAP for transducin Gαt, it potentiates the GAP activity of RGS9 by increasing its affinity for transducin-α (50). Similarly, co-expression of PDE6C with Pγ alone does not yield functional PDE6, but Pγ potentiates the chaperone activity of AIPL1.
The co-expression system developed in this study makes it possible to address many unresolved questions about structure-function relationships of PDE6 and AIPL1, to delineate the mechanisms of related retinal diseases, and to screen for pathogenic mutations among the protein variants. As proof of principle, we investigated the roles of the FKBP and TPR domains in the AIPL1 chaperone function and screened a panel of LCA-linked mutants for their ability to fold PDE6. We found that both domains of AIPL1 play unique roles in the folding of PDE6 and that their functions cannot be replicated by the corresponding domains of AIP despite their high sequence conservation. The exclusive role of AIPL1FKBP includes its binding to the prenylated C termini of PDE6 (Fig. 4, C and D). The TPR domains of AIPL1 and AIP bind the HSP90 C-terminal signature sequence MEVEED similarly to well known interactions of TPR domain proteins with HSP90 (49, 51–53). Thus, AIPL1TPR fulfills its unique role in PDE6 folding via a mechanism that is distinct from “generic” binding of HSP90 to TPR domains.
Our screen of LCA-linked AIPL1 mutants for the ability to chaperone PDE6 reveals similar proportions of pathogenic and seemingly benign variants. When experimental approaches are unavailable, the potential pathogenicity of mutant proteins is often evaluated in silico, using mutation analysis software. Our experimental results are consistent with the consensus predictions from three programs: PolyPhen-2 (54), SIFT (55), and PMut (56), that AIPL1 mutants V71F, W72S, C89R, and C239R are pathological but V96I, T114I, and Y134F are likely benign (Table 1). However, our data for other mutants are not consistent with predictions. Whereas computational analysis predicts that R38C and K53W are damaging to AIPL1 structure and/or function, these mutants retained the ability to chaperone PDE6C in our analysis. Conversely, although the G262S mutation is clearly not harmful to the protein itself, its splicing in vitro is altered, and thus it may be pathogenic (24). Thus, our results demonstrate the utility of the PDE6C expression system for validation of pathogenicity of AIPL1 variants.
TABLE 1.
Predictions from in silico analysis of AIPL1 mutants investigated in this study
| PolyPhen | SIFT | PMut | Chaperone activity | |
|---|---|---|---|---|
| R38C | Probably damaging | Damaging | Pathological | Yes |
| R53W | Probably damaging | Damaging | Pathological | Yes |
| V71F | Probably damaging | Damaging | Pathological | No |
| W72S | Probably damaging | Damaging | Pathological | No |
| C89R | Probably damaging | Damaging | Pathological | No |
| V96I | Benign | Tolerated | Neutral | Yes |
| T114I | Benign | Tolerated | Pathological | Yes |
| Y134F | Possibly damaging | Tolerated | Neutral | Yes |
| C239R | Probably damaging | Damaging | Pathological | No |
| G262S | Possibly damaging | Tolerated | Neutral | Yes |
Mapping of the mutations to the model structure of AIPL1 shows that residues corresponding to the benign variants are surface-exposed and/or situated in the flexible “insert” region that links the last two β-strands in the core FKBP domain (Fig. 1B) (32). In contrast, pathogenic mutations V71F, W72S, and C89R involve residues that are embedded in the core FKBP domain and may alter or destabilize the FKBP fold (Fig. 1B). This idea is supported by the finding that mutants W72S and C89R formed aggregates in COS7 cells (Fig. 6). Problems that have plagued the field are that a high degree of polymorphism in AIPL1 has made it difficult to reliably establish disease causation and that the consequences of many PDE6 mutations linked to retinitis pigmentosa and autosomal recessive achromatopsia are unknown (4, 23, 57). Our heterologous expression system, with its simplicity and sensitivity, offers a potentially effective solution for overcoming these hurdles in validating the pathogenicity of AIPL1 and PDE6 mutations, a step that will be required for the development of patient-specific therapies.
Experimental Procedures
Plasmids/Cloning
DNA sequence encoding the full-length mouse AIPL1 was PCR-amplified from pET15b vector harboring mouse AIPL1 gene (32) using 5′ primer with BamHI site and 3′ primer that encodes an HA tag and an XbaI site. The PCR product was then cloned into pcDNA3.1(+) (Invitrogen). Mouse AIP DNA was PCR-amplified from mouse retina cDNA and cloned into either the NdeI/XhoI sites of the pET15b vector (Novagen), for expression as a His6-tagged protein in E. coli, or the HindIII/XhoI sites of pcDNA3.1(+), for expression as an HA-tagged protein in cultured HEK293T cells. Mutations were introduced into AIPL1 using the standard QuikChange site-directed mutagenesis protocol. The mouse AIPL1FKBP-AIPTPR chimera was generated in pcDNA3.1 by replacing residues 162–328 of AIPL1 with residues 163–330 of AIP in a two-step PCR procedure. In the first step, DNA sequence encoding AIP163–330 was amplified from the AIP template using a forward hybrid primer containing the joined AIPL1 and AIP sequences and a reverse primer containing an HA tag and XbaI site. In the second PCR amplification, this PCR product was used as reverse primer with the AIPL1 template, and a forward primer containing the HindIII site. A similar two-step PCR protocol was used to generate mouse AIPFKBP-AIPL1TPR chimera composed of AIP1–162 and AIPL1162–328. DNA coding the FLAG-tagged human PDE6C was PCR-amplified from the PDE6C transgene (31) and cloned into pcDNA3.1(+) using BamHI/EcoRV sites. DNA coding cone Pγ was PCR-amplified from mouse retina cDNA and cloned into the pFIV3.2CAGmcswtIRESeGFP vector using BsrGI and ClaI sites such that the Pγ sequence was fused to the C terminus of EGFP, with the EGFP sequence located downstream of an IRES. Subsequently, the EGFP-Pγ and IRES-EGFP-Pγ sequences were PCR-amplified and cloned into the NotI/XbaI sites of the pcDNA3.1(+) and pcDNA3.1-FLAG-PDE6C plasmids, respectively. The sequences of all constructs were verified by automated DNA sequencing at the UI DNA Core Facility.
Cell Culture and Fractionation of Cell Lysates
HEK293T or COS7 cells were cultured and maintained in DMEM containing 10% FBS (Gibco). Cells were transfected with human PDE6C alone (2 μg) or co-transfected with mouse Pγ or mouse AIPL1 (1 μg each) alone or both Pγ and AIPL1 (0.7 μg each) plasmids using FuGene6 (Promega) according to the manufacturer's instructions. Transfected cells were collected 48 h post-transfection. Cell lysates, prepared in 20 mm Tris-HCl buffer (pH 7.5) containing 120 mm KCl and 1 mm MgCl2 (buffer A) were analyzed by Western blotting for protein expression and assayed for PDE activity. For immunofluorescence, 48h post-transfection, the cells were seeded onto poly-d-lysine-coated (0.1 mg/ml) 4-well chambered glass slide and allowed to grow for additional 24 h before fixation with 4% formaldehyde.
For membrane fractionation, PDE6C, AIPL1, and Pγ co-transfected HEK293T cell lysate was prepared in buffer A and centrifuged at 125,000 × g for 30 min at 4 °C in a Beckman Optima TLX Ultracentrifuge. Pellet thus obtained was further resuspended in hypotonic buffer (5 mm Tris-HCl, pH 7.5, 1 mm MgCl2) and centrifuged at 125,000 × g for 30 min at 4 °C to obtain hypotonic supernatant. The extracted pellet was resuspended in the same buffer and homogenized in a 1.5-ml tube using a disposable pestle.
Protein Expression in E. coli and Purification
His6-tagged mouse AIPL1 and AIP, as well as mutant forms of these, were expressed in BL21-codon plus E. coli cells by induction with 100 μm isopropyl1-thio-β-d-galactopyranoside at 16 °C overnight. Cell pellets were sonicated on ice (five 30-s pulses) in 50 mm Tris-HCl buffer (pH 8) containing 100 mm NaCl, 50 mm l-arginine, 50 mm l-glutamic acid, 10% glycerol, 10 mm DTT (buffer B), and EDTA-free mini protease inhibitor mixture (Roche Applied Science). His6-tagged AIPL1 protein and mutant forms were purified over nickel-nitrilotriacetic acid resin (Novagen), using buffer B containing 250 mm imidazole for elution. Nickel-nitrilotriacetic acid affinity-purified proteins were additionally purified by gel filtration chromatography on a HiLoad 16/600 Superdex 75 column (GE Healthcare) equilibrated against 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 50 mm l-arginine, 50 mm l-glutamic acid, 5% glycerol, and 5 mm DTT.
Western Blotting
Proteins separated by 4–12% SDS-PAGE were transferred to a nitrocellulose membrane using iBlot Western blotting kit (Invitrogen) and analyzed using mouse monoclonal anti-FLAG antibody (Sigma), (1:2000 dilution), mouse anti-HA (BioLegend) (1:1000 dilution), and mouse anti-EGFP (Sigma) (1:2000) primary antibodies. The antibody-antigen complexes were detected using horseradish peroxidase-conjugated goat anti-mouse (1:10000 dilution) secondary antibody and enhanced chemiluminescence reagents obtained from GE Healthcare.
Immunofluorescence
HEK293T or COS7 cells on chambered glass slides were washed once with PBS and fixed for 15 min in 4% formaldehyde in PBS at 25 °C. The cells were then washed once in PBS and permeabilized in 0.1% Triton X-100 in PBS for 5 min at 25 °C. Permeabilized cells were washed three times for 5 min each in PBS, blocked for 60 min in blocking solution (1% BSA in 1× PBS), and then incubated at 4 °C overnight with either mouse anti-FLAG, rabbit anti-PDE6C (31) or mouse anti-HA antibodies diluted (1:2000) in the blocking solution. After washing in PBS three times for 5 min each, the cells were incubated in the dark for 1 h in Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 568-conjugated goat anti-rabbit (Life Technologies) or Alexa Fluor 647-conjugated anti-mouse (ThermoFisher) secondary antibodies (1:5000) diluted in the blocking solution. The cells were washed three times for 5 min each in PBS, and the nuclei were counterstained with To-Pro-3 (1:1000) (ThermoFisher) for 30 min in the dark at 25 °C. The cells were mounted using Vectashield mounting medium (Vector Laboratories) and imaged using Plan-Neofluar 40×/1.3 oil lens and a LSM 510 confocal microscope (Zeiss).
PDE6 Activity Assay
cGMP hydrolysis was measured in cell lysates obtained from HEK293T cells 48 h post-transfection. Where indicated, samples were treated with 0.1 mg/ml tosylphenylalanyl chloromethyl ketone-Trypsin (Sigma) on ice for 10 min to selectively degrade Pγ, after which trypsin was inhibited with the addition of 10-fold excess of soybean trypsin inhibitor (Sigma) and incubation for 5 min at 25 °C. Cell lysates (protein concentration, 5–10 mg/ml) were diluted 4–600 fold into 40 μl (final volume) of 20 mm Tris-HCl (pH 7.5) buffer containing 120 mm NaCl, 2 mm MgSO4, 1 mm 2-mercaptoethanol, 0.1 unit of bacterial alkaline phosphatase, 10 μm [3H]cGMP (100,000 cpm) (Perkin-Elmer) for 5–10 min at 37 °C. The reaction was stopped by the addition of AG1-X2 cation exchange resin (0.5 ml of 20% bed volume suspension). Samples were incubated for 6 min at 25 °C with occasional mixing and spun at 10,000 × g for 3 min. 0.25 ml of the supernatant was removed for counting in a scintillation counter.
Fluorescence Assay
A peptide corresponding to the C terminus of mouse PDE6A (GAPASKSC, Cys farnesylated and carboxymethylated) was custom made, labeled with 7-amino-4-methylcoumarin-3-acetyl)amino) hexanoic acid succinimidyl ester (AMCA), and purified as previously described (49). A FRET assay testing the binding of PDE6A peptide probe (AMCA-CtPDE6A-farnesyl) to AIPL1 and AIP was performed on an F-2500 fluorescence spectrophotometer (Hitachi) in 1 ml of 100 mm HEPES (pH 7.3) with excitation at 280 nm and emission at 440 nm. AMCA-CtPDE6A-farnesyl served as the acceptor of AIPL1 Trp fluorescence. The concentrations of AMCA-CtPDE6A-farnesyl, AIPL1, and AIP were determined using ϵ350 =19,000, ϵ280 = 57,870, and ϵ280 = 37400 m−1 cm−1, respectively. The data were fitted with the equation for binding with ligand depletion using the Prism software (GraphPad) as previously described (32). Kd is expressed as the mean ± S.E. for at least three independent measurements.
In Silico Analysis
AIPL1 variants were examined using three mutation analysis programs: SIFT (55), PolyPhen-2 (54), and PMut (56).
Author Contributions
K. N. G. and N. O. A. designed the research; K. N. G., K. B., R. P. Y., and N. O. A. performed the research; K. N. G., K. B., R. P. Y., and N. O. A. analyzed the data; and K. N. G. and N. O. A. wrote the paper.
Acknowledgments
We thank Dr. A. Majumder for cloning of the mouse cone Pγ-subunit and Christine Blaumueller for critical reading of the manuscript.
This work was supported by National Institutes of Health Grant EY-10843 (to N. O. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PDE6
- photoreceptor phosphodiesterase-6
- LCA
- Leber congenital amaurosis
- FKBP
- FK506-binding protein
- TPR
- tetratricopeptide repeat
- AMCA
- 7-amino-4-methylcoumarin-3-acetyl) amino) hexanoic acid, succinimidyl ester
- IRES
- internal ribosome entry site.
References
- 1. Fu Y., and Yau K. W. (2007) Phototransduction in mouse rods and cones. Pflugers Arch. 454, 805–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arshavsky V. Y., and Burns M. E. (2014) Current understanding of signal amplification in phototransduction. Cell. Logist. 4, e29390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McLaughlin M. E., Ehrhart T. L., Berson E. L., and Dryja T. P. (1995) Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 92, 3249–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dryja T. P., Rucinski D. E., Chen S. H., and Berson E. L. (1999) Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 40, 1859–1865 [PubMed] [Google Scholar]
- 5. Dvir L., Srour G., Abu-Ras R., Miller B., Shalev S. A., and Ben-Yosef T. (2010) Autosomal-recessive early-onset retinitis pigmentosa caused by a mutation in PDE6G, the gene encoding the gamma subunit of rod cGMP phosphodiesterase. Am. J. Hum. Genet. 87, 258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gal A., Orth U., Baehr W., Schwinger E., and Rosenberg T. (1994) Heterozygous missense mutation in the rod cGMP phosphodiesterase β-subunit gene in autosomal dominant stationary night blindness. Nat. Genet. 7, 64–68 [DOI] [PubMed] [Google Scholar]
- 7. Chang B., Grau T., Dangel S., Hurd R., Jurklies B., Sener E. C., Andreasson S., Dollfus H., Baumann B., Bolz S., Artemyev N., Kohl S., Heckenlively J., and Wissinger B. (2009) A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc. Natl. Acad. Sci. U.S.A. 106, 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiadens A. A., den Hollander A. I., Roosing S., Nabuurs S. B., Zekveld-Vroon R. C., Collin R. W., De Baere E., Koenekoop R. K., van Schooneveld M. J., Strom T. M., van Lith-Verhoeven J. J., Lotery A. J., van Moll-Ramirez N., Leroy B. P., van den Born L. I., et al. (2009) Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am. J. Hum. Genet. 85, 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grau T., Artemyev N. O., Rosenberg T., Dollfus H., Haugen O. H., Cumhur Sener E., Jurklies B., Andreasson S., Kernstock C., Larsen M., Zrenner E., Wissinger B., and Kohl S. (2011) Decreased catalytic activity and altered activation properties of PDE6C mutants associated with autosomal recessive achromatopsia. Hum. Mol. Genet. 20, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohl S., Coppieters F., Meire F., Schaich S., Roosing S., Brennenstuhl C., Bolz S., van Genderen M. M., Riemslag F. C., European Retinal Disease Consortium, Lukowski R., den Hollander A. I., Cremers F. P., De Baere E., Hoyng C. B., et al. (2012) A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am. J. Hum. Genet. 91, 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sohocki M. M., Bowne S. J., Sullivan L. S., Blackshaw S., Cepko C. L., Payne A. M., Bhattacharya S. S., Khaliq S., Qasim Mehdi S., Birch D. G., Harrison W. R., Elder F. F., Heckenlively J. R., and Daiger S. P. (2000) Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat. Genet. 24, 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. den Hollander A. I., Roepman R., Koenekoop R. K., and Cremers F. P. (2008) Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 27, 391–419 [DOI] [PubMed] [Google Scholar]
- 13. Koenekoop R. K. (2004) An overview of Leber congenital amaurosis: a model to understand human retinal development. Surv. Ophthalmol. 49, 379–398 [DOI] [PubMed] [Google Scholar]
- 14. Ramamurthy V., Niemi G. A., Reh T. A., and Hurley J. B. (2004) Leber congenital amaurosis linked to AIPL1: a mouse model reveals destabilization of cGMP phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 101, 13897–13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X., Bulgakov O. V., Wen X. H., Woodruff M. L., Pawlyk B., Yang J., Fain G. L., Sandberg M. A., Makino C. L., and Li T. (2004) AIPL1, the protein that is defective in Leber congenital amaurosis, is essential for the biosynthesis of retinal rod cGMP phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 101, 13903–13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van der Spuy J., and Cheetham M. E. (2006) The chaperone function of the LCA protein AIPL1: AIPL1 chaperone function. Adv. Exp. Med. Biol. 572, 471–476 [DOI] [PubMed] [Google Scholar]
- 17. Schwartz M. L., Hurley J. B., and Ramamurthy V. (2006) Biochemical function of the LCA linked protein, aryl hydrocarbon receptor interacting protein like-1 (AIPL1): role of AIPL1 in retina. Adv. Exp. Med. Biol. 572, 89–94 [DOI] [PubMed] [Google Scholar]
- 18. Trivellin G., and Korbonits M. (2011) AIP and its interacting partners. J. Endocrinol. 210, 137–155 [DOI] [PubMed] [Google Scholar]
- 19. Granovsky A. E., Natochin M., McEntaffer R. L., Haik T. L., Francis S. H., Corbin J. D., and Artemyev N. O. (1998) Probing domain functions of chimeric PDE6α′/PDE5 cGMP-phosphodiesterase. J. Biol. Chem. 273, 24485–24490 [DOI] [PubMed] [Google Scholar]
- 20. Qin N., and Baehr W. (1994) Expression and mutagenesis of mouse rod photoreceptor cGMP phosphodiesterase. J. Biol. Chem. 269, 3265–3271 [PubMed] [Google Scholar]
- 21. Muradov H., Boyd K. K., and Artemyev N. O. (2006) Analysis of PDE6 function using chimeric PDE5/6 catalytic domains. Vision research 46, 860–868 [DOI] [PubMed] [Google Scholar]
- 22. Cahill K. B., Quade J. H., Carleton K. L., and Cote R. H. (2012) Identification of amino acid residues responsible for the selectivity of tadalafil binding to two closely related phosphodiesterases, PDE5 and PDE6. J. Biol. Chem. 287, 41406–41416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan M. H., Mackay D. S., Cowing J., Tran H. V., Smith A. J., Wright G. A., Dev-Borman A., Henderson R. H., Moradi P., Russell-Eggitt I., MacLaren R. E., Robson A. G., Cheetham M. E., Thompson D. A., Webster A. R., et al. (2012) Leber congenital amaurosis associated with AIPL1: challenges in ascribing disease causation, clinical findings, and implications for gene therapy. PLoS One 7, e32330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bellingham J., Davidson A. E., Aboshiha J., Simonelli F., Bainbridge J. W., Michaelides M., and van der Spuy J. (2015) Investigation of aberrant splicing induced by AIPL1 variations as a cause of Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 56, 7784–7793 [DOI] [PubMed] [Google Scholar]
- 25. Hurley J. B., and Stryer L. (1982) Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J. Biol. Chem. 257, 11094–11099 [PubMed] [Google Scholar]
- 26. Ovchinnikov Yu A., Lipkin V. M., Kumarev V. P., Gubanov V. V., Khramtsov N. V., Akhmedov N. B., Zagranichny V. E., and Muradov K. G. (1986) Cyclic GMP phosphodiesterase from cattle retina. Amino acid sequence of the gamma-subunit and nucleotide sequence of the corresponding cDNA. FEBS Lett. 204, 288–292 [DOI] [PubMed] [Google Scholar]
- 27. Gillespie P. G., and Beavo J. A. (1988) Characterization of a bovine cone photoreceptor phosphodiesterase purified by cyclic GMP-Sepharose chromatography. J. Biol. Chem. 263, 8133–8141 [PubMed] [Google Scholar]
- 28. Muradov H., Boyd K. K., Haeri M., Kerov V., Knox B. E., and Artemyev N. O. (2009) Characterization of human cone phosphodiesterase-6 ectopically expressed in Xenopus laevis rods. J. Biol. Chem. 284, 32662–32669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anant J. S., Ong O. C., Xie H. Y., Clarke S., O'Brien P. J., and Fung B. K. (1992) In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J. Biol. Chem. 267, 687–690 [PubMed] [Google Scholar]
- 30. Baehr W., Devlin M. J., and Applebury M. L. (1979) Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J. Biol. Chem. 254, 11669–11677 [PubMed] [Google Scholar]
- 31. Majumder A., Pahlberg J., Muradov H., Boyd K. K., Sampath A. P., and Artemyev N. O. (2015) Exchange of cone for rod phosphodiesterase 6 catalytic subunits in rod photoreceptors mimics in part features of light adaptation. J. Neurosci. 35, 9225–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Majumder A., Gopalakrishna K. N., Cheguru P., Gakhar L., and Artemyev N. O. (2013) Interaction of aryl hydrocarbon receptor-interacting protein-like 1 with the farnesyl moiety. J. Biol. Chem. 288, 21320–21328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sohocki M. M., Perrault I., Leroy B. P., Payne A. M., Dharmaraj S., Bhattacharya S. S., Kaplan J., Maumenee I. H., Koenekoop R., Meire F. M., Birch D. G., Heckenlively J. R., and Daiger S. P. (2000) Prevalence of AIPL1 mutations in inherited retinal degenerative disease. Mol. Genet. Metab. 70, 142–150 [DOI] [PubMed] [Google Scholar]
- 34. Dharmaraj S., Leroy B. P., Sohocki M. M., Koenekoop R. K., Perrault I., Anwar K., Khaliq S., Devi R. S., Birch D. G., De Pool E., Izquierdo N., Van Maldergem L., Ismail M., Payne A. M., Holder G. E., et al. (2004) The phenotype of Leber congenital amaurosis in patients with AIPL1 mutations. Arch. Ophthalmol. 122, 1029–1037 [DOI] [PubMed] [Google Scholar]
- 35. Galvin J. A., Fishman G. A., Stone E. M., and Koenekoop R. K. (2005) Evaluation of genotype-phenotype associations in leber congenital amaurosis. Retina 25, 919–929 [DOI] [PubMed] [Google Scholar]
- 36. Stone E. M. (2007) Leber congenital amaurosis: a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson Memorial Lecture. Am. J. Ophthalmol. 144, 791–811 [DOI] [PubMed] [Google Scholar]
- 37. Conti M., and Beavo J. (2007) Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- 38. Cote R. H. (2004) Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int. J. Impot. Res. 16, S28–S33 [DOI] [PubMed] [Google Scholar]
- 39. Arshavsky V. Y., Lamb T. D., and Pugh E. N. Jr. (2002) G proteins and phototransduction. Annu. Rev. Physiol. 64, 153–187 [DOI] [PubMed] [Google Scholar]
- 40. Kirschman L. T., Kolandaivelu S., Frederick J. M., Dang L., Goldberg A. F., Baehr W., and Ramamurthy V. (2010) The Leber congenital amaurosis protein, AIPL1, is needed for the viability and functioning of cone photoreceptor cells. Hum. Mol. Genet. 19, 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsang S. H., Gouras P., Yamashita C. K., Kjeldbye H., Fisher J., Farber D. B., and Goff S. P. (1996) Retinal degeneration in mice lacking the gamma subunit of the rod cGMP phosphodiesterase. Science 272, 1026–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farber D. B., and Lolley R. N. (1974) Cyclic guanosine monophosphate: elevation in degenerating photoreceptor cells of the C3H mouse retina. Science 186, 449–451 [DOI] [PubMed] [Google Scholar]
- 43. Bowes C., Li T., Danciger M., Baxter L. C., Applebury M. L., and Farber D. B. (1990) Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature 347, 677–680 [DOI] [PubMed] [Google Scholar]
- 44. Pittler S. J., and Baehr W. (1991) Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc. Natl. Acad. Sci. U.S.A. 88, 8322–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pandit J., Forman M. D., Fennell K. F., Dillman K. S., and Menniti F. S. (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barren B., Gakhar L., Muradov H., Boyd K. K., Ramaswamy S., and Artemyev N. O. (2009) Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. EMBO J. 28, 3613–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeng-Elmore X., Gao X. Z., Pellarin R., Schneidman-Duhovny D., Zhang X. J., Kozacka K. A., Tang Y., Sali A., Chalkley R. J., Cote R. H., and Chu F. (2014) Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J. Mol. Biol. 426, 3713–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Z., He F., Constantine R., Baker M. L., Baehr W., Schmid M. F., Wensel T. G., and Agosto M. A. (2015) Domain organization and conformational plasticity of the G protein effector, PDE6. J. Biol. Chem. 290, 12833–12843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yadav R. P., Majumder A., Gakhar L., and Artemyev N. O. (2015) Extended conformation of the proline-rich domain of human aryl hydrocarbon receptor-interacting protein-like 1: implications for retina disease. J. Neurochem. 135, 165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skiba N. P., Hopp J. A., and Arshavsky V. Y. (2000) The effector enzyme regulates the duration of G protein signaling in vertebrate photoreceptors by increasing the affinity between transducin and RGS protein. J. Biol. Chem. 275, 32716–32720 [DOI] [PubMed] [Google Scholar]
- 51. Hidalgo-de-Quintana J., Evans R. J., Cheetham M. E., and van der Spuy J. (2008) The Leber congenital amaurosis protein AIPL1 functions as part of a chaperone heterocomplex. Invest. Ophthalmol. Vis. Sci. 49, 2878–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morgan R. M., Hernández-Ramírez L. C., Trivellin G., Zhou L., Roe S. M., Korbonits M., and Prodromou C. (2012) Structure of the TPR domain of AIP: lack of client protein interaction with the C-terminal alpha-7 helix of the TPR domain of AIP is sufficient for pituitary adenoma predisposition. PLoS One 7, e53339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taipale M., Jarosz D. F., and Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 [DOI] [PubMed] [Google Scholar]
- 54. Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., and Sunyaev S. R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sim N. L., Kumar P., Hu J., Henikoff S., Schneider G., and Ng P. C. (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ferrer-Costa C., Gelpí J. L., Zamakola L., Parraga I., de la Cruz X., and Orozco M. (2005) PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21, 3176–3178 [DOI] [PubMed] [Google Scholar]
- 57. Cheguru P., Majumder A., and Artemyev N. O. (2015) Distinct patterns of compartmentalization and proteolytic stability of PDE6C mutants linked to achromatopsia. Mol. Cell. Neurosci. 64, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]