Abstract
Glycosylation of the Notch receptor is essential for its activity and serves as an important modulator of signaling. Three major forms of O-glycosylation are predicted to occur at consensus sites within the epidermal growth factor-like repeats in the extracellular domain of the receptor: O-fucosylation, O-glucosylation, and O-GlcNAcylation. We have performed comprehensive mass spectral analyses of these three types of O-glycosylation on Drosophila Notch produced in S2 cells and identified peptides containing all 22 predicted O-fucose sites, all 18 predicted O-glucose sites, and all 18 putative O-GlcNAc sites. Using semiquantitative mass spectral methods, we have evaluated the occupancy and relative amounts of glycans at each site. The majority of the O-fucose sites were modified to high stoichiometries. Upon expression of the β3-N-acetylglucosaminyltransferase Fringe with Notch, we observed varying degrees of elongation beyond O-fucose monosaccharide, indicating that Fringe preferentially modifies certain sites more than others. Rumi modified O-glucose sites to high stoichiometries, although elongation of the O-glucose was site-specific. Although the current putative consensus sequence for O-GlcNAcylation predicts 18 O-GlcNAc sites on Notch, we only observed apparent O-GlcNAc modification at five sites. In addition, we performed mass spectral analysis on endogenous Notch purified from Drosophila embryos and found that the glycosylation states were similar to those found on Notch from S2 cells. These data provide foundational information for future studies investigating the mechanisms of how O-glycosylation regulates Notch activity.
Keywords: glycosylation, glycosyltransferase, mass spectrometry (MS), Notch receptor, O-linked N-acetylglucosamine (O-GlcNAc), EGF repeat, Fringe, O-linked fucose, O-linked glucose
Introduction
The Notch signaling pathway is evolutionarily conserved in all metazoans and controls a number of critical developmental processes (1–4). Consequently, defects in Notch signaling are linked to several diseases in humans (5). These diseases include various types of cancer, such as T-cell acute lymphoblastic leukemia, which was the first human disease to be linked to Notch signaling (6), and a number of developmental disorders, including Alagille syndrome (7–10) and CADASIL (11). Nonetheless, Notch signaling was originally characterized in Drosophila (12, 13). In flies, the Notch receptor is canonically activated upon binding to either of two ligands, Delta or Serrate. The extracellular domain (ECD)3 of Drosophila Notch consists largely of 36 tandem epidermal growth factor-like (EGF) repeats, which comprise the ligand binding domain of the receptor (14, 15). Each EGF repeat is ∼40 amino acids in length and has six conserved cysteines, forming three disulfide bonds. EGF repeats are found in a wide variety of both cell surface and secreted proteins, including all Notch homologs, as well as the Notch ligands. In addition, EGF repeats can bear a variety of post- and co-translational modifications, including glycosylation.
There are currently three forms of O-linked glycosylation reported on Notch, O-fucose, O-glucose, and O-GlcNAc, all of which occur at consensus sequences within the EGF repeats. The consensus sequence for O-fucosylation exists at a site located between the second and third conserved cysteine residues on a serine or threonine (C2XXXX(S/T)C3) (16) and is catalyzed by O-fucosyltransferase 1 (Ofut1 in flies, POFUT1 in mammals) (17). Loss of Ofut1/Pofut1 in flies or mice results in Notch-null phenotypes, indicating that O-fucosylation is essential for Notch function (18–20). The β3GlcNAc-transferases of the Fringe family can extend O-fucose in flies and mammals (21) and most notably play important roles in modulating Notch-ligand binding (22–25). In mammals, the GlcNAcβ1-3Fuc-O-Ser/Thr disaccharide can be further elongated to a tetrasaccharide: Siaα2–6Galβ1–4GlcNAcβ1–3Fuc-O-Ser/Thr (26). In flies, O-fucose on Notch has not been seen elongated past the GlcNAcβ1–3Fuc-O-Ser/Thr disaccharide by galactosyltransferases or sialyltransferases (23). However, in an O-glycomics analysis of total protein extracts from fly embryos, Aoki et al. (27) reported a novel GlcNAcβ1–3(GlcAβ1–4)fucitol glycan. Interestingly, this trisaccharide was observed in Fringe-rich tissues during embryonic and larval stages of fly development when Notch is known to function (27). Whether this O-fucose trisaccharide occurs on Notch remains to be resolved.
Notch EGF repeats are O-glucosylated by protein O-glucosyltransferase 1 (Rumi in flies, POGLUT1 in mammals) at a serine within a consensus sequence between the first and second conserved cysteine (C1XSX(A/P)C2) (28, 29). O-Glucosylation is also essential for Notch function in flies because loss of rumi in flies results in temperature-sensitive Notch-null phenotypes (29, 30). Loss of mouse Poglut1 also results in a severe decrease in NOTCH1 signaling in embryos and several cell lines (30, 31), although POGLUT1 does not seem to be essential for NOTCH2 signaling during liver development (32). O-Glucose can also be extended to Xylα1–3Glcβ-O-Ser by glucoside α3-xylosyltransferases (Shams in flies and GXYLT1 and GXYLT2 in mammals) (33, 34). This disaccharide can be further extended to a trisaccharide by the addition of a terminal xylose, generating Xylα1–3Xylα1–3Glcβ-O-Ser, by xyloside α3-xylosyltransferase (XXYLT1) (35). We have previously observed the O-glucose trisaccharide as the predominant O-glucose glycoform on NOTCH1 expressed in mammalian cells, whereas O-glucose has been observed in mono-, di-, and trisaccharide forms on Drosophila Notch, depending on the EGF repeat examined (28, 33).
The third type of O-glycosylation that occurs on EGF repeats is O-GlcNAcylation, which is mediated by the EGF domain-specific O-GlcNAc transferase (Eogt in flies, EOGT1 in mammals) (36, 37). EOGT is localized to the endoplasmic reticulum and modifies secreted and membrane proteins, making it distinct from the nuclear and cytoplasmic O-GlcNAc transferase (36). Although the function of O-GlcNAc on Notch remains largely unknown, recent studies show that mutations in EOGT and Notch signaling pathway proteins are linked to Adams-Oliver syndrome (38–40). The current putative consensus sequence for O-GlcNAc exists on a serine or threonine between the fifth and sixth conserved cysteine (C5XXGX(T/S)GXXC6) (41). Of note, whereas Matsuura et al. and others have reported the presence of O-GlcNAc on Notch (36, 37, 41, 42), like the other types of O-glycosylation, no data regarding how efficiently the sites on Notch are modified have been provided.
The role of O-glycosylation on the EGF repeats of Notch has been studied intensely in recent years, especially the mechanisms of how Fringe modulates Notch-ligand binding (16, 43). However, to gain additional insight into how these essential modifications impart functionality to the receptor, it is critical to first establish whether the predicted sites of O-glycosylation are modified as well as to determine the relative amounts of the various glycoforms of O-glycosylation at each of those sites. We have previously shown Notch to be O-fucosylated and O-glucosylated at a subset of predicted sites, using protein expressed in S2 cells (23, 29, 33). Here we present comprehensive site-mapping data of O-fucose, O-glucose, and O-GlcNAc glycans on EGF repeats 1–36 from Drosophila Notch produced in the presence or absence of Fringe in S2 cells. Using semiquantitative mass spectral approaches, we examine site occupancy and efficiency of elongation at each site. In addition, we performed an initial analysis of the glycosylation state of endogenous Notch immunopurified from Drosophila embryos.
Results
Drosophila Notch Is Extensively Modified with O-Glycosylation in S2 Cells
Using the current consensus sequences for O-glycosylation on EGF repeats, the Drosophila Notch ECD is predicted to have 22 O-fucose sites, 18 O-glucose sites, and 18 O-GlcNAc sites (Fig. 1A). To study the O-glycans on Notch, we used an established protocol for purifying protein from conditioned culture medium and preparing protein for mass spectral analyses, which includes reduction/alkylation, protease digestion, and analysis by nano-LC-MS/MS (44). By using multiple proteases, such as trypsin, chymotrypsin, and V8, we observed peptides covering about 90% of the sequence from EGF repeats 1–36 (Fig. 1B). We were therefore able to provide a comprehensive summary of the O-glycosylation on Notch.
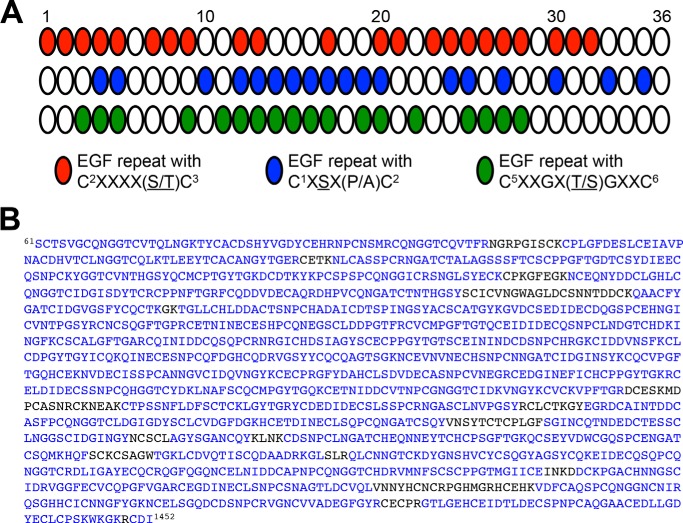
FIGURE 1.
Total coverage from mass spectral analyses of Notch, including all predicted O-glycosylation sites. A, schematic representation of the predicted sites of O-fucosylation (red), O-glucosylation (blue), and O-GlcNAcylation (green) found on EGF repeats 1–36 from the ECD of Drosophila Notch based on current consensus sequences. White ovals, EGF repeats that do not contain any consensus sequence for O-glycosylation. B, the protein sequence of Drosophila Notch expressed for mass spectral analyses includes amino acids 61–1452. Blue letters, residues covered from peptides identified from online database searches (using Matrix Science Mascot) and neutral loss searches. Total coverage is ∼90%.
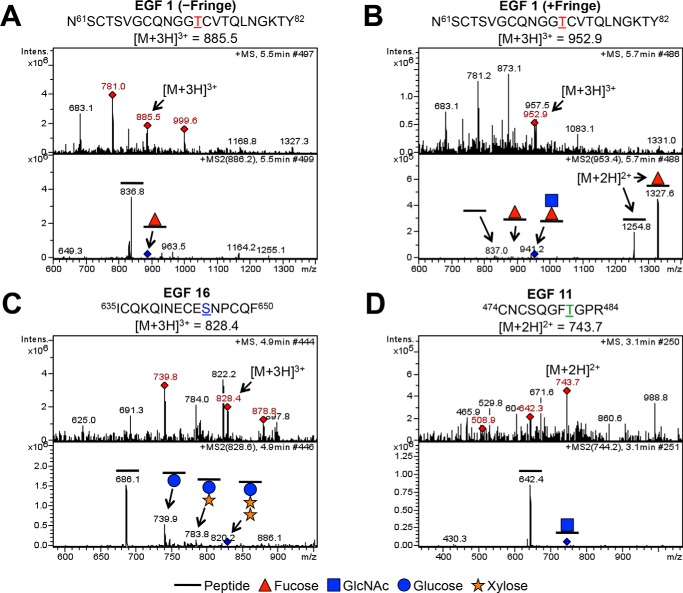
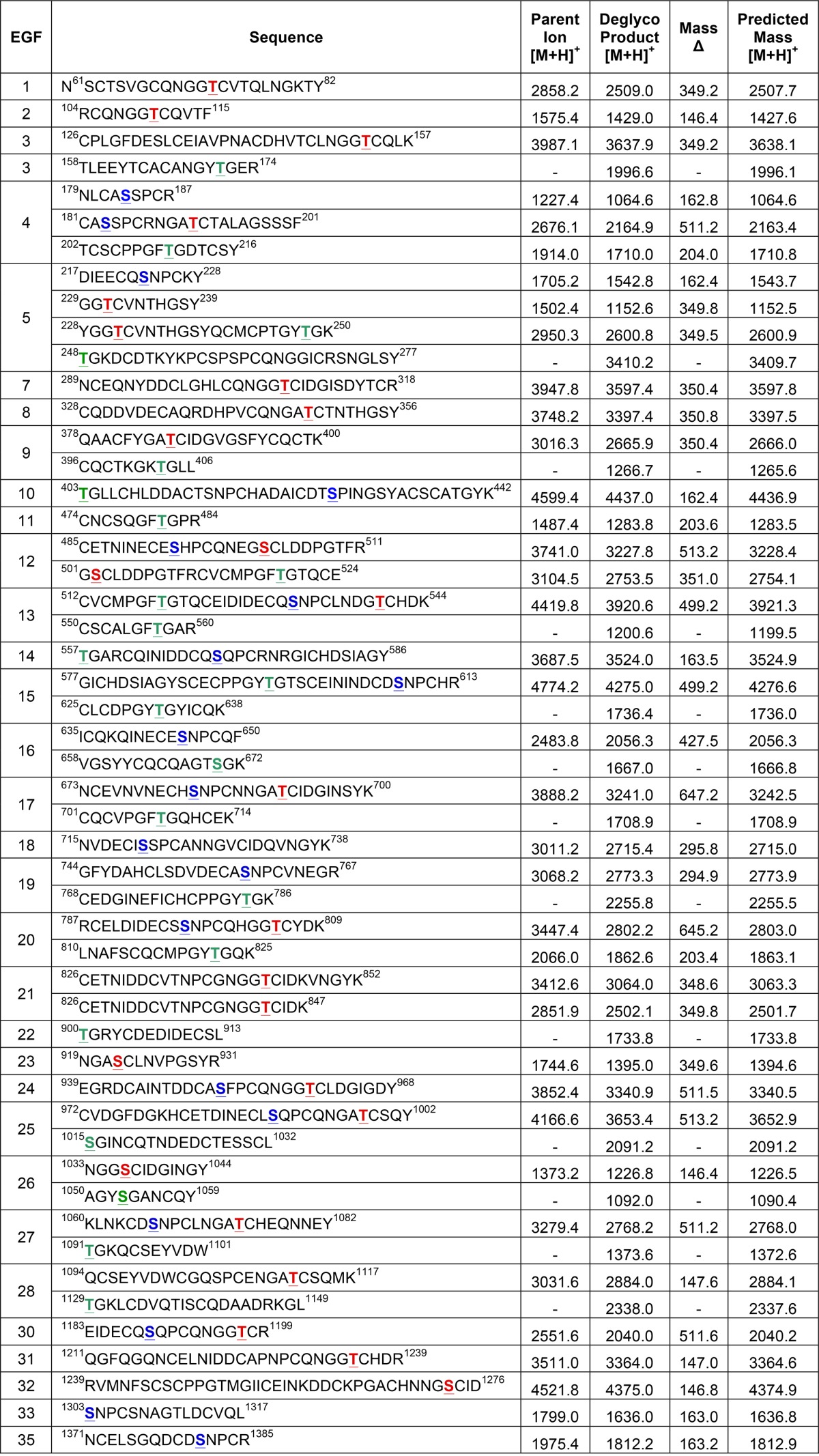
Examples of spectra for each form of O-glycosylation are shown in Fig. 2. In each case, the top panel shows an MS spectrum, where red diamonds represent the ions chosen for fragmentation, and the bottom panel shows the MS2 spectrum of a particular parent ion (Fig. 2, A–D). In the MS2 spectra, we observe the neutral losses of sugars from peptides. For instance, in the absence of Fringe, we observed the peptide 61NSCTSVGCQNGGTCVTQLNGKTY82 from EGF1 modified with an O-fucose monosaccharide (Fig. 2A, −Fringe). We identified the same peptide generated in the presence of Fringe, now modified with a GlcNAc-fucose disaccharide, indicating Fringe modification of the O-fucose (Fig. 2B, +Fringe). An example of O-glucosylation is seen on a peptide from EGF16, 635ICQKQINECESNPCQF650 (Fig. 2C). The MS2 spectra revealed neutral losses, showing that the peptide was modified with a Xyl-Xyl-Glc trisaccharide, which we have observed previously (33). Last, we also observed neutral losses consistent with O-GlcNAcylation at several sites. One example of an unreported site of O-GlcNAcylation was seen on the peptide 474CNCSQGFTGPR484 from EGF11 (Fig. 2D). Ultimately, from our mass spectral analyses, we can observe all three types of O-glycosylation on peptides generated from EGF1 to -36 from Drosophila Notch purified from S2 cells. Spectra for all sites identified are shown in supplemental Fig. S3 and summarized in Table 1.
FIGURE 2.
O-Fucosylation, O-glucosylation, and O-GlcNAcylation of EGF repeats 1–36 from Notch ECD. Examples of mass spectra for the three major types of O-glycosylation are shown (A–D). Top panels, MS spectra; red diamonds, ions chosen for fragmentation. Parent ions corresponding to specified glycosylated peptides (A, EGF1 m/z = 885.5; B, EGF1 m/z = 952.9; C, EGF16 m/z = 828.4; D, EGF11 m/z = 743.7) are fragmented to produce MS2 spectra, shown in the bottom panels. Other ions in the MS spectra are from co-eluting material. Blue diamonds in MS2 spectra indicate the position of the parent ion. A, mass spectra showing the peptide 61NSCTSVGCQNGGTCVTQLNGKTY82 from EGF1 in the +3 charge state. The peptide generated in the absence of Fringe shows a neutral loss of a fucose. B, mass spectra showing the same peptide from EGF1 in the +3 charge state, now generated in the presence of Fringe, shows sequential neutral losses of GlcNAc and fucose. Loss of fucose from the same peptide in the +2 charge state is seen to the right of the parent ion in the MS2 spectrum. A and B, asparagine residue before amino acid 61 comes from the vector and is not part of the Notch sequence. C, mass spectra showing the peptide 635ICQKQINECESNPCQF650 from EGF16 in the +3 charge state. Sequential neutral loss of two xyloses and a glucose is seen in the MS2 spectra. D, mass spectra showing the peptide 474CNCSQGFTGPR484 from EGF11 in the +2 charge state. A neutral loss consistent with that of a GlcNAc is seen in the MS2 spectra. Spectra analyzing O-glucosylation and O-GlcNAcylation were taken from Notch generated in the absence of Fringe. Red triangle, fucose (deoxyhexose); blue circle, glucose (hexose). Orange star, xylose (pentose). Blue square, GlcNAc (HexNAc).
TABLE 1.
O-Glycosylated peptides from EGF 1–36 of Drosophila Notch
EGF repeats 1–36 were produced in S2 cells co-transfected with or without Fringe. Peptides were generated from protease digestions with trypsin, chymotrypsin, or V8. Certain peptides were also identified from endogenous Notch purified from Drosophila embryos. Spectra corresponding to each peptide are provided in Fig. S3. Predicted O-glycosylation sites are underlined, and are colored red for O-fucose, blue for O-glucose or green for O-GlcNAc. Parent ions of the most glycosylated form of each peptide found in the MS/MS data are listed, with the corresponding observed deglycosylated peptide. Peptides only seen as unmodified in the MS/MS data are listed only as the Deglycosylated Product. Masses of all ions were converted to the singly charged state [M + H]+. Predicted masses <1,500 Da were calculated with monoisotopic masses, while peptides >1,500 Da were calculated as average masses.
Comparison of Extracted Ion Chromatograms and Multiple-reaction Monitoring as Semiquantitative Approaches to Analyzing Peptide Glycoforms
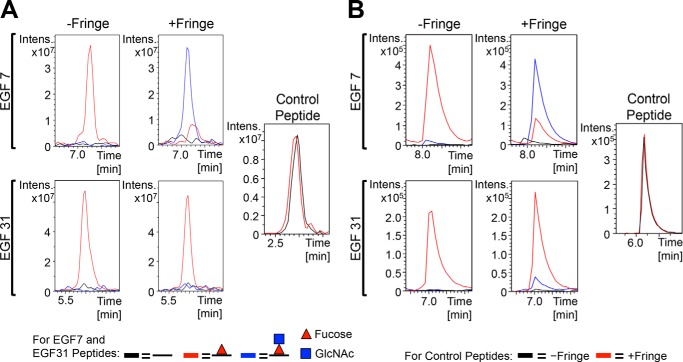
To compare the relative levels of different glycoforms found at each O-glycosylation site more quantitatively, we compared two label-free mass spectral methods: extracted ion chromatograms (EICs) and multiple-reaction monitoring (MRM) (Fig. 3). EICs depict a plot of the intensity of a specified parent ion monitored over time. In contrast, instead of monitoring parent ions over time, MRM follows the intensity of a specified transition ion, which is generated upon fragmentation of a chosen parent ion. Therefore, for each technique, the data were searched for ions corresponding to peptides modified with different glycoforms, and the relative intensities were compared with each other in the same chromatograms. Fig. 3 provides a comparison between EICs and MRM for the O-fucose modification at EGF7 and EGF31 produced in the presence and absence of Fringe. In the absence of Fringe, both EGF7 and EGF31 were modified with O-fucose monosaccharide at high stoichiometry, represented by the red line, with very little unmodified peptide (Fig. 3, A and B, −Fringe panels). There was also no GlcNAc-fucose disaccharide detected, consistent with the inability to detect endogenous Fringe activity in S2 cells (21). However, in the presence of Fringe, we saw that both the EICs and MRM chromatograms show that EGF7 now becomes modified with GlcNAc-fucose disaccharide, represented by the increased intensity of the blue line, whereas EGF31 mostly remained as the fucose monosaccharide glycoform (Fig. 3, A and B, +Fringe panels). Interestingly, the intensities of the fucosylated peptides from −Fringe and +Fringe samples were comparable, and the intensities of unglycosylated control peptides from each sample were the same, indicating that Fringe elongation did not affect ionization efficiency of these peptides. We performed similar analyses for other O-fucose sites, and overall, we found that EIC and MRM data support each other when comparing different glycoforms of fucosylation (compare Fig. 4 with supplemental Fig. S1). Both methods are appropriate for monitoring relative amounts of peptide glycoforms; however, each has advantages and disadvantages. For instance, MRM chromatograms have a reduced signal/noise ratio. However, the increased complexity of the fragmentation pattern of peptides bearing more than one O-glycan significantly complicates the MRM analysis. Therefore, we decided to use EICs for the remaining analyses.
FIGURE 3.
Comparison of extracted ion chromatograms and multiple-reaction monitoring for semiquantitative analysis of glycosylation. EICs (A) and MRM chromatograms (B) show the relative levels of unmodified (black line), O-fucose monosaccharide (red line), and O-fucose disaccharide (blue line) glycoforms of peptides 289NCEQNYDDCLGHLCQNGGTCIDGISDYTCR318 from EGF7 and 1211QGFQGQNCELNIDDCAPNPCQNGGTCHDR1239 from EGF31. Internal unglycosylated control peptides used were 272SNGLSYECK280 from EGF6 (A) and 1200DLIGAYECQCR1210 from EGF30 (B) from samples generated in the absence of Fringe (black line) or presence of Fringe (red line). Lists of the ions used for each glycoform in the EICs and MRM chromatograms are provided in supplemental Fig. S3.
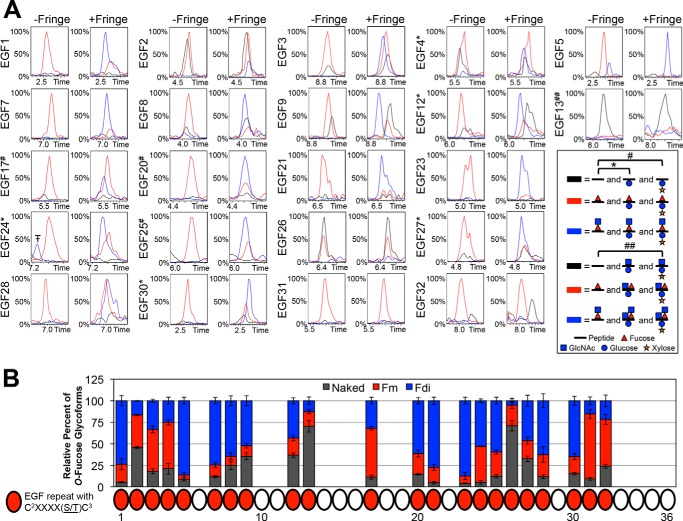
FIGURE 4.
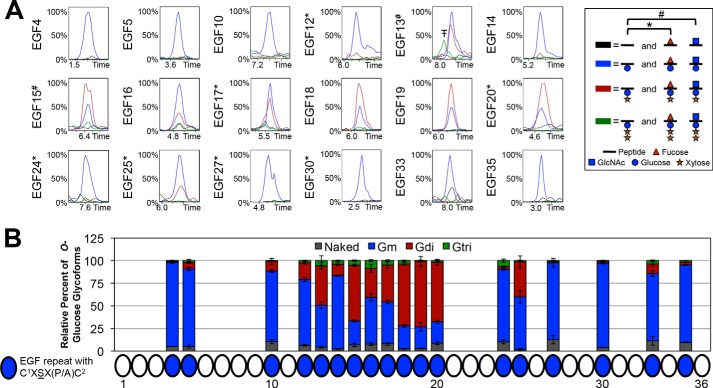
O-Fucosylation occurs at high stoichiometry, but Fringe elongation is site-specific. A, representative EICs showing the relative intensities of unmodified (black line), O-fucose monosaccharide (red line), and O-fucose disaccharide (blue line) glycoforms of peptides containing each predicted O-fucose site in the absence of Fringe (−Fringe) and presence of Fringe (+Fringe). Peptides searched in each EIC are listed in Table 1, and corresponding spectra are shown in supplemental Fig. S3. EICs of O-fucose sites represented by peptides also containing an O-glucose site include searches for O-glucose monosaccharide (*) and O-glucose disaccharide (#). The peptide corresponding to the EGF13 O-fucose site also contained an O-glucose and O-GlcNAc site (##). There is a small amount of O-fucose disaccharide observed on EGF5 in the absence of Fringe, suggesting that there are low levels of endogenous Fringe activity in S2 cells, and EGF5 is the most sensitive to that activity. Additional combinations of O-fucose, O-glucose, and O-GlcNAc glycoforms were searched; however, none were present in significant amounts, and therefore, they are not included in the chromatograms shown. Time is represented in minutes. Red triangle, fucose (deoxyhexose). Blue circle, glucose (hexose). Orange star, xylose (pentose). Blue square, GlcNAc (HexNAc). The peak indicated byT–corresponds to a contaminating ion with the same parent ion mass as the ion of interest, within the specified mass precision range of ±0.5 Da used in the EIC. B, peak areas of each O-fucose glycoform were quantified and represented as a percentage of the total peak area from each EIC (see “Experimental Procedures” for more information). Error bars, S.E.
O-Fucosylation Occurs at High Stoichiometry, but Fringe Elongation Is Site-specific
We then used these methods to investigate the modifications at every predicted O-glycosylation site in the ECD of Notch, beginning with O-fucosylation. We identified peptides containing O-fucose sites from all of the 22 EGF repeats containing the consensus sequence for O-fucosylation (Fig. 1A, Table 1, and supplemental Fig. S3). We generated EICs of naked peptides (unmodified; black line), fucose monosaccharide (red line), and GlcNAc-fucose disaccharide (blue line) glycoforms for each site and compared their relative amounts from samples of Notch produced in the absence of Fringe (Fig. 4A, −Fringe panels). The majority of the O-fucose sites were modified to high stoichiometries in the absence of Fringe (Fig. 4A, −Fringe panels), indicating that Ofut1 modified those EGF repeats with high efficiency. Some sites were O-fucosylated with small amounts of unmodified peptide (EGF3, -4, -8, -9, -12, -27, and -32). EGF2 and -26 had lower levels of O-fucosylation, whereas EGF13 appeared to be the least efficiently modified site, as seen in the −Fringe EICs.
We then co-expressed Notch with Fringe and reexamined EICs for the O-fucosylated peptides (Fig. 4A, +Fringe panels). Some sites showed a more complete conversion of fucose monosaccharide to disaccharide with Fringe co-expression (EGF1, -5, -7, -21, and -23). Several other sites showed varying amounts of monosaccharide remaining with the appearance of disaccharide (EGF3, -4, -8, -9, -12, -17, -20, -24, -25, -27, -28, and -30). Last, there were sites that appeared to be poorly modified by Fringe, showing very little or no apparent conversion of fucose monosaccharide to disaccharide in the EICs (EGF2, -13, -26, -31, and -32). These results revealed that Fringe modified certain O-fucose sites more efficiently than others. To more clearly represent the overall specificity of Fringe elongation at each site, peak areas corresponding to each O-fucose glycoform were quantified within EICs of triplicate samples co-expressed with Fringe (Fig. 4B). Quantifying peak areas in chromatograms provided a simpler and more quantitative summary of the extent of O-fucosylation and Fringe elongation at each site.
Drosophila Notch Is Modified with O-Glucose Mono-, Di-, and Trisaccharide in S2 Cells
In previous studies, we briefly investigated O-glucosylation of Drosophila Notch using similar mass spectral methods, and O-glucose monosaccharide, disaccharide, and trisaccharide were observed modifying certain EGF repeats (29, 33). However, no data regarding the relative levels of each of those O-glucose glycoforms was reported. Here, we have identified peptides containing all 18 sites predicted to be O-glucosylated based on the consensus sequence (Fig. 1A, Table 1, and supplemental Fig. S3). EICs were generated to compare naked peptides (unmodified; black line), glucose monosaccharide (blue line), xylose-glucose disaccharide (green line), and xylose-xylose-glucose trisaccharide (maroon line) glycoforms for each site (Fig. 5A). EICs showed that Rumi modified each O-glucose site to high stoichiometries. Several sites were modified with Xyl-Glc disaccharide in various amounts, although most notably in the region from EGF15 to EGF20, and on EGF13 and EGF25. Although O-glucose trisaccharide was observed on EGF16 and EGF18 previously (33), EICs showed that the trisaccharide glycoform is not the major modification at those or any other sites. To provide a similar summary as Fringe elongation of O-fucose sites, peak areas corresponding to each O-glucose glycoform were quantified and compared together for each O-glucose site (Fig. 5B). Overall, EICs revealed that whereas the addition of O-glucose occurred at high stoichiometries, elongation of O-glucose was site-specific and partial in S2 cells.
FIGURE 5.
O-glucosylation occurs at high stoichiometry, but elongation is limited. A, representative EICs showing the relative intensities of unmodified (black line), O-glucose monosaccharide (blue line), O-glucose disaccharide (maroon line), and O-glucose trisaccharide (green line) glycoforms of peptides containing each predicted O-glucose site. List of peptides searched in each EIC are in Table 1, and corresponding spectra are in supplemental Fig. S3. EICs of O-glucose sites represented by peptides also containing an O-fucose site include searches for O-fucose monosaccharide (*). Peptide containing the O-glucose site on EGF13 also contains an O-GlcNAc site, so the EIC includes searches for O-GlcNAc (#). Additional combinations of O-fucose, O-glucose, and O-GlcNAc glycoforms were searched; however, none were present in significant amounts, and therefore, they are not included in the chromatograms shown. Time is represented in minutes. Red triangle, fucose (deoxyhexose). Blue circle, glucose (hexose). Orange star, xylose (pentose). Blue square, GlcNAc (HexNAc). The peak indicated byT–corresponds to a contaminating ion with the same parent ion mass as the ion of interest, within the specified mass precision range of ±0.5 Da used in the EIC. B, peak areas of each O-glucose glycoform were quantified and represented as a percentage of the total peak area from each EIC (see “Experimental Procedures” for more information). Error bars, S.E.
Drosophila Notch Shows Glycosylation Consistent with the Presence of O-GlcNAc on EGF Repeats
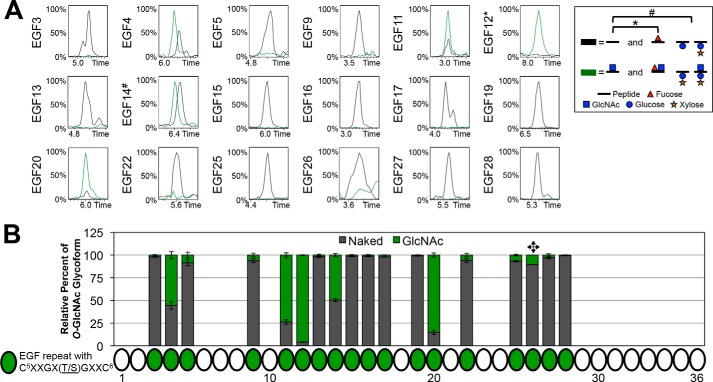
We have examined all 18 predicted O-GlcNAcylation sites based on the current putative consensus sequence (Fig. 1A, Table 1, and supplemental Fig. S3). Whereas previous studies have detected O-GlcNAc on EGF20 of Notch using mass spectral methods and other techniques (36, 37, 41, 42), here we confirm the presence of a HexNAc on EGF20 and several other EGF repeats. Although we cannot differentiate GlcNAc from GalNAc using these methods, the localization of the HexNAc to peptides containing O-GlcNAcylation sites suggests a GlcNAc modification. Additional support comes from the fact that none of the HexNAc-modified peptides in our data could be detected with an additional hexose (Galβ1,3GalNAc is the most abundant O-GalNAc species in flies (27)). EICs compared naked peptides (unmodified; black line) and modifications consistent with that of an O-GlcNAc (green line) (Fig. 6A). Interestingly, only five sites showed an O-GlcNAc modification; none of the other predicted sites appeared to be modified. In addition to EGF20, we observed that EGF4, -11, -12, and -14 were also modified, although EGF4 and EGF14 were modified to a lesser extent. A summary of the O-GlcNAcylation at each predicted site is shown from the quantified peak areas from the EICs (Fig. 6B). Quantification of the EICs indicated that EGF5, -9, -25, and -26 could contain small amounts of modification, although we could not confirm the presence of O-GlcNAc at these sites in any MS2 spectra (Table 1 and supplemental Fig. S3).
FIGURE 6.
O-GlcNAc appears present on EGF4, EGF11, EGF12, EGF14, and EGF20.
A, representative EICs showing the relative intensities of unmodified (black line) and O-GlcNAc (green line) for each predicted O-GlcNAc site. Peptides searched in each EIC are listed in Table 1, and corresponding spectra are shown in supplemental Fig. S3. The peptide containing the O-GlcNAc site on EGF12 also contains an O-fucose site, so the EIC includes searches for O-fucose (*). The peptide containing the EGF14 O-GlcNAc site also contains an O-glucose site, and the EIC includes searches for O-glucose mono- and disaccharide (#). Additional combinations of O-fucose, O-glucose, and O-GlcNAc glycoforms were searched; however, none were present in significant amounts, and therefore, they are not included in the chromatograms shown. Time is represented in minutes. Red triangle, fucose (deoxyhexose). Blue circle, glucose (hexose). Orange star, xylose (pentose). Blue square, GlcNAc (HexNAc). B, peak areas of each O-GlcNAc glycoform were quantified and represented as a percentage of the total peak area from each EIC (see “Experimental Procedures” for more information). Quantification of only one EIC from a sample of Notch produced in the presence of Fringe was performed for the EGF26 O-GlcNAc site ( ). Error bars, S.E.
). Error bars, S.E.
Endogenous Notch from Drosophila Embryos Has O-Glycosylation Similar to That of Notch Produced from S2 Cells
Although using EGF1 to -36 of Drosophila Notch generated from S2 cells provides an excellent experimental system for generating protein for mass spectral studies, one possible caveat is that protein made in a cell culture system may have glycosylation patterns different from those of Notch produced in vivo. Therefore, we generated a Notch genomic transgene, Notchgt-FH, in which the expression of Notch is driven by the endogenous promoter and enhancers. To facilitate purification, the transgene also contains a triple FLAG tag and a double HA tag after the EGF repeats and before the LNR repeats in the ECD of the Notch protein (Fig. 7A). To determine whether the transgene was functional, we performed gene dosage and rescue experiments. As reported previously, removing one copy of Notch results in wing margin loss and wing vein thickening in adult N+/− females (45) (Fig. 7B, arrow and arrowhead, respectively). These phenotypes were rescued upon the addition of one copy of the Notchgt-FH transgene (Fig. 7C). Adding an extra copy of Notch results in additional wing vein material in the adult wings called the Confluens phenotype (45, 46). Because N+/−; Notchgt-FH/+ females did not exhibit the Confluens phenotype (Fig. 7C), the level of Notch expressed from the Notchgt-FH transgene should be similar to that expressed from an endogenous copy of Notch. Drosophila Notch resides on the X chromosome, and males harboring a null allele of Notch are embryonic lethal (47). However, one copy of Notchgt-FH rescued the lethality of Notch-null males. The rescued adult males did not exhibit any morphological defects associated with decreased Notch signaling, as evidenced by normal leg joints (Fig. 7D, arrows), normal bristle pattern in the thorax (Fig. 7E), and normal wings (Fig. 7F). Together, these observations indicate that the Notchgt-FH is functional in vivo and behaves similarly to an endogenous copy of Notch.
FIGURE 7.
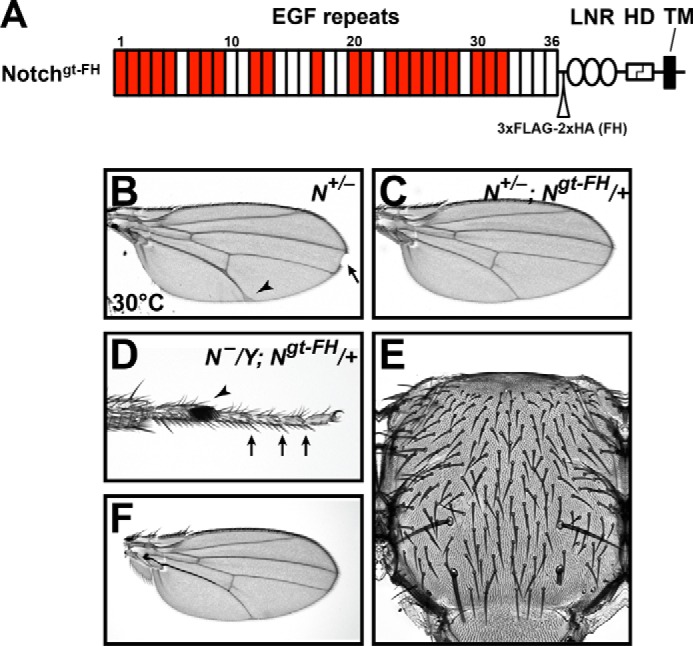
A Notch transgene, Ngt-FH, mimics an endogenous copy of Notch. A, diagram of the Ngt-FH ECD, showing 36 EGF repeats, three LNR domains, the heterodimerization (HD) domain, and transmembrane (TM) domain. A triple FLAG tag and a double HA tag were inserted after the EGF repeats and before the LNR repeats. EGF repeats containing an O-fucosylation consensus sequence are colored red. B, a Notch haploinsufficient female wing showing thickened veins (arrowhead) and wing margin loss (arrow). These phenotypes could be rescued with one copy of Ngt-FH (C). D–F, phenotypes exhibited by a Notch-null hemizygous male rescued from embryonic lethality to adulthood with one copy of Ngt-FH. Note the normal leg joints (arrows in D), normal bristle pattern on the thorax (E), and normal wings (F). The arrowhead in D marks the sex comb, which is a male-specific feature of the Drosophila legs.
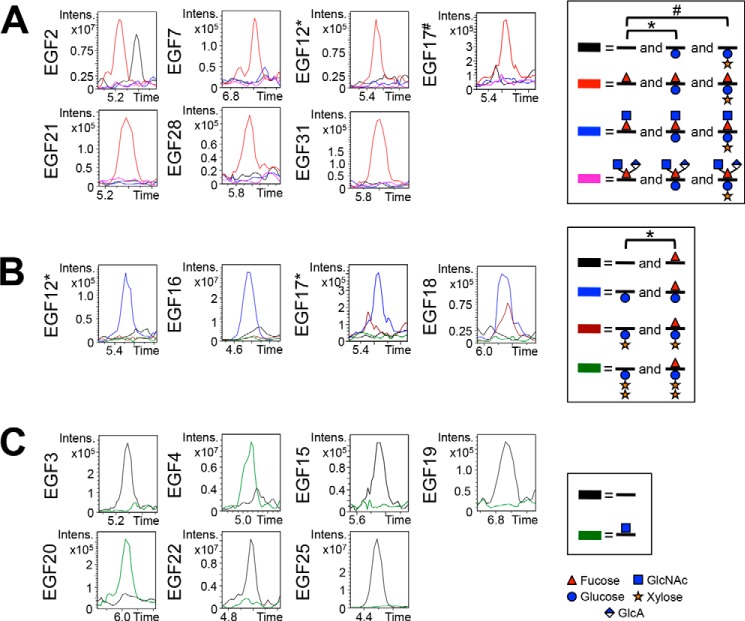
To examine whether the O-glycosylation patterns we observed on Notch made in S2 cells were similar to that of endogenous Notch, we performed mass spectral analysis of Notch purified from Drosophila in the mid-embryonic stage. Although ions were not detected for all O-glycosylation sites due to limited material and co-purified contaminating proteins that were not present with the S2 cell-derived protein, we did observe O-glycosylation at several EGF repeats (Fig. 8). Peptides containing O-fucose sites were detected with essentially no elongation past the fucose monosaccharide glycoform (Fig. 8A). This result is consistent with the fact that Fringe is not expressed at a high level during the mid-embryonic stage (48, 49). However, because Aoki et al. (27) proposed that a novel branched trisaccharide GlcNAcβ1–3(GlcAβ1–4)Fuc glycan could be modifying Notch protein in Drosophila embryos at this stage, we performed EIC searches for this glycoform. From the EICs, we did not observe the O-fucose trisaccharide glycoform as a major species at any of the O-fucose sites that we were able to obtain from the endogenous Notch protein (Fig. 8A, pink line). We cannot rule out the presence of the trisaccharide at other sites.
FIGURE 8.
O-Glycosylation on endogenous Notch is similar to that seen from S2 cells. EICs showing O-fucosylation (A), O-glucosylation (B), and O-GlcNAcylation (C) of peptides from several EGF repeats from endogenous Notch protein purified from Drosophila. To more directly compare the O-glycosylation of endogenous Notch with Notch generated in S2 cells, only peptides analyzed from S2 cells were also analyzed from endogenous Notch (Table 1 and supplemental Fig. S3). Similar combinations of O-fucose, O-glucose, and O-GlcNAc glycoforms were seen modifying Notch produced in S2 cells. A, EICs showing the relative levels of unmodified (black line), O-fucose monosaccharide (red line), O-fucose disaccharide (blue line), and the novel O-fucose trisaccharide (pink line) for each predicted O-fucose site. EICs of O-fucose sites represented by peptides also containing an O-glucose site include searches for O-glucose monosaccharide (*) and O-glucose disaccharide (#). B, EICs showing the relative levels of unmodified (black line), O-glucose monosaccharide (blue line), O-glucose disaccharide (maroon line), and O-glucose trisaccharide (green line) for each predicted O-glucose site. Peptides searched in each EIC are listed in Table 1, and corresponding spectra are shown in supplemental Fig. S3. EICs of O-glucose sites represented by peptides also containing an O-fucose site include searches for O-fucose monosaccharide (*). C, EICs showing the relative levels of unmodified (black line) and O-GlcNAc (green line) for each predicted O-GlcNAc site. A–C, time is represented in minutes. Red triangle, fucose (deoxyhexose). Blue circle, glucose (hexose). Orange star, xylose (pentose). Blue square, GlcNAc (HexNAc). Half-blue diamond, GlcA.
We also detected several peptides containing O-glucose sites and observed that O-glucosylation occurred at high stoichiometries with some extension to the xylose-glucose disaccharide glycoform. Although EGF17 appeared to have slightly less O-glucose disaccharide, the data were comparable with the data obtained from S2 cells (Fig. 8B; compare with Fig. 5A). Several peptides containing predicted O-GlcNAc sites were also detected. EGF4 and 20 appeared to be modified; however, EGF4 seemed somewhat more modified than we had observed in S2 cells (Fig. 8C; compare with Fig. 6A). These slight differences may be due to the fact that the S2 samples were averages of biological triplicates, whereas the in vivo samples were not. Other sites remained unmodified (Fig. 8C). Overall, these data provide evidence that the O-glycosylation on Notch made in S2 cells mimics that found on Notch endogenously produced from embryos.
Discussion
Notch is an important signaling receptor that is involved in many developmental processes and whose activity is modulated by glycosylation of the EGF repeats in the ECD. Although a few prior publications have identified some sites of O-glycosylation on the ECD, this is the first comprehensive report that includes data showing site occupancy and the relative levels of glycan extension at each site. We performed mass spectral analyses of EGF repeats 1–36 from Drosophila Notch expressed and purified from S2 cells and utilized EICs to obtain semiquantitative data on the relative amounts of various glycoforms at each O-fucose, O-glucose, and O-GlcNAc site. Moreover, we report the O-glycan modifications of the Notch protein expressed in Drosophila embryos. To our knowledge, this is the first analysis of the O-glycosylation on the Notch protein expressed at nearly endogenous levels in an intact animal. Our data confirm that the O-glycosylation profiles of Notch overexpressed in S2 cells in this and previous studies faithfully represent the in vivo O-glycosylation of Notch.
We analyzed each of the 22 O-fucose sites predicted to be modified by Ofut1, and we observed that the majority of sites were modified to high stoichiometries (Fig. 4A). Interestingly, EGF repeats 2, 13, and 26 had the least amount of fucosylation. However, comparing the residues that compose the O-fucose consensus sequences in these EGF repeats did not seem to provide specific amino acids predictive of low O-fucosylation efficiency (supplemental Fig. S2A). Although there is a unique aspartate two residues away from the modified threonine in EGF13, EGF12 has a glutamate in the same position and is more efficiently modified by Ofut1. The predicted modified residue in EGF26 is a serine, which could presumably be a poorer substrate for Ofut1. However, EGF12, -23, and -32 also have serine residues. In fact, other than the serine, the residues in the consensus sequence of EGF26 are very similar to the sequence in the subsequent and more modified EGF repeat, EGF27. Thus, the C2XXXX(S/T)C3 still accurately predicts more than 95% of the O-fucosylation sites. The specific factors that reduce the efficiency of O-fucosylation of EGF13 in the Notch ECD by Ofut1 remain unclear.
Although we know that Fringe is a critical modulator of Notch activity, we do not know specifically how Fringe modification of O-fucose enhances Notch interactions with Delta and at the same time reduces interactions with Serrate. In our mass spectral analyses, we were particularly interested in determining which O-fucose sites were modified by Fringe because those sites should play important roles in the effects Fringe has on Notch. Recent structural studies on the ligand binding domain of mammalian Notch1 show that the O-fucose glycans on EGF12 directly interact with ligands upon binding (22, 50). From the EIC analysis, we observed that Fringe elongates some sites more efficiently than others. Elongation varied from little on EGF2, -26, -31, or -32 to almost complete modification of O-fucose monosaccharide to disaccharide on EGF5, -7, -21, and -23. No specific residues within individual EGF repeats appear to distinctly correlate with Fringe elongation (supplemental Fig. S2B). Of note, EGF6, -22, and -29 are well conserved non-calcium-binding EGF repeats within Notch (15), and interestingly, the most Fringe-elongated sites surround these EGF repeats. The distribution of calcium-binding EGF repeats is well conserved across several species (15), and calcium-binding EGF repeats have more rigid structures than non-calcium-binding EGF repeats (51). It is possible that the predicted flexibility of those regions of the Notch ECD allow for more efficient modification by Fringe.
Aoki et al. (27) found a novel GlcAβ1–4(GlcNAcβ1–3)Fuc branched trisaccharide in an O-glycomics study of protein extracts from fly embryos. This trisaccharide was significantly enriched in the dorsal compartment of the third instar wing imaginal discs (27), where Fringe is expressed. Accordingly, the authors proposed that this novel glycan may modify Notch, given that a Fringe-elongated O-fucose can serve as a substrate for the generation of this branched trisaccharide and because of the correlation between its tissue distribution and Fringe expression. Using our established mass spectral methods for analyzing O-glycosylation, we did not observe such an O-fucose trisaccharide on endogenous Notch isolated from embryos. Our in vivo data from embryos suggest that the above-mentioned branched O-fucose trisaccharide is either on other proteins found in embryo extracts or on sites that we have not yet mapped on endogenous Notch. Isolation of endogenous Notch from later stages of fly development using our immunoprecipitation methodology would allow us to determine whether this glycan exists on Notch at later developmental stages (e.g. in third instar imaginal discs).
We have previously mapped several O-glucose sites on Drosophila Notch (29, 33). Here we provide data on the efficiency of O-glucosylation at individual sites. We were able to detect peptides from each of the 18 predicted O-glucosylated EGF repeats of Drosophila Notch. We observed O-glucosylation at high stoichiometries at each site, confirming the accuracy of the C1XSX(P/A)C2 consensus sequence. Nonetheless, elongation by xylose was limited to certain sites. Although no obvious patterns of residues within the consensus sequence correlate with extension of the O-glucose (supplemental Fig. S2C), previous studies have shown that certain amino acids within the O-glucose consensus sequence can affect the elongation (52). In addition, it appears that the elongated sites coincide with a stretch of several tandem calcium-binding EGF repeats (15), suggesting that the rigidity of that region could enhance xylosylation. Although determining the efficiency of glucosylation and xylosylation provides additional insight to investigating the role of O-glucosylation on Notch activity, there are still unanswered questions. The exact molecular mechanisms of how O-glucosylation promotes Notch activity (29, 45, 53) and xylosylation of the O-glucose inhibits Notch activity (33) are yet to be determined.
Analysis of the embryonic phenotypes of the Eogt mutant in flies indicates that O-GlcNAcylation is not essential for Notch signaling during Drosophila embryogenesis (36). However, additional experiments have uncovered a dosage-sensitive genetic interaction between several components of the Notch signaling pathway and Eogt knockdown adult flies (54). Moreover, mutations in human EOGT have been reported in an autosomal recessive form of a human disease called Adams-Oliver syndrome (55, 56). Notably, mutations in several Notch pathway components cause an autosomal dominant form of the same disease (39, 40, 57–59). Together, these reports provide strong evidence that O-GlcNAcylation might modulate Notch signaling. As the third major type of O-glycosylation on Notch, we mapped the 18 predicted O-GlcNAc sites. Surprisingly, only four sites appeared to be modified with O-GlcNAc. This suggests that the current consensus sequence should be revised to more accurately predict sites of modification. Based on the residues found in the consensus sequences from EGF4, -11, -12, and -20 from Drosophila Notch, it is possible that threonines are preferred over serines as the modified residue and that only phenylalanine and tyrosine aromatic residues can be tolerated adjacent to the hydroxyl residue (C5XXG(F/Y)TGXXC6) (supplemental Fig. S2D). Also, the modified sites have a high prevalence of proline or glutamine immediately before the first glycine (C5X(P/Q)G(F/Y)TGXXC6). However, previous studies have found other EGF repeats with contradictory sequences to be modified with O-GlcNAc (41), so ultimately, it is possible that something other than the residues within the putative consensus sequence determine O-GlcNAcylation. Additionally, like the other types of O-glycosylation, more work needs to be done to better understand the specific mechanisms of how these modifications affect Notch activity.
Experimental Procedures
Expression Constructs
The constructs expressing EGF repeats 1–36 from the Drosophila Notch ECD with a C-terminal 3XFLAG tag (N-3XFLAG) and Drosophila Fringe with a C-terminal His6 tag (Fng), were generous gifts from Dr. Kenneth Irvine (Rutgers University) and were described previously (60). Construction of these plasmids, transfection of Drosophila S2 cells, and purification of the resulting proteins from medium has been described (21, 23, 60). pMTHy empty vector, used as a control for transfections, was a generous gift from Dr. J. Peter Gergen (Stony Brook University).
Cell Culture and Protein Purification
S2 cells were co-transfected with either 10 μg each of N-3XFLAG and pMTHy empty vector or 10 μg each of N-3XFLAG and Fng, using the calcium phosphate transfection method to generate unmodified or in vivo Fringe-elongated Notch protein. Transfections were performed based on protocols from the Calcium Phosphate Transfection Kit (Invitrogen), although the buffers used were made in-laboratory. Cells were cultured in Schneider's Drosophila medium (Lonza) supplemented with 10% heat-inactivated FBS and 1% penicillin-streptomycin.
For purification of N-3XFLAG (with or without Fng), expression in transfected S2 cells was induced by the addition of 0.7 mm CuSO4 for 3 days. Conditioned medium was cleared by centrifugation and incubated with anti-FLAG beads (Sigma) overnight at 4 °C with rotation. Purified protein was eluted from beads using 3XFLAG peptide (Sigma) as per the manufacturer's instructions. To remove excess 3XFLAG peptide, samples were passed through Zeba desalting columns (Thermo Fisher Scientific).
Analysis of Glycopeptides by Nano-LC-Electrospray Ionization-MS
Purified Notch protein was reduced, alkylated, and subjected to in-gel digestions with either trypsin, chymotrypsin, or V8 protease and analyzed by nano-liquid chromatography/tandem mass spectrometry using an Agilent 6340 ion trap mass spectrometer with a nano-HPLC CHIP cube interface autosampler as reported previously (28, 44). Constant neutral loss searches as well as generation of extracted ion chromatograms and multiple-reaction monitoring chromatograms were performed as reported previously using the Data Analysis software from Agilent (44, 61). All O-glucosylation and O-GlcNAcylation site mapping was performed from samples generated in the absence of Fringe. Online database searches were conducted using Matrix Science Mascot, with search parameters allowing two missed cleavages and carbamidomethyl as a fixed modification of cysteines. Amino acid numbering of all Notch peptides is based on the sequence from Uniprot accession number P07207.
Quantification of Extracted Ion Chromatograms
Specific ions corresponding to the respective glycoforms of peptides containing consensus sequences were searched from the MS data to generate the EICs. Each trace was then processed with a Gaussian smoothing algorithm, and background values across the chromatogram were subtracted from each trace using the Data Analysis software. The area under each chromatogram trace was quantified from the same time range for each glycoform in the same chromatogram. Quantifications of chromatograms were obtained from mass spectral data of protein purified from three separate transfections of N-3XFLAG (exceptions include the analyses of the O-fucose site from EGF32, which used protein from two separate transfections and one technical replicate of the same preparation, and the O-GlcNAc site from EGF26, which used protein from a single transfection). Areas were averaged, and the percentage of total peak area was calculated to represent the relative amounts of each glycoform for one site of glycosylation.
Generation of Notchgt-FH Transgenic Flies and Fly Genetics
The Notch genomic transgene with a 3XFLAG-2XHA (FH) epitope tag was generated by gap repair mutagenesis as described previously (45, 62). The FH sequence was inserted between EGF36 and the LNR repeats into the Notch-attB-P[acman]-ApR construct (45). The precise location and sequence of the tag is as follows: 1465GSDYKDHDGDYKDHDIDYKDDDDKLYPYDVPDYAGYPYDVPDYARS1466. The numbered amino acids mark the Notch amino acids (Uniprot accession number P07207) between which the tag was inserted; 3XFLAG and 2XHA are underlined. ΦC31-mediated integration (63) was used to integrate the NotchFH-attB-P[acman]-ApR construct into the VK22 docking site (64) in the fly genome and to generate the PBac{Notchgt-FH}VK22 strain, which will be called Notchgt-FH hereafter. Other stocks used in this study are the Notch-null allele N55e11/FM7 and the vas-int-ZH-2A; attVK22 stock (63, 64) (Bloomington Drosophila Stock Center). The genetic interaction and rescue crosses were performed at 30 °C. Dissection, mounting, and image acquisition for the characterization of adult fly tissues were performed as described previously (29). Images were processed with Adobe Photoshop CS2 and were assembled in Adobe Illustrator CS2.
Immunoprecipitation of Endogenous Notch from Notchgt-FH Embryos
7.0 g of Notchgt-FH embryos were collected, aged to 16 h, and stored at −80 °C until 20 min before lysis. Embryos were lysed in TBS, pH 7.4, 1.0% Nonidet P-40, and 0.2% PMSF supplemented with Protease Inhibitor Mixture (EDTA-free, Roche Applied Science) using a probe sonicator in short pulses over a 5-min incubation on ice. Lysate was incubated on ice for an additional 10 min with vortexing and then cleared by centrifugation. The resulting supernatant was collected, followed by adding additional lysis buffer to the remaining insoluble pellet for another 10-min incubation on ice with vortexing. The second lysate was centrifuged and pooled with the previous lysate. Cleared lysates were filtered using a 0.22-μm filter. Approximately 85 ml of filtered lysate were incubated with 900 μl of EZ-view red anti-FLAG beads (Sigma) on a rotator at 4 °C for 12 h. Following centrifugation, beads were washed with 0.1% Nonidet P-40 in TBS and then subjected to either a harsher ionic wash of 1.0 m NaCl in TBS or a detergent wash of 0.1% SDS in TBS. After centrifuging at 10,000 rpm at 4 °C for 1 min, protein was eluted using 3XFLAG peptide (Sigma) at 250 ng/μl.
Author Contributions
B. M. H. and N. A. R. designed, coordinated, and analyzed the data in Figs. 1–6 and 8 and Table 1 and wrote the first draft of the manuscript. H. M. provided technical assistance for the data in Fig. 8. J. L. and H. J.-N. designed, coordinated, and analyzed the data in Fig. 7. R. S. H. conceived and coordinated the study and assisted in data analysis. All authors participated in editing the manuscript.
Supplementary Material
Acknowledgments
We thank Rodrigo Fernandez-Valdivia and Yi-Dong Li for discussions and technical assistance with generation of the Notchgt-FH transgene. We thank the Bloomington Drosophila Stock Center (National Institutes of Health Grant P40OD018537) for fly stocks. We also thank Spyros Artavanis-Tsakonas and Kazuya Hori for sharing unpublished data indicating that the insertion of epitope tags between EGF repeats and LNR repeats does not disrupt the function of the Drosophila Notch in vivo. We thank the Haltiwanger laboratory for comments on the manuscript.
This work was supported in part by NIGMS, National Institutes of Health, Grants R01GM084135 (to H. J.-N.) and R01GM061126 (to R. S. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S3.
- ECD
- extracellular domain
- EGF
- epidermal growth factor-like
- Fuc
- fucose
- HexNAc
- N-acetylhexosamine
- Sia
- sialic acid
- GlcA
- glucuronic acid
- Xyl
- xylose
- EOGT
- EGF domain-specific O-GlcNAc transferase
- EIC
- extracted ion chromatogram
- MRM
- multiple-reaction monitoring.
References
- 1. Artavanis-Tsakonas S., Rand M. D., and Lake R. J. (1999) Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2. Hori K., Sen A., and Artavanis-Tsakonas S. (2013) Notch signaling at a glance. J. Cell Sci. 126, 2135–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kopan R., and Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lai E. C. (2004) Notch signaling: control of cell communication and cell fate. Development 131, 965–973 [DOI] [PubMed] [Google Scholar]
- 5. Penton A. L., Leonard L. D., and Spinner N. B. (2012) Notch signaling in human development and disease. Semin. Cell Dev. Biol. 23, 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., Smith S. D., and Sklar J. (1991) TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 [DOI] [PubMed] [Google Scholar]
- 7. Li L., Krantz I. D., Deng Y., Genin A., Banta A. B., Collins C. C., Qi M., Trask B. J., Kuo W. L., Cochran J., Costa T., Pierpont M. E., Rand E. B., Piccoli D. A., Hood L., and Spinner N. B. (1997) Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16, 243–251 [DOI] [PubMed] [Google Scholar]
- 8. Kamath B. M., Bauer R. C., Loomes K. M., Chao G., Gerfen J., Hutchinson A., Hardikar W., Hirschfield G., Jara P., Krantz I. D., Lapunzina P., Leonard L., Ling S., Ng V. L., Hoang P. L., et al. (2012) NOTCH2 mutations in Alagille syndrome. J. Med. Genet. 49, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oda T., Elkahloun A. G., Pike B. L., Okajima K., Krantz I. D., Genin A., Piccoli D. A., Meltzer P. S., Spinner N. B., Collins F. S., and Chandrasekharappa S. C. (1997) Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16, 235–242 [DOI] [PubMed] [Google Scholar]
- 10. McDaniell R., Warthen D. M., Sanchez-Lara P. A., Pai A., Krantz I. D., Piccoli D. A., and Spinner N. B. (2006) NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am. J. Hum. Genet. 79, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P., Alamowitch S., Domenga V., Cécillion M., Marechal E., Maciazek J., Vayssiere C., Cruaud C., Cabanis E. A., Ruchoux M. M., et al. (1996) Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383, 707–710 [DOI] [PubMed] [Google Scholar]
- 12. Wharton K. A., Johansen K. M., Xu T., and Artavanis-Tsakonas S. (1985) Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 43, 567–581 [DOI] [PubMed] [Google Scholar]
- 13. Dexter J. S. (1914) The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am. Naturalist 48, 712–758 [Google Scholar]
- 14. Rebay I., Fleming R. J., Fehon R. G., Cherbas L., Cherbas P., and Artavanis-Tsakonas S. (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67, 687–699 [DOI] [PubMed] [Google Scholar]
- 15. Xu A., Lei L., and Irvine K. D. (2005) Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J. Biol. Chem. 280, 30158–30165 [DOI] [PubMed] [Google Scholar]
- 16. Rana N. A., and Haltiwanger R. S. (2011) Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 21, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Shao L., Shi S., Harris R. J., Spellman M. W., Stanley P., and Haltiwanger R. S. (2001) Modification of epidermal growth factor-like repeats with O-fucose: molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J. Biol. Chem. 276, 40338–40345 [DOI] [PubMed] [Google Scholar]
- 18. Okajima T., and Irvine K. D. (2002) Regulation of notch signaling by O-linked fucose. Cell 111, 893–904 [DOI] [PubMed] [Google Scholar]
- 19. Shi S., and Stanley P. (2003) Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 100, 5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasamura T., Sasaki N., Miyashita F., Nakao S., Ishikawa H. O., Ito M., Kitagawa M., Harigaya K., Spana E., Bilder D., Perrimon N., and Matsuno K. (2003) neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development 130, 4785–4795 [DOI] [PubMed] [Google Scholar]
- 21. Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., and Vogt T. F. (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 22. Taylor P., Takeuchi H., Sheppard D., Chillakuri C., Lea S. M., Haltiwanger R. S., and Handford P. A. (2014) Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc. Natl. Acad. Sci. U.S.A. 111, 7290–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu A., Haines N., Dlugosz M., Rana N. A., Takeuchi H., Haltiwanger R. S., and Irvine K. D. (2007) In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. J. Biol. Chem. 282, 35153–35162 [DOI] [PubMed] [Google Scholar]
- 24. LeBon L., Lee T. V., Sprinzak D., Jafar-Nejad H., and Elowitz M. B. (2014) Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states. Elife 3, e02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panin V. M., Papayannopoulos V., Wilson R., and Irvine K. D. (1997) Fringe modulates Notch-ligand interactions. Nature 387, 908–912 [DOI] [PubMed] [Google Scholar]
- 26. Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., and Haltiwanger R. S. (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 275, 9604–9611 [DOI] [PubMed] [Google Scholar]
- 27. Aoki K., Porterfield M., Lee S. S., Dong B., Nguyen K., McGlamry K. H., and Tiemeyer M. (2008) The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J. Biol. Chem. 283, 30385–30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rana N. A., Nita-Lazar A., Takeuchi H., Kakuda S., Luther K. B., and Haltiwanger R. S. (2011) O-Glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J. Biol. Chem. 286, 31623–31637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Acar M., Jafar-Nejad H., Takeuchi H., Rajan A., Ibrani D., Rana N. A., Pan H., Haltiwanger R. S., and Bellen H. J. (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fernandez-Valdivia R., Takeuchi H., Samarghandi A., Lopez M., Leonardi J., Haltiwanger R. S., and Jafar-Nejad H. (2011) Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramkumar N., Harvey B. M., Lee J. D., Alcorn H. L., Silva-Gagliardi N. F., McGlade C. J., Bestor T. H., Wijnholds J., Haltiwanger R. S., and Anderson K. V. (2015) Protein O-glucosyltransferase 1 (POGLUT1) promotes mouse gastrulation through modification of the apical polarity protein CRUMBS2. PLoS Genet. 11, e1005551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thakurdas S. M., Lopez M. F., Kakuda S., Fernandez-Valdivia R., Zarrin-Khameh N., Haltiwanger R. S., and Jafar-Nejad H. (2016) Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi). Hepatology 63, 550–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee T. V., Sethi M. K., Leonardi J., Rana N. A., Buettner F. F., Haltiwanger R. S., Bakker H., and Jafar-Nejad H. (2013) Negative regulation of notch signaling by xylose. PLoS Genet. 9, e1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sethi M. K., Buettner F. F., Krylov V. B., Takeuchi H., Nifantiev N. E., Haltiwanger R. S., Gerardy-Schahn R., and Bakker H. (2010) Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J. Biol. Chem. 285, 1582–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sethi M. K., Buettner F. F., Ashikov A., Krylov V. B., Takeuchi H., Nifantiev N. E., Haltiwanger R. S., Gerardy-Schahn R., and Bakker H. (2012) Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. J. Biol. Chem. 287, 2739–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakaidani Y., Nomura T., Matsuura A., Ito M., Suzuki E., Murakami K., Nadano D., Matsuda T., Furukawa K., and Okajima T. (2011) O-Linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2, 583. [DOI] [PubMed] [Google Scholar]
- 37. Sakaidani Y., Ichiyanagi N., Saito C., Nomura T., Ito M., Nishio Y., Nadano D., Matsuda T., Furukawa K., and Okajima T. (2012) O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem. Biophys. Res. Commun. 419, 14–19 [DOI] [PubMed] [Google Scholar]
- 38. Ogawa M., Sawaguchi S., Kawai T., Nadano D., Matsuda T., Yagi H., Kato K., Furukawa K., and Okajima T. (2015) Impaired O-linked N-acetylglucosaminylation in the endoplasmic reticulum by mutated epidermal growth factor (EGF) domain-specific O-linked N-acetylglucosamine transferase found in Adams-Oliver syndrome. J. Biol. Chem. 290, 2137–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meester J. A., Southgate L., Stittrich A. B., Venselaar H., Beekmans S. J., den Hollander N., Bijlsma E. K., Helderman-van den Enden A., Verheij J. B., Glusman G., Roach J. C., Lehman A., Patel M. S., de Vries B. B., Ruivenkamp C., Itin P., Prescott K., Clarke S., Trembath R., Zenker M., Sukalo M., Van Laer L., Loeys B., and Wuyts W. (2015) Heterozygous loss-of-function mutations in DLL4 cause Adams-Oliver syndrome. Am. J. Hum. Genet. 97, 475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stittrich A. B., Lehman A., Bodian D. L., Ashworth J., Zong Z., Li H., Lam P., Khromykh A., Iyer R. K., Vockley J. G., Baveja R., Silva E. S., Dixon J., Leon E. L., Solomon B. D., et al. (2014) Mutations in NOTCH1 cause Adams-Oliver syndrome. Am. J. Hum. Genet. 95, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G. 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., and Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., and Okajima T. (2008) O-Linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 43. Takeuchi H., and Haltiwanger R. S. (2014) Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 453, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kakuda S., and Haltiwanger R. S. (2014) Analyzing the posttranslational modification status of Notch using mass spectrometry. Methods Mol. Biol. 1187, 209–221 [DOI] [PubMed] [Google Scholar]
- 45. Leonardi J., Fernandez-Valdivia R., Li Y. D., Simcox A. A., and Jafar-Nejad H. (2011) Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development 138, 3569–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Welshons W. J. (1971) Genetic basis for two types of recessive lethality at the notch locus of Drosophila. Genetics 68, 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Artavanis-Tsakonas S., Muskavitch M. A., and Yedvobnick B. (1983) Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 80, 1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S. E., Richards S., Ashburner M., Hartenstein V., Celniker S. E., and Rubin G. M. (2002) Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3, RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., Hartenstein V., Celniker S. E., and Rubin G. M. (2007) Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8, R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luca V. C., Jude K. M., Pierce N. W., Nachury M. V., Fischer S., and Garcia K. C. (2015) Structural biology: structural basis for Notch1 engagement of Delta-like 4. Science 347, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hambleton S., Valeyev N. V., Muranyi A., Knott V., Werner J. M., McMichael A. J., Handford P. A., and Downing A. K. (2004) Structural and functional properties of the human notch-1 ligand binding region. Structure 12, 2173–2183 [DOI] [PubMed] [Google Scholar]
- 52. Takeuchi H., Kantharia J., Sethi M. K., Bakker H., and Haltiwanger R. S. (2012) Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. J. Biol. Chem. 287, 33934–33944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perdigoto C. N., Schweisguth F., and Bardin A. J. (2011) Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 138, 4585–4595 [DOI] [PubMed] [Google Scholar]
- 54. Müller R., Jenny A., and Stanley P. (2013) The EGF repeat-specific O-GlcNAc-transferase Eogt interacts with notch signaling and pyrimidine metabolism pathways in Drosophila. PLoS One 8, e62835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shaheen R., Aglan M., Keppler-Noreuil K., Faqeih E., Ansari S., Horton K., Ashour A., Zaki M. S., Al-Zahrani F., Cueto-González A. M., Abdel-Salam G., Temtamy S., and Alkuraya F. S. (2013) Mutations in EOGT confirm the genetic heterogeneity of autosomal-recessive Adams-Oliver syndrome. Am. J. Hum. Genet. 92, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen I., Silberstein E., Perez Y., Landau D., Elbedour K., Langer Y., Kadir R., Volodarsky M., Sivan S., Narkis G., and Birk O. S. (2014) Autosomal recessive Adams-Oliver syndrome caused by homozygous mutation in EOGT, encoding an EGF domain-specific O-GlcNAc transferase. Eur. J. Hum. Genet. 22, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aminkeng F. (2015) DLL4 loss-of-function heterozygous mutations cause Adams-Oliver syndrome. Clin. Genet. 88, 532. [DOI] [PubMed] [Google Scholar]
- 58. Hassed S. J., Wiley G. B., Wang S., Lee J. Y., Li S., Xu W., Zhao Z. J., Mulvihill J. J., Robertson J., Warner J., and Gaffney P. M. (2012) RBPJ mutations identified in two families affected by Adams-Oliver syndrome. Am. J. Hum. Genet. 91, 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Southgate L., Sukalo M., Karountzos A. S., Taylor E. J., Collinson C. S., Ruddy D., Snape K. M., Dallapiccola B., Tolmie J. L., Joss S., Brancati F., Digilio M. C., Graul-Neumann L. M., Salviati L., Coerdt W., et al. (2015) Haploinsufficiency of the NOTCH1 receptor as a cause of Adams-Oliver syndrome with variable cardiac anomalies. Circ. Cardiovasc. Genet. 8, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okajima T., Xu A., and Irvine K. D. (2003) Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J. Biol. Chem. 278, 42340–42345 [DOI] [PubMed] [Google Scholar]
- 61. Yamamoto S., Charng W. L., Rana N. A., Kakuda S., Jaiswal M., Bayat V., Xiong B., Zhang K., Sandoval H., David G., Wang H., Haltiwanger R. S., and Bellen H. J. (2012) A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 338, 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leonardi J., and Jafar-Nejad H. (2014) Structure-function analysis of Drosophila Notch using genomic rescue transgenes. Methods Mol. Biol. 1187, 29–46 [DOI] [PubMed] [Google Scholar]
- 63. Bischof J., Maeda R. K., Hediger M., Karch F., and Basler K. (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Venken K. J., He Y., Hoskins R. A., and Bellen H. J. (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.