Abstract
CCAAT/enhancer-binding protein (C/EBPα) can appoint mouse bone marrow (MBM) cells to the osteoclast (OC) lineage for osteoclastogenesis. However, whether C/EBPα is also involved in OC differentiation and activity is unknown. Here we demonstrated that C/EBPα overexpression in MBM cells can promote OC differentiation and strongly induce the expression of the OC genes encoding the nuclear factor of activated T-cells, c1 (NFATc1), cathepsin K (Cstk), and tartrate-resistant acid phosphatase 5 (TRAP) with receptor activator of NF-κB ligand-evoked OC lineage priming. Furthermore, while investigating the specific stage of OC differentiation that is regulated by C/EBPα, our gene overexpression studies revealed that, although C/EBPα plays a stronger role in the early stage of OC differentiation, it is also involved in the later stage. Accordingly, C/EBPα knockdown drastically inhibits osteoclastogenesis and markedly abrogates the expression of NFATc1, Cstk, and TRAP during OC differentiation. Consistently, C/EBPα silencing revealed that, although lack of C/EBPα affects all stages of OC differentiation, it has more impact on the early stage. Importantly, we showed that ectopic expression of rat C/EBPα restores osteoclastogenesis in C/EBPα-depleted MBM cells. Furthermore, our subsequent functional assays showed that C/EBPα exhibits a dispensable role on actin ring formation by mature OCs but is critically involved in bone resorption by stimulating extracellular acidification and regulating cell survival. We revealed that C/EBPα is important for receptor activator of NF-κB ligand-induced Akt activation, which is crucial for OC survival. Collectively, these results indicate that C/EBPα functions throughout osteoclastogenesis as well as in OC function. This study provides additional understanding of the roles of C/EBPα in OC biology.
Keywords: Akt PKB, bone, CCAAT-enhancer-binding protein (C/EBP), differentiation, osteoclast, transcription factor
Introduction
C/EBPα3 is a transcription factor of the C/EBP family of transcription factors, and members of this family share a conserved leucine zipper dimerization domain (1). C/EBPα is critical for hematopoiesis and granulopoiesis in particular through induction of myeloid lineage-specific genes and regulation of the cell cycle for cell differentiation (2, 3). Thereby, C/EBPα can couple cell lineage commitment to terminal cell differentiation (4, 5). This is underscored by studies demonstrating that global deletion of the C/EBPα gene in mice (C/EBPα−/− mice) causes early death and a lack of mature granulocytes aside from other issues related to defective homeostasis (6) (7). Consistently, conditional deletion of the C/EBPα gene in adult mice impedes the differentiation of granulocytes, leading to an increase in myeloblasts (8). As a result, mutations that affect C/EBPα expression and/or function have been shown to be strongly associated with certain types of acute myeloid leukemia in humans (4, 9, 10). Notably, we have recently revealed a novel role for C/EBPα in osteoclastogenesis by mediating the commitment of OC precursors from the hematopoietic cell lineage into the OC lineage (11).
OCs are polykaryon bone-resorbing cells that are critical for skeletal development and bone homeostasis (12). Moreover, OCs are implicated in the pathogenesis and morbidity of numerous bone diseases, including periodontitis, rheumatoid arthritis, and post-menopausal osteoporosis (12, 13). OCs differentiate from cells of the monocyte/macrophage linage upon stimulation by M-CSF and RANKL (14). Although M-CSF mainly promotes the proliferation of OC precursors, RANKL is responsible for OC differentiation and is also crucial for the survival and activity of mature OCs (15, 16). Specifically, binding of RANKL to its receptor, RANK, on the cell surface of OC progenitors transduces intracellular signaling, leading to activation of many critical OC transcription factors, including NFATc1, the master regulator of OC differentiation (17, 18). In fact, NFATc1 is essential for the induction of numerous OC markers, including Cstk, an OC-specific gene, during osteoclastogenesis (17, 19, 20, 21).
To elucidate the molecular events by which specific factors mediate the commitment of OC precursors into the OC lineage for osteoclastogenesis, we have recently mapped the critical cis-regulatory element in the Cstk promoter and identified C/EBPα as its critical cis-regulatory element-binding protein (11). We revealed that C/EBPα is highly expressed in OCs. Furthermore, our forced expression studies revealed that C/EBPα can appoint MBM cells, widely used as primary OC precursors, into the OC lineage and thereby up-regulate OC genes independently of RANKL, indicating that C/EBPα can induce OC lineage priming. Consistent with this notion, we have reported that newborn C/EBPα−/− mice display a severe osteopetrotic phenotype from impaired OC development. However, it remains unclear whether the defective OC formation observed in this previous study stems from a defect in OC lineage commitment or whether it is the result of the requirement for C/EBPα in other stages of osteoclastogenesis.
In this study, we investigated the roles of C/EBPα in OC differentiation and function in vitro by utilizing both gain-of-function and loss-of-function strategies. This study has not only expanded our understanding of the roles of C/EBPα in OC biology but has also further supported the potential of C/EBPα as a promising therapeutic target for bone loss stemming from bone disorders of excessive OC formation and/or activity.
Experimental Procedures
Chemicals and Biological Reagents
All chemicals were purchased from Sigma. Synthetic oligonucleotides were obtained from Life Technologies. Recombinant mouse RANKL (catalog no. 462-TEC) and M-CSF (catalog no. 416-ML) were from R&D Systems. Anti-FLAG antibody (catalog no. F1804-1 mg) was purchased from Sigma. Anti-C/EBPα (catalog no. SC-61) and β-actin (catalog no. SC-81178) antibodies were from Santa Cruz Biotechnology. Anti-Akt (catalog no. 4685S) and anti-pAkt (catalog no. 2965S) were from Cell Signaling Technology.
Construct Generation
The pMX-puro-3×FLAG vector was engineered by cloning a synthesized 3×FLAG oligonucleotide into the pMX-puro vector (22). The pMX-puro-3×FLAG-p42C/EBPα (FLAG-C/EBPα) and pMX-puro-3×FLAG-p30C/EBPα (rat C/EBPα) constructs were prepared by first amplifying mouse p42C/EBPα and rat p30C/EBPα cDNAs from the pSport6-C/EBPα (Addgene) and pcDNA3.1-ratC/EBPα (Addgene) vectors, respectively. The amplified cDNAs were then subcloned in-frame with the 3×FLAG sequence into the pMx-puro-3×FLAG vector. The constructs were confirmed by sequencing. The pMX-puro-GFP vector was generated in a previous study (23).
Retroviral/Lentiviral Infection of MBM Cells
For retroviral infection, the retrovirus 293GPG packaging cell line was cultured in DMEM supplemented with 10% heat-inactivated FBS, G418, tetracycline, penicillin/streptomycin, and puromycin as described previously (24). 293GPG cells were then transiently transfected with pMX retroviral constructs using the calcium phosphate precipitation method (25). The virus supernatant was collected at days 2, 3, and 4 after transfection. For lentiviral transfection, C/EBPα shRNA or scramble shRNA lentiviral constructs from Sigma and packaging vectors were co-transfected into 293T cells using the calcium phosphate precipitation method (25, 26). The virus supernatant was harvested 60 h after transfection. The virus supernatant was then utilized to infect MBM cells for osteoclastogenesis assays.
In Vitro Osteoclastogenesis Assays
MBM cells were isolated from long bones of 4- to 6-week-old C57BL/6 mice as described previously (23, 27). Briefly, MBM cells (5 × 104 cells/well) were cultured in 24-well culture dishes in α-minimal essential medium supplemented with 10% heat-inactivated FBS and M-CSF (20 ng/ml) for 48 h. Some cells were submitted to osteoclastogenesis assays as indicated in individual experiments. Other cells were infected with virus in the presence of M-CSF (10 ng/ml) and Polybrene for 24 h before being submitted to osteoclastogenesis assays (23). For rescue osteoclastogenesis assays, MBM cells were first infected with a lentivirus for 24 h to deplete C/EBPα and then infected with a retrovirus encoding rat C/EBPα for 24 h to rescue C/EBPα expression as described previously (23, 28). At the end of the osteoclastogenesis assays, all cells were stained for TRAP activity using a leukocyte acid phosphatase kit (catalog no. 387-A, Sigma) according to the instructions of the manufacturer. Assay quantification was carried by counting or accessing the size of the multinucleated TRAP-positive cells (more than three nuclei) in representative areas. The experiments involving mice were approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Actin Ring and Acridine Orange (AO) Staining
MBM cells (5 × 104 cells/well) were treated as indicated in individual experiments. Actin ring staining was performed as described previously (23, 29). Briefly, cells were fixed with 4% paraformaldehyde for 10 min and then treated with 0.1% Triton X-100 in PBS for 10 min before staining with Alexa Fluor 488 phalloidin in PBS for 20 min. AO staining was also performed as described previously (29). Cells were incubated with 5 μg/ml AO solution in α-minimal essential medium at 37 °C for 15 min and then washed with PBS. Cells were imaged using a light-emitting diode fluorescence microscope (DM3000, Leica), and a representative image from each assay is shown.
In Vitro Bone Resorption Assays
Bone resorption analysis was carried as described previously (30, 31). Briefly, MBM cells (5 × 104 cells/well) seeded on bovine cortical bone slices were treated as indicated in individual experiments. Bone slices were then collected, and cells were removed with 0.25 m ammonium hydroxide and mechanical agitation. Bone resorption pits were imaged by a FEI Co. QuantaTM 650 FEG SEM (30) at the University of Alabama at Birmingham School of Engineering or stained with hematoxylin as described previously (31). A representative area from each assay was shown. Data were quantified by measuring the percent resorbed areas in three random areas using ImageJ software from the National Institutes of Health.
Western Blotting Analysis
Western blotting analysis was performed as described previously (32). Briefly, protein lysate was prepared and submitted to gel electrophoresis. Membranes were washed, and enhanced chemiluminescence detection was carried using Luminata Forte HRP substrate from Millipore. Membranes were visualized using a C-DiGit® blot scanner and Image Studio software from Li-Cor.
Quantitative Real-time PCR (qPCR) Analysis
qPCR analysis was carried out as described previously (11). Briefly, MBM cells were treated as indicated in individual experiments, and total RNA was collected using TRIzol reagent (Life Technologies). 1 μg of total RNA was used for cDNA synthesis by reverse transcription using SuperScript® VILOTM Master Mix (Life Technologies) according to the instructions of the manufacturer. qPCR reactions were carried using Fast SYBR® Green Master Mix reagent (Life Technologies). PCR conditions and primer sequences are available upon request. Hypoxanthine-guanine phosphoribosyl transferase (Hprt) was used as an endogenous control for normalization.
Statistical Analysis
Data are reported as mean ± S.D. Statistical significance was assessed using Student's t test. p > 0.05 was considered significant.
Results
C/EBPα Overexpression Can Mediate OC Differentiation and Strongly Induce Gene Expression with RANKL-evoked Lineage Priming
We have recently shown that forced expression of C/EBPα can reprogram MBM cells into OC-like cells and thereby primes these cells into the OC lineage for osteoclastogenesis in the absence of RANKL (11). To further examine the role of C/EBPα in OCs, we first wanted to confirm this previous finding by using the 293GPG retroviral system for gene expression (24). As expected, C/EBPα overexpression in MBM cells could initiate osteoclastogenesis by generating TRAP-positive mononucleated cells independently of RANKL compared with a GFP control (supplemental Fig. 1A). Various studies have demonstrated that RANKL mediates OC formation by inducing the expression of various genes, including NFATc1, Cstk, and TRAP (21, 33, 34). To confirm the ability of C/EBPα to mediate OC lineage priming, we examined its ability to induce the expression of the aforementioned OC genes independently of RANKL. C/EBPα overexpression in MBM cells significantly up-regulated the expression of NFATc1, TRAP, and Cstk without RANKL stimulation (supplemental Fig. 1B). These results replicate our previous finding and thus confirm that C/EBPα can mediate OC lineage commitment in the absence of RANKL.
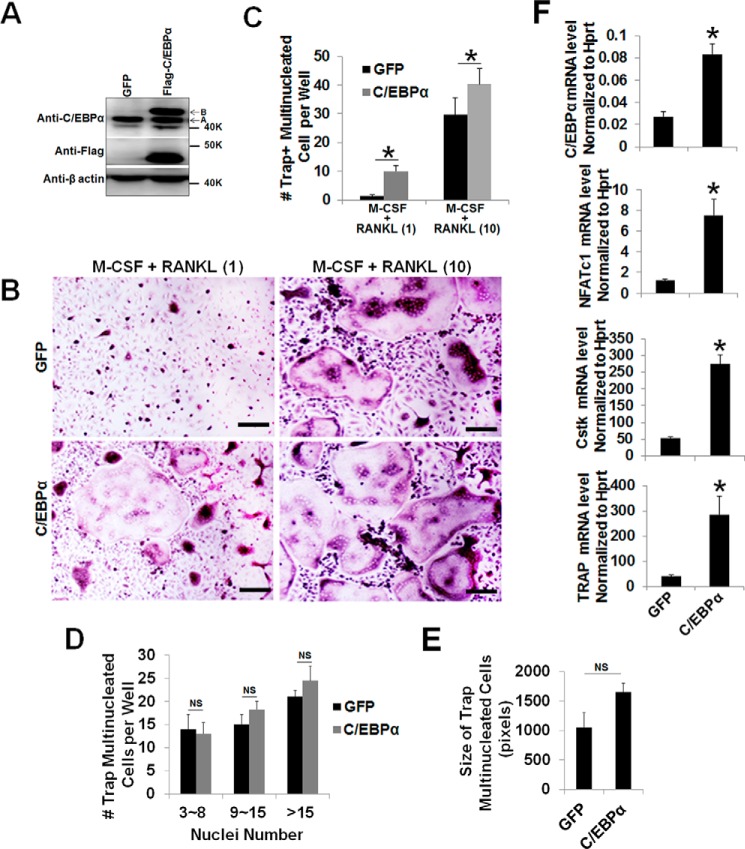
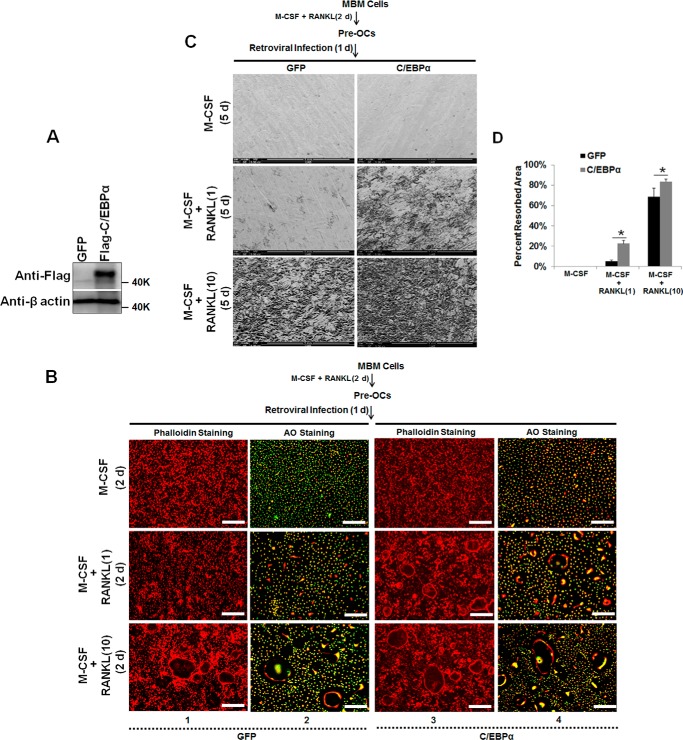
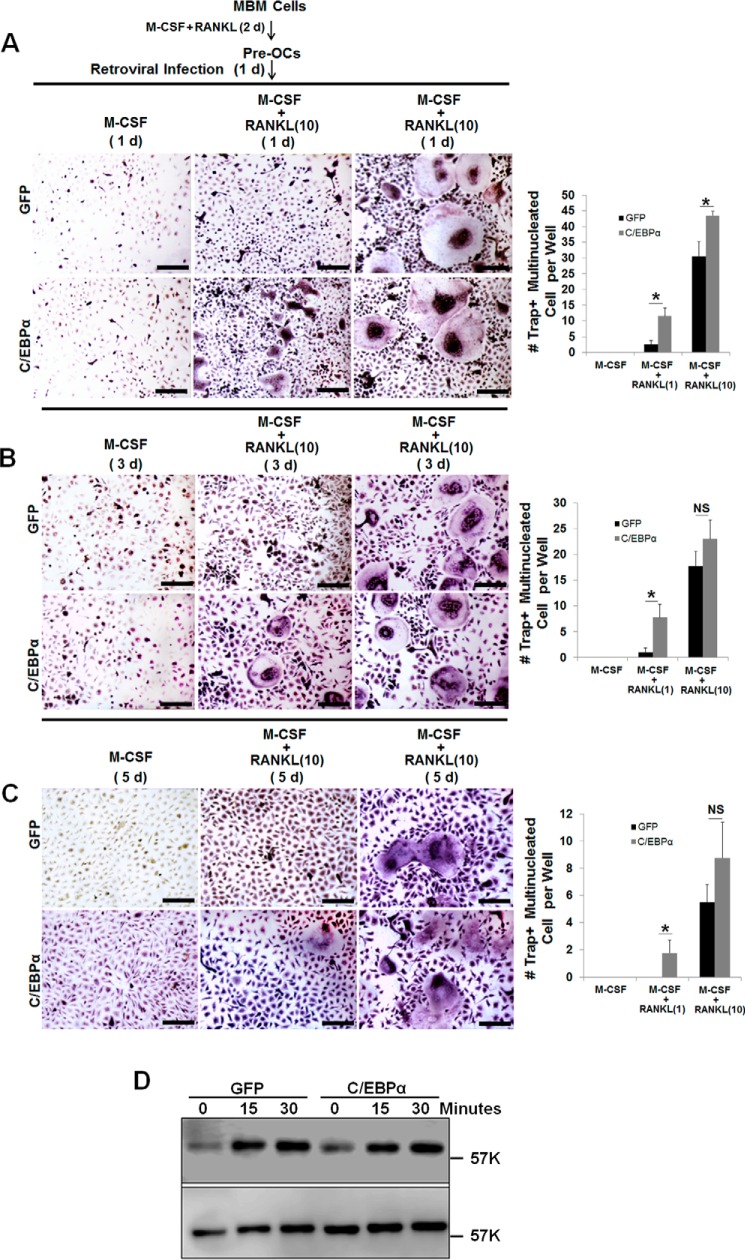
Next we focused on OC differentiation to investigate whether C/EBPα is also required beyond the lineage priming stage of osteoclastogenesis. It was reported that, although treatment of MBM cells with permissive levels of RANKL is unable to promote OC differentiation, these RANKL doses are sufficient to commit MBM cells into the OC lineage for TNF-α- and IL-1-mediated OC differentiation (23, 35–37). Hence, we utilized this strategy to examine the role of C/EBPα in OC differentiation. The permissive levels of RANKL required for OC lineage commitment is about one-tenth of the optimal RANKL dosage (10 ng/ml) utilized in standard in vitro osteoclastogenesis assays involving M-CSF and RANKL (11). Hence, we overexpressed C/EBPα in the presence of 1 ng/ml RANKL to promote OC differentiation with RANKL-induced lineage commitment (Fig. 1). MBM cells overexpressing C/EBPα or expressing the GFP control, as confirmed by Western blotting analysis (Fig. 1A), were submitted to osteoclastogenesis assays (Fig. 1B). Although MBM cells expressing GFP or overexpressing C/EBPα formed numerous OCs with M-CSF and 10 ng/ml RANKL, the C/EBPα overexpressers generated significantly more OCs than the GFP controls (Fig. 1C). Interestingly, only the cells overexpressing C/EBPα, but not the GFP expressers, could promote OC differentiation with permissive RANKL doses, indicating that C/EBPα overexpression can stimulate OC differentiation (Fig. 1, B and C). Furthermore, our data showed that C/EBPα overexpression did not induce a significant increase in OC size compared with the control cells (Fig. 1, D and E). To elucidate the molecular basis of the role of C/EBPα in OC differentiation, we examined the ability of permissive RANKL levels to induce the expression of the aforementioned OC genes in the C/EBPα overexpressers. Data showed that permissive RANKL dosages drastically up-regulated the expression of NFATc1, Cstk, and TRAP in MBM cells overexpressing C/EBPα compared with the GFP controls during OC differentiation (Fig. 1F).
FIGURE 1.
C/EBPα overexpression mediates OC differentiation and strongly induces gene expression with RANKL-evoked lineage commitment. A, MBM cells expressing the GFP control (GFP) or FLAG-C/EBPα were treated with M-CSF alone (10 ng/ml) for 4 days and then submitted to Western blotting analysis using β-actin as a loading control. A, endogenous C/EBPα; B, FLAG-C/EBPα. B, MBM cells expressing GFP or FLAG-C/EBPα (C/EBPα) were cultured with M-CSF (10 ng/ml) plus RANKL (1 ng/ml) or M-CSF (10 ng/ml) plus RANKL (10 ng/ml) for 4 days. The cultures were then stained for TRAP activity. Scale bars = 200 μm. C, quantification of B for the number of TRAP-positive multinucleated cells in at least three independent experiments. D and E, quantification of OC size for B via the number of nuclei (D) and size (E) of TRAP-positive multinucleated cells in at least three independent experiments. F, MBM cells expressing GFP or C/EBPα were stimulated with M-CSF (10 ng/ml) plus RANKL (1 ng/ml) for 3 days. Gene expression was assessed by qPCR using Hprt as a loading control from three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
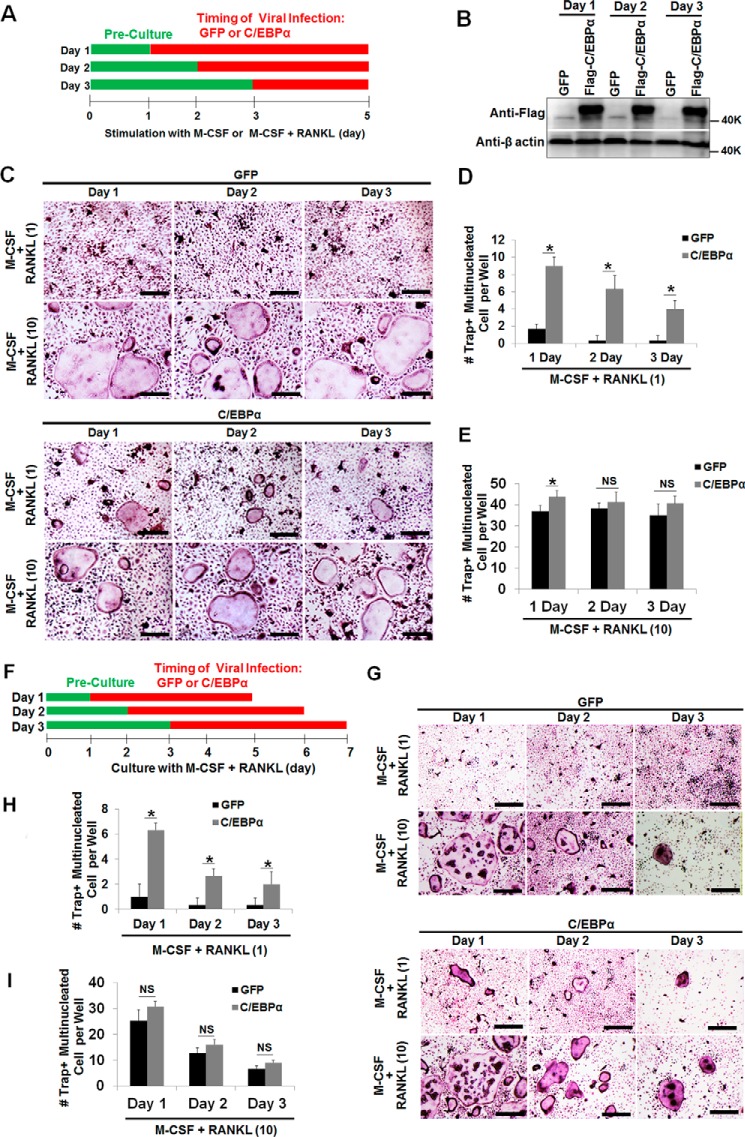
Given this new finding regarding the role of C/EBPα in OC differentiation (Fig. 1), we then sought to isolate the specific stage of OC differentiation that is regulated by C/EBPα (Fig. 2). Standard osteoclastogenesis assays require the stimulation of OC precursors with M-CSF and RANKL for 4–5 days (11, 23, 27, 36), so we infected MBM cells with a virus encoding the C/EBPα gene or GFP control in different stages of osteoclastogenesis to investigate the effects of C/EBPα overexpression in OC differentiation (Fig. 2A). Previous studies showed that treatment of MBM cells with M-CSF and RANKL for 12–24 h can commit these cells to the OC lineage for osteoclastogenesis (38, 39), so we overexpressed the C/EBPα gene on day 1, 2, or 3 of osteoclastogenesis to investigate its role in OC differentiation. C/EBPα overexpression on day 1, 2, or 3 of osteoclastogenesis, as confirmed by Western blotting analysis (Fig. 2B), could induce OC differentiation with permissive RANKL dosages (Fig. 2, C, bottom row versus center row, and D) compared with GFP controls. However, we noticed that C/EBPα played a stronger role in the early stage (day 1) than later stage (day 2 or 3) of osteoclastogenesis, as substantiated by OC numbers (Fig. 2C). Consistently, culture of the C/EBPα overexpressers with M-CSF and 10 ng/ml RANKL generated more OCs from cells in which C/EBPα was overexpressed in the early stage of osteoclastogenesis than from cells in which C/EBPα was overexpressed in the later stage (Fig. 2, C, bottom row, and E). In line with our previous data (11), we found that C/EBPα overexpression on day 0 of osteoclastogenesis gave rise to more OCs than its overexpression on either day 1, 2, or 3 (data not shown), further confirming the role of C/EBPα in OC lineage priming in addition to its role in OC differentiation. However, the stronger effect of C/EBPα overexpression in the early stage of OC differentiation compared with the later stage may result from the different duration of C/EBPα overexpression in these osteoclastogenesis assays. Therefore, in a modified experiment, we cultured all cells for 4 days after C/EBPα overexpression in the different stages of OC differentiation (Fig. 2F). Our results confirmed that C/EBPα overexpression in the early stage of osteoclastogenesis exhibited a stronger role in OC differentiation compared with GFP controls (Fig. 2, G–I). Moreover, C/EBPα overexpressers treated with M-CSF and 10 ng/ml RANKL generated more OCs than the GFP control cells (Fig. 2, E and I). Taken together, these results show that C/EBPα can promote OC differentiation by strongly inducing gene expression from RANKL-primed MBM cells.
FIGURE 2.
C/EBPα overexpression plays a stronger role in the early stage of OC differentiation but is also important for the later stage. A, schematic of the research strategy for B–E. B, MBM cells were infected with a retrovirus encoding the GFP control (GFP) or FLAG-C/EBPα (C/EBPα) on day 1, 2, or 3 of osteoclastogenesis in the presence of M-CSF alone (10 ng/ml) for 24 h. The cultures were then continued for 5 days with M-CSF alone. Gene expression was examined by Western blotting using β-actin as a loading control. C, the same set of assays as in A was repeated, but cells were infected with a retrovirus for 24 h while undergoing treatment with M-CSF (10 ng/ml) plus RANKL (1 ng/ml) or M-CSF (10 ng/ml) plus RANKL (10 ng/ml) for 5 days. D and E, quantification of the assays in C for M-CSF + RANKL (1 ng/ml, D) or M-CSF + RANKL (10 ng/ml, E) in at least three independent experiments. F, schematic of the research strategy for G–I. G, MBM cells were infected with a retrovirus encoding the GFP control or C/EBPα on day 1, 2, or 3 of osteoclastogenesis with M-CSF (10 ng/ml) and RANKL (1 ng/ml) or M-CSF (10 ng/ml) plus RANKL (10 ng/ml) for 24 h. Following the infection, all cells were cultured with M-CSF (10 ng/ml) and RANKL (1 ng/ml) or M-CSF (10 ng/ml) plus RANKL (10 ng/ml) for 4 days. The cultures in C and G were stained for TRAP activity. H–I, quantification of the assays in G for M-CSF + RANKL (1 ng/ml, H) or M-CSF + RANKL (10 ng/ml, I) from at least three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show averages ± S.D. *, p < 0.05; NS, not significant. Scale bars = 200 μm.
C/EBPα Silencing Inhibits OC Differentiation and Markedly Abrogates Gene Expression
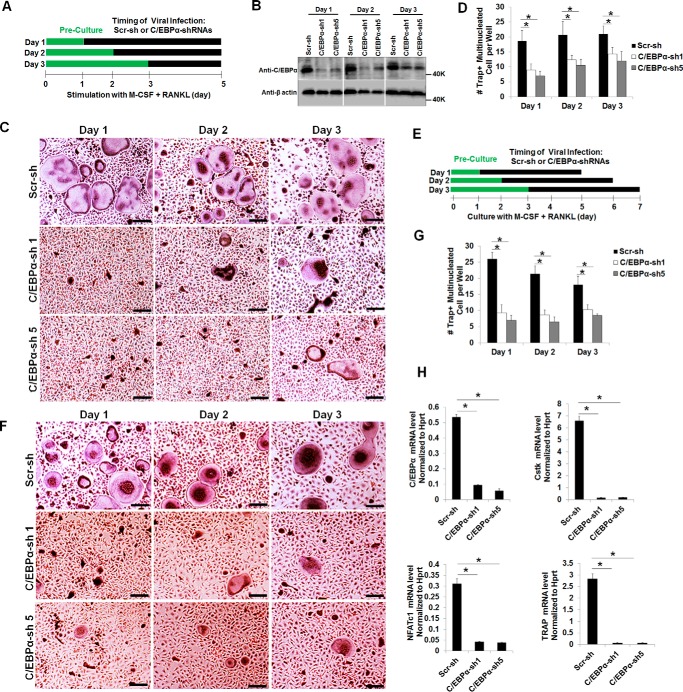
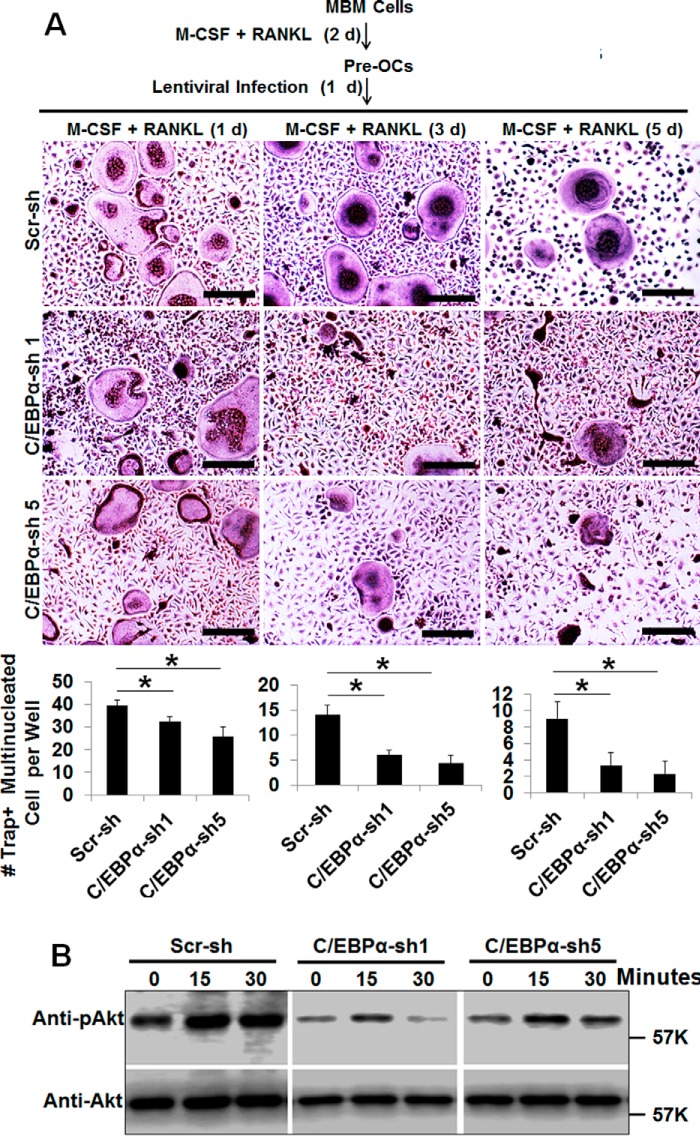
To further investigate the role of C/EBPα in OC differentiation, we utilized a loss-of-function strategy to examine the impact of C/EBPα silencing on osteoclastogenesis (supplemental Fig. 2 and Fig. 3). We were able to knock down the C/EBPα gene using three of five C/EBPα shRNA constructs (C/EBPα shRNA 1, 3, and 5) that were purchased from Sigma (supplemental Fig. 2, A and B). The ability of MBM cells expressing C/EBPα shRNA 1, 3, or 5 to promote osteoclastogenesis was drastically inhibited compared with scramble controls (supplemental Fig. 2, C and D). Notably, MBM cells expressing C/EBPα shRNA 2 or 4, which were unable to suppress C/EBPα expression (supplemental Fig. 2, A and B), failed to inhibit osteoclastogenesis (supplemental Fig. 2, C and D), confirming the specificity of C/EBPα shRNA constructs 1, 3, and 5 in inhibiting osteoclastogenesis. Because C/EBPα shRNA constructs 1 and 5 can drastically inhibit osteoclastogenesis (supplemental Fig. 2C), these two shRNA constructs were chosen to further examine the role of C/EBPα in OC differentiation (Fig. 3, A–D). Similar to C/EBPα overexpression in different stages of OC differentiation (Fig. 2), we infected MBM cells with a virus encoding C/EBPα shRNAs or the scramble control on day 1, 2, or 3 of osteoclastogenesis (Fig. 3) to examine the impact of C/EBPα silencing on OC differentiation. We found that C/EBPα silencing in the early stage (day 1) of osteoclastogenesis had a stronger inhibitory effect on OC formation than the later stage (day 2 or 3) (Fig. 3, B–D), confirming that C/EBPα plays a stronger role in the early stage of osteoclastogenesis. Nevertheless, the stronger effect of C/EBPα silencing in the early stage of osteoclastogenesis compared with the later stage may result from the different duration of C/EBPα silencing in these osteoclastogenesis assays. Therefore, in a modified experiment, we cultured all cells for 4 days after C/EBPα silencing in the different stages of OC differentiation (Fig. 3E). Our results further confirmed that C/EBPα silencing in the early stage of osteoclastogenesis exhibited a stronger role in OC differentiation compared with the scramble control (Fig. 3, F and G). Furthermore, our gene expression analysis showed that C/EBPα depletion drastically abrogated the expression of NFATc1, Cstk, and TRAP during OC differentiation (Fig. 3H). The results show that C/EBPα silencing impedes OC differentiation by blunting gene expression.
FIGURE 3.
C/EBPα silencing has a stronger impact in the early stage of OC differentiation and drastically suppresses gene expression. A, schematic of the research strategy for B–D. B, MBM cells were infected with a lentivirus encoding scramble shRNA control (Scr-sh) or C/EBPα shRNA constructs 1 or 5 (C/EBPα-sh 1 or 5) on day 1, 2, or 3 of osteoclastogenesis in the presence of M-CSF (10 ng/ml) for 24 h. The cultures were then continued for 5 days with M-CSF alone. Gene expression was examined by Western blotting using β-actin as a loading control. C, the same set of assays as in B were repeated, but cells were infected with a lentivirus for 24 h while undergoing treatment with M-CSF (10 ng/ml) plus RANKL (10 ng/ml) for 5 days. D, quantification for C from at least three independent experiments. E, schematic of the research strategy for F and G. F, MBM cells were infected with a lentivirus encoding Scr-sh or C/EBPα-sh 1 or 5 on day 1, 2, or 3 of osteoclastogenesis with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 24 h. Following the infection, all cells were cultured with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 4 days. The cultures in C and F were then stained for TRAP activity. G, quantification for F from three independent experiments. H, MBM cells expressing Scr-sh or C/EBPα-sh 1 or 5 were stimulated with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 3 days. Gene expression was assessed by qPCR using Hprt as a loading control in three independent experiments. Error bars show averages ± S.D. *, p < 0.05. Scale bars = 200 μm.
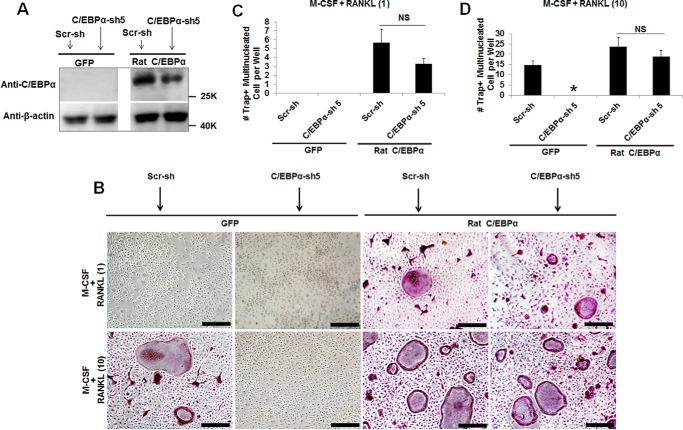
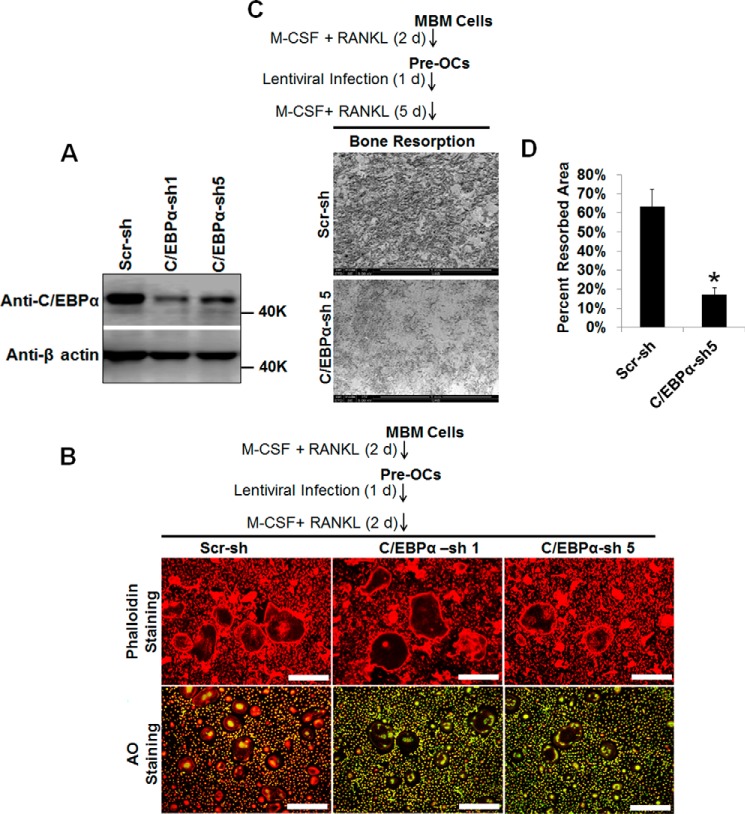
Ectopic Expression of Rat C/EBPα Can Restore Osteoclastogenesis in C/EBPα-depleted MBM Cells
The C/EBPα gene is highly homologous between mouse and rat and has two natural isoforms: the fully translated protein (p42) and a shorter protein (p30) that lacks the first 117 amino acids at the N terminus (40). To confirm the role of C/EBPα in OC differentiation, we rescued C/EBPα expression after shRNA-induced gene silencing to investigate its impact on OC differentiation (Fig. 4). Because the region targeted by C/EBPα shRNA construct 5 is located around nucleotides 98–130 of the 5′ end of C/EBPα cDNA, we used rat p30C/EBPα cDNA, which lacks this region, to rescue C/EBPα expression after gene silencing (Fig. 4A). Although C/EBPα depletion followed by GFP expression failed to promote osteoclastogenesis with 10 ng/ml RANKL, ectopic expression of rat C/EBPα in C/EBPα-depleted cells submitted to the same treatment could restore osteoclastogenesis (Fig. 4, B and D). As expected, expression of GFP or rat C/EBPα in MBM cells expressing scramble shRNA controls could mediate osteoclastogenesis with 10 ng/ml RANKL (Fig. 4, B–D). Importantly, only MBM cells doubly expressing either C/EBPα shRNA and rat C/EBPα or scramble shRNA and rat C/EBPα could mediate osteoclastogenesis with permissive RANKL doses but not MBM cells expressing C/EBPα shRNA and GFP control or scramble shRNA and the GFP control (Fig. 4, B and C). These results confirm that C/EBPα is critical for OC differentiation.
FIGURE 4.
Ectopic expression of rat C/EBPα restores osteoclastogenesis in C/EBPα-depleted MBM cells. A, MBM cells were first infected with a virus encoding a scramble shRNA control (Scr-sh) or C/EBPα shRNA construct 5 (C/EBPα-sh 5) for 24 h and then infected with a virus encoding the GFP control (GFP) or FLAG-rat p30C/EBPα (rat C/EBPα) for 24 h. Doubly infected cells were then treated with M-CSF (10 ng/ml) for 3 days before being submitted to Western blotting analysis using β-actin as a loading control. B, the same set of assays as in A was repeated, but after the double infection, cells were treated with M-CSF (10 ng/ml) plus RANKL (1 ng/ml) or M-CSF (10 ng/ml) plus RANKL (10 ng/ml) for 4 days. Cultures were then stained for TRAP activity. Scale bars = 200 μm. C and D, quantification of the assays in B for M-CSF + RANKL (1 ng/ml, C) or M-CSF + RANKL (10 ng/ml, D) are shown from three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show mean ± S.D. *, p < 0.05; NS, not significant.
C/EBPα Is Important for OC Activity
Upon finding that C/EBPα is also involved in OC differentiation aside from its previously identified role in OC lineage commitment (11), we investigated whether C/EBPα is still required beyond OC differentiation by examining its role in OC function. Toward this end, MBM cells were first differentiated into pre-OCs and then infected with a retrovirus to overexpress the C/EBPα gene (Fig. 5A), which was then continued with RANKL stimulation to form mature OCs for examination of actin ring formation, a critical feature of mature OCs, and extracellular acidification, critical for OC activity (Fig. 5B) and to promote bone resorption by mature OCs (Fig. 5, C and D, and supplemental Fig. 3). As negative controls, GFP expressers or C/EBPα overexpressers cultured with M-CSF alone remained mononucleated and did not form any actin ring (Fig. 5B, columns 1 versus 3, top row). Interestingly, C/EBPα overexpressers cultured with M-CSF alone showed extracellular acidification by mononucleated cells as compared with GFP controls (Fig. 5B, columns 2 versus 4, top row). As expected, GFP expressers or C/EBPα overexpressers treated with 10 ng/ml RANKL formed numerous OCs assessed by actin ring formation (Fig. 5B, column 1 versus column 3, bottom row) and extracellular acidification (41, 42) (Fig. 5B, column 2 versus column 4, bottom row). Notably, although C/EBPα overexpressers treated with permissive RANKL doses formed many Ocs, as substantiated by actin ring formation (Fig. 5B, column 3, center row) and extracellular acidification (Fig. 5B, column 4, center row), GFP expressers submitted to the same treatment remained mononucleated and formed no actin ring (Fig. 5B, column 1, center row) but showed little extracellular acidification by mononucleated cells (Fig. 5B, column 2, center row). These results indicate that C/EBPα overexpression does not influence actin formation but can stimulate extracellular acidification in mature OCs. Consistent with the role of C/EBPα in extracellular acidification, we found that C/EBPα overexpression significantly promoted bone resorption with 10 ng/ml RANKL, as assessed by SEM analysis (Fig. 5, C, bottom row, and D) or hematoxylin staining of the bone resorption pits (supplemental Fig. 3). Moreover, although GFP expressers treated with permissive RANKL doses barely induced bone resorption, C/EBPα overexpressers submitted to this treatment strongly induced bone resorption (Fig. 5, C, center row, and D, and supplemental Fig. 3).
FIGURE 5.
C/EBPα overexpression does not modulate actin ring formation but stimulates bone resorption by OCs. A, MBM cells were first treated with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 2 days (d) before infection with a retrovirus encoding the GFP control (GFP) or FLAG-C/EBPα (C/EBPα) for 1 day. The treatments were then continued for 2 more days. Gene expression was examined by Western blotting using β-actin as a loading control. B, the same set of assays as in A was repeated, and some cells were stained with Alexa Fluor 488-phalloidin (Phalloidin Staining) for actin ring analysis, and others were submitted to AO staining (AO Staining) for analysis of extracellular acidification in three independent experiments. Scale bars = 200 μm. C, the same set of experiments as in A was carried out with cells seeded on bone slices, but after infection, the cells were cultured for 5 more days before SEM analysis of the bone resorption pits. D, quantification for the bone resorption assays shown in C from three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show mean ± S.D. *, p < 0.05.
To confirm this finding, we turned to our loss-of-function strategy to examine the effect of C/EBPα depletion on OC activity (Fig. 6 and supplemental Fig. 4). MBM cells were differentiated into pre-OCs and then infected with a virus to ablate C/EBPα expression (Fig. 6A), which was then continued with RANKL stimulation to generate mature OCs for examination of actin ring formation and extracellular acidification (Fig. 6B) or to promote bone resorption by mature OCs (Fig. 6, C and D, and supplemental Fig. 4). C/EBPα silencing gave rise to fewer OCs than scramble controls but did not affect actin ring formation (Fig. 6B, left column). Fascinatingly, C/EBPα silencing markedly affected extracellular acidification by mature OCs, as assessed by AO staining (Fig. 6B). Moreover, C/EBPα depletion drastically abrogated bone resorption by OCs, as assessed by SEM analysis (Fig. 6, C and D) and hematoxylin staining of the bone resorption pits (supplemental Fig. 4). Taken together, these findings indicate that, although C/EBPα is dispensable for the actin ring, it is crucially involved in extracellular acidification and bone resorption by mature OCs.
FIGURE 6.
C/EBPα silencing does not affect actin ring formation but attenuates bone resorption by OCs. A, MBM cells were first treated with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 2 days before infection with a lentivirus encoding scramble shRNA control (Scr-sh) or C/EBPα shRNA construct 1 or 5 (C/EBPα-sh 1 or 5) for 1 day (d). The treatments were then continued for 2 more days. Gene expression was examined by Western blotting using β-actin as loading control. B, the same set of assays as in A was repeated, and some cells were stained with Alexa Fluor 488-phalloidin (Phalloidin Staining) for actin ring analysis in three independent experiments. The other cells were submitted to AO staining (AO Staining) for analysis of extracellular acidification in three independent experiments. Scale bars = 200 μm. C, the same set of experiments as in A was carried out with cells seeded on bone slices, but after infection, the cells were cultured for 5 more days before SEM analysis of the bone resorption pits. D, quantification for the bone resorption assays shown in C from three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show mean ± S.D. *, p < 0.05.
C/EBPα Can Regulate Cell Survival
Given the drastic impact of C/EBPα silencing on osteoclastic bone resorption (Fig. 6 and supplemental Fig. 4), we reasoned that C/EBPα might also regulate cell survival aside from its role in extracellular acidification. This is consistent with the notion that an increase in OC survival by C/EBPα may also promote OC activity independently or in concert with an increase in extracellular acidification. In addressing this notion, MBM cells were first differentiated into pre-OCs and then infected with a virus to overexpress C/EBPα, which was followed by osteoclastogenic treatments to complete a 4-, 6-, and 8-day culture for examination of OC survival by assessing OC numbers (Fig. 7, A–C). We found that C/EBPα overexpression significantly enhanced OC survival from culture with M-CSF and 10 ng/ml RANKL after 4 days of culture (Fig. 7A) but only slightly enhanced OC survival after 6 or 8 days of culture (Fig. 7, B and C) compared with GFP controls. Interestingly, we noticed that, although C/EBPα overexpression generated few OCs with permissive RANKL doses that survived through 8 days of culture, GFP controls formed no OCs, further confirming the role of C/EBPα in OC differentiation (Fig. 7, A–C). Numerous studies have reported that RANKL can promote OC survival by activating the Akt signaling pathway in OC precursors (43–47). Hence, we examined the ability of C/EBPα overexpression to activate this pathway in MBM cells. However, the ability of C/EBPα overexpression to stimulate RANKL-induced Akt activation in MBM cells overexpressing the C/EBPα gene was similar to that of GFP controls (Fig. 7D), indicating that C/EBPα overexpression does not modulate RANKL-induced Akt activation to promote cell survival.
FIGURE 7.
Analysis of the role of C/EBPα overexpression in OC survival. A–C, C/EBPα overexpression slightly enhances OC survival. MBM cells were first treated with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 2 days (d) before infection with a retrovirus encoding the GFP control (GFP) or FLAG-C/EBPα (C/EBPα) for 1 day in the presence of M-CSF (10 ng/ml) alone, M-CSF (10 ng/ml) plus RANKL (1 ng/ml), or M-CSF (10 ng/ml) plus RANKL (10 ng/ml). The cultures were then continued to complete a 4-day (A), 6-day (B), and 8-day culture (C). The cultures were stained for TRAP activity. Quantification is shown in the right panels from at least three independent experiments. Scale bars = 200 μm. D, C/EBPα overexpression does not enhance RANKL-induced Akt activation in MBM cells. MBM cells expressing GFP or overexpressing C/EBPα were treated with RANKL (10 ng/ml) as indicated. The activation of the Akt pathway was assessed by Western blotting as phosphorylation of Akt using total Akt as a loading control in three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show mean ± S.D. *, p < 0.05; NS, not significant.
Finally, we repeated these survival experiments using the loss-of-function strategy by silencing the C/EBPα gene (Fig. 8). Fascinatingly, C/EBPα depletion drastically attenuated OC survival after 4, 6, and 8 days of culture, as assessed by a decrease in OC numbers (Fig. 8A). Consistently, C/EBPα ablation suppressed Akt activation by RANKL in MBM cells (Fig. 8B). Collectively, the data show that C/EBPα is critical for OC activity by also regulating cell survival.
FIGURE 8.
Analysis of the role of C/EBPα silencing in OC survival. A, C/EBPα silencing attenuates OC survival. MBM cells were first treated with M-CSF (10 ng/ml) and RANKL (10 ng/ml) for 2 days (d) before infection with a lentivirus encoding scramble shRNA control (Scr-sh) or C/EBPα shRNA construct 1 or 5 (C/EBPα-sh 1 or 5) for 1 day in the presence M-CSF (10 ng/ml) plus RANKL (10 ng/ml). The cultures were then continued to complete a 4-, 6-, and 8-day culture. The cultures were stained for TRAP activity. Quantification is shown in the bottom panel from three independent experiments. Scale bars = 200 μm. B, C/EBPα silencing attenuates RANKL-induced Akt activation in MBM cells. MBM cells expressing Scr-sh or C/EBPα-sh 1 or 5 were treated with RANKL (10 ng/ml) as indicated. The activation of the Akt pathway was assessed by Western blotting as phosphorylation of Akt (pAkt) using total Akt (Akt) as a loading control in three independent experiments. The numbers in parentheses show concentration in nanograms per milliliter. Error bars show mean ± S.D. *, p < 0.05.
Discussion
We have previously reported that C/EBPα is essential for OC lineage commitment, but its roles in terminal osteoclastogenesis and OC activity remain unresolved. This study was aimed at addressing these critical issues.
C/EBPα Can Mediate OC Differentiation in Precommitted MBM Cells
The C/EBPα protein was initially identified based on its ability to interact with gene promoters (1) and has two natural isoforms. p42C/EBPα has two transcription activation domains (TAD1 and TAD2) at the N terminus and a basic leucine zipper domain at the C terminus. p30C/EBPα lacks the first 117 amino acids of the N terminus, which include the TAD1 region (48). Although the TAD1 and TAD2 domains can recruit co-activators and remodeling complex, the basic leucine zipper domain is important for protein interaction and DNA binding for gene expression (40). In addressing the role of C/EBPα in OC differentiation, we first utilized a gain-of-function strategy to overexpress p42C/EBPα in MBM cells in the presence of permissive RANKL levels to promote OC differentiation. We adopted this strategy to investigate whether C/EBPα is required beyond the lineage priming of osteoclastogenesis. We showed that C/EBPα overexpression can induce OC differentiation. In deciphering the specific stage of osteoclastogenesis that is regulated by C/EBPα, we showed that C/EBPα functions throughout osteoclastogenesis but plays a more critical role in the early stage. This finding is consistent with our previous report that further supports the role of C/EBPα in lineage commitment while also elucidating its novel role in OC differentiation (11). In addressing the molecular basis of the role of C/EBPα overexpression in OC differentiation, we showed that C/EBPα overexpression can drastically promote the expression of OC genes with permissive levels of RANKL.
C/EBPα Silencing Abrogates OC Differentiation
Moreover, our C/EBPα silencing studies not only reinforce our findings that C/EBPα functions throughout OC differentiation but also supports that C/EBPα is more critical for the early stage of OC differentiation. In confirming the molecular basis of the requirement of C/EBPα in OC differentiation, C/EBPα ablation drastically abrogates the expression of the OC gene during differentiation. Finally, we confirmed the role of C/EBPα in OC differentiation by rescuing osteoclastogenesis in C/EBPα-depleted cells via ectopic expression of rat p30C/EBPα. We noted that rat p30C/EBPα only partially mediates osteoclastogenesis compared with mouse p42C/EBPα (data not shown). The inability of rat p30C/EBPα to fully mediate osteoclastogenesis in these assays stems from the lack of the TAD1 region (49, 50). Nonetheless, rat p30C/EBPα retains the TAD2 domain, which can compensate for the loss of the TAD1 domain by functioning with the basic leucine zipper region to promote cell differentiation (40). Notably, the ability of rat p30C/EBPα to restore osteoclastogenesis in our assays is consistent with another study reporting that p30C/EBPα can also promote adipocyte differentiation (48).
C/EBPα Promotes OC Activity by Stimulating Extracellular Acidification and Regulating Cell Survival
We showed that, although C/EBPα plays no overt role in actin ring formation, C/EBPα is critical for extracellular acidification (29), indicating that C/EBPα is crucial for osteoclastic bone resorption. Although our gene expression analyses suggest that the ability of C/EBPα to activate OC genes may account for its role in bone resorption, we suspected that C/EBPα might also promote OC function by other mechanisms. Hence, we have previously reported that C/EBPα does not play any roles in the activation of the NF-κB and p38 signaling pathways in OC precursors, which are important for OC survival (51–54). We thus focused on investigating whether C/EBPα could activate the Akt signaling pathway in OC precursors, which has also been reported to be a strong regulator of OC survival (54–57). We revealed that C/EBPα can induce Akt activation to promote OC survival (58, 59). Importantly, this finding also suggests that a threshold level of C/EBPα may be required for effective Akt activation and that a drastic increase in C/EBPα expression may not substantially enhance Akt activation by RANKL. Hence, C/EBPα can modulate OC activity by inducing gene expression, stimulating extracellular acidification, and regulating cell survival. However, it remains to be investigated how C/EBPα can stimulate extracellular acidification in OCs. We speculate that C/EBPα may modulate the expression of specific genes, such as carbonic anhydrase II, vacuolar H+-ATPase and chloride channel ClC-7, which are known to play roles in OC acidification.
In summary, our data reveal that C/EBPα is critical for OC differentiation and activity aside from its previously reported role in OC lineage priming (11). It is likely that C/EBPα functions in a complex with different proteins at different stages of osteoclastogenesis to exert its actions. We anticipate that identification of the partners with which C/EBPα interacts will be crucial to enhance our understanding of its roles in OCs. Therefore, future studies are warranted to elucidate the molecular mechanism(s) through which C/EBPα functions, potentially in concert with other factors, in OCs.
Author Contributions
J. J., W. C., and Y. P. L. designed the study, carried out the experiments, analyzed the data, and prepared the manuscript. X. F. provided the pMX-puro and pMX-GFP retroviral constructs as well as the 293GPG retroviral packaging cells and analyzed the data. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the University of Alabama at Birmingham Center for Metabolic Bone Disease (P30 AR046031) for assistance. We also appreciate the support of the SEM Core at the University of Alabama at Birmingham School of Engineering.
This work was supported by National Institutes of Health Grants R01-AR-044741, R01DE023813, and AR-055307 (to Y. P. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. 1–4.
- C/EBPα
- CCAAT/enhancer-binding protein α
- OC
- osteoclast
- RANKL
- receptor activator of NF-κB ligand
- MBM
- mouse bone marrow
- TRAP
- tartrate-resistant acid phosphatase 5
- AO
- acridine orange
- qPCR
- quantitative PCR
- Hprt
- hypoxanthine-guanine phosphoribosyl transferase
- TAD
- transcription activation domain
- SEM
- scanning electron microscopy.
References
- 1. Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., and McKnight S. L. (1988) Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 2, 786–800 [DOI] [PubMed] [Google Scholar]
- 2. Nerlov C. (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 17, 318–324 [DOI] [PubMed] [Google Scholar]
- 3. Ramji D. P., and Foka P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M. L., Dayaram T., Owens B. M., Shigematsu H., Levantini E., Huettner C. S., Lekstrom-Himes J. A., Akashi K., and Tenen D. G. (2004) Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP α. Immunity 21, 853–863 [DOI] [PubMed] [Google Scholar]
- 5. Mueller B. U., and Pabst T. (2006) C/EBPα and the pathophysiology of acute myeloid leukemia. Curr. Opin. Hematol. 13, 7–14 [DOI] [PubMed] [Google Scholar]
- 6. Zhang D. E., Zhang P., Wang N. D., Hetherington C. J., Darlington G. J., and Tenen D. G. (1997) Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang N. D., Finegold M. J., Bradley A., Ou C. N., Abdelsayed S. V., Wilde M. D., Taylor L. R., Wilson D. R., and Darlington G. J. (1995) Impaired energy homeostasis in C/EBP α knockout mice. Science 269, 1108–1112 [DOI] [PubMed] [Google Scholar]
- 8. Ye M., Zhang H., Amabile G., Yang H., Staber P. B., Zhang P., Levantini E., Alberich-Jordà M., Zhang J., Kawasaki A., and Tenen D. G. (2013) C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol. 15, 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Porse B. T., Bryder D., Theilgaard-Mönch K., Hasemann M. S., Anderson K., Damgaard I., Jacobsen S. E., and Nerlov C. (2005) Loss of C/EBP α cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J. Exp. Med. 202, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roe J. S., and Vakoc C. R. (2014) C/EBPα: critical at the origin of leukemic transformation. J. Exp. Med. 211, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W., Zhu G., Hao L., Wu M., Ci H., and Li Y. P. (2013) C/EBPα regulates osteoclast lineage commitment. Proc. Natl. Acad. Sci. U.S.A. 110, 7294–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng X., and McDonald J. M. (2011) Disorders of bone remodeling. Annu. Rev. Pathol. 6, 121–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Souza P. P., and Lerner U. H. (2013) The role of cytokines in inflammatory bone loss. Immunol. Invest. 42, 555–622 [DOI] [PubMed] [Google Scholar]
- 14. Boyle W. J., Simonet W. S., and Lacey D. L. (2003) Osteoclast differentiation and activation. Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 15. Teitelbaum S. L. (2000) Bone resorption by osteoclasts. Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 16. Horowitz M. C., Xi Y., Wilson K., and Kacena M. A. (2001) Control of osteoclastogenesis and bone resorption by members of the TNF family of receptors and ligands. Cytokine Growth Factor Rev. 12, 9–18 [DOI] [PubMed] [Google Scholar]
- 17. Asagiri M., and Takayanagi H. (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 18. Karsenty G., and Wagner E. F. (2002) Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2, 389–406 [DOI] [PubMed] [Google Scholar]
- 19. Li Y. P., Alexander M., Wucherpfennig A. L., Yelick P., Chen W., and Stashenko P. (1995) Cloning and complete coding sequence of a novel human cathepsin expressed in giant cells of osteoclastomas. J. Bone Miner Res. 10, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 20. Chen W., Yang S., Abe Y., Li M., Wang Y., Shao J., Li E., and Li Y. P. (2007) Novel pycnodysostosis mouse model uncovers cathepsin K function as a potential regulator of osteoclast apoptosis and senescence. Hum. Mol. Genet 16, 410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takayanagi H. (2007) The role of NFAT in osteoclast formation. Ann. N.Y. Acad. Sci. 1116, 227–237 [DOI] [PubMed] [Google Scholar]
- 22. Onishi M., Nosaka T., Misawa K., Mui A. L., Gorman D., McMahon M., Miyajima A., and Kitamura T. (1998) Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18, 3871–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jules J., Shi Z., Liu J., Xu D., Wang S., and Feng X. (2010) Receptor activator of NF-κB (RANK) cytoplasmic IVVY535–538 motif plays an essential role in tumor necrosis factor-α (TNF)-mediated osteoclastogenesis. J. Biol. Chem. 285, 37427–37435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ory D. S., Neugeboren B. A., and Mulligan R. C. (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakoda T., Kasahara N., Kedes L., and Ohyanagi M. (2007) Calcium phosphate coprecipitation greatly enhances transduction of cardiac myocytes and vascular smooth muscle cells by lentivirus vectors. Exp. Clin. Cardiol. 12, 133–138 [PMC free article] [PubMed] [Google Scholar]
- 26. Graham F. L., Smiley J., Russell W. C., and Nairn R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36, 59–74 [DOI] [PubMed] [Google Scholar]
- 27. Yang S., and Li Y. P. (2007) RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev. 21, 1803–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jules J., Wang S., Shi Z., Liu J., Wei S., and Feng X. (2015) The IVVY motif and tumor necrosis factor receptor-associated factor (TRAF) sites in the cytoplasmic domain of the receptor activator of nuclear factor κB (RANK) cooperate to induce osteoclastogenesis. J. Biol. Chem. 290, 23738–23750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang D. Q., Feng S., Chen W., Zhao H., Paulson C., and Li Y. P. (2012) V-ATPase subunit ATP6AP1 (Ac45) regulates osteoclast differentiation, extracellular acidification, lysosomal trafficking, and protease exocytosis in osteoclast-mediated bone resorption. J. Bone Miner. Res. 27, 1695–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang S., Hao L., McConnell M., Zhou X., Wang M., Zhang Y., Mountz J. D., Reddy M., Eleazer P. D., Li Y. P., and Chen W. (2013) Inhibition of Rgs10 expression prevents immune cell infiltration in bacteria-induced inflammatory lesions and osteoclast-mediated bone destruction. Bone Res. 1, 267–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura T., Takahashi N., Akatsu T., Sasaki T., Udagawa N., Tanaka S., and Suda T. (1993) New resorption assay with mouse osteoclast-like multinucleated cells formed in vitro. J. Bone Miner Res. 8, 953–960 [DOI] [PubMed] [Google Scholar]
- 32. Chen W., Ma J., Zhu G., Jules J., Wu M., McConnell M., Tian F., Paulson C., Zhou X., Wang L., and Li Y. P. (2014) Cbfβ deletion in mice recapitulates cleidocranial dysplasia and reveals multiple functions of Cbfβ required for skeletal development. Proc. Natl. Acad. Sci. U.S.A. 111, 8482–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cappellen D., Luong-Nguyen N. H., Bongiovanni S., Grenet O., Wanke C., and Susa M. (2002) Transcriptional program of mouse osteoclast differentiation governed by the macrophage colony-stimulating factor and the ligand for the receptor activator of NFκ B. J. Biol. Chem. 277, 21971–21982 [DOI] [PubMed] [Google Scholar]
- 34. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., and Taniguchi T. (2002) Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 35. Lam J., Takeshita S., Barker J. E., Kanagawa O., Ross F. P., and Teitelbaum S. L. (2000) TNF-α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Invest. 106, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei S., Kitaura H., Zhou P., Ross F. P., and Teitelbaum S. L. (2005) IL-1 mediates TNF-induced osteoclastogenesis. J. Clin. Invest. 115, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jules J., Zhang P., Ashley J. W., Wei S., Shi Z., Liu J., Michalek S. M., and Feng X. (2012) Molecular basis of requirement of receptor activator of nuclear factor κB signaling for interleukin 1-mediated osteoclastogenesis. J. Biol. Chem. 287, 15728–15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taguchi Y., Gohda J., Koga T., Takayanagi H., and Inoue J. (2009) A unique domain in RANK is required for Gab2 and PLCγ2 binding to establish osteoclastogenic signals. Genes Cells 14, 1331–1345 [DOI] [PubMed] [Google Scholar]
- 39. Xu D., Wang S., Liu W., Liu J., and Feng X. (2006) A novel receptor activator of NF-κB (RANK) cytoplasmic motif plays an essential role in osteoclastogenesis by committing macrophages to the osteoclast lineage. J. Biol. Chem. 281, 4678–4690 [DOI] [PubMed] [Google Scholar]
- 40. Reckzeh K., and Cammenga J. (2010) Molecular mechanisms underlying deregulation of C/EBPα in acute myeloid leukemia. Int. J. Hematol. 91, 557–568 [DOI] [PubMed] [Google Scholar]
- 41. Li Y. P., Chen W., Liang Y., Li E., and Stashenko P. (1999) Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat. Genet. 23, 447–451 [DOI] [PubMed] [Google Scholar]
- 42. Li Y. P., Chen W., and Stashenko P. (1996) Molecular cloning and characterization of a putative novel human osteoclast-specific 116-kDa vacuolar proton pump subunit. Biochem. Biophys. Res. Commun. 218, 813–821 [DOI] [PubMed] [Google Scholar]
- 43. Liu W., Wang S., Wei S., Sun L., and Feng X. (2005) Receptor activator of NF-κB (RANK) cytoplasmic motif, 369PFQEP373, plays a predominant role in osteoclast survival in part by activating Akt/PKB and its downstream effector AFX/FOXO4. J. Biol. Chem. 280, 43064–43072 [DOI] [PubMed] [Google Scholar]
- 44. Fuller K., Wong B., Fox S., Choi Y., and Chambers T. J. (1998) TRANCE is necessary and sufficient for osteoblast-mediated activation of bone resorption in osteoclasts. J. Exp. Med. 188, 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wong B. R., Besser D., Kim N., Arron J. R., Vologodskaia M., Hanafusa H., and Choi Y. (1999) TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol. Cell 4, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 46. Murakami T., Yamamoto M., Ono K., Nishikawa M., Nagata N., Motoyoshi K., and Akatsu T. (1998) Transforming growth factor-β1 increases mRNA levels of osteoclastogenesis inhibitory factor in osteoblastic/stromal cells and inhibits the survival of murine osteoclast-like cells. Biochem. Biophys. Res. Commun. 252, 747–752 [DOI] [PubMed] [Google Scholar]
- 47. Zhao Q., Shao J., Chen W., and Li Y. P. (2007) Osteoclast differentiation and gene regulation. Front Biosci. 12, 2519–2529 [DOI] [PubMed] [Google Scholar]
- 48. Lin F. T., MacDougald O. A., Diehl A. M., and Lane M. D. (1993) A 30-kDa alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. U.S.A. 90, 9606–9610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nerlov C., and Ziff E. B. (1995) CCAAT/enhancer binding protein α amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 14, 4318–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H., Iakova P., Wilde M., Welm A., Goode T., Roesler W. J., and Timchenko N. A. (2001) C/EBPα arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8, 817–828 [DOI] [PubMed] [Google Scholar]
- 51. Gao B., Chen W., Hao L., Zhu G., Feng S., Ci H., Zhou X., Stashenko P., and Li Y. P. (2013) Inhibiting periapical lesions through AAV-RNAi silencing of cathepsin K. J. Dent. Res. 92, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boyce B. F., Xiu Y., Li J., Xing L., and Yao Z. (2015) NF-κB-mediated regulation of osteoclastogenesis. Endocrinol. Metab. (Seoul) 30, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tai T. W., Su F. C., Chen C. Y., Jou I. M., and Lin C. F. (2014) Activation of p38 MAPK-regulated Bcl-xL signaling increases survival against zoledronic acid-induced apoptosis in osteoclast precursors. Bone 67, 166–174 [DOI] [PubMed] [Google Scholar]
- 54. Gingery A., Bradley E. W., Pederson L., Ruan M., Horwood N. J., and Oursler M. J. (2008) TGF-β coordinately activates TAK1/MEK/AKT/NFκB and SMAD pathways to promote osteoclast survival. Exp. Cell Res. 314, 2725–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cicek M., Vrabel A., Sturchio C., Pederson L., Hawse J. R., Subramaniam M., Spelsberg T. C., and Oursler M. J. (2011) TGF-β inducible early gene 1 regulates osteoclast differentiation and survival by mediating the NFATc1, AKT, and MEK/ERK signaling pathways. PLoS ONE 6, e17522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kwak H. B., Sun H. M., Ha H., Lee J. H., Kim H. N., and Lee Z. H. (2008) AG490, a Jak2-specific inhibitor, induces osteoclast survival by activating the Akt and ERK signaling pathways. Mol. Cells 26, 436–442 [PubMed] [Google Scholar]
- 57. Hu J. P., Nishishita K., Sakai E., Yoshida H., Kato Y., Tsukuba T., and Okamoto K. (2008) Berberine inhibits RANKL-induced osteoclast formation and survival through suppressing the NF-κB and Akt pathways. Eur. J. Pharmacol. 580, 70–79 [DOI] [PubMed] [Google Scholar]
- 58. Qu X., Zhai Z., Liu X., Li H., Ouyang Z., Wu C., Liu G., Fan Q., Tang T., Qin A., and Dai K. (2014) Dioscin inhibits osteoclast differentiation and bone resorption though down-regulating the Akt signaling cascades. Biochem. Biophys. Res. Commun. 443, 658–665 [DOI] [PubMed] [Google Scholar]
- 59. Tu Q., Zhang J., Dong L. Q., Saunders E., Luo E., Tang J., and Chen J. (2011) Adiponectin inhibits osteoclastogenesis and bone resorption via APPL1-mediated suppression of Akt1. J. Biol. Chem. 286, 12542–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.