Abstract
Motivation: Animals from worms and insects to birds and mammals show distinct body plans; however, the embryonic development of diverse body plans with tissues and organs within is controlled by a surprisingly few signaling pathways. It is well recognized that combinatorial use of and dynamic interactions among signaling pathways follow specific logic to control complex and accurate developmental signaling and patterning, but it remains elusive what such logic is, or even, what it looks like.
Results: We have developed a computational model for Drosophila eye development with innovated methods to reveal how interactions among multiple pathways control the dynamically generated hexagonal array of R8 cells. We obtained two novel findings. First, the coupling between the long-range inductive signals produced by the proneural Hh signaling and the short-range restrictive signals produced by the antineural Notch and EGFR signaling is essential for generating accurately spaced R8s. Second, the spatiotemporal orders of key signaling events reveal a robust pattern of lateral inhibition conducted by Ato-coordinated Notch and EGFR signaling to collectively determine R8 patterning. This pattern, stipulating the orders of signaling and comparable to the protocols of communication, may help decipher the well-appreciated but poorly defined logic of developmental signaling.
Availability and implementation: The model is available upon request.
Contact: hao.zhu@ymail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
In developing cells, cascades of signaling events are believed to follow specific logic to make several evolutionarily conserved signaling pathways control the patterning of numerous phenotypically distinct body plans. At the genomic level, the DNA sequence encodes rich logic that accurately determines the conditions each gene is turned ON and OFF (Istrail and Davidson, 2005). Above this level, it is argued that ‘the heart of the matter (developmental signaling) is not so much the individual molecules involved, but more the flow of information and the logic of the system they participate in’ (Lawrence and Struhl, 1996), but this logic has never been clearly defined. Such logic is essential for ultimately deciphering morphogenesis and morphological evolution (Carroll, 2008). Integrating protein-protein interactions into complicated maps simply makes dynamic signaling deceptively complex without revealing spatiotemporal information of interactions (Guruharsha et al., 2011). Many researchers build models and compute molecular concentrations, and upon which infer signaling events and their spatiotemporal orders in cells. But such inference is difficult, error-prone and inaccurate in nature, especially when crosstalk occurs dynamically among pathways in rapidly patterning cells.
Drosophila eye development, controlled by signaling among equipotential cells and producing a striking cellular pattern, has been used to uncover the logic of developmental signaling (Freeman, 1997). The patterning of about 800 hexagonally arrayed photoreceptor 8 (R8) cells is the first step of eye development controlled by signaling among proneural genes, antineural genes and conserved signaling pathways. These genes and pathways together form multiple spatiotemporally closed feedbacks, controlling the accurate and moving process of R8 patterning.
Despite intensive studies of these proneural genes, antineural genes and Hh/Notch/EGFR/Dpp/Wnt pathways in R8 patterning in the past three decades (excellently reviewed in (Frankfort and Mardon, 2002)), the quantitative and dynamic picture of the self-sustained signaling that drives the morphogenetic furrow (MF) movement and R8 patterning remains absent. Intensive crosstalk among these genes and pathways makes it difficult to unveil the picture experimentally. Instead, to integrate experimental findings into a computational model to examine how the fine-grained and accurate organogenesis is performed is valuable for unfolding the spatiotemporally orchestrated signaling process. Recently, Lubensky et al. (2011) developed a concise model containing four equations to simulate the striped pattern of R8 specification, upon which they suggested that the dynamics of positive induction play a central role in the selection of certain cells as R8s and that R8s are defined before the appearance of the complete group of proneural cells. Their results are quite different from what experimental studies revealed, and no signaling events in R8 patterning were examined (Lubensky et al., 2011). In fact, in conventional computational studies, the logic of signaling has never been explicitly defined and addressed.
Here, we describe a model consisting of the master proneural gene ato (abbreviations of genes are in Supplementary Table S1) and key components (ligands, receptors and effectors) in the Hh, EGFR, Notch and Dpp pathways. The model combines the differential equation formulation and the handling of signaling events, aiming to define and decipher the logic of developmental signaling that controls R8 patterning in particular, and other morphogenetic events in general. We obtained two novel findings. First, the coupling between the long-range inductive signals produced by the proneural Hh signaling and the short-range restrictive signals produced by the antineural Notch and EGFR signaling is essential for generating accurately spaced R8s. Second, the spatiotemporal orders of key signaling events reveal a robust pattern of lateral inhibition conducted by Ato-coordinated Notch and EGFR signaling to collectively determine R8 patterning. These findings, unreported before, help unveil the logic of developmental signaling, and may make computational (and even experimental) researchers further examine the order and disorder of cell signaling.
2 Materials and methods
2.1 Model formulation and simulation
Upon intensive literature review we integrated well-accepted experimental findings on Drosophila eye development into a concise model that covers key components in the Hh/Notch/EGFR/Dpp pathways (Fig. 1). To make the model in a reasonable size, the complex MAP kinase pathway, for example, is represented by only one reaction from EGF receptor to the activated MAPK. In the 2-dimensional cell space (50 × 50 and 100 × 100) each cell is a system of 42 mathematical equations, and dX/dt means dX[i,j]/dt with [i,j] indicating cell’s position. Initial conditions of equations provide the initial signals to drive signaling (Fig. 1). Equations in all cells were solved simultaneously using the second-order Runge-Kutta method with adaptive time steps. We adopted very simple initial conditions (Supplementary Methods), confirming that the model does not rely on specific initial conditions. Since this is a non-dimensionalized model, time period does not indicate true time. When new R8s are determined, equations in old R8s (actually in all cells) are still being solved, with molecular concentrations at stable levels and some events continually occurring to make the R8 fate maintained. Nonlinearity of gene expression and molecular interactions is described by Hill functions in equations. Parameters were determined upon (i) experimental evidence, (ii) deep exploration of constraints among interacting molecules, (iii) some general properties of mRNAs and proteins and (iv) tuning the model to produce the correct R8 arrays.
Fig. 1.
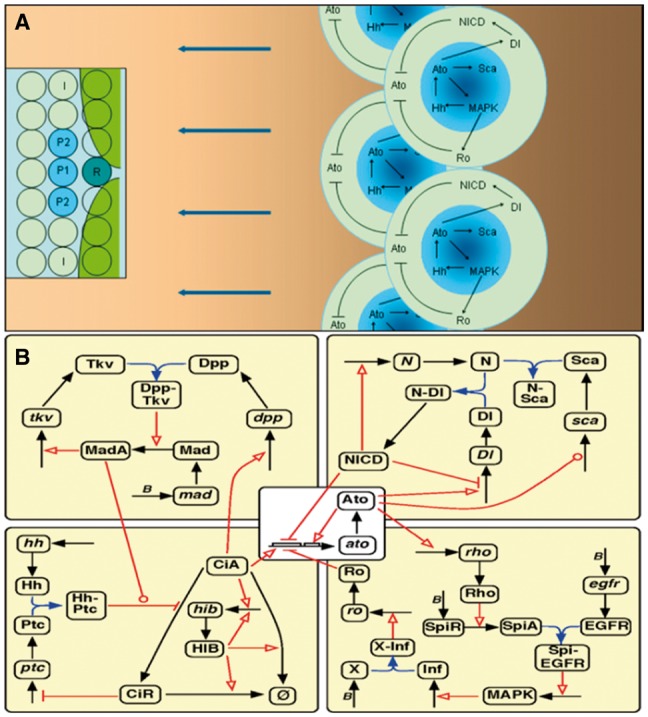
Ato-coordinated Hh, Dpp, Notch and EGFR signaling during R8 patterning. (A) Hh secretion and diffusion drives the anterior (leftward) move of Hh signaling and R8 patterning. Anteriorly diffused Hh causes CiA accumulation, which activates ato in cells anterior to the inhibitory Ro domain (green areas in inset) produced by the last column of (posterior) R8s. Ato induces sca in proneural cells (the dark center of shadowed blue areas) and delta in more cells (shadowed blue areas) to restrain and activate Notch signaling respectively, and induces rho in R8 precursors to activate EGFR/MAPK signaling. EGFR signaling first activates ro in non-R8 cells and later hh in R2/R5 cells. Thus, the Ato-EGFR-Hh-Ato feedback drives the posterior-to-anterior (right-to-left) moving of Hh signaling and R8 fate determination. In the inset R, P1, P2 and I indicate the R8 precursor, proneural cells and intervening cells. (B) The static (canonical) signaling network in each cell. Black and blue arrows indicate the production/degradation of mRNAs/proteins and binding between proteins. Red links indicate proteins’ regulatory functions, those with arrows and bars indicating positive and negative regulation (including transcriptional activation and inhibition), and those with circles indicating more complex regulation. Specifically, that from Ato to sca indicates first activation then inhibition, and that from MadA (Supplementary Table S1) to the red link of HhPtc (Hh-bound Patched) indicates that MadA enhances HhPtc’s function. Letter B indicates basal expression. Hh, Dpp, Sca, SpiA and Inf diffuse into cell. Hh diffuses several ommatidia anteriorly (see panel A) to bind to Ptc and prevents CiA from degrading to the negative CiR. Accumulated CiA induces ato and dpp expression in cells within and anterior to the MF, but is degraded by HIB in cells posterior to the MF. HIB expression is induced by CiA but quickly reaches self-activation, forming a CiA-HIB-CiA negative feedback. Ato induces Rho directly (which represses ro) and Ro indirectly via Rho and EGFR signaling and a diffusible inhibitory factor (Inf, with X as its putative receptor) downstream to EGFR
2.2 Definition and capture of signaling events
When protein A’s concentration reaches the half-maximal activation/repression coefficient in the Hill function describing how A nonlinearly activates/represses B (a protein or a gene), A sends the message activation or repression to B, which is captured as A_Act_B or A_Rep_B. Upon experimental studies, ligand–receptor binding shows unclear or lesser nonlinearity than transcriptional activation/repression and protein activation/repression by phosphorylation. Lacking clear evidence whether ligand–receptor binding happens at particular concentrations, binding events may not be defined optimally. For an event determined upon several other events, it is defined upon the logic relationship of the related events (Supplementary Methods). Some events, such as Ato_Act_hh_0_p1, represent several steps of signaling occurred in or between cells. Defined events (Supplementary Table S2) are captured in each and every cell during simulations. Definition and capture of events and numeric solution of equations were facilitated by a program (available upon request) developed for multi-cellular modeling (Zhu et al., 2005).
3 Results
3.1 The long-range Hh signaling and the short-range Notch and EGFR signaling form the core system for R8 patterning
As multiple genes and pathways participate in R8 patterning, our first question is what comprises the core signaling system. According to experimental studies, Wnt signaling is dispensable for R8 patterning in eye discs central area. Upon literature review and the examination of different model components, we found that the master proneural gene ato and about 20 genes and their products in the Hh/Notch/EGFR/Dpp pathways form the most parsimonious model of R8 patterning (Fig. 1). Ato and the Hh/EGFR/Notch pathways suffice to generate R8 arrays even when parameters vary considerably. Since the anteriorly diffused Hh (secreted by R2/R5 cells in patterned ommatidia) first activates the positive feedback between CiA accumulation and Ptc expression and then allows the increased CiA to activate ato expression (Chen et al., 1999; Strutt and Mlodzik, 1996), the long-range Hh signaling triggers the proneural fate in most cells in a band of cells ahead of the MF. Subsequently, Ato triggers the short-range Notch and EGFR signaling in these cells to spatially restrict ato expression firstly into proneural cells and finally into R8 cells (NICD and Ro inhibit ato expression) (Fig. 1). These simulated processes match experimental observations very well (Supplementary Fig. S2). Simulations suggest that, to dynamically generate the subtle R8 pattern, Ato activity is essential to spatiotemporally coordinate the long-range and short-range signaling systems (Supplementary Fig. S1). Dpp signaling is reported to promote CiA accumulation, and thus, to accelerate MF movement (Fu and Baker, 2003). Simulations suggest that Dpp signaling has two opposite effects—while MadA promotes CiA accumulation, it also negatively affects the process, because an expanded CiA domain also causes an expanded Ptc domain which would significantly increase Hh consumption. This finding may explain why Dpp signaling only slightly accelerates MF movement.
3.2 Signaling events elucidate discrete steps of cell fate determination
Next we addressed the properties of the core signaling system, especially, the generated signaling events. It is experimentally observed that ato is widely up-regulated initially in all cells in the MF, then down-regulated firstly in intervening cells (I cells) and finally in all proneural cells except R8 (Frankfort and Mardon, 2002) (Fig. 1), but the timing and order of signaling events remain unknown. Simulations reveal how R8 fate is determined by two positive events, CiA_Act_ato and Ato_Act_ato, together with two negative events, NICD_Rep_ato and Ro_Rep_ato. We examined these events in cells during different columns of R8 patterning. Computed molecular concentrations and captured signaling events show that ato is induced earliest by CiA_Act_ato in the R8 precursor (which is closest to the sources of Hh but is beyond the action of Ro_Rep_ato produced by cells posterior to the MF), and then in proneural cells (Fig. 1; Supplementary Fig. S1). Sca, induced by high Ato, binds to Notch on the R8 precursor and blocks Notch signaling within. Delta, widely induced by ato expression, binds to Notch on neighboring cells and activates Notch signaling in these cells (Powell et al., 2001). The low Sca in the P1/P2/I cells, diffused from the R8 precursor and/or produced locally, does not effectively block Notch signaling and prevent NICD_Rep_ato in these cells. Later, since the I cells are beyond the scope of SpiA but within the scope of the inhibitory factor Inf (Baonza et al., 2001), there is Ro_Rep_ato but no EgfrB_Act_MAPK in these cells. The spatiotemporally ordered events of Notch/EGFR signaling in these cells, in the background of the Hh gradient and the Ro domain, provide a mechanistic explanation for computed molecular concentrations and experimentally observed cell fate determination. Notably, while molecular concentrations look the same in different columns of R8, signaling events (especially events of Dpp and Hh signaling) show variations (Figs 2 and 3), which reveal a noticeable aspect of developmental signaling and patterning. Simulations suggest that, for correct R8 spacing, an adequate time interval between EGFR and Hh activation is required to allow EGFR signaling and Ro distribution to become stable when a new round of ato expression is activated.
Fig. 2.
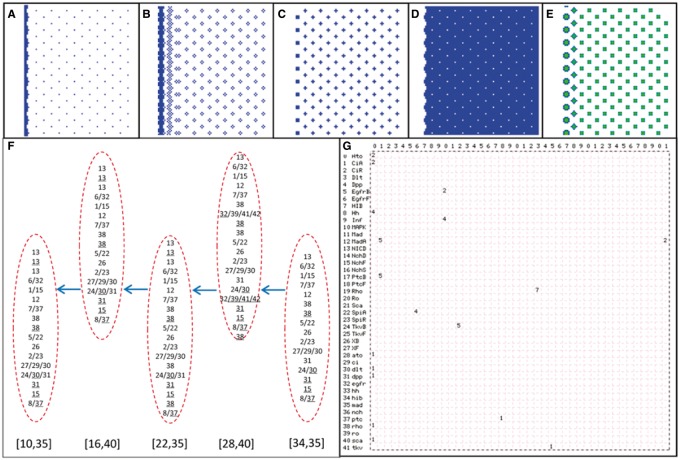
Simulation results. (A–D) The distribution of event Ato_Act_dlt, NICD_Rep_ato, EgfrB_Act_MAPK and Ro_Rep_ato at a time step in the 100 × 100 cell space. (E) The distribution of MAPK concentration at a time step in the 100 × 100 cell space. (F) The sequences of events in some R8s in different columns of ommatidia. Arrows indicate the movement of the MF. [34,35], [28,40], [22,35], [16,40] and [10,35] indicate the position of five R8s. (G) Events occurring at a time step form an emergent network; a stable network formed by continually occurring events indicates an attractor of signaling. Numbers in the top row, as in the left column, indicate the 42 molecules (Supplementary Table S1), and numbers in the lattice indicate events, including expression (Exp), binding (Bid), ubiquitination (Ubi), activation (Act) (Supplementary Table 2)
Fig. 3.
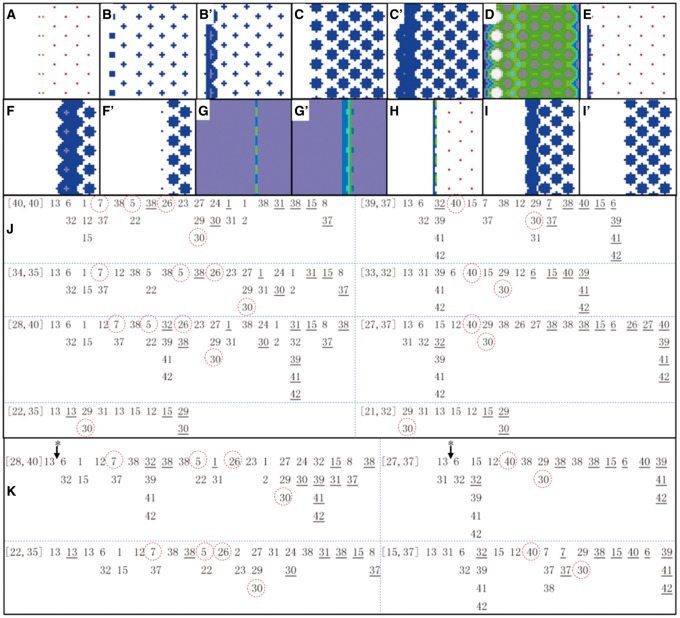
Impact of changed parameters and changed events on R8 patterning. (A–E) are distributions of Rho, Sca, SpiA, XB and Ato concentration, respectively, at some time steps under trNICD_ato = 0.003→0.0045. For comparisons, (B′) and (C′) are distributions of Sca and SpiA concentration at the same time steps under the default condition. This change of trNICD_ato causes ectopic (more posterior) ato expression (indicated by ectopic Rho at t = 588 in (A)), influences Notch signaling (indicated by Sca at t = 786 in (B)) and influences EGFR signaling (indicated by SpiA at t = 786 in (C)). New R8s are generated, but with a defect (indicated by XB and Ato at t = 860 in (D) and (E)). (F–I) are distributions of SpiA, CiA, ato and SpiA concentration at different time steps under taAto_rho = 0.35→0.175. For comparisons, (F′), (G′) and (I′) are distributions of SpiA, CiA and SpiA at the same time steps under the default condition. This change of taAto_rho activates rho expression and EGFR signaling earlier (indicated by SpiA at t = 307 in (F)). The change also influences Hh signaling via Ato (indicated by CiA at t = 371 in (G)), arrests R8 generation (indicated by ato at t = 426 in (H)) and influences EGFR signaling (indicated by SpiA at t = 475 in (I)). (J) The sequences of some events in the case taAto-rho = 0.35→0.175. [40,40] and [39,37] indicates cell position; events (as numbered in Supplementary Table S2, and ordered left-to-right) without and with an underline indicate the initiation and stop of the events; circled events are critical ones and follow rigorous orders. In R8 cells ([40,40], [34,35], [28,40], [22,35] at the left side) the order of events appears the same as that under the default condition (compare the marked 7, 5, 26, 30 with those in Fig. 2), but in the I cells ([39,37], [33,32], [27,37], [21,32] at the right side), the occurrence of the event NICD_Rep_ato is gradually postponed and the activation of ro becomes relatively earlier (see the marked 40, 30 in these cells). (K) When taAto_rho = 0.35→0.175 is corrected at t = 400 (indicated by * with an arrow), all columns of R8 are generated, but a correction made at t = 450 is not successful, because it is too late to correct the subsequent EGFR signaling and ro expression. Events 7, 5, 26, 30 shown in the two R8 cells ([28,40] and [22,35]), and events 40, 30 shown in the two I cells ([27,37] and [15,37]), follow the correct order, but the timing and their positions in the event sequences are different
3.3 Notch/EGFR signaling forms concerted lateral inhibition
Notch signaling is ubiquitously used in embryogenesis by cells that acquire (or are about to acquire) a particular fate to stop their neighbors from acquiring the same fate. Notch and Delta signaling thus implements a lateral inhibition mechanism that acts between directly connecting cells, which has been examined intensively (Collier, 1996; Webb and Owen, 2004). How inhibition happens in cells not directly connected is less examined and poorly understood. While EGFR signaling regulates Ato levels in cells within an ommatidium (Chen and Chien, 1999), it is difficult to determine experimentally the relative order of Ato_Act_ato and Ato_Act_rho, or whether rho is activated before a cell obtains the R8 fate, and therefore, when EGFR signaling begins to regulate R8 patterning (Spencer et al., 1998). Simulations indicate that Ato_Act_rho should happen before Ato_Act_ato (taAto_rho = 0.35 and taAto_ato = 0.45, the parameter setting taAto_rho < taAto_ato enables Ato_Act_rho to happen earlier than Ato_Act_ato) and that increasing or decreasing taAto_rho by 50% (which would cause a delayed or precocious Ato_Act_rho) makes R8 patterning fail after 3 or 4 rounds of R8 patterning. These results suggest that Notch and EGFR signaling, activated by Ato at different time points (taAto_delta = 0.01 and taAto_rho = 0.35), forms concerted lateral inhibition to regulate competitive R8 determination (Baonza et al., 2001; Dokucu et al., 1996; Powell et al., 2001). Given that Ro is activated by EGFR via a hypothesized diffusible inhibitory factor (Baonza et al., 2001), NICD_Rep_ato and Ro_Rep_ato act in different spatial scope and temporal stage (Ro_Rep_ato occurs slightly later in more cells, including in R8 precursors for a short time, see Figs 2 and 3), with a proper timing, on ato. Since Ato-activated Notch and EGFR signaling also acts in the development of chordotonal sense organ precursors (Zur Lage and Jarman, 2000), it is sensible to infer that they combine to make robust lateral inhibition in a large group of cells for competitive cell fate determination.
3.4 R8 patterning is robust to changes in some signaling events
Since the spatiotemporal restraint of ato expression by Notch and EGFR signaling is essential for R8 determination, we compared the impact of changed Ato_Act_rho and NICD_Rep_ato, two events in the R8 cells, on R8 determination. We made the two events happen earlier and later by decreasing and increasing their controlling parameters taAto_rho and trNICD_ato by 50%. Notably, the changed NICD_Rep_ato did not affect R8 patterning (trNICD_ato = 0.003→0.0015), or just made the fifth column of R8 slightly more posterior (closer to the previous column of R8s) (trNICD_ato = 0.003→0.0045) (Fig. 3A–E). Similarly, the changed Ato_Act_rho did not affect R8 patterning until the forth column of R8 (taAto_rho = 0.35→0.175) (Fig. 3F–I). In this case, EGFR and Notch signaling was seen to become gradually, instead of immediately, decoupled (Fig. 3J). R8 patterning of fifth to eighth columns can be recovered when taAto_rho = 0.35→0.175 is corrected (akin to rescue of a mutation in ato) at an early time (t = 400), but cannot if taAto_rho = 0.35→0.175 is corrected at a late time (t = 450). The corrected parameter changes the timing and order of Notch/EGFR signaling events (Fig. 3K).
Robust developmental patterning can be generated by properties of signaling such as positive and negative feedbacks, and by redundancy in signaling as evidenced by multiple antineural genes involved in R8 patterning. So far, robustness of a computational model is measured by the model’s sensitivity to changes in parameters (von Dassow et al., 2000). Nevertheless, changes in parameters do not necessarily, or do not in a simple way, cause changes in signaling events (Zhu and Mao, 2015). To examine the sensitivity of R8 patterning to changes in signaling events, we captured events in successive rounds of R8 patterning. The results indicate that signaling events show round-to-round variations and the correct R8 pattern was generated robustly despite significant changes in the timing, order and occurrence of some events (Fig. 2). As described above, the model can also buffer the impact of changed parameters to delay the occurrence of changed or erroneous events. These results, for the first time, reveal a new dimension of robustness of developmental signaling, which is impressive in R8 patterning and may be ubiquitous in developmental patterning.
3.5 The spatiotemporal orders of key events show a specific pattern of signaling in cells
Developmental patterning is controlled by an array of tissue-specific selector genes together with a few evolutionarily conserved signaling pathways (Affolter and Mann, 2001). In developmental biology, a fundamental question is to what extent does developmental signaling share intrinsic commons. To explore the properties of the Ato-coordinated Hh, EGFR and Notch signaling, apart from taAto_rho, taAto_ato and taAto_delta, we further increased and decreased the value of 39 other parameters by 50% and examined the impact of these changes on R8 patterning. In many cases (40 out of 78) R8 patterning remained successful. In the cases where R8 patterning was wrong, changes often did not cause immediate failure of R8 patterning; instead, one or two columns of R8 were generated before failure occurred (14 out of 38). These indicate that the Hh/Notch/EGFR/Dpp signaling as modeled here is rather robust against parameter changes. We then analyzed the sequences of signaling events produced by the 78 simulations. In all cases, as long as a column of R8 was generated, events of Notch/EGFR signaling in the R8 show a specific order—Ato_Act_sca, Ato_Act_rho, EgfrB_Act_MAPK and XB_Ato_ro. Moreover, as long as the order of these events occurred in a cell, the cell became R8. The correspondence between the R8 fate and the order of these events, regardless of changes in the timing, order and occurrence of other events caused by parameter changes, may reveal an important property of the Ato-coordinated Hh/Notch/EGFR/Dpp signaling in R8 patterning. Such orders of signaling in and between cells are comparable to protocols of communication (such as TCP/IP) in and between computers; both stipulate orderly transmission of information.
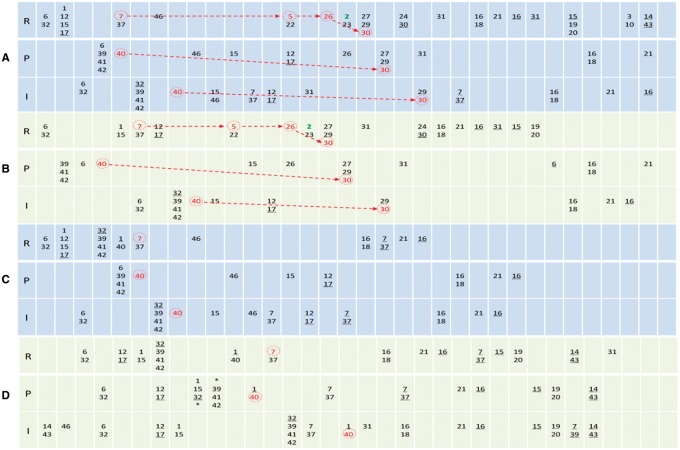
In the R8 cells, as CiA_Act_ato is not repressed by Notch and EGFR signaling, the self-sustained Ato expression (event 2) occurs at the correct time following event 7, 5 and 26 and marks R8 fate (Fig. 4). In the proneural cell P1 and the intervening cell I, key events of Notch/EGFR also show specific orders. Unique to the P1 cell is the order NICD_Rep_ato, EgfrB_Act_MAPK and XB_Ato_ro, lacking Ato_Act_sca and Ato_Act_rho; and unique to the I cell is the order NICD_Rep_ato and XB_Ato_ro, further lacking EgfrB_Act_MAPK (Fig. 4). Generally, in non-R8 cells where signaling is less constrained sequences of events are more varied. In failed or wrong R8 patterning, these orders are disrupted in these cells, indicating that concerted Notch/EGFR signaling follows a specific spatiotemporal pattern in multiple cells. From these results we hypothesize that such orders and patterns may intrinsically characterize the concerted lateral inhibition conducted by Notch/EGFR signaling, for R8 patterning in particular and for competitive cell fate determination in general (Culi et al., 2001; Hajnal et al., 1997).
Fig. 4.
The sequences of events in an R8 cell, a P1 cell and an I cell during R8 patterning. In all panels, R, P and I indicate the R8 cell, the P1 cell and the I cell (see inset in Fig. 1). Events are numbered as in Supplementary Table S2 and ordered left-to-right. Columns help display the order of events but do not contain time information. Multiple numbers in a unit (and those with ‘*’ in two units) indicate concurrent events. Events without and with an underline indicate the initiation and stop of the events. Red numbers indicate key Notch/EGFR signaling events. The self-sustained Ato expression (event 2) follows event 7, 5 and 26 and marks the R8 fate. Notch and EGFR signaling, by repressing Ato expression in other cells, allows Ato_Exp_ato to occur only in R8 cells. Red events, together with event 2, are key events that make a cell R8, and they show specific orders in these cells in successful R8 patterning (A, B), but do not in unsuccessful R8 patterning (C, D). In non-R8 cells (and failed R8 cells), sequences of events vary more significantly as signaling is less constrained (note that the fate of these cells is undetermined at the moment of R8 determination). (A) With the default parameter setting, R8 patterning is correct. (B) With taAto_delta = 0.005, R8 patterning is correct. (C) With taXB_ro = 0.00825, R8 patterning fails. (D) With baseptc = 0.06, R8 patterning fails
4. Discussion
4.1 A note on parameter settings and captured events
We chose Drosophila eye development to explore the logic of developmental signaling because Drosophila eye development is the most studied, best characterized and remarkably accurate developmental patterning process, no other process provides more or better data for systematic and quantitative investigations targeting coordinated pathway interactions. It allows us to determine parameters upon abundant experimental reports and to explore vast constraints among parameters (Supplementary Methods). For such a complex model, the chance of accidentally producing the subtle R8 array with parameters in unreasonable ranges is extremely small. Our conclusions are upon signaling events under varied orders but not molecular concentrations, and are therefore independent of specific parameter values. For a small model it may be preferable to build it upon a specifically designed experiment with parameters being accurately measured; but for a large one it is only feasible to build it upon abundant experimental findings with parameters being reasonably estimated and rigorously tested.
A question is whether it is meaningful, or possible, that event sequences in different R8 cells are somewhat different. Functionally redundant signaling components and feedbacks, parallel activities of multiple pathways, intrinsic stochasticity of gene expression, fluctuation of molecular concentrations and tissue specific environments all make it possible for molecular interaction to generate different event sequences in different cells, including cells obtaining the same fate at different times and different places. Li et al., using a completely different method, revealed that a yeast cell can undergo different state transition pathways to fall into an attractor and the pathway is robust against small perturbations (Li et al., 2004); this equals to that a cell undergoes different molecular interaction events to reach a stable state (including to enter into its final fate as we examine here). Also comparable to their finding that it is very unlikely for a sequence of events to deviate from the cell-cycle pathway, we explicitly demonstrate that the order of key events is highly robust. Thus, events and orders of events may be better than molecule concentrations for unveiling properties of signaling.
It is well justified to define and model nonlinear molecular interactions (such as protein activation and repression by phosphorylation) as a series of events (Naldi et al., 2009) or logic-like operations (Li et al., 2004), but to what extent ligand–receptor binding is nonlinear is less clear. The way we define ligand–receptor binding may not be optimal, which may influence not only the timing, duration and occurrence of binding events per se but also event sequences as a whole. This may, to some extent, explain the variation of inessential events in silico and in vivo. Another question is whether changes of events (for example, a delayed or absent A_Act_B) observed in simulations occur in vivo. In the model, if a protein’s concentration does not reach the defined threshold for it to interact with its target, the defined event does not occur; this does not necessarily mean that the event would be absent in vivo. Nevertheless, upon the widely accepted mass-action rule, the absence of A_Act_B due to low concentration of A indicates that A_Act_B would occur for a shorter period in vivo and thereby influence the sequence and order of related events. Thus, simulated sequences and orders of events indeed reasonably reflect in vivo situations.
4.2 Potential missing regulators of Notch signaling
Ato induces the expression of two Notch ligands, the membrane-tethered Delta and the diffusible Sca. Delta/Notch binding produces NICD but Sca/Notch binding, with a higher affinity, just stabilizes Notch (Baker and Yu, 1998; Lee et al., 1996; Powell et al., 2001). As Sca is only observed in proneural cells, it is suggested that cells expressing high levels of Ato express Sca to repress Notch signaling in proneural cells (Powell et al., 2001). We tried a range of values for the two parameters (taAto_delta and taAto_sca) that control the activation of delta and sca by Ato. A relatively small taAto_sca resulted in timely Sca expression, but in a domain that was too broad; a relatively large taAto_sca resulted in Sca expression in a correctly confined domain, but too late. Neither agrees with the experimentally observed timely and confined Sca distribution. To check if a large Sca/Notch binding affinity would make Sca distribution correctly confined, different kDlN and kScaN (the parameters reflecting Notch/Delta and Notch/Sca binding affinity) were tried. A large kScaN alone was unable to fully prevent Delta/Notch binding and ato repression in proneural cells and, since Sca is diffusible, a very large kScaN resulted in repression of Notch lateral inhibition in intervening cells.
Since Delta/Notch binding was not adequately blocked by Sca in proneural cells in the model, ato down regulation by NICD (event NICD_Rep_ato) happened simultaneously in both proneural and intervening cells (Figs 3 and 4) (experimental observations are that ato down regulation happens first in intervening cells). A question is, if this probably flawed Notch signaling is due to sca expression being solely controlled by Ato, what could be the extra regulators of Sca? No evidence highlighting any co-regulators within the Notch pathway has been reported. It is reported that EGFR activation promotes Delta expression (Tsuda et al., 2002), and it is speculated that MAPK activity in proneural cells could help antagonize Notch lateral inhibition (Jones and Moses, 2004). However, as our simulations show, the EGFR/MAPK system acts too late to participate in Sca regulation. Whether, apart from the Notch and EGFR pathways, there are components in other pathways that participate in the timely regulation of Sca is an open question. Nevertheless, the model indicates that even with the flaw in Notch signaling, R8 patterning is still very robust, resistant to many parameter changes.
4.3 Concerted EGFR and Notch signaling
Notch signaling is involved in diverse biological processes to generate NICD to activate a range of target genes. A specific and well characterized aspect of Notch signaling is lateral inhibition, by which a cell that obtains or is about to obtain a particular fate inhibits its neighboring cells from obtaining the same fate. In this situation NICD generation is maintained and amplified for a period of time by Notch signaling via a negative feedback in which NICD, together with other factors, enhances Notch expression and represses Delta expression to force cells to adopt different fates (Collier, 1996; Heitzler, 1991).
EGFR is reported to act antagonistically with Notch-mediated lateral inhibition to promote proneural genes in proneural clusters (Culi et al., 2001). Nevertheless, Drosophila eye provides an example showing that, by activating the downstream Ro via an inhibitory factor (Baonza et al., 2001; Spencer et al., 1998), it also acts synergistically with Notch to repress proneural genes in intervening cells. By examining how the model responds to changed parameters, we find that EGFR signaling, by activating diffusible Ro, influences R8 patterning more significantly than Notch signaling (the latter functions at smaller scale) (Fig. 3). Given that Notch and EGFR can be tissue-specifically activated by multiple proneural genes, including ato, achaete and scute, we postulate that, besides Ro (an eye-specific homeobox gene (Saint et al., 1988)), other tissue-specific inhibitory EGFR downstream targets should also exist, which enable EGFR signaling to play specific roles in different cells, antagonistically or synergistically with Notch. Indeed, other pathways also work synergistically instead of independently for information processing during development (Hayward et al., 2008). Moreover, simulations suggest that the proposed inhibitory factor in the eye, produced dependent on EGFR signaling, may have a half-life comparable to that of PtcB and a diffusion scope comparable to that of Spitz.
This study predicts that the key events of Notch and EGFR signalling follow a specific and robust spatiotemporal order in R8 and its neighbouring cells. This spatiotemporal order also enables EGFR signaling and Ro expression to be correctly and stably distributed in cells when a new round of ato expression and R8 generation is activated. Notably, compared with events in R8s, events in other cells are not only fewer but also more varied, which is reasonable because the fate of these cells is undetermined.
4.4 A new dimension of robustness of developmental patterning
Glypicans are reported to play important roles in regulating the movement of diffusible molecules and their gradients in cells by buffering their concentration fluctuations (Kreuger et al., 2004; Lin, 2004). Since no glypican is included in the model, when a failed or ectopic R8 occurs, it considerably affects the concentration of Hh, Inf and other diffusible proteins in cells and impair the following R8 patterning. Despite the absence of glypicans and the flaw in Notch signaling, the model still shows impressive robustness against changes in parameters and in signaling events. The robustness of R8 patterning, examined for the first time upon captured signaling events, reveals a new dimension of robustness of developmental signaling and patterning. Different from such mechanisms as feedbacks and redundant components in signaling pathways, this dimension of robustness is determined by the occurrence and order of essential signaling events (Zhu and Mao, 2015), whereas the occurrence, timing and order of other events can be varied without visibly influencing R8 patterning. This property may widely exist in developmental signaling and patterning.
4.5 The logic of developmental signaling
‘Timing is everything’ in developmental signaling has been well recognized, but details of how critically timing influences signaling are poorly uncovered. More fundamental than timing is the logic of developmental signaling, which is frequently discussed but never clearly defined. A case in point is Notch pathway, which is remarkably simple but performs exceedingly complex signaling, making it important to reveal how Notch signaling intrinsically follows specific logic (or protocols) when interacting with other pathways. Our simulations show that for R8 patterning controlled jointly by the inductive Hh signaling and restrictive Notch/EGFR signaling, the timing of some events is critical but of others is less important, upon which key and inessential events can be determined. Moreover, sequences of key events and their spatiotemporal orders specify a clear pattern of intracellular and intercellular signaling required for orderly transmission of information. We do not assume that every single event is correct, but believe that as a whole such patterns, concrete and tangible for a definition of logic and comparable to protocols of communication (such as the TCP/IP protocol for communication between computer programs), more fundamentally characterize developmental signaling than gradual or abrupt changes of molecular concentrations. We postulate that the revealed pattern may also dominate concerted lateral inhibition during many processes of competitive cell fate determination and positively answer the question of ‘whether pattern formation in other animals uses all or some of the same logical steps’ (Lawrence and Struhl, 1996).
Supplementary Material
Funding
This work was supported by UK Engineering and Physical Sciences Research Council (EP/C539044, EP/C539052) and National Science Foundation of China (31071165).
Conflict of Interest: none declared.
References
- Affolter M., Mann R. (2001) Legs, eyes, or wings-Selectors and signals make the difference. Science, 292, 1080–1081. [DOI] [PubMed] [Google Scholar]
- Baker N.E., Yu S.Y. (1998) The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech. Dev., 74, 3–14. [DOI] [PubMed] [Google Scholar]
- Baonza A. et al. (2001) A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol., 11, 396–404. [DOI] [PubMed] [Google Scholar]
- Carroll S.B. (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell, 134, 25–36. [DOI] [PubMed] [Google Scholar]
- Chen C.H. et al. (1999) Nuclear trafficking of cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell, 98, 305–316. [DOI] [PubMed] [Google Scholar]
- Chen C.K., Chien C.T. (1999) Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc. Natl. Acad. Sci. U. S. A., 96, 5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J.R. et al. (1996) Pattern formation by lateral inhibition with feedback: a mathematical model of Delta-Notch intercellular signalling. J. Theor. Biol., 183, 429–446. [DOI] [PubMed] [Google Scholar]
- Culi J.E. et al. (2001) The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development, 128, 299–308. [DOI] [PubMed] [Google Scholar]
- Dokucu M.E. et al. (1996) Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development, 122, 4139–4147. [DOI] [PubMed] [Google Scholar]
- Frankfort B.J., Mardon G. (2002) R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development, 129, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Freeman M. (1997) Cell determination strategies in the Drosophila eye. Development, 124, 261–270. [DOI] [PubMed] [Google Scholar]
- Fu W., Baker N.E. (2003) Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development, 130, 5229–5239. [DOI] [PubMed] [Google Scholar]
- Guruharsha K.G. et al. (2011) A protein complex network of Drosophila melanogaster. Cell, 147, 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal A. et al. (1997) Inhibition of C. elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev., 11, 2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward P. et al. (2008) Wnt/Notch signalling and information processing during development. Development, 135, 411–424. [DOI] [PubMed] [Google Scholar]
- Heitzler P. Simpson,P. (1991) The choice of cell fate in the epidermis of Drosophila. Cell, 64, 1083–1092. [DOI] [PubMed] [Google Scholar]
- Istrail S., Davidson E.H. (2005) Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. U. S. A., 102, 4954–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C., Moses K. (2004) Cell-cycle regulation and cell-type specification in the developing Drosophila compound eye. Semin. Cell Dev. Biol., 15, 75–81. [DOI] [PubMed] [Google Scholar]
- Kreuger J. et al. (2004) Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev. Cell, 7, 503–512. [DOI] [PubMed] [Google Scholar]
- Lawrence P.A., Struhl G. (1996) Morphogens, compartments, and pattern: lessons from Drosophila? Cell, 85, 951–961. [DOI] [PubMed] [Google Scholar]
- Lee E.C. et al. (1996) The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol. Cell Biol., 16, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. et al. (2004) The yeast cell-cycle network is robustly designed. Proc. Natl. Acad. Sci. U. S. A., 101, 4781–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. (2004) Functions of heparan sulfate proteoglycans in cell signaling during development. Development, 131, 6009–6021. [DOI] [PubMed] [Google Scholar]
- Lubensky D.K. et al. (2011) A dynamical model of ommatidial crystal formation. Proc. Natl. Acad. Sci. U. S. A., 108, 11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldi A. et al. (2009) Logical modelling of regulatory networks with GINsim 2.3. Bio. Syst., 97, 134–139. [DOI] [PubMed] [Google Scholar]
- Powell P.A. et al. (2001) Scabrous complexes with Notch to mediate boundary formation. Nature, 409, 626–630. [DOI] [PubMed] [Google Scholar]
- Saint R. et al. (1988) Pattern formation in the developing eye of Drosophila melanogaster is regulated by the homeo-box gene, rough. Nature, 334, 151–154. [DOI] [PubMed] [Google Scholar]
- Spencer S.A. et al. (1998) Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development, 125, 4777–4790. [DOI] [PubMed] [Google Scholar]
- Strutt D.I., Mlodzik M. (1996) The regulation of hedgehog and decapentaplegic during Drosophila eye imaginal disc development. Mech. Dev., 58, 39–50. [DOI] [PubMed] [Google Scholar]
- Tsuda L. et al. (2002) An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell, 110, 625–637. [DOI] [PubMed] [Google Scholar]
- von Dassow G. et al. (2000) The segment polarity network is a robust developmental module. Nature, 406, 188–192. [DOI] [PubMed] [Google Scholar]
- Webb S.D., Owen M.R. (2004) Oscillations and patterns in spatially discrete models for developmental intercellular signalling. J. Math. Biol., 48, 444–476. [DOI] [PubMed] [Google Scholar]
- Zhu H., Mao Y. (2015) Robustness of cell cycle control and flexible orders of signaling events. Sci. Rep., 5, 14627.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. et al. (2005) Cellular automata with object-oriented features for parallel molecular network modeling. IEEE Trans. Nanobioscience, 4, 141–148. [DOI] [PubMed] [Google Scholar]
- Zur Lage P., Jarman A.P. (2000) Antagonism of EGFR and Notch signaling in the reiterative recruitment of Drosophila adult chordotonal sense organ precursors. Nature, 406, 188–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.