Abstract
Fibroblast growth factor 21 (Fgf21) has emerged as a potential plasma marker to diagnose non-alcoholic fatty liver disease (NAFLD). To study the molecular processes underlying the association of plasma Fgf21 with NAFLD, we explored the liver transcriptome data of a mild NAFLD model of aging C57BL/6J mice at 12, 24, and 28 months of age. The plasma Fgf21 level significantly correlated with intrahepatic triglyceride content. At the molecular level, elevated plasma Fgf21 levels were associated with dysregulated metabolic and cancer-related pathways. The up-regulated Fgf21 levels in NAFLD were implied to be a protective response against the NAFLD-induced adverse effects, e.g. lipotoxicity, oxidative stress and endoplasmic reticulum stress. An in vivo PPARα challenge demonstrated the dysregulation of PPARα signalling in the presence of NAFLD, which resulted in a stochastically increasing hepatic expression of Fgf21. Notably, elevated plasma Fgf21 was associated with declining expression of Klb, Fgf21’s crucial co-receptor, which suggests a resistance to Fgf21. Therefore, although liver fat accumulation is a benign stage of NAFLD, the elevated plasma Fgf21 likely indicated vulnerability to metabolic stressors that may contribute towards progression to end-stage NAFLD. In conclusion, plasma levels of Fgf21 reflect liver fat accumulation and dysregulation of metabolic pathways in the liver.
Non-alcoholic fatty liver disease (NAFLD) has been recognised as a hepatic manifestation of metabolic syndrome. NAFLD covers a spectrum of liver injuries ranging from fat accumulation in the liver (steatosis) to the more severe condition of steatohepatitis (NASH). Considering that NAFLD is currently the most common liver disorder, prevalence of which has been reported to be 20–40% in the US1,2, the population would benefit from a diagnosis from the early stage of NAFLD. Intervention as simple as weight management through diet and exercise is the most effective way leading to reduced liver fat, NASH remission, and also reduction of fibrosis3. However, at present a population-based screening tool for NAFLD is still lacking. Serum screening of liver enzymes and a liver ultrasound technique have been employed in screenings and clinical studies4, but these procedures are, for different reasons, suboptimal. A number of studies have pointed out that the blood-screening test of the commonly analysed liver enzymes, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), γ-glutamyl-transpeptidase (γ-GT) and albumin, poorly diagnose NAFLD5,6,7,8. The ultrasound technique has better accuracy than the blood screening test, but is also suboptimal due to the low sensitivity of this method9. Magnetic resonance imaging and spectroscopy has a higher sensitivity, but this technique requires specific and expensive instruments, which limits its measurement availability. Diagnosis on liver biopsies is the most accurate way to determine the presence of NAFLD, but this procedure is highly invasive and not suitable for population-based screening. Hence, new accurate and non-invasive measures are required for the diagnosis of NAFLD.
Recently, fibroblast growth factor 21 (FGF21 (Fgf21 in mice)) has emerged as a potential diagnostic marker for NAFLD. Serum FGF21 is found to be elevated in NAFLD patients, as compared to healthy subjects, and correlates with hepatic fat content and the degree of liver steatosis4,10,11,12. Moreover, the performance of FGF21 has been examined in a 3-year prospective study in China and high serum FGF21 was found to be a determinant of NAFLD, showing an area under curve of receiver operating characteristic (AUROC) of 0.81613. Serum FGF21 has also been reported to increase the accuracy of non-alcoholic steatohepatitis (NASH) diagnosis using cytokeratin-18 fragment (CK-18)14. Therefore, plasma/serum FGF21 seems a promising diagnostic marker for an accurate and non-invasive diagnosis of NAFLD.
FGF21 has multiple metabolic functions, regulating energy homeostasis, glucose-lipid metabolism and insulin sensitivity13. However, it is currently unclear which metabolic functions of FGF21 underlie the association of plasma FGF21 level and NAFLD. In the present study, we aim to identify the putative molecular mechanisms that underlie the association of plasma FGF21 level with NAFLD. A complicating factor for a human study in this field is that the accurate assessment of NAFLD would require liver biopsies from healthy subjects, which is ethically undesirable. Therefore, we employed a cohort of aging mice to investigate the association between NAFLD, which was determined by IHTG level, and plasma Fgf21. The use of a cohort of aging mice, which consists of four different age time points, enables us to investigate whether plasma Fgf21 can act as a biomarker at different age time points. NAFLD has been reported in subjects of all ages, but particularly in middle to old age (40–65 years old in human)15, which indicates that NAFLD develops over many years. To create a mice cohort that simulates the slow onset of NAFLD, we included a medium-fat diet (MF; 25E% from fat) as a diet that induces the development of NAFLD. The energy contribution from fat in this diet group is considered mild, compared to other NAFLD-inducing dietary interventions that commonly acutely stimulate NAFLD pathologies within several weeks of feeding by applying a high-fat diet (45E% from fat)16,17. In addition to the MF diet, we introduced a normal diet (10E% from fat) as the control (C) group and a calorie restriction diet (CR; 30E% reduced feeding compared to control group) as a diet regimen that prevents NAFLD. Then, we investigated whether the use of plasma Fgf21 as marker for NAFLD in our mice cohort is comparable to what has previously been reported for humans. We next searched for the biological processes underlying the association of plasma Fgf21 with NAFLD by performing microarray analysis on liver mRNA by using two approaches. First, we searched for pathways and upstream regulators associated with plasma Fgf21 by performing a gene co-expression network analysis using weighted gene co-expression network analysis (WGCNA)18. In the second approach, we investigated the major transcriptional difference in NAFLD by performing gene set enrichment analysis (GSEA), and then determining the relevance of Fgf21 in the NAFLD-related pathways. To further investigate PPARα signalling as one of the dysregulated pathways, we examined the PPARα response to its agonist at the gene expression level. Since we found that some of the mice of 6 months of age displayed a high level of plasma Fgf21 in the absence of elevated IHTG levels, we also explored the microarray data to search for the functions of the genes associated with the elevated Fgf21 plasma level without accumulation of IHTG.
Materials and Methods
Ethics statement
The animal experiment was approved by the Local Committee for Care and Use of Laboratory Animals at Wageningen University (code number: drs-2010151b) and performed in accordance with the institutional and national guidelines for the care and use of animals.
Mice aging study
The mice aging study was a part of the IDEAL mice aging cohort that has been described in detail previously19,20. Briefly, male C57BL/6J mice (age of 7 weeks) were purchased from Janvier (Cedex, France) and were housed in pairs of two in the light and temperature (20 °C)-controlled animal facility of Wageningen University (12-hour light/dark cycle, light on at 04.00). The mice were acclimated for 2 weeks, receiving standard AIN-93G (Research Diet Services, Wijk bij Duurstede, The Netherlands) upon arrival. All mice were provided with ad libitum access to water. The study design is presented in Supplementary Fig. S1. The diet intervention started at the age of 9 weeks. The mice were housed individually and randomly distributed into three intervention groups: 1) control diet (C, 10E% fat, n = 89) receiving AIN-93W diet ad libitum; 2) calorie restricted diet (CR, n = 117) receiving AIN-93W-CR in portions containing 70E% of the mean energy intake compared to the mice on the control diet; 3) medium fat diet (MF, 25E% fat, n = 127) receiving AIN-93W-MF ad libitum. AIN-93W-CR contains an increased concentration of vitamins and minerals in order to feed these mice the same concentrations of micronutrients as the mice receiving the AIN-93W diet and to avoid malnutrition. Portion sizes for the mice on the CR were based on food intake of mice on the control diet and adjusted every 6 months. The rations were provided each day at 15.30, 30 minutes before the light was switched off. The complete diet compositions are listed in Supplementary Table S1 (Research Diet Services, Wijk bij Duurstede, The Netherlands).
The mice were culled at the age of 6, 12, 24 and 28 months. At each sacrifice, 12–18 mice of each intervention group were sacrificed between 14.00–17.00 on consecutive days (the remaining mice stayed in the experiment and were evaluated at older ages). Mice were paired per dietary intervention group according to body weight at sacrifice, so that mock and PPARα agonist Wy-14,643 (Wy) treatment were provided to mice with similar body weight. Prior to sacrifice each mouse was first fasted for 4 hours, after which they received an intragastric gavage of either solvent (0.5% carboxymethyl cellulose) or Wy dispersed in solvent (160 mg Wy/kg body weight) and were fasted again for another 6 hours. Body weight, liver weight, IHTG and 4-hydroxyproline were measured in both the mock- and Wy-treated animals. Plasma Fgf21 and liver microarray analysis were performed for mock-treated animals only, as Wy substance is a PPARα agonist potentially affecting Fgf21 expression. At the sacrifice, the mice were sedated with isoflurane (1.5%) in a mixture of nitrous oxide (70%) and oxygen (30%). Blood samples were collected by cardiac puncture, which was followed by neck dislocation. Weight of various organs was measured and, subsequently, organs/tissues were snap-frozen and stored at −80 °C until further molecular/biochemical analysis.
Measurement of hepatic steatosis and fibrosis
Intrahepatic triglyceride (IHTG) content was determined in 5% liver homogenates prepared in buffer containing 250 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl (pH 7.5), using the triglyceride Liquicolor Monoreagent (Instruchemie, Delfzijl, The Netherlands), according to manufacturer’s instruction. The IHTG level was applied as the diagnosis standard of NAFLD using the 5% or 50 mg TG per gram liver criterion from Kleiner’s scoring21. Liver fibrosis is represented by 4-hydroxyproline content measurement in the liver. The 4-hydroxyproline analysis was performed as previously described in Hillebrandt et al.22.
Measurement of plasma Fgf21 level and other plasma markers
The plasma Fgf21 concentration was determined using Rat/Mouse FGF-21 ELISA kits (Milipore, cat #EZRMFGF21-26K), according to the manufacturer’s instructions. Plasma insulin was measured using a Mouse Adipokine (MADKMAG-71K) kit (Millipore, Billerica, MA, USA), according to the manufacturer’s instructions. The cytokeratin-18 plasma concentration was measured using Mouse Cytokeratin 18-M30 ELISA kit (Cusabio, Hubei, China).
RNA isolation
Total RNA from liver, tibialis anterior muscle, colon scrapings and epidydimal white adipose tissue were isolated using TRIzol reagent (Invitrogen, Breda, The Netherlands), according to the manufacturer’s instructions. The RNA was treated with DNAse and purified on columns using the RNeasy microkit (Qiagen, Venlo, The Netherlands). RNA concentration was measured on a NanoDrop ND-1000 UV–vis spectrophotometer (Isogen, Maarsen, The Netherlands) and RNA integrity was checked on an Agilent 2100 Bioanalyzer (Agilent Technologies, Amsterdam, The Netherlands) with 6000 Nano Chips, according to the manufacturer’s instructions. RNA was judged as suitable only if samples showed intact bands of 18S and 28S ribosomal RNA subunits, displayed no chromosomal peaks or RNA degradation products and had a RNA integrity number (RIN) above 8.0.
Microarray hybridization
Hybridization, washing and scanning of Affymetrix GeneChip Mouse Gene 1.1 ST arrays were performed according to standard Affymetrix protocols as described previously19,23. Microarray analysis was performed in MADMAX, a pipeline for statistical analysis of microarray data24. Arrays were normalized using the Robust Multiarray Average method25,26. Probe sets were defined according to Dai et al.27. In this method probes are assigned to unique gene identifiers, in this case Entrez IDs. The probes on the Gene 1.1 ST arrays represent 21,225 Entrez IDs. For the analysis, only genes having 1) an inter-quartile range of >0.1 and 2) an intensity value of >20 on at least five arrays were taken into account, which resulted in 15,885 genes in the dataset. Array data have been submitted to the Gene Expression Omnibus, with accession number GSE84495.
cDNA synthesis and real-time quantitative PCR
The microarray data was validated by real-time quantitative PCR (Q-PCR). For each individual sample, single-stranded complementary DNA was synthesized from 1 μg of total RNA using the First Strand cDNA Synthesis kit (Thermo Scientific, Landsmeer, The Netherlands), following the supplier’s protocol. Q-PCR was performed using SensiMix SYBR No-ROX kit (Bioline, Alphen aan de Rijn, The Netherlands) and a CFX384 thermal cycler (Bio-Rad, Veenendaal, The Netherlands). The following thermal cycling conditions were used: 2 min at 94 °C, followed by 40 cycles of 94 °C for 15 s and 60 °C for 45 s. PCR reactions to validate Fgf21 expression were performed in duplicate and all samples were normalized to Rplp0 expression. Primer sequences were retrieved from the online PrimerBank database28 and the sequences of the primers used are listed in Supplementary Table S2.
Statistical analysis
Data were analysed with GraphPad Prism 5.04. The data were expressed as mean ± standard error mean. Comparison between two groups was performed using student t-test, whereas comparison between 3 or more groups was performed using ANOVA. Correlation between two parameters was presented as Pearson correlation coefficient (r) and p-value. A p-value of <0.05 was considered significant. The receiver operating characteristic (ROC) curve analysis was carried out and the area under the ROC curves (AUROCs) were calculated to represent their performance to predict NAFLD. Optimal cut-off points were calculated for sensitivity and specificity reference (Youden Index).
Hepatic transcriptomics data analysis
For the microarray data analysis, differentially expressed probe sets were identified by using linear models (library limma) and the intensity-based moderated t-statistic (IBMT) method was applied29,30. Resulting log2 intensities and p-values were used for further descriptive bioinformatic analysis of the data. Gene set enrichment analysis (GSEA; http://www.broad.mit.edu/gsea/) was performed in MADMAX24. Gene sets with a false discovery rate (FDR) q-value of <0.01 were considered significantly enriched.
Gene co-expression networks (modules) were constructed using the blockwiseModules R function in Weighted Gene Co-expression Network Analysis (WGCNA)18. WGCNA uses a network distance coupled with hierarchical clustering and dynamic tree cutting to define modules as branches of a cluster tree. Gene modules, which summarize the main patterns of variation, are defined in an unbiased fashion and denoted by colors. The first principal component represents the summary of the module and is referred to as the module eigengene (ME). MEs were then related to plasma Fgf21 level and other NAFLD-related traits. This approach avoids the multiple testing from thousands of individual transcripts to only a number of modules. To explore the functional pathways and predicted upstream regulators of the gene modules, Ingenuity pathway analysis (IPA; Ingenuity® Systems) was used.
Results
Control and medium-fat diet groups developed NAFLD at middle and old age
The dietary interventions exerted pronounced effects on the mice’s body and liver weight, as shown in Fig. 1a,b, respectively. During aging, the MF-exposed animals gained the highest body and liver weight, while the CR-fed animals were the leanest. The weight gain was accompanied by an increase in IHTG content (Fig. 1c). While the CR-fed animals only showed a modest increase over time, the C- and MF-fed animals displayed an elevated IHTG level starting at middle-age, i.e. at 12 months. Liver fibrosis, which was represented by the measurement of 4-hydroxyproline (4-HP) content in the liver (Fig. 1d), showed to increase at old age (24 and 28 month) in the MF diet group.
Figure 1. Physiological changes induced by the different dietary interventions at the age of 6, 12, 24, and 28 months.
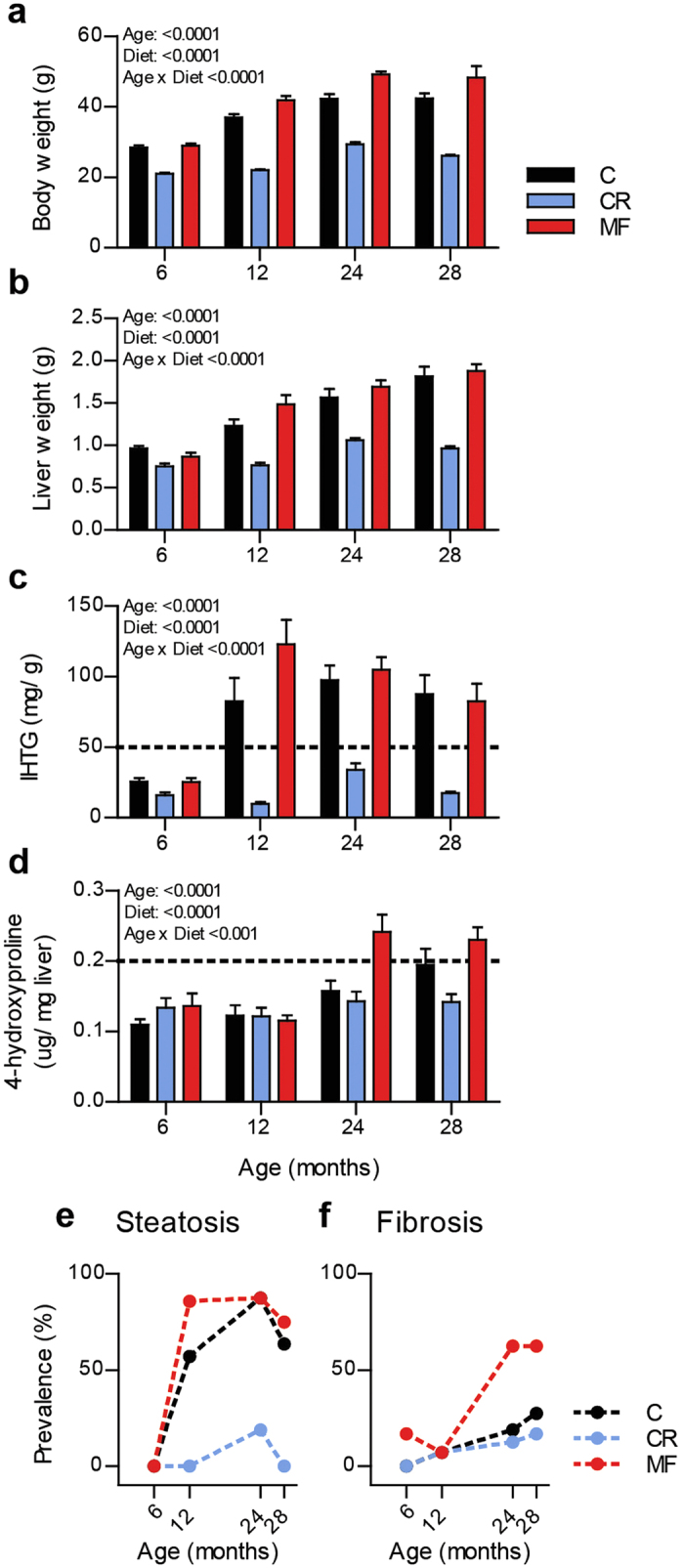
Body weight (a) liver weight (b) and IHTG content (c) dramatically increased over time, except for the CR-fed animals which stayed lean. Significance (p-value) of age, diet and interaction were evaluated using two-way ANOVA. Error bars represent s.e.m. Prevalence of hepatic steatosis (e) and fibrosis (f) in the mice aging cohort at different age time points.
The IHTG level was applied as the diagnosis standard of hepatic steatosis using the 5% or 50 mg TG per gram liver criterion from Kleiner’s scoring21. The prevalence of hepatic steatosis development in the cohort is depicted in Fig. 1e. At the age of 6 months, none of the mice in any of the diet groups developed hepatic steatosis, indicating a healthy liver condition at young/mature adult age. Hepatic steatosis development became visible in 12-month old mice exposed to C and MF diets, reaching up to 85.7% in the MF diet group. At the 24-month time point, the prevalence of hepatic steatosis increased considerably in the control group. At this age over 80% in the C and MF diet groups displayed steatosis (Fig. 1e) but at the age of 28 months, a slight decrease of prevalence was observed.
Since a criterion of 4-HP level for hepatic fibrosis diagnosis has not been clearly defined, we adapted the level of 4-HP that was reported by Fuchs et al. to be associated with extensive portal fibrosis (equivalent with Ishak fibrosis scoring stage 2–3)31. A level of >0.200 μg of 4-HP per mg liver was applied as an indication of liver fibrosis. Increasing prevalence of hepatic fibrosis was only pronounced in the MF intervention group at 24 and 28 months of age (Fig. 1f). These results showed that hepatic steatosis occurred in both the C and MF intervention groups at the middle-age time point, while hepatic fibrosis in particularly developed in the MF diet group at an old age. On the other hand, the CR-fed animals were protected from developing hepatic steatosis and fibrosis.
Plasma Fgf21 reflected the hepatic fat accumulation at middle and old age, but not at mature adult age
The results presented in Fig. 2a show that plasma Fgf21 levels of the C- and MF-exposed animals were higher than that of the CR-exposed animals at all ages. Noticeably, plasma Fgf21 levels were particularly high at 6 months of age in the C and CR diet groups. Next, to examine a specific diet- or age-related effect on plasma Fgf21 levels, the correlation with IHTG content was carried out separately by diet and age. The CR diet group displayed low IHTG and plasma Fgf21 concentration, while the C and MF groups showed some variation. However, overall we did not observe significant correlation in any of the different diet groups (Fig. 2b). On the other hand, the comparison by age indicated that, except for the 6-month-old mice, the correlations at different ages were comparable (Fig. 2c). Some of the young mice, independent of their diet types, exhibited high plasma Fgf21 concentrations despite their low IHTG levels. This observation implies that the plasma Fgf21 concentration reflects the hepatic fat accumulation at middle-age (12 months) and old age (24 and 28 months), which results in an overall correlation coefficient of 0.52 (p < 0.0001), but performs differently at younger age (Fig. 2d).
Figure 2. Plasma Fgf21 reflected the hepatic fat accumulation at middle and old age, but not at mature adult age.
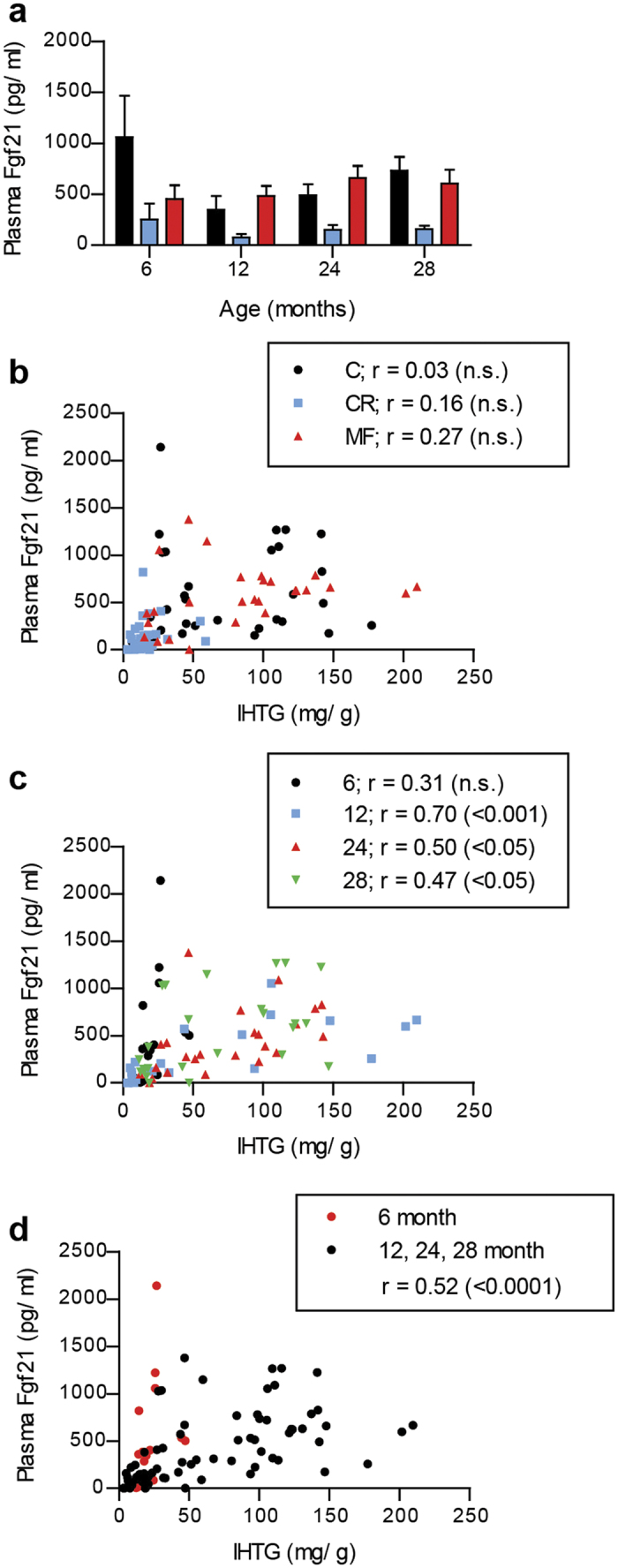
(a) Different plasma Fgf21 levels induced by the dietary interventions at the age of 6, 12, 24, and 28 months. Correlation of IHTG and plasma Fgf21 for different dietary interventions (b) and ages (c). (d) Plasma Fgf21 was positively correlated with IHTG levels at older ages, while the young 6-month-old mice had elevated plasma Fgf21, despite their low IHTG levels (in red symbols). r values were calculated with Pearson’s correlations and their significance are indicated in the parentheses.
Plasma Fgf21 levels reflected liver Fgf21 expression
The gene expression levels of Fgf21 in the liver were obtained from the microarray data and revealed that, similar to the plasma Fgf21 levels, the C- and MF-exposed animals displayed higher expression levels of Fgf21 (Fig. 3a). Notably, the higher levels of plasma Fgf21 at the earliest time point of 6 months were also reflected in the Fgf21 liver expression level. The results presented in Fig. 3b show that the plasma Fgf21 levels significantly correlated to hepatic Fgf21 expression levels obtained by microarray analysis (r = 0.63, p < 0.0001). This result was confirmed by Q-PCR analysis (r = 0.71, p < 0.0001, Supplementary Fig. S2a). These observations indicate that the plasma Fgf21 concentration reflects the Fgf21 mRNA levels in liver tissue. In addition, we examined whether plasma Fgf21 increased with body or liver weight. Correlation analyses confirmed that plasma Fgf21 had significant positive correlation with body and liver weights (Supplementary Fig. S2b,c). As Fgf21 has been reported to be expressed abundantly, not only in the liver, but also in other tissue types, we also examined whether the plasma Fgf21 concentrations reflect the Fgf21 expression levels in epidydimal white adipose tissue (eWAT), tibialis anterior muscle and colon tissue. The results presented in Fig. 3c–e show that plasma Fgf21 levels was strongly associated with the liver expression, compared to the other tissues. The expression levels of Fgf21 in the muscle and colon were extremely low (Fig. 3d,e). The correlation of plasma Fgf21 and expression in eWAT was significant, however, the levels of expression in the eWAT were much lower compared to those in the liver tissue (Fig. 3c).
Figure 3. Plasma Fgf21 levels were strongly reflected by the expression of Fgf21 in the liver.
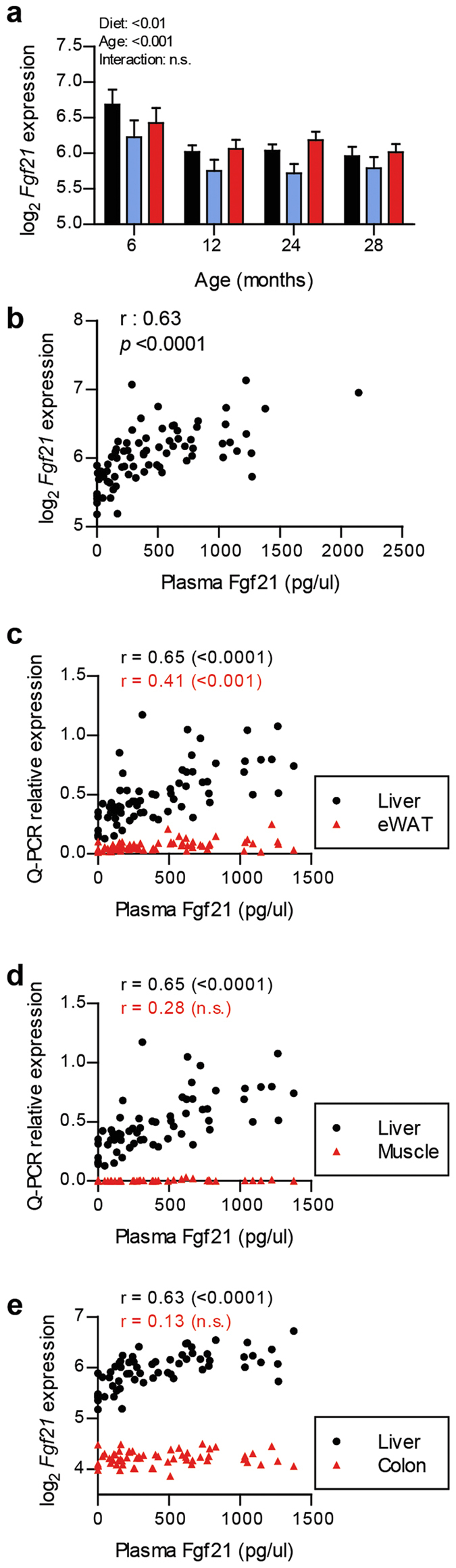
(a) Different expression levels of Fgf21 in the liver induced by the dietary interventions and age. (b) Significant positive correlation between plasma Fgf21 and Fgf21 expression in the liver. The association between plasma and expression levels of Fgf21 was also compared in other tissue types, (c) epidydimal white adipose tissue (eWAT), (d) muscle, and (e) colon tissue. r values were calculated with Pearson’s correlations and their significance are indicated in the parentheses.
To examine whether plasma Fgf21 in mice indicates hepatic fat accumulation as observed in humans, the mice were divided into two groups: those that either developed or did not develop NAFLD according to the IHTG criterion of higher or lower than 50 mg TG per gram liver from Kleiner’s scoring21. This resulted in 53 animals without NAFLD, which largely consisted of young or CR-fed animals, and 36 animals with NAFLD, which mostly consisted of older animals or under the C or MF dietary regimen. The average IHTG contents of the animals with and without NAFLD were 111.1 and 22.2 mg TG per gram liver, respectively. The characteristics of animals with and without NAFLD are summarized in Supplementary Table S3, showing that the animals with NAFLD had a significantly heavier body weight, larger epididymal fat depot, enlarged liver, and lower liver 4-HP content. The levels of fasting plasma insulin and the liver injury marker alanine aminotransferase (ALT) were elevated in the group of animals with NAFLD.
Subsequently, AUROC analysis was performed using optimal cut-off points determined by the Youden Index (plasma Fgf21 >222.0 pg/ml). The results summarized in Table 1 show that the sensitivity was remarkably high at 91.4%, while the specificity was low (57.1%). This resulted in an AUROC of 0.77 (Supplementary Fig. S3a) and an accuracy of 71.4% (positive predictive value/PPV of 60.4% and negative predictive value/NPV of 90.3%). Since plasma Fgf21 appears to perform differently at the age of 6 months and a previous study of obese children indicated that plasma Fgf21 did not provide additional value in predicting NAFLD32, we also tested the Fgf21 performance in older animals only. When the 6 months old animals were excluded, the AUROC and specificity of Fgf21 were improved to 0.84 (Supplementary Fig. S3b) and 73.5%, while the sensitivity remained high (88.6%). In addition, the accuracy was improved to 81.2% (PPV of 77.5% and NPV of 86.2%). Diagnosis using more markers commonly yield a better outcome, so we applied combinations of plasma Fgf21 with other plasma or trait markers for NAFLD, i.e. plasma ALT, plasma CK-18 and body weight. The performance accuracy described in Table 2 revealed that the plasma Fgf21 was best combined with plasma ALT (accuracy of 88.4%) or body weight (87.0%), while the combination with plasma CK-18 (81.2%) did not improve the performance of Fgf21.
Table 1. Overall performance of plasma Fgf21 levels for the diagnosis of NAFLD.
| AUROC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|
| All time points | |||||
| 0.77 (0.67–0.87) | 91.4 | 57.1 | 60.4 | 90.3 | 71.4 |
| Without 6 months old | |||||
| 0.84 (0.74–0.94) | 88.6 | 73.5 | 77.5 | 86.2 | 81.2 |
Table 2. NAFLD diagnosis performance of plasma Fgf21 in combination with other markers.
| Fgf21 in combination with | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Plasma ALT | 80.0 | 97.1 | 96.6 | 82.5 | 88.4 |
| Plasma CK-18 | 82.9 | 79.4 | 80.6 | 81.8 | 81.2 |
| Body weight | 80.0 | 94.1 | 93.3 | 82.1 | 87.0 |
The gene module strongly correlated with the plasma Fgf21 levels contained steatosis- and cancer-related genes
To explore the molecular mechanism underlying the involvement of Fgf21 in NAFLD development, microarray analysis was performed on mRNA isolated from the livers of the 12, 24, and 28-month old animals. In the search for the molecular processes underlying the association of plasma Fgf21 with NAFLD, the microarray data were analysed by a two-step approach: 1) gene co-expression network analysis using WGCNA18 and 2) functional pathways and up-stream regulator analysis were determined by applying Ingenuity pathway analysis (IPA) on the co-expression network.
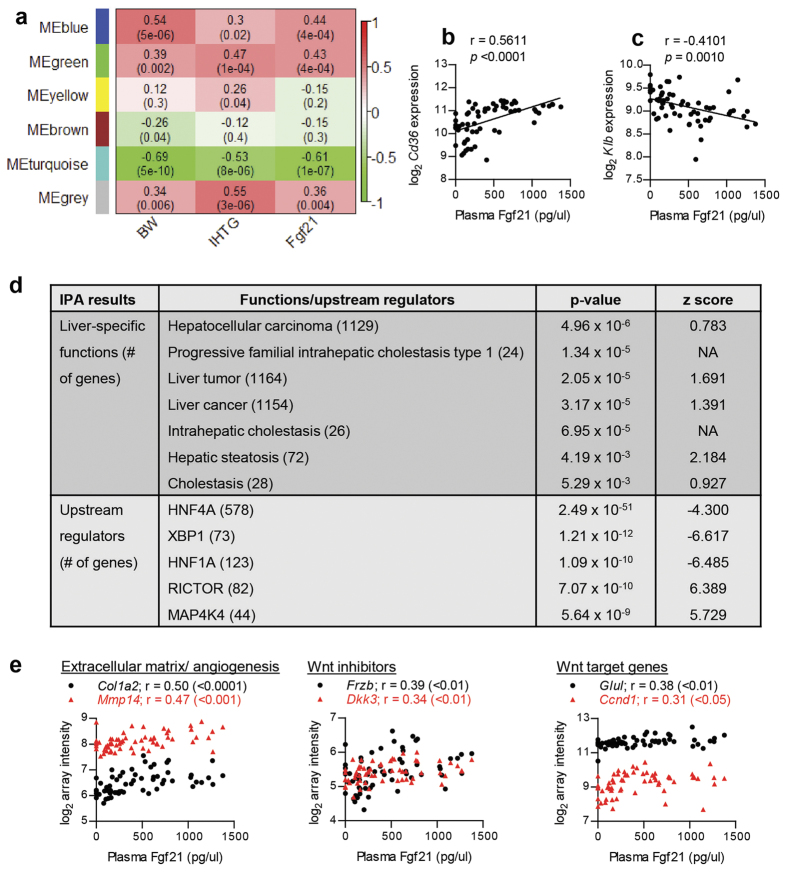
The WGCNA results presented in Fig. 4a, show that 5 modules were created from the hepatic transcriptome data and that 3 of them significantly correlated with plasma Fgf21 (p < 0.05; module grey denotes background genes outside of modules). The module displaying the most significant correlation with plasma Fgf21 was MEturquoise (r = −0.61). Interestingly, cluster differentiation 36 (Cd36), a fatty acid transporter gene involved in steatosis development, was identified as the top regulated gene in this module. A correlation analysis between plasma Fgf21 and Cd36 expression levels showed a positive significant association (Fig. 4b). Intriguingly, beta-Klotho gene (Klb), a co-receptor component that is required for Fgf21 metabolic activity33,34, was included in this module. The expression of Klb decreased with the increase of plasma Fgf21 (Fig. 4c). In addition, strong inverse correlations with Klb expression were also observed for body weight and IHTG (Supplementary Fig. S4a). We also examined plasma Fgf21 correlation with the liver gene expression levels of the 4 members of the Fgf receptor family and observed a significant negative correlation with Fgfr2 and Fgfr4 expression, similarly to Klb (Supplementary Fig. S4b).
Figure 4. Liver biological processes associated with plasma Fgf21 level.
(a) Heatmap depicting the correlation between gene modules (in rows) and phenotypes (in columns). The top values in each cell represents the correlation coefficient between the module and phenotype with the correlation p-value in parentheses. Red and green color represents positive and negative correlation, respectively. Correlation between plasma Fgf21 levels and hepatic expressions of Cd36 (b) and Klb (c). (d) Biological processes and regulators associated with MEturquoise, which is the most significant modules correlated with plasma Fgf21. Significant liver-specific functions and upstream regulators are reported in p-values and z-scores. Positive and negative z-score represent predicted activation and inhibition, respectively. (e) Correlation between plasma Fgf21 and expression levels of hepatocellular carcinoma-related genes within MEturqouise, which included genes related to extracellular matrix formation and angiogenesis (Col1a2 and Mmp14), inhibition of Wnt signalling (Frzb and Dkk3), and downstream target of Wnt signalling (Glul and Ccnd1). r values were calculated with Pearson’s correlations and their significance are indicated in the parentheses.
Next, we investigated which biological processes are represented by the genes in the MEturquoise module, by using IPA. Figure 4d shows the liver-specific functions with p-value < 0.01 and the 5 most significant regulators identified by IPA, which were ranked by p-value. Predicted activation/inhibition z-scores are displayed when available. The liver-specific functions associated with this module included hepatic steatosis and cholestasis, but the most significant function was the hepatocellular carcinoma (HCC). A number of genes related to HCC functions were identified in this module, including collagen type I, alpha 2 (Col1a2), matrix metallopeptidase 14 (Mmp14), frizzled-related protein (Frzb), dickkopf WNT signalling pathway inhibitor 3 (Dkk3), glutamate-ammonia ligase (Glul), and cyclin D1 (Ccnd1). The functions of these genes include extracellular matrix formation/angiogenesis (Col1a2 and Mmp14), inhibitors of Wnt signalling (Frzb and Dkk3), and regulation of Wnt target genes (Glul and Ccnd1). The expression levels of these genes were significantly correlated with plasma Fgf21 levels (Fig. 4e), demonstrating that plasma Fgf21 level is associated with HCC-related signalling. Moreover, the identification of predicted upstream regulators further confirmed the association of this module with hepatocellular carcinoma (Fig. 4d): predicted inhibition of hepatocyte nuclear factor 4α and 1α (HNF4A and HNF1A), both are tumor suppressor regulators, while rapamycin-insensitive companion of mTOR (RICTOR) and mitogen-activated protein 4 kinase 4 (MAP4K4), factors involved in cancer development, were activated. X-box binding protein (XBP1) was also identified as one of the top upstream regulators.
The elevated plasma Fgf21 levels in young animals without accumulation of IHTG were related to the up-regulation of lipid metabolism by PPARα, PPARGC1α and PPARγ
To elucidate the functions of the genes associated with the elevated Fgf21 plasma level at young age without accumulation of IHTG, WGCNA analysis was performed with the inclusion of the 6-month-old animals. For this purpose, we searched for a gene module that was significantly associated with plasma Fgf21, but not with IHTG, and we found that module MEgreen (1286 genes) fulfilled this criterion (Supplementary Fig. S5a). This module has a significant correlation with Fgf21 (r = 0.44, p < 0.0001), but not with IHTG (r = 0.004, p = 1). To screen for the genes strongly correlated with the plasma Fgf21, we filtered for the genes with a correlation coefficient larger than 0.4. IPA of biological functions revealed that these genes play a role in lipid metabolism (top 3 functions/regulators are listed in Supplementary Fig. S5b) and IPA identified peroxisome proliferator-activated receptor α (PPARα), peroxisome proliferator-activated receptor γ coactivator 1-α (PPARGC1α), and PPARγ as the predicted upstream regulators.
NRF2 and PPARα targets, pathways differentially up-regulated by NAFLD, were linked to Fgf21
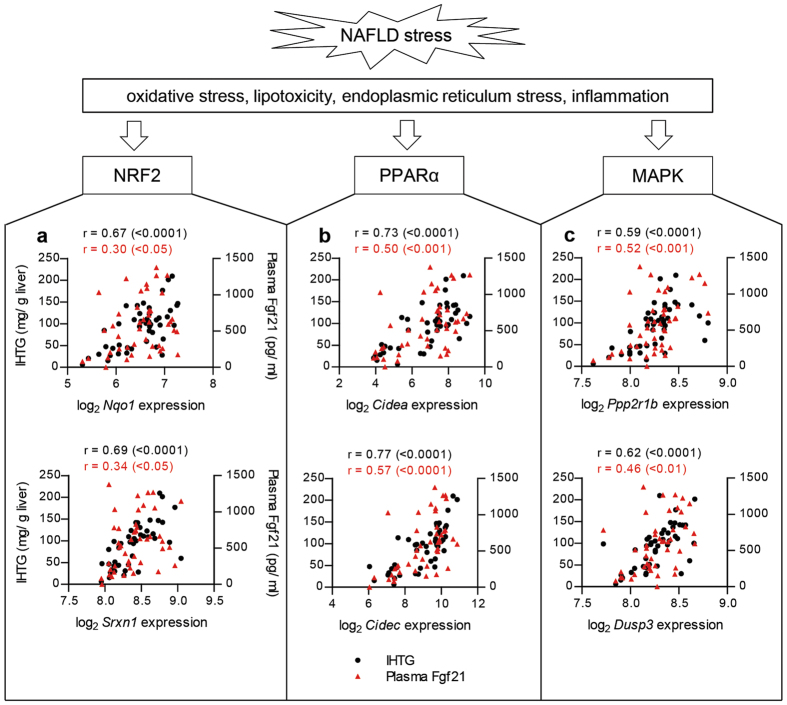
Next, to determine the relevance of Fgf21 in the NAFLD-related pathways, we first identified the pathways differentially regulated in the animals with NAFLD by performing GSEA. Then, we explored the differentially regulated pathways in NAFLD for their link to Fgf21. The gene expression data of 12, 24, and 28-month old animals were analysed with the exclusion of the CR group, since the latter group has a markedly different gene expression profile19. Forty-five animals were included in the microarray analysis (32 and 13 animals, with and without NAFLD, respectively). The GSEA results (based on FDR q value < 0.01), presented in Tables 3 and 4, revealed that 18 and 12 pathways were up- and down-regulated in NAFLD, respectively (lists of pathways with FDR q value < 0.05 is available in Supplementary Tables S4 and S5). The up-regulated pathways were dominated by pathways related to oxidative stress (nuclear factor (erythroid-derived 2)-like 2 or NRF2 targets, glutathione metabolism), energy and lipid metabolism (PPARα targets, oxidative phosphorylation and electron transport chain and fatty acid metabolism). The down-regulated pathways included various complement cascades pathways. Interestingly, NRF2 and PPARα targets, the 2 most significantly enriched up-regulated pathways, contained genes that have been previously identified for their strong correlation with IHTG35: NAD(P)H dehydrogenase quinone 1 (Nqo1), sulfiredoxin 1 (Srxn1), cell death-inducing DFFA-like effector a (Cidea) and c (Cidec). Genes in the core enrichment of NRF2 and PPARα targets are listed in Supplementary Tables S6 and S7. The gene expression levels of these genes were analysed for their correlation with IHTG, which revealed highly significant correlations (p < 0.0001; Fig. 5a,b). Although not as strong, the expression levels of these genes were also significantly correlated with the plasma Fgf21 levels. Thus, the differentially regulated PPARα and NRF2 target genes were pointed out to be the link between plasma Fgf21 levels and IHTG content during NAFLD.
Table 3. List of the significantly enriched up-regulated pathways in NAFLD.
| Enriched up-regulated pathways | NES* | FDR q-value |
|---|---|---|
| NRF2 TARGETS | 2.617 | 0.00000 |
| PPARA TARGETS | 2.404 | 0.00000 |
| WP1248 OXIDATIVE PHOSPHORYLATION | 2.328 | 0.00000 |
| WP295 ELECTRON TRANSPORT CHAIN | 2.299 | 0.00068 |
| KEGG OXIDATIVE PHOSPHORYLATION | 2.260 | 0.00081 |
| KEGG LYSOSOME | 2.223 | 0.00090 |
| WP1269 FATTY ACID BETA OXIDATION | 2.208 | 0.00097 |
| KEGG FATTY ACID ELONGATION | 2.180 | 0.00135 |
| KEGG FATTY ACID DEGRADATION | 2.146 | 0.00210 |
| MITOCHONDRIAL TRANSLATION | 2.113 | 0.00337 |
| MITOCHONDRIAL TRANSLATION TERMINATION | 2.102 | 0.00344 |
| MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 2.079 | 0.00418 |
| KEGG GLUTATHIONE METABOLISM | 2.052 | 0.00625 |
| RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | 2.048 | 0.00629 |
| SPHINGOLIPID METABOLISM | 2.032 | 0.00775 |
| AQUAPORIN MEDIATED TRANSPORT | 2.025 | 0.00786 |
| KEGG SYNAPTIC VESICLE CYCLE | 2.020 | 0.00795 |
| MITOCHONDRIAL TRANSLATION INITIATION | 2.003 | 0.00937 |
*Normalised enrichment score (NES); a statistical test for gene set enrichment.
Table 4. List of the significantly enriched down-regulated pathways in NAFLD.
| Enriched down-regulated pathways | NES | FDR q-value |
|---|---|---|
| WP449 COMPLEMENT AND COAGULATION CASCADES | −2.496 | 0.00000 |
| KEGG COMPLEMENT AND COAGULATION CASCADES | −2.496 | 0.00000 |
| KEGG SELENOCOMPOUND METABOLISM | −2.229 | 0.00018 |
| FORMATION OF FIBRIN CLOT CLOTTING CASCADE | −2.272 | 0.00024 |
| WP200 COMPLEMENT ACTIVATION CLASSICAL PATHWAY | −2.219 | 0.00028 |
| BIOC INTRINSICPATHWAY | −2.081 | 0.00213 |
| COMMON PATHWAY | 2.086 | 0.00226 |
| REGULATION OF COMPLEMENT CASCADE | −2.066 | 0.00304 |
| COMPLEMENT CASCADE | −2.023 | 0.00428 |
| AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | −1.995 | 0.00606 |
| WP460 BLOOD CLOTTING CASCADE | −1.962 | 0.00857 |
| INTRINSIC PATHWAY | −1.963 | 0.00928 |
Figure 5. The differentially regulated pathways in NAFLD were reflected by plasma Fgf21 levels.
(a) Stress induced by NAFLD may activate NRF2, PPARα and MAPK, and the expression of their target genes. The expression levels of the NRF2 (b) PPARα (c) and MAPK (d) target genes were strongly correlated with IHTG and more modestly with plasma Fgf21 levels (in black and red color, respectively). r values were calculated with Pearson’s correlations and their significance are indicated in the parentheses.
Notably, MAPK targets was also among the pathways enriched in the NAFLD-differentially regulated pathway (full list of genes in core enrichment of MAPK targets in Supplementary Table S8). This is in line with the finding of MAP4K4 activation in Fig. 4d.. The top MAPK target genes, protein phosphatase 2, regulatory subunit A, beta (Ppp2r1b) and dual specificity phosphatase 3 (Dusp3), showed significant correlations to both IHTG content and plasma Fgf21 levels (Fig. 5c). Therefore, this signifies the association of plasma Fgf21 with liver cancer-related signalling.
Gene expression response to PPARα activation demonstrates its dysregulation in NAFLD
Based on the essential role of PPARα in lipid homeostasis, we further investigated whether the dysregulation of PPARα in NAFLD extends to an altered response when the system is challenged. To examine the response of PPARα, prior to each sacrifice the PPARα agonist Wy-14,643 (Wy) substance was administered to half of the mice of each intervention group, while the other half of the group received mock treatment. A number of PPARα target genes, including Fgf2136,37, were analysed by Q-PCR, namely monoacylglycerol O-acyltransferase 1 (Mogat1), G0/G1 switch 2 (G0s2), acyl-CoA thioesterase 3 (Acot3), hydroxymethyl glutary coenzyme A reductase (Hmgcr) (Fig. 6a–e). These genes were selected to represent different functions regulated by PPARα (lipogenesis: Mogat1; lipolysis: G0s2; fatty acid oxidation: Acot3; cholesterol metabolism: Hmgcr), As shown in Fig. 6a, Wy treatment led to induced hepatic Fgf21 expression and the 2-way ANOVA test indicated an interaction between NAFLD and the Wy response. Intriguingly, animals with NAFLD exhibited an augmented response to the treatment. Similarly to Fgf21 expression, the induction of Mogat1 and Hmgcr expression levels were stronger in the animals with NAFLD (Fig. 6b,e). However, not all genes demonstrated the stochastic response to Wy treatment. G0s2 and Acot3 expression levels were similarly up-regulated regardless of the presence of NAFLD (Fig. 6c,d). The expression of Pparα itself was also examined and the results in Fig. 6f show that the stimulation of PPARα did not differ between the animals with and without NAFLD. Although NAFLD presence did not alter the response to Wy treatment of all PPARα target genes, the expression profiles of Fgf21, Mogat1 and Hmgcr underscore the dysregulation PPARα signalling pathway in NAFLD.
Figure 6. The response to PPARα challenge test showed that the presence of NAFLD partially altered the response at the gene expression levels.
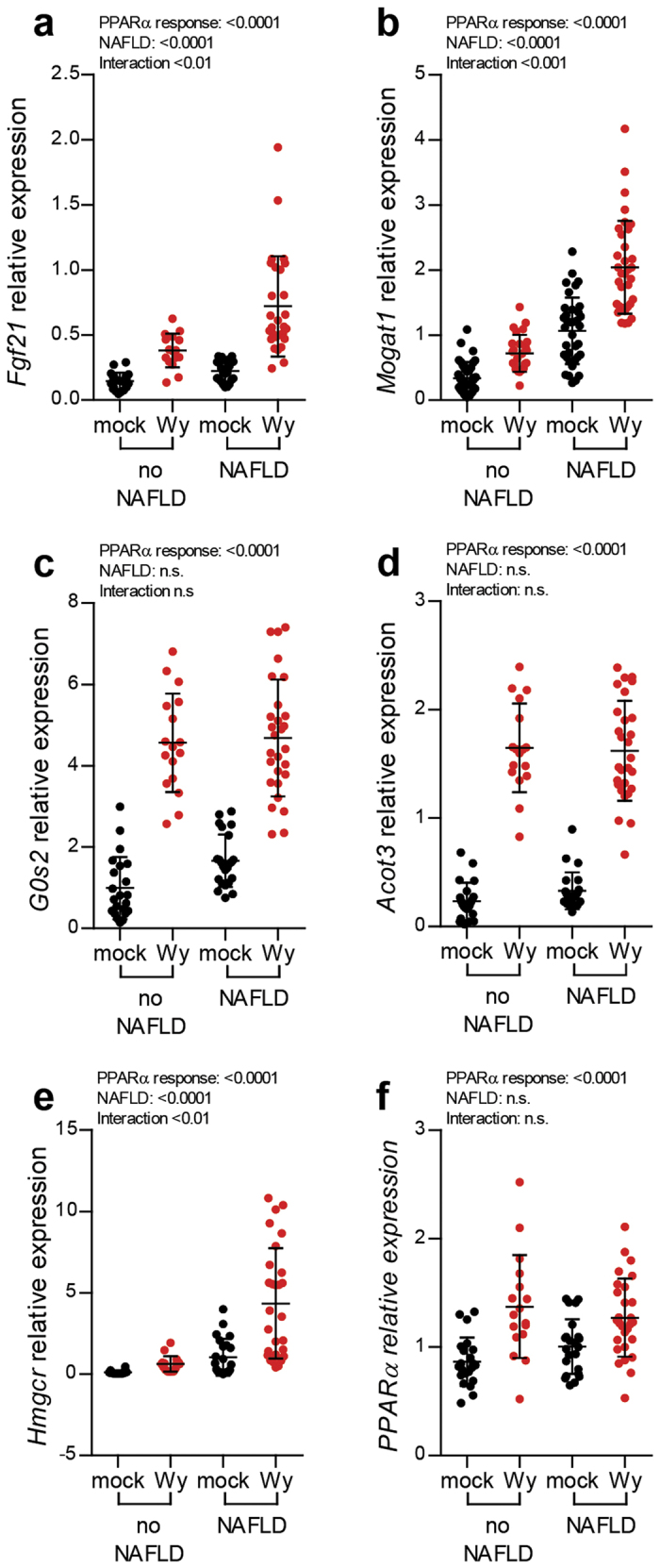
(a) Fgf21; (b) Mogat1; (c) G0s2; (d) Acot3; (e) Hmgcr; and (f) Pparα. The effect of PPARα agonist and NAFLD, as well as any interaction between them, were analysed using two-way ANOVA.
Discussion
In our mouse aging cohort, NAFLD development started at middle-age in Control (C)- and medium-fat (MF)-exposed mice, but not in the calorie restricted (CR)-fed animals, which stayed lean over time. The prevalence of NAFLD in the ad libitum C- and MF-fed groups increased during aging, which reflects weight gain and aging as risk factors for developing NAFLD38. In this study, we applied a long-term exposure to a less extreme diet compared to previous studies16,17 by using a 25E% medium-fat diet to mimic the slow onset of NAFLD in the human population, which did not induce severe NAFLD. We assessed this by analysing the liver fibrosis marker, 4-hydroxyproline content, and observed a lower prevalence of liver fibrosis, compared to liver steatosis or benign NAFLD.
The analysis of plasma Fgf21 levels in the different intervention groups and ages suggests that there is an age-effect on plasma Fgf21 levels, although we did not observe a diet-dependent effect on plasma Fgf21 levels. We found that at the age of 6 months, some of the mice displayed a high level of plasma Fgf21 in the absence of elevated IHTG levels. A previous study in children has also revealed a lack of correlation between serum FGF21 and NAFLD parameters39, moreover another study showed an inverse correlation between FGF21 and hepatic damage40. The addition of serum FGF21 in a NAFLD diagnostic model for children and adolescent also failed to improve the diagnostic performance32. An age of 6 months in mice is equivalent to mature adult age (~30 years old in human)41 and does not correspond to a developmental period in childhood. However, both mice and human data support either an absence of correlation or different correlation between plasma Fgf21 and NAFLD in younger age groups. Overall, our results reveal that, with the exception of the 6-month-old animals, plasma Fgf21 levels significantly correlated with IHTG content and performed well as a plasma marker for NAFLD diagnosis. This is in agreement with the results of previous studies in humans10,11.
The pathogenesis of NAFLD is attributed to a multi-hit process that includes lipotoxicity, oxidative stress and endoplasmic reticulum (ER) stress. Liver fat accumulation involves excess fatty acid supply to the liver, which triggers fatty acid oxidation and, consequently, oxidative stress from microsomal enzymes and ER stress. We found that NRF2 and PPARα targets were the most significantly enriched up-regulated pathways in the animals with NAFLD. NRF2 acts as a protective measure against oxidative stress, by producing antioxidant proteins. Meanwhile, PPARα activation is crucial for maintaining the homeostasis of fatty acid metabolism by increasing mitochondrial β-oxidation, thereby reducing the potential for fatty acid-induced lipotoxicity42. In addition, the WGCNA followed by pathway analysis pointed XBP1 out as one of the predicted upstream regulators of Fgf21. The elevated expression of FGF21 has been described as a counteractive mechanism for ER stress, by modulating lipid metabolism43,44,45. Therefore, the up-regulation of Fgf21 in NAFLD appears to be a simultaneous protection against lipotoxicity, oxidative stress and ER stress in NAFLD.
Intriguingly, we found that plasma Fgf21 levels at young age were related to fatty acid oxidation (L-carnitine shuttle and β-oxidation), which also corresponds to PPARα activation. While this seems to be contradictory with the idea of PPARα activation as a protective measure for fatty liver, both mouse and human studies have reported that the increased mitochondrial activity and β-oxidation do not necessarily reflect an efficient electron transport chain46,47,48. The authors found that the electron transport chain in subjects with hepatic steatosis and/or obesity is inefficient46,47,48. Therefore, it is worthwhile noting that, despite the up-regulated fatty acid oxidation in both young animals without NAFLD and old animals with NAFLD, the up-regulation in old animals with NAFLD measurement might lead to perturbing consequences, such as hepatic oxidative stress.
The in vivo PPARα challenge performed in this study provides a novel insight into the ability of maintaining metabolic homeostasis. A dynamic measurement by using system perturbation or challenge tests are likely more valuable to define metabolic health or resilience, compared to more static measurements49. By performing the PPARα agonist treatment, we demonstrate that the PPARα response at the transcriptional level is partially altered in the presence of NAFLD. It appears that liver fat is the burden of the liver’s plasticity of lipid metabolism. A similar notion was reported by Hyotylainen and co-workers, showing that high liver fat markedly hampers the ability of the liver to adaptively regulate metabolism to meet the excessive demands on basic liver functions. As a consequence, individuals with NAFLD may be more vulnerable to various metabolic stressors on the liver50. This underlines that, although hepatic steatosis is considered benign (first hit), when the metabolic system faces a challenge (second/multiple hits), the ability to maintain or regain homeostasis might have been compromised.
Furthermore, we revealed the association between plasma Fgf21 and transcriptional changes related to hepatocellular carcinoma (HCC) development. The predicted regulators (HNF1A, HNF4A, RICTOR, MAP4K4), functional pathways and genes (Wnt target genes Ccnd1 and Glul) associated with plasma Fgf21 suggest dysregulations of metabolic and proliferative pathways51,52, which are characteristics of a benign hepatocellular tumor. Although these dysregulations alone were not sufficient to induce carcinogenesis, it likely increases the susceptibility to HCC development. In line with this hypothesis, a pre-malignant stage in NAFLD has been shown to denote stress, inflammation and even apoptosis, which pre-condition and initiate pro-oncogenic signals53. This is supported by the evidence that both liver expression and circulating levels of FGF21 are increased in patients with hepatitis, cirrhosis and hepatocarcinoma54. Therefore, the population would likely benefit from the use of plasma Fgf21 as a biomarker of an early stage of NAFLD.
Although the association between plasma Fgf21 and HCC-related function is not as strong as its performance in reflecting the benign NAFLD stage, plasma Fgf21 might be beneficial in improving the performance of biomarkers for advanced stages of NAFLD. This has been suggested in the study of a NASH biomarker, that for the purpose of defining the stages of NASH, plasma CK-18 performs better than FGF21, but adding FGF21 to the CK-18 model significantly improved the performance14. We evaluated the combined analysis of plasma Fgf21 and CK-18 to detect NAFLD in our study, but it did not result in a higher accuracy compared to merely Fgf21 analysis. A plausible explanation for this observation is that our mice cohort modelled a rather mild NAFLD development, so that the addition of plasma CK-18, which represents advanced stages of NAFLD, did not effectively improve the analysis.
In this study, we discovered that the expression level of Klb, a critical co-receptor of the FGF receptors, was negatively correlated with plasma Fgf21, IHTG and body weight. In addition, the liver expression levels of the FGF receptors Fgfr2 and Fgfr4 showed similar patterns, although the declining expression levels were not as strong as observed for Klb. FGFR2 and FGFR4 proteins, along with FGFR1, have been shown to form a transmembrane complex with β-klotho to mediate the effects of FGF21 in adipocytes55. Although the type of FGF receptor that forms a complex with β-klotho protein in the liver is still unclear, this observation underlines the growing notion that metabolic system might develop a resistance to mediate the beneficial effect of Fgf21. The Fgf21 resistance due to its co-receptor alteration is an essential issue to be addressed during the further development of FGF21 as a novel pharmacological agent for metabolic diseases56. Both human and mice studies have reported increased FGF21 gene expression or circulating protein levels with obesity and/or metabolic syndrome57,58,59. One of these studies also demonstrated that the diet-induced obese mice with an elevated endogenous level of Fgf21 responded poorly to acute exogenous Fgf21 administration59. Since Klb plays a critical role in mediating Fgf21’s metabolic activity34,60, the declining Klb expression that occurs over a long-term obesity and/or hepatic steatosis development might result in Fgf21 resistance. Therefore, in order to assess the possibility of resistance to FGF21 treatment, the consequence of the down-regulation of critical FGF21 receptors/co-receptors in the liver on sensitivity to endo- and exogenous FGF21 warrants further investigation. It is worthwhile noting that, in the acute induction of obesity in mouse model, Klb expression was not altered in the obese state59.
Taken together, in this study, we demonstrate that plasma Fgf21 levels strongly reflects liver fat accumulation, confirming its potential as NAFLD marker. However, this association is age-dependent and does not apply at the age of 6 months in the C57BL/6J mice. The molecular link between plasma Fgf21 and IHTG levels was associated with dysregulation of both metabolic and cancer-related pathways. The up-regulated Fgf21 levels in NAFLD appears to be a measure to maintain homeostasis against the adverse effects in NAFLD, e.g. lipotoxicity, oxidative stress and endoplasmic reticulum stress. The elevated plasma Fgf21 is also associated with declining expression of Klb, its crucial co-receptor, which suggests a resistance to Fgf21. Therefore, although liver fat accumulation is a benign stage of NAFLD, the liver is likely more vulnerable to metabolic stressors and progress to end-stage liver disease. The in vivo PPARα challenge further demonstrates the dysregulation of PPARα signalling in the presence of NAFLD, which results in a stochastically increasing hepatic expression of Fgf21. In conclusion, Fgf21 plasma levels reflect liver fat accumulation and dysregulation of metabolic pathways at a transcriptional level in the liver of C57BL/6J mice.
Additional Information
How to cite this article: Rusli, F. et al. Fibroblast growth factor 21 reflects liver fat accumulation and dysregulation of signalling pathways in the liver of C57BL/6J mice. Sci. Rep. 6, 30484; doi: 10.1038/srep30484 (2016).
Supplementary Material
Acknowledgments
This work was financially supported by European Union’s Seventh Framework Programme (FP7/2007–2011) IDEAL-aging under grant agreement no. 259679.
Footnotes
Author Contributions F.R. and J.D. conceived the study and designed the experiment with input from W.T.S. and M.B. F.R., E.A. and C.L. performed the experiments and data analysis. M.V.B and E.B.vd.A. supervised the data analysis. F.R., J.D. and E.A. wrote the manuscript with contributions from all authors. M.M., M.B. and W.T.S. supervised the entire project.
References
- Lazo M. et al. Prevalence of Nonalcoholic Fatty Liver Disease in the United States: The Third National Health and Nutrition Examination Survey, 1988–1994. American Journal of Epidemiology 178, 38–45, doi: 10.1093/aje/kws448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satapathy S. K. & Sanyal A. J. Epidemiology and Natural History of Nonalcoholic Fatty Liver Disease. Semin Liver Dis 35, 221–235, doi: 10.1055/s-0035-1562943 (2015). [DOI] [PubMed] [Google Scholar]
- Marchesini G. & Petta S. & dale Grave, R. Diet, Weight Loss, and Liver Health in NAFLD: Pathophysiology, Evidence and Practice. Hepatology, doi: 10.1002/hep.28392 (2015). [DOI] [PubMed] [Google Scholar]
- Yan H. et al. Circulating fibroblast growth factor 21 levels are closely associated with hepatic fat content: a cross-sectional study. PloS one 6, e24895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano E., Tagawa K., Yamaoka K. & Mori M. Test validity of periodic liver function tests in a population of Japanese male bank employees. Journal of clinical epidemiology 54, 945–951, doi: 10.1016/s0895-4356(01)00355-9 (2001). [DOI] [PubMed] [Google Scholar]
- Mofrad P. et al. Clinical and histologic spectrum of nonalcoholic fatty liver disease associated with normal ALT values. Hepatology 37, 1286–1292 (2003). [DOI] [PubMed] [Google Scholar]
- Angulo P. Nonalcoholic fatty liver disease. New England Journal of Medicine 346, 1221–1231 (2002). [DOI] [PubMed] [Google Scholar]
- Chalasani N. et al. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. The American journal of gastroenterology 107, 811–826, doi: 10.1038/ajg.2012.128 (2012). [DOI] [PubMed] [Google Scholar]
- Saadeh S. et al. The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology 123, 745–750, doi: 10.1053/gast.2002.35354 (2002). [DOI] [PubMed] [Google Scholar]
- Dushay J. et al. Increased fibroblast growth factor 21 in obesity and nonalcoholic fatty liver disease. Gastroenterology 139, 456–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Fibroblast growth factor 21 levels are increased in nonalcoholic fatty liver disease patients and are correlated with hepatic triglyceride. Journal of Hepatology 53, 934–940, doi: 10.1016/j.jhep.2010.05.018 (2010). [DOI] [PubMed] [Google Scholar]
- Yilmaz Y. et al. Increased serum FGF21 levels in patients with nonalcoholic fatty liver disease. European journal of clinical investigation 40, 887–892, (2010). [DOI] [PubMed] [Google Scholar]
- Li H., Zhang J. & Jia W. Fibroblast growth factor 21: a novel metabolic regulator from pharmacology to physiology. Frontiers of Medicine 7, 25–30, doi: 10.1007/s11684-013-0244-8 (2013). [DOI] [PubMed] [Google Scholar]
- Shen J. et al. Non-invasive diagnosis of non-alcoholic steatohepatitis by combined serum biomarkers. Journal of Hepatology 56, 1363–1370, doi: 10.1016/j.jhep.2011.12.025 (2012). [DOI] [PubMed] [Google Scholar]
- Bellentani S., Scaglioni F., Marino M. & Bedogni G. Epidemiology of Non-Alcoholic Fatty Liver Disease. Dig Dis 28, 155–161 (2010). [DOI] [PubMed] [Google Scholar]
- Duval C. et al. Adipose Tissue Dysfunction Signals Progression of Hepatic Steatosis Towards Nonalcoholic Steatohepatitis in C57Bl/6 Mice. Diabetes 59, 3181–3191, doi: 10.2337/db10-0224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner R., Schölmerich J. & Bollheimer L. C. High-fat Diets: Modeling the Metabolic Disorders of Human Obesity in Rodents. Obesity 15, 798–808, doi: 10.1038/oby.2007.608 (2007). [DOI] [PubMed] [Google Scholar]
- Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559, doi: 10.1186/1471-2105-9-559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusli F. et al. A weekly alternating diet between caloric restriction and medium fat protects the liver from fatty liver development in middle-aged C57BL/6J mice. Molecular nutrition & food research 59, 533–543, doi: 10.1002/mnfr.201400621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Norren K. et al. Behavioural changes are a major contributing factor in the reduction of sarcopenia in caloric-restricted ageing mice. Journal of Cachexia, Sarcopenia and Muscle 6, 253–268, doi: 10.1002/jcsm.12024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner D. E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005). [DOI] [PubMed] [Google Scholar]
- Hillebrandt S. et al. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat Genet 37, 835–843 (2005). [DOI] [PubMed] [Google Scholar]
- Steegenga W. T. et al. Sexually dimorphic characteristics of the small intestine and colon of prepubescent C57BL/6 mice. Biology of Sex Differences 5, 1–17, doi: 10.1186/s13293-014-0011-9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. et al. MADMAX - Management and analysis database for multiple ~omics experiments. Journal of integrative bioinformatics 8, 160, doi: 10.2390/biecoll-jib-2011-160 (2011). [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Åstrand M. & Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193, doi: 10.1093/bioinformatics/19.2.185 (2003). [DOI] [PubMed] [Google Scholar]
- Irizarry R. A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research 31, e15, doi: 10.1093/nar/gng015 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Research 33, e175, doi: 10.1093/nar/gni179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. & Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Research 31, e154, doi: 10.1093/nar/gng154 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor M. A. et al. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics 7, doi: 10.1186/1471-2105-7-538 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. C. et al. Molecular MRI of collagen to diagnose and stage liver fibrosis. Journal of Hepatology 59, 992–998, doi: 10.1016/j.jhep.2013.06.026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot B. G. et al. Accuracy of prediction scores and novel biomarkers for predicting nonalcoholic fatty liver disease in obese children. Obesity 21, 583–590 (2013). [DOI] [PubMed] [Google Scholar]
- Ogawa Y. et al. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proceedings of the National Academy of Sciences of the United States of America 104, 7432–7437, doi: 10.1073/pnas.0701600104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. et al. βKlotho Is Required for Fibroblast Growth Factor 21 Effects on Growth and Metabolism. Cell Metabolism 16, 387–393, doi: 10.1016/j.cmet.2012.08.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S. T. et al. The genetic architecture of NAFLD among inbred strains of mice. eLife 4, e05607, doi: 10.7554/eLife.05607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badman M. K. et al. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell metabolism 5, 426–437 (2007). [DOI] [PubMed] [Google Scholar]
- Inagaki T. et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell metabolism 5, 415–425 (2007). [DOI] [PubMed] [Google Scholar]
- Duseja A. & Chalasani N. Epidemiology and risk factors of nonalcoholic fatty liver disease (NAFLD). Hepatology International 7, 755–764 (2013). [DOI] [PubMed] [Google Scholar]
- Reinehr T., Woelfle J., Wunsch R. & Roth C. L. Fibroblast growth factor 21 (FGF-21) and its relation to obesity, metabolic syndrome, and nonalcoholic fatty liver in children: a longitudinal analysis. The Journal of Clinical Endocrinology & Metabolism 97, 2143–2150 (2012). [DOI] [PubMed] [Google Scholar]
- Alisi A. et al. Association between serum atypical fibroblast growth factors 21 and 19 and pediatric nonalcoholic fatty liver disease. PloS one 8, e67160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K. et al. In The Mouse in Biomedical Research (Second Edition) 637–672 (Academic Press, 2007). [Google Scholar]
- Berson A. et al. Steatohepatitis-inducing drugs cause mitochondrial dysfunction and lipid peroxidation in rat hepatocytes. Gastroenterology 114, 764–774, doi: 10.1016/s0016-5085(98)70590-6 (1998). [DOI] [PubMed] [Google Scholar]
- Schaap F. G., Kremer A. E., Lamers W. H., Jansen P. L. & Gaemers I. C. Fibroblast growth factor 21 is induced by endoplasmic reticulum stress. Biochimie 95, 692–699 (2013). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. Fibroblast growth factor 21 participates in adaptation to endoplasmic reticulum stress and attenuates obesity-induced hepatic metabolic stress. Diabetologia 58, 809–818, doi: 10.1007/s00125-014-3475-6 (2015). [DOI] [PubMed] [Google Scholar]
- Jiang S. et al. Fibroblast Growth Factor 21 Is Regulated by the IRE1α-XBP1 Branch of the Unfolded Protein Response and Counteracts Endoplasmic Reticulum Stress-induced Hepatic Steatosis. J. Biol. Chem. 289, 29751–29765, doi: 10.1074/jbc.M114.565960 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunny N. E., Parks, Elizabeth J., Browning, Jeffrey D. & Burgess, Shawn C. Excessive Hepatic Mitochondrial TCA Cycle and Gluconeogenesis in Humans with Nonalcoholic Fatty Liver Disease. Cell Metabolism 14, 804–810, doi: 10.1016/j.cmet.2011.11.004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satapati S. et al. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. Journal of Lipid Research 53, 1080–1092, doi: 10.1194/jlr.M023382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliaki C. et al. Adaptation of Hepatic Mitochondrial Function in Humans with Non-Alcoholic Fatty Liver Is Lost in Steatohepatitis. Cell Metabolism 21, 739–746, doi: 10.1016/j.cmet.2015.04.004 (2015). [DOI] [PubMed] [Google Scholar]
- van Ommen B., van der Greef J., Ordovas J. M. & Daniel H. Phenotypic flexibility as key factor in the human nutrition and health relationship. Genes & Nutrition 9, 1–9, doi: 10.1007/s12263-014-0423-5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyotylainen T. et al. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat Commun 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D. T. et al. Control of Pancreas and Liver Gene Expression by HNF Transcription Factors. Science 303, 1378–1381, doi: 10.1126/science.1089769 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault J. C., Bioulac–Sage P. & Zucman–Rossi J. Hepatocellular Benign Tumors—From Molecular Classification to Personalized Clinical Care. Gastroenterology 144, 888–902, doi: 10.1053/j.gastro.2013.02.032 (2013). [DOI] [PubMed] [Google Scholar]
- Stickel F. & Hellerbrand C. Non-alcoholic fatty liver disease as a risk factor for hepatocellular carcinoma: mechanisms and implications. Gut, doi: 10.1136/gut.2009.199661 (2010). [DOI] [PubMed] [Google Scholar]
- Yang C. et al. Activation of Liver FGF21 in hepatocarcinogenesis and during hepatic stress. BMC gastroenterology 13, 67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A. et al. FGF‐21/FGF‐21 receptor interaction and activation is determined by βKlotho. Journal of cellular physiology 215, 1–7 (2008). [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A. & DiMarchi R. FGF21 Revolutions: Recent Advances Illuminating FGF21 Biology and Medicinal Properties. Trends in endocrinology and metabolism: TEM 26, 608–617, doi: 10.1016/j.tem.2015.09.007 (2015). [DOI] [PubMed] [Google Scholar]
- Muise E. S. et al. Adipose Fibroblast Growth Factor 21 Is Up-Regulated by Peroxisome Proliferator-Activated Receptor γ and Altered Metabolic States. Molecular Pharmacology 74, 403–412, doi: 10.1124/mol.108.044826 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Serum FGF21 Levels Are Increased in Obesity and Are Independently Associated With the Metabolic Syndrome in Humans. Diabetes 57, 1246–1253, doi: 10.2337/db07-1476 (2008). [DOI] [PubMed] [Google Scholar]
- Fisher f. M. et al. Obesity Is a Fibroblast Growth Factor 21 (FGF21)-Resistant State. Diabetes 59, 2781–2789, doi: 10.2337/db10-0193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y. et al. βKlotho is required for metabolic activity of fibroblast growth factor 21. Proceedings of the National Academy of Sciences 104, 7432–7437, doi: 10.1073/pnas.0701600104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.