Abstract
Alterations in the gut microbiota play a crucial role in host physiology and metabolism; however, the molecular pathways underlying these changes in diet-induced obesity are unclear. Mechanistic target of rapamycin (mTOR) signaling pathway is associated with metabolic disorders such as obesity and type 2 diabetes (T2D). Therefore, we examined whether changes in the regulation of mTOR signaling induced by diet (a high-fat diet [HFD] or normal-chow diet) and/or therapeutics (resveratrol [a specific inhibitor of mTOR complex 1] or rapamycin [an inhibitor of both mTOR complex 1 and 2]) altered the composition of the gut microbiota in mice. Oral administration of resveratrol prevented glucose intolerance and fat accumulation in HFD-fed mice, whereas rapamycin significantly impaired glucose tolerance and exacerbated intestinal inflammation. The abundance of Lactococcus, Clostridium XI, Oscillibacter, and Hydrogenoanaerobacterium increased under the HFD condition; however, the abundance of these species declined after resveratrol treatment. Conversely, the abundance of unclassified Marinilabiliaceae and Turicibacter decreased in response to a HFD or rapamycin. Taken together, these results demonstrated that changes in the composition of intestinal microbiota induced by changes in mTOR activity correlate with obese and diabetic phenotypes.
Obesity is a major risk factor for various chronic diseases, including type 2 diabetes (T2D), cardiovascular disease, hypertension, non-alcoholic fatty liver disease and cancer1. The fundamental cause of obesity is an imbalance between energy intake from foods and energy expenditure through basal metabolism, physical activity and thermogenesis2. Since the basal metabolism rate (BMR) accounts for about 60–75% of the total energy expenditure2, a low BMR per unit of body weight is one of the risk factors for obesity3. Moreover, energy balance is influenced by complex interactions between genetic, environmental and psychosocial factors4. With respect to energy intake, changes in gastrointestinal (GI) motility contribute to obesity by regulating not only the digestive efficiency but also appetite and satiety5. Interestingly, recent studies suggest that gut microbiota play an important role in energy harvest and obesity via interactions with GI motility6,7.
The composition of the gut microbiota is influenced by the genetic background, immune status, age, sex and (especially) diet of the host8. Although a high-fat diet (HFD) alters the composition of the intestinal microbiota9, recent studies show that the gut microbiota themselves promote obesity and a diabetic phenotype10,11. By contrast, several species of intestinal microbe have a beneficial effect on obesity and obesity-related metabolic disorders via their ability to modulate immune homeostasis12,13. We recently demonstrated that oral administration of the mucin-degrading bacterium Akkermansia muciniphila, which is markedly more abundant in metformin-treated HFD-fed mice than in HFD-fed control mice, improved glucose tolerance and alleviated adipose tissue inflammation in diet-induced obese (DIO) and diabetic mice by inducing proliferation of mucin-producing intestinal goblet cells and adipose tissue-resident Foxp3+ regulatory T cells14. Also, previous studies revealed that compositional changes in the gut microbiota resulting from genetic ablation of antimicrobial peptides such as defensin and RegIII-gamma could affect host physiology15,16. Even though studies have shown consistently that gut microbiota regulate host metabolism and immune status, both the molecular mechanisms by which energy-rich diets alter the gut microbiota, and the interactions between the host and the gut microbiota that maintain metabolic homeostasis, remain unclear.
Mechanistic target of rapamycin (mTOR) is a central regulator of energy storage and consumption, and is implicated in deleterious states such as cancer, metabolic diseases and ageing17. Because over-activation of mTOR complex 1 (mTORC1) signaling by excessive energy intake plays a crucial role in metabolic disorders17, mTOR inhibitors such as rapamycin (23,27-epoxy-3H-pyrido[2,1-c][1,4]oxaazacyclohentriacontine), metformin (1,1-dimethylbiguanide hydrochloride) and resveratrol (3,4′,5-trihydroxy-trans-stilbene) are used to treat those suffering from obesity and T2D14,18,19. Both metformin and resveratrol activate AMP-activated protein kinase (AMPK), which in turn inhibits mTORC114,18. Although rapamycin is also thought to inhibit mTORC1-S6 kinase 1 (S6K1) activation, long-term treatment with rapamycin also inactivates mTOR complex 2 (mTORC2) and Akt (Ser473), leading to impaired glucose homeostasis and insulin activity19. Several studies have shown increased activation of AMPK in the liver, muscle and colon tissue of germ-free mice that are resistant to HFD-induced obesity, suggesting that the intestinal microbiota may modulate AMPK activation in the host20,21. These findings raise the possibility that changes in mTOR signaling may have both direct and indirect effects on the intestinal microbiota, which in turn would contribute to obese and diabetic phenotypes.
Here, we examined the effects of resveratrol and rapamycin on mTOR signaling, metabolic marker expression, and the composition of the gut microbiota in both normal and DIO mice. The results show that resveratrol and rapamycin have marked effects not only on the mTOR signaling activity and metabolic marker expression but also on the composition of gut microbiota, suggesting that specific microbial groups correlate with the pathophysiological phenotypes associated with obesity and T2D.
Results
Resveratrol, but not rapamycin, mitigates HFD-induced obesity
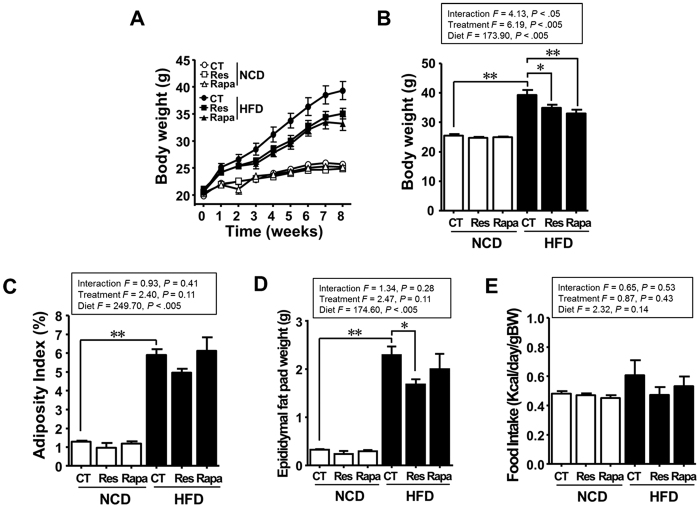
As expected, the body weight (BW) and fat deposition of HFD-fed mice were higher than those of normal-chow diet (NCD)-fed mice (P < 0.005; Fig. 1). The BW and epididymal fat pad weight of the HFD-fed mice treated with resveratrol (HFD-Res) were significantly lower than those of the HFD-fed mice without any additional treatment (HFD-CT) (P < 0.05; Fig. 1A–C). By contrast, epididymal fat pad weight in the HFD-fed mice treated with rapamycin (HFD-Rapa) was not lower than that in the HFD-CT mice (P = 0.60), although BW was markedly lower than that in the HFD-Res mice (P < 0.005). There were no marked differences in the amount of food consumed by the animals in the control and treatment groups (Fig. 1E). Thus, resveratrol mitigated both BW gain and fat deposition. However, although rapamycin mitigated weight gain in HFD-fed mice, it had no effect on fat mass.
Figure 1. Resveratrol, but not rapamycin, mitigates HFD-induced obesity.
(A) Effects of resveratrol or rapamycin treatment on temporal changes in the body weight (BW) of NCD- or HFD-fed mice over 8 weeks. (B) BW, (C) adiposity index (AI), and (D) epididymal fat pad weight measured after 8 weeks of resveratrol or rapamycin treatment (E) Food intake (FI) by the resveratrol- or rapamycin-treated groups. Data are expressed as the mean ± SEM (n = 5 per group). F- and p-values are from two-way ANOVA after Bonferroni’s post hoc test. *P < 0.05, **P < 0.005.
Resveratrol improves, whereas rapamycin impairs, glucose homeostasis
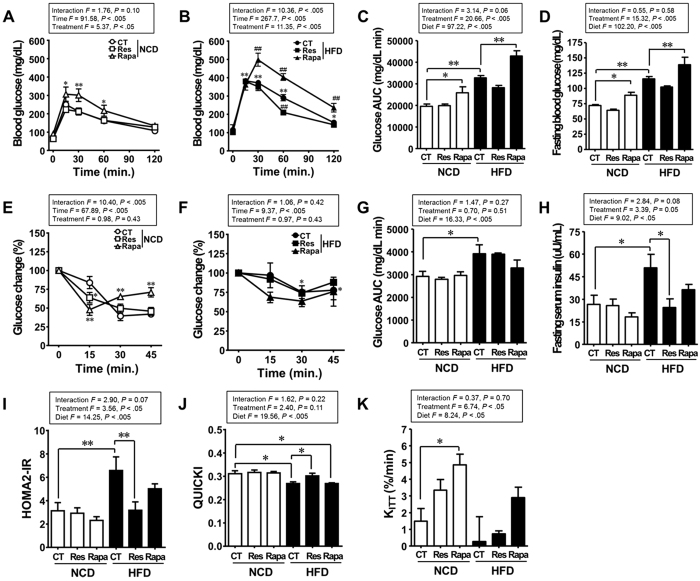
To determine whether resveratrol or rapamycin affect glucose homeostasis, we performed the glucose tolerance test (GTT) and insulin tolerance test (ITT) at the end of the treatment period. As expected, compared with the NCD-fed mice without any additional treatment (NCD-CT), the HFD-CT group showed an increase in the area under the curve (AUC) during GTT and ITT, along with an increase in fasting blood glucose and fasting serum insulin levels (Fig. 2A–H). The homeostatic model assessment 2 (HOMA2) model and quantitative insulin sensitivity check index (QUICKI) are measures of insulin resistance and insulin sensitivity, respectively, and are derived from fasting glucose and insulin levels22,23. Here, we found that the HOMA2-insulin resistance (IR) index was significantly higher and QUICKI significantly lower in the HFD-CT group than in the NCD-CT group (Fig. 2I,J). These results indicate that a HFD impairs glucose tolerance and induces insulin resistance. When compared with the HFD-CT group, the HFD-Res group showed significantly improved glucose intolerance and insulin resistance, both of which were confirmed by a reduction in the blood glucose level at 60 min. during GTT, reduced levels of fasting serum insulin, a lower HOMA2-IR index and a higher QUICKI index (Fig. 2B,H–J). Unexpectedly, chronic rapamycin treatment led to impaired glucose homeostasis, including in the NCD-fed mice. There was a marked increase in glucose intolerance (AUC of the GTT) and fasting blood glucose level in the rapamycin-treated mice compared with the control mice (Fig. 2A–D). However, the rate constant for plasma glucose disappearance (KITT) during ITT was higher in the NCD-fed mice treated with rapamycin (NCD-Rapa) than in the NCD-CT mice (Fig. 2K). Taken together, these results suggest that chronic rapamycin treatment is associated with impaired insulin synthesis or secretion, but not with insulin resistance. Thus, resveratrol treatment prevented hyperinsulinaemia and insulin resistance in DIO mice, whereas rapamycin triggered glucose intolerance and disrupted pancreatic beta cell function in both healthy and DIO mice.
Figure 2. Resveratrol improves, whereas rapamycin impairs, glucose homeostasis.
(A,B) Blood glucose levels (repeated measures two-way ANOVA after Bonferroni’s post hoc test, F = 13.88; P < 0.005 for interaction, F = 186.0; P < 0.005 for time, F = 92.14; P < 0.005 for diet, *P < 0.05; **P < 0.005 compared with NCD-CT, #P < 0.05; ##P < 0.005 compared with HFD-CT) and (C) area under the curve (AUC) during the glucose tolerance test (GTT) (n = 5 per group). (D) Fasting blood glucose levels (n = 10 per group). (E,F) Blood glucose levels (repeated measures two-way ANOVA after Bonferroni’s post hoc test, F = 3.33; P = 0.06 for interaction, F = 19.65; P < 0.005 for time, F = 5.63; P = 0.08 for diet, *P < 0.05; **P < 0.005 compared with NCD-CT) and (G) AUC during the insulin tolerance test (ITT) (n = 3 per group). (H) Fasting serum insulin levels (n = 5 per group). (I) HOMA2 indices and (J) QUICKI were calculated from fasting glucose and insulin levels (n = 5 per group). (K) The rate constant for plasma glucose disappearance (KITT) during the insulin tolerance test (ITT) (n = 3 per group). Mice were overnight-fasted (16 h) before the GTT and ITT. Data are expressed as the mean ± SEM. F- and p-values are from two-way ANOVA after Bonferroni’s post hoc test. *P < 0.05, **P < 0.005.
Resveratrol ameliorates, whereas rapamycin exacerbates, intestinal inflammation
We next examined the effects of resveratrol and rapamycin on gut inflammation in NCD or HFD-fed mice by measuring faecal lipocalin-2 (LCN-2), a sensitive biomarkers for low-grade intestinal inflammation24, and colon length. The concentration of faecal LCN-2 in HFD-CT mice was significantly higher than that in NCD-CT mice (P < 0.05; Supplementary Fig. S2A). Moreover, higher levels of LCN-2 were observed in HFD-Rapa mice than in HFD-CT mice (P < 0.05). Furthermore, the length of the colon in HFD-Rapa mice was significantly shorter than that in NCD-CT mice (P < 0.05; Supplementary Fig. S2B), and the levels of faecal LCN-2 correlated negatively with colon length (r = −0.92; P < 0.005) (Supplementary Fig. S2C). These observations indicate that rapamycin aggravates intestinal inflammation in DIO mice.
Resveratrol specifically inhibits mTORC1, whereas rapamycin inhibits both mTORC1 and 2
The ratio of phosphorylated S6 (Ser235/236) to total S6 protein (p-S6/total S6) was significantly higher, and the ratio of phosphorylated Akt1 (Ser473) to total Akt1 (p-Akt1/total Akt1) lower, in HFD-CT mice than in NCD-CT mice (Supplementary Fig. S3A,B). These results suggest that a HFD activates mTORC1. By contrast, the p-S6/total S6 ratio was lower, and the p-Akt1/total Akt1 ratio and cAMP level were higher, in HFD-Res mice than in HFD-CT mice (Supplementary Fig. S3C), indicating that resveratrol inhibits mTORC1 and activates mTORC2 in HFD-fed mice. By contrast, chronic rapamycin treatment led to reduced p-S6/total S6 and p-Akt1/total Akt1 ratios, regardless of diet, demonstrating that rapamycin suppresses the activity of both mTORC1 and 2.
Effect of resveratrol and rapamycin on the gut microbiota
To ascertain the effects of differential regulation of mTOR activity on the gut microbiota, we analysed the sequences of bacterial 16S rRNA gene amplicons using 454 pyrosequencing technology. After quality control processing, we obtained 215,932 high-quality sequences, with an average of 7,198 (±358) reads per sample. Compared with NCD-CT, we observed a significant reduction in the operational taxonomic units (OTUs) and bacterial alpha diversity indices in the HFD-fed groups (Supplementary Table S1). However, there was no significant difference in the alpha diversity indices between resveratrol- and rapamycin-treated groups. Although the relative abundance of Firmicutes (P < 0.005) was higher, and that of Bacteroidetes lower (P < 0.005), in HFD-CT mice than in NCD-CT mice, resveratrol and rapamycin treatment under NCD or HFD feeding conditions did not significantly affect the faecal Firmicutes/Bacteroidetes (F/B) ratio (Supplementary Fig. S4).
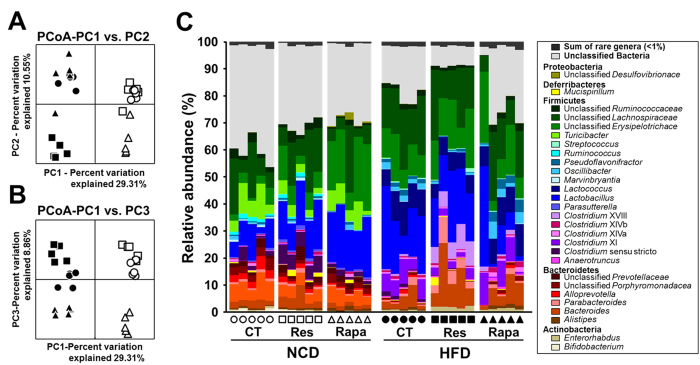
Using principle coordinate analysis (PCoA) based on unweighted UniFrac distances, we next compared the composition of the gut microbiota in the diet and treatment groups. The PC1 axis of the PCoA clearly separated the gut bacterial community according to dietary type (Fig. 3A). Furthermore, each resveratrol- or rapamycin-treated group formed a distinct cluster from the control groups along the PC3 axis (Fig. 3B), suggesting that resveratrol or rapamycin has differential effects on gut microbial communities in NCD- and HFD-fed mice.
Figure 3. Changes in the faecal bacterial community following resveratrol or rapamycin treatment.
Bacterial communities were clustered using unweighted UniFrac distance-based principal coordinates analysis (PCoA). (A) Principal coordinate (PC) 1 versus PC2 and (B) PC1 versus PC3. The percentage variation in the plotted PC is indicated on the axes. (C) Bar charts showing the relative abundance (%) of different bacterial genera in the different diet and treatment groups. Each group of mice is represented by a different symbol or bar on the x axis of the graph, and each spot or column indicates one sample (n = 5 per group).
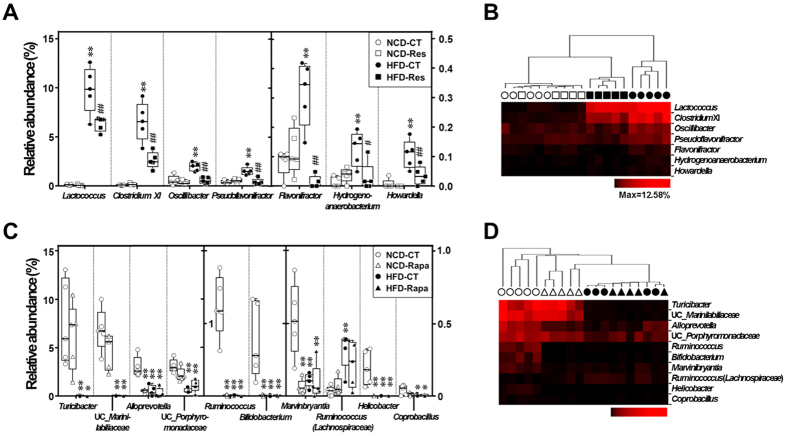
To determine whether resveratrol or rapamycin induce more specific changes in the gut bacterial taxa, we performed a nearest shrunken centroid (NSC) analysis. Statistical analysis of variance (ANOVA) and NSC analyses revealed that changes in the abundance of 17 taxa accounted for the observed changes in the gut microbiota induced by diet and resveratrol or rapamycin treatment, which suggests a correlation between the antidiabetic effect of resveratrol or diabetic effect of rapamycin and specific subsets of gut bacteria. The relative abundances of Lactococcus, Clostridium XI, Oscillibacter, Pseudoflavonifractor, Flavonifractor, Hydrogenoanaerobacterium and Howardella were significantly higher in HFD-fed mice, and resveratrol treatment reversed these HFD-induced changes in bacterial abundance (Figs 3C and 4A). Furthermore, hierarchical clustering showed that the bacterial profiles of HFD-Res mice resembled more those of NCD-fed mice than those of HFD-CT mice (Fig. 4B). By contrast, rapamycin changed the relative abundances of Turicibacter, unclassified Marinilabiliaceae, Alloprevotella, unclassified Porphyromonadaceae, Ruminococcus, Bifidobacterium, Marvinbryantia, Ruminococcus (Lachnospiraceae), Helicobacter, and Coprobacillus to those observed in HFD-fed mice (Figs 3C and 4C). With the exception of Ruminococcus (Lachnospiraceae), these bacteria were more abundant in NCD-fed mice than in HFD-fed or rapamycin-treated mice. The results were confirmed by hierarchical clustering analysis, which revealed that the bacterial profiles in NCD-Rapa mice clustered more closely with those in HFD-fed mice than with those in NCD-CT mice (Fig. 4d). Thus, resveratrol prevented changes in the relative abundance of specific HFD-induced bacteria, resulting in bacterial levels similar to those seen in NCD-fed mice, while rapamycin contributed to HFD-induced changes in gut microbiota.
Figure 4.
Marked differences in the relative abundance of gut bacterial genera in the different diet and treatment groups. Effect of (A,B) resveratrol or (C,D) rapamycin on the relative abundance of 7 or 10 bacterial genera, and hierarchical clustering analysis of these bacterial profiles based on the Manhattan distance, were examined in NCD- and HFD-fed mice. Data are expressed as the mean ± SEM (n = 5 per groups). F- and p-values are from two-way ANOVA after Bonferroni’s post hoc test (Supplementary Table S2). *P < 0.05 and **P < 0.005 compared with NCD-CT. #P < 0.05 and ##P < 0.005 compared with HFD-CT.
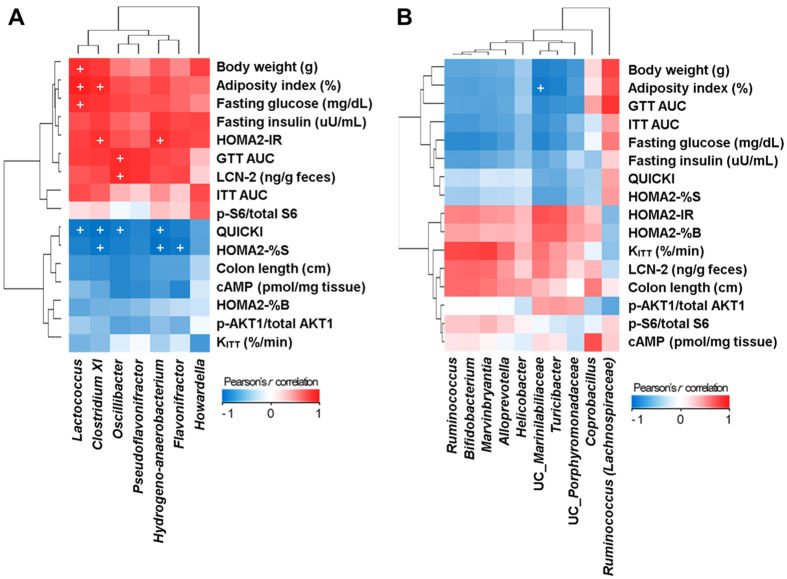
With this in mind, we performed linear regression analysis to examine for a possible connection between the abundance of specific gut bacteria and host metabolic parameters. Despite the small sample size, Pearson’s correlation analysis showed that several metabolic parameters correlated with the abundance of specific populations of gut bacteria (Fig. 5). Specifically, the relative abundances of five taxa (Lactococcus, Clostridium XI, Oscillibacter, Hydrogenoanaerobacterium and Flavonifractor) were higher in HFD-fed mice and were reduced by resveratrol treatment. These abundances correlated positively with biomarkers for metabolic syndrome (e.g., BW, AI, AUC during GTT, fasting blood glucose level, the HOMA2-IR index, and faecal LCN-2 levels), and correlated negatively with biomarkers for insulin sensitivity (e.g., QUICKI and HOMA2-%S indices). Conversly, the relative abundance of unclassified Marinilabiliaceae which were lower in HFD-fed and rapamycin-treated mice, correlated negatively with AI. Therefore, HFD and rapamycin not only contribute to the mTOR signaling activity and the host diabetic phenotype, but also influence the composition of the gut microbiota.
Figure 5.
Pearson’s r correlation coefficients heat maps showing the association between metabolic markers and the abundance of specific bacterial genera after (A) resveratrol or (B) rapamycin treatment. Given the large number of correlation tests performed, a significance threshold of P < 0.005 was used, which is indicated by ‘+’.
Discussion
We previously showed that oral administration of mucin-degrading Akkermansia muciniphila, which is markedly more abundant in metformin-treated HFD-fed mice, resulted in improved glucose homeostasis and reduced adipose tissue inflammation via its ability to induce goblet cells in the intestine and regulatory T cells in adipose tissue14. Since metformin is a key regulator of mTOR signaling, the present study examined whether the mechanisms underlying host central energy metabolism, which are controlled by differential regulation of the mTOR pathway, involve changes in the composition of gut microbiota.
Consistent with the results of a previous study18, we found that resveratrol protected HFD-fed mice against glucose intolerance, hyperinsulinaemia, fat deposition and BW gain. These effects seem to be attributed to the various mechanisms of resveratrol, including the inhibition of mTORC1 signaling pathway. Conversely, prolonged rapamycin treatment disrupted glucose homeostasis and pancreatic beta cell function in both NCD and HFD-fed mice by inhibiting mTORC1 and mTORC2 and preventing Akt activation19. Consistent with our results, recent studies indicate that mTOR signaling is important not only for glucose and lipid metabolism25 but also for the regulation of gut barrier function26 and immune homeostasis via its effects on immune cell profiles27 and cytokine production28, both of which may play an essential role in controlling gut microbiota and affect the pathogenesis of obesity and diabetes. In addition, over-activation of mTORC1 signaling via TSC2 inactivation suppresses the differentiation of intestinal goblet and Paneth cells29, both of which might contribute to dysbiosis of gut microbiota by reducing the production of mucus and antimicrobial peptides, respectively16,30. Interestingly, chronic rapamycin treatment has been suggested to aggravate intestinal inflammation in diet-induced obese mice. Recent various studies have confirmed that rapamycin treatment contributes to inflammation by promoting the expression of pro-inflammatory cytokines, such as interleukin-12 (IL-12), IL-6, IL-1β and tumor necrosis factor-α, and by inhibiting of IL-10 expression in immune cells31,32,33. These pro-inflammatory responses have been attributed to the activation of the nuclear factor-kB (NF-kB) and/or forkhead box O1 (FoxO1) or the inhibition of signal transducer and activator of transcription 3 (STAT3). In particular, mTORC2 signaling is crucial for regulating immune status because inactivity of Akt (a downstream target of mTORC2 signaling) leads to impairment of FoxO1 phosphorylation34. Impairment of FoxO1 phosphorylation promotes not only gluconeogenesis in the liver, protein catabolism in muscle, and apoptosis in pancreatic beta cells, but also inflammation of adipose tissue by increasing TLR4-mediated signaling in mature macrophages34. Macrophages in adipose tissue play an important role in the pathogenesis of insulin resistance and obesity via their ability to produce pro-inflammatory cytokines35. Furthermore, FoxO1 directly regulates the production of antimicrobial peptides by binding to the regulatory region of the antimicrobial peptide gene promoter34, thereby modulating the composition of the gut microbial community16. Here, we showed that the prolonged rapamycin treatment is associated with obese and diabetic phenotypes, whereas resveratrol treatment is associated with improved metabolic biomarker profiles in DIO mice.
We also found that the pathophysiology induced by changes in the regulation of the mTOR pathway were associated with a marked shift in the composition of gut microbiota. When considering the recent studies showing that dysbiosis of the gut microbiota directly affects host metabolism36,37, the changes in gut microbiota by resveratrol or rapamycin treatment may have the potential to affect DIO and diabetes. We also found that the relative abundances of 17 bacterial taxa were changed significantly after resveratrol or rapamycin treatment. Among these, Lactococcus, Clostridium XI, Oscillibacter, and Hydrogenoanaerobacterium were most strongly associated with obese and diabetic phenotypes. The proportions of these taxa correlated significantly and positively with BW, AI, glucose intolerance, insulin resistance and intestinal inflammatory marker gene expression. Several studies have highlighted a link between the abundance of these taxa and HFD-induced obesity14,38,39; however, little is known about their physiological roles. Qiao et al.38 showed that an increase in the Lactococcus population in Peyer’s patches is associated with an obesity-prone phenotype, and was positively correlated with the levels of pro-inflammatory cytokines such as interleukin (IL)-6 and TNF-α, but negatively associated with the levels of anti-inflammatory cytokines such as IL-10. Moreover, mice fed a NCD or a HFD with 30% caloric-restriction showed a significant reduction in the abundance of Lactococcus40. Our recent study also demonstrated that HFD-fed mice orally administered with metformin had lower abundances of Lactococcus and Hydrogenoanaerobacterium14. Yoshimoto et al.41 showed that the proportion of Clostridium XI is markedly higher in HFD-fed mice and is associated with higher levels of deoxycolic acid, a known carcinogen of the colon and liver41,42. Oscillibacter is positively correlated with gut permeability43, which can influence adiposity and systemic inflammation in obese prone donors and their GF recipients fed a HF diet39. In addition, Methanobrevibacter smithii, the predominant microbe in the obese human gut44, utilizes hydrogen for methanogenesis and increases energy uptake efficiency by interacting with hydrogen-producing Hydrogenoanaerobacterium45. Based on these results, we suggest that activating the mTORC2 signaling pathway followed by mTORC1 inhibition with resveratrol suppresses the growth of obesity-associated gut microbiota, such as Lactococcus, Clostridium XI, Oscillibacter, and Hydrogenoanaerobacterium.
In conclusion, the results presented herein demonstrate that changes in the composition of the gut microbiota caused by treatment of resveratrol and rapamycin are correlated with alterations in BW, fat deposition, insulin resistance, and intestinal inflammation in DIO mice. Although the identities of specific HFD-related molecules that enrich or diminish certain populations of gut microbes remain to be identified, mTOR signaling would appear to be a key component of the regulation of the composition of gut microbiota in DIO mice.
Methods
Animals
Four-week-old male C57BL/6J mice were purchased from Japan SLC, Inc. (Haruno Production Facility, Japan) and maintained in groups of no more than five mice per cage at the animal facility at Kyung Hee University. Mice were housed under specific pathogen-free conditions at 48 ± 6% relative humidity and temperature- and light-controlled conditions (25 ± 1 °C; 14 hr light/10 hr dark cycle) with free access to food and water. After 1 week of acclimation, mice were fed either a NCD (12.41% kcal from fat, 24.52% kcal from protein, 63.07% kcal from carbohydrate; #38057; Purina Korea, Inc., Seoul, South Korea; Supplementary Table S3) or a HFD (60% kcal from fat, 20% kcal from protein, 20% kcal from carbohydrate; #D12492; Research Diets Inc., New Brunswick, USA; Supplementary Table S4). During the 8 week study, male C57BL/6J mice were divided into six groups (5-week-old; n = 5 per group) as follows: (1) an NCD without any additional treatment (NCD-CT); (2) an NCD plus resveratrol (NCD-Res); (3) an NCD plus rapamycin (NCD-Rapa); (4) a HFD without any additional treatment (HFD-CT); (5) a HFD plus resveratrol (HFD-Res); or (6) a HFD plus rapamycin (HFD-Rapa). The resveratrol-treated (NCD-Res and HFD-Res) mice received 200 mg/kg/day of resveratrol (ChromaDex, Inc., Irvine, CA) and the rapamycin-treated (NCD-Rapa and HFD-Rapa) mice received 3 mg/kg/day of rapamycin (Enzo Life Sciences, Inc., Farmingdale, NY) by oral gavage (ethanol-dissolved stock solution diluted in phosphate buffered saline) throughout the experimental period (5 days per week). Control mice were gavaged with phosphate buffered saline alone by the same person. Food intake was manually monitored per cage during a four-week period. All animal experiments were approved by and performed in accordance with the guideline of the committee for care and use of laboratory animals of College of Pharmacy, Kyung Hee University (KHP-2013-08-2-R1).
Samples collection
Faecal samples were freshly collected after 8 weeks of resveratrol or rapamycin treatment, and then stored at −80 °C. At the end of the treatment period, the mice were anesthetized using isoflurane (2-chloro-2-(difluoromethoxy)-1,1,1-trifluoro-ethane) after overnight-fasting (16 hr). Livers, intestines, epididymal fat pads and blood samples were rapidly collected, washed briefly in PBS and stored at −80 °C until processing. Epididymal fat pads were weighted for the calculation of the AI (g epididymal fat pads weight/g BW·100). Serum was separated from blood using Microtrainer™ tubes (BD, Franklin Lakes, NJ) for insulin analyses.
Analysis of glucose homeostasis
A GTT or ITT was performed at the end of the treatment period. Overnight-fasted mice received a glucose load (1.5 g/kg BW) by oral gavage or an insulin load (0.75 U/kg BW) by intraperitoneal injection. A blood glucometer (Accu-Check Performa, Roche) was used to measure blood glucose levels both before and after glucose or insulin loading. The updated homeostatic model assessment (HOMA2), which includes HOMA2-IR (insulin resistance), HOMA2-%B (pancreatic beta cell function) and HOMA2-%S (insulin sensitivity) indices, was used to calculate QUICKI and KITT as described previously23,46,47. The HOMA2 model was calculated using fasting glucose and fasting insulin levels measured before sacrifice and before glucose loading for the GTT.
Enzyme-linked immunosorbent assay
To evaluate activation of both mTORC1 and 2 signaling complexes, liver samples were examined using a Mouse/Rat cAMP Parameter Assay Kit (R&D Systems Inc., Minneapolis, MN) for cellular cAMP level and PathScan sandwich ELISA Kits (Cell Signaling Technology, Beverly, MA) for total S6, phosphorylated S6 (Ser235/236), and total Akt and phosphorylated Akt1 (Ser473). To examine Akt1 phosphorylation, 0.5 U of insulin per kg body weight were injected 10 minutes before blood collection. Serum insulin concentrations were measured using Mouse Insulin ELISA kit AKRIN-011T (Shibayagi, Gunma, Japan). To assess intestinal inflammation, the level of faecal LCN-2 was measured using a Mouse Lipocalin-2/NGAL Quantikine ELISA Kit (R&D Systems Inc., Minneapolis, MN) as described by Chassaing et al.24. All ELISAs were performed according to the manufacturers’ protocols.
DNA extraction, bacterial 16S rRNA gene amplification and 454 pyrosequencing
Metagenomic DNA was extracted from faecal samples (0.02 g per sample) using the repeated bead beating plus column method as previously described14. The V1 and V2 hyper-variable regions of the bacterial 16S rRNA gene were amplified from each extracted DNA sample using barcode primers14. Five replicated PCR products per sample were pooled and purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA). The pooled DNA was sequenced using 454 pyrosequencing GS FLX Titanium (Roche 454 Life Sciences, Branford, CT). Sequencing was performed by Macrogen (Seoul, Korea).
Analysis of bacterial 16S rRNA gene sequences and community comparison
Initial processing of sequence data, quality control, phylotype binning, and taxonomic alignment of raw sequencing reads were performed using Quantitative Insight into Microbial Ecology (QIIME) software (package 1. 8. 0)48. The following parameters were used for quality filtering: minimum/maximum length = 200/1000; no ambiguous bases; no primer mismatches; average quality score >25; and homopolymer run <6 nucleotides. The reverse primers were trimmed away from denoised sequences analysed using the Denoiser algorithm49. OTUs with 97% sequence identity were clustered using UCLUST software and the Greengenes core set as a reference sequence database50. A representative sequence was chosen for each phylotype and aligned against the Greengenes core set using PyNAST48. ChimeraSlayer was then used to remove potentially chimeric sequences from the aligned representative sequences51. Taxonomic classification of representative sequences from each OTU was performed using the RDP classifier, with a minimal confidence of 60%. FastTree52 was then used to build a phylogenetic tree based on the aligned sequences. UniFrac-based beta-diversity was visualized after PCoA53. Over- or under-represented bacterial genera within a given category (diet, treatment, or diet-treatment combinations) were determined using the NSC method54. A heat map was made and hierarchical cluster analysis was performed by applying the Manhattan distance method to the processed and normalized data using PermutMatrix software55.
Statistical analysis
Data were expressed as the mean ± SEM. All statistical analyses and Pearson’s correlation coefficients were performed using GraphPad Prism software (version 6.0; GraphPad Software, SD, USA). Pearson’s correlation coefficient heat maps were visualized using Excel and PowerPoint (version 2010; Microsoft Corporation, WA, USA). In experiments comparing multiple groups, differences were analysed by two-way ANOVA followed by Bonferroni’s post-hoc test. GTT and ITT were analysed using a repeated measure two-way ANOVA with both time and group as sources of variation. P values < 0.05 were regarded as significant (*P < 0.05 and **P < 0.005).
Additional Information
How to cite this article: Jung, M.-J. et al. Chronic Repression of mTOR Complex 2 Induces Changes in the Gut Microbiota of Diet-induced Obese Mice. Sci. Rep. 6, 30887; doi: 10.1038/srep30887 (2016).
Supplementary Material
Acknowledgments
This work was supported by a grant from the Mid-Career Researcher Program (2011-0028854) through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (MEST) and by the Strategic Initiative for Microbiomes (as part of the (multi-ministerial) Genome Technology to Business Translation Program) in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Footnotes
Author Contributions M.-J.J. helped perform the experiments, analysed the data, presented the results, and prepared the manuscript. J.L. performed the majority of experiments and prepared the manuscript. M.-S.K. helped analyze the data and revised the manuscript. D.-W.H. helped perform the experiments. N.-R.S. revised the manuscript. J.-H.Y. revised the manuscript. P.S.K. helped perform the experiments. T.W.W. revised the manuscript. J.-W.B. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analyses.
References
- Allison D. B., Fontaine K. R., Manson J. E., Stevens J. & VanItallie T. B. Annual deaths attributable to obesity in the United States. JAMA 282, 1530–1538 (1999). [DOI] [PubMed] [Google Scholar]
- Genton L., Melzer K. & Pichard C. Energy and macronutrient requirements for physical fitness in exercising subjects. Clin Nutr 29, 413–423 (2010). [DOI] [PubMed] [Google Scholar]
- Horgan G. & Stubbs J. Predicting basal metabolic rate in the obese is difficult. Eur J Clin Nutr 57, 335–340 (2003). [DOI] [PubMed] [Google Scholar]
- Spiegelman B. M. & Flier J. S. Obesity and the regulation of energy balance. Cell 104, 531–543 (2001). [DOI] [PubMed] [Google Scholar]
- Badman M. K. & Flier J. S. The gut and energy balance: visceral allies in the obesity wars. Science 307, 1909–1914 (2005). [DOI] [PubMed] [Google Scholar]
- Quigley E. M. Microflora modulation of motility. J Neurogastroenterol Motil 17, 140–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara G. et al. Interactions between commensal bacteria and gut sensorimotor function in health and disease. Am J Gastroenterol 100, 2560–2568 (2005). [DOI] [PubMed] [Google Scholar]
- Tremaroli V. & Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249 (2012). [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Bäckhed F., Fulton L. & Gordon J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101, 15718–15723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1131 (2006). [DOI] [PubMed] [Google Scholar]
- Atarashi K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L. & Kasper D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008). [DOI] [PubMed] [Google Scholar]
- Shin N.-R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2013). [DOI] [PubMed] [Google Scholar]
- Vaishnava S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.-H. et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782 (2008). [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A. & Sabatini D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12, 21–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-J. et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148, 421–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming D. W. et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Manchester J. K., Semenkovich C. F. & Gordon J. I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 104, 979–984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D. R. et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 13, 517–526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A. et al. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metabol 85, 2402–2410 (2000). [DOI] [PubMed] [Google Scholar]
- Levy J. C., Matthews D. R. & Hermans M. P. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care 21, 2191–2192 (1998). [DOI] [PubMed] [Google Scholar]
- Chassaing B. et al. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PloS one 7, e44328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. Fat cell–specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes 59, 1397–1406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. et al. Loss of enteral nutrition in a mouse model results in intestinal epithelial barrier dysfunction. Ann N Y Acad Sci 1258, 71–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A. W., Turnquist H. R. & Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol 9, 324–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-j. et al. Resveratrol pre-treatment reduces early inflammatory responses induced by status epilepticus via mTOR signaling. Brain Res 1492, 122–129 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou Y., Rychahou P., Wang Q., Weiss H. & Evers B. TSC2/mTORC1 signaling controls Paneth and goblet cell differentiation in the intestinal epithelium. Cell Death Dis 6, e1631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F. et al. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PloS one 9, e85254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T. et al. Inhibition of mTOR blocks the anti-inflammatory effects of glucocorticoids in myeloid immune cells. Blood 117, 4273–4283 (2011). [DOI] [PubMed] [Google Scholar]
- Brown J., Wang H., Suttles J., Graves D. T. & Martin M. Mammalian target of rapamycin complex 2 (mTORC2) negatively regulates Toll-like receptor 4-mediated inflammatory response via FoxO1. J Biol Chem 286, 44295–44305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T. et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29, 565–577 (2008). [DOI] [PubMed] [Google Scholar]
- Fan W. et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J 29, 4223–4236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky J. M. & Glass C. K. Macrophages, inflammation, and insulin resistance. Annu rev physiol 72, 219–246 (2010). [DOI] [PubMed] [Google Scholar]
- Caricilli A. M. et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol 9, e1001212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kimura I. et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun 4, 1829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Sun J., Xie Z., Shi Y. & Le G. Propensity to high-fat diet-induced obesity in mice is associated with the indigenous opportunistic bacteria on the interior of Peyer’s patches. J Clin Biochem Nutr 55, 120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca F. A. et al. Replication of obesity and associated signaling pathways through transfer of microbiota from obese prone rat. Diabetes, 63, 1624–1636 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun 4, 2163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). [DOI] [PubMed] [Google Scholar]
- McGarr S. E., Ridlon J. M. & Hylemon P. B. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol 39, 98–109 (2005). [PubMed] [Google Scholar]
- Lam Y. Y. et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PloS one 7, e34233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R. et al. Methane and hydrogen positivity on breath test is associated with greater body mass index and body fat. J Clin Endocrinol Metab 98, E698–E702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L. & Dong X. Hydrogenoanaerobacterium saccharovorans gen. nov., sp. nov., isolated from H2-producing UASB granules. Int J Syst Evol Microbiol 59, 295–299 (2009). [DOI] [PubMed] [Google Scholar]
- Katz A. et al. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab 85, 2402–2410 (2000). [DOI] [PubMed] [Google Scholar]
- Bonora E. et al. Estimates of in vivo insulin action in man: Comparison of insulin tolerance tests with euglycemic and hyperglycemic glucose clamp studies. J Clin Endocrinol Metab 68, 374–378 (1989). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J. & Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7, 668–669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2640–2641 (2010). [DOI] [PubMed] [Google Scholar]
- Haas B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21, 494–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. & Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O. et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA 108 Suppl 1, 4592–4598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraux G. & Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21, 1280–1281 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.