Abstract
In insects, trehalose serves as the main sugar component of haemolymph. Trehalose is also recognized as a mediator of desiccation survival due to its proposed ability to stabilize membranes and proteins. Although the physiological role of trehalose in insects has been documented for decades, genetic evidence to support the importance of trehalose metabolism remains incomplete. We here show on the basis of genetic and biochemical evidence that the trehalose synthesis enzyme Tps1 is solely responsible for the de novo synthesis of trehalose in Drosophila. Conversely, a lack of the gene for the trehalose hydrolyzing enzyme Treh causes an accumulation of trehalose that is lethal during the pupal period, as is observed with Tps1 mutants. Lack of either Tps1 or Treh results in a significant reduction in circulating glucose, suggesting that the maintenance of glucose levels requires a continuous turnover of trehalose. Furthermore, changes in trehalose levels are positively correlated with the haemolymph water volume. In addition, both Tps1 and Treh mutant larvae exhibit a high lethality after desiccation stress. These results demonstrate that the regulation of trehalose metabolism is essential for normal development, body water homeostasis, and desiccation tolerance in Drosophila.
Organisms live in variable environments, and therefore adaptive strategies have evolved to cope with adverse environmental conditions such as starvation, low oxygen levels, and lack of water. Approximately 65 to 70% of our body weight consists of water, with one-third of the total being extracellular fluid. Energy obtained from the metabolic breakdown of nutrients such as sugar is utilized for various growth and maintenance processes. Water is not only the medium for enzymatic and chemical reactions in the body; it also moves nutrients and oxygen through the circulatory system. Therefore, maintenance of systemic water homeostasis and fluid balance is critical in most organisms, including humans, to tolerate changes in osmolarity and prevent desiccation1,2,3.
Trehalose is a non-reducing disaccharide that is present in a broad range of organisms including bacteria, yeast, fungi, plants, and invertebrates4,5. In insects, trehalose serves as the main sugar component of haemolymph. It is synthesized in the fat body, an organ analogous to the mammalian liver, and released into the haemolymph6,7,8. Trehalose has been recognized to be an important osmoprotectant and a mediator of desiccation tolerance because of its stability and inert chemical properties9,10,11,12,13. Many of these roles have been documented in the budding yeast Saccharomyces cerevisiae, where trehalose is proposed to play a protective role by functioning as a chemical chaperone, which prevents protein denaturation and aggregation and influences protein folding through trehalose-protein interactions14,15. In addition, trehalose contributes to the preservation of membrane phospholipids organization through the packing of lipid acyl chains9,10,16,17. In the sleeping chironomid Polypedilum vanderplanki, trehalose accumulates, making up as much as 20% of the dry body mass, and is thought to replace water in tissues to successfully achieve an anhydrobiotic state10,13. Although trehalose accumulation is not the only mechanisms involved in desiccation tolerance, it has been demonstrated to be an important factor for desiccation tolerance based on genetic evidence in diverse multicellular organisms such as the nematode C. elegans16, the mosquito Anopheles gambiae18, and the fruit fly Drosophila melanogaster19. The physiological role of trehalose in insects has been proposed for decades. However, genetic evidence to independently validate the importance of trehalose metabolism and the role of trehalose as a chemical chaperone remains incomplete.
Trehalose is synthesized by the trehalose synthesis enzyme Tps1 and is hydrolyzed by trehalase. Two trehalase genes, a soluble form called Tre-1 and a membrane-bound form called Tre-2 that has a transmembrane domain at the C-terminus, have been identified in many insects, including the silkworm Bombyx mori20,21, the bamboo borer Omphisa fuscidentalis22, the mired bug Apolygus lucorum23, and the whitefly Bemisia tabaci24. Although differences in expression pattern of the two trehalase genes have been reported in several insects, the functional significance of each trehalase being distributed in different locations remains unknown.
We previously reported that larvae lacking trehalose exhibit diet-dependent phenotypes relating to growth and survival in the genetically tractable organism, D. melanogaster25. In this manuscript, we describe the molecular characterization of the genes responsible for trehalose metabolism, Tps1 and Treh. We further determined the importance of each isoform of Treh by generating isoform-specific mutants. These genetic mutants allow us to directly assess the physiological role of trehalose during development. We demonstrate the importance of trehalose metabolism for the maintenance of free glucose levels. In addition, defects in trehalose metabolism affect water homeostasis and desiccation tolerance.
Results
Tps1 is solely responsible for the de novo synthesis of trehalose in Drosophila
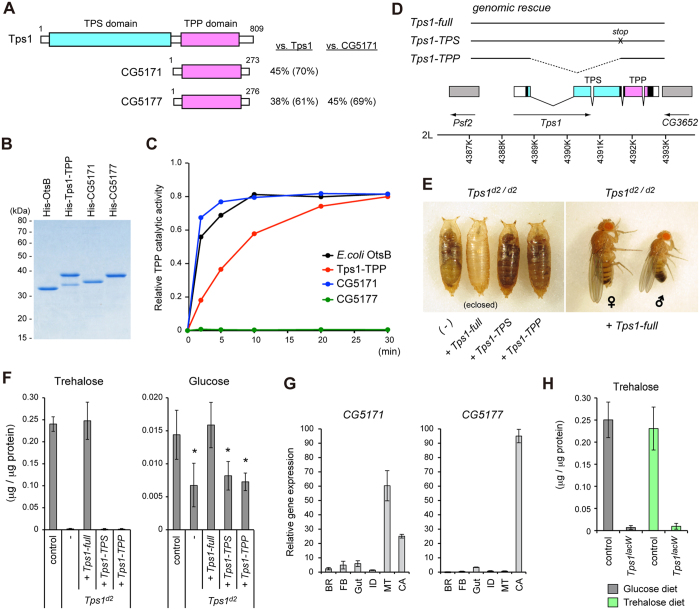
The gene for the trehalose synthesis enzyme, Tps1, is exclusively expressed in the fat body25. We have previously shown that the knockdown of Tps1 in the fat body fully recapitulates the ubiquitous knockdown phenotype25. Tps1 has two functionally distinct catalytic domains4,5. The N-terminal TPS (trehalose-6-phosphate (T6P) synthase) domain catalyzes the production of T6P using glucose-6-phosphate and UDP-glucose. The C-terminal TPP (T6P phosphatase) domain then dephosphorylates T6P to generate trehalose. We failed to detect genes other than Tps1 in the Drosophila genome that possess a TPS domain, whereas there are two uncharacterized genes containing only a TPP domain: CG5171 and CG5177 (Fig. 1A). Similarly, there are multiple genes in C. elegans, plants, and bacteria that contain only a TPP domain in addition to Tps15,26,27,28. It has been shown that the TPP domains of Tps1 in some plant species have lost their catalytic activity29. These observations raise a question about the necessity of the C-terminal TPP domain in the Tps1 gene product in Drosophila.
Figure 1. Tps1 is solely responsible for the de novo synthesis of trehalose in Drosophila.
(A) Domain structure of Tps1 and the TPP-domain containing proteins, CG5171 and CG5177. Numbers refer to amino acids. Identities and similarities of amino acids between proteins are shown on the right. (B) Recombinant His-tagged proteins used in the assay were analyzed by CBB staining. (C) Relative TPP activity was analyzed by the incubation of a recombinant enzyme and the substrate trehalose-6-phosphate. (D) Schematic representation of the Tps1 locus and molecular nature of the genomic rescue constructs. The protein-coding regions and untranslated regions are represented by filled boxes and open boxes, respectively. The transcript regions of the neighboring genes are represented by grey boxes. (E) Both the TPS and TPP domains are required to rescue the Tps1 mutant phenotype from pupal lethality. (F) Trehalose and glucose levels were analyzed in late third instar larvae of the indicated genotypes. (G) The expression patterns of CG5171 and CG5177 were analyzed by qRT-PCR in mid third instar larvae. BR, Central nervous system; FB, Fat body; ID, Imaginal discs; MT, Malpighian tubule; CA, Carcass. Relative gene expression levels in each tissue are shown. Total values are set to 100. (H) The food containing trehalose instead of glucose did not rescue the trehalose levels in the Tps1 mutant wandering larvae. All the values are means and SD (n = 6 [F,H] or n = 3 [G]). Statistical significance is determined by two-tailed Student’s t-test (P < 0.05).
To address this issue, we directly assessed the phosphatase activity of the TPP domain of Drosophila Tps1 in vitro. A bacterially expressed Tps1-TPP was purified, and its enzymatic activity was compared with those of CG5171, CG5177, and an E. coli gene product, OtsB, which possesses the TPP domain (Fig. 1B). We found that recombinant Tps1-TPP and CG5171, but not CG5177, dephosphorylated T6P under our experimental conditions (Fig. 1C). Time-course analyses revealed that Tps1-TPP showed a lower activity than CG5171 and OtsB in vitro. The enzymatic activity of Tps1 may be post-translationally regulated via mechanisms such as phosphorylation and methylation, as has been demonstrated in yeast and bacteria4,30.
If CG5171 can substitute for the function of TPP in Tps1, the TPS domain of Tps1 may be sufficient to rescue the mutant phenotype in Tps1 null mutants. To further confirm the necessity of the TPS and TPP domains of Tps1 in vivo, we generated genomic-rescue constructs with a full-length Tps1 gene, a Tps1 gene with a stop codon before the TPP domain, and a Tps1 gene lacking the TPS domain (Fig. 1D). One-copy of the full-length Tps1 genomic construct completely rescued the lethality of the Tps1 mutants, whereas neither the TPS domain nor the TPP domain of Tps1 rescued the mutant phenotype (Fig. 1E). Consistently, the reductions of the trehalose and glucose levels in the Tps1 mutants were not rescued by the TPS domain nor the TPP domain of Tps1 (Fig. 1F). These results indicate that the TPS and TPP domains in Tps1 are both required for trehalose synthesis in vivo. We observed that CG5171 was mainly expressed in the Malpighian tubules and the components of the carcass, which included body wall muscles, but not in the fat body (Fig. 1G). The difference in the expressing tissues likely explains the in vivo requirement of the TPP domain of Tps1. Furthermore, feeding trehalose to the flies failed to restore trehalose levels or rescue pupal lethality in the Tps1 mutants (Fig. 1H), suggesting that trehalose is not directly supplied from the food through the gut in flies. Alternatively, the absorption rate of trehalose through the gut may be low, and therefore dietary trehalose is not enough to restore the normal levels of circulating trehalose. Taken together, these results indicate that Tps1 is solely responsible for the de novo synthesis of trehalose in Drosophila.
Treh mutations are lethal in the pupal period
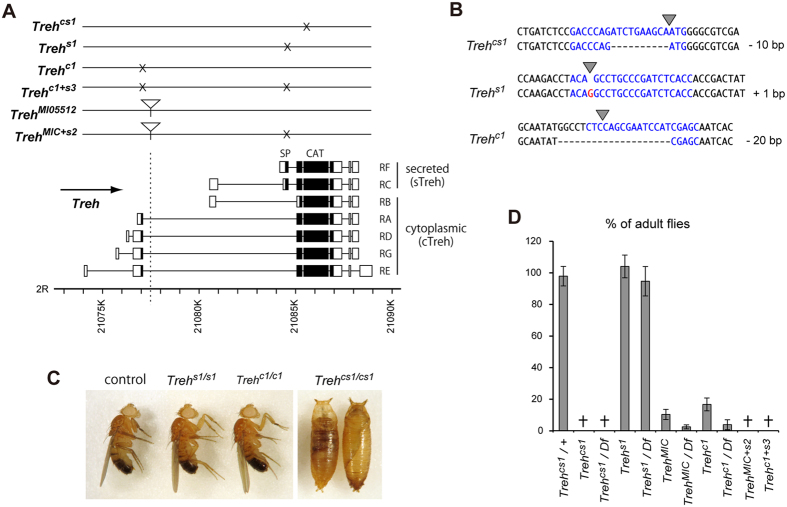
We next examined the functional significance of the trehalose hydrolyzing enzyme. In the Drosophila genome, there are two genes that possess a catalytic domain for trehalose hydrolysis: Treh and CG6262. Because CG6262 is dominantly expressed in adult testis (Flybase), Treh is thought to be the main enzyme that catalyzes trehalose during development. To determine whether Treh plays an essential role in trehalose metabolism and development, we generated Treh mutants using CRISPR/Cas9 technology. We isolated several Treh mutant strains with frameshift deletions/insertions at the section of the catalytic domain nearest to the N-terminal using three distinct single-guide RNAs (sgRNAs). Homozygous mutants of the isolated frameshift mutants generated from each sgRNA construct survived the larval period but displayed complete lethality during pupal period. Hereafter, we used one of the deletion alleles of Treh, named Trehcs1 (Fig. 2A). Trehcs1 lacks 10 bp, resulting in an unusual transcript with a premature stop codon and a lack of the entire catalytic domain (Fig. 2B). Transheterozygotes of Trehcs1 with a deficiency allele lacking the Treh locus survived the larval period and died at the pupal stage (Fig. 2C,D), suggesting that the Trehcs1 allele is functionally null. Taken together, these results indicate that Treh is essential for pupal development, and the Treh mutants display lethality at a stage identical to that of the Tps1 mutants that we have previously reported on ref. 25.
Figure 2. Generation and characterization of the Treh mutants in Drosophila.
(A) Schematic representation of the Treh locus and the molecular nature of the mutants. Protein-coding regions and untranslated regions are represented by filled boxes and open boxes, respectively. The P-element insertion sites are marked with an inverted triangle. Each transcript is based on information from FlyBase. (B) Sequences of sgRNA target sites and the deletion/insertion of Treh mutants. The 20 bp target sequence corresponding to each target site is indicated in blue and the cleavage sites of Cas9 are shown as triangles. (C) Trehcs1 homozygous mutants were lethal at the pupal period. Trehs1 mutant adult and Trehc1 escaping adult are shown. (D) Lethality of the Treh mutants. Percentages of the indicated mutant adults were determined based on the ratio of heterozygous mutants in each vial. All the values are means and SD (n > 6 [D]).
Cytoplasmic Treh rather than secreted Treh is critical for normal development
Treh in Drosophila is thought to be produced in two different forms via variations in alternative splicing (Flybase): a putative secreted form (sTreh), with a signal peptide at the N-terminus, and a cytoplasmic form (cTreh), without a signal peptide (Fig. 2A). Both sTreh and cTreh share the same catalytic domain for trehalose hydrolysis. Both sTreh and cTreh transcripts are initiated at several different transcriptional start sites, and therefore various transcripts (5 corresponding to cTreh and 2 corresponding to sTreh) are described in the Flybase. To examine the functional importance of cTreh and sTreh, we next examined the mutant phenotype of an available Minos-transposon in the Treh locus (MI05112, hereafter named TrehMIC). The transposon is inserted in the first intron of the coding region of cTreh (Fig. 2A). The TrehMIC mutants displayed lethality in the pharate adult stage. However, adult escapers were observed under regular rearing conditions (Fig. 2D). The number of adult escapers was significantly reduced in TrehMIC transheterozygotes with the deficiency allele, suggesting that TrehMIC is a hypomorphic allele.
Because TrehMIC likely affects the major cTreh transcripts, including RA, RD, RG, and RE, but not RB, and the sTreh transcripts, TrehMIC is likely to be a strong allele of cTreh. It is possible that cTreh functions redundantly with sTreh. To address this possibility, we used the CRISPR/Cas9 technique to generate sTreh-specific mutants by introducing small deletions at the region within the signal peptide sequence (Fig. 2A). All isolated mutants with frameshift deletions/insertions displayed homozygous viability, were fertile, and had no visible lethality during development. Hereafter, we used one of the deletion allele of sTreh, named Trehs1, which has a frameshift insertion of 1 bp (Fig. 2B,C). Trehs1 transheterozygotes with the deficiency allele also exhibited no lethality (Fig. 2D), indicating that sTreh is dispensable for successful development. Then, we introduced a similar frameshift deletion/insertion at the signal peptide sequence in the TrehMIC allele using the identical sgRNA construct (Fig. 2A). Homozygous TrehMIC mutants having a small deletion within the sTreh transcripts (hereafter named TrehMIC+s2) displayed complete lethality during pupal period (Fig. 2D). To further validate these results, we also introduced a small deletion at the first exon of the major cTreh transcripts (hereafter named Trehc1). Homozygous Trehc1 mutants displayed pupal lethality, with some adult escapers and with results similar to those observed with the TrehMIC allele. The introduction of a small deletion at the signal peptide sequence on the Trehc1 allele (hereafter named Trehc1+s3) exhibited complete pupal lethality (Fig. 2A,D). These results indicate that cTreh and sTreh function redundantly, although the function of cTreh, rather than sTreh, is dominant within the Treh gene locus.
Treh is required for the maintenance of glucose levels and starvation tolerance
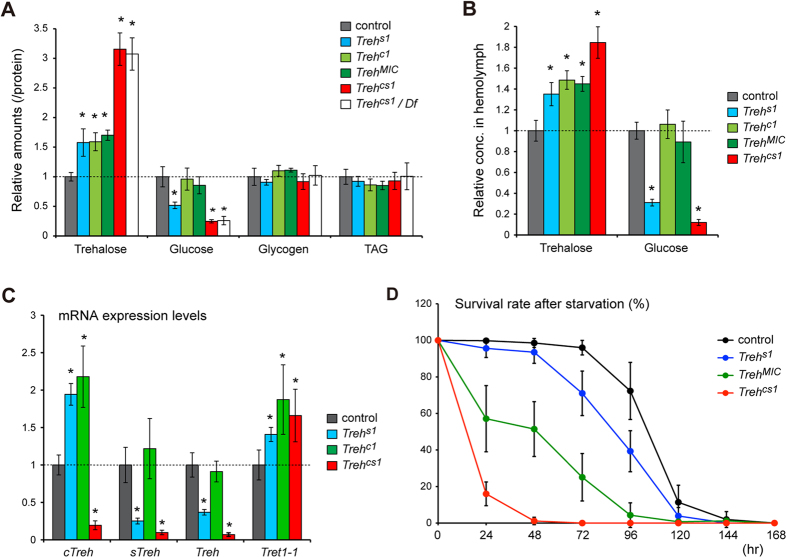
To further characterize the Treh mutants, we next analyzed trehalose levels. Trehcs1 mutant larvae exhibited significant increase in trehalose at the late third instar larval stage (Fig. 3A). Furthermore, trehalose concentrations in the circulating haemolymph of the Trehcs1 mutants were also up-regulated, although the degree of increase was less drastic when compared with the increases in the whole larvae (Fig. 3B). Consistent with the observed lethality of the mutations, TrehMIC and Trehc1 mutant larvae displayed a partial increase in trehalose levels when compared with the increases in the Trehcs1 mutants. We next examined glycogen and triacylglycerol (TAG) levels to understand the metabolic consequences of defective trehalose catabolism in flies; however, the levels were not significantly altered in the Treh mutants (Fig. 3A). Conversely, glucose levels in the Trehcs1 mutants were significantly lower. Trehs1 mutants had increased trehalose levels, similar to cTreh mutants, whereas glucose levels were reduced only in sTreh mutants and not in cTreh mutants. Circulating glucose in the haemolymph was also decreased in Trehcs1 and Trehs1 mutants (Fig. 3B). The reduction of glucose levels in Treh mutants is reminiscent of the Tps1 mutant phenotype (Fig. 1F)25, suggesting that the maintenance of glucose levels requires a continuous turnover of trehalose, including its synthesis and breakdown. Specifically, sTreh rather than cTreh plays an important role in regulating the levels of free glucose. Nevertheless, given that sTreh mutants displayed no lethality, severe dysregulation of glucose levels in the haemolymph does not appear to be directly related to pupal lethality in Trehcs1 mutants.
Figure 3. Treh mutations affect trehalose and glucose levels in Drosophila.
(A) Treh mutations increase trehalose level in late third instar larvae. Each value was normalized by protein levels and further normalized according to the level in the control larvae. (B) Trehalose and glucose concentrations in the haemolymph were analyzed in late third instar larvae of the indicated genotypes. (C) Treh transcript levels were analyzed by qRT-PCR at the mid third instar stage of the indicated genotypes. (D) Treh mutants exhibited lethality under starvation conditions. Early third instar larvae were transferred to a vial containing 0.8% agar in PBS, and the number of surviving larvae were counted at the indicated time points. Statistical significance is determined by two-tailed Student’s t-test (P < 0.01). All the values are means and SD (n > 4 [A,B,D] or n = 3 [C]).
Our results demonstrate that Trehcs1 mutants exhibit a significant increase in trehalose levels in the whole larvae, which is most likely not additive between cTreh and sTreh mutants. These observations raise a possibility of a compensatory mechanism between cTreh and sTreh. We observed that Treh transcript levels were significantly reduced in Trehcs1 mutants based on a qRT-PCR analysis with primer sets that amplified the regions outside the small deletions (Fig. 3C). More specifically, sTreh transcripts were significantly down-regulated in Trehs1 mutants, suggesting positive feedback regulation in response to its own enzyme activity. On the other hand, the expression of cTreh was up-regulated in both Trehs1 and Trehc1 mutants, suggesting negative feedback regulation at the level of transcription and/or depending on the stability of mRNA to compensate for a defect in trehalose hydrolysis. Expression of the trehalose transporter Tret1-1 was also up-regulated in Treh mutants, suggesting that feedback regulation co-operates not only in Treh but also at the level of trehalose transport. The compensatory up-regulation of cTreh and Tret1-1 may in part account for the viability of the Trehs1 mutants.
To understand the role of Treh under conditions of dietary stress, we analyzed the starvation tolerance of Treh mutants. The control larvae survived for 4 days on a water-only diet, whereas almost all Trehcs1 mutant larvae died 2 days after the initiation of starvation conditions (Fig. 3D). TrehMIC mutants exhibited a weaker but still apparent lethality under starvation conditions, whereas Trehs1 mutants showed only a slight reduction in survival rate when compared with the control larvae. Furthermore, Trehcs1 mutants failed to grow to pupae under feeding conditions that lacked dietary glucose (data not shown), which is similar to the results for Tps1 mutants25. These results suggest that Treh mutants are sensitive to the levels of dietary glucose for their survival and larval growth. Because the reliance on dietary glucose for survival and growth in Treh mutant larvae is consistent with the results observed in Tps1 mutants25, the observed phenotypes related to starvation and low-dietary glucose levels are due to defects in trehalose metabolism rather than to an over-accumulation of trehalose.
Treh is responsible for trehalose catabolism in vivo and in vitro
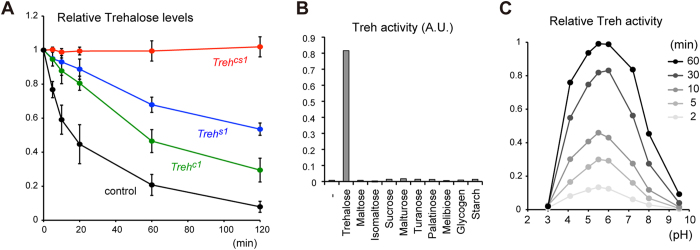
We next measured the trehalase activity in larval homogenates to directly test the role of Treh in trehalose hydrolysis. Wild-type homogenates exhibited an enzyme activity with the hydrolysis of trehalose occurring in a time-dependent manner (Fig. 4A). On the other hand, Trehcs1 mutant larvae had no detectable trehalose hydrolysis activity, indicating that Treh is solely responsible for the hydrolysis of trehalose at this developmental stage. Consistent with the functional redundancy between sTreh and cTreh, sTreh and cTreh mutants showed a partial trehalose hydrolysis activity under these conditions. To further characterize the Treh gene product, we produced recombinant His-tagged cTreh proteins in E. coli. The recombinant cTreh exhibited a specific breakdown of trehalose to produce glucose in vitro (Fig. 4B). The activity of His-cTreh was observed in vitro between pH 4.0 and pH 8.0, with a maximum activity at pH 5.5 ± 0.5 (Fig. 4C). Because the catalytic domain of Treh is shared between sTreh and cTreh, these results indicate that Treh is responsible for trehalose catabolism in vivo and in vitro.
Figure 4. Treh is responsible for trehalose catabolism in vivo and in vitro.
(A) Trehalase activity was measured in larval homogenates. Relative levels of trehalose are shown for each mutant. (B) Recombinant His-tagged Drosophila Treh specifically hydrolyzed trehalose in vitro. (C) Trehalase activity was observed in a broad range of pH conditions. Relative Treh activity as determined by the hydrolysis of trehalose is shown over time. All the values are means and SD (n = 3 [A]).
Changes in trehalose levels influence the water levels in the haemolymph
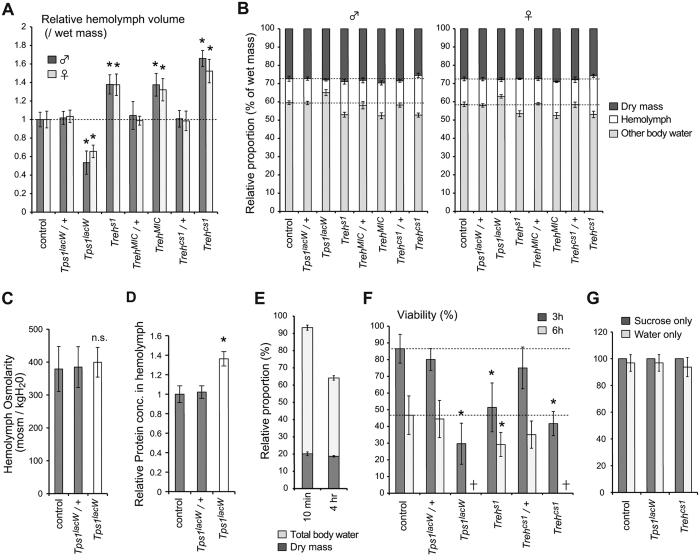
During the course of experiments, we noticed that the volume of haemolymph was considerably higher in Treh mutants than in the control larvae. In contrast, Tps1 mutants appeared to retain less haemolymph based on the difficulty in collecting larval haemolymph from these mutants. These observations regarding haemolymph volume were confirmed by quantifying the weight of the haemolymph (Fig. 5A). The increase in haemolymph water volume explains why the changes in trehalose concentrations in the haemolymph of Trehcs1 mutants were less drastic than the increase in trehalose detected in the whole larvae. Body water is the primary determinant of the osmolality of the extracellular fluid, and trehalose is known to be an organic osmoprotectant in several organisms5,9,10,11,12. Therefore, alterations of trehalose levels likely affect osmolality, thereby changing the water content of the haemolymph. To further examine the mutant phenotype, we next examined whether defects in trehalose metabolism altered water content at the whole-organism level. Tps1 mutants did not exhibit a significant decrease in the percentage of water in whole larvae (Fig. 5B). These results suggest that Tps1 mutants retain more intracellular fluid at the expense of the reduction of water in the haemolymph. Because of the size difference between males and females at the wandering stage, we analyzed the haemolymph volume and dry mass separately in males and females. We found that the relative proportion of dry mass and haemolymph volume per wet mass was relatively comparable between sexes. Our results suggest that changes in the trehalose levels affect body water homeostasis equally in males and females. We also analyzed the trehalose and glucose levels in males and females and found no significant differences between sexes (data not shown).
Figure 5. Trehalose metabolism affects haemolymph water volume and desiccation tolerance.
(A) Tps1 and Treh mutations affect the volume of haemolymph at the wandering stage. The weights of haemolymph in the indicated genotypes were measured and normalized according to wet weight. Heterozygous mutants are included to validate the accuracy of the measurements. (B) Relative proportions of haemolymph volume, dry weight, and the predicted water content in tissues are shown. (C) Tps1 mutants exhibited no change in haemolymph osmolarity. (D) Protein concentrations in the haemolymph were analyzed in late third instar larvae of the indicated genotypes. (E) Desiccation reduced water contents in early third instar larvae. Relative proportions of water content and dry weight were normalized to the values of wet weight before desiccation. (F) Survival rates after desiccation and rehydration. Early third instar larvae were used to test 3 hr or 6 hr of desiccation followed by rehydration. (G) Survival rates under sucrose or water-only dietary conditions. Early third instar larvae were kept under each condition for 12 hours. Statistical significance is determined by two-tailed Student’s t-test (P < 0.01). All the values are means and SD (n > 6 [A–D] or n > 3 [E–G]).
Importantly, we failed to detect a decrease in osmotic pressure in the haemolymph of Tps1 mutants (Fig. 5C), even though these mutants retained less haemolymph than the controls. Tps1 mutants showed a higher concentration of total protein in their haemolymph (Fig. 5D). These results support the possibility that the reduction of haemolymph water volume in Tps1 mutants is the result of alterations in osmotic pressure. Conversely, Treh mutants showed a significant decrease in tissue water content instead of an increase in haemolymph volume (Fig. 5B). Furthermore, Trehcs1 mutants displayed a slight reduction in solute levels relative to their water content, suggesting a relative increase in total body water. Taken together, these results indicate that trehalose levels are positively correlated with extracellular fluid volume due to alterations in osmotic pressure.
Importance of trehalose metabolism in desiccation tolerance
A strong relationship has been observed between haemolymph volume, carbohydrate levels, and survival during desiccation in a D. melanogaster population selected for desiccation resistance31. Because the genetic manipulation of trehalose levels affects water levels in haemolymph, we next examined the sensitivity of Tps1 and Treh mutants to desiccation stress. During the desiccation process, larvae gradually stop moving and reduce their body size. An approximately 35% reduction in water contents in whole larvae was observed 4 hours after desiccation under our experimental conditions (Fig. 5E). In contrast, dry weight was reduced by 7% during that period. More than 80% of the wild type early 3rd instar larvae recovered with rehydration after 3 hours of desiccation (Fig. 5F). Approximately 40% recovered after 5 hours, and 15% recovered after 7 hours of desiccation. Under these conditions, Tps1 mutants exhibited a higher lethality than the heterozygous mutants and wild-type larvae. Similarly, Trehcs1 mutants exhibited a higher lethality rate, similar to that in Tps1 mutants, after rehydration. We showed that both Tps1 and Treh mutants are sensitive under starvation conditions (Fig. 3D)25. To confirm whether the observed lethality is due to the desiccation stress rather than starvation stress, we analyzed the starvation phenotype for a short period. More than 90% of mutant larvae survived under water-only or sucrose-only dietary conditions for 12 hours (Fig. 5G), indicating the specific phenotype relates to desiccation but not starvation under these conditions.
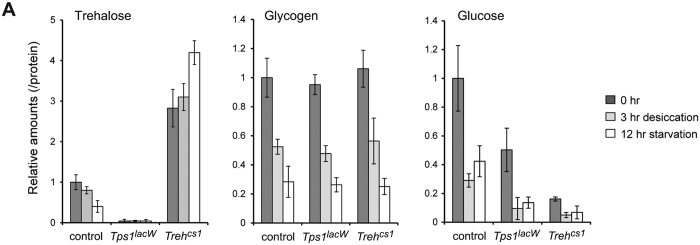
To further understand the importance of trehalose metabolism under stress conditions, we analyzed the sugar levels in Tps1 and Treh mutants following desiccation and starvation stress. We found that Treh mutants further increased the amount of trehalose after 12 hours of starvation, but not after 3 hours of desiccation (Fig. 6), indicating the active production of trehalose by Tps1 following starvation stress. Glycogen and glucose were significantly reduced after 3 hours of desiccation, whereas trehalose was slightly reduced. In addition, free glucose was more reduced after 3 hours of desiccation than it was after 12 hours of starvation. Tps1 and Treh mutants retained less glucose than did the control. Both desiccation and starvation further reduced the glucose levels in Tps1 and Treh mutants. Conversely, the changes in the glycogen levels in Tps1 and Treh mutants were almost comparable to those observed in the control. These results raise the possibility that Tps1 and Treh mutants are sensitive following desiccation stress due to a reduction in the free glucose level in addition to the defects in body water homeostasis. Taken together, these results suggest that the physiological role of the trehalose pathway is closely related to metabolic regulation and that desiccation tolerance is not attributable simply to increased concentrations of trehalose.
Figure 6. Changes in sugar levels following desiccation and starvation stress.
(A) The amounts of trehalose, glycogen, and glucose were analyzed in early third-instar larvae following desiccation and starvation stress. Each value was normalized by protein levels and further normalized according to the level in the control larvae. All the values are means and SD (n > 3).
Discussion
Trehalose is synthesized in a two-step enzymatic reaction. In this study, we showed that both the TPS and the TPP domain in Tps1 are required for the de novo synthesis of trehalose in Drosophila. Although the function of the TPP domain proteins CG5171 and CG5177 remains unknown, the TPP domain of Tps1 is functional and solely involved in trehalose synthesis in the fat body. Interestingly, T6P is not just a precursor of trehalose in the biosynthetic pathway, it also controls the flux of glycolysis as a signaling molecule in yeast and plants32,33. In addition, T6P acts as an endogenous inhibitor of SNF1-related protein kinase in plants and controls metabolism and growth in response to starvation34. Glucose is directly or indirectly sensed in several ways to control cellular energy metabolism35,36,37. Likewise, it is possible that a metabolite such as T6P and/or trehalose functions as a signaling molecule in addition to serving as an energy source. T6P can be detected in adult haemolymph at a relatively low level in Drosophila38. It will be interesting to investigate the function of T6P in response to environmental changes in Drosophila.
We found that cTreh, and not sTreh, is critical for normal development, although to some extent, sTreh and cTreh function redundantly. The trehalose transporters in Drosophila, Tret1-1 and Tret1-2, are expressed in both the fat body and in several peripheral tissues25,39. Therefore, it is likely that cTreh hydrolyzes trehalose within cells. In contrast, sTreh plays an important role for the maintenance of glucose levels in the haemolymph and in body as a whole. However, defects in cTreh have little effect on circulating glucose levels. These observations suggest that the hydrolysis of trehalose in circulating haemolymph directly influences the levels of free glucose. Furthermore, the co-existence of the enzyme and its substrate in the haemolymph has been suggested as a possible regulatory mechanism of trehalase, along with inhibitory proteins22,40,41. Therefore, it is thought that sTreh activity is post-translationally regulated by unknown mechanisms. Our results also suggest that the expression of sTreh is positively regulated by its own activity, while cTreh expression is regulated in a compensatory manner. Nevertheless, the reduction of free glucose levels in Treh mutants does not directly correlate with the observed lethality of the mutation. It has been reported that soluble and membrane-bound trehalase exhibit dynamic changes in expression patterns during metamorphosis20,22,42. Consistently with these findings, the amount of trehalose is more rapidly decreased than that of glycogen and TAG during metamorphosis25. Further analysis will be required to understand a precise role of trehalose during the pupal period.
Depletion of body water leads to a deleterious effect on organisms. Our results demonstrate that the haemolymph sugar trehalose significantly influences the volume of haemolymph and thereby is important for regulating the intracellular fluid. Although trehalose is thought to play a protective role under desiccating conditions, our results suggest that trehalose metabolism rather than or in addition to the presence of trehalose itself is critical for desiccation tolerance in Drosophila larvae. Consistent with this hypothesis, the importance of trehalose metabolism for desiccation tolerance has been documented in yeasts43. It is also important to consider direct versus indirect effects of trehalose on biological systems44.
The excretory system, composed of the kidneys in mammals and of the Malpighian tubules and the hindgut in insects, functions to maintain systemic water homeostasis45,46. Ion and water balance in insects is regulated by the balance between excretion by the Malpighian tubules and absorption by the hindgut and rectum47. The regulation of cellular ion and water homeostasis in the Malpighian tubules has been proposed to be a key physiological mechanism underlying stress tolerance, such as resistance to desiccation47,48,49. In the brown planthopper, Malpighian tubules function in the reabsorption of trehalose by a proton-dependent trehalose transporter50. The Malpighian tubules in several insect species exhibit high expression levels of trehalase23,42. Consistent with this, we previously reported that cTreh is highly expressed in the Malpighian tubules during the larval period25. However, this expression was almost completely down-regulated at the white pupal stage (data not shown). In addition to the reabsorption of trehalose, the high level expression of cTreh in the Malpighian tubules suggests the importance of trehalose as an energy source for maintaining organ function in the tubules, and it may indirectly contribute to water homeostasis and osmoregulation. Of note, mammals lack the gene Tps1 but retain trehalase. Trehalase is highly expressed in the kidney brush border membranes, although the function and physiological relevance of this expression are still not clear5. In this sense, it will be interesting to investigate the local requirements of Treh in the Malpighian tubules in relation to body water homeostasis.
Materials and Methods
Drosophila strains
Drosophila melanogaster flies were reared on a standard agar-yeast-cornmeal medium at 25 °C. Details of the composition of the food have been described previously25. For trehalose feeding experiments, 10 g glucose in the food was substituted for 10 g trehalose (Hayashibara Co.) per 100 ml. All experiments were conducted under non-crowded conditions. No yeast paste was added to the fly tubes for any of the experiments. The strain w1118 was used as a control. Tps1lacW and Tps1d2 have been described previously25. Mi{MIC}TrehMI05512 and Df(2R)Exel6072 (a deficiency strain with a deleted genomic region including the Treh locus) were obtained from the Bloomington Drosophila Stock Center. y2 cho2 v1; attP40{nos-Cas9}/CyO and y2 cho2 v1, P{nos-Cas9, y+, v+}/FM7c, KrGal4, UAS-GFP were obtained from the National Institute of Genetics Drosophila Stock Center. Mi{MIC}TrehMI05512 and Df(2R)Exel6072 were back-crossed three times with the y− w− and w− strain, respectively, and were used for the experiments. The back-crossed Mi{MIC}TrehMI05512 was renamed as TrehMIC.
Generation of the Treh mutants
Generation of the Treh allele was carried out using the CRISPR/Cas9 system with the pBFv-U6.2 vector51. Sense and antisense oligonucleotides corresponding to sgRNA target sequences were annealed and inserted into BbsI-digested pBFv-U6.2 vectors. Treh sgRNA vectors were injected into embryos carrying attP2 and nos-phiC31 and the transgenic strains were generated (BestGene, Inc). The nos-Cas9-based gene targeting was carried out as previously described51. Independent isogenized strains for each sgRNA construct were established. Indel mutations were analyzed via genome DNA extraction and PCR amplification of the DNA fragment including the target site, followed by sequence analysis. We isolated several frameshift mutations for Treh by using three different sgRNA that target the catalytic domain of Treh. All of the isolated Treh mutants are pupal-lethal. Likewise, we isolated several frameshift mutations for sTreh by using three different sgRNAs that target the region within the signal peptide. All of the isolated sTreh mutants are viable and fertile. We isolated three frameshift mutations for cTreh by using one sgRNA, and all of them are pupa-lethal with some escapers. We chose 1 strain for each target site for further analyses and renamed them Trehcs1, Trehs1, and Trehc1. TrehMIC and Trehc1 mutants were further crossed with flies carrying sTreh sgRNA and nos-Cas9 to introduce indel mutations in sTreh.
qRT-PCR analysis
qRT-PCR analyses was done as described previously52. The primers used to detect sTreh, Tret1-1, and rp49 levels are described previously25. The primers used are as follows: Treh (common) sense primer: 5′-TGGGCACCGATGCAGTACATCCTG-3′, Treh (common) antisense primer: 5′-CCGAACTCATCGGCGTTGTACTTC-3′, cTreh sense primer: 5′-ATACGGCAGTGATCAAATCGAGTG-3′, cTreh antisense primer: 5′-TGCGACAAAGACTGTTGTTTCCTG-3′, CG5171 sense primer: 5′-CTTCGGAGATCTGCACAAAGTTCG-3′, CG5171 antisense primer: 5′-AAGCACTCTCATGGCATCTTCATCG-3′, CG5177 sense primer: 5′-CAAGTTGAAGGCCAAGCTGATTGC-3′, CG5177 antisense primer: 5′-CATAGACGATCTTCAGGTTCTTGG-3′.
Plasmid construction
The cDNAs encoding Tps1, CG5171, CG5177, and Treh were cloned by RT-PCR using sequenced strains obtained from the Bloomington Stock Center. The DNA fragments of the Tps1 genomic region were cloned by genomic PCR using the same strains. The cDNA encoding OtsB was cloned by genomic PCR using the E. coli strain DH5a. All PCR fragments were validated by DNA sequencing. Tps1 genomic fragments were subcloned into pCaSpeR4 vectors. Transformants were obtained using a standard injection method (BestGene, Inc). We analyzed 7 independent transformants inserted into the third chromosome for each rescue construct and obtained the same results. For bacterial expression, pET28a vectors containing an N-terminal His-tag (Novagen) were used.
Protein production and in vitro assays
His-tagged proteins were produced in BL21(DE3) cells, purified on Ni-NTA agarose (QIAGEN), and dialyzed in PBS. The reaction for TPP was carried out in a 30 μl assay mixture containing 20 mM Na2HPO4 (pH 7.4), 2 mM NaH2PO4, 1.8 mM KH2PO4, 132 mM NaCl, 2.7 mM KCl, 5 mM MgCl2, 0.2 mg/ml BSA, 0.5 mM T6P (SIGMA), and 0.02 nmol of recombinant enzyme. Reactions were started with the enzyme at 30 °C and stopped after appropriate times by placing the tubes at 100 °C for 5 min. After cooling to room temperature, the amounts of trehalose were determined using a glucose assay kit (SIGMA) after the treatment with trehalase (SIGMA). Net activities were calculated by subtracting the values without treatment with trehalase.
The reaction for Treh was carried out in a 15 μl assay mixture containing 5 μg sugar and 0.12 pmol of His-cTreh in PBS. After an incubation at 30 °C for 2 hours, the amounts of glucose were determined using a glucose assay kit (SIGMA). For the measurement of Treh activity, the reaction was carried out in a 20 μl assay mixture containing 5 μg trehalose and 0.01 pmol His-cTreh under various buffer conditions as follows; pH 3.0 (20 mM ammonium formate buffer), pH 4.1, pH 5.0, and pH 5.5 (20 mM sodium acetate buffer), pH 6.0 and pH 7.2 (20 mM phosphate buffer), pH 8.0 and pH 9.5 (20 mM ammonium bicarbonate buffer). After the incubation at 30 °C for the indicated period, the reaction was stopped by heat inactivation at 90 °C for 5 min. The amounts of glucose were determined using a glucose assay kit (SIGMA). Because of the excess amount of the glucose assay solution relative to the volume of reaction mixture, the detection of glucose was not affected by the different pH conditions (data not shown).
Measurement of protein, TAG, and sugar levels
Measurement of protein, TAG, trehalose, glycogen, and glucose was performed as described previously25. Haemolymph sample preparation was also performed as previously25,53. Osmolality was measured using a Micro-Osmometer (Vogel).
Starvation assay
Starvation assay was performed as described previously53,54.
Trehalase activity assay
Two late third instar larvae were rinsed in PBS and homogenized on ice in 100 μl of 10 mM ammonium acetate buffer (pH 5.0) containing 0.1% TritonX-100, 2.5 mM EDTA, and Complete Protease inhibitor (Roche). Five μl of the homogenates was mixed on ice with 10 μl of substrate solution (200 ng/μl trehalose and 20 ng/μl mannitol-1-13C (SIGMA) as an internal control in 10 mM ammonium acetate, pH 5.0). The reactions were started at 30 °C and stopped after appropriate times by placing the tubes at 90 °C for 5 min. After cooling to room temperature, assay reactions were mixed with 85 μl of acetonitrile, cleared by centrifugation, and 20 μl of the supernatant was diluted with 20 μl H2O. The amounts of trehalose were quantified by LC-MS/MS.
Quantification of trehalose and glucose by LC-MS/MS
Chromatographic separation was performed on an ACQUITY BEH Amide column (100 mm × 2.1 mm, 1.7 mm particles, Waters) in combination with a VanGuard precolumn (5 mm × 2.1 mm, 1.7 mm particles) using an Acquity UPLC H-Class System (Waters). Elution was performed at 30 °C under isocratic conditions (0.3 mL/min, 70% acetonitrile and 30% 10 mM ammonium bicarbonate, pH 10.0). The mass spectrometric analysis was performed using a Xevo TQD triple quadrupole mass spectrometer (Waters) coupled with an electro-spray ionization source in the negative ion mode. The MRM transitions of m/z 341.2 >119 and m/z 182.1 >88.9 were used to quantify trehalose and mannitol-13C, respectively. Analytical conditions were optimized using standards solution. Sample concentrations were calculated from the standard curve obtained from serial dilution of each standard. The amounts of trehalose were normalized to the levels of mannitol-1-13C and further normalized to the levels at time 0 to determine the relative hydrolysis rate in each mutant. TPP activity in trehalose synthesis is Mg2+ dependent, whereas the Treh activity is not affected by the addition of EDTA (data not shown). Therefore, we assumed that there was no de novo production of trehalose during the incubation period. For the quantification of glucose in the haemolymph, the MRM transition of m/z 179.1 >89.0 was used to detect glucose under conditions identical to those described above.
Measurement of haemolymph volume and desiccation tolerance
Developmental staging and measurement of wet weight were performed as previously described52,53. The procedures for the measurement of haemolymph volume and the desiccation assay were slightly modified based on previous studies31,55. For the estimation of haemolymph volume, late third instar larvae were collected, rinsed in PBS, and dried on a Kimwipe. After measuring the wet weight of 8–10 pooled larvae, their cuticle were carefully torn to release the haemolymph on a Parafilm membrane. The released haemolymph from individual larvae was immediately absorbed with a small piece of Kimwipe. The weight of haemolymph was determined by subtracting the weight of the Kimwipe in 1.5 ml tube before and after haemolymph absorption. For measuring dry weight, the wet weight of 8–10 pooled larvae was first measured in 1.5 ml tubes. After being heated at 100 °C for 2 hours, the dry weight of the pooled larvae was measured. Total body water was estimated based on the reduction in mass after dehydration. The proportions of dry weight, total body water, and haemolymph volume were calculated by normalizing them to the values for wet weight.
For the desiccation experiments, early third instar larvae were collected. They were rinsed in PBS and dried carefully on a Kimwipe. The individual larvae were placed in 48 well plates. The plates were covered by a Kimwipe and kept under 10–15% relative humidity in a desiccation chamber containing silica gel for the indicated times. Because of the difficulty in judging the survival rate after desiccation, larvae were rehydrated with a 5% sucrose solution in PBS for 12 hours. The survival rate of larvae was judged based on the movement of the mouth hook under a stereomicroscope.
Additional Information
How to cite this article: Yoshida, M. et al. Molecular characterization of Tps1 and Treh genes in Drosophila and their role in body water homeostasis. Sci. Rep. 6, 30582; doi: 10.1038/srep30582 (2016).
Acknowledgments
We thank the Bloomington Drosophila Stock Center and the National Institute of Genetics Drosophila Stock Center for fly stocks; T. Yamada, K. Higuchi, and N. Nishimura for technical help; members of Fly labs in RIKEN CDB for their valuable support and discussion; K. Banzai and K. Hironaka for comments on the manuscript. This work was supported in part by Scientific Research Grants from MEXT (to T.N.).
Footnotes
Author Contributions M.Y., H.M. and T.N. designed the experiments. M.Y., H.M., H.K. and T.N. performed the experiments and analyzed the data. T.N. wrote the manuscript. All authors reviewed the manuscript.
References
- Jéquier E. & Constant F. Water as an essential nutrient: the physiological basis of hydration. Eur J Clin Nutr 64, 115–123 (2010). [DOI] [PubMed] [Google Scholar]
- Adler S. M. & Verbalis J. G. Disorders of body water homeostasis in critical illness. Endocrinol Metab Clin North Am 35, 873–894 (2006). [DOI] [PubMed] [Google Scholar]
- Stanhewicz A. E. & Kenney W. L. Determinants of water and sodium intake and output. Nutr Rev 73, 73–82 (2015). [DOI] [PubMed] [Google Scholar]
- Becker A., Schlöder P., Steele J. E. & Wegener G. The regulation of trehalose metabolism in insects. Experientia 52, 433–439 (1996). [DOI] [PubMed] [Google Scholar]
- Elbein A. D., Pan Y. T., Pastuszak I. & Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–17RR (2003). [DOI] [PubMed] [Google Scholar]
- Wyatt G. R. & Kalf G. F. The chemistry of insect haemolymph. Trehalose and other carbohydrate. J Gen Physiol 40, 833–846 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candy D. J. & Kilby B. A. Site and mode of trehalose biosynthesis in the locust. Nature 183, 1594–1595 (1959). [DOI] [PubMed] [Google Scholar]
- Murphy T. A. & Wyatt G. R. The enzymes of glycogen and trehalose synthesis in silk moth fat body. J Biol Chem 240, 1500–1508 (1965). [PubMed] [Google Scholar]
- Crowe J. H., Carpenter J. F. & Crowe L. M. The role of vitrification in anhydrobiosis. Annu Rev Physiol 60, 73–103 (1998). [DOI] [PubMed] [Google Scholar]
- Cornette R. & Kikawada T. The induction of anhydrobiosis in the sleeping chironomid: current status of our knowledge. IUBMB Life 63, 419–429 (2011). [DOI] [PubMed] [Google Scholar]
- Thorat L. J., Gaikwad S. M. & Nath B. B. Trehalose as an indicator of desiccation stress in Drosophila melanogaster larvae: a potential marker of anhydrobiosis. Biochem Biophys Res Commun 419, 638–642 (2012). [DOI] [PubMed] [Google Scholar]
- Shukla E., Thorat L. J., Nath B. B. & Gaikwad S. M. Insect trehalase: physiological significance and potential applications. Glycobiology 25, 357–367 (2015). [DOI] [PubMed] [Google Scholar]
- Mitsumasu K. et al. Enzymatic control of anhydrobiosis-related accumulation of trehalose in the sleeping chironomid, Polypedilum vanderplanki. FEBS J 277, 4215–4228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia H. & Koshland D. E. Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol 24, 2758–2766 (2014). [DOI] [PubMed] [Google Scholar]
- Tapia H., Young L., Fox D., Bertozzi C. R. & Koshland D. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci USA 112, 6122–6127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkut C. et al. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21, 1331–1336 (2011). [DOI] [PubMed] [Google Scholar]
- Erkut C. & Kurzchalia T. V. The C. elegans dauer larva as a paradigm to study metabolic suppression and desiccation tolerance. Planta 242, 389–396 (2015). [DOI] [PubMed] [Google Scholar]
- Liu K., Dong Y., Huang Y., Rasgon J. L. & Agre P. Impact of trehalose transporter knockdown on Anopheles gambiae stress adaptation and susceptibility to Plasmodium falciparum infection. Proc Natl Acad Sci USA 110, 17504–17509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorat L., Mani K. P., Thangaraj P., Chatterjee S. & Nath B. B. Downregulation of dTps1 in Drosophila melanogaster larvae confirms involvement of trehalose in redox regulation following desiccation. Cell Stress Chaperones 21, 285–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsumasu K., Azuma M., Niimi T., Yamashita O. & Yaginuma T. Changes in the expression of soluble and integral-membrane trehalases in the midgut during metamorphosis in Bombyx mori. Zoolog Sci 25, 693–698 (2008). [DOI] [PubMed] [Google Scholar]
- Mitsumasu K., Azuma M., Niimi T., Yamashita O. & Yaginuma T. Membrane-penetrating trehalase from silkworm Bombyx mori. Molecular cloning and localization in larval midgut. Insect Mol Biol 14, 501–508 (2005). [DOI] [PubMed] [Google Scholar]
- Tatun N., Singtripop T. & Sakurai S. Dual control of midgut trehalase activity by 20-hydroxyecdysone and an inhibitory factor in the bamboo borer Omphisa fuscidentalis Hampson. J Insect Physiol 54, 351–357 (2008). [DOI] [PubMed] [Google Scholar]
- Tan Y. et al. Molecular characterization of soluble and membrane-bound trehalases in the cotton mirid bug, Apolygus lucorum. Arch Insect Biochem Physiol 86, 107–121 (2014). [DOI] [PubMed] [Google Scholar]
- Wang J., He W. B., Su Y. L., Bing X. L. & Liu S. S. Molecular characterization of soluble and membrane-bound trehalases of the whitefly, Bemisia tabaci. Arch Insect Biochem Physiol 85, 216–233 (2014). [DOI] [PubMed] [Google Scholar]
- Matsuda H., Yamada T., Yoshida M. & Nishimura T. Flies without trehalose. J Biol Chem 290, 1244–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormish J. D. & McGhee J. D. The C. elegans lethal gut-obstructed gob-1 gene is trehalose-6-phosphate phosphatase. Dev Biol 287, 35–47 (2005). [DOI] [PubMed] [Google Scholar]
- Zang B., Li H., Li W., Deng X. W. & Wang X. Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol Biol 76, 507–522 (2011). [DOI] [PubMed] [Google Scholar]
- Yang H. L., Liu Y. J., Wang C. L. & Zeng Q. Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS One 7, e42438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesteene L., Ramon M., Le Roy K., Van Dijck P. & Rolland F. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol Plant 3, 406–419 (2010). [DOI] [PubMed] [Google Scholar]
- Sengupta S. et al. Purification, characterization, sequencing and molecular cloning of a novel cysteine methyltransferase that regulates trehalose-6-phosphate synthase from Saccharomyces cerevisiae. Biochim Biophys Acta 1840, 1861–1871 (2014). [DOI] [PubMed] [Google Scholar]
- Folk D. G. & Bradley T. J. Evolved patterns and rates of water loss and ion regulation in laboratory-selected populations of Drosophila melanogaster. J Exp Biol 206, 2779–2786 (2003). [DOI] [PubMed] [Google Scholar]
- Gonçalves P. & Planta R. J. Starting up yeast glycolysis. Trends Microbiol 6, 314–319 (1998). [DOI] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D. & Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol 59, 417–441 (2008). [DOI] [PubMed] [Google Scholar]
- Schluepmann H., Berke L. & Sanchez-Perez G. F. Metabolism control over growth: a case for trehalose-6-phosphate in plants. J Exp Bot 63, 3379–3390 (2012). [DOI] [PubMed] [Google Scholar]
- Rutter G. A., Da Silva Xavier G. & Leclerc I. Roles of 5′-AMP-activated protein kinase (AMPK) in mammalian glucose homoeostasis. Biochem J 375, 1–16 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havula E. & Hietakangas V. Glucose sensing by ChREBP/MondoA-Mlx transcription factors. Semin Cell Dev Biol 23, 640–647 (2012). [DOI] [PubMed] [Google Scholar]
- Oosterveer M. H. & Schoonjans K. Hepatic glucose sensing and integrative pathways in the liver. Cell Mol Life Sci 71, 1453–1467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata F. et al. Necrosis-driven systemic immune response alters SAM metabolism through the FOXO-GNMT axis. Cell Rep 7, 821–833 (2014). [DOI] [PubMed] [Google Scholar]
- Kikawada T. et al. Trehalose transporter 1, a facilitated and high-capacity trehalose transporter, allows exogenous trehalose uptake into cells. Proc Natl Acad Sci USA 104, 11585–11590 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y., Jahagirdar A. P., Yaguchi M. & Downer R. G. Purification and characterization of trehalase inhibitor from haemolymph of the American cockroach, Periplaneta americana. J Biol Chem 264, 16165–16169 (1989). [PubMed] [Google Scholar]
- de Mesquita J. F., Paschoalin V. M. & Panek A. D. Modulation of trehalase activity in Saccharomyces cerevisiae by an intrinsic protein. Biochim Biophys Acta 1334, 233–239 (1997). [DOI] [PubMed] [Google Scholar]
- Chen J. et al. Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA interference. PLoS One 5, e10133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney P. A., Schieler A., Chen J. C., Rabinowitz J. D. & Botstein D. Characterizing the in vivo role of trehalose in Saccharomyces cerevisiae using the AGT1 transporter. Proc Natl Acad Sci USA 112, 6116–6121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean M., Teste M. A., François J. M. & Parrou J. L. Yeast tolerance to various stresses relies on the trehalose-6P synthase (Tps1) protein, not on trehalose. J Biol Chem 290, 16177–16190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gibbs A. G. & Matzkin L. M. Evolution of water balance in the genus Drosophila. J Exp Biol 204, 2331–2338 (2001). [DOI] [PubMed] [Google Scholar]
- Chown S. L., Sørensen J. G. & Terblanche J. S. Water loss in insects: An environmental change perspective. J Insect Physiol 57, 1070–1084 (2011). [DOI] [PubMed] [Google Scholar]
- Folk D. G., Han C. & Bradley T. J. Water acquisition and partitioning in Drosophila melanogaster: effects of selection for desiccation-resistance. J Exp Biol 204, 3323–3331 (2001). [DOI] [PubMed] [Google Scholar]
- Davies S. A. et al. Cell signalling mechanisms for insect stress tolerance. J Exp Biol 217, 119–128 (2014). [DOI] [PubMed] [Google Scholar]
- Albers M. A. & Bradley T. J. Osmotic regulation in adult Drosophila melanogaster during dehydration and rehydration. J Exp Biol 207, 2313–2321 (2004). [DOI] [PubMed] [Google Scholar]
- Kikuta S., Hagiwara-Komoda Y., Noda H. & Kikawada T. A novel member of the trehalose transporter family functions as an h(+)-dependent trehalose transporter in the reabsorption of trehalose in malpighian tubules. Front Physiol 3, 290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S. & Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Nishimori Y. & Nishimura T. Conserved role for the Dachshund protein with Drosophila Pax6 homolog Eyeless in insulin expression. Proc Natl Acad Sci USA 109, 2406–2411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N. et al. A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev 27, 87–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N. & Nishimura T. Signaling from glia and cholinergic neurons controls nutrient-dependent production of an insulin-like peptide for Drosophila body growth. Dev Cell 35, 295–310 (2015). [DOI] [PubMed] [Google Scholar]
- Luan Z., Quigley C. & Li H. S. The putative Na+/Cl−-dependent neurotransmitter/osmolyte transporter inebriated in the Drosophila hindgut is essential for the maintenance of systemic water homeostasis. Sci Rep 5, 7993 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]