Abstract
Previous studies have demonstrated that gestational diabetes mellitus (GDM) and Type 2 diabetes mellitus (T2D) share common genetic polymorphisms. We conducted meta-analysis and subgroup analysis of all available variants and determined the effects of confounding and experimental components on the genetic association of GDM. Any case-controlled or cohort studies with genotype distribution compared GDM cases with controls were included. In total, 28 articles including 8,204 cases and 15,221 controls for 6 polymorphisms were studied. rs10830963(MTNR1B), rs7903146(TCF7L2), and rs1801278(IRS1) were significantly associated with the increased GDM risk. The association of rs4402960(IGF2BP2) and rs1800629(TNF-α) was significant only when the studies with control allele frequency deviation and publication bias were excluded. Further subgroup analysis showed the risk alleles of rs7903146(TCF7L2) and rs1801282(PPARG) were significantly associated with the GDM risk only in Asian, but not in Caucasian population. The OGTT test using 100 g, but not 75 g; and genotype detection by other assays, but not Taqman method, were also significantly associated with increased GDM risk in rs1801278(IRS1) and rs7903146(TCF7L2). Overall GDM was associated with rs10830963(MTNR1B), rs7903146(TCF7L2), and rs1801278(IRS1), but only rs7903146(TCF7L2) and rs1801282(PPARG) were significant in Asian populations. While rs1801278(IRS1) and rs7903146(TCF7L2) were significantly affected by OGTT protocol and genotyping methods.
Gestational diabetes mellitus (GDM), one of the most common pregnancy complications, is defined as the onset or first recognition of glucose intolerance during pregnancy1. Adverse pregnancy outcomes of GDM impact on both mothers and their offspring during and after pregnancy2. Pregnant women complicated with GDM tend to have increased risk of miscarriage, hypertensive disorders, macrosomia, operative delivery and postpartum hemorrhage; and 2.6% to 70% will develop diabetes mellitus 28 years later3. The offspring of these women are associated with large for gestational age, premature birth, neonatal respiratory distress syndrome, hypoglycemia, and also impaired glucose metabolism in early age4.
Globally, the prevalence of GDM varies in population and ethnicity from 1% to 14%7. GDM affects 5% to 6% of all pregnancies in the U.S.A.5; <5% in South Korea, South Africa and United Kingdom; <10% in Italy and Australia; and nearly 20% in Bermuda and Nepal4. GDM received increasing attention globally due to its continuous increase in prevalence, particular in developing countries, including China, India and Africa6. For example, the prevalence of GDM was increased from 2.3% in 1999 to 6.8% in 2008 in northern China7.
GDM and Type 2 diabetes mellitus (T2D) have similar pathogenesis with impaired insulin secretion and increased insulin resistance8. Previous studies demonstrated that GDM and T2D share common genetic polymorphisms, with similar magnitude of the effect sizes on the same risk alleles9. Recent meta-analysis demonstrated association of individual genetic variants with the risk of GDM10. However, most analyses were limited by only examining single or small numbers of genetic variants, thus sample sizes were not sufficient11. The interaction between genetic polymorphisms and other confounding risk factors; and the effects of study populations, demographic characteristics and detection methods were commonly neglected12,13. In this study, we conducted a large scale meta-analysis and further subgroup analysis of most GDM associated genetic variants whose pathogenesis is similar to those of T2D in order to confirm its genetic association and dissect the effects of ethnicity, sample size, OGTT criteria, maternal age, parity, BMI and genotyping methods on the association.
Methods
Literature search
All recent genetic association studies published in recent years (2004 to 2015) were searched in Medline, Evidence Based Medicine (EBM), Pubmed and Web of Science (ISI) for studies published in English and in the WanFang and China National Knowledge Infrastructure (CNKI) database for literature published in Chinese. Keywords in the searches included “gestational diabetes mellitus”, “GDM”, and “genetic”. In addition, the references cited by the original studies, reference lists, conference proceedings, and Google scholar were also searched. Articles published in language other than English and Chinese were not included in this analysis.
Eligibility criteria
Inclusion criteria included any case-controlled or cohort studies with genotype distribution information in both GDM cases and controls. Exclusion criteria included review papers, commentary articles, publication in other languages, studies of postpartum diabetes mellitus, duplicated reports and case series. Genetic studies with no information on genetic polymorphisms, incomplete genotyping data and family trio studies were also excluded. For meta-analysis, a genetic variant reported by less than 3 independent studies and meta-analysis papers were further excluded.
Study selection and validity assessment
Data extraction was independently completed by two reviewers (L.W. and L.C.) from all eligible publications. If a consensus could not be reached, a third reviewer (C.C.W.) settled the discrepancy. If any outstanding queries remain, the corresponding authors of the publication were contacted for data clarification. When the data were not available in the published articles or a further clarification was needed, we contacted the authors by e-mail.
Data extraction and Statistical analysis
Studied variants and its molecular functions in T2D/GDM were recorded. Information was extracted from each selected study, including names of the authors, publication year, study ethnicity, sample size and the definition of cases and controls, the diagnostic criterion for GDM used in the study, the mean age and BMI of cases and controls, genotyping methods, and the frequency of risk alleles were defined according to genotypes. Pooled odds ratio (OR) and 95% confidence intervals (CI) were calculated by allele models of the selected studies on the same variant. Significance levels were determined by Z test. Forest plots were used to demonstrate effect sizes and their confidence intervals (CI)14. Heterogeneity amongst the studies of the genetic variants was assessed by I2 statistic which reflected the heterogeneity proportion in the total variation of effect size. If I2 > 50%, random-effects model (REM) was used. While I2 ≤ 50%, fixed-effects model (FEM) was used to determine the significant heterogeneity15. Hardy-Weinberg equilibrium (HWE) was tested to assess the stability of allele and genotype frequencies in the study population of each study. χ2 test by Fisher’s exact method was used to estimate whether there was significant deviation from HWE among the control group. Significant association was repeatedly determined by either no, HWE only, or combined HWE and Funnel outlier exclusion. All statistical analyses were carried out using Stata software version 12.1 (Stata Corporation, College Station, TX, USA). The significance level was set as 0.05, except Cochran’s Q test for heterogeneity as 0.1.
Subgroup analysis
Effects of different confounding factors of GDM, demographic characteristics of the study populations, diagnostic criteria of GDM, and study methodology of the genetic association studies were tested by subgroup analysis. For the confounding factors, maternal age (<35 vs ≥35), previous history of GDM (yes vs. no) and BMI before and during pregnancy (<25 vs ≥25) were compared. For demographic data, parity (nullpara vs multipara) and gestational age (2nd trimester vs 3rd trimester) were compared. For the diagnosis of GDM, diagnostic criteria using different approaches (75 g OGTT vs 100 g OGTT) were compared. For study methods, genotyping methods (Taqman vs others), study ethnicity (Asian vs Caucasian) and sample sizes (small vs large) were also compared. Sample size was defined as small or large according to calculation using an online sample size estimator (OSSE) on the included papers for each variant (Supplementary Table 1). The confounding and experimental factors of a genetic variant in less than 3 independent studies were excluded due to insufficient data for subgroup analysis. Pooled odds ratios (ORs) and 95% confidence intervals (CI) in each subgroup were calculated as above. χ2 test by Cochran Q model was used to detect the heterogeneity between subgroups.
Publication bias of the included studies
Funnel plots were employed to check the potential publication bias due to significant deviation in the CI distribution from the other studies16. Egger’s test was used to assess the possibility of publication bias by outlier distribution of the standard error of log OR.
Sensitivity analysis of the meta-analysis
To test the robustness of uncertainty in the meta-analysis, sensitivity analyses were employed.
Results
Characteristics of the included studies
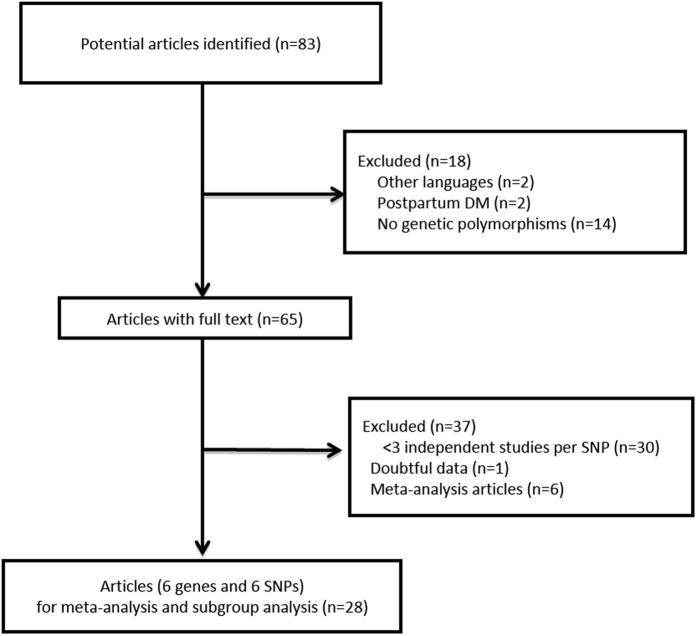
Literature search identified 83 potential articles, including 75 from PubMed and ISI (2004–2015) and 8 from the WanFang database (2004–2015). After abstracts were screened, 18 articles were excluded: 2 published in other languages (Polish and Czech), 2 studies which included postpartum DM, 14 which had no information on genetic polymorphisms. In addition, 37 articles were excluded for the different reasons as listed: 30 articles whereby the reported genetic variants have been studied in less than 3 independent studies for appropriate meta-analysis, 6 which were published meta-analysis and 1 which had doubtful data with potential reversed wild-type allele numbers and risk allele numbers but could not be verified by contacting the author17. One study18 included for its control subjects both men and women with average age more than 60-year-old and no evidence of DM. In addition the frequency of polymorphism associated with DM is low, therefore this study was regarded as eligible for inclusion. In total, 28 articles (7 papers from China, 8 papers from other regions of Asia and 13 papers from Western countries) were included for the meta-analysis and subgroup-analysis. All included studies were case-controlled studies, including 8,204 cases and 15,221 controls for 6 loci (Fig. 1). Of the 6 loci, Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), Melatonin receptor 1B (MTNR1B), Transcription factor 7-like 2 (TCF7L2) are involved in insulin secretion19,20,21; Insulin receptor substrate 1 (IRS1) and Peroxisome proliferator-activated receptor gamma (PPARG) are involved in insulin resistance22,23; while Tumor necrosis factor alpha (TNF-α) is involved in inflammation in DM and/or GDM pathogenesis (Supplementary Table 2)24. Though three variants (rs4402960 in IGF2BP2, rs10830963 in MTNR1B and rs7903146 in TCF7L2) are located at intron regions, these causal variants do modulate gene function and impact on downstream transcriptional machinery25. Another three variants (rs1801278 in IRS1, rs1801282 in PPARG and rs1800629 in TNF-α) are located at exon regions. Study characteristics, detailed genotype information and allele distribution in each included study are summarized in Supplementary Tables 3 and 4, respectively. Results of meta-analysis and sub-group analysis in each SNP are detailed as below. Sensitivity analysis did not identify any significant uncertainty in the analysis.
Figure 1. Flowchart of the systematic search methodology.
Meta-analysis
Variants involved in insulin secretion
IGF2BP2 (rs4402960) There were 3 articles which studied the T allele of rs4402960 but meta-analysis did not identify any significant association of the variant with the increased risk of GDM (pooled OR 1.12, 95% CI 0.89–1.40, P = 0.353, Table 1 and Supplementary Figure 1). There was significant heterogeneity across the studies (I2 = 68.4%; p = 0.042). There was no significant HWE variation (Supplementary Table 4), but there was potential publication bias (Supplementary Figure 3). After the outlier19 from the funnel plot analysis was removed, the association between the variant and GDM was significant (pooled OR 1.22, 95% CI 1.09–1.36; p < 0.001, Table 1 and Supplementary Figure 3) and heterogeneity was not significant (I2 = 0%; p = 0.637). There were not enough studies for subgroup analysis.
Table 1. Associations between genetic variants and GDM risk.
| Gene | Variant | Minor allele | Exclusion | Number of studies | Sample size (cases/controls) | OR(95% CI) | P (Z)a | Heterogeneity |
|
|---|---|---|---|---|---|---|---|---|---|
| I2 | P (Q)b | ||||||||
| IGF2BP2 | rs4402960 | T | Noc | 3 | 1688/1712 | 1.12 (0.89, 1.40) | 0.353 | 68.4% | 0.042 |
| HWE & Funnele | 2 | 1593/1671 | 1.22 (1.09, 1.36) | <0.001 | 0% | 0.637 | |||
| MTNR1B | rs10830963 | G | Noc | 7 | 2705/4325 | 1.31 (1.18, 1.47) | <0.001 | 44.2% | 0.097 |
| HWEd | 5 | 2548/3022 | 1.28 (1.14, 1.44) | <0.001 | 50.3% | 0.090 | |||
| TCF7L2 | rs7903146 | T | Noc | 9 | 3206/6334 | 1.41 (1.16, 1.72) | 0.001 | 80.8% | <0.001 |
| HWEd | 7 | 2127/2871 | 1.40 (1.03, 1.90) | 0.032 | 85.1% | <0.001 | |||
| HWE & Funnele | 6 | 1866/2495 | 1.57 (1.38, 1.79) | <0.001 | 9% | 0.359 | |||
| IRS1 | rs1801278 | T | Noc | 5 | 1307/1973 | 1.53 (1.08, 2.15) | 0.015 | 49.6% | 0.094 |
| PPARG | rs1801282 | G | Noc | 10 | 2929/6969 | 0.89 (0.75, 1.05) | 0.154 | 30.8% | 0.162 |
| HWEd | 8 | 2544/3015 | 0.86 (0.68, 1.08) | 0.181 | 44.0% | 0.085 | |||
| HWE & Funnele | 7 | 2365/2835 | 0.95 (0.80, 1.13) | 0.587 | 16.7% | 0.302 | |||
| TNF-α | rs1800629 | A | Noc | 3 | 196/181 | 1.38 (0.37, 5.16) | 0.633 | 85.4% | 0.001 |
| HWEd | 2 | 86/79 | 2.69 (1.28, 5.68) | 0.009 | 24.5% | 0.25 | |||
aP value for Z test.
bP value for the Cochran χ2 Q test.
cNo exclusion of studies for meta-analysis.
dStudies after removing articles deviated from Hardy-Weinberg equilibrium (HWE) in control (P < 0.05) for meta-analysis.
eStudies after removing articles deviated from Hardy-Weinberg equilibrium in control (P < 0.05) and outliers in funnel plot (Funnel) for meta-analysis.
MTNR1B (rs10830963) There were 7 eligible studies which examined the rs10830963 polymorphism. Meta-analysis showed that the G allele was significantly associated with increased risk of GDM (pooled OR 1.31, 95% CI 1.18–1.47, p < 0.001; Heterogeneity I2 44.2%, p = 0.097; Table 1, Supplementary Figure 1). There was significant departure from HWE in 2 studies26,27 (Supplementary Table 4) and the significant association was maintained when these studies were excluded (pooled OR 1.28, 95% CI 1.14–1.44, p < 0.001; Heterogeneity I2 50.3%, p = 0.09; Table 1, Supplementary Figure 2). There was no publication bias under the funnel plot analysis (Supplementary Figure 3). There were enough studies for subgroup analysis for ethnicity, OGTT criteria, genotyping methods and sample size only. Studies looking at ethnicity found the risk allele was significantly associated with increased risk of GDM both in Asian populations (pooled OR 1.23, 95% CI 1.10–1.38, p < 0.001; I2 37.7%, p = 0.186) and Caucasian populations (pooled OR 1.49, 95% CI 1.22–1.82, p < 0.001, Table 2). Studies either used 75 g or 100 g in OGTT for GDM (pooled OR 1.38, 95% CI 1.20–1.57, p < 0.001 and pooled OR 1.20, 95% CI 1.01–1.44, p = 0.043, respectively; Table 2); studies used Taqman assay or other assays as genotype detection method (pooled OR 1.28, 95% CI 1.08–1.51, p = 0.004 and pooled OR 1.29, 95% CI 1.08–1.54, p = 0.005, respectively; Table 2); and studies with both larger sample size (≥336) and smaller sample size (<336) (pooled OR 1.27, 95% CI 1.12–1.44, p < 0.001 and pooled OR 1.53, 95% CI 1.01–2.32, p = 0.047, respectively; Table 2) were significantly associated between the risk variant and GDM. But only studies included subjects with high mean pre-pregnancy BMI (≥25), but not low mean pre-pregnancy BMI (<25), were significantly associated between the risk variant and GDM (pooled OR 1.24, 95% CI 1.02–1.51, p = 0.033, Table 2).
Table 2. Subgroup analysis of genetic variants in GDM (with HWE excluded only).
| Subgroup | Number of studies | Number of cases | Number of controls | OR(95% CI) | P (Z)a | Heterogeneity |
|
|---|---|---|---|---|---|---|---|
| I2 | P (Q)b | ||||||
| MTNR1B rs10830963 | |||||||
| Ethnicity | |||||||
| Asian | 4 | 2090 | 2600 | 1.23 (1.10, 1.38) | <0.001 | 37.7% | 0.186 |
| Caucasian | 1 | 458 | 422 | 1.49 (1.22, 1.82) | <0.001 | NA | NA |
| OGTT | |||||||
| 75 g | 3 | 895 | 993 | 1.38 (1.20, 1.57) | <0.001 | 0.0% | 0.375 |
| 100 g | 2 | 1653 | 2029 | 1.20 (1.01, 1.44) | 0.043 | 72.6% | 0.056 |
| Genotype method | |||||||
| Taqman assay | 3 | 2111 | 2451 | 1.28 (1.08, 1.51) | 0.004 | 72.2% | 0.027 |
| Others | 2 | 437 | 571 | 1.29 (1.08, 1.54) | 0.005 | 0.0% | 0.375 |
| Sample size | |||||||
| small | 1 | 87 | 91 | 1.53 (1.01, 2.32) | 0.047 | NA | NA |
| large | 4 | 2461 | 2931 | 1.27 (1.12, 1.44) | <0.001 | 58.4% | 0.066 |
| Pre-BMI | |||||||
| <25 | 2 | 812 | 1130 | 1.22 (0.9, 1.65) | 0.202 | 54.2% | 0.14 |
| ≥25 | 1 | 350 | 480 | 1.24 (1.02, 1.51) | 0.033 | NA | NA |
| TCF7L2 rs7903146 | |||||||
| Ethnicity | |||||||
| Asian | 3 | 1008 | 801 | 1.58 (1.12, 2.23) | 0.009 | 39.1% | 0.194 |
| Caucasian | 4 | 1119 | 2070 | 1.32 (0.86, 2.03) | 0.212 | 91.4% | 0.212 |
| OGTT | |||||||
| 75 g | 5 | 1219 | 2170 | 1.43 (0.97, 2.09) | 0.07 | 89.7% | <0.001 |
| 100 g | 2 | 908 | 701 | 1.36 (0.94, 1.97) | 0.100 | 12.3% | 0.286 |
| Genotype method | |||||||
| Taqman assay | 4 | 1839 | 2590 | 1.24 (0.82, 1.87) | 0.306 | 90.1% | <0.001 |
| Others | 3 | 288 | 281 | 1.73 (1.20, 2.50) | 0.003 | 49.8% | 0.136 |
| IRS1 rs1801278 | |||||||
| Ethnicity | |||||||
| Asian | 2 | 262 | 400 | 2.15 (0.80, 5.74) | 0.128 | 50.3% | 0.156 |
| Caucasian | 3 | 1045 | 1573 | 1.41 (0.99, 2.01) | 0.059 | 56.3% | 0.101 |
| OGTT | |||||||
| 75 g | 2 | 736 | 1296 | 1.31 (0.82, 2.08) | 0.261 | 70.3% | 0.067 |
| 100 g | 3 | 571 | 677 | 1.88 (1.21, 2.94) | 0.005 | 6.8% | 0.342 |
| Genotype method | |||||||
| Taqman assay | 1 | 588 | 1189 | 1.04 (0.75, 1.45) | 0.805 | NA | NA |
| Others | 4 | 719 | 784 | 1.77 (1.33, 2.35) | <0.001 | 0.0% | 0.515 |
| PPARG rs1801282 | |||||||
| Ethnicity | |||||||
| Asian | 5 | 1259 | 1126 | 0.72 (0.56, 0.93) | 0.011 | 24.8% | 0.256 |
| Caucasian | 3 | 1285 | 1889 | 1.07 (0.91, 1.18) | 0.611 | 0.0% | 0.474 |
| OGTT | |||||||
| 75 g | 5 | 1519 | 2242 | 0.98 (0.86, 1.12) | 0.812 | 51.7% | 0.082 |
| 100 g | 3 | 1025 | 773 | 0.80 (0.53, 1.21) | 0.294 | 24.3% | 0.267 |
| Genotype method | |||||||
| Taqman assay | 2 | 1505 | 1864 | 0.95 (0.71, 1.26) | 0.706 | 55.9% | 0.132 |
| Others | 6 | 1039 | 1151 | 0.87 (0.71, 1.05) | 0.141 | 45.3% | 0.104 |
aP values in subgroups.
bP values for Cochran’s Q statistic test used to assess the heterogeneity.
NA not available.
TCF7L2 (rs7903146) There were 9 published articles which studied the rs7903146 polymorphism. Meta-analysis showed that the T allele was significantly associated with increased risk of GDM (pooled OR 1.41, 95% CI 1.16–1.72, p = 0.001; Heterogeneity I2 80.8%, p < 0.001; Table 1 and Supplementary Figure 1). HWE was significant in 2 studies28,29 (Supplementary Table 4), the significant association was maintained when those studies were removed (pooled OR 1.40, 95% CI 1.03–1.90, p = 0.032; Heterogeneity I2 85.1%, p < 0.001 Table 1 and Supplementary Figure 2). Funnel plot analysis indicated potential publication bias in 1 study30 (Supplementary Figure 3), the association was still significant when this study was excluded (pooled OR 1.57, 95% CI 1.38–1.79, p < 0.001; Heterogeneity I2 9%, p = 0.359; Table 1 and Supplementary Figure 4). Only ethnicity, OGTT criteria and genotyping methods have enough studies for further subgroup analysis. Comparing studies in different ethnicity showed the risk allele was significantly associated with the increased risk of GDM in the Asian population only (pooled OR 1.58, 95% CI 1.12–2.23, p = 0.009; I2 39.1%, p = 0.194), but not in Caucasian population (pooled OR 1.32, 95% CI 0.86–2.03, p = 0.212; I2 91.4%, p = 0.212); Table 2). Studies which either used 75 g or 100 g in OGTT for GDM diagnosis had no statistical significant association between the risk alleles and GDM (Table 2). If studies with potential publication bias were not included in subgroup analysis, the association became significant in Caucasian population (pooled OR 1.55, 95% CI 1.35–1.79, p < 0.001; heterogeneity I2 6.3%, p = 0.344) and GDM diagnosis using 75 g OGTT (pooled OR 1.62, 95% CI 1.39–1.89, heterogeneity I2 19.8%, p = 0.291) only (Supplementary Table 5). Studies which used methods other than Taqman assay as genotype detection were also significant (Table 2).
Variants involved in insulin resistance
IRS1 (rs1801278) Five eligible articles studied rs1801278 polymorphism. Meta-analysis showed that the T allele was significantly associated with the increased risk of GDM (pooled OR 1.53, 95% CI 1.08–2.15, p = 0.015; Heterogeneity I2 49.6%, p = 0.094; Table 1 and Supplementary Figure 1). There were no deviation of HWE and no publication bias (Supplementary Table 4 and Supplementary Figure 3). Only ethnicity, OGTT criteria and genotyping methods had enough studies for subgroup analysis. Subgroup analysis according to ethnicity showed the risk allele was not significantly associated with GDM in Asian population or Caucasian population (Table 2). Only studies which used 100 g in OGTT for GDM diagnosis had significant association in GDM risk (pooled OR 1.88, 95% CI 1.21–2.94, p = 0.005; Table 2), while studies which used 75 g in OGTT for GDM diagnosis had no significance association (pooled OR 1.31, 95% CI 0.82–2.08, p = 0.261; Table 2). Studies used method other than Taqman assay as genotype detection (pooled OR 1.77, 95% CI 1.33–2.35, p < 0.001) were also significant associated between the risk alleles and GDM (Table 2).
PPARG (rs1801282) There were 10 included articles which studied the rs1801282 polymorphism. Meta-analysis showed that the G allele was not significantly associated with the increased risk of GDM (pooled OR 0.89, 95% CI 0.75–1.05, p = 0.154; Heterogeneity I2 30.8%, p = 0.162; Table 1 and Supplementary Figure 1). HWE was significant in 2 studies29,31 (Supplementary Table 4), the significant association was not maintained when these studies with excluded (pooled OR 0.86, 95% CI 0.68–1.08, p = 0.181; Heterogeneity I2 44%, p = 0.085; Table 1 and Supplementary Figure 2). Funnel plot analysis indicated potential publication bias in 1 study32 (Supplementary Figure 3), there was no significant association even when this study was also excluded (pooled OR 0.95, 95% CI 0.80–1.13, p0.587; Heterogeneity I2 16.7%, p = 0.302; Table 1 and Supplementary Figure 4). Again only ethnicity, OGTT criteria and genotyping methods have enough studies for subgroup analysis. Subgroup analysis on ethnicity showed the risk allele was significantly associated with decreased risk of GDM in Asian populations (pooled OR 0.72, 95% CI 0.56–0.93, p = 0.011; I2 24.8%, p = 0.256), but not in Caucasian populations (pooled OR 1.07, 95% CI 0.91–1.18, p = 0.611; I2 0%, p = 0.474; Table 2). Studies which used either 75 g or 100 g in OGTT for GDM diagnosis had no significant association between the risk alleles and GDM (Table 2). If studies with potential publication bias were not included, the above significant association was not maintained (Supplementary Table 5). Studies used methods either Taqman assay or other assays as genotype detection were also not significant (Table 2).
Variant involved in inflammation
TNF-α (rs1800629) There were 3 included articles which studied the rs1800629 polymorphism. Meta-analysis showed no significant association between rs1800629 and GDM risk (pooled OR 1.38, 95% CI 0.37–5.16); P = 0.633 Table 1 and Supplementary Figure 1). HWE was significant in 1 study24 (Supplementary Table 4), the association of rs1800629 in GDM risk become significant when this study with excluded (pooled OR 2.69, 95% CI 1.28–5.68; p = 0.009, Table 1 and Supplementary Figure 2). There was no potential publication bias (Supplementary Figure 3) and there were not enough studies for subgroup analysis.
Discussion
Previous meta-analysis identified few genetic polymorphism associated with GDM10,11. However, these studies have certain limitations, such as single or few variants per each study, insufficient sample sizes, study heterogeneity and neglected confounding factors such as maternal age, BMI, diabetic history, etc. In our current study, we conducted a larger-scale meta-analysis and reduced the study heterogeneity to confirm the association between the risk alleles and GDM. In addition, we further carried out subgroup-analysis of ethnicity, OGTT criteria, genotyping methods and sample size in order to obtain more in-depth interpretation. Our meta-analysis included the alleles in IGF2BP2, MTNR1B, TCF7L2, IRS1, TNF-α, and PPARG genes, and showed the risk alleles of MTNR1B, TCF7L2 and IRS1, but not PPARG, were significantly associated with increased risk of GDM with or without study heterogeneity, HWE derivation and publication bias. The results confirmed previous meta-analyses that the risk allele of rs7903146 in TCF7L233 and rs1801278 in IRS1 and rs10830963 in MTNR1B were associated with increased risk of GDM34, while there was no significant association between rs1801282 polymorphism in PPARG and GDM risk35. However, the risk alleles of IGF2BP2 and TNF-α were significantly associated with increased risk of GDM only after studies which failed HWE were excluded, which had not been identified in previous studies11, indicating its potential association in GDM.
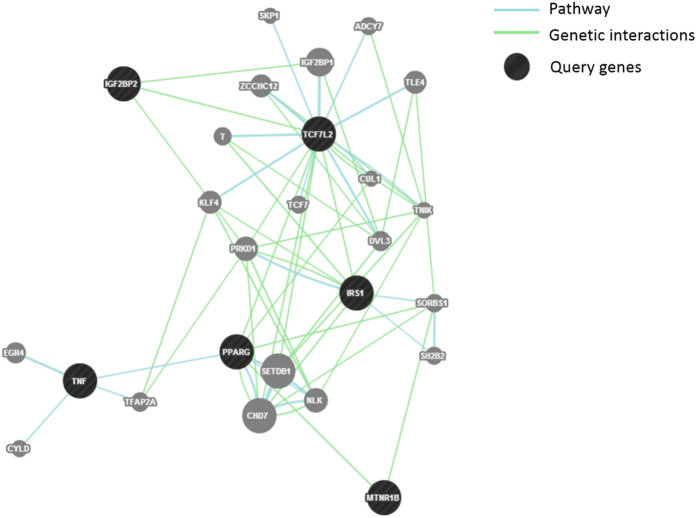
The underlying mechanism of GDM include impaired β-cell function, and decreased insulin sensitivity Women with GDM had a higher risk of developing diabetes than those with normal blood glucose after pregnancy36. It has been suggested that GDM and T2DM may share common pathogenic pathways37. GDM is a multifactorial metabolic disorder in which loci interact with environmental factors as well as family history and obesity38. Some studies examined the association of genetic polymorphism between GDM and T2D and supported the hypothesis that both had common genetic background18,29,39. In our study, only MTNR1B40 and TCF7L241 involved in impaired insulin secretion, and IRS142 involved in insulin resistance in T2D were significantly associated with GDM. It further supports impaired insulin secretion and insulin resistance may involve in the pathogenesis of GDM as in T2D. By using Gene Interaction Network (GeneMENIA), TNF-α and PPARG showed direct link to each other under category of gene pathway. It indicates the direct gene-gene interactions between TNF-α and PPARG (Fig. 2). Lin et al. confirm the interactions by radiation hybrid genotyping in the mammalian genome43. Although the association between TNF-α and PPARG has been shown, their direct genetic interaction in GDM is still unclear. Obesity and insulin resistance are associated with inflammatory factors and play a pivotal role in the development of T2D and GDM44. TNF-α, as a pro-inflammatory cytokine, can activate some signaling pathways to inhibit the insulin activity45. However there are not enough studies of TNF-α to show the relationship between BMI and GDM. On the other hand, TCF7L2 showed direct link with IGF2BP2, IRS1 and PPARG, but indirectly with MTNR1B, indicating the potential genetic association between TCF7L2 and other genes important for T2D and/or GDM. Gene-gene interactions would provide possibility for future genetic research.
Figure 2. Gene pathway and genetic interactions of the SNPs in GDM.
Prevalence of GDM in Asian countries is now higher than that in other countries6. In our subgroup-analysis, only 2 study populations, Asian and Caucasian, could be compared. There were not enough studies included other populations for the comparison. The results showed that TCF7L2 (rs7903146) and PPARG (rs1801282) were significantly associated with increased/decreased risk of GDM in Asian population, mostly from China, but not in Caucasian population. All 7 previous genetic studies shows the significant association of TCF7L2 (rs7903146) in Caucasian populations were positive in 6 out 7 studies with highest OR as 2.0446, but negative in 1 study30 with the lowest OR as 0.69. In contrast, genetic studies shows the association in Asian populations were positive in 2 out of 3 studies with highest OR as 2.0647 but negative in 1 study48 with OR as 1.08. For all 5 previous genetic studies show the negative association of PPARG (rs1801282) in Caucasian populations but only 1 out of 5 studies in Asian population were positive32. Here our subgroup results suggest underlying ethnic predisposition of TCF7L2 and PPARG in the development of GDM in Asian population only, but larger scale study will be necessary to confirm the results. In addition, larger pre-pregnancy BMI (≥25) has significant association in MTNR1B (rs10830963) with GDM, indicating the potential effects of obesity on the role of MTNR1B mutation in GDM risk10. Apart from ethnicity and obesity, genotype and OGTT test methods also affect the genetic association of the risk alleles and GDM. Genotyping in TCF7L2 (rs7903146) and IRS1 (rs1801278) were significantly associated with increased risk of GDM in other methods, including PCR-RFLP, rather than Taqman assay. PCR-RFLP is a rapid, simple and inexpensive genotyping technique that exploits variations in homologous DNA sequences but poor in conditions of compound heterozygous mutation49 and non-coding regions that the restriction enzymes may not work properly50. In our study no studied variants were compound heterozygous mutation but rs7903146 was a non-coding allele. The identified significant association may imply potential error in the mutation detection method by PCR-RFLP method, the results should be validated by Sanger sequencing, the gold standard of the genotyping. On the other hand, the OGTT test using 100 g, but not 75 g, was significantly associated with increased GDM risk in IRS-1 (rs1801278) only. One-step 75 g OGTT was recommended by The international Association of Diabetes and Pregnancy Study Groups (IADPSG) while two-step 100 g OGTT was recommended by the National Diabetes Data Group (NDDG) and Carpenter and Coustan for GDM diagnosis5. Compared with 100 g OGTT, 75 g OGTT would increase the prevalence of GDM twofold to threefold if using the Carpenter and Coustan’s diagnostic criteria. The 100 g OGTT might increase the sensitivity to detect the genetic polymorphism in IRS-1. IRS-1 is involved in insulin resistance, how different glucose loading and threshold of abnormal plasma glucose levels affect the insulin resistance remained to elucidate.
Although this present study is comprehensive, included more variants for meta-analysis and carried out subgroup-analysis, there were several limitations. Firstly, more accurate analysis using the adjusted data by age and BMI would be preferable to direct use of unadjusted raw data. Secondly, environmental factors including cigarette smoking, alcohol consumption, and dietary habit are important confounding factors which should preferably be considered and recorded in genetic studies. Thirdly, due to the limitation of included studies with small sample sizes, the odd ratio are small and the results will be seriously affected by any studies removed due to significant heterogeneity, HWE and publication bias. Since a strict minimal number of studies was required for meta-analysis, some of important variants of CDKAL151, KCNJ1137and GCK52could not be included. A genome-wide association study has demonstrated that genetic variants in CDKAL1 gene and near MTNR1B are associated with GDM in Korean women (p < 5.0 × 10−8), however there is no other genome-wide association study available to extract the variant information for present meta-analysis. The genetic association of these variants with GDM should also be included in future if sufficient association studies become available.
Overall GDM was associated with rs10830963, rs7903146, and rs1801278, but only rs7903146 and rs1801282 were significant in Asian populations. While rs1801278 and rs7903146 were significantly affected by OGTT protocol and genotyping methods. In summary, the present study dissected the effects of subgroup information on the genetic association of GDM, which not only confirmed the genetic association for some of the variants, but also to explore the effects of ethnicity, diagnosis criteria, genotype methods and BMI on the results. These factors should be considered in the association between the genetic variants and the risk of GDM in future genetic studies.
Additional Information
How to cite this article: Wu, L. et al. Genetic variants associated with gestational diabetes mellitus: a meta-analysis and subgroup analysis. Sci. Rep. 6, 30539; doi: 10.1038/srep30539 (2016).
Supplementary Material
Acknowledgments
This work was partially supported by the Hong Kong Obstetrician and Gynaecologist Trust Fund 2016.
Footnotes
Author Contributions L.W. and C.C.W. drafted the original manuscript, L.W. and L.C. performed statistic analysis, L.W., W.H.T. and R.C.W.M. revised the manuscript and approved the final version.
References
- American Diabetes, A. Diagnosis and classification of diabetes mellitus. Diabetes care 32 Suppl 1, S62–67, doi: 10.2337/dc09-S062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille J. & Grant A. M. The genetics of gestational diabetes mellitus: evidence for relationship with type 2 diabetes mellitus. Genetics in medicine: official journal of the American College of Medical Genetics 10, 240–250, doi: 10.1097/GIM.0b013e31816b8710 (2008). [DOI] [PubMed] [Google Scholar]
- Kim C., Newton K. M. & Knopp R. H. Gestational diabetes and the incidence of type 2 diabetes - A systematic review. Diabetes care 25, 1862–1868, doi: DOI 10.2337/diacare.25.10.1862 (2002). [DOI] [PubMed] [Google Scholar]
- Kampmann U. et al. Gestational diabetes: A clinical update. World J Diabetes 6, 1065–1072, doi: 10.4239/wjd.v6.i8.1065 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDorsten J. P. et al. National Institutes of Health consensus development conference statement: diagnosing gestational diabetes mellitus, March 4–6, 2013. Obstet Gynecol 122, 358–369, doi: 10.1097/AOG.0b013e31829c3e64 (2013). [DOI] [PubMed] [Google Scholar]
- Tutino G. E. et al. Diabetes and pregnancy: perspectives from Asia. Diabetic medicine: a journal of the British Diabetic Association 31, 302–318, doi: 10.1111/dme.12396 (2014). [DOI] [PubMed] [Google Scholar]
- Leng J. et al. Prevalence of gestational diabetes mellitus and its risk factors in Chinese pregnant women: a prospective population-based study in Tianjin, China. Plos one 10, e0121029, doi: 10.1371/journal.pone.0121029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl C. Etiology and pathogenesis of gestational diabetes. Diabetes care 21 Suppl 2, B19–26 (1998). [PubMed] [Google Scholar]
- Huopio H. et al. Association of risk variants for type 2 diabetes and hyperglycemia with gestational diabetes. European journal of endocrinology/European Federation of Endocrine Societies 169, 291–297, doi: 10.1530/EJE-13-0286 (2013). [DOI] [PubMed] [Google Scholar]
- Mao H., Li Q. & Gao S. Meta-analysis of the relationship between common type 2 diabetes risk gene variants with gestational diabetes mellitus. Plos one 7, e45882, doi: 10.1371/journal.pone.0045882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Human reproduction update 19, 376–390, doi: 10.1093/humupd/dmt013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz N., Nachum Z. & Green M. S. The prevalence of gestational diabetes mellitus recurrence-effect of ethnicity and parity: a metaanalysis. American journal of obstetrics and gynecology, doi: 10.1016/j.ajog.2015.03.011 (2015). [DOI] [PubMed] [Google Scholar]
- Chong Y. S. et al. Ethnic differences translate to inadequacy of high-risk screening for gestational diabetes mellitus in an Asian population: a cohort study. BMC pregnancy and childbirth 14, 345, doi: 10.1186/1471-2393-14-345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J. L., Zhong W. Z., Zheng M. H. & Chen F. Meta—Analysis in Stata. The Journa]of Evidence—Based Medicine 7, 363–368 (2007). [Google Scholar]
- Wang D., Mou Z. Y., Zhai J. X., Zong H. X. & Zhao X. D. Application of Stata software to test heterogeneity in Meta—analysis method. Chinese Journal of Epidemiology 29, 726–729 (2008). [PubMed] [Google Scholar]
- Wang D., Mou Z. Y., Zhai J. X., Zong H. X. & Zhao X. D. Study on Stata software in investigating publication bias in meta-analysis. Modern Preventive Medicine 35, 2819–2822 (2008). [Google Scholar]
- Nor Khatijah Mohd Aris et al. An Analysis of Targeted Single Nucleotide Polymorphisms for the Risk Prediction of Gestational Diabetes Mellitus in a Cohort of Malaysian Patients. Asia-Pacific Journal of Molecular Medicine, 1–8 (2011). [Google Scholar]
- Cho Y. M. et al. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia 52, 253–261, doi: 10.1007/s00125-008-1196-4 (2009). [DOI] [PubMed] [Google Scholar]
- Chon S. J. et al. Association of variants in PPARgamma(2), IGF2BP2, and KCNQ1 with a susceptibility to gestational diabetes mellitus in a Korean population. Yonsei medical journal 54, 352–357, doi: 10.3349/ymj.2013.54.2.352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y. et al. Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC medical genetics 12, 82, doi: 10.1186/1471-2350-12-82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K. et al. Transcription factor 7-like 2 gene polymorphisms and gestational diabetes mellitus. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet 25, 1783–1786, doi: 10.3109/14767058.2012.663831 (2012). [DOI] [PubMed] [Google Scholar]
- Alharbi K. K., Khan I. A., Abotalib Z. & Al-Hakeem M. M. Insulin receptor substrate-1 (IRS-1) Gly927Arg: correlation with gestational diabetes mellitus in Saudi women. BioMed research international 2014, 146495, doi: 10.1155/2014/146495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tok E. C. et al. PPAR-gamma2 Pro12Ala polymorphism is associated with weight gain in women with gestational diabetes mellitus. European journal of obstetrics, gynecology, and reproductive biology 129, 25–30, doi: 10.1016/j.ejogrb.2006.03.016 (2006). [DOI] [PubMed] [Google Scholar]
- Montazeri S., Nalliah S. & Radhakrishnan A. K. Is there a genetic variation association in the IL-10 and TNF alpha promoter gene with gestational diabetes mellitus? Hereditas 147, 94–102, doi: 10.1111/j.1601-5223.2009.02134.x (2010). [DOI] [PubMed] [Google Scholar]
- Xia Q., Deliard S., Yuan C. X., Johnson M. E. & Grant S. F. Characterization of the transcriptional machinery bound across the widely presumed type 2 diabetes causal variant, rs7903146, within TCF7L2. European journal of human genetics: EJHG 23, 103–109, doi: 10.1038/ejhg.2014.48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuebe A. M. et al. Maternal genotype and gestational diabetes. American journal of perinatology 31, 69–76, doi: 10.1055/s-0033-1334451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassi M. et al. The rs10830963 variant of melatonin receptor MTNR1B is associated with increased risk for gestational diabetes mellitus in a Greek population. Horm-Int J Endocrino 11, 70–76 (2012). [DOI] [PubMed] [Google Scholar]
- Papadopoulou A. et al. Gestational diabetes mellitus is associated with TCF7L2 gene polymorphisms independent of HLA-DQB1*0602 genotypes and islet cell autoantibodies. Diabetic medicine: a journal of the British Diabetic Association 28, 1018–1027, doi: 10.1111/j.1464-5491.2011.03359.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauenborg J. et al. Common type 2 diabetes risk gene variants associate with gestational diabetes. The Journal of clinical endocrinology and metabolism 94, 145–150, doi: 10.1210/jc.2008-1336 (2009). [DOI] [PubMed] [Google Scholar]
- Vcelak J. et al. T2D risk haplotypes of the TCF7L2 gene in the Czech population sample: the association with free fatty acids composition. Physiological research/Academia Scientiarum Bohemoslovaca 61, 229–240 (2012). [DOI] [PubMed] [Google Scholar]
- Heude B. et al. Association of the Pro12Ala and C1431T variants of PPARgamma and their haplotypes with susceptibility to gestational diabetes. The Journal of clinical endocrinology and metabolism 96, E1656–1660, doi: 10.1210/jc.2011-0381 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu Y. & Wu C. Y. Relationship between Pr012Ala polymorphism in peroxisome proliferators-activated receptor gamma 2 gene and gestationul diabetes mellitus. The Journal of Practical Medicine 25, 3 (2009). [Google Scholar]
- Kang S., Xie Z. & Zhang D. Association of the rs7903146 polymorphism in transcription factor 7-like 2 (TCF7L2) gene with gestational diabetes mellitus: a meta-analysis. Gynecological endocrinology: the official journal of the International Society of Gynecological Endocrinology 29, 873–877, doi: 10.3109/09513590.2013.813469 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sun C. M., Hu X. Q. & Zhao ,. Y. Relationship between melatonin receptor 1B and insulin receptor substrate 1 polymorphisms with gestational diabetes mellitus: a systematic review and meta-analysis. Scientific reports 4, 6113, doi: 10.1038/srep06113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang C., Li X., Huang Z. & Qian J. Quantitative assessment of the influence of PPARG P12A polymorphism on gestational diabetes mellitus risk. Molecular biology reports 40, 811–817, doi: 10.1007/s11033-012-2119-5 (2013). [DOI] [PubMed] [Google Scholar]
- Bao W. et al. Long-term risk of type 2 diabetes mellitus in relation to BMI and weight change among women with a history of gestational diabetes mellitus: a prospective cohort study. Diabetologia 58, 1212–1219, doi: 10.1007/s00125-015-3537-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaat N. et al. Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia 48, 2544–2551, doi: 10.1007/s00125-005-0035-0 (2005). [DOI] [PubMed] [Google Scholar]
- Cho G. J. et al. Secular Trends of Gestational Diabetes Mellitus and Changes in Its Risk Factors. PloS one 10, e0136017, doi: 10.1371/journal.pone.0136017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappa K. I. et al. Gestational diabetes mellitus shares polymorphisms of genes associated with insulin resistance and type 2 diabetes in the Greek population. Gynecological endocrinology: the official journal of the International Society of Gynecological Endocrinology 27, 267–272, doi: 10.3109/09513590.2010.490609 (2011). [DOI] [PubMed] [Google Scholar]
- Salman M. et al. MTNR1B gene polymorphisms and susceptibility to Type 2 Diabetes: A pilot study in South Indians. Gene 566, 189–193, doi: 10.1016/j.gene.2015.04.064 (2015). [DOI] [PubMed] [Google Scholar]
- Zhou Y. et al. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet 23, 6419–6431, doi: 10.1093/hmg/ddu359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. et al. Impairment of insulin receptor substrate 1 signaling by insulin resistance inhibits neurite outgrowth and aggravates neuronal cell death. Neuroscience 301, 26–38, doi: 10.1016/j.neuroscience.2015.05.072 (2015). [DOI] [PubMed] [Google Scholar]
- Lin A., Wang R. T., Ahn S., Park C. C. & Smith D. J. A genome-wide map of human genetic interactions inferred from radiation hybrid genotypes. Genome research 20, 1122–1132, doi: 10.1101/gr.104216.109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G. S. Inflammation and metabolic disorders. Nature 444, 860–867, doi: 10.1038/nature05485 (2006). [DOI] [PubMed] [Google Scholar]
- Altinova A. E. et al. Circulating concentrations of adiponectin and tumor necrosis factor-alpha in gestational diabetes mellitus. Gynecological endocrinology: the official journal of the International Society of Gynecological Endocrinology 23, 161–165, doi: 10.1080/09513590701227960 (2007). [DOI] [PubMed] [Google Scholar]
- Ekelund M. et al. Genetic prediction of postpartum diabetes in women with gestational diabetes mellitus. Diabetes research and clinical practice 97, 394–398, doi: 10.1016/j.diabres.2012.04.020 (2012). [DOI] [PubMed] [Google Scholar]
- Xiling S., Cai Q. H., Zou M. Y. & Shen Y. S. Correlation between TCF7L2 gene polymorphism and genetic susceptibility in women with gestational diabetes mellitus. Chinese Journal of Obstetrics and Gynecology 49, 6 (2014). [PubMed] [Google Scholar]
- Rizk N., Rooshenas A. A, E, F. & Shaat N. The Associations of Transcription Factor 7-like 2 [TCF7L2] Gene with Gestational Diabetes Mellitus in State of Qatar. Bloomsbury Qatar Foundation Journals. Available from: http://www.qscience.com/doi/pdfplus/ 10.5339/qfarf.2011.bmp8. (2011). [DOI] [Google Scholar]
- Jin Y. W., Qu Y. J., Wang H., Bai J. L. & Song F. Limitation of PCR-RFLP method for the detection of genetic mutations in spinal muscular atrophy. Chinese journal of Medical Genetics 29, 4 (2012). [DOI] [PubMed] [Google Scholar]
- Pulvirenti A., Solieri L., De Vero L. & Giudici P. Limitations on the use of polymerase chain reaction–restriction fragment length polymorphism analysis of the rDNA NTS2 region for the taxonomic classification of the species Saccharomyces cerevisiae. Can J Microbiol 51, 759–764, doi: 10.1139/w05-062 (2005). [DOI] [PubMed] [Google Scholar]
- Kwak S. H. et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61, 531–541, doi: 10.2337/db11-1034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freathy R. M. et al. Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study: common genetic variants in GCK and TCF7L2 are associated with fasting and postchallenge glucose levels in pregnancy and with the new consensus definition of gestational diabetes mellitus from the International Association of Diabetes and Pregnancy Study Groups. Diabetes 59, 2682–2689, doi: 10.2337/db10-0177 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.