Abstract
Ruminant animals contribute significantly to the global value of agriculture and rely on a complex microbial community for efficient digestion. However, little is known of how this microbial-host relationship develops and is maintained. To begin to address this, we have determined the ability of three Bifidobacterium species isolated from the faeces of newborn calves to grow on carbohydrates typical of a newborn ruminant diet. Genome sequences have been determined for these bacteria with analysis of the genomes providing insights into the host association and identification of several genes that may mediate interactions with the ruminant gastrointestinal tract. The present study provides a starting point from which we can define the role of potential beneficial microbes in the nutrition of young ruminants and begin to influence the interactions between the microbiota and the host. The differences observed in genomic content hint at niche partitioning among the bifidobacterial species analysed and the different strategies they employ to successfully adapt to this habitat.
Ruminant animals such as cattle and sheep contribute significantly to the global value of agriculture by supporting the livelihoods and food security of almost a billion people worldwide1. Ruminants are foregut fermenters that have evolved an efficient digestive system, which relies on a complex microbial community to ferment plant fibre and provide fermentation end-products and other nutrients for growth of the animal2. This reliance develops at birth where vertical transmission of microbes from the mother is considered a pivotal route for microbiota establishment in newborns3. Currently, little is known of the microbial-host interactions that occur during a ruminants’ early life and whether these interactions influence the lifetime performance and health of the animal.
Newborn ruminants (pre-ruminants) have a physically and metabolically underdeveloped rumen (fore-stomach) which means they are naturally dependent on milk during their early stages of life, and gradually transition and adapt to solid feed during which time the rumen develops4. The provision of milk to the neonate has considerable implications in the development of the infant intestinal microbiota. Milk selects for a highly adapted intestinal microbiota, dominated by bifidobacteria. Several constituents of milk such as oligosaccharides and glycoconjugates are known to selectively enrich for bifidobacteria5. These microbes have been shown to have a versatile and important role in human infant gut development ranging from stimulation and maintenance of the intestinal mucosal barrier and its immune response, to prevention of the attachment of pathogens and production of a range of beneficial metabolic substrates6,7,8. However, the on-farm management of pre-ruminants, particularly dairy calves, has traditionally focused on separating the newborn from their dams and restricting the amount of milk, or milk replacer, offered in order to encourage solid feed intake and accelerate weaning and rumen development4. This means calves do not have extended access to the nutritional and additional benefits that milk provides, and it remains to be seen whether this strategy has a negative impact on the animal’s development and subsequent performance. Recent research has shown that increased nutrient intake from milk, or milk replacer, during the pre-weaning period positively impacts lactation milk yield and lifetime performance of the animal9.
Pre-ruminants are often susceptible to a number of microbial pathogens during their first months of life which can severely affect growth efficiency and overall productivity4. Use of Bifidobacterium strains as direct-fed microbials (DFMs) are an attractive possibility that may provide beneficial effects for young livestock during a time when the risks of morbidity and mortality are high. There are several studies describing the isolation of bifidobacteria from young ruminants and some limited reports on their use as probiotics, which highlight their influence on immune function and their ability to reduce the incidence of diarrhea in young calves10,11. However, there is no research investigating the role of ruminant-derived bifidobacteria on ruminant gut development or whether there are beneficial microbial interactions occurring within the host ruminant during this early life phase. To begin to address these questions we have sequenced the genomes of three Bifidobacterium species isolated from the faeces of newborn calves and examined the ability of these organisms to grow on specific carbohydrates typical of the newborn ruminant diet.
Results and Discussion
Phylogenetic analysis
Three strains of Bifidobacterium were isolated from freshly expelled calf faecal samples during the milk-feeding period (1 to 5 days old). A phylogenetic analysis based on the 16S rRNA gene sequences of the type strains of the recognized species of the genus Bifidobacterium and the isolated strains from this study was performed (Fig. S1). This analysis indicated that the isolated strains were members of three different Bifidobacterium species; Bifidobacterium choerinum AGR2158, Bifidobacterium pseudolongum subsp. globosum AGR2145, and Bifidobacterium longum subsp. suis AGR2137. AGR2145 and AGR2158 were isolated from the same animal. B. choerinum was originally described as a species by Scardovi et al.12. It is considered an autochthonous Bifidobacterium species of the pig that is well adapted to the gut of pre-weaned piglets. However, B. choerinum has also been isolated from young ruminant faeces and from sewage10,12,13. Members of the B. pseudolongum species are typically isolated from the gastrointestinal tract with the subspecies B. pseudolongum subsp. globosum mainly found in the rumen and intestine of ruminants14. B. longum subsp. suis was defined as a subspecies by Mattarelli et al.15 alongside the two other subspecies B. longum subsp. infantis and B. longum subsp. longum. B. longum subsp. suis is commonly isolated from the faeces of piglets but has also been isolated from the faeces of young ruminants13. While members of the Bifidobacterium genus can be readily cultured from young ruminant sources, knowledge of their apparent abundance in the microbiome of the young ruminant has been limited. This has been a consequence of selected DNA extraction techniques, inadequate primer choice or a combination of both as evidenced by analysis of the human infant gut microbiota16,17. The strains isolated in this study are phylogenetically closely related to the Bifidobacterium lineages (B. longum, B. animalis, B. breve,) which are used and/or have high potential as probiotics in human infants18 (Fig. S1). This highlights their possible application as beneficial organisms for the development of the intestinal microbiota of pre-ruminants.
General genome characteristics
The genome features of the isolated strains and their comparison against relevant type strains of the genus Bifidobacterium are listed in Table 1. As expected, based on their phylogeny (Fig. S1) the genome sequences of the calf isolates AGR2145 and AGR2158 are more similar to each other than to AGR2137. Both genomes display a higher guanine and cytosine (G + C) content, 63.30% and 64.10% respectively, than that of the AGR2137 genome, 59.86%. These G + C percentages are consistent with what has been observed for the relevant type strains (Table 1)19. Additionally, both AGR2145 and AGR2158 display a more similar codon and amino acid usage profile than that of AGR2137 (Fig. S2). The three genomes range in size between 1.9 and 2.2 Mb which is within the range (1.73–3.25 Mb) reported for genome sequences of the genus Bifidobacterium19. Functional assignment of the calf bifidobacterial ORFeomes based on the Clusters of Orthologous Groups (COG) database indicate a functional designation for, on average, 67% of the total number of predicted protein-coding genes for each genome. A comparative analysis of the ORFeomes indicates a conserved set of 1,142 gene families shared between the three Bifidobacterium species examined. (Fig. 1a). COG categories were assigned to these 1,142 gene families. This analysis highlighted the comparable metabolism between the three calf isolates with 35% of the conserved gene families being assigned to metabolism, 23% to information storage and processing, 15% to cellular processes and processing and 20% to the poorly characterized category (Fig. S3). Genes unique to each of the calf genomes were examined and are discussed throughout the text (Figs 1 and S3). These genes may serve as targets for functional studies to elucidate the different strategies each of the calf bifidobacterial species employ to successfully adapt, co-exist, and survive within the pre-ruminant gastrointestinal tract. ORFeomes of the relevant type (sub) species were also compared against each of the relevant calf ORFeomes (Fig. 1b–d) to identify genes distinct to the calf isolates. As expected, the majority of these distinct genes (on average 67%) were assigned no functional annotation. Those assigned a functional category are summarised in Fig. S4 with the COG carbohydrate transport and metabolism category being the most abundant for the genomes of AGR2145 and AGR2137.
Table 1. General features of the Bifidobacterium strains used in this study.
| Organism | B. longum subsp. suis | B. longum subsp. suis | B. longum subsp. infantis | B. longum subsp. longum | B. pseudolongum subsp. globosum | B. pseudolongum subsp. globosum | B. pseudolongum subsp. pseudolongum | B. choerinum | B. choerinum |
|---|---|---|---|---|---|---|---|---|---|
| Strain | AGR2137 | LMG21814T | ATCC15697T | LMG13197T | AGR2145 | LMG11569T | LMG11571T | AGR2158 | LMG10510T |
| Isolation source | Calf faeces | Pig faeces | Infant intestine | Adult intestine | Calf faeces | Bovine rumen | Pig faeces | Calf faeces | Piglet faeces |
| Status | Draft | Draft | Complete | Draft | Draft | Draft | Draft | Draft | Draft |
| Number of DNA scaffolds | 49 | 36 | 1 | 8 | 30 | 26 | 11 | 19 | 20 |
| Genome size (bp) | 2,270,377 | 2,335,832 | 2,832,748 | 2,384,703 | 1,992,204 | 1,935,255 | 1,898,683 | 2,190,080 | 2.096,121 |
| G + C content (%) | 59.86 | 59.96 | 59.86 | 60.33 | 63.30 | 63.39 | 63.06 | 64.10 | 65.53 |
| Number of protein-coding genes | 1,897 | 1,999 | 2,486 | 1,924 | 1,654 | 1,598 | 1,551 | 1742 | 1,684 |
| Number of genes assigned to COGs | 1,250 | 1,253 | 1,396 | 1,235 | 1,119 | 1,089 | 1,072 | 1,143 | 1107 |
| Number of genes with signal peptides | 85 | 57 | 133 | 67 | 88 | 48 | 48 | 100 | 52 |
| Number of genes with transmembrane helices | 497 | 517 | 627 | 509 | 441 | 417 | 416 | 467 | 448 |
| Genbank accession no. | ATWX00000000 | JGZA00000000 | CP001095 | JGYZ00000000 | ATWW00000000 | JGZG00000000 | JGZH00000000 | AUJM00000000.1 | JGYU00000000 |
Figure 1. Genomic diversity of the calf bifidobacterial species.

Panel a displays a Venn diagram of homologues shared between the three calf genome sequences. Panel b, c, & d show a Venn diagram representation of each of the respective calf genome sequences compared against the relevant type strain from the Bifidobacterium genus.
The availability of the genome sequences of members of the genus Bifidobacterium allowed a Functional Genome Distribution (FGD) tree to be constructed20 (Fig. 2). In contrast to an evolutionary phylogeny, FGD analyzes the functional relationship between microbes based on their predicted ORFeomes. This approach takes into account genotype adaptations which might render organisms more similar to each other than what their respective evolutionary heritage would indicate. Based on this analysis, the three genomes examined cluster closely with their respective type strains. The FGD tree is largely comparable with the phylogenetic clusters identified in the supertree described for the genus of Bifidobacterium19. Both AGR2145 and AGR2158 fall within the B. pseudolongum phylogenetic group and AGR2137 within the B. longum phylogenetic group. Analysis of potential horizontal gene transfer (HGT) events in the calf genome sequences identified percentages of alien genes, compared to the total number of ORFs. This analysis revealed 13.4% (AGR2137), 9.1% (AGR2145) and 7.8% (AGR21258) respectively, of the calf ORFeomes are predicted to have undergone HGT. Predicting the donors of these putative alien genes (Table S1) indicated a preferential origin from members of the Alphaproteobacteria for AGR2145 and AGR2158 while other members of the Actinobacteria were the preferential origin for AGR2137. Analysis of the COG assignment of these predicted alien genes, excluding genes with no known function, is summarised in Fig. S5.
Figure 2. Functional genome distribution of the Bifidobacterium genus.
All strains are colour-coded according to their isolation source and species with a use and/or high potential as probiotics in human infants, as detailed by Di Gioia et al.18, are underlined.
Genomic features unique to the B. choerinum AGR2158 genome include a putative type VII/WXG100 secretion system and a prophage. A cluster of 15 open reading frames (ORFs) was identified in the genome of AGR2158 that represents a type VII/WXG100 secretion system (Fig. S6). This denotes the first report of such as system in the Bifidobacterium genus. However, comparative analysis has revealed a similar cluster of genes can also be found in the genome of B. dentium Bd1 (Fig. S6, Genbank accession number CP001750). Type VII/WXG100 secretion systems are known to be present in several Gram-positive organisms. They were initially identified in Mycobacterium tuberculosis and although type VII/WXG100 secretion systems are thought to play a role in bacterial pathogenesis in some organisms21 they have also been associated with other cellular functions22. Their role in bifidobacteria remains unknown. A 41 kb prophage (G606DRAFT_1765-1820) has been identified in the genome of AGR2158 (Fig. S7) which is different from the 56 kb prophage identified in the type strain of this species (LMG 10510)23. The modular organization of the AGR2158 prophage is similar to that reported for other Bifidobacterium prophages24,25.
Genomics of carbohydrate utilization
Carbohydrate metabolism and transport is a major activity for bifidobacteria26,27 and genes involved in this functional category make up 9.5% (AGR2158), 11.8% (AGR2145) and 12.4% (AGR2137) of the calf Bifidobacterium genomes. The ability of the three strains to grow on various carbohydrates is shown in Table 2, and the genes predicted to encode proteins associated with carbohydrate metabolism and transport are shown in Table S2.
Table 2. Carbohydrate utilisation by the three Bifidobacterium strains.
| Carbohydrate | AGR2137 | AGR2145 | AGR2158 |
|---|---|---|---|
| Arabinose | + | + | − |
| Fructose | + | − | − |
| Galactose | + | + | + |
| Glucose | + | + | + |
| N-acetyl glucosamine | − | − | − |
| Xylose | + | + | − |
| Cellobiose | − | − | − |
| Lactose | + | + | + |
| Maltose | + | + | + |
| Melibiose | + | + | + |
| Sucrose | + | − | + |
| 3′-sialyl lactose | − | − | − |
| 6′-sialyl lactose | − | − | − |
| Raffinose | + | + | + |
| Fructo-oligosaccharides | + | + | + |
| Galacto-oligosaccharides | + | + | + |
| Inulin | − | − | − |
| Xylo-oligosaccharides | − | + | + |
Milk and host-derived carbohydrates
Studies of bifidobacteria from human infants have shown how strains of B. bifidum and B. longum subsp. infantis are able to metabolize human milk oligosaccharides28 and the glycan component of host mucins29. It is expected that bifidobacteria from young ruminants will be exposed to comparable nutrient sources, but this has not been investigated. Bovine milk and colostrum contain a diversity of free oligosaccharides, but their concentration is much lower than in human milk23. Bovine milk also contains several glycoproteins which are substituted with a range of N- and O-linked glycans30. The carbohydrate composition of calf intestinal mucin has been determined31 and is made up predominantly of galactose, N-acetylglucosamine, and N-acetylgalactosamine, with lower amounts of fucose, mannose and sialic acids. Ruminants also secrete large amounts of saliva which contains numerous N-linked glycoproteins32 but their carbohydrate composition is not known.
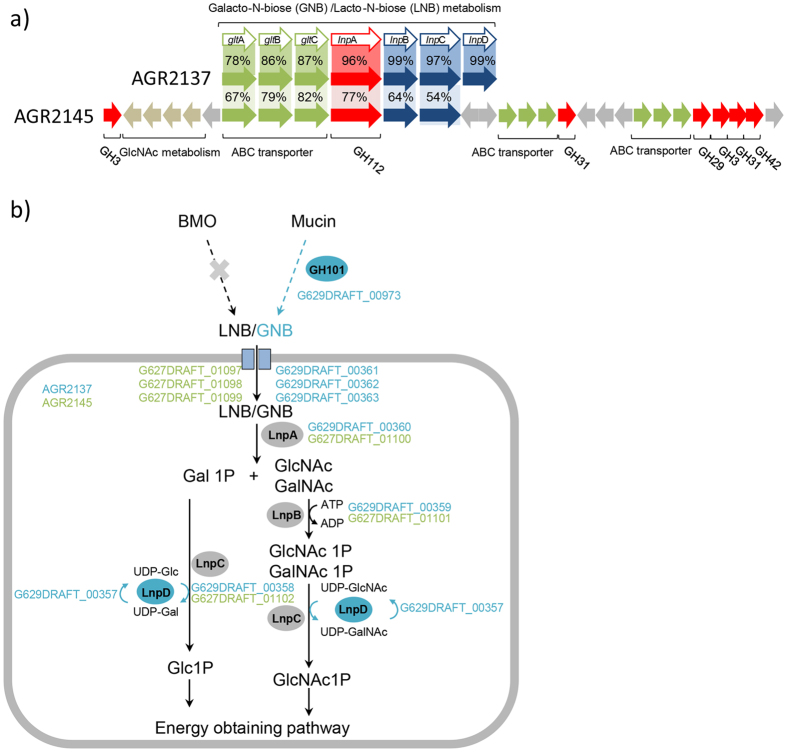
Lactose is the predominant carbohydrate in bovine milk and is able to be fermented by the three calf Bifidobacterium strains investigated (Table 2). Most bovine milk oligosaccharides are sialylated, but none of the calf strains were able to ferment 3′- or 6′-sialyl lactose, or had sialidase genes in their genome sequence. However, other bacteria in the gut may be able to initiate oligosaccharide breakdown and thus cross-feed the bifidobacterial strains33. Other bacteria isolated from the same samples as the three Bifidobacterium strains have also been genome sequenced and two of these bacteria (Ruminococcus gnavus AGR2154 and Dorea longicatena AGR2136) encode sialidases. The metabolism of lacto-N-biose (LNB) derived from milk oligosaccharides and galacto-N-biose (GNB) derived from mucin occurs via a common pathway and has been well studied in infant-derived bifidobacteria. AGR2137 encodes the gene cluster (Fig. 3a,b) for the complete LNB/GNB metabolism pathway as described in B. longum34. B. pseudolongum strains have previously been reported to ferment lacto-N-biose35 and AGR2145 encodes a similar gene cluster to that found in B. longum strains (G629DRAFT_00357-00363). However, in AGR2145 the galE gene homolog (lnpD) is missing and the LNB/GNB genes form part of a novel 29 gene cluster ((G627DRAFT_01091-01120) which includes genes for two other ABC transporters and several glycoside hydrolases (Fig. 2a,b, Table S2). This gene cluster is not present in the genome of the type strain of B. pseudolongum subsp. globosum (DSM 20092T). The LNB/GNB metabolism genes are not part of the AGR2158 genome.
Figure 3. The galacto-N-biose (GNB)/lacto-N-biose (LNB) pathway found in bifidobacteria.
(a) The gene cluster encoding the pathway was adapted from Kitaoka et al.28. Using the B. longum JCM1217 gene cluster (BLLJ_1620-1626)) as a reference (Genbank accession number NC_015067) the corresponding gene clusters are shown for AGR2137 (G629DRAFT_00357-00363) and AGR2145 (G627DRAFT_01097-01102). BLASTP percent identities are shown. Only genes predicted to be involved in the LNB/GNB gene cluster are shown for AGR2137 while additional carbohydrate utilising genes surrounding the LNB/GNB gene cluster are shown for AGR2145 (G627DRAFT_01091-01120). This diagram is not drawn to scale. (b) Schematic representation of the pathway. GalNAc1P, N-acetylgalactosamine 1 phosphate; Gal1P, galactose 1-phosphate; GlcNAc1P, N-acetylglucosamine 1 phosphate, Glc1P, glucose 1-phosphate. Locus tags of genes from AGR2137 are shown in blue, while those from AGR2145 are shown in green.
AGR2137 encodes homologues of glycoside hydrolases that have been shown to be involved in the metabolism of oligosaccharides derived from host mucins (Table S2). These include a GH101 family endo-alpha-N-acetylgalactosaminidase36 that cleaves GNB from gastroduodenal mucin, and an GH129 family alpha-N-acetylgalactosaminidase37 that removes N-acetylgalactosamine from intestinal mucin.
Plant carbohydrates
The diet of forage fed ruminants is rich in structural carbohydrates including cellulose, hemicellulose and pectin, storage polysaccharides such as starch and water-soluble fructans and raffinose family oligosaccharides. Consequently, young ruminants are exposed to a variety of plant carbohydrates as they transition to having a fully functional rumen.
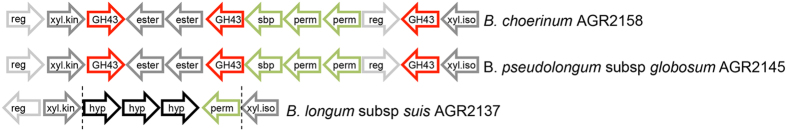
Hemicellulose is a major component of plant cell walls and its degradation results in the accumulation of xylo- and arabinoxylo-oligosaccharides (XOS, AXOS) that are selectively fermented by bifidobacteria. An AXOS utilization locus, bounded by xylose isomerase (xylA) and xylulokinase (xylB) genes, has been identified by transcriptional analysis in B. animalis subsp. lactis38 and a homologue of this locus is found in both AGR2145 and AGR2158 (Fig. 4). This locus includes the gene for a highly conserved ABC transporter substrate-binding protein which mediates the uptake of a range of XOS and AXOS oligomers39 (Table S2). In AGR2137 although the xylA and xylB genes are present, the other AXOS utilization locus genes have been lost (Fig. 4) and thus this strain is unable to ferment XOS (Table 2). Two of the calf Bifidobacterium strains are also able to grow on arabinose and xylose (Table 2). AGR2137 has genes for a pentose ABC transporter (G629DRAFT_00299-00304), while AGR2145 has a pentose ABC transporter as part of a larger gene cluster including a GH51 family alpha-L-arabinofuranosidase and genes for arabinose metabolism (G627DRAFT_00515-0000526). The pentose transporter has been lost from AGR2158 although a fragment of the permease gene remains (G606DRAFT_00824).
Figure 4. Genomic content and organization of XOS utilisation gene clusters.
Gene functions are coloured as follows: glycoside hydrolases (GH) in red, carbohydrate esterases (ester), xylose isomerase (xyl.iso) and xylulose kinase (xul.kin) all in dark grey, ABC transporter substrate binding proteins (SBPs) and permeases (perm) in green, transcriptional regulators (reg) in light grey, hypothetical proteins (hypo) in black.
AGR2137 also encodes the gene cluster that mediates the utilization of galactans derived from pectin in several B. breve and B. longum strains (G629DRAFT_01106-01111)40. This locus contains the gene for a secreted GH53 family arabinogalactan endo-1,4-beta-galactosidase that acts on galacto-oligosaccharides (GOS) and releases galactotriose which is transported into the cell and further broken down by a GH42 family beta-galactosidase40.
B. pseudolongum strains have been shown to ferment starch, amylopectin and pullulan41, and both AGR2145 and AGR2158 encode several signal peptide-containing GH13 family glycoside hydrolases which may mediate the breakdown of dextran, pullulan, starch and other glucans. All three strains can ferment maltose (Table 2) and each encodes a maltose ABC transporter (COG2182) associated with GH13 and GH77 glycoside hydrolases (Table S2). The three calf strains ferment fructo-oligosaccharides (FOS, Table 2) and encode a three gene operon similar to that shown to be involved in fructo-oligosaccharide breakdown in B. breve and B. longum42. A sucrose utilization operon containing a GH13 sucrose phosphorylase/transglycosylase has been characterized in B. longum43 and is present in AGR2137 and AGR2158. However, AGR2145 is unable to use sucrose (Table 2) and the operon in this strain (G627DRAFT_00811-00815) contains an inserted toxin-antitoxin gene cassette (Table S2).
Melibiose and raffinose are found in a wide variety of plants and a cluster of genes containing the alpha-galactosidase necessary to metabolize these sugars has been characterized in bifidobacteria44. AGR2145 and AGR2158 have a gene cluster similar to that found in B. animalis strains37,44, with genes encoding two GH36 and one GH13 glycoside hydrolases, while AGR2137 has a different gene arrangement with only one GH36.
Microbial-host interactions
During its lifetime, a ruminant will continuously encounter microorganisms that range from those essential for health and productivity to those causing disease, hampering productivity. Consequently, the ruminant’s immune system must learn to distinguish between beneficial and pathogenic microbes, and understanding how this is mediated will be important to improve the lifetime performance of the animal.
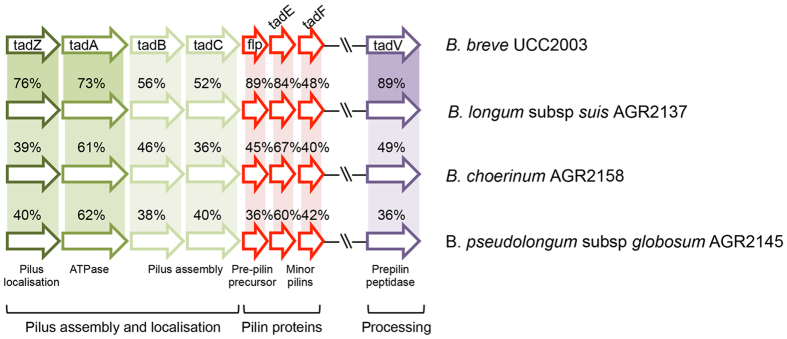
Microbial surface appendages are considered important for microbial-host interactions, and several species of bifidobacteria are known to produce pili-like structures45. Research with B. breve UCC2003 identified a type IVb tight adherence (Tad) pilus-encoding gene cluster which is essential for efficient in vivo murine gut colonization, and supports the concept of a pili-mediated host colonization and persistence mechanism for bifidobacteria8. The genomes of the three calf strains all contain a homologous tad locus (Fig. 5) suggesting that type IVb pili may play a role as a host colonization factor in the gastrointestinal tract of young ruminants. Members of the Bifidobacteriaceae are also known to elaborate pili via a sortase-assembly mechanism. Sortase dependent pili of B. bifidum PRL2010 have been shown to modulate bacterium-host interactions46,47. Analysis of the genomes of AGR2158 and AGR2145 has identified two sortases in each genome that cluster with genes encoding proteins with predicted cell wall sorting signals, and which may represent pilus loci (Fig. S7). Further investigation will be necessary to determine any involvement in host interactions.
Figure 5. Schematic representation of the tad locus.
Gene names and the functions of the encoded proteins are indicated. Figure adapted from O’Connell Motherway et al.8.
Exopolysaccharides (EPS) are considered surface molecules that act as mediators of cross-talk between microbes and their host. Several beneficial activities have been attributed to EPS synthesized by Bifidobacterium species including modulation of the host’s immune system, formation of a protective physical barrier, modulation of the intestinal microbiota, and antagonism against pathogens48. The genomes of AGR2145 and AGR2158 each encode two EPS gene clusters (Table S3) which correspond to the relatively conserved eps3 and eps4 gene clusters which have been described as being common to strains of B. pseudolongum, B. choerinum and B. animalis isolated from the gastrointestinal tract of animals49. The EPS genes were not assembled in the AGR2137 genome (Table S3), but rhamnose biosynthesis genes which are a common feature of bifidobacterial EPS gene clusters are present49. Future work will be required to confirm EPS production in the strains analysed.
Bile salts play an important role in the host’s defence against ingested microorganisms50, and constitute a physiological barrier, which bifidobacteria need to overcome in order to survive and colonize within the gastrointestinal tract of the young ruminant. Specific bile resistance mechanisms reported in bifidobacteria include bile salt hydrolysis51 and bile efflux52,53. Bile salt hydrolases (BSHs) also referred to as cholylglycine hydrolases catalyse the hydrolysis of glycine- and/or taurine-conjugated bile salts into amino acid residues and free bile acids. The genomes of all three calf isolates contain a bsh gene (AGR2137, G629DRAFT_01360; AGR2145, G627_00122; AGR2158, G606DRAFT_1516). Genetic organisation around the bsh gene is analogous to that reported for B. animalis (AGR2145 and AGR2158) or B. longum51 (AGR2137). Efflux pumps are a common detoxification mechanism employed by bacteria and their role in bile tolerance has been demonstrated for B. breve and B. longum52,53. The AGR2137 genome has a homologous bile efflux pump gene (G629DRAFT_00578) and transporter gene (G629DRAFT_00581) suggesting a common mechanism of bile resistance in these species.
Fukuda et al.7 identified a ‘probiotic’ carbohydrate ABC transporter in three Bifidobacterium strains that correlated with the ability of these strains to protect against E. coli O157-induced death in mice. Expression of these transporter genes was highly induced by fructose, and it was proposed that fructose metabolism enables these cells to produce sufficient acetate to inhibit E. coli. This ABC transporter is characterized by the presence of genes assigned to functional categories COG1129, COG1172 and COG1879. The same gene cluster is found in AGR2137 (G629DRAFT_00719-00722), and this was the only calf strain able to use fructose (Table 2).
Conclusion
This study has examined the genomes of three species of Bifidobacterium isolated from newborn calves, and focused on the genetic strategies these bacteria use for carbohydrate metabolism and host colonization and persistence. Dietary carbohydrates and host-derived glycans are the main energy sources for bifidobacteria and all three strains have the ability to utilize a range of carbohydrates that are likely to be available in the calf gastrointestinal environment. However, B. longum subsp. suis AGR2137 seems better adapted to the utilization of host glycans, while B. pseudolongum subp. globosum AGR2145 may have a more developed repertoire of genes allowing for the utilization of milk oligosaccharides, and B. choerinum AGR2158 looks to be restricted to the utilisation of plant polysaccharides. These differences in carbohydrate preference, and the presence of additional carbohydrate metabolism genes (such as sialidases) in other bacteria from the same environment, highlight the cooperative nature of polysaccharide metabolism and cross-feeding in gut environments. Analysis of the genomes has also provided insights into the association of these bifidobacterial species with their hosts and has resulted in the identification of several genes with a potential role in these processes. AGR2137 appears to be more similar in its predicted host-microbial processes to those described for the human infant related bifidobacterial species. The differences in genomic content hint at niche partitioning among the three different bifidobacteria analysed and give a first indication of the different strategies they employ to successfully colonize and survive in the pre-ruminant gastrointestinal habitat.
Methods
Isolation and 16S rRNA gene sequencing
Freshly voided faecal samples were taken from calves (1 to 3 days old) using a sterile wooden spatula and placed in a pre-weighed plastic container. Samples were weighed again, resuspended in 1 ml MS buffer and serially diluted in MS buffer under anaerobic conditions. MS buffer contained 40 ml mineral solution 1 (6 g/L K2HPO4), 40 ml mineral solution 2 (6 g/L KH2PO4, 6 g/L (NH4)2SO4, 12 g/L NaCl, 2.55 g/L MgSO4.7H2O, 1.69 g/L CaCl2.2H2O), 500 μl resazurin (0.1% w/v) and distilled water to 1 L. The solution was boiled and cooled under O2-free N2 and pH adjusted to 6.9, and then 0.5 g cysteine.HCl and 3 g Na2CO3 were added. The serially diluted faecal suspensions (90 μl volumes) were plated in duplicate onto bifidobacterial selective medium made according to the method of Leuscher et al.54 and incubated at 37 °C in an anaerobic glove box (95% CO2, 5% H2). Plates were inspected after 24 and 48 hrs and individual colonies were purified by repeated streaking onto fresh agar plates. Colony purity was confirmed by Gram stain and microscopic examination. Bacterial strains were stored at −85 °C on de Man, Rogosa, Sharpe (MRS, Oxoid) or Reinorced Clostridial Medium (RCM, Oxoid) agar slopes.
Full length amplification and sequencing of the 16S rRNA gene from the calf bifidobacterial isolates was undertaken using the bifF11 (5′-AGG GTT CGA TTC TGG CTC AGG), bifF782 (5′-GAT TAG ATA CCC TGG TAG TCC), bifR711 (5′-TTC CCG ATA TCT ACA CAT TCC), and bifR1543 (5′-AGG TGA TCC AGC CGC ACC TTC C) primers, designed to match conserved regions of the bifidobacterial full length 16S rRNA gene determined using an alignment of twenty diverse species generated in MEGA655. PCR amplicons were generated using the KAPA Readymix HiFi HotStart ReadyMix according to the manufacturer’s instructions. After column purification (Qiagen PCR purification kit) PCR products were Sanger sequenced (Massey Genome Service, Massey University, New Zealand) and full length contiguous sequences assembled in Geneious (version 7.0).
Carbohydrate growth assays
Bifidobacterial cultures were routinely cultured in MRS broth supplemented with 0.05% (w/v/) cysteine HCl. MRS media was dissolved in distilled water, boiled and then cooled on ice under a stream of O2-free CO2. Once cooled the cysteine HCl was added and aliquots were dispensed into Hungate tubes gassed with CO2. For carbohydrate utilisation a semi-synthetic medium based on MRS was used that was supplemented with carbohydrate (1% w/v) immediately before inoculation. The carbohydrates added were glucose, fructose, galactose, xylose, arabinose, N-acetyl galactosamine (Sigma), sucrose, cellobiose, lactose, melibiose (Sigma), maltose, 3′ or 6′ sialyl lactose (Carbosynth, UK), raffinose (Sigma), inulin (average DP ≥23, Orafti HP, Beneo Orafti, Belgium), galacto-oligosaccharide (Neo Cremar Co., Ltd., South Korea), fructo-oligosaccharide (DP 2–8, Orafti P95, Beneo Orafti, Belgium), xylo-oligosaccharide (DP 2–8, Shandong Longlive Biotechnology Co., Ltd., China). Each carbohydrate was made as a 10% (w/v/) stock solution in distilled water, and sterilised by filtration through a 0.22 μm syringe filter into a rubber-stoppered serum bottle that had been flushed with O2-free N2 and autoclaved. Stationary phase bifidobacterial cultures (100 μl) were inoculated aseptically into carbohydrate-supplemented broth using a sterile syringe filled with O2-free CO2 and the optical density at 600nm recorded immediately. Carbohydrate utilisation was assessed in triplicate with A600 measurements recorded over a period of 24–36 hours. Without carbohydrate supplementation, the semi-synthetic medium was unable to sustain bacterial growth above an A600 of ~0.2.
Genomic DNA preparation
Cultures of Bifidobacterium strains AGR2137, AGR2145 and AGR2158 were grown in Reinforced Clostridial Medium (RCM) at 37 °C under anaerobic conditions. DNA for sequencing was obtained using the Qiagen Genomic-tip kit and following the manufacturer’s instructions for the 500/g size extraction.
Genome sequencing and sequence analysis
The draft genomes of strains AGR2145, AGR2158 and AGR2137 were generated at the Joint Genome Institute using Illumina technology. For each of the genomes, an Illumina standard shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform. Annotation and analysis of the genomes was performed using a combination of the Integrated Microbial Genomes (IMG) Expert Review system56 and a manual curation using a GAMOLA/ARTEMIS software suite as described previously57. To identify conserved gene families, the program OrthoMCL (v1.4) was used using default parameters58. To identify genes that may have been acquired by horizontal gene transfer (HGT), the results from Alienhunter59 and COLOMBO (v3.8) implemented with the program SIGI-HMM60 were merged. Alienhunter was run with default parameters, while COLOMBO was run with a sensitivity value of 0.4. To generate the FGD tree, draft and completed Bifidobacterium (type strains) genomes19 in FASTA format were downloaded from the National Center for Biotechnology (NCBI). Draft genomes were concatenated using a non-bleeding spacer sequence and gene models were created for all genomes using an updated version of GAMOLA57. Resulting Genbank files were then subjected to a Functional Genome Distribution (FGD) analysis20 and the calculated distance matrix was imported into Molecular Evolutionary Genetics Analysis version 6 (MEGA6)55. The functional distribution was visualized using the unweighted pair group method with arithmetic mean (UPGMA).
Additional Information
How to cite this article: Kelly, W. J. et al. Genomic analysis of three Bifidobacterium species isolated from the calf gastrointestinal tract. Sci. Rep. 6, 30768; doi: 10.1038/srep30768 (2016).
Supplementary Material
Acknowledgments
This work was supported by AgResearch PreSeed Contract No. A19573. The Hungate1000 project is funded by the New Zealand Government in support of the Livestock Research Group of the Global Research Alliance on Agricultural Greenhouse Gases. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. The authors would like to thank Stephen and Mary Barr for access to their farm and support of this research.
Footnotes
Author Contributions This study was designed by W.J.K., A.L.C. and S.C.L. Experimental research was carried out by ACL, S.C.La, R.P., K.H.T., D.E.O. and S.C.L. Data analysis was performed by W.J.K., A.LC., E.A., N.S. and T.W. W.J.K., A.L.C. and S.C.L. wrote the manuscripts. All authors approved the manuscript before submission.
References
- FAO. The second report on the state of the World’s Animal Genetic Resources for Food and Agriculture (eds Scherf, B.D. & Pilling, D.) FAO Commission on Genetic Resources for Food and Agriculture Assessments, Rome (2015) Available at http://www.fao.org/3/a-i4787e/index.html (Accessed: 19th May 2016).
- Clauss M., Hume I. D. & Hummel J. Evolutionary adaptations of ruminants and their potential relevance for modern production systems. Animal 4, 979–992 (2010). [DOI] [PubMed] [Google Scholar]
- Milani C. et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 81, 7078–7086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A. et al. Invited review: Transitioning from milk to solid feed in dairy heifers. J. Dairy Sci. 99, 885–902 (2016). [DOI] [PubMed] [Google Scholar]
- Pacheco A. R., Barile D., Underwood M. A. & Mills D. A. The impact of the milk glycobiome on the neonate gut microbiota. Annu. Rev. Anim. Biosci. 3, 419–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy S. C., Higgins D. G., Fitzgerald G. F. & van Sinderen D. Getting better with bifidobacteria. J. Appl. Microbiol. 98, 1303–1315 (2005). [DOI] [PubMed] [Google Scholar]
- Fukuda S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011). [DOI] [PubMed] [Google Scholar]
- O’Connell Motherway M. et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. USA 108, 11217–11222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberon F. & van Amburgh M. E. Lactation Biology Symposium: The effect of nutrient intake from milk or milk replacer of preweaned dairy calves on lactation milk yield as adults: a meta-analysis of current data. J. Anim. Sci. 91, 706–712 (2013). [DOI] [PubMed] [Google Scholar]
- Bunešová V., Vlková E., Killer J., Rada V. & Ročková S. Identification of Bifidobacterium strains from faeces of lambs. Small Ruminant Res. 105, 355–360 (2012). [Google Scholar]
- Signorini M. L. et al. Impact of probiotic administration on the health and fecal microbiota of young calves: a meta-analysis of randomized controlled trials of lactic acid bacteria. Res. Vet. Sci. 93, 250–258 (2012). [DOI] [PubMed] [Google Scholar]
- Scardovi V., Trovatelli L. D., Biavati B. & Zani G. Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucleic acid homology relationships. Int. J. Syst. Bacteriol. 29, 291–311 (1979). [Google Scholar]
- Vlková E. et al. Survival of Bifidobacteria administrated to calves. Folia. Microbiol. 55, 390–392 (2010). [DOI] [PubMed] [Google Scholar]
- Yaeshima T., Fujisawa T. & Mitsuoka T. Bifidobacterium globosum, subjective synonym of Bifidobacterium pseudolongum, and description of Bifidobacterium pseudolongum subsp. pseudolongum comb. nov. and Bifidobacterium pseudolongum subsp. globosum comb. nov. System Appl. Microbiol. 15, 380–385 (1992). [Google Scholar]
- Mattarelli P., Bonaparte C.,. Pot B. & Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int. J. Syst. Evol. Microbiol. 58, 767–772 (2008). [DOI] [PubMed] [Google Scholar]
- Walker A. W. et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C. et al. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS. Microbiol. Ecol. 90, 493–503 (2014). [DOI] [PubMed] [Google Scholar]
- Di Gioia D., Aloisio I., Mazzola G. & Biavati B. Bifidobacteria: their impact on gut microbiota composition and their applications as probiotics in infants. Appl. Microbiol. Biotechnol. 98, 563–577 (2014). [DOI] [PubMed] [Google Scholar]
- Milani. et al. Genomic encyclopaedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 80, 6290–6302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann E. Tracing lifestyle adaptation in prokaryotic genomes. Front Microbiol. 3, 48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah A. M. et al. Type VII secretion–mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891 (2007). [DOI] [PubMed] [Google Scholar]
- Huppert L. A. et al. The ESX system in Bacillus subtilis mediates protein secretion. PLoS One 9, e96267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht S. et al. A comparative study of free oligosaccharides in the milk of domestic animals. Br. J. Nutr. 111, 1313–1328 (2014). [DOI] [PubMed] [Google Scholar]
- Lugli G. A. et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol. doi: 10.1111/1462-2920.13154 (2015). [DOI] [PubMed] [Google Scholar]
- Ventura M. et al. Prophage-like elements in bifidobacteria: Insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl. Environ. Microbiol. 71, 8692–8705 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C. et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 82, 980–991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki et al. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int. J. Genomics 2015, 567809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv. Nutr. 3, 422S–429S (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F. et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA 107, 19514–19519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan N., Kane M., Joshi L. & Hickey R. M. Structural and functional characteristics of bovine milk protein glycosylation. Glycobiology 24, 220–236 (2014). [DOI] [PubMed] [Google Scholar]
- Montagne L., Toullec R. & Lallès J. P. Calf intestinal mucin: isolation, partial characterization, and measurement in ileal digesta with an enzyme-linked immunosorbent assay. J. Dairy Sci. 83, 507–517 (2000). [DOI] [PubMed] [Google Scholar]
- Ang C.-S. et al. Global survey of the bovine salivary proteome: integrating multidimensional prefractionation, targeted, and glycocapture strategies. J. Proteome Res. 10, 5059–5069 (2011). [DOI] [PubMed] [Google Scholar]
- Milani C. et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 5, 15782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto M. & Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl. Environ. Microbiol. 73, 6444–6449 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J.-Z. et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl. Environ. Microbiol. 76, 54–59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R. et al. Crystallographic and mutational analyses of substrate recognition of endo-α-acetylgalactosaminidase from Bifidobacterium longum. J. Biochem. 146, 389–398 (2009). [DOI] [PubMed] [Google Scholar]
- Kiyohara M. et al. α-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J. Biol. Chem. 287, 693–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. M. et al. Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 14, 312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejby M. et al. Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp. lactis Bl-04. Mol. Microbiol. 90, 1100–1112 (2013). [DOI] [PubMed] [Google Scholar]
- O’Connell Motherway M., Kinsella M., Fitzgerald G. F. & van Sinderen D. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb. Biotechnol. 6, 67–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S. M., Fitzgerald G. F. & van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72, 5289–5296 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F. et al. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl. Environ. Microbiol. 76, 3206–3219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. et al. Cloning and expression of sucrose phosphorylase gene from Bifidobacterium longum in E. coli and characterization of the recombinant enzyme. Biotechnol. Lett. 25, 1211–1217 (2003). [DOI] [PubMed] [Google Scholar]
- O’Connell K. J. et al. Metabolism of four α-glycosidic linkage-containing oligosaccharides by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 79, 6280–6292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroni E. et al. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb. Cell Fact. 10, Suppl 1 S16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F. et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. USA 110, 11151–11156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F. et al. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol. Lett. 357, 23–33 (2014). [DOI] [PubMed] [Google Scholar]
- Hidalgo-Cantabrana C. et al. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl. Environ. Microbiol. 80, 9–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario. et al. Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol. Ecol. 92, fiw056 (2016). [DOI] [PubMed] [Google Scholar]
- Begley M., Gahan C. G. & Hill C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 (2005). [DOI] [PubMed] [Google Scholar]
- Tanaka H., Hashiba H., Kok J. & Mierau I. Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 2502–2512 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueimonde M., Garrigues C., van Sinderen D., de los Reyes-Gavilán C. G. & Margolles A. Bile-inducible efflux transporter from Bifidobacterium longum NCC2705, conferring bile resistance. Appl. Environ. Microbiol. 75, 3153–3160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz L. et al. A bile-inducible membrane protein mediates bifidobacterial bile resistance. Microb. Biotechnol. 5, 523–535 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuscher R. G., Bew J., Simpson P., Ross P. R. & Stanton C. A collaborative study of a method for the enumeration of probiotic bifidobacteria in animal feed. Int. J. Food. Microbiol. 83, 161–170 (2003). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. Supporting community annotation and user collaboration in the integrated microbial genomes (IMG) system. BMC Genomics 17, 307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy S. C. et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5, e8926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G. S. & Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22, 2196–2203 (2006). [DOI] [PubMed] [Google Scholar]
- Waack. et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7, 142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.