Abstract
The ability to assemble molecules into supramolecular architectures of controllable size and symmetry is a long sought after goal of nanotechnology and material engineering. Proteins are particularly attractive for molecular assembly due to their inherent molecular recognition and self-assembly capabilities. Advances in the computational prediction of protein folding and quaternary assembly have enabled the design of proteins that self-assemble into complex yet predictable shapes. These protein nanostructures are opening new possibilities in biomaterials, metabolic engineering, molecular delivery, tissue engineering, and a plethora of nanomaterials. Images of protein constructs assembled from simpler structures draw comparison to characters of calligraphy. In both cases, elaborate designs emerge from basic subunits, resulting in the translation of form into function with a high degree of artistry.
Short abstract
Controlled self-assembly of proteins is being exploited in the emerging field of protein nanotechnology to create nanoscale structures. Simple protein subunits assembled into complex designs give rise to functional materials and devices.
Introduction
The intricate and ordered complexes that proteins adopt in nature is central to many biological processes, ranging from cellular scaffolding provided by cytoskeletal proteins to the encapsulation of nucleic acids in viral capsids. Exploiting this remarkable fidelity and precision in self-assembly is highly attractive for the fabrication of structurally defined materials with nanometer dimensions. Researchers have spent considerable effort attempting to mimic nature to sculpt proteins into structural templates and devices. Early attempts focused on repurposing naturally occurring protein nanostructures such as viral capsids.1 However, these top-down approaches are limited in terms of engineerability and versatility. Modern nanobiotechnology aims to build structures from the ground up, creating assemblies not found in nature, which can be functionalized and used in a diverse range of applications from nanoelectronics and energy, to biomedicine and the environment. Indeed, nascent efforts to control the assembly of proteins into precise shapes and patterns can be thought of in similar terms as the development of letters or characters to comprise an alphabet, with the former activity directed toward communication and the latter toward new technological capabilities. Both, however, reflect an intrinsic artistry.
As with all self-assembling systems, the final structure of a biopolymer, such as a protein, is encoded by interactions of the material’s components defined by their properties and order within the linear polymer.2 Therefore, the engineering of biopolymers seeks to exploit sequence–structure relationships to drive the folding and assembly of specific, well-ordered shapes. This has been strikingly illustrated by DNA nanotechnology,3 in which strands of DNA are designed to fold by complementary Watson–Crick base pairing into a variety of structures including lattices, tubes, tiles, and bricks.4,5 Moving beyond the creation of simple shapes, the functional use of DNA nanotechnology has been impressively demonstrated by the production of nanoscale cages for therapeutic drug encapsulation,6 molecular machines,7 and biocomputing nanorobots.8 However, real-world application of DNA nanostructures is in its infancy and requires expensive and laborious chemical synthesis that limits technological applications.9
The creation of protein nanostructures has lagged behind due to the greater complexity of their structures. Proteins are composed of amino acids that vary in their electrostatic charge and hydrophobicity, which results in difficult-to-predict cooperative and long-range interactions. While nature has had millennia of trial-and-error to evolve proteins to fold into specific shapes, the engineering of proteins has proven to be challenging due to incomplete understanding of how a sequence of amino acids determines a protein’s three-dimensional shape, and of how multiple proteins assemble into complex arrangements. The complexity of proteins, however, has advantages in terms of chemistry and molecular recognition, as well as architecture. Proteins are capable of performing catalytic reactions, of interacting with both organic and inorganic molecules, and of assembling into intricate structures, all of which are ideal attributes for advanced materials. Furthermore, mature technology exists for the efficient and economical production of recombinant proteins in a range of microbial hosts.10
Recent improvements in the prediction of protein folding11 and protein–protein interactions12,13 indicate that the design of protein nanostructures is becoming increasingly feasible. This has occurred in part through advances in computational power that enable increasingly sophisticated algorithms that accurately model protein structure. Critical to the success of this molecular modeling has been the crystallography-solved protein structures submitted to the Protein Data Bank (PDB). This constantly growing database of over 100,000 protein structures has also revealed that many proteins in nature are oligomeric in structure, and assemble either with themselves or with other proteins into complexes. Using a combination of computational design and exploitation of natural protein assemblies, researchers are seeking to create novel protein shapes. In many ways, this engineering of protein assembly is reminiscent of the masterful strokes of a calligrapher who joins simple lines into complex patterns (Figure 1). As with calligraphy, these protein materials are more than just the sum of their parts, as the specific dimensions and topology of the structures yield unique properties. Therefore, an ultimate goal of protein nanotechnology is the ability to reliably create specific protein characters without error, which can subsequently be functionalized and combined for diverse applications. Extending the calligraphy analogy one step further, much like combining meaningless letters into a meaningful narrative, a set of protein characters could create the potential for vast combinations that produce functions far beyond the properties of the characters themselves.
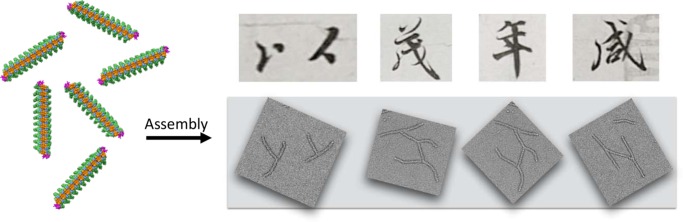
Figure 1.
Assembly of proteins into nanostructured shapes. Simple filamentous structures can be assembled together into aesthetically ordered patterns as shown by transmission electron microscopy. The interlocking shapes produced conjure the aesthetic of “protein calligraphy”, as shown by the comparison to characters of Chinese calligraphy.
Scripting Novel Protein Nanostructures
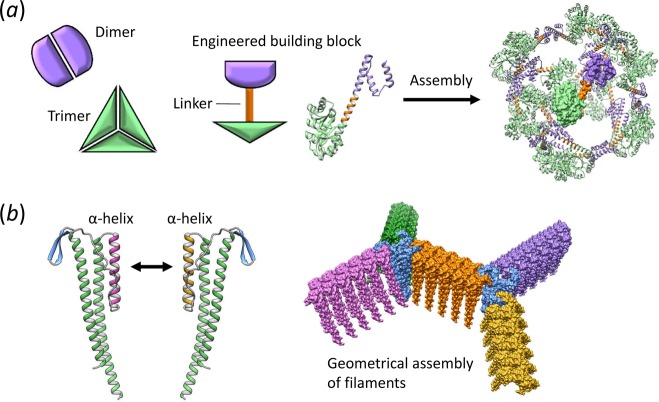
Pioneering approaches to create novel protein nanostructures relied on naturally occurring oligomerization domains to guide the assembly process.14 Many proteins have evolved to self-associate through noncovalent oligomeric interactions to form dimers or higher-order oligomers.12 Domains that associate two at a time (dimers) or three at a time (trimers) are particularly common in nature, and are a rich source of raw building materials for creating self-assembling objects. An elegant approach for building protein nanostructures joins two different oligomerization domains by a semirigid linker into a single fusion protein (Figure 2A). Each domain in the fusion protein has a strong tendency to associate with other copies of itself. As a consequence of this design, many identical copies of the fusion protein self-assemble into filaments or symmetrical cagelike objects, depending on the arrangement of oligomerization domains.14,15 A recent demonstration of this technique involved joining dimerization and trimerization domains together (Figure 2A), which assembled into a homogeneous 24-subunit porous cube with a 23 nm diameter and large internal volume.16 Furthermore, varying the angle or symmetry between the oligomerization domains enables control over the cube’s dimensions17,15 or the creation of two-dimensional lattices over extended surface areas.18,19
Figure 2.
Strategies for engineering self-assembling protein nanostructures. (a) The fusion of two different naturally occurring oligomeric protein domains by a semirigid linker for the creation of building blocks that self-assemble into cagelike structures. (b) Design of de novo coiled coils to attach multiple protein filaments together into complex geometries.
The assembly of protein nanostructures is not limited to using naturally occurring oligomerization domains as improvements in our understanding of the rules governing protein–protein interactions12 are facilitating more direct strategies for designing large protein assemblies.13 Molecular modeling software is increasingly able to predict protein–protein interactions,20,21 which make it possible to design proteins to form novel contact interfaces for the creation of nanostructures. Using this strategy, cagelike structures have recently been created with atomic level accuracy.22,23 This was achieved by redesigning the interface between protein domains to provide the energetic forces required to drive the assembly process, as well as orient the proteins into the geometry of the desired structure. Importantly, the final structures of the 24-subunit protein assemblies—either tetrahedrons or octahedrons—were in close agreement with the computational design. The engineering of new protein–protein interfaces is not without challenges, however, as current methods have a low success rate, which requires many individual designs to be experimentally evaluated in order to find a functional interaction.24 Therefore, the long-term goal is to have reliable computational methods for designing protein assemblies without experimental trial-and-error, but this will require deeper insights into the nature of protein–protein interactions.12
The next level of sophistication in protein nanostructure engineering is increasingly focused on creating small modular domains composed of de novo designed secondary structure that assemble with specific partners into larger assemblies. This methodology avoids having to redesign the entire contact interface of a protein, and instead imparts modularity to control assembly of individual proteins. One approach that has had notable success seeks to use the simple but well-understood structural elements of coiled coils to create complex shapes. Coiled-coil domains are intertwined helical sequences that associate together by inter- and intramolecular protein–protein interactions into elongated bundles.25 These protein domains are attractive building blocks as the rules governing coiled-coil assembly have been thoroughly characterized,26,27 which has facilitated the rational and computational design of artificial coiled coils.28,29
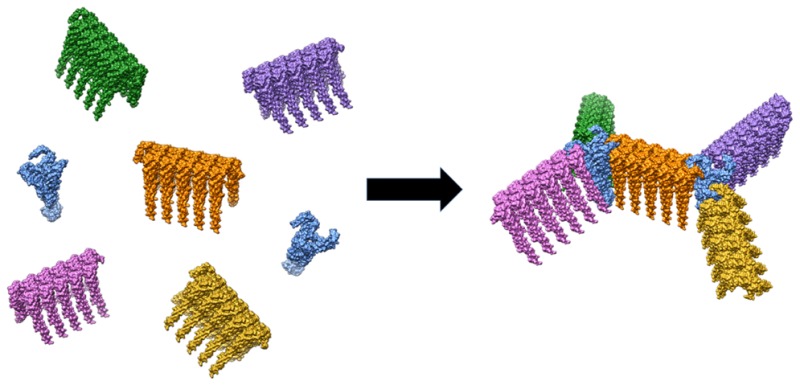
Engineering coiled-coil interaction specificity has enabled a variety of protein nanostructures to be created, including fibers,30,31 tubes,32 cages,33,34 and responsive hydrogels.35 Our recent paper demonstrated that the natural protein–protein interface of a filamentous protein can be redesigned using coiled coils to impart specificity and drive filament assembly into multifaceted structures.36 In this approach, modular connector proteins were created by replacing one of the two contact interfaces in the filament subunit with opposing helical domains that associate together as tight heterodimeric coiled coils (Figure 2B). The resulting pair of connector proteins bound to each other with high specificity while also incorporating into nascent filaments. Furthermore, the addition of a trimerization domain into one of the engineered subunits enabled the creation of a three-way connector that assembles with filaments containing the opposing helical sequence into geometrically ordered shapes such as pinwheels (Figure 1). These protein shapes were ideal templates for building nanomaterials that included highly conductive nanowires,36 and demonstrate that the calligraphy of proteins is maturing beyond the proof of principle stage and moving toward application in solving a host of challenging problems.
Spatial Organization of Enzymes
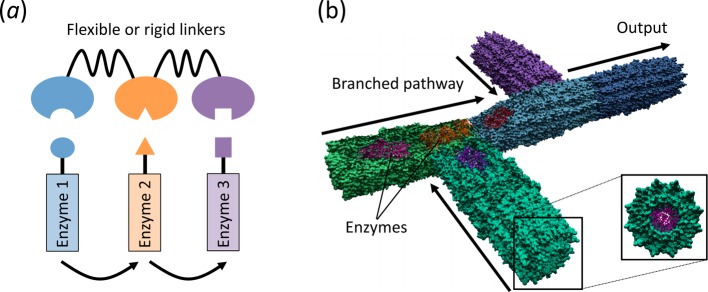
The controlled assembly of multiple proteins has found application in the construction of novel enzymatic pathways for metabolic engineering, which are promising alternatives to synthetic chemistry for cheaply and renewably producing molecules of value. Biopolymer scaffolds such as the hypothetical protein example in Figure 2B could prove ideal for the alignment of multiple enzymes to facilitate substrate channelling, a process whereby the reaction product of one enzyme is transferred directly from the enzyme’s active site into the active site of the next enzyme of a multistep reaction sequence. This can accelerate the rate of metabolic reactions37,38 and reduce diversion of substrate/intermediates into competing pathways that lead to undesirable side reactions.39,40 An emerging strategy has been to use protein scaffolds composed of a series of protein–protein interaction domains that bind to specific peptide sequences (Figure 3A). Using this approach, enzymes fused to cognate peptide sequences are attached along a protein or nucleic acid scaffold in ordered arrangements for sequential enzymatic reactions.37,40,41 The natural modularity of this system enables stoichiometric control over the enzymes and reaction fluxes, resulting in improved efficiency of a variety of enzymatic pathways.42 These scaffold systems lack physical barriers and work by concentrating intermediates before diffusion into the environment can occur.38 The actual mechanism for the enhanced metabolic flux observed by positioning enzymes in close proximity is at present unclear. The distance between the active sites of adjacent enzymes is most likely too large for an intermediate produced by one enzyme to be processed by an adjacent enzyme before diffusing in solution. Instead, recent molecular modeling suggests that the clustering of enzymes into large agglomerates improves the probability that the intermediate will be processed by a downstream enzyme.38
Figure 3.
Spatial organization of enzymes for substrate channelling. (a) Synthetic protein scaffold to position enzymes for modular control over metabolic pathways. Adapted from reference (40). (b) Proposed scheme for creating a protein pipeline to integrate multiple-enzyme pathways for sequential catalytic reactions. Enzymes are encapsulated within tubular proteins that are assembled together, which enables direct diffusion of intermediates between active sites.
A more direct strategy for spatial isolation of enzymes would be to encapsulate them within an engineered protein cage, which has the inherent advantages of isolating reactive intermediates from competing or incompatible processes.43 Initial attempts to encapsulate enzymes exploited naturally occurring protein containers such as virus-like particles44 and bacterial microcompartments.45,46 Engineered protein cages (Figure 2A) are also promising for enzyme encapsulation; however, practical application of enzyme encapsulation will require methods to be developed for the regulation of metabolite entry and exit, and ways in which to localize enzymes into the cage. If these problems can be solved, then elegant enzyme cascades can be envisaged whereby individual enzymes are positioned in a series of protein containers that prevent metabolic intermediates from diffusing away before reacting with sequential enzymes (Figure 3B). These containers could ultimately be assembled into networks, allowing multiple inputs into branched metabolic pathways, with intermediates reacted upon in a precise and ordered manner, a long sought after goal in metabolic engineering.
Nanomaterial Scaffolds
Beyond generating 2D structures to serve as templates, assembling organized protein nanostructures has advantages over traditional processing methods for the fabrication of nanoscale devices. In particular, mature technology exists to evolve peptide sequences for the recognition and binding of nearly any conceivable material.47 Judicious selection of binding peptides combined with an appropriate protein nanostructure enables the incorporation of functional materials into regular patterns with nanometer precision. The almost encyclopedic design of binding peptides has facilitated the creation of a diverse range of materials including conductive metallic nanowires,1 lithium ion batteries,48 and carbon nanotube solar cells.49 Many of these materials, particular in the area of nanoelectronics, are beginning to move out of the laboratory and into real-world application. Furthermore, the natural ability of many protein domains to bind to specific nucleic acid sequences, for example, zinc fingers, has enabled the creation of protein–nucleic acid coassembling nanomaterials.41,50 These hybrid materials have the potential to meld the highly ordered assembly of DNA nanotechnology with the functionality of proteins.
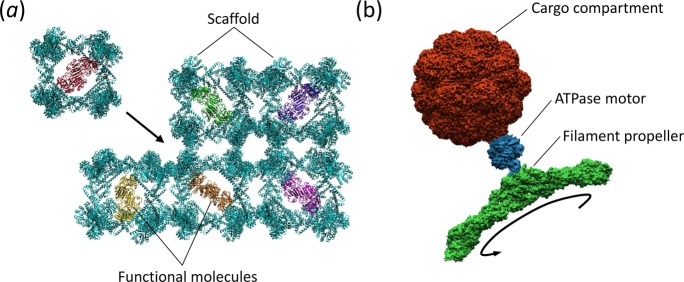
In addition to the templation of functional materials upon the surface of protein nanostructures, the internal cavity of protein cages or containers is ideal for the compartmentalization of functional molecules (e.g., Figure 2A). Protein nanostructures with larger internal openings could be useful as reaction vessels or as vehicles for delivering cargoes, including as agents for nanomedicine. Indeed, functionalization of protein delivery vectors with subcellular targeting signals can facilitate the active transport and delivery of therapeutic drugs to specific locations within mammalian cells.51 These multicomponent delivery vectors have virus-like functionality, and mimic the ability of viruses to target and enter specific cells, with the release of their drug/DNA cargo triggered by a decrease in the pH of the endosome/lysozyme.52 The assembly of multiple cages (Figure 4A) would also facilitate the creation of structures with high porosity.16 Highly porous materials, as exemplified by metal–organic frameworks (MOFs),53 have been examined for applications in catalysis, gas storage, and biomimetic mineralization;54 presumably protein nanostructures could find similar roles, especially as scaffolds for three-dimensional tissue culture.
Figure 4.
Future directions for the creation of functional protein nanostructures and materials. (a) Embedding of functional molecules within large-scale protein frameworks. (b) An example of a nanorobot capable of encapsulating a cargo within a compartment and transportation using an F1-ATPase molecular motor attached to a filament nanopropeller.
It is important to note, however, that many proteins have specific environmental limitations, such as a narrow range of structural stability, which may render them impractical for harsh material fabrication conditions. Therefore, of particular interest is the use of ultrastable proteins that retain their native structure at extremes of pH or temperature or in the presence of chemical denaturants.55 The continual discovery of extremophilic organisms that thrive under adverse environmental conditions is providing new source material as well as insights into protein stability and will enable the creation of ultrastable protein materials.56,57
Combining elements of structural biology, mechanical engineering, and materials science, the design and fabrication of biological nanomachines ranging from levers and rotors to motor-driven assemblies is advancing rapidly in scope and complexity. Proposed applications of such systems include sorting and transporting nanoscale cargo;58 driving and accelerating self-assembly processes of nanostructures;59 mixing and pumping fluids;60 and various other mechanical maneuvers.61 Many of these engineered protein machines exploit naturally occurring molecular motors; however, de novo design of proteins is progressing to a point whereby novel functions can be created such as a recently described metal ion transporter that actively moves Zn2+ and Co2+ across lipid membranes.62An ultimate goal, which bridges science fiction with emerging reality, is the construction of biobased or bioinspired “nanobots” that are capable of performing myriad mechanical and analytical tasks in a multitude of environments. Recently, DNA nanotechnology has made large strides in the creation of nanobots for the transport and delivery of therapeutic agents;58 however, despite numerous proof-of-concept examples of biological moving parts and simple devices, protein nanobots have yet to advance to the same level of sophistication. Well-known examples of biological rotary and linear-motion motors in nature include F1-ATPase63 and kinesin,64 which have been incorporated into a variety of nanoscale mechanical constructs. A surface-immobilized F1-ATPase motor, for example, has been used to rotate an attached filamentous protein through the efficient conversion of chemical energy.60 Improvements in the assembly of proteins into ordered shapes should enable the next level of nanobots to be created, whereby a motor such as an F1-ATPase is able to drive a nanoscale protein propeller and transport a cargo encapsulated within a protein compartment (Figure 4B). Combined with targeting peptides and environmental responsiveness, these nanobots could be programmed to seek out and interact with or destroy specific targets, e.g., cancer cells.
Protein Calligraphy: Form and Function
Overall, this is an exciting time in the development of protein nanostructures. The building of protein complexes has improved to the point where highly ordered structures can be designed and built in a customizable manner; next-generation genome sequencing and crystallography are rapidly expanding the database of known protein structures; the dramatic reduction in the cost and time required to synthesize genes65 and build protein expression constructs66 greatly accelerates prototyping of new protein designs; and near-atomic resolution imaging by cryo-electron microscopy enables the structure of large protein assemblies to be resolved.67 Taken together, this confluence of scientific advances will enable “protein calligraphy” to move from “writing” single characters to crafting the equivalent of sentences, i.e., rational assemblages of shapes that can translate abstract matter into powerful devices, even convey information, for creating function and solving problems.
Acknowledgments
The authors acknowledge support from the Air Force Office of Scientific Research (FA9550-14-1-0026).
The authors declare no competing financial interest.
References
- Mao C.; Solis D. J.; Reiss B. D.; Kottmann S. T.; Sweeney R. Y.; Hayhurst A.; Georgiou G.; Iverson B.; Belcher A. M. Virus-Based Toolkit for the Directed Synthesis of Magnetic and Semiconducting Nanowires. Science 2004, 303, 213–217. 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- Perrier S. Polymer Folding: ABC of Molecular Origami. Nat. Chem. 2011, 3, 194–196. 10.1038/nchem.995. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Ke Y.; Ong L. L.; Shih W. M.; Yin P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science 2012, 338, 1177–1183. 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson E.; Mohammed A.; Gardell J.; Masich S.; Czeizler E.; Orponen P.; Högberg B. DNA Rendering of Polyhedral Meshes at the Nanoscale. Nature 2015, 523, 441–444. 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- Edwardson T. G. W.; Carneiro K. M. M.; McLaughlin C. K.; Serpell C. J.; Sleiman H. F. Site-Specific Positioning of Dendritic Alkyl Chains on DNA Cages Enables Their Geometry-Dependent Self-Assembly. Nat. Chem. 2013, 5, 868–875. 10.1038/nchem.1745. [DOI] [PubMed] [Google Scholar]
- Wickham S. F. J.; Bath J.; Katsuda Y.; Endo M.; Hidaka K.; Sugiyama H.; Turberfield A. J. A DNA-Based Molecular Motor That Can Navigate a Network of Tracks. Nat. Nanotechnol. 2012, 7, 169–173. 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- Amir Y.; Ben-Ishay E.; Levner D.; Ittah S.; Abu-Horowitz A.; Bachelet I. Universal Computing by DNA Origami Robots in a Living Animal. Nat. Nanotechnol. 2014, 9, 353–357. 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro A. V.; Han D.; Shih W. M.; Yan H. Challenges and Opportunities for Structural DNA Nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772. 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräslund S.; Nordlund P.; Weigelt J.; Bray J.; Gileadi O.; Knapp S.; Oppermann U.; Arrowsmith C.; Hui R.; Ming J.; et al. Protein Production and Purification. Nat. Methods 2008, 5, 135–146. 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A.; MacCallum J. L. The Protein-Folding Problem, 50 Years on. Science 2012, 338, 1042–1046. 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- Ahnert S. E.; Marsh J. A.; Hernández H.; Robinson C. V.; Teichmann S. A. Principles of Assembly Reveal a Periodic Table of Protein Complexes. Science 2015, 350, aaa2245. 10.1126/science.aaa2245. [DOI] [PubMed] [Google Scholar]
- Grueninger D.; Treiber N.; Ziegler M. O. P.; Koetter J. W. A.; Schulze M.-S.; Schulz G. E. Designed Protein-Protein Association. Science 2008, 319, 206–209. 10.1126/science.1150421. [DOI] [PubMed] [Google Scholar]
- Padilla J. E.; Colovos C.; Yeates T. O. Nanohedra: Using Symmetry to Design Self Assembling Protein Cages, Layers, Crystals, and Filaments. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 2217–2221. 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.-T.; Tsai K.-L.; Sawaya M. R.; Asturias F. J.; Yeates T. O. Structure and Flexibility of Nanoscale Protein Cages Designed by Symmetric Self-Assembly. J. Am. Chem. Soc. 2013, 135, 7738–7743. 10.1021/ja402277f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.-T.; Reading E.; Hura G. L.; Tsai K.-L.; Laganowsky A.; Asturias F. J.; Tainer J. A.; Robinson C. V.; Yeates T. O. Structure of a Designed Protein Cage That Self-Assembles into a Highly Porous Cube. Nat. Chem. 2014, 6, 1065–1071. 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.-T.; Cascio D.; Yeates T. O. Structure of a 16-Nm Cage Designed by Using Protein Oligomers. Science 2012, 336, 1129. 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- Sinclair J. C.; Davies K. M.; Vénien-Bryan C.; Noble M. E. M. Generation of Protein Lattices by Fusing Proteins with Matching Rotational Symmetry. Nat. Nanotechnol. 2011, 6, 558–562. 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- Gonen S.; DiMaio F.; Gonen T.; Baker D. Design of Ordered Two-Dimensional Arrays Mediated by Noncovalent Protein-Protein Interfaces. Science 2015, 348, 1365–1368. 10.1126/science.aaa9897. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A.; Tyka M.; Lewis S. M.; Lange O. F.; Thompson J.; Jacak R.; Kaufman K.; Renfrew P. D.; Smith C. A.; Sheffler W.; et al. ROSETTA3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. Methods Enzymol. 2011, 487, 545–574. 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F.; Leaver-Fay A.; Bradley P.; Baker D.; André I. Modeling Symmetric Macromolecular Structures in Rosetta3. PLoS One 2011, 6, e20450. 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N. P.; Sheffler W.; Sawaya M. R.; Vollmar B. S.; Sumida J. P.; André I.; Gonen T.; Yeates T. O.; Baker D. Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy. Science 2012, 336, 1171–1174. 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N. P.; Bale J. B.; Sheffler W.; McNamara D. E.; Gonen S.; Gonen T.; Yeates T. O.; Baker D. Accurate Design of Co-Assembling Multi-Component Protein Nanomaterials. Nature 2014, 510, 103–108. 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman S. J.; Whitehead T. A.; Ekiert D. C.; Dreyfus C.; Corn J. E.; Strauch E.-M.; Wilson I. A.; Baker D. Computational Design of Proteins Targeting the Conserved Stem Region of Influenza Hemagglutinin. Science 2011, 332, 816–821. 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolovic B.; Danial M.; Klok H.-A. Coiled Coils: Attractive Protein Folding Motifs for the Fabrication of Self-Assembled, Responsive and Bioactive Materials. Chem. Soc. Rev. 2010, 39, 3541–3575. 10.1039/b914339b. [DOI] [PubMed] [Google Scholar]
- Moutevelis E.; Woolfson D. N. A Periodic Table of Coiled-Coil Protein Structures. J. Mol. Biol. 2009, 385, 726–732. 10.1016/j.jmb.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Wood C. W.; Bruning M.; Ibarra A. Á.; Bartlett G. J.; Thomson A. R.; Sessions R. B.; Brady R. L.; Woolfson D. N. CCBuilder: An Interactive Web-Based Tool for Building, Designing and Assessing Coiled-Coil Protein Assemblies. Bioinformatics 2014, 30, 3029–3035. 10.1093/bioinformatics/btu502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley E. H. C.; Sessions R. B.; Thomson A. R.; Woolfson D. N. Designed Alpha-Helical Tectons for Constructing Multicomponent Synthetic Biological Systems. J. Am. Chem. Soc. 2009, 131, 928–930. 10.1021/ja804231a. [DOI] [PubMed] [Google Scholar]
- Thomson A. R.; Wood C. W.; Burton A. J.; Bartlett G. J.; Sessions R. B.; Brady R. L.; Woolfson D. N. Computational Design of Water-Soluble α-Helical Barrels. Science 2014, 346, 485–488. 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- Ryadnov M. G.; Woolfson D. N. Engineering the Morphology of a Self-Assembling Protein Fibre. Nat. Mater. 2003, 2, 329–332. 10.1038/nmat885. [DOI] [PubMed] [Google Scholar]
- Boyle A. L.; Bromley E. H. C.; Bartlett G. J.; Sessions R. B.; Sharp T. H.; Williams C. L.; Curmi P. M. G.; Forde N. R.; Linke H.; Woolfson D. N. Squaring the Circle in Peptide Assembly: From Fibers to Discrete Nanostructures by de Novo Design. J. Am. Chem. Soc. 2012, 134, 15457–15467. 10.1021/ja3053943. [DOI] [PubMed] [Google Scholar]
- Ueda M.; Makino A.; Imai T.; Sugiyama J.; Kimura S. Rational Design of Peptide Nanotubes for Varying Diameters and Lengths. J. Pept. Sci. 2011, 17, 94–99. 10.1002/psc.1304. [DOI] [PubMed] [Google Scholar]
- Gradišar H.; Božič S.; Doles T.; Vengust D.; Hafner-Bratkovič I.; Mertelj A.; Webb B.; Šali A.; Klavžar S.; Jerala R. Design of a Single-Chain Polypeptide Tetrahedron Assembled from Coiled-Coil Segments. Nat. Chem. Biol. 2013, 9, 362–366. 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. M.; Harniman R. L.; Barnes F. R. H.; Boyle A. L.; Collins A.; Mantell J.; Sharp T. H.; Antognozzi M.; Booth P. J.; Linden N.; et al. Self-Assembling Cages from Coiled-Coil Peptide Modules. Science 2013, 340, 595–599. 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banwell E. F.; Abelardo E. S.; Adams D. J.; Birchall M. A.; Corrigan A.; Donald A. M.; Kirkland M.; Serpell L. C.; Butler M. F.; Woolfson D. N. Rational Design and Application of Responsive α-Helical Peptide Hydrogels. Nat. Mater. 2009, 8, 596–600. 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. J.; Giger L.; Kim S. S.; Naik R. R.; Clark D. S. Geometrical Assembly of Ultrastable Protein Templates for Nanomaterials. Nat. Commun. 2016, 7, 11771. 10.1038/ncomms11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilner O. I.; Weizmann Y.; Gill R.; Lioubashevski O.; Freeman R.; Willner I. Enzyme Cascades Activated on Topologically Programmed DNA Scaffolds. Nat. Nanotechnol. 2009, 4, 249–254. 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- Castellana M.; Wilson M. Z.; Xu Y.; Joshi P.; Cristea I. M.; Rabinowitz J. D.; Gitai Z.; Wingreen N. S. Enzyme Clustering Accelerates Processing of Intermediates through Metabolic Channeling. Nat. Biotechnol. 2014, 32, 1011–1018. 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornscheuer U. T.; Huisman G. W.; Kazlauskas R. J.; Lutz S.; Moore J. C.; Robins K. Engineering the Third Wave of Biocatalysis. Nature 2012, 485, 185–194. 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- Dueber J. E.; Wu G. C.; Malmirchegini G. R.; Moon T. S.; Petzold C. J.; Ullal A. V.; Prather K. L. J.; Keasling J. D. Synthetic Protein Scaffolds Provide Modular Control over Metabolic Flux. Nat. Biotechnol. 2009, 27, 753–759. 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Delebecque C. J.; Lindner A. B.; Silver P. A.; Aldaye F. A. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science 2011, 333, 470–474. 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- Whitaker W. R.; Davis S. A.; Arkin A. P.; Dueber J. E. Engineering Robust Control of Two-Component System Phosphotransfer Using Modular Scaffolds. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 18090–18095. 10.1073/pnas.1209230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. C.; Patterson D. P.; Saboda K. N.; Edwards E. J.; Miettinen H. M.; Basu G.; Thielges M. C.; Douglas T. Self-Assembling Biomolecular Catalysts for Hydrogen Production. Nat. Chem. 2016, 8, 179–185. 10.1038/nchem.2416. [DOI] [PubMed] [Google Scholar]
- Patterson D. P.; Schwarz B.; Waters R. S.; Gedeon T.; Douglas T. Encapsulation of an Enzyme Cascade within the Bacteriophage P22 Virus-Like Particle. ACS Chem. Biol. 2014, 9, 359–365. 10.1021/cb4006529. [DOI] [PubMed] [Google Scholar]
- Wörsdörfer B.; Woycechowsky K. J.; Hilvert D. Directed Evolution of a Protein Container. Science 2011, 331, 589–592. 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- Kerfeld C. A.; Erbilgin O. Bacterial Microcompartments and the Modular Construction of Microbial Metabolism. Trends Microbiol. 2015, 23, 22–34. 10.1016/j.tim.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Care A.; Bergquist P. L.; Sunna A. Solid-Binding Peptides: Smart Tools for Nanobiotechnology. Trends Biotechnol. 2015, 33, 259–268. 10.1016/j.tibtech.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Lee Y. J.; Yi H.; Kim W.-J.; Kang K.; Yun D. S.; Strano M. S.; Ceder G.; Belcher A. M. Fabricating Genetically Engineered High-Power Lithium-Ion Batteries Using Multiple Virus Genes. Science 2009, 324, 1051–1055. 10.1126/science.1171541. [DOI] [PubMed] [Google Scholar]
- Dang X.; Yi H.; Ham M.-H.; Qi J.; Yun D. S.; Ladewski R.; Strano M. S.; Hammond P. T.; Belcher A. M. Virus-Templated Self-Assembled Single-Walled Carbon Nanotubes for Highly Efficient Electron Collection in Photovoltaic Devices. Nat. Nanotechnol. 2011, 6, 377–384. 10.1038/nnano.2011.50. [DOI] [PubMed] [Google Scholar]
- Mou Y.; Yu J.-Y.; Wannier T. M.; Guo C.-L.; Mayo S. L. Computational Design of Co-Assembling Protein-DNA Nanowires. Nature 2015, 525, 230–233. 10.1038/nature14874. [DOI] [PubMed] [Google Scholar]
- Glover D. J.; Ng S. M.; Mechler A.; Martin L. L.; Jans D. A. Multifunctional Protein Nanocarriers for Targeted Nuclear Gene Delivery in Nondividing Cells. FASEB J. 2009, 23, 2996–3006. 10.1096/fj.09-131425. [DOI] [PubMed] [Google Scholar]
- Glover D. J. Artificial Viruses: Exploiting Viral Trafficking for Therapeutics. Infect. Disord.: Drug Targets 2012, 12, 68–80. 10.2174/187152612798995000. [DOI] [PubMed] [Google Scholar]
- Zhou H.-C. “J.”; Kitagawa S. Metal–Organic Frameworks (MOFs). Chem. Soc. Rev. 2014, 43, 5415–5418. 10.1039/C4CS90059F. [DOI] [PubMed] [Google Scholar]
- Liang K.; Ricco R.; Doherty C. M.; Styles M. J.; Bell S.; Kirby N.; Mudie S.; Haylock D.; Hill A. J.; Doonan C. J.; et al. Biomimetic Mineralization of Metal-Organic Frameworks as Protective Coatings for Biomacromolecules. Nat. Commun. 2015, 6, 7240. 10.1038/ncomms8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszka M. J.; Clark M. E.; Schneider E.; Clark D. S. Nature versus Nurture: Developing Enzymes That Function under Extreme Conditions. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 77–102. 10.1146/annurev-chembioeng-061010-114239. [DOI] [PubMed] [Google Scholar]
- Glover D. J.; Giger L.; Kim J. R.; Clark D. S. Engineering Protein Filaments with Enhanced Thermostability for Nanomaterials. Biotechnol. J. 2013, 8, 228–236. 10.1002/biot.201200009. [DOI] [PubMed] [Google Scholar]
- Glover D. J.; Clark D. S. Oligomeric Assembly Is Required for Chaperone Activity of the Filamentous γ-Prefoldin. FEBS J. 2015, 282, 2985–2997. 10.1111/febs.13341. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Lewandowski B.; Bo G. D.; Ward J. W.; Papmeyer M.; Kuschel S.; Aldegunde M. J.; Gramlich P. M. E.; Heckmann D.; Goldup S. M.; D’Souza D. M.; et al. Sequence-Specific Peptide Synthesis by an Artificial Small-Molecule Machine. Science 2013, 339, 189–193. 10.1126/science.1229753. [DOI] [PubMed] [Google Scholar]
- Soong R. K.; Bachand G. D.; Neves H. P.; Olkhovets A. G.; Craighead H. G.; Montemagno C. D. Powering an Inorganic Nanodevice with a Biomolecular Motor. Science 2000, 290, 1555–1558. 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- Goel A.; Vogel V. Harnessing Biological Motors to Engineer Systems for Nanoscale Transport and Assembly. Nat. Nanotechnol. 2008, 3, 465–475. 10.1038/nnano.2008.190. [DOI] [PubMed] [Google Scholar]
- Joh N. H.; Wang T.; Bhate M. P.; Acharya R.; Wu Y.; Grabe M.; Hong M.; Grigoryan G.; DeGrado W. F. De Novo Design of a Transmembrane Zn2+-Transporting Four-Helix Bundle. Science 2014, 346, 1520–1524. 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji H.; Yasuda R.; Yoshida M.; Kinosita K. Direct Observation of the Rotation of F1-ATPase. Nature 1997, 386, 299–302. 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- Wollman A. J. M.; Sanchez-Cano C.; Carstairs H. M. J.; Cross R. A.; Turberfield A. J. Transport and Self-Organization across Different Length Scales Powered by Motor Proteins and Programmed by DNA. Nat. Nanotechnol. 2014, 9, 44–47. 10.1038/nnano.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S.; Church G. M. Large-Scale de Novo DNA Synthesis: Technologies and Applications. Nat. Methods 2014, 11, 499–507. 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini A.; Storch M.; Baldwin G. S.; Ellis T. Bricks and Blueprints: Methods and Standards for DNA Assembly. Nat. Rev. Mol. Cell Biol. 2015, 16, 568–576. 10.1038/nrm4014. [DOI] [PubMed] [Google Scholar]
- Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 2015, 161, 450–457. 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]