Abstract
Therapeutic targeting of membrane-associated viral proteins is complicated by the challenge of investigating their enzymatic activities in the native membrane-bound state. To permit functional characterization of these proteins, we hypothesized that the supported lipid bilayer (SLB) can support in situ reconstitution of membrane-associated viral protein complexes. As proof-of-principle, we selected the hepatitis C virus (HCV) NS5B polymerase which is essential for HCV genome replication, and determined that the SLB platform enables functional reconstitution of membrane protein activity. Quartz crystal microbalance with dissipation (QCM-D) monitoring enabled label-free detection of full-length NS5B membrane association, its interaction with replicase subunits NS3, NS5A, and template RNA, and most importantly its RNA synthesis activity. This latter activity could be inhibited by the addition of candidate small molecule drugs. Collectively, our results demonstrate that the SLB platform can support functional studies of membrane-associated viral proteins engaged in critical biological activities.
Short abstract
Biomimetic supported lipid bilayers provide a surface-sensitive measurement platform to reconstitute functional viral replication complexes and evaluate the performance of drug inhibitors.
Introduction
Biological membranes support a wide range of macromolecular interactions and are critical for cellular homeostasis and protection.1−4 Membrane-associated protein complexes also perform essential functions during the genome replication of many viral pathogens. For example, the formation of a membrane-associated replication complex, composed of viral proteins and replicating RNA, is a hallmark of all positive-strand RNA viruses.5−8 Despite the biological importance of these complexes, there is a lack of robust, quantitative tools to perform functional analysis of membrane-associated viral proteins in their native state. Beyond the resultant challenges for studying key molecular details of viral replicase complex assembly and function, this technical hurdle also limits the ability to discover and characterize inhibitors that bind to and interfere with components of membrane-associated protein complexes.
To address these challenges, we hypothesized that the supported lipid bilayer (SLB) might be an excellent platform to host membrane-associated proteins involved in viral replication. Indeed, SLBs and related model membrane platforms9−11 (e.g., tethered lipid bilayer, adsorbed vesicles) have enabled the investigation of various classes of membrane-associated proteins, including transmembrane proteins,12 anchored proteins,13 and interfacial enzymes.14 Formed by the self-assembly of lipid vesicles upon interaction with certain planar solid surfaces, SLBs are robust and offer a well-characterized membranous setting upon which to study dynamic biological interactions.15,16 We were particularly interested in integrating the SLB platform together with the quartz crystal microbalance with dissipation (QCM-D) nanomass sensor. The technique enables real-time, quantitative, and label-free monitoring of macromolecular interactions at solid–liquid interfaces,17 and has been previously utilized for measuring bacterial polymerase kinetics with a surface-attached oligonucleotide configuration that involves transiently bound polymerase.18,19 The development of a measurement platform to investigate polymerase reactions at membrane interfaces remains an outstanding goal, and such measurement capabilities have never been applied to study viral replication complexes. Within this context, we further hypothesized that QCM-D monitoring of an SLB platform would enable functional characterization of viral protein enzymatic activity, particularly that related to genome replication.
To test these hypotheses, we selected the hepatitis C virus (HCV) as a model system. HCV is a single-strand, positive sense RNA virus that belongs to the Hepacivirus genus of the Flaviviridae family. HCV infection affects approximately 150 million individuals globally.20 Current treatment options for HCV have improved, yet remain suboptimal for many patients.21 The core enzyme of the HCV replicase complex, the NS5B RNA-dependent RNA polymerase, is required for virus replication in vivo(22) and is a prime target in the development of the most effective current and future therapies.21
HCV forms its characteristic replicase complex—consisting of viral nonstructural proteins, RNA template, and host factors—in association with an endoplasmic reticulum (ER)-derived, virally induced, membrane structure termed the “membranous web”23 that supports NS5B function.24,25In vitro studies of NS5B polymerase have, for the most part, employed the catalytic core of the protein, the so-called NS5B-ΔC21, devoid of 21 hydrophobic amino acid residues from the C-terminus that are used for membrane anchorage. This is because of NS5B-ΔC21’s greater solubility, ease of purification, and greater activity in solution as compared to the full-length version (NS5B-FL).26,27 In spite of the advantages that NS5B-ΔC21 confers for in vitro analysis, studies on NS5B-FL are important because its hydrophobic tail is necessary for full functionality of the enzyme in vivo, including membrane association and likely also the mediation of critical protein–protein or protein–RNA interactions. Hence, in vitro studies that examine NS5B-FL are crucial to elucidate its complete mechanism of action and will provide additional opportunities for drug discovery research directed toward blockage of its function. To date, however, the reconstitution of NS5B-FL in a membranous environment has not been replicated in order to restore polymerase function.
In this study, we report that the SLB platform is able to successfully host assembly of a functional HCV replicase system composed of membrane-associated NS5B-FL, either alone or with known replicase complex components such as the HCV NS3 and NS5A proteins, and template RNA that is capable of robust RNA synthesis in vitro. Importantly, QCM-D monitoring enabled label-free detection of NS5B-FL membrane association, its binding to replicase complex protein and RNA components, and the direct sensing and characterization of NS5B-FL RNA polymerase activity as well as its inhibition by a candidate small molecule drug. We also demonstrate that this system could identify a single nucleotide change in the HCV template RNA that can abolish functional RNA polymerase activity, while still allowing RNA binding and translation of the viral RNA intact. Taken together, this study describes the creation of the first in vitro system for examining NS5B-FL RNA binding and polymerase activity in association with a lipid membrane. We envision that the technology presented herein will find broad application for studying the assembly and function of a wide range of viral protein complexes, and for developing potential therapeutic inhibitors of such complexes.
Results
In Vitro Activity of NS5B Isoforms
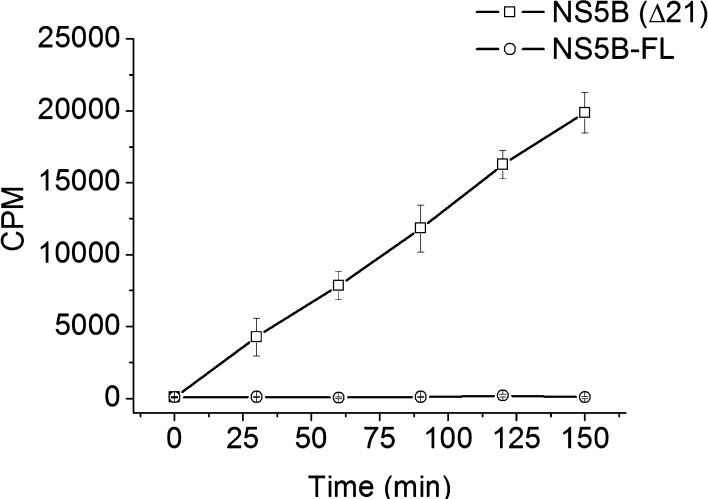
We first measured the RdRp activity of NS5B-ΔC21 and NS5B-FL in solution using the 377 nucleotide RNA template (the 3′ terminus of the viral negative strand RNA, termed “cIRES”) as described in Klumpp et al.28 (see Supporting Information for detailed protocols of protein purifications, gel characterizations (Figure S1), and assay conditions). NS5B-ΔC21 demonstrated polymerase activity with an apparent steady state kcat value of 0.10 ± 0.0 min–1, which was in good agreement with literature values (Figure 1).29 Under the same assay conditions, however, RNA polymerase activity was not detectable with NS5B-FL. Crystallographic structural studies using NS5B-ΔC21 protein and other deletion mutants have suggested that in solution the C-terminus of NS5B can fold back into the active site of the polymerase and interfere with RNA polymerase activity.30,31 Consistent with the crystal structure information and the very low enzyme activity of NS5B-FL in solution, modeling of the full C-terminus of NS5B in the absence of its natural membrane target predicts an energetically favorable conformation of the hydrophobic tail wherein the latter is folded back and positioned into the active site (see proposed model in Figure S2). Based on these observations, we investigated whether the cell membrane-mimicking SLB platform could restore NS5B-FL function by supporting membrane association of its C-terminus.
Figure 1.
Biochemical activities of full-length (FL) and truncated (Δ21) HCV NS5B. In vitro polymerase activity of HCV NS5B protein in solution, measured as incorporation of radiolabeled nucleosides into larger RNA molecules. Enzyme activity is shown as CPM (counts per minute) of trichloroacetic acid precipitated, labeled RNA. NS5B-ΔC21 without the 21 amino acid C-terminal hydrophobic tail demonstrates time-dependent enzymatic activity. By contrast, NS5B-FL activity is undetectable under the same assay conditions.
NS5B-FL Membrane Association
As such, an SLB platform was self-assembled on a silicon oxide coated quartz crystal by the vesicle fusion process.15 To briefly describe the process, unilamellar lipid vesicles consisting of zwitterionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid are allowed to adsorb on the hydrophilic silicon oxide surface. Upon reaching a critical surface coverage, the vesicles spontaneously rupture and reassemble to form a planar bilayer.16 On the quartz crystal microbalance with dissipation (QCM-D) system, mass adsorption onto the silicon oxide surface causes a proportional decrease in the resonance frequency of the oscillating quartz crystal, which allows for a sensitive and direct quantification of the bound mass for rigid films (Figure S3A). The exact response of the crystal’s oscillation behavior, captured by the dissipation function, is related to the degree of viscoelasticity of the absorbed mass (Figure S3B,C).16,32 When lipid vesicles adsorb to the quartz crystal substrate beyond a critical surface coverage, there is a characteristic vesicle rupturing signature with two-step kinetics.16 The vesicles first adsorb to the substrate and remain intact until reaching a maximum frequency change of −40 Hz, which reflects the adsorption of lipid and hydrodynamically coupled solvent mass on the sensor platform. After reaching this maximum value, the frequency signal increases as the result of vesicle rupture and subsequent release of encapsulated solvent and excess lipid molecules. The final resonance frequency change of −24 Hz indicates the formation of an SLB (Figure S3D).33 New SLBs were generated de novo for each experiment.
We then tested if the SLB platform can support NS5B-FL binding. Following a buffer wash to stabilize the SLB, NS5B-FL protein was next added and the frequency signal further decreased to −62 Hz, indicating a 38 Hz saturating signal equivalent of NS5B-FL associated with the SLB within 15 min (Figure S2B, Figure 2A, see arrow labeled “NS5B-FL”). The observed change in resonance frequency upon addition of NS5B to the SLB platform (ΔfNS5B) can be converted into the mass of bound NS5B-FL by the Sauerbrey equation34 as follows:
| 1 |
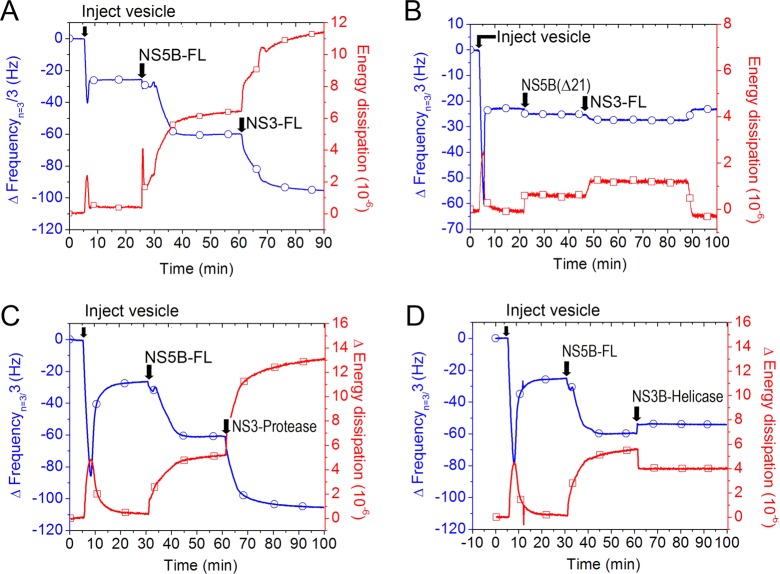
Figure 2.
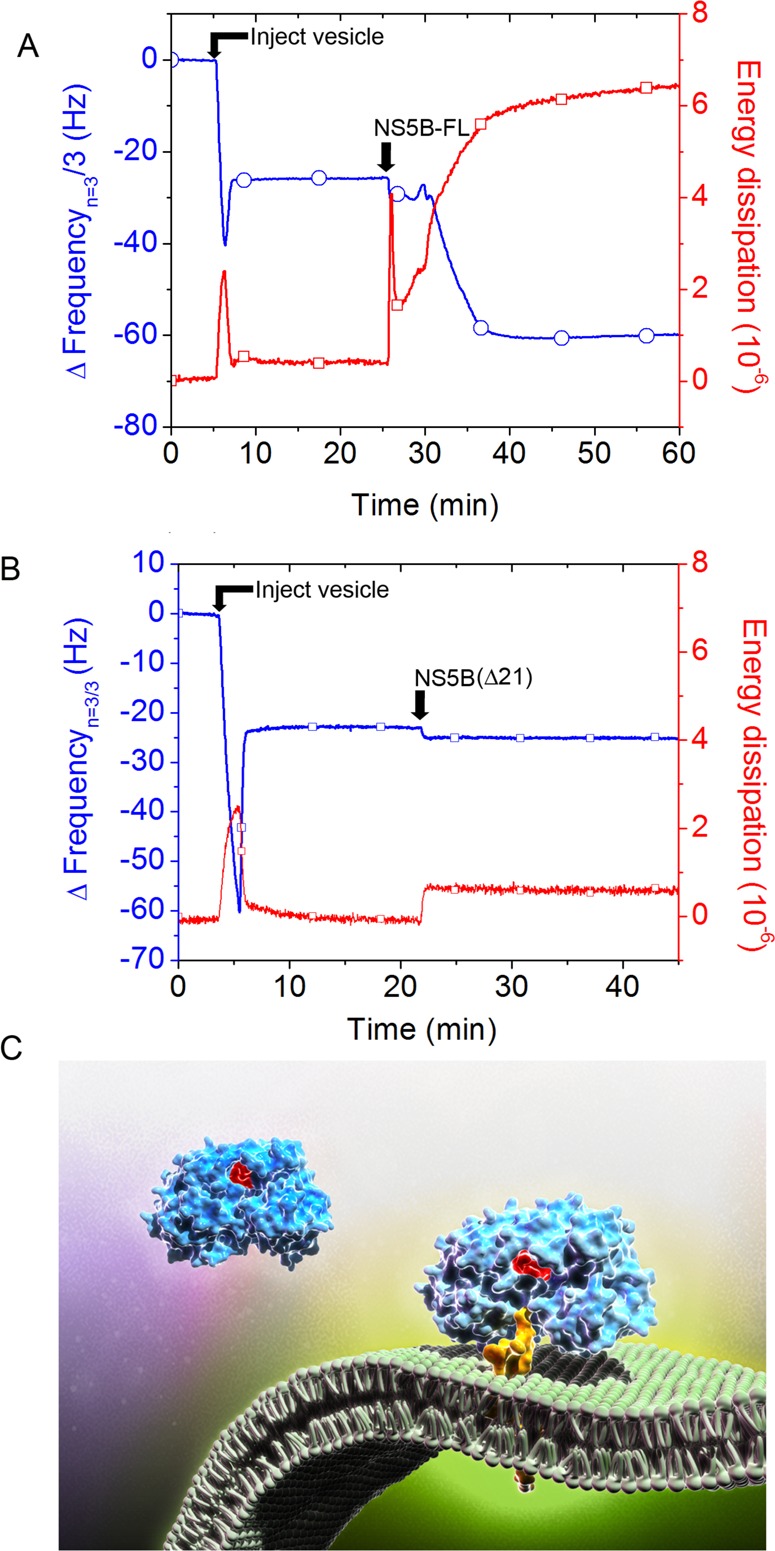
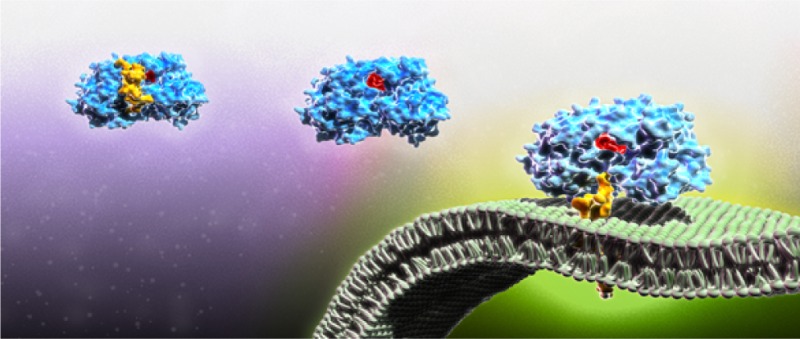
HCV NS5B protein C-terminal tail mediates binding to the planar lipid bilayer membrane. (A) QCM-D frequency (blue) and energy dissipation (red) tracings, indicating lipid bilayer formation signature after injection of lipid vesicles, followed by injection of NS5B-FL and binding to the planar lipid bilayer membrane as indicated by frequency reduction, which correlates with increasing mass on the quartz crystal. Typically, we used the third overtone value (15 MHz) for frequency (n = 3), and present frequency data with fundamental overtone value (5 MHz), Δfn/n. (B) Apparent minimal binding of the NS5B-ΔC21 protein, which is reversible upon buffer wash (not shown). (C) Illustration of proposed NS5B-ΔC21 and NS5B-FL interaction with the bilayer membrane. Active site residues are colored red, and C-terminal domain is colored yellow. The NS5B-ΔC21 protein lacks the transmembrane domain. Thus, its minimal apparent binding signal is caused by nonspecific physiosorption. By contrast, the 21 amino acid C-terminal hydrophobic tail of the NS5B-FL protein mediates binding by preferential insertion into the bilayer membrane with hydrophobic interactions being the primary driving force.
Using eq 1 and the observed frequency change, we estimated that, upon reaching binding saturation, there are ∼6 × 1012 NS5B molecules bound per cm2 of active QCM-D sensing area. Similar calculations permitted us to determine the number of individual molecules bound to the sensor surface during each subsequent step of replicase complex assembly (Table S2). This analytical approach allowed us to compare the relative effects of different subunit factors on replicase-mediated polymerization by calculating “apparent” kinetic values, although it should be stressed that absolute determination of kinetic values would likely require more complex hydrodynamic modeling or utilization of an alternative label-free, surface-sensitive measurement technique. These factors along with consideration of the possible measurement uncertainties arising from the conversion of changes of resonance frequency into molecular mass for different classes of biomolecules are discussed below.
In contrast to the NS5B-FL case, the QCM-D responses demonstrated that NS5B-ΔC21 does not bind to the SLB platform (Figure 2B, see arrow labeled “NS5B-ΔC21”). The NS5B-ΔC21 mutant was unable to bind the SLB platform regardless of whether the His-tag label was present at the N- or C-terminus (Figure S4). The minor frequency and dissipation changes were removed by a buffer wash, indicating the absence of specific binding. To confirm the membrane-associating domain, we also investigated membrane binding of a 21 amino acid peptide corresponding to the NS5B C-terminus on the SLB platform. Concentration-dependent binding kinetics were observed, and the kinetic signature of frequency and dissipation changes were consistent with membrane penetration35 (Figure S5). Collectively, these data demonstrate that NS5B-FL protein can specifically bind to a lipid membrane and that this binding interaction is mediated by membrane penetration of its C-terminal 21 amino acid tail.
NS5B-FL Interaction with NS3 Protein
Based on this platform, we next sought to determine if we could detect the binding interaction of the NS5B-FL with another replicase complex protein, NS3. Indeed, in HCV infected cells, NS5B is part of a multicomponent complex of RNA and proteins that together presumably specify and regulate the requisite functions for viral genome replication. Figure 3A shows the addition of full-length NS3 to membrane-bound NS5B-FL. NS3 binding was indicated by a −33 Hz frequency shift and concomitant increase in dissipation of 8 × 10–6. As a control, we also determined that NS3 did not bind to the SLB platform itself (Figure S6) or when it was added after NS5B-ΔC21 (Figure 3B). As the frequency change is directly dependent on the mass added, and because there was no binding of NS3 in the absence of NS5B-FL, the individual frequency changes from the sequential binding of NS5B-FL and NS3 are indicative of the stoichiometry of NS5B-NS3 binding. Quantitative estimates of relative protein mass addition to the SLB platform indicated one-to-one stoichiometric binding between full-length NS3 and NS5B-FL (Figure 3A).
Figure 3.
Assembly of the membrane-associated HCV NS3–NS5B complex requires the NS3 protease domain. (A) Full length NS3 protein binds to membrane-associated NS5B-FL protein. (B) NS3 does not bind to membranes in the absence of NS5B-FL. To demonstrate this, we first injected NS5B-ΔC21, followed by injection of NS3. Only minor changes in frequency and dissipation are apparent, and they are reversible by buffer wash. For A and B, QCM-D monitoring was as in Figure 2. (C) Addition of the protease domain fragment of NS3 results in significant binding to membrane associated NS5B-FL, whereas (D) no significant binding of the NS3 helicase domain fragment to NS5B-FL is observed.
We also used this method to determine the interaction domain of NS3 with NS5B. As shown in Figure 3C, the NS3 protease domain showed significant binding to membrane-associated NS5B-FL. The kinetics of binding of the NS3 protease domain was similar to that of full-length NS3. In contrast, the NS3 helicase domain did not bind to NS5B (Figure 3D). Taken together, these data support a direct and stoichiometric binding of NS3 protein to membrane-associated NS5B-FL that is mediated through the NS3 protease domain. Beyond replicase complex assembly, we next sought to determine whether the SLB platform could restore NS5B-FL enzyme activity.
Membrane Association Restores NS5B-FL Polymerase Activity
To assess restoration of NS5B-FL polymerase activity, we first investigated if the increase in mass from RNA binding to NS5B could be measured. We used cIRES RNA, derived from the HCV antigenome, as template RNA in this study. This RNA corresponds to the last 378 nucleotides at the 3′ end of the HCV negative strand RNA. This RNA sequence is required to initiate the RNA polymerization reaction for production of progeny plus strand RNA genomes. Addition of cIRES template RNA to membrane-bound NS5B was associated with a reduction in resonance frequency, indicating RNA binding to NS5B-FL in an ∼1:5 molar ratio (Figure S7C).
Based on this protein–RNA complex, we next determined if the reconstituted NS5B-FL was catalytically active. RNA synthesis results from nucleotide incorporation that can be sensed as mass addition by QCM-D monitoring. Strikingly, when nucleoside triphosphates (NTPs) were added to the membrane-bound NS5B–RNA complex, a continuous decrease in resonance frequency was observed, consistent with mass addition from nucleotide incorporation and RNA synthesis (Figure 4A, Figure S7C). By contrast, there was no effect on resonance frequency in the absence of NTP (Figure 4A) or when fewer than all four native NTP species were present in the reaction (Figure S8). Therefore, the decline in resonance frequency was dependent on the presence of all four native NTPs and consistent with RNA synthesis activity.
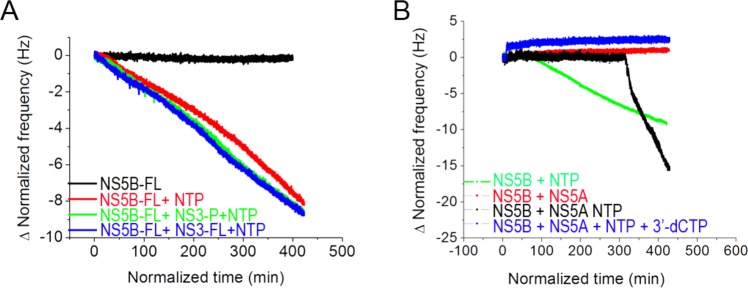
Figure 4.
RNA-dependent, RNA polymerase activity of the membrane-associated HCV replicase complex. NS5B-FL was immobilized on the lipid bilayer membrane as shown in previous figures. NS3 or NS5A proteins were added as indicated. cIRES RNA template was bound as shown in Figure S7. Subsequent addition of ribonucleoside triphosphates (NTPs) resulted in continuous reduction of oscillation frequency, consistent with template-directed RNA strand synthesis. (A) Effect of NS3 on NS5B polymerase activity. QCM-D resonance frequency response as a function of time following addition of NTPs to membrane-associated NS5B-FL + cIRES RNA template (red), membrane-associated NS5B-FL + full-length NS3 + cIRES RNA template (blue), or membrane-associated NS5B-FL + NS3 protease domain + cIRES RNA template (green). A negative control reaction consisting of membrane-associated NS5B-FL + cIRES RNA template without addition of NTPs, wherein no RNA synthesis occurred, is also shown (black). (B) Effect of NS5A on NS5B polymerase activity. QCM-D resonance frequency response as a function of time following addition of NTPs to membrane-associated NS5B-FL + cIRES RNA template, without (green) or with (black) added NS5A. Reactions performed without NTPs (red) or with chain-terminating inhibitor, 3′-dCTP, added together with the NTPs (blue). Each NTP and inhibitor had a concentration of 500 μM.
Beyond qualitative assessment of the QCM-D kinetic traces, we sought to quantitatively measure the effects of replicase subunits NS3 and NS5B on polymerase activity. Therefore, we calculated apparent kcat based on the time-dependent QCM-D resonance frequency response upon the addition of NTPs to initiate the RNA polymerization reaction. The rate of RNA polymerization was measured as a function of the change in resonance frequency over time (Δf/Δtime), allowing for a direct assessment of the absolute amount of RNA product synthesized over this time interval as well as monitoring of the RNA polymerization kinetics. With regard to the latter, eq 1 and the corresponding molecular weight of NMP were applied to obtain the following:
| 2 |
Taken together, the results from eqs 1 and 2 were combined in eq 3 shown below to determine the apparent catalytic rate constant, kcat (see Tables S1 and S2 for complete data sets of calculated values).
| 3 |
Based on this strategy, we determined the apparent kcat of NS5B-FL to be 0.13 ± 0.02 min–1 (Figure 4, Table S1, and Figure S7C). The RNA polymerase activity was similar for the membrane-bound NS5B complexed with NS3 (0.12 ± 0.03 min–1) (Figure 4A, Table S1, and Figure S7D). In both cases, we observed linear polymerization kinetics and similar specific activity.
In contrast to NS3, we found that addition of another HCV replicase subunit, NS5A, had a profound effect on NS5B polymerase activity. Full-length NS5A carries an N-terminal amphipathic peptide that independently interacts with bilayer membranes.36−38 In order to determine if there is a direct interaction between NS5B and NS5A, we used an N-terminal deletion mutant of NS5A in this study. Whereas full-length NS5A binds the membrane bilayer on its own (Figure S9A), the truncated version of NS5A used for the polymerase assay (NS5A-Δ32, which is devoid of NS5A’s N-terminal amphipathic helix membrane-anchoring segment), as expected, does not (Figure S9B). This NS5A bound to membrane-bound NS5B–RNA complex with a 1:2 stoichiometry, and this complex was active in RNA synthesis. The rate of mass increase after the addition of NTPs to the NS5B–NS5A–RNA complex was significantly higher than that obtained in the absence of NS5A (Figure 4B, compare black and green lines, respectively). The apparent kcat of the NS5A containing complex was 0.83 ± 0.33 min–1 (Figure 4B, Table S2, and Figure S7E), which translates into an ∼6-fold increase in polymerase activity as compared to NS5B alone (Figure 4B). As controls, there was no change in oscillation frequency when NTPs were omitted from the reaction (Figure 4B, red trace) or when the chain terminating inhibitor 3′-dCTP was added to the reaction (Figure 4B, blue trace). NS5A could associate with NS5B on the platform independent of cIRES RNA (Figure S9C). Interestingly, there was a substantial NS5A-dependent lag time of approximately 350 min after the addition of NS5A to the membrane-bound NS5B–RNA complex, before RNA synthesis initiated spontaneously (Figure 4B and Figure S7E). The presence of a lag phase may indicate a requirement for substantial conformational rearrangements on the bilayer membrane to form an optimally active NS5A–NS5B–RNA complex that, once properly configured, exhibits remarkably enhanced RdRp activity. The NS5A-stimulated polymerization appeared to be more efficient if NS5A was added after (Figure S7F), as opposed to before (Figure S7E), interaction of NS5B with its RNA template.
To further characterize the nature of the synthesized RNA product, we extracted the total RNA directly off of the QCM quartz crystals and analyzed the RNA products by Northern blots using strand-specific probes (Figure S10). A single distinct ∼200 nucleotide long plus strand RNA product is produced off of the 378 nucleotide minus strand RNA (cIRES) template. Given the increase in absolute mass post addition of nucleotides to initiate the polymerase reaction, these results indicate that, at the very least, on average every enzyme bound template is actively transcribed to generate the ∼200 nucleotide long plus strand RNA product. Interestingly, there appears to be a strong pause site at about the midpoint of the minus strand RNA template. While addition of NS5A was associated with an observed increased apparent elongation rate (Figure S7F), the newly synthesized product still ended at the same position (Figure S10).
We further tested the feasibility of this platform for NS5B drug development. A small molecule chain-terminating polymerase inhibitor, 3′-deoxycytidine triphosphate (3′-dCTP), could completely stop the reaction and stabilize the resonance frequency (Figure 4B, Figure S7G).
To explore the potential of this system to assess the activity of chemotypes beyond nucleoside analogue inhibitors, we also determined the effect on NS5B activity of two non-nucleoside allosteric inhibitors that bind at different sites on the NS5B enzyme (HCV-796 binds the palm site II, and VX-222 binds the thumb site II; see Figure S11 for chemical structures).39,40 As shown in Figure 5, when nucleoside triphosphates (NTPs) were added to the membrane-bound NS5B–RNA complex along with vehicle control, a continuous decrease in resonance frequency was observed, consistent with mass addition from nucleotide incorporation and RNA synthesis (red trace). In contrast, in the presence of HCV-796 (turquoise trace), or VX-222 (green trace), significant inhibition of anti-NS5B activity was observed, approaching that seen with 3′-dCTP (pink trace).
Figure 5.
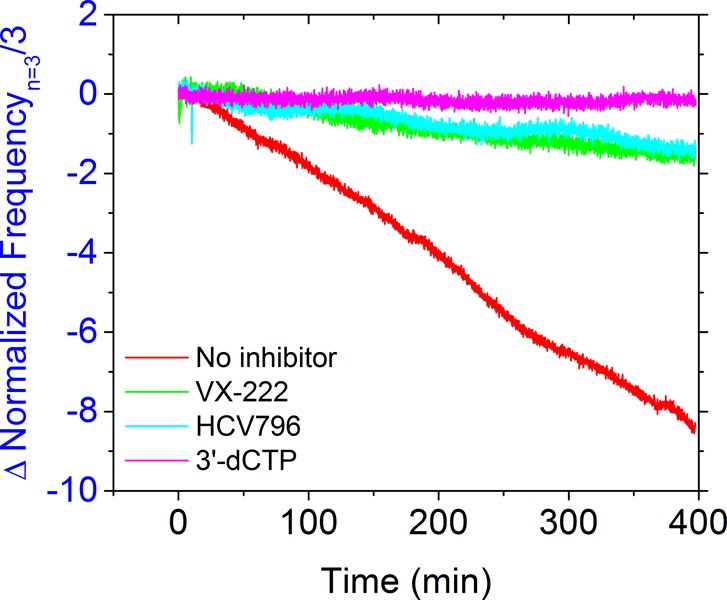
RNA-dependent, RNA polymerase activity of the membrane-associated HCV replicase complex in the presence of different chemotype inhibitors. NS5B-FL was immobilized on the lipid bilayer membrane as shown in previous figures. cIRES RNA template was bound followed by subsequent addition of ribonucleoside triphosphates (NTPs) in the presence of either vehicle control (red), the non-nucleoside inhibitor HCV-796 (turquoise), the non-nucleoside inhibitor VX-222 (green), or the nucleoside analogue inhibitor 3′-dCTP (pink). Each NTP and inhibitor had a concentration of 500 μM. The resulting effects on template-directed RNA strand synthesis as measured by continuous reduction of oscillation frequency were recorded as in Figure 4.
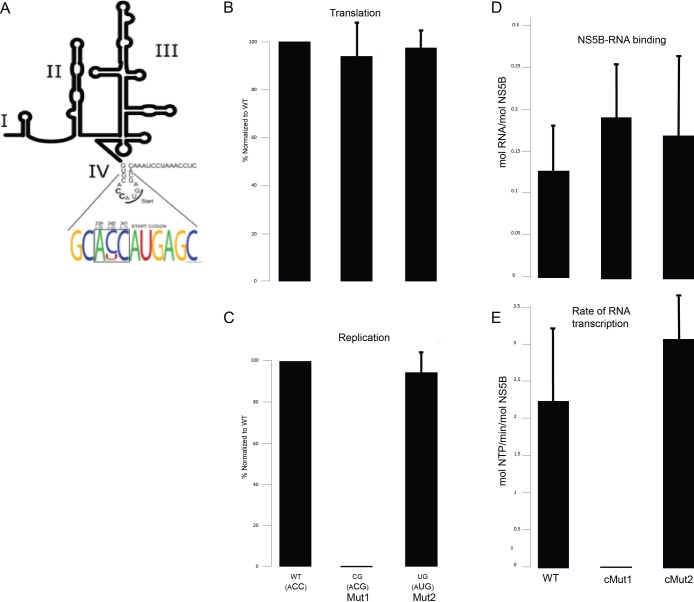
Finally, in addition to using this replicase system to better understand the chemical reactions mediated by the protein components of a replication complex, we also sought to leverage the unique capabilities of this replicase system to probe the role of the RNA template in the replication cycle. The SLB system allows for a simple and direct fashion to investigate the specific sequence motifs within HCV RNA that are important for RNA recognition, binding, and polymerase activity, respectively. As an example, we focused on the segment of HCV RNA that constitutes the HCV internal ribosome entrance site, or IRES. The latter is responsible for cap-independent viral translation. The AUG start codon for translation initiation is located in domain IV of the IRES.41 The three nucleotides preceding this start site (ACC) are part of a Kozak consensus sequence, a translation initiation motif.42 In a Kozak sequence, position −3 from the AUG codon requires a purine, but positions −2 and −1 can be other nucleotides (nt) and still maintain efficient translation.42 Given the high mutation rate of HCV, our observation of a high degree of conservation found at positions −2 and −1 in a large collection of HCV isolates (Figure 6A) led us to hypothesize that positions −2 and −1 might have a role in aspects of the HCV life cycle other than translation, such as viral replication. To test this hypothesis, we performed an extensive mutagenesis study of the ACC triplet preceding the AUG start codon. One interesting mutant (Mut1, encoding “ACG”) demonstrated that while the positions −2 and −1 can be mutated from their highly conserved nature without a significant effect on viral translation (Figure 6B), this had a profound effect on viral RNA replication (Figure 6C). Interestingly, this impairment could be compensated by altering the nucleotide at position −2 (Mut2, encoding “AUG”) (Figures 6B and 6C). When the complementary nucleotide changes at these positions (“cMut1” and “cMut2”, respectively) were introduced into the corresponding 3′ terminus of the negative strand (i.e., the cIRES) template of our replicase system, there was no effect on the ability of NS5B to bind these mutant templates (Figure 6D). As shown in Figure 6E, however, there were profound differential effects on the polymerization reaction. In particular, while the replication defective cMut1 was highly impaired in NS5B catalyzed RNA synthesis, the cMut2 compensatory mutant restored wild-type level of NS5B activity. Together, these data reveal that domain IV of the HCV IRES harbors signals that regulate viral RNA replication independent of translation, most likely via affecting polymerization activity on the corresponding 3′ terminus of the negative strand.
Figure 6.
Membrane-associated HCV replicase assays with mutant RNA templates identifies transcriptional defect associated with impaired HCV RNA genome replication. (A) Weblogo analysis indicating a high degree of conservation at positions −2 and −1 from the AUG translational start site codon in the Kozak consensus sequence in the plus (+) RNA strand of a large collection of HCV isolates. (B) Effect of mutation at positions −2 and −1 to CG (Mut1) or UG (Mut2) on HCV translation. (C) Effect of mutation at positions −2 and −1 to CG (Mut1) or UG (Mut2) on HCV RNA genome replication. (D) NS5B-FL was immobilized on lipid bilayer membranes as shown in previous figures. Wild type or mutant (cMut1 or cMut2, which contain mutations in the minus (−) strand cIRES RNA complementary to Mut1 and Mut2, respectively) cIRES RNA templates were bound as shown in Figure S7C. (E) RNA-dependent RNA transcription activity (mol NTP/min/mol NS5B) observed upon subsequent addition of ribonucleoside triphosphates (NTPs), as in Figure S7.
Discussion
The experimental strategy presented herein offers several advances over existing methods to interrogate the molecular mechanisms which drive self-assembly and function of membrane-associated viral protein complexes. As a test case, we have focused on the HCV replicase complex, although many of the exemplified principles are broadly applicable to other such complexes. First, the SLB platform provides a cell membrane mimicking environment upon which the NS5B-FL protein can function, as is the case in its natural biological environment. Indeed, we found that our NS5B-FL not only displays robust RNA polymerase activity in this assay format but is only functional in this more natural membrane-associated context. The commonly used truncated version of HCV polymerase, NS5B-ΔC21, could not bind to the bilayer membrane and was only active in solution. This finding is in good agreement with past reports where NS5B-FL was found to be either inactive43,44 or poorly active with only minute amounts of RNA synthesized in a relatively long incubation time and requiring specific buffer conditions including detergent, salt, and glycerol to solubilize the NS5B protein.22,45,46
We demonstrate here that, in terms of sensing capabilities, the QCM-D biosensor permits sequential label-free detection of SLB platform self-assembly, NS5B-FL membrane association, kinetics and stoichiometry of NS5B-FL interactions with other proteins (such as NS3 and NS5A) and RNA (e.g., the cIRES RNA template), RNA-dependent RNA polymerase activity, and the latter’s disruption by a small molecule inhibitor. Moreover, these individual steps can be dissected, thus permitting identification of the roles of different components which can lead to a more complete understanding of functional interactions within a membrane associated protein–RNA complex. Importantly, this analysis can be quantified since real-time binding kinetics are monitored. In this initial report, we have elected to use the Sauerbrey relationship for converting the QCM-D frequency shifts into the adsorbed molar number of bound proteins or synthesized RNA stands. While this relationship is quite accurate for rigid films, its application can lead to underestimations of the effective mass for viscoelastic films such as adsorbed protein layers47 and oligonucleotides.48 As such, the kinetics values obtained in this work are best viewed comparatively at this stage, and there is opportunity to apply more complex models to analyze such data. At the same time, the best approach to tackle this problem remains outstanding as even classical viscoelastic models likely do not properly capture the hydrodynamic behavior of discrete, membrane-associated viral replication complexes.49
It should also be emphasized that the QCM-D technique is an acoustic sensor which measures the hydrodynamically coupled mass of an adsorbate, including the bound biomolecules and associated solvent. Due to the coupled solvent, the actual, “dry” mass of biomolecule can be much lower and depends on the types of biomolecule which entrap different amounts of water.50 The dry mass of protein can be 1.3- to 3-times overestimated by QCM-D analysis, while the dry mass of nucleic acids can be overestimated by up to 10 times.51 In the current work, the kinetic values are treated as apparent values while correction factors could in principle be applied to estimate absolute values although it would not affect the main conclusions of this work. Nevertheless, it should be emphasized that there is uncertainty caused by the fact that, for QCM-D measurement data, conversion of changes in resonance frequency into molecular mass varies for different biomolecular systems, and this caveat should be taken into consideration when comparing quantitative values reported here with future studies.
With ongoing advances to form SLBs on formerly intractable substrates such as gold52 and aluminum oxide,53 there is also potential to explore optical biosensor techniques that are insensitive to hydration mass. Above all, it should be emphasized that the present study demonstrates that the SLB system offers unparalleled advantages over solution-based assays for dissecting the macromolecular interactions involved in viral replicase complex assembly and function, and furthermore the platform is compatible with different surface-sensitive measurement techniques. More detailed studies will be aimed at determining if multiple NS5B molecules could bind per RNA molecule, or if only a fraction of membrane-bound NS5B was reconstituted into a conformation that allowed RNA binding. Indeed, evidence for functional NS5B oligomerization has been published previously.25,54,55
Nonetheless, based on the present format, the ability of the QCM-D system to follow an RNA synthesis reaction in real time allows kinetic analyses of the reaction. It is important to note that the calculated apparent kcat of 0.13 ± 0.02 min–1 for NS5B-FL represents a minimum lower estimate, based on the assumption that 100% of the membrane associated NS5B is active and that all NS5B molecules equally contribute to RNA synthesis. For example, as the ratio of RNA template to NS5B on the bilayer membrane was ∼1:5, not all membrane-bound NS5B may be contributing to the polymerization reaction, and a proportion of the protein may be primarily serving a structural role.
In terms of replicase subunit effects, NS5A exhibited marked stimulatory activity on NS5B polymerase activity, but this effect initiated only after a significant lag time of approximately 350 min. This is consistent with the results of Quezada and Kane,56 where NS5A was shown to stimulate NS5B’s activity during the elongation phase. In addition, the complex assembled on the lipid bilayer was able to synthesize plus strand RNA (off of the minus strand cIRES template) which was previously not observed.56 Interestingly, the lag time suggests that NS5A may induce a conformational rearrangement of the NS5B–RNA complex on the bilayer membrane. This could be associated with changing oligomeric states of NS5A and/or NS5B proteins to form the fully functional replicase complex. Such a process might be accelerated by another HCV replicase subunit (supplied by the virus or host).
Characterization of the nature of the synthesized RNA product (Figure S10) revealed that a single distinct ∼200 nucleotide long plus strand RNA product is produced off of the 378 nucleotide minus strand RNA (cIRES) template. These results provide additional confirmation that the change in QCM signal seen upon addition of nucleotides to the complex of membrane-bound NS5B and cIRES negative strand RNA template indeed represents accumulation of newly synthesized RNA. Given the increase in absolute mass upon initiation of the polymerase reaction, we calculate that, at the very least, on average, every enzyme bound negative strand template is actively transcribed to generate the ∼200 nucleotide long plus strand RNA product, indicating a remarkable (∼100%) efficiency.
Further studies will aim at addition of other subunits and their interaction on the membrane platform and host factor(s) that are currently absent in our minimal component in vitro assay. Moreover, our currently configured format offers an ideal assay system for identifying such factors. This system is also ideal for evaluating the RNA template requirements of NS5B. For example, as shown in Figure 6, using this assay system with wild-type and mutant RNA templates allowed us to determine that the mechanism whereby a recently discovered mutation found to be lethal for HCV replication involves specific defects in virus RNA transcription.
Finally, in addition to providing a new platform to investigate key molecular details and mechanisms involved in membrane-associated HCV replicase activity and complex assembly, our results (Figure 5) provide proof-of-concept that candidate small molecule inhibitors of these processes can be evaluated using the membrane-associated authentic NS5B-FL replicase system described here. Beyond the HCV model pathogen, there is great potential for this technology to validate novel targets in other pathogens that are similarly dependent on membrane-associated viral replicase complexes (e.g., HAV, Dengue, JEV) and, more broadly, targets residing in other biologically important membrane-associated complexes that collectively might lead to the development of novel classes of therapeutics.
Materials and Methods
Expression of Recombinant Proteins
Recombinant full length NS3 (NS3-FL, BK strain, a.a. 1–631), NS3 helicase domain (BK strain, a.a 182–631), NS3 protease domain (BK strain, a.a. 1–181), truncated NS5A (NS5A-Δ32, BK strain, a.a. 33–447), truncated NS5B (NS5B-Δ21, BK strain, a.a. 1–570), and truncated NS5B (NS5B-Δ21, BK strain, a.a. 1–570) were expressed in Escherichia coli BL21(DE3) cells. A single colony was used to inoculate TB medium at 37 °C. Protein expression was induced with 0.5 mM IPTG at OD600 between 0.6 and 0.8. Cells were incubated for 18 h at 20 °C and harvested by centrifugation at 4700g for 20 min at 4 °C. Recombinant baculovirus expressing full length NS5B (NS5B-FL, BK-strain, a.a. 1–591) was generated and used to infect Sf9 insect cells at an MOI of 0.5 and harvested after 72 h via centrifugation at 10000g for 10 min.
Purification of Recombinant Proteins
NS3-FL pellets were resuspended, lysed, loaded onto a nickel column (Qiagen), washed, and eluted. Gradient peaks were collected, dialyzed, centrifuged, and loaded on a heparin–sepharose HP column (GE). Peak fractions were pooled and stored at −80 °C. NS3 protease, NS3 helicase, NS5A-Δ32, pellets were handled as described above, except that a Superdex 75 column was used instead of heparin–sepharose HP. NS5B-Δ21 was purified using the following three successive columns: nickel, SP Sepharose, and Superdex 200. NS5B-FL was batch-purified using nickel resin (Qiagen). See Supporting Information for additional details.
Small Unilamellar Vesicle Preparation
Small unilamellar vesicles of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids, Alabaster, AL, USA) were prepared by the extrusion method. For planar lipid bilayer formation and NS5B membrane association studies, we used a Tris buffer composed of 10 mM Tris [pH 7.5], 150 mM NaCl, and 1 mM ethylene diamine tetraacetic acid (EDTA) in 18.2 MΩ cm Milli-Q water (MilliPore, Oregon, USA). Extruded unilamellar vesicles were prepared in the following manner: Lipid films were prepared by first drying the as-supplied lipids dissolved in chloroform under a gentle stream of nitrogen at room temperature. Then, the resulting lipid film was stored under vacuum for at least 5 h in order to remove residual chloroform. Multilamellar vesicles were prepared by first swelling the lipid film in aqueous solution and then vortexing periodically for 5 min. The resulting multilamellar vesicles were subsequently sized by a miniextruder (Avanti Polar Lipids, Alabaster, AL, USA) through polycarbonate membranes with nominal 100, 50, and 30 nm pores. Vesicles were generally prepared at a nominal lipid concentration of ∼5 mg mL–1 and subsequently diluted before experiments. Vesicles were generally used within 1 h of preparation. A DEPC-treated aqueous solution (Ambion Inc.) consisting of 40 mM Tris-HCl (pH 8.0), 40 mM NaCl, 4 mM MgCl2, and 4 mM DTT was used for RNA binding and replicase assays.
Dynamic Light Scattering (DLS)
The effective diameter of the vesicles was measured using dynamic light scattering with a Brookhaven 90 Plus Particle Analyzer (Brookhaven Instruments Corporation, Holtsville, NY) at 25 °C in order to verify the correct unilamellar size distribution for prompting bilayer self-assembly. Dynamic light scattering (DLS) was performed by a 90 Plus particle size analyzer, and the results were analyzed by digital autocorrelator software (Brookhaven Instruments Corporation, New York, USA). All measurements were taken at a scattering angle of 90° where the reflection effect is minimized. All autocorrelation functions were also analyzed by CONTIN and non-negatively constrained least squares (NNLS) algorithms to check for multimodal distributions.
Quartz Crystal Microbalance with Dissipation Monitoring (QCM-D)
Adsorption kinetics and the viscoelastic properties of the adsorbed layer were studied using a Q-Sense E4 (Q-Sense AB, Gothenburg, Sweden) equipped with a flow pump, as previously described (refs (9) and (32)). The crystal was initially driven near its resonance frequency as indicated by a maximum in the current. To capture the characteristic dissipation, the drive circuit was short-circuited and the exponential decay of the crystal oscillation was recorded and analyzed, yielding the frequency and dissipation changes at 5, 15, 25, 35, 45, 55, and 65 MHz. The temperature of the Q-Sense cell was set at 24.0 °C and accurately controlled by a Peltier element in the cell with fluctuations smaller than ±0.05 °C. Most experiments were repeated at least three times with a standard deviation of less than 5%. Each QCM crystal was treated with oxygen plasma at ∼80 W for ∼5 min prior to use (March Plasmod Plasma Etcher, March Instruments, Concord, CA, USA).
Sequence Alignment and Weblogo Analysis
An alignment of the nucleotides surrounding the AUG start codon in domain IV of the HCV IRES was performed using all 471 sequences from the Los Alamos HCV Sequence Database. Weblogo (http://weblogo.berkeley.edu/) was used to generate the sequence alignment display in Figure 6A.
Plasmids and in Vitro RNA Transcription
Standard recombinant DNA technology was used to construct and purify all plasmids. The plasmid FL-J6/JFH-5′C19Rluc2AUbi that consists of the full-length HCV genome and expresses Renilla luciferase was a gift from Charles M. Rice.57 Site-directed mutagenesis was performed using the Stratagene QuikChange Lightning Kit (Agilent Technologies). All mutations were introduced into this plasmid, and the plasmids were analyzed by automated DNA sequencing. In vitro RNA transcription was carried out with the T7MEGAscript kit (Ambion) according to the manufacturer’s protocol. The integrity of the RNA and its concentration were confirmed by 1% agarose gel electrophoresis and ethidium bromide staining.
Cell Cultures and Transfection
Huh 7 cells were maintained in DMEM (Gibco) supplemented with 1% l-glutamine (Gibco), 1% penicillin, 1% streptomycin (Gibco), 1× nonessential amino acids (Gibco) and 10% FBS (Omega Scientific). Cells were seeded in a 6-well plate at 106 cells/well 1 day prior to transfection. Transfection was performed the next day using Lipofectamine 2000 (Invitrogen) and 2.5 μg of HCV RNA. Four hours post transfection, cells were assessed for viability using Alamar Blue assay. Aliquots of equal numbers of cells transfected with individual mutants were subjected to Luciferase assays to assess translation. 15,000 transfected cells per well were plated into 96-well plates. 48 h later, cells were subjected to viability and luciferase assays to assess replication (see below).
HCV Translation and Genome Replication Assays
Both viral translation and viral replication were determined using a Renilla luciferase assay (Promega). Briefly, either 4 or 48 h post transfection, after assessing viability, cells were washed with ice-cold PBS. For the 4 h time point, 60,000 cells were lysed with 20 μL of ice-cold Renilla lysis buffer (Promega) and loaded into a well of a 96-well plate. For the 48 h time point, 20 μL of ice-cold Renilla lysis buffer was added to each well of a 96-well plate. 100 μL of the Renilla luciferase assay buffer containing assay substrate was injected and luciferase activity measured using a plate reader. All experiments were done at least three times, each time with at least quadruplicates.
Acknowledgments
We thank Biing Y. Lin and Sam S. Lee for generously supplying the VX-222 compound. The authors wish to acknowledge support from the National Research Foundation (NRF -NRFF2011-01) and the National Medical Research Council (NMRC/CBRG/0005/2012) (to N.-J.C.); Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (to J.S.G.); NIH RO1 DK064223, RO1 AI087917, RO1 AI099245, and U19 AI109662; N.J.C. is a recipient of an American Liver Foundation Postdoctoral Fellowship Award and a Global Roche Postdoctoral Fellowship. R.J.H. is supported by NIH Graduate Training Grant 5T32AI007328 and a Stanford BIO-X Interdisciplinary Graduate Fellowship. E.A.P. is supported by F30DK099017, the HHMI Medical Student Research Fellowship, the Paul and Daisy Soros Foundation, and the Stanford Medical Scientist Training Program.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.6b00112.
Characterization and experimental details (PDF)
Author Present Address
§ Nanyang Technological University, Singapore.
Author Contributions
∥ N.-J.C. and E.A.P. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Conner S. D.; Schmid S. L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Simons K.; Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Singer S. J.; Nicolson G. L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- Steyer J. A.; Almers W. A real-time view of life within 100 nm of the plasma membrane. Nat. Rev. Mol. Cell Biol. 2001, 2, 268–275. 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- Bienz K.; Egger D.; Rasser Y.; Bossart W. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology 1980, 100, 390–399. 10.1016/0042-6822(80)90530-9. [DOI] [PubMed] [Google Scholar]
- Chu P. W.; Westaway E. G. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 1992, 125, 177–191. 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- Froshauer S.; Kartenbeck J.; Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988, 107, 2075–2086. 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy D. A.; Giddings T. H. Jr.; Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000, 74, 8953–8965. 10.1128/JVI.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R. P.; Bérat R.; Brisson A. R. Formation of solid-supported lipid bilayers: an integrated view. Langmuir 2006, 22, 3497–3505. 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- Chan Y.-H. M.; Boxer S. G. Model membrane systems and their applications. Curr. Opin. Chem. Biol. 2007, 11, 581–587. 10.1016/j.cbpa.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J. A.; Knoll W.; Cho N.-J. Biotechnology applications of tethered lipid bilayer membranes. Materials 2012, 5, 2637–2657. 10.3390/ma5122637. [DOI] [Google Scholar]
- Salafsky J.; Groves J. T.; Boxer S. G. Architecture and function of membrane proteins in planar supported bilayers: a study with photosynthetic reaction centers. Biochemistry 1996, 35, 14773–14781. 10.1021/bi961432i. [DOI] [PubMed] [Google Scholar]
- Granéli A.; Rydström J.; Kasemo B.; Höök F. Formation of supported lipid bilayer membranes on SiO2 from proteoliposomes containing transmembrane proteins. Langmuir 2003, 19, 842–850. 10.1021/la026231w. [DOI] [Google Scholar]
- Jackman J. A.; Cho N.-J.; Duran R. S.; Frank C. W. Interfacial binding dynamics of bee venom phospholipase A2 investigated by dynamic light scattering and quartz crystal microbalance. Langmuir 2010, 26, 4103–4112. 10.1021/la903117x. [DOI] [PubMed] [Google Scholar]
- Cho N. J.; Frank C. W.; Kasemo B.; Hook F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat. Protoc. 2010, 5, 1096–1106. 10.1038/nprot.2010.65. [DOI] [PubMed] [Google Scholar]
- Keller C. A.; Kasemo B. Surface specific kinetics of lipid vesicle adsorption measured with a quartz crystal microbalance. Biophys. J. 1998, 75, 1397–1402. 10.1016/S0006-3495(98)74057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodahl M.; Kasemo B. A simple setup to simultaneously measure the resonant frequency and the absolute dissipation factor of a quartz crystal microbalance. Rev. Sci. Instrum. 1996, 67, 3238–3241. 10.1063/1.1147494. [DOI] [Google Scholar]
- Niikura K.; Matsuno H.; Okahata Y. Direct monitoring of DNA polymerase reactions on a quartz-crystal microbalance. J. Am. Chem. Soc. 1998, 120, 8537–8538. 10.1021/ja980967r. [DOI] [PubMed] [Google Scholar]
- Stengel G.; Höök F.; Knoll W. Viscoelastic modeling of template-directed DNA synthesis. Anal. Chem. 2005, 77, 3709–3714. 10.1021/ac048302x. [DOI] [PubMed] [Google Scholar]
- World Health Organization, Hepatitis C-Global Prevalence (Update); 1999; pp 425–427. [Google Scholar]
- Asselah T.; Boyer N.; Saadoun D.; Martinot-Peignoux M.; Marcellin P. Direct-acting antivirals for the treatment of hepatitis C virus infection: optimizing current IFN-free treatment and future perspectives. Liver Int. 2016, 36, 47–57. 10.1111/liv.13027. [DOI] [PubMed] [Google Scholar]
- Behrens S. E.; Tomei L.; De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996, 15, 12–22. [PMC free article] [PubMed] [Google Scholar]
- Wolk B.; Buchele B.; Moradpour D.; Rice C. M. A dynamic view of hepatitis C virus replication complexes. J. Virol. 2008, 82 (21), 10519–10531. 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D.; Aligo J.; Xu C.; Park W. S.; Koc H.; Heo W. D.; Konan K. V. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 2010, 398 (1), 21–37. 10.1016/j.virol.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinkert D.; Bartenschlager R.; Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005, 79, 13594–13605. 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E.; Wright-Minogue J.; Fang J. W.; Baroudy B. M.; Lau J. Y.; Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 1999, 73, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T.; Kaneko S.; Shirota Y.; Qin W.; Nomura T.; Kobayashi K.; Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 1998, 273, 15479–15486. 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- Klumpp K.; Leveque V.; Le Pogam S.; Ma H.; Jiang W. R.; Kang H.; Granycome C.; Singer M.; Laxton C.; Hang J. Q.; Sarma K.; Smith D. B.; Heindl D.; Hobbs C. J.; Merrett J. H.; Symons J.; Cammack N.; Martin J. A.; Devos R.; Najera I. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 2006, 281, 3793–3799. 10.1074/jbc.M510195200. [DOI] [PubMed] [Google Scholar]
- Johnson R. B.; Sun X. L.; Hockman M. A.; Villarreal E. C.; Wakulchik M.; Wang Q. M. Specificity and mechanism analysis of hepatitis C virus RNA-dependent RNA polymerase. Arch. Biochem. Biophys. 2000, 377, 129–134. 10.1006/abbi.2000.1749. [DOI] [PubMed] [Google Scholar]
- Adachi T.; Ago H.; Habuka N.; Okuda K.; Komatsu M.; Ikeda S.; Yatsunami K. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta, Proteins Proteomics 2002, 1601, 38–48. 10.1016/S1570-9639(02)00433-8. [DOI] [PubMed] [Google Scholar]
- Leveque V. J.; Johnson R. B.; Parsons S.; Ren J.; Xie C.; Zhang F.; Wang Q. M. Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis. J. Virol. 2003, 77, 9020–9028. 10.1128/JVI.77.16.9020-9028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodahl M.; Hook F.; Fredriksson C.; Keller C. A.; Krozer A.; Brzezinski P.; Voinova M.; Kasemo B. Simultaneous frequency and dissipation factor QCM measurements of biomolecular adsorption and cell adhesion. Faraday Discuss. 1997, 107, 229–246. 10.1039/a703137h. [DOI] [PubMed] [Google Scholar]
- Cho N. J.; Frank C. W.; Kasemo B.; Hook F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat. Protoc. 2010, 5, 1096–1106. 10.1038/nprot.2010.65. [DOI] [PubMed] [Google Scholar]
- Sauerbrey G. Use of quartz vibration for weighing thin films on a microbalance. Eur. Phys. J. A 1959, 155, 206–212. 10.1007/BF01337937. [DOI] [Google Scholar]
- McCubbin G. A.; Praporski S.; Piantavigna S.; Knappe D.; Hoffmann R.; Bowie J. H.; Separovic F.; Martin L. L. QCM-D fingerprinting of membrane-active peptides. Eur. Biophys. J. 2011, 40, 437–446. 10.1007/s00249-010-0652-5. [DOI] [PubMed] [Google Scholar]
- Brass V.; Pal Z.; Sapay N.; Deleage G.; Blum H. E.; Penin F.; Moradpour D. Conserved determinants for membrane association of nonstructural protein 5A from hepatitis C virus and related viruses. J. Virol. 2007, 81, 2745–2757. 10.1128/JVI.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N. J.; Cheong K. H.; Lee C.; Frank C. W.; Glenn J. S. Binding dynamics of hepatitis C virus’ NS5A amphipathic peptide to cell and model membranes. J. Virol. 2007, 81, 6682–6689. 10.1128/JVI.02783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar M.; Cheong K. H.; Liu P.; Greenberg H. B.; Rice C. M.; Glenn J. S. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 2003, 77, 6055–6061. 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W.; Jiao P.; Liu H.; Yao X. Molecular modeling and residue interaction network studies on the mechanism of binding and resistance of the HCV NS5B polymerase mutants to VX-222 and ANA598. Antiviral Res. 2014, 104, 40–51. 10.1016/j.antiviral.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Eltahla A. A.; Luciani F.; White P. A.; Lloyd A. R.; Bull R. A. Inhibitors of the Hepatitis C Virus Polymerase; Mode of Action and Resistance. Viruses 2015, 7, 5206–5224. 10.3390/v7102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M.; Brown E. A.; Lemon S. M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 1996, 2, 955–968. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Al R. H.; Xie Y.; Wang Y.; Hagedorn C. H. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 1998, 53, 141–149. 10.1016/S0168-1702(97)00147-0. [DOI] [PubMed] [Google Scholar]
- Luo G.; Hamatake R. K.; Mathis D. M.; Racela J.; Rigat K. L.; Lemm J.; Colonno R. J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 2000, 74, 851–863. 10.1128/JVI.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Francesco R.; Behrens S. E.; Tomei L.; Altamura S.; Jiricny J. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 1996, 275, 58–67. 10.1016/S0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- Lohmann V.; Korner F.; Herian U.; Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 1997, 71, 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höök F.; Kasemo B.; Nylander T.; Fant C.; Sott K.; Elwing H. Variations in coupled water, viscoelastic properties, and film thickness of a Mefp-1 protein film during adsorption and cross-linking: a quartz crystal microbalance with dissipation monitoring, ellipsometry, and surface plasmon resonance study. Anal. Chem. 2001, 73, 5796–5804. 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
- Larsson C.; Rodahl M.; Höök F. Characterization of DNA immobilization and subsequent hybridization on a 2D arrangement of streptavidin on a biotin-modified lipid bilayer supported on SiO2. Anal. Chem. 2003, 75, 5080–5087. 10.1021/ac034269n. [DOI] [PubMed] [Google Scholar]
- Reviakine I.; Johannsmann D.; Richter R. P. Hearing what you cannot see and visualizing what you hear: interpreting quartz crystal microbalance data from solvated interfaces. Anal. Chem. 2011, 83, 8838–8848. 10.1021/ac201778h. [DOI] [PubMed] [Google Scholar]
- Bingen P.; Wang G.; Steinmetz N. F.; Rodahl M.; Richter R. P. Solvation effects in the quartz crystal microbalance with dissipation monitoring response to biomolecular adsorption. A phenomenological approach. Anal. Chem. 2008, 80, 8880–8890. 10.1021/ac8011686. [DOI] [PubMed] [Google Scholar]
- Stengel G.Real-time monitoring of DNA hybridization and replication using optical and acoustic biosensors; Max-Planck Institute for Polymer Research: 2004. [Google Scholar]
- Tabaei S. R.; Choi J.-H.; Haw Zan G.; Zhdanov V. P.; Cho N.-J. Solvent-assisted lipid bilayer formation on silicon dioxide and gold. Langmuir 2014, 30, 10363–10373. 10.1021/la501534f. [DOI] [PubMed] [Google Scholar]
- Jackman J. A.; Tabaei S. R.; Zhao Z.; Yorulmaz S.; Cho N.-J. Self-assembly formation of lipid bilayer coatings on bare aluminum oxide: overcoming the force of interfacial water. ACS Appl. Mater. Interfaces 2015, 7 (1), 959–968. 10.1021/am507651h. [DOI] [PubMed] [Google Scholar]
- Clemente-Casares P.; Lopez-Jimenez A. J.; Bellon-Echeverria I.; Encinar J. A.; Martinez-Alfaro E.; Perez-Flores R.; Mas A. De novo polymerase activity and oligomerization of hepatitis C virus RNA-dependent RNA-polymerases from genotypes 1 to 5. PLoS One 2011, 6, e18515. 10.1371/journal.pone.0018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. M.; Hockman M. A.; Staschke K.; Johnson R. B.; Case K. A.; Lu J.; Parsons S.; Zhang F.; Rathnachalam R.; Kirkegaard K.; Colacino J. M. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002, 76, 3865–3872. 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada E. M.; Kane C. M. The Hepatitis C Virus NS5A Stimulates NS5B During In Vitro RNA Synthesis in a Template Specific Manner. Open Biochem. J. 2009, 3, 39–48. 10.2174/1874091X00903010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne D. M.; Jones C. T.; Evans M. J.; Lindenbach B. D.; McKeating J. A.; Rice C. M. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 2006, 80, 1734–1741. 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.