Abstract
Integrin adhesion receptors mediate the adhesion of blood cells, such as leukocytes, to other cells, such as endothelial cells. Integrins also are critical for anchorage of hematopoietic precursors to the extracellular matrix. Blood cells can dynamically regulate the affinities of integrins for their ligands (“activation”), an event central to their functions. Here we review recent progress in understanding the mechanisms of integrin activation with a focus on the functions of blood cells. We discuss how talin binding to the integrin β cytoplasmic domain, in conjunction with the plasma membrane, induces long-range allosteric rearrangements that lead to integrin activation. Second, we review our understanding of how signaling events, particularly those involving Rap1 small guanosine triphosphate (GTP)hydrolases, can regulate the talin–integrin interaction and resulting activation. Third, we review recent findings that highlight the role of the Rap1-GTP-interacting adapter molecule (RIAM), encoded by the APBB1IP gene, in leukocyte integrin activation and consequently in leukocyte trafficking.
Introduction
Circulating blood cells patrol the body to guard against pathogens and maintain homeostasis. The adhesive interactions of leukocytes with the blood vessel walls are transient and dynamic and serve a wide range of immune functions.1 Similarly, platelets do not stably adhere to the intact vessel wall; however, when the endothelium is disrupted, a rapid interaction between circulating platelets and the vessel leads to the formation of a platelet–leukocyte–fibrin aggregate that mediates hemostasis.2 Members of the integrin adhesion receptor family play important roles in these processes. Integrins are type 1 transmembrane receptors that mediate cell–cell and cell–extracellular matrix adhesion.3 Each integrin heterodimer contains 1 α and 1 β subunit. In vertebrates, 18 α and 8 β subunits can form 24 integrins with characteristic tissue distributions and distinct ligand binding specificities.3,4 Leukocyte integrins include αLβ2 (LFA-1, CD11a/CD18), α4β1 (VLA-4), and αMβ2 (Mac-1, CD11b/CD18),5,6 whereas integrin αIIbβ3 (GPIIb–IIIa) is by far the most abundant platelet integrin.7 Integrins sense chemical and physical properties of extracellular matrix and control signal transduction pathways that regulate cell adhesion, proliferation, differentiation, and apoptosis.3 Mutations of integrin genes can lead to blood diseases such as leukocyte adhesion deficiency I syndrome,8,9 due to mutations in the gene encoding integrin β2, or Glanzmann thrombasthenia,10 due to mutations in the genes encoding αIIb or β3.
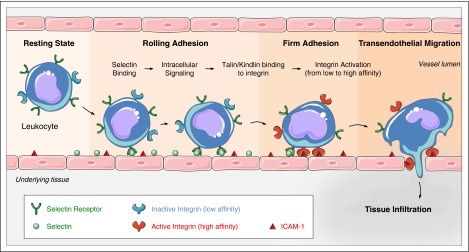
Integrins in blood cells are usually in a low-affinity state until agonist stimulation induces a high-affinity form, a process operationally defined as “integrin activation.”6,11-13 On platelet stimulation by agonists such as thrombin or collagen, activated αIIbβ3 binds fibrinogen, fibronectin, and von Willebrand factor to enable both stable adherence to the vessel wall and platelet aggregation.3 Similarly, when rolling leukocytes are stimulated by agonists such as chemokines, the activated β2 integrins bind to endothelial ICAMs to enable leukocyte arrest and subsequent extravasation during inflammatory responses (Figure 1).3 The capacity of intracellular signaling pathways to induce such changes in integrin conformation and affinity has been termed inside-out signaling to contrast with outside-in signaling that occurs when ligand binding to the extracellular domain initiates intracellular signals. Binding of talin to the integrin β cytoplasmic tail is a critical final step in the process of integrin activation.14-17 Here, we discuss our current understanding of the molecular mechanisms underlying integrin activation in blood cells, focusing on regulation of recruitment of talin to integrins and the structural basis of talin-induced activation.
Figure 1.
Leukocyte trafficking depends on integrin activation. Leukocytes in flowing blood are captured by and roll via selectin ligand interactions. Selectin engagement signals talin-mediated integrin activation that leads to integrin extension and slowed rolling. Activation of the leukocytes, usually through G-protein–coupled receptors culminates in kindlin and talin-mediated arrest and subsequent leukocyte transmigration. Graphical art has been adapted from Les Laboratoires Servier © 2016.
Talin induces integrin activation
Regulation of integrin transmembrane signaling
The effects of deletion of talin in mouse blood cells have unequivocally confirmed its importance in integrin activation in platelets18,19 and leukocytes20 as first predicted from model systems.21,22 Moreover, structural studies23-25 have enabled construction of mice with point mutations in integrin β326 or talin27,28 that block αIIbβ3 activation and therefore inhibit platelet aggregation, hemostasis, and thrombosis. Similarly, the talin1(L325R) mutation that specifically blocks the capacity of talin to activate integrins impairs the capacity of rolling neutrophils to arrest and consequently inhibits renal reperfusion injury.29 These data firmly establish the importance of talin binding to the integrin β cytoplasmic domain in activation and focus attention on the mechanisms by talin causes activation.30
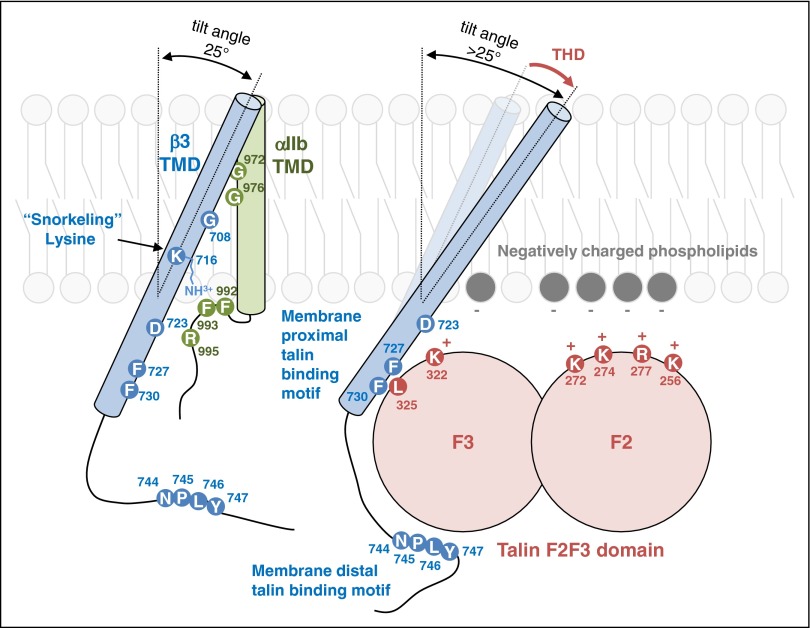
Both site-directed31-33 and random34 mutagenesis combined with Förster resonance energy transfer35 suggested that intersubunit interactions of the integrin α and β cytoplasmic and transmembrane domains constrain integrins in a low-affinity “off” state. In consequence, disruption of these α–β interactions is a central event in activation. Structural studies showed that integrin β3 transmembrane domain (TMD) is tilted ∼25° in a model lipid bilayer.36,37 The β3 tilt angle is maintained by “snorkeling,”38 the positioning of the positively charged end of lysine side chain (K716 in case of integrin β3) in the negatively charged phosphate head group of lipids near the membrane–water interface (Figure 2).37,39 This tilt allows helix packing interaction between αIIb and β3 TMDs mediated by glycine residues αIIb(G972), αIIb(G976), and β3(G708) (Figure 2). Changes in the tilt angle caused by loss of the snorkeling lysine residue β3(K716E)39 or TMD-shortening mutation β3(L712R)34 can disrupt the αIIb-β3 TMD interaction and cause activation of the integrin. Similarly, mutation of the paralogous snorkeling residue in β1A(K732) can also disrupt α5β1 interaction and cause α5β1 activation.39 Thus, changing the tilt angle may be a general way to control integrin activation,34 and the role of the TMD tilt in regulating other integrins, such as α4β7, warrants future investigation.
Figure 2.
A model of talin-dependent integrin activation. The tilt angle of β3 TMD is maintained by the interaction between the positively charged end of K716 and negatively charged-phosphate head group. TMD packing interaction of integrin αIIb and β3 is mediated through glycine residues (G972, G976 in αIIb and G708 in β3) in the middle of the TMDs. The TMD interaction is also stabilized by electrostatic interaction between D723 and R995. The membrane distal region of β3 cytoplasmic tail interacts with talin through an NPXY motif, which is part of the primary talin-binding site. The membrane proximal region of β3 cytoplasmic tail interacts with talin via hydrophobic interaction between two Phe residues (F727 and F730) in β3 and L325 in talin. The positively charged residues (K322, K272, K274, R277, and K256) on the surface of talin can interact with lipid bilayer. Talin binding to the β3 membrane proximal region and lipid bilayer increases the tilt angle of β3 TMD thereby disrupting the helical packing interaction of the TMD leading to their rearrangement and resulting long range conformational change in the integrin. The amino acid residues of integrin αIIb (blue), β3 (green), and talin1 (red) are numbered.
Talin head domain contains a FERM (band 4.1, ezrin, radixin, and moesin) domain composed of 4 subdomains: F0, F1, F2, and F3. Ectopic expression of the talin head induces activation.21,40 Furthermore, it can activate purified integrin αIIbβ3 embedded in an artificial lipid bilayer as assessed by both ligand binding affinity and conformational rearrangement.16 The binding of talin to sites in the integrin β cytoplasmic tail, and in combination with binding to negatively charged membrane lipids, activates integrins αIIbβ3 in cells and can change the tilt angle of β3 TMD (Figure 2).41,42 Furthermore, insertion of a flexible kink in the β3 TMD prevents transmission of tilting across the membrane and blocks integrin activation.41,42 Thus, the capacity of talin to change the topology of β3 TMD disrupts the αIIb-β3 TMD interaction, thereby activating integrin αIIbβ3 (Figure 2).
Is talin dispensable for integrin activation?
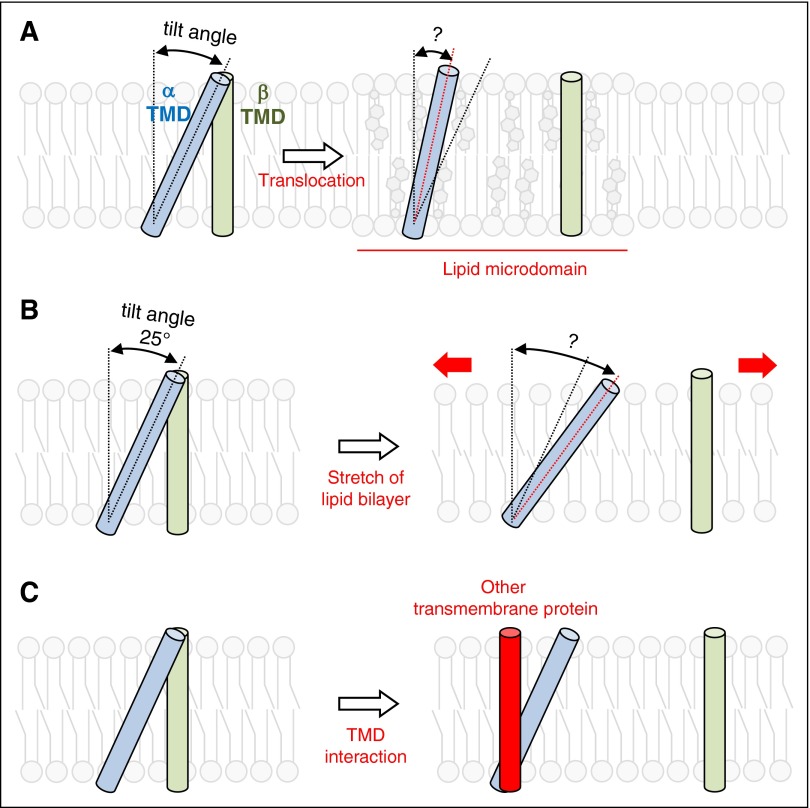
Mutations in integrins or talin that prevent their interaction block the activation of blood cell integrins. In principle, other mechanisms exist to disrupt the integrin αβ TMD interaction or directly alter the conformation of the extracellular domain and could lead to integrin activation. For example, localization of activated integrins to lipid microdomains (Figure 3A)43 occurs in leukocytes.44,45 Such microdomains are enriched in rigid and planar-shaped cholesterol molecules and unsaturated lipids, and the integrin β TMD tilt angle may be altered due to the altered membrane properties in these microdomains. In addition, mechanical stretch of the lipid bilayer, which could be caused by the forces experienced by a rolling leukocyte or platelet, could deform the membrane (Figure 3B), changing thickness, curvature, or surface tension.46 These changes in membrane properties could alter TMD topologies, TMD helix–helix interaction, and thus activities of membrane proteins.47-52 Indeed, a previous study showed that the affinity of integrin αvβ3 is increased by stretching cells, although the increased affinity was ascribed to stretch-induced activation of phosphatidylinositol 3-kinase.53 It would be of interest to test whether membrane stretch or altered lipid composition can change integrin β TMD topologies to mediate talin-independent integrin activation. Finally, the interaction of integrin TMDs with other TMD can activate integrins (Figure 3C) as elegantly demonstrated by the Bennett and DeGrado groups.54,55 When integrins are clustered, intermolecular TMD interactions between integrins themselves could compete with the intramolecular α-β TMD interaction. Indeed, homomeric (α-α or β-β) TMD interactions between integrins54 and more recently heteromeric (α-β) TMD interaction56 have been proposed to cause integrin activation by disrupting the intramolecular α-β TMD interaction.
Figure 3.
Hypothetical models of talin-independent integrin activation. (A) Translocation of integrins to lipid microdomains, distinct regions of the plasma membrane enriched in cholesterols and unsaturated lipids, can change the tilt angle of β3 TMD and disrupt the TMD interaction. (B) Stretch of lipid bilayer may change the properties of lipid bilayers, leading to the change in TMD tilt angle and disruption of the α-β TMD interaction. (C) When one of TMDs in the integrin molecule interacts with other TMDs, the α-β TMD interaction in the integrin can be disrupted. The disrupted α-β TMD interactions can then activate integrins.
Cooperation between talin and kindlins in integrin activation
The strength of integrin-mediated cell adhesion is determined by the affinity state of each integrin and the number of integrin–ligand interactions at the site of adhesion.57 Kindlins, a family of FERM domain-containing proteins, bind to integrin β cytoplasmic tails and play essential roles in the integrin-mediated cell–extracellular matrix interaction.58,59 When kindlin-2 is depleted in Chinese hamster ovary cells ectopically expressing integrin αIIbβ3, talin-induced ligand binding to integrins is inhibited, establishing that talin is insufficient for full activation when kindlin-2 is absent in these cells.60 Kindlin-3 is the dominant kindlin in blood cells and landmark studies established that this kindlin is required for efficient binding of multivalent ligands to platelets and leukocytes.61 Furthermore, kindlin 3 mutations results in life-threatening platelet and leukocyte dysfunction in humans with leukocyte adhesion deficiency type 3 (also known as LAD1v).61-64 In addition to their role in increasing ligand binding to integrins, kindlins can also enhance the ability of integrins to transmit biochemical signals into cells,61 and recent study suggests that kindlins can promote the clustering of talin-activated integrins, thereby increasing the binding of multivalent ligands, such as fibrinogen.65 Because integrin clustering is important in the capacity of integrins to signal into cells, kindlin-mediated clustering can contribute to “outside-in” integrin signaling. Readers are referred to other reviews for a more detailed description of kindlins and their role comparison with that of talin in integrin functions.58,59,66
Rap proteins integrate adhesive stimuli
Rap proteins
Whereas talin and kindlins play critical roles in the final events in integrin-mediated adhesion, how their interactions with the integrins are regulated are only partially understood. Although Rap1-independent pathways of integrin activation clearly exist,67 Rap1 guanosine triphosphate (GTP) hydrolases (GTPases) are perhaps the most completely studied relays of signals from cell surface receptors to integrin activation. Rap proteins are small GTPases of the Ras family.68 In mammals, the Rap GTPases are encoded by 2 Rap1 genes, Rap1A and Rap1B, whose protein products are 95% identical in sequence, and 2 Rap2 genes encoding proteins that share 60% identity with Rap1. Whereas Rap1A is expressed ubiquitously, Rap1B is the predominant isoform and the most abundant Ras family member in platelets. Rap1 proteins are targeted to lipid membranes by covalently attachment of a geranylgeranyl lipid to the terminal cysteine during Rap1 synthesis and transport through the Golgi.69 In many cell types, Rap1 associates with specific endosomal compartment to serve as a pool of membrane that can be rapidly mobilized and translocate to the cell surface during cellular stimulation.69,70 Accumulating evidence indicates that Rap1 is a physiologic activator of integrins, playing important roles in the regulation of a variety of integrin-dependent cellular functions.71 Here, we focus on description of direct Rap1 regulators and regulation of integrin function in blood cells. Readers are referred to other reviews for a more complete listing of Rap1 signaling and functions in hematopoietic cells.72,73
Roles of Rap GTPases in integrin-mediated functions of blood cells
The physiologic relevance of Rap1 function in integrins was first reported in lymphocytes isolated from mice lacking a functional Rap1A gene.74 Rap1A-deficient T and B cells have diminished adhesive capacity to ICAM-1 and fibronectin substrates.74 Furthermore, LFA-1 polarization on the surface of activated T cells is impaired in Rap1A knockout cells74; however, these defects do not result in hematopoietic or cell homing abnormalities. Transgenic mice that constitutively express an active Rap1A mutant (Rap1A-V12) within the T-cell lineage exhibit strong T-cell adhesion via β1 and β2 integrins.75 Rap1B is the dominant Rap1 isoform in B cells where it is crucial for B-cell development, marginal zone B-cell maturation, and T-dependent humoral responses.76 Additionally, absence of Rap1B leads to reduced B-cell adhesion, chemotaxis and in vivo homing.
The Rap1B knockout mice have a mild bleeding defect due to abnormal platelet function.77 In vitro aggregation of Rap1B-null platelets is reduced in response to stimulation with both G-protein–coupled receptor (GPCR)-linked and GPCR-independent agonists due to impaired, but not abrogated, integrin αIIbβ3 activation. Moreover, Rap1B-null platelets are protected from thrombosis.77 Platelets also express Rap2B that associates with αIIbβ3 and translocates to the actin cytoskeleton on platelet stimulation.78
Several Rap1 effectors including regulator for cell adhesion and polarization enriched in lymphoid tissues (RAPL),79 Rap1-GTP-interacting adaptor molecule (RIAM),80 and Ras associated and diluted domains (RADIL)81 can link Rap1 to integrin activation or avidity in various cell types. The connection between talin and RAPL or RADIL is unknown; thus, in the section below, we focus on the mechanisms of RIAM-mediated regulation of integrin activation.
Regulatory mechanisms of Rap1
Rap1, like other small G proteins, cycles between guanosine diphosphate (GDP)-bound inactive and guanosine triphosphate (GTP)-bound active forms to act as timed molecular switches that can precisely control cellular processes.68 Conversion from the GDP-bound form to the GTP-bound form is mediated by guanine nucleotide exchange factors (GEFs) that facilitate the release of GDP and binding to GTP. On activation by GTP binding, Rap1 undergoes a conformational change that exposes an effector-binding loop, enabling the recruitment of a variety of effectors. Rap1 signaling is terminated by the hydrolysis of bound GTP to GDP through specific GTPase-activating proteins (GAPs).
Several Rap1 GEFs have been identified, enabling Rap1 to respond to a diverse set of stimuli including chemokine signaling through GPCRs, signaling downstream of antigen receptors (T-cell receptors [TCRs] and B-cell receptors), and cell adhesion molecules.5,71,73 A critical step on the path to integrin activation is the proper localization and activation of Rap1 GEFs. C3G (Crk SH3-domain-binding guanine nucleotide releasing factor, also known as RAPGEF1) is the first Rap1 GEF to be identified.82 C3G regulates Rap1 activation induced by tyrosine kinases and plays an important role in activation of β1 and β2 integrins in many hematopoietic cell types. Moreover, studies using transgenic mice with specific C3G overexpression in megakaryocytes revealed higher activation and aggregation of transgenic platelets compared with wild-type animals; in contrast, platelets expressing a C3G mutant with a deletion in the GEF domain showed impaired activation and aggregation.83 C3G activation is mostly regulated by translocation to the plasma membrane through its interaction with SH3 domains of Crk family of adapter proteins.84,85 Ligand activation of a variety of receptors such as cytokine receptors for interleukin-3 or erythropoietin86, the TCR,87 and the B-cell receptor88 nucleates sites of tyrosine phosphorylation at the plasma membrane. The SH2 domain in Crk enables translocation of the Crk/C3G complex to these foci of tyrosine phosphorylation to mediate integrin activation.84,85 Calcium and diacylglycerol (CalDAG)-GEFs comprise another family of GEFs that responds to calcium and diacylglycerol (DAG).89 Four CalDAG-GEF gene products have been identified. CalDAG-GEFI (RasGRP2) is a Rap GEF expressed in the brain and hematopoietic cells. CalDAG-GEFIII (RasGRP3) is expressed predominantly in B cells. CalDAG-GEFII (RasGRP1) is a Ras regulator preferentially expressed in T cells. RasGRP4 is a Ras activator highly expressed in mast cells. In Jurkat T cells, TCR-mediated activation of Rap1 depends on phospholipase C-γ, and this activity is likely to be mediated by CalDAG-GEFI, which is required to activate LFA-1.90 Compelling in vivo evidence for the role of Rap1 in integrin function come from mice deficient in CalDAG-GEFI.91,92 These mice exhibit defective Rap1 and β1, β2, and β3 integrin activation resulting in a combination of defects in leukocyte and platelet functions. In patients, a point mutation in the CalDAG-GEFI gene impairs αIIbβ3 integrin inside-out and outside-in signaling resulting in platelet dysfunction associated with severe bleeding.93 Another family of Rap1GEFs includes members of Epac family.94 Although Epac1 is expressed in most tissues with relatively low levels in leukocytes, Epac2 is predominantly expressed in the brain and in the adrenal gland.95 Binding of cAMP activates Epac GTP exchange activity.94 The Epac1/Rap1 signaling pathway promotes red blood cell adhesion to laminin.96 Activation of Epac1 in monocytes enhances adhesion to endothelial cells and promotes cell polarization and chemotaxis.97 PDZ-GEF1/2 characterized by the presence of a PSD-95/DlgA/ZO-1 (PDZ) domain98 and dedicators of cytokinesis protein 4 (DOCK4)99 are other classes of Rap1/2 activators, but information on their expression and functional role in blood cells is currently limited.
In contrast to the molecular diversity of GEFs, only 2 families of Rap1-specific GAPs have been identified. The first consists of Rap1GAP100 and its splice variant Rap1GAPII.101 Their activity and translocation to the plasma membrane are regulated by direct interaction with Gα proteins.101 The second family of Rap1-specific GAPs encompasses the structurally related proteins signal-induced proliferation-associated protein-1 (SPA-1).102 SPA-1 is characterized by the presence of a PDZ domain that mediates interaction with membrane-associated proteins. The expression patterns of SPA-1 and Rap1GAP are quite distinct and tend to be segregate in human tissues.103 SPA-1 is predominantly expressed in the lymphoid tissues in the absence of Rap1GAP, whereas tissues such as brain, kidney, and pancreas strongly express Rap1GAP with little SPA-1 expression. SPA-1 is the principal GAP for Rap1 in hematopoietic progenitors. Although the dependence on integrin signaling is unknown, null mutant mice of the SPA-1 gene develop a spectrum of myeloid disorders.104 Rap1GAPII is the only known GAP of Rap1 in platelets105; however, its role in platelet function remains unknown.
RIAM relays Rap1 signaling to talin
Biochemical signaling through RIAM
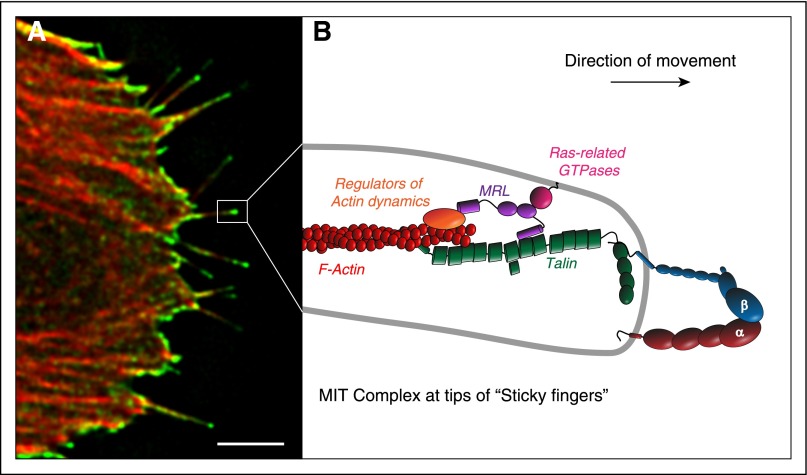
RIAM was identified as a Rap1-binding protein important for leukocyte integrin activation.80 RIAM induces β1 and β2 integrin-mediated adhesion in Jurkat T cells and is required for Rap1-dependent adhesion in these cells. RIAM contains Ras association (RA) and pleckstrin homology (PH) domains and proline-rich regions, which are defining features of the Mig-10/RIAM/Lamellipodin (MRL) family of adapter proteins. Recruitment of RIAM to the plasma membrane is mediated through its RA and PH domains that are both required for lymphocyte adhesion.106 Several critical adaptor molecules in T cells are necessary to translate TCR engagement to integrin activation, particularly 55-kDa src kinase-associated phosphoprotein (SKAP-55) and the adhesion and degranulation-promoting adapter protein (ADAP). RIAM constitutively interacts with the SKAP-55/ADAP module to promote the membrane targeting of the RIAM-Rap1 module on antigen stimulation of T cells to facilitate LFA-1 integrin activation.107 Furthermore, studies using synthetic approach in combination with model nonhematopoietic cells reported that Rap1 mediates integrin activation by forming a complex containing talin in combination with RIAM, which targets talin to integrin.108 Mapping studies identified short amphipathic helices in the RIAM N terminus that bind talin, and joining those helical peptides to the membrane targeting sequences of Rap1 led to a minimized Rap1-RIAM module that is sufficient to recruit talin to activate integrins.109 Thus, RIAM functions as a scaffold that in effect connects the membrane targeting sequences in Rap1 to talin, thereby recruiting talin to the plasma membrane and activating integrins.30,109 In adherent cells, RIAM is most abundant at the cell edge and lamellipodium and is then subsequently reduced in mature adhesions due to direct competition with vinculin for binding sites on talin.110 Vinculin stabilizes adhesions, increasing their ability to transmit forces, whereas RIAM promotes lamellipodial protrusion.110 The transition of integrin-based adhesions from drivers of lamellipodial protrusion to stable focal adhesions delineates a molecular switch in adhesion maturation. The protrusive activity of RIAM is likely mediated by modulation of the actin cytoskeleton through binding to both profilin and Ena/VASP (enabled/vasodilator-stimulated phosphoprotein).80 Overexpression of RIAM induces actin polymerization and cell spreading and lamellipodia formation. In contrast, RIAM knockdown cells have reduced content of polymerized actin.80 RIAM, or its paralogue, Lamellipodin (Lpd), forms a complex with talin and integrin that is enriched at the tip of growing actin filament in lamellipodial and filopodial protrusions (Figure 4).111 In this complex, the N terminus of the MRL protein binds and recruits talin to the plasma membrane to induce integrin activation, and the C terminus of MRL protein increases processive actin polymerization in part by recruiting ENA/VASP, thereby propelling the movement of the “sticky fingers” (Figure 4). Future work is needed to assess the role of the RIAM-driven sticky fingers in leukocyte adhesion and migration during immune responses. Although there is strong evidence that RIAM serves integrin inside-out signaling, its capacity to regulate actin dynamics portends a potential contribution to signaling that follows adhesion (outside-in signaling); indeed, such a role has been documented in β1 integrin outside-in signaling in melanoma cells.112 Future studies in hematopoietic cells are clearly warranted.
Figure 4.
The MRL protein-integrin-talin (MIT) complex forms the molecular basis of sticky fingers that direct cell migration. RIAM and its paralogue, lamellipodin, are the mammalian MRL proteins. (A) The MIT complex, visualized by bimolecular fluorescence complementation (green) between RIAM and integrin αIIbβ3, is enriched at the tips of growing actin filaments (red) in protruding regions of migrating cells. (B) Schematic representation of the MIT complex at the tips of sticky fingers. The N terminus of the MRL protein (RIAM or Lamellipodin) binds talin, thereby enabling its recruitment to the integrin to induce activation. The C terminus of MRL proteins increases processive actin polymerization in part by recruiting ENA/VASP and activators of the ARP2/3 complex to drive the rapid translocation of the integrins. Together, these 2 biochemical functions of the MRL proteins result in the formation of the sticky fingers at the cell edge that direct protrusion during cell migration. Scale bar: 5 μm. Adapted from Lagarrigue et al.111
Biological functions of RIAM
RIAM is abundant in hematopoietic cells, and RIAM knockdown blocks agonist-mediated αIIbβ3 activation in primary mouse megakaryocytes.30 Despite in vitro evidence for an important role of the ternary Rap1/RIAM/talin module in αIIbβ3 integrin activation, genetic deletion of the RIAM gene in mice does not affect development, homeostasis, or platelet integrin functions.113-115 This unexpected finding challenges the view of RIAM as key regulator of integrin activation and indicates that RIAM-independent mechanisms exist for Rap1 to mediate its effects on platelet integrin function. RIAM levels are low in platelets, whereas Rap1, talin1, and integrins are highly expressed in platelets,116 implying the existence of alternative Rap effectors that can regulate talin in platelets. Lamellipodin is not expressed in platelets, thereby excluding a compensatory mechanism.113 The fact that RIAM is dispensable for platelet integrin activation also raises the question whether integrin functions in other hematopoietic cells are compromised in the RIAM-null mice. Recent studies have answered this important question.114,115 Work from Fässler's group highlighted the essential role of RIAM in the activation of β2 integrin in neutrophils, macrophages, and T cells.114 By contrast, β1 and β3 integrin functions are only partially affected by the absence of RIAM in these leukocytes.114 Interestingly, RIAM deficiency has a less pronounced effect on leukocyte β2 integrin function than talin1 deficiency,114 emphasizing the existence of RIAM-independent mechanisms, leading to talin recruitment to integrins. Importantly, RIAM-deficient mice exhibit significant leukocytosis associated with leukocyte adhesion deficiency and impaired leukocyte extravasation.114 At the same time, Philips' group reported that the Rap1/RIAM module is essential for efficient lymphocyte adhesion to ICAM-1 and VCAM-1 and for proper trafficking of B and T cells to secondary lymphoid organs.115 Accordingly, they showed that RIAM is required for a normal humoral response to a T-dependent antigen. Furthermore, RIAM-deficient macrophages exhibit impaired adhesion and spreading on ICAM-1,114 thereby complementing previous studies indicating that RIAM is actively involved in complement-dependent phagocytosis mediated by Rap1 by recruiting talin to integrin αMβ2 in the human myeloid cell lines HL-60 and THP-1.117 Although there are alternative RIAM-independent pathways, these studies bring RIAM to the forefront of integrin regulation in leukocytes.114,115 These findings underscore the fact that the molecular mechanisms leading to talin recruitment to integrins and resulting integrin activation operate in both a cell type– and integrin-specific manner.
Translational implications
Leukocyte recruitment into a tissue is a hallmark of inflammation. Due to their critical role in leukocyte recruitment, leukocyte integrins have emerged as potential therapeutic targets in several inflammatory diseases, and some antileukocyte integrin therapeutics have reached the clinic.118,119 Similarly, the essential role of αIIbβ3 in platelet aggregation and thrombus formation has led to αIIbβ3 antagonists used to treat cardiovascular diseases.7,119 That said, mechanism-based toxicities, ascribable to the effects of complete blockade of integrins, have limited the utility of such therapies.7,118,119
Although much has been learned about how talin and kindlins enable integrin functions, there is a need to decipher the sequence of signal transduction events from adhesive stimuli to talin and kindlin recruitment to integrins. The function of Rap proteins as hubs in integrin inside-out signaling is widely accepted, but their connection with talin remains incompletely characterized. Recent studies established a role for RIAM in relaying signals downstream of Rap1 in leukocytes.114,115 RIAM-deficient mice exhibit significant leukocytosis associated with a loss of β2 integrin activation resulting in impaired leukocyte adhesion to inflamed vessels and accumulation in the circulation.114,115 Additional work is required to determine whether inhibiting RIAM can ameliorate diseases that depend on leukocyte integrin functions, eg, psoriasis and inflammatory bowel disease.118,119 Because RIAM-deficient mice are viable, fertile, and healthy,113-115 amelioration of disease models would portend a potential therapeutic benefit in blocking RIAM or the signaling events that enable its functions. Reverse genetic experiments (mutating a gene and analyzing resulting phenotype) in mice revealed that RIAM is dispensable for platelet function and associated with mild neutrophil and lymphocyte function defects.113-115 These studies strongly suggest that forward genetics of humans (identifying a phenotype and tying it to a mutated gene) will uncover patients with loss-of-function mutations in the RIAM gene associated with a mild leukocyte adhesion deficiency syndrome and normal platelet function. Therefore, the future of RIAM research is exciting, promising a new class of therapeutic targets with the potential to impact inflammatory human diseases without compromising platelet function. Moreover, the dispensability of RIAM for platelet function leaves open the question of how Rap1 regulates integrin activation in platelets.113-115
Acknowledgments
The authors thank Alexandre R. Gingras for carefully reading the manuscript. The authors apologize to those whose studies could not be cited owing to space limitations.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea and funded by Ministry of Education grant NRF-2013R1A1A1007773 (C.K.). Work from the Ginsberg laboratory was funded by grants from the National Institutes of Health, and F.L. was a postdoctoral fellow of the American Heart Association (13POST16950078).
Authorship
Contribution: F.L. wrote the review and C.K. and M.H.G. wrote and edited the review.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark H. Ginsberg, Department of Medicine, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093; e-mail: mhginsberg@ucsd.edu; or Chungho Kim, Department of Life Sciences, Korea University, 145 Anam-Ro, Seongbuk-Gu, Seoul 136-701, South Korea; e-mail: chungho@korea.ac.kr.
References
- 1.Vestweber D. How leukocytes cross the vascular endothelium. Nat Rev Immunol. 2015;15(11):692–704. doi: 10.1038/nri3908. [DOI] [PubMed] [Google Scholar]
- 2.Davì G, Patrono C. Platelet activation and atherothrombosis. N Engl J Med. 2007;357(24):2482–2494. doi: 10.1056/NEJMra071014. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 4.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119(Pt 19):3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogg N, Patzak I, Willenbrock F. The insider’s guide to leukocyte integrin signalling and function. Nat Rev Immunol. 2011;11(6):416–426. doi: 10.1038/nri2986. [DOI] [PubMed] [Google Scholar]
- 6.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bledzka K, Smyth SS, Plow EF. Integrin αIIbβ3: from discovery to efficacious therapeutic target. Circ Res. 2013;112(8):1189–1200. doi: 10.1161/CIRCRESAHA.112.300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnaout MA, Dana N, Gupta SK, Tenen DG, Fathallah DM. Point mutations impairing cell surface expression of the common beta subunit (CD18) in a patient with leukocyte adhesion molecule (Leu-CAM) deficiency. J Clin Invest. 1990;85(3):977–981. doi: 10.1172/JCI114529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wardlaw AJ, Hibbs ML, Stacker SA, Springer TA. Distinct mutations in two patients with leukocyte adhesion deficiency and their functional correlates. J Exp Med. 1990;172(1):335–345. doi: 10.1084/jem.172.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buitrago L, Rendon A, Liang Y, et al. ThromboGenomics Consortium. αIIbβ3 variants defined by next-generation sequencing: predicting variants likely to cause Glanzmann thrombasthenia. Proc Natl Acad Sci USA. 2015;112(15):E1898–E1907. doi: 10.1073/pnas.1422238112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24(1):107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110(5):599–11. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 13.Xiong JP, Stehle T, Diefenbach B, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294(5541):339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11(4):288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011;27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 16.Ye F, Hu G, Taylor D, et al. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188(1):157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye F, Lagarrigue F, Ginsberg MH. SnapShot: Talin and the modular nature of the integrin adhesome. Cell. 2014;156(6):1340-1341. [DOI] [PMC free article] [PubMed]
- 18.Nieswandt B, Moser M, Pleines I, et al. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J Exp Med. 2007;204(13):3113–3118. doi: 10.1084/jem.20071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrich BG, Marchese P, Ruggeri ZM, et al. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J Exp Med. 2007;204(13):3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefort CT, Rossaint J, Moser M, et al. Distinct roles for talin-1 and kindlin-3 in LFA-1 extension and affinity regulation. Blood. 2012;119(18):4275–4282. doi: 10.1182/blood-2011-08-373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274(40):28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 22.Tadokoro S, Shattil SJ, Eto K, et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302(5642):103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 23.Anthis NJ, Wegener KL, Ye F, et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28(22):3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Alvarez B, de Pereda JM, Calderwood DA, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11(1):49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 25.Wegener KL, Partridge AW, Han J, et al. Structural basis of integrin activation by talin. Cell. 2007;128(1):171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 26.Petrich BG, Fogelstrand P, Partridge AW, et al. The antithrombotic potential of selective blockade of talin-dependent integrin alpha IIb beta 3 (platelet GPIIb-IIIa) activation. J Clin Invest. 2007;117(8):2250–2259. doi: 10.1172/JCI31024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haling JR, Monkley SJ, Critchley DR, Petrich BG. Talin-dependent integrin activation is required for fibrin clot retraction by platelets. Blood. 2011;117(5):1719–1722. doi: 10.1182/blood-2010-09-305433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefanini L, Ye F, Snider AK, et al. A talin mutant that impairs talin-integrin binding in platelets decelerates αIIbβ3 activation without pathological bleeding. Blood. 2014;123(17):2722–2731. doi: 10.1182/blood-2013-12-543363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yago T, Petrich BG, Zhang N, et al. Blocking neutrophil integrin activation prevents ischemia-reperfusion injury. J Exp Med. 2015;212(8):1267–1281. doi: 10.1084/jem.20142358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe N, Bodin L, Pandey M, et al. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J Cell Biol. 2008;181(7):1211–1222. doi: 10.1083/jcb.200803094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger BW, Kulp DW, Span LM, et al. Consensus motif for integrin transmembrane helix association. Proc Natl Acad Sci USA. 2010;107(2):703–708. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes PE, Diaz-Gonzalez F, Leong L, et al. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J Biol Chem. 1996;271(12):6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 33.Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2(6):e153. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain helix packing stabilizes integrin alphaIIbbeta3 in the low affinity state. J Biol Chem. 2005;280(8):7294–7300. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- 35.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301(5640):1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 36.Lau TL, Partridge AW, Ginsberg MH, Ulmer TS. Structure of the integrin beta3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry. 2008;47(13):4008–4016. doi: 10.1021/bi800107a. [DOI] [PubMed] [Google Scholar]
- 37.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28(9):1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 2003;544(1-3):69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 39.Kim C, Schmidt T, Cho EG, Ye F, Ulmer TS, Ginsberg MH. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2011;481(7380):209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calderwood DA, Yan B, de Pereda JM, et al. The phosphotyrosine binding-like domain of talin activates integrins. J Biol Chem. 2002;277(24):21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 41.Kalli AC, Campbell ID, Sansom MS. Multiscale simulations suggest a mechanism for integrin inside-out activation. Proc Natl Acad Sci USA. 2011;108(29):11890–11895. doi: 10.1073/pnas.1104505108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim C, Ye F, Hu X, Ginsberg MH. Talin activates integrins by altering the topology of the β transmembrane domain. J Cell Biol. 2012;197(5):605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 44.Leitinger B, Hogg N. The involvement of lipid rafts in the regulation of integrin function. J Cell Sci. 2002;115(Pt 5):963–972. doi: 10.1242/jcs.115.5.963. [DOI] [PubMed] [Google Scholar]
- 45.Decker L, ffrench-Constant C. Lipid rafts and integrin activation regulate oligodendrocyte survival. J Neurosci. 2004;24(15):3816–3825. doi: 10.1523/JNEUROSCI.5725-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamill OP, Martinac B. Molecular basis of mechanotransduction in living cells. Physiol Rev. 2001;81(2):685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- 47.Anishkin A, Loukin SH, Teng J, Kung C. Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc Natl Acad Sci USA. 2014;111(22):7898–7905. doi: 10.1073/pnas.1313364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berrier C, Pozza A, de Lacroix de Lavalette A, et al. The purified mechanosensitive channel TREK-1 is directly sensitive to membrane tension. J Biol Chem. 2013;288(38):27307–27314. doi: 10.1074/jbc.M113.478321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brohawn SG, Su Z, MacKinnon R. Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc Natl Acad Sci USA. 2014;111(9):3614–3619. doi: 10.1073/pnas.1320768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cymer F, Veerappan A, Schneider D. Transmembrane helix-helix interactions are modulated by the sequence context and by lipid bilayer properties. Biochim Biophys Acta. 2012;1818(4):963–973. doi: 10.1016/j.bbamem.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 51.Nomura T, Cranfield CG, Deplazes E, et al. Differential effects of lipids and lyso-lipids on the mechanosensitivity of the mechanosensitive channels MscL and MscS. Proc Natl Acad Sci USA. 2012;109(22):8770–8775. doi: 10.1073/pnas.1200051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459(7245):379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katsumi A, Naoe T, Matsushita T, Kaibuchi K, Schwartz MA. Integrin activation and matrix binding mediate cellular responses to mechanical stretch. J Biol Chem. 2005;280(17):16546–16549. doi: 10.1074/jbc.C400455200. [DOI] [PubMed] [Google Scholar]
- 54.Li R, Mitra N, Gratkowski H, et al. Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science. 2003;300(5620):795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 55.Yin H, Litvinov RI, Vilaire G, et al. Activation of platelet alphaIIbbeta3 by an exogenous peptide corresponding to the transmembrane domain of alphaIIb. J Biol Chem. 2006;281(48):36732–36741. doi: 10.1074/jbc.M605877200. [DOI] [PubMed] [Google Scholar]
- 56.Ye F, Kim SJ, Kim C. Intermolecular transmembrane domain interactions activate integrin αIIbβ3. J Biol Chem. 2014;289(26):18507–18513. doi: 10.1074/jbc.M113.541888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carman CV, Springer TA. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr Opin Cell Biol. 2003;15(5):547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Karaköse E, Schiller HB, Fässler R. The kindlins at a glance. J Cell Sci. 2010;123(Pt 14):2353–2356. doi: 10.1242/jcs.064600. [DOI] [PubMed] [Google Scholar]
- 59.Plow EF, Qin J, Byzova T. Kindling the flame of integrin activation and function with kindlins. Curr Opin Hematol. 2009;16(5):323–328. doi: 10.1097/MOH.0b013e32832ea389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ussar S, Moser M, Widmaier M, et al. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 2008;4(12):e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14(3):325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 62.Kuijpers TW, van de Vijver E, Weterman MA, et al. LAD-1/variant syndrome is caused by mutations in FERMT3. Blood. 2009;113(19):4740–4746. doi: 10.1182/blood-2008-10-182154. [DOI] [PubMed] [Google Scholar]
- 63.Malinin NL, Zhang L, Choi J, et al. A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat Med. 2009;15(3):313–318. doi: 10.1038/nm.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svensson L, Howarth K, McDowall A, et al. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat Med. 2009;15(3):306–312. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye F, Petrich BG, Anekal P, et al. The mechanism of kindlin-mediated activation of integrin αIIbβ3. Curr Biol. 2013;23(22):2288–2295. doi: 10.1016/j.cub.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye F, Snider AK, Ginsberg MH. Talin and kindlin: the one-two punch in integrin activation. Front Med. 2014;8(1):6–16. doi: 10.1007/s11684-014-0317-3. [DOI] [PubMed] [Google Scholar]
- 67.Cifuni SM, Wagner DD, Bergmeier W. CalDAG-GEFI and protein kinase C represent alternative pathways leading to activation of integrin alphaIIbbeta3 in platelets. Blood. 2008;112(5):1696–1703. doi: 10.1182/blood-2008-02-139733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bos JL. Ras-like GTPases. Biochim Biophys Acta. 1997;1333(2):M19–M31. doi: 10.1016/s0304-419x(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 69.Bivona TG, Wiener HH, Ahearn IM, Silletti J, Chiu VK, Philips MR. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J Cell Biol. 2004;164(3):461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mor A, Wynne JP, Ahearn IM, Dustin ML, Du G, Philips MR. Phospholipase D1 regulates lymphocyte adhesion via upregulation of Rap1 at the plasma membrane. Mol Cell Biol. 2009;29(12):3297–3306. doi: 10.1128/MCB.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001;2(5):369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 72.Stork PJ, Dillon TJ. Multiple roles of Rap1 in hematopoietic cells: complementary versus antagonistic functions. Blood. 2005;106(9):2952–2961. doi: 10.1182/blood-2005-03-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheele JS, Marks RE, Boss GR. Signaling by small GTPases in the immune system. Immunol Rev. 2007;218:92–101. doi: 10.1111/j.1600-065X.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- 74.Duchniewicz M, Zemojtel T, Kolanczyk M, Grossmann S, Scheele JS, Zwartkruis FJ. Rap1A-deficient T and B cells show impaired integrin-mediated cell adhesion. Mol Cell Biol. 2006;26(2):643–653. doi: 10.1128/MCB.26.2.643-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sebzda E, Bracke M, Tugal T, Hogg N, Cantrell DA. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat Immunol. 2002;3(3):251–258. doi: 10.1038/ni765. [DOI] [PubMed] [Google Scholar]
- 76.Chu H, Awasthi A, White GC, II, Chrzanowska-Wodnicka M, Malarkannan S. Rap1b regulates B cell development, homing, and T cell-dependent humoral immunity. J Immunol. 2008;181(5):3373–3383. doi: 10.4049/jimmunol.181.5.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC., II Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005;115(3):680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Torti M, Ramaschi G, Sinigaglia F, Lapetina EG, Balduini C. Glycoprotein IIb-IIIa and the translocation of Rap2B to the platelet cytoskeleton. Proc Natl Acad Sci USA. 1994;91(10):4239–4243. doi: 10.1073/pnas.91.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4(8):741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 80.Lafuente EM, van Puijenbroek AA, Krause M, et al. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7(4):585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 81.Liu L, Aerbajinai W, Ahmed SM, Rodgers GP, Angers S, Parent CA. Radil controls neutrophil adhesion and motility through β2-integrin activation. Mol Biol Cell. 2012;23(24):4751–4765. doi: 10.1091/mbc.E12-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gotoh T, Hattori S, Nakamura S, et al. Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol Cell Biol. 1995;15(12):6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gutiérrez-Herrero S, Maia V, Gutiérrez-Berzal J, et al. C3G transgenic mouse models with specific expression in platelets reveal a new role for C3G in platelet clotting through its GEF activity. Biochim Biophys Acta. 2012;1823(8):1366–1377. doi: 10.1016/j.bbamcr.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 84.Ichiba T, Kuraishi Y, Sakai O, et al. Enhancement of guanine-nucleotide exchange activity of C3G for Rap1 by the expression of Crk, CrkL, and Grb2. J Biol Chem. 1997;272(35):22215–22220. doi: 10.1074/jbc.272.35.22215. [DOI] [PubMed] [Google Scholar]
- 85.Arai A, Nosaka Y, Kohsaka H, Miyasaka N, Miura O. CrkL activates integrin-mediated hematopoietic cell adhesion through the guanine nucleotide exchange factor C3G. Blood. 1999;93(11):3713–3722. [PubMed] [Google Scholar]
- 86.Arai A, Nosaka Y, Kanda E, Yamamoto K, Miyasaka N, Miura O. Rap1 is activated by erythropoietin or interleukin-3 and is involved in regulation of beta1 integrin-mediated hematopoietic cell adhesion. J Biol Chem. 2001;276(13):10453–10462. doi: 10.1074/jbc.M004627200. [DOI] [PubMed] [Google Scholar]
- 87.Nolz JC, Nacusi LP, Segovis CM, et al. The WAVE2 complex regulates T cell receptor signaling to integrins via Abl- and CrkL-C3G-mediated activation of Rap1. J Cell Biol. 2008;182(6):1231–1244. doi: 10.1083/jcb.200801121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smit L, van der Horst G, Borst J. Sos, Vav, and C3G participate in B cell receptor-induced signaling pathways and differentially associate with Shc-Grb2, Crk, and Crk-L adaptors. J Biol Chem. 1996;271(15):8564–8569. doi: 10.1074/jbc.271.15.8564. [DOI] [PubMed] [Google Scholar]
- 89.Quilliam LA, Rebhun JF, Castro AF. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog Nucleic Acid Res Mol Biol. 2002;71:391–444. doi: 10.1016/s0079-6603(02)71047-7. [DOI] [PubMed] [Google Scholar]
- 90.Katagiri K, Shimonaka M, Kinashi T. Rap1-mediated lymphocyte function-associated antigen-1 activation by the T cell antigen receptor is dependent on phospholipase C-gamma1. J Biol Chem. 2004;279(12):11875–11881. doi: 10.1074/jbc.M310717200. [DOI] [PubMed] [Google Scholar]
- 91.Bergmeier W, Goerge T, Wang HW, et al. Mice lacking the signaling molecule CalDAG-GEFI represent a model for leukocyte adhesion deficiency type III. J Clin Invest. 2007;117(6):1699–1707. doi: 10.1172/JCI30575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crittenden JR, Bergmeier W, Zhang Y, et al. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat Med. 2004;10(9):982–986. doi: 10.1038/nm1098. [DOI] [PubMed] [Google Scholar]
- 93.Canault M, Ghalloussi D, Grosdidier C, et al. Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding. J Exp Med. 2014;211(7):1349–1362. doi: 10.1084/jem.20130477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Rooij J, Zwartkruis FJ, Verheijen MH, et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396(6710):474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 95.Kawasaki H, Springett GM, Mochizuki N, et al. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282(5397):2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 96.Murphy MM, Zayed MA, Evans A, et al. Role of Rap1 in promoting sickle red blood cell adhesion to laminin via BCAM/LU. Blood. 2005;105(8):3322–3329. doi: 10.1182/blood-2004-07-2881. [DOI] [PubMed] [Google Scholar]
- 97.Lorenowicz MJ, van Gils J, de Boer M, Hordijk PL, Fernandez-Borja M. Epac1-Rap1 signaling regulates monocyte adhesion and chemotaxis. J Leukoc Biol. 2006;80(6):1542–1552. doi: 10.1189/jlb.0506357. [DOI] [PubMed] [Google Scholar]
- 98.Kuiperij HB, de Rooij J, Rehmann H, et al. Characterisation of PDZ-GEFs, a family of guanine nucleotide exchange factors specific for Rap1 and Rap2. Biochim Biophys Acta. 2003;1593(2-3):141–149. doi: 10.1016/s0167-4889(02)00365-8. [DOI] [PubMed] [Google Scholar]
- 99.Yajnik V, Paulding C, Sordella R, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112(5):673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 100.Polakis PG, Rubinfeld B, Evans T, McCormick F. Purification of a plasma membrane-associated GTPase-activating protein specific for rap1/Krev-1 from HL60 cells. Proc Natl Acad Sci USA. 1991;88(1):239–243. doi: 10.1073/pnas.88.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mochizuki N, Ohba Y, Kiyokawa E, et al. Activation of the ERK/MAPK pathway by an isoform of rap1GAP associated with G alpha(i). Nature. 1999;400(6747):891–894. doi: 10.1038/23738. [DOI] [PubMed] [Google Scholar]
- 102.Hattori M, Tsukamoto N, Nur-e-Kamal MS, et al. Molecular cloning of a novel mitogen-inducible nuclear protein with a Ran GTPase-activating domain that affects cell cycle progression. Mol Cell Biol. 1995;15(1):552–560. doi: 10.1128/mcb.15.1.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kurachi H, Wada Y, Tsukamoto N, et al. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2. Segregate expression profiles from a rap1GAP gene product. J Biol Chem. 1997;272(44):28081–28088. doi: 10.1074/jbc.272.44.28081. [DOI] [PubMed] [Google Scholar]
- 104.Ishida D, Kometani K, Yang H, et al. Myeloproliferative stem cell disorders by deregulated Rap1 activation in SPA-1-deficient mice. Cancer Cell. 2003;4(1):55–65. doi: 10.1016/s1535-6108(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 105.Schultess J, Danielewski O, Smolenski AP. Rap1GAP2 is a new GTPase-activating protein of Rap1 expressed in human platelets. Blood. 2005;105(8):3185–3192. doi: 10.1182/blood-2004-09-3605. [DOI] [PubMed] [Google Scholar]
- 106.Wynne JP, Wu J, Su W, et al. Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector. J Cell Biol. 2012;199(2):317–330. doi: 10.1083/jcb.201201157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ménasché G, Kliche S, Chen EJ, Stradal TE, Schraven B, Koretzky G. RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation. Mol Cell Biol. 2007;27(11):4070–4081. doi: 10.1128/MCB.02011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han J, Lim CJ, Watanabe N, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16(18):1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 109.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem. 2009;284(8):5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee HS, Anekal P, Lim CJ, Liu CC, Ginsberg MH. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Mol Biol Cell. 2013;24(9):1354–1362. doi: 10.1091/mbc.E12-09-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lagarrigue F, Vikas Anekal P, Lee HS, et al. A RIAM/lamellipodin-talin-integrin complex forms the tip of sticky fingers that guide cell migration. Nat Commun. 2015;6:8492. doi: 10.1038/ncomms9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coló GP, Lafuente EM, Teixidó J. The MRL proteins: adapting cell adhesion, migration and growth. Eur J Cell Biol. 2012;91(11-12):861–868. doi: 10.1016/j.ejcb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 113.Stritt S, Wolf K, Lorenz V, et al. Rap1-GTP-interacting adaptor molecule (RIAM) is dispensable for platelet integrin activation and function in mice. Blood. 2015;125(2):219–222. doi: 10.1182/blood-2014-08-597542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klapproth S, Sperandio M, Pinheiro EM, et al. Loss of the Rap-1 effector RIAM results in leukocyte adhesion deficiency due to impaired beta2 integrin function in mice. Blood. 2015;126(25):2704–2712. doi: 10.1182/blood-2015-05-647453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Su W, Wynne J, Pinheiro EM, et al. Rap1 and its effector RIAM are required for lymphocyte trafficking. Blood. 2015;126(25):2695–2703. doi: 10.1182/blood-2015-05-644104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zeiler M, Moser M, Mann M. Copy number analysis of the murine platelet proteome spanning the complete abundance range. Mol Cell Proteomics. 2014;13(12):3435–3445. doi: 10.1074/mcp.M114.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Medraño-Fernandez I, Reyes R, Olazabal I, et al. RIAM (Rap1-interacting adaptor molecule) regulates complement-dependent phagocytosis. Cell Mol Life Sci. 2013;70(13):2395–2410. doi: 10.1007/s00018-013-1268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mitroulis I, Alexaki VI, Kourtzelis I, Ziogas A, Hajishengallis G, Chavakis T. Leukocyte integrins: role in leukocyte recruitment and as therapeutic targets in inflammatory disease. Pharmacol Ther. 2015;147:123–135. doi: 10.1016/j.pharmthera.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yonekawa K, Harlan JM. Targeting leukocyte integrins in human diseases. J Leukoc Biol. 2005;77(2):129–140. doi: 10.1189/jlb.0804460. [DOI] [PubMed] [Google Scholar]