Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Low level of total CD79b protein impairs BCR assembly in CLL samples.
IL-4 rescues CD79b protein and sIgM and BCR signaling in CLL samples.
Abstract
Chronic lymphocytic leukemia (CLL) cells express poor levels of surface immunoglobulin (sIg), and many are minimally activated or anergic in response to B-cell receptor (BCR) crosslinking in vitro. Paradoxically, CLL cells in patients are highly activated through BCR signaling and expand in proliferation centers, suggesting that the function of sIg signaling is rescued. Here, we find that, compared with normal naïve B cells, CLL cells express a low level of total CD79b protein but normal levels of CD79a and IgM protein. Association of both CD79a and CD79b to IgM is markedly reduced. We further find that interleukin-4 (IL-4) markedly rescues CD79b and sIgM protein in CLL samples. These changes significantly enhance signaling in response to BCR crosslinking. Furthermore, we find that these changes are more pronounced in immunoglobulin heavy chain variable (IGHV)-unmutated CLL cells than IGHV-mutated CLL cells. The results described herein reveal that reduced sIgM is due to low expression of total CD79b protein in CLL cells. IL-4 substantially restores CD79b protein expression, sIgM expression, and BCR signaling.
Introduction
Reduced expression of surface immunoglobulin (sIg) on the cell surface is a hallmark of chronic lymphocytic leukemia (CLL) cells.1,2 Based on the mutation status of the immunoglobulin heavy chain variable (IGHV) region gene, CLL diseases are divided into 2 major subsets.3 In general, patients with IGHV-mutated CLL (M-CLL) cells follow a milder disease course than IGHV-unmutated CLL (U-CLL), and IGHV mutation status is a critical prognostic marker in CLL disease.4,5 However, the reasons for different clinical courses between mutated and unmutated populations remain unclear.
Peripheral CLL cells express poor levels of sIg, and many of them are minimally activated or anergic in response to BCR crosslinking in vitro.6 Paradoxically, BCR plays an important role for the selection of normal B cells into the leukemic state and the subsequent proliferation of CLL cells posttransformation.7-9 CLL cells express a skewed BCR repertoire illustrated by preferential IGHV usage such as IGHV1-69,3 and 30% of CLL cells express “stereotyped BCRs.”10-13 These features indicate that CLL cells experience antigen selection. Recent evidence demonstrates that proliferation of CLL cells in tissues accounts for a daily birth rate of 0.1% to 2% of the entire clone.14 CLL samples exhibit intraclonal heterogeneity. CXCR4dimCD5hi cells, recent lymphoid tissue emigrants, exhibit more proliferation than the CXCR4hiCD5dim fraction that represents a resting subpopulation15 and the vigorously proliferating CLL cells express higher sIgM than resting CLL cells.16 Gene expression profile data17 show that BCR signaling is predominantly activated in CLL cells isolated from proliferating areas within lymphatic tissues. Moreover, sustained BCR signaling is critical for CLL survival.18 BCR signaling has been considered as a target for therapeutic purposes. Recent therapeutic breakthroughs have been achieved by using the BTK inhibitor ibrutinib and the PI3Kδ inhibitor idelalesib in CLL patients.19,20 These findings suggest that sIg expression on CLL samples is restored in proliferation centers. However, the molecular mechanism for BCR rescuing remains unknown.
CLL cells express a higher level of interleukin-4 (IL-4) receptor than normal naïve B cells.21 IL-4–expressing T cells are expanded in CLL patients, and a higher proportion of IL-4–expressing T cells is evident in patients with aggressive disease.22 In proliferation centers, CLL cells are in intimate contact with T cells that express abundant IL-4.23,24 In vitro, IL-4 promotes CLL cell survival.25 In line with these features, injection of recombinant IL-4 resulted in an increased number of CLL B cells in some clinical trials.26 These data suggest that IL-4 is closely associated with and may play a key pathogenic role in CLL disease.
Protein synthesis occurs in the endoplasmic reticulum where proteins undergo modification such as N-glycosylation and folding. Only the correctly assembled protein complexes can pass into the Golgi apparatus where protein complexes obtain further modifications and additions, and achieve mature form.27 The BCR complex is comprised of one dimeric Ig, and one heterodimer of CD79a and CD79b.28 The latter is a prerequisite for, and sufficient to promote, maturation and membrane transport of IgM.29,30 CLL cells exhibit glycosylation and folding defects of the IgM and CD79a chains,31 which results in impaired BCR assembly and reduced sIgM in CLL samples. Our recent studies32-34 have shown that IL-4 reprograms sIgM expression and substantially enhances anti–Ig-initiated B-cell activation in mouse B cells. In the present study, we find that CLL cells express a low level of total CD79b protein. Uncoupled CD79a and CD79b expression impedes assembly of IgM-BCR complexes. IL-4 markedly rescues CD79b and sIgM expression that enhance CLL cell activation in response to BCR crosslinking.
Materials and methods
Human samples
Heparinized venous blood was collected from untreated patients with CLL after informed consent. U-CLLs were chosen randomly from a group that expressed IGHV with <2.0% mutation. M-CLLs were randomly chosen from a group that had IGHV with >2.0% mutation. Cases included in this study comprised only those expressing sIgM. Normal control peripheral blood was obtained from healthy donors. The Institutional Review Board of Northwell Health approved these studies, which were also conducted in accordance with the Declaration of Helsinki.
CLL cell culture and coculture
The human marrow stromal cell line 5 (HS-5) was maintained in RPMI 1640 supplemented with 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.2, 2 mM glutamine, 1% penicillin/streptomycin, and 10% fetal calf serum. For coculture experiments, HS-5 cells were seeded at 3 × 105 cells/mL and incubated overnight. Non-adherent HS-5 cells were washed out with fetal calf serum free medium, and CLL cells were seeded onto HS-5 monolayers at 2 × 106 cells per mL. This coculture strategy is used in the whole study unless specifically stated otherwise.
Flow cytometry and cell sorting
Cocultured CLL cells were incubated with varying combinations of the following antibodies: V-450–anti-CD19, phycoerythrin-CY7-CD5, allophycocyanin (APC)–anti-IgM, or APC–anti-IgD. For viable CLL cell sorting, cocultured cells were harvested, washed with phosphate-buffered saline twice, and then stained with anti-human CD19 and CD5 antibodies. CLL cells were easily separated from HS-5 cells based on their size. Viable CD19+CD5+ CLL cells were separated from dead CLL cells, based on staining with propidium iodide. For normal naïve B-cell sorting, healthy peripheral blood mononuclear cells were incubated with the following antibodies: phycoerythrin-CD20, fluorescein isothiocyanate-CD5, and APC-CD27. CD20+CD5+CD27− B cells were sorted as normal controls.
CLL cell activation
Sorted viable CLL cells were rested in medium for 3 hours followed by a stimulation with F(ab′)2 goat anti-human IgM antibody (10 μg/mL) for 0, 5, or 15 minutes. The anergic CLL samples refer to those that respond to BCR crosslinking by increasing <10% of basal phosphorylated extracellular signal-regulated kinase (pERK).
Quantitative reverse transcription polymerase chain reaction (PCR)
Total RNA was obtained from CLL samples. Complementary DNA was prepared using Avian myeloblastosis virus reverse transcriptase (Roche Applied Sciences). CD79b-specific primers (forward: CCTATCTTCCTGCTGCTGGA; reverse: CATAGGTGGCTGTCTGGTCA) and actin-specific primers (forward: ACTCTTCCAGCCTTCCTTCC; reverse: CGTACAGGTCTTT GCGGATG) were used for real-time PCR. CD79b messenger RNA (mRNA) expression was assessed and normalized to actin.
Immunoblot analysis and immunoprecipitation
For immunoblot analysis, CLL cells were lysed in NP40 lysis buffer and equal amounts of protein for each condition (15-30 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation followed by immunoblot. For immunoprecipitation, CLL cells were lysed in digitonin lysis buffer containing 1% digitonin, 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and 1× Roche complete protease inhibitor tablets. The cell extracts were precleared followed by incubation with goat anti-human IgM polyclonal antibody overnight at 4°C. Subsequently, protein A/G UltraLink resin was added and incubated for 1 hour at 4°C. Immunoprecipitated proteins were washed with 1% digitonin lysis buffer 3 times and were subjected to immunoblot.
Antibodies and reagents
Affinity-purified F(ab′)2 fragments of polyclonal goat anti-human IgM (anti-Ig) antibody and horseradish peroxidase-conjugated mouse anti-rabbit monoclonal antibody were obtained from Jackson ImmunoResearch Laboratories. Neutralizing anti-human IL-4 antibody was obtained from BioLegend. Propidium iodide and fluorescently labeled mouse anti-human monoclonal antibodies against CD19, CD5, IgM, CXCR4, CD27, CD20, and IgD were obtained from BD Pharmingen. Polyclonal anti-pERK, anti-phosphorylated spleen tyrosine kinase (pSYK), anti-extracellular signal-regulated kinase (ERK), and anti-SYK antibodies were obtained from Cell Signaling Technology. Monoclonal anti-CD79a and anti-CD79b were obtained from Abcam. Endo H was obtained from New England Biolabs. Recombinant human IL-4, IL-13, IL-6, IL-1, IL-21, and CD40L were obtained from PeproTech.
Statistical analysis
The Student t test was used for statistical analysis; results with P < .05 were considered significant.
Results
CLL cells express a low level of total CD79b protein
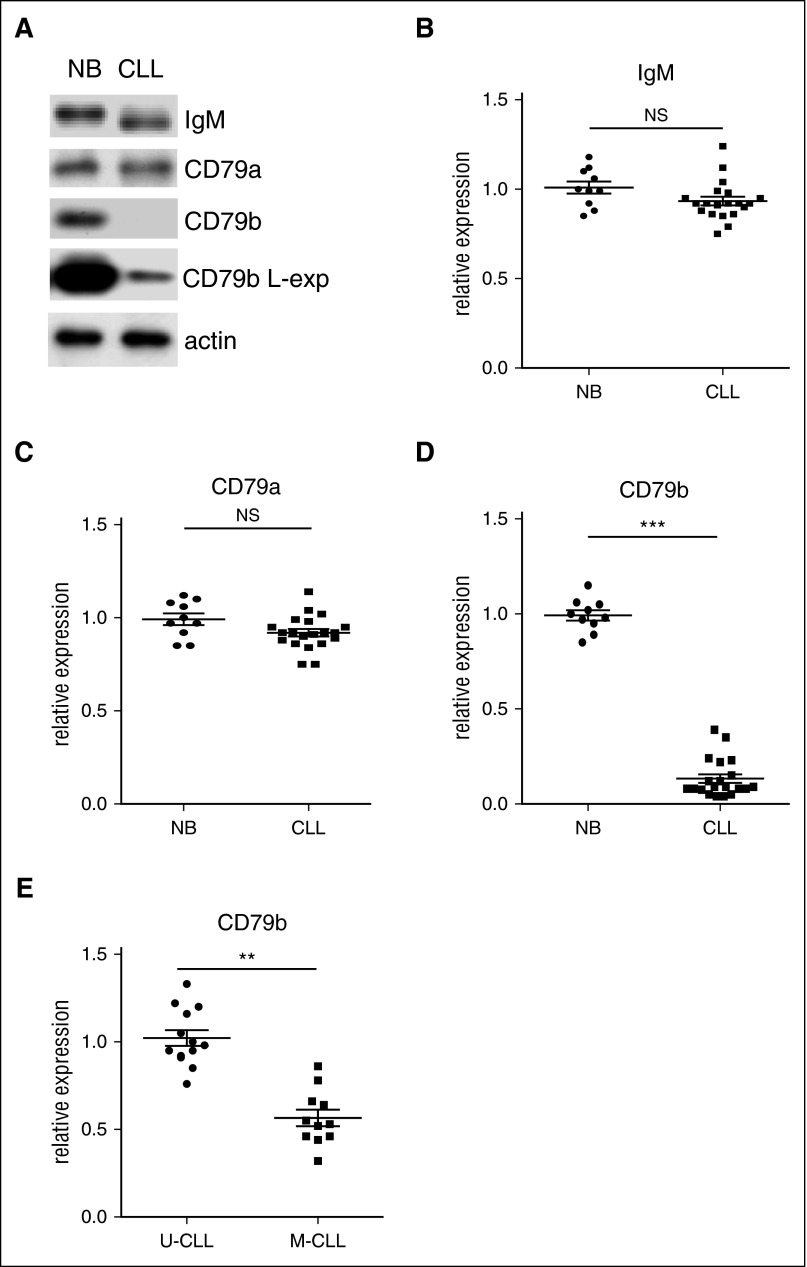
To examine the molecular mechanism for reduced sIg in CLL samples, total protein expression of IgM, CD79a, and CD79b was examined by immunoblot. Compared with normal naïve B cells, CLL cells (Table 1)35 express a low level of total CD79b protein (Figure 1A,D; decrease ≈12.5 ± 2.6-fold [P = .0024]), and M-CLL samples express less total CD79b protein than U-CLL samples (Figure 1E; decrease ≈1.8 ± 0.45-fold [P = .0048]). In contrast, CLL cells express relatively normal total amounts of CD79a and IgM protein (Figure 1A-C).
Table 1.
Clinical feathers of the CLL patients
| CLL sample ID | RAI score* | Treatment | Median length of disease course (mo) |
|---|---|---|---|
| CLL0614 | N/D | N | >130 |
| CLL0920 | 0 | N | >95 |
| CLL1016 | III | N | 46 |
| CLL1082 | 0 | N | >84 |
| CLL1083 | 0 | N | >83 |
| CLL1090 | 0 | N | >90 |
| CLL1104 | I | N | >82 |
| CLL1148 | 0 | N | >82 |
| CLL1189 | 0 | N | >99 |
| CLL1217 | 0 | N | >72 |
| CLL1232 | 0 | N | 64 |
| CLL1251 | II | N | >135 |
| CLL1274 | I | N | >81 |
| CLL1300 | 0 | N | >88 |
| CLL1309 | 0 | N | >96 |
| CLL1453 | I | N | >305 |
| CLL1512 | I | N | >94 |
| CLL1524 | I | N | >56 |
| CLL1539 | 0 | N | >130 |
| CLL1566 | 0 | N | >46 |
| CLL1570 | N/D | N | >224 |
| CLL1606 | 0 | N | >52 |
| CLL1625 | N/D | N | >58 |
| CLL1656 | I | N | >81 |
| CLL1731 | 0 | N | >26 |
N, no; N/D, not determined.
According to criteria previously described by Rai et al.35
Figure 1.
CLL cells express a low level of total CD79b protein. (A-B) Total IgM, (A,C) CD79a, and (A,D) CD79b protein in human normal naive B-cell samples (n = 10) and CLL cell samples (n = 25) were examined by immunoblot. Membranes were stripped and reprobed with anti-actin antibody as a loading control. The results in (A) show 1 representative case, and l-exp represents a longer exposure. The results in (B-D) represent relative expression of total IgM, CD79a, and CD79b protein in CLL samples (lines represent mean ± standard error of the mean [SEM] of 25 samples), with total IgM, CD79a, and CD79b protein expression in human normal naïve B-cell samples set at “1.0” (lines represent mean ± SEM of 10 samples). The results in (E) represent the relative expression of total CD79b protein in U-CLL samples vs M-CLL samples with total CD79b protein expression in U-CLL samples set at “1.0” (lines represent mean ± SEM of 11 samples). **P < .01; ***P < .005. NB, naïve B cell; NS, not significant.
Assembly of the IgM-BCR complex is impaired in CLL cells
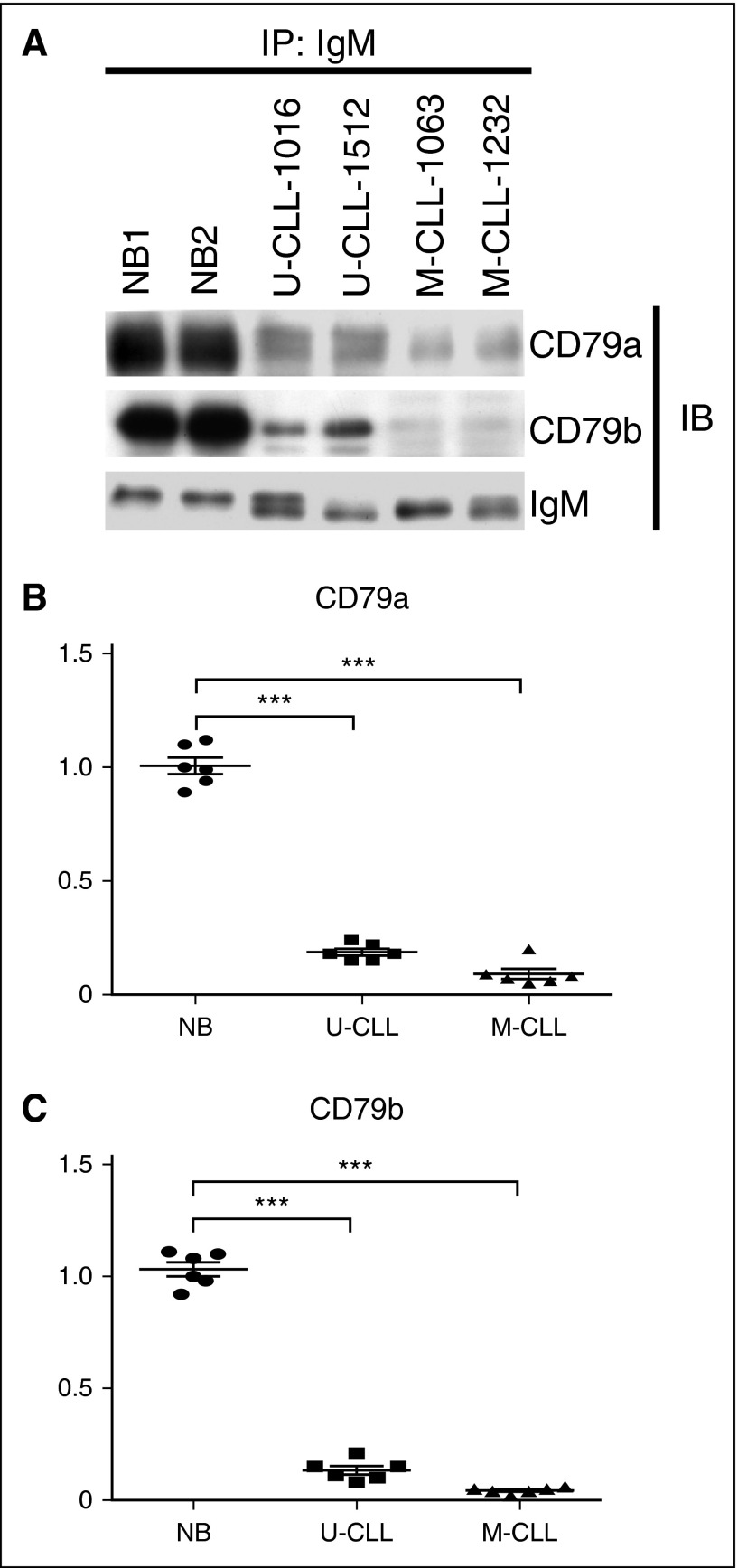
The levels of CD79a and CD79b proteins are uncoupled in CLL samples, suggesting that assembly of IgM-BCR complexes may be impaired. To examine this, CLL cells, as well as normal naïve B cells, were lysed in digitonin buffer, which does not disassociate elements of the BCR complexes, and total IgM was immunoprecipitated. In normal naïve B cells, because CD79a and CD79b form heterodimers that are associated with IgM, large amounts of CD79a and CD79b proteins were detected by immunoblot in immunoprecipitates. In contrast, because CLL cells express a low level of total CD79b protein, very little CD79b protein was detected by immunoblot in immunoprecipitates (Figure 2A,C; decrease ≈5.3 ± 1.6-fold [P = .0048] in U-CLL samples and 8.9 ± 3.2-fold [P = .0033] in M-CLL samples). CLL cells express uncoupled CD79a and CD79b protein, and a large amount of CD79a fails to form heterodimers with CD79b. Thus, although CLL cells express normal total CD79a protein, IgM-associated CD79a is markedly reduced (Figure 2A-B; decrease ≈6.2 ± 1.8-fold [P = .0039] in U-CLL samples and 7.5 ± 2.4-fold [P = .0025] in M-CLL samples).
Figure 2.
Impaired assembly of IgM with CD79a and CD79b in CLL samples. (A) Human normal naïve B-cell samples (n = 6), U-CLL cell samples (n = 6), and M-CLL cell samples (n = 6) were extracted in digitonin buffer. IgM protein was immunoprecipitated, and IgM-associated CD79a and CD79b proteins were examined by immunoblot. Membranes were stripped and reprobed with anti-IgM antibody as a loading control. The results in (B-C) represent the relative association of CD79a and CD79b to IgM in U-CLL and M-CLL cell samples (lines represent mean ± SEM of 6 samples), with total IgM-associated CD79a or CD79b protein in human normal naïve B-cell samples set at “1” (lines represent mean ± SEM of 6 samples). The values of IgM-associated CD79a and CD79b are normalized to anti–IgM-precipitated IgM in all samples. The results in (A) show 1 representative experiment with 2 samples of each population. ***P < .005. IB, immunoblot; IP, immunoprecipitated; NB, naïve B cell.
IL-4 rescues CD79b protein expression in CLL cells
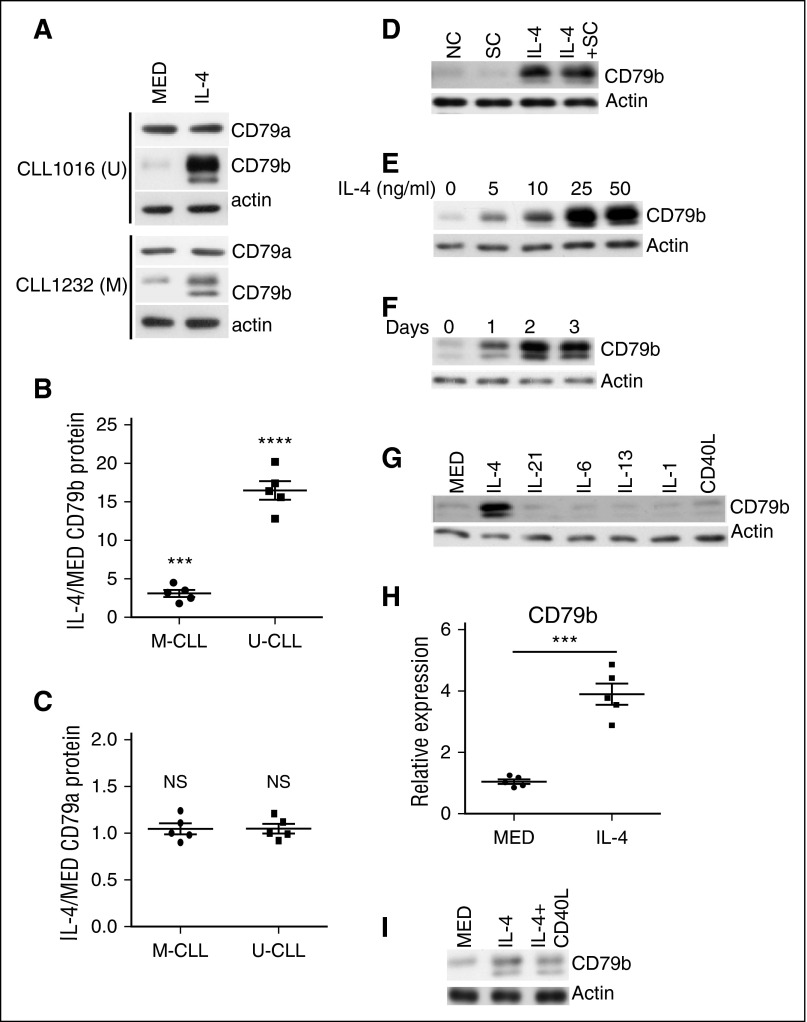
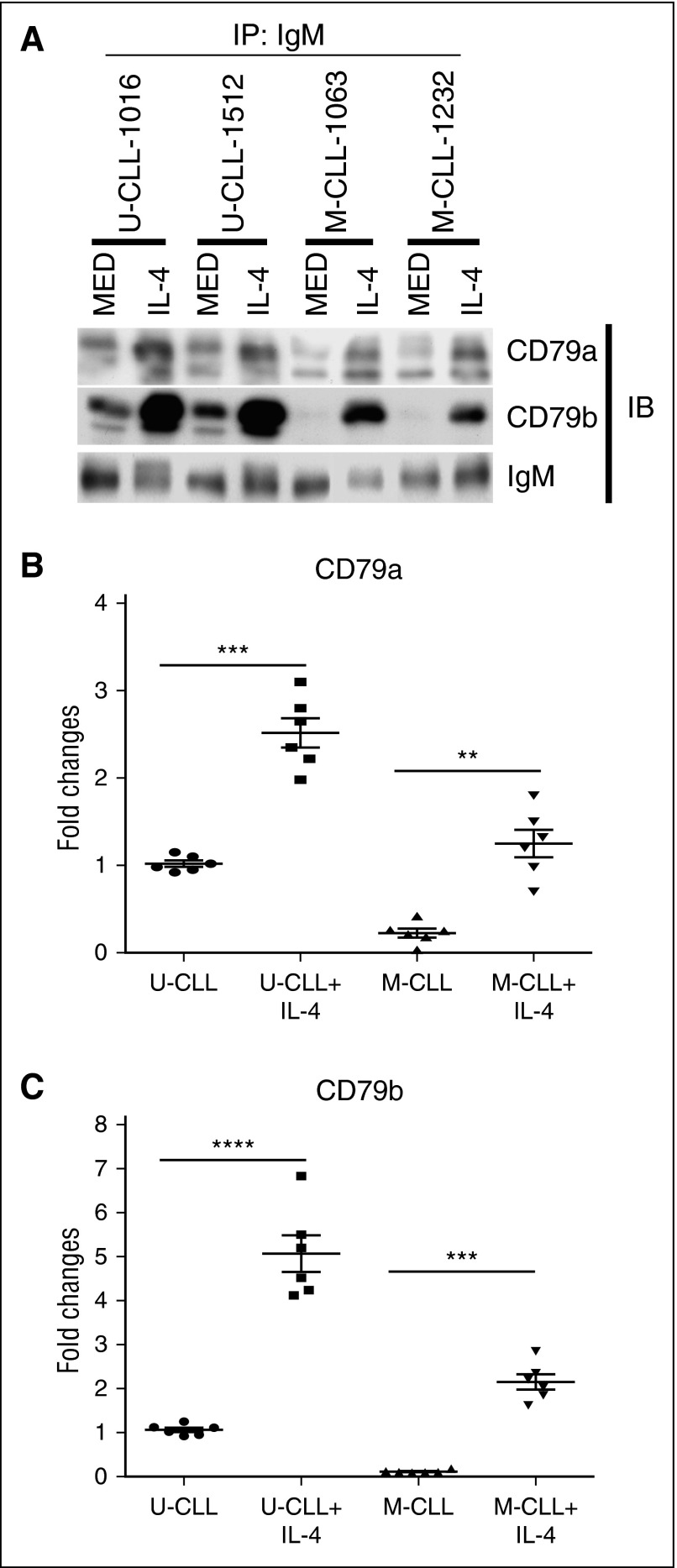
We showed that IL-4 upregulates CD79b protein expression in mouse primary B cells,32 suggesting that IL-4 might be a potential microenvironmental factor rescuing CD79b protein expression in CLL cells. To test this, CLL cells were cocultured with a human stromal cell line, HS-5, in the presence or absence (as a negative control) of IL-4 for 48 hours. Coculture with HS-5 cells prevents CLL cell death in vitro.36 After coculture, viable CLL (CD5+CD19+) cells were sorted, and total protein of CD79a and CD79b was evaluated by immunoblot. IL-4 markedly increases CD79b protein expression in CLL samples (Figure 3A-B). In contrast, IL-4 does not significantly change CD79a protein expression (Figure 3A,C). When comparing responses of U-CLL vs M-CLL to IL-4, unexpectedly, we find that IL-4 restores much more CD79b protein in U-CLL cells than it does in M-CLL cells (Figure 3A-B; increase ≈16.5 ± 3.4-fold [P = .00078] in U-CLL samples and 3.5 ± 1.4-fold [P = .0038] in M-CLL samples).
Figure 3.
IL-4 rescues total CD79b protein expression in CLL cells. (A) U-CLL cells (top) and M-CLL cells (bottom) were cocultured in the presence of IL-4 or the absence (MED) of IL-4 (25 ng/mL) for 48 hours. CD79a and CD79b protein in viable CLL cells was examined by immunoblot. (B-C) Summary of results representing fold change in CD79a and CD79b protein expression, respectively. Lines represent mean ± SEM of 5 samples. Each spot represents an individual sample. (D) CLL samples were NC, or cocultured in the absence (SC) or presence with IL-4 (25 ng/mL) (IL-4 + SC), or cultured with IL-4 (25 ng/mL) (IL-4) alone for 48 hours. CD79b protein in viable CLL cells was examined by immunoblot. (E) CLL samples were cocultured with IL-4 at 0, 5, 10, 25, or 50 ng/mL for 48 hours. CD79b protein in viable CLL cells was examined by immunoblot. (F) CLL samples were cocultured with IL-4 (25 ng/mL) for 0, 24, 48, or 72 hours. CD79b protein in viable CLL cells was examined by immunoblot. (G) CLL samples were cocultured in the absence (MED) or presence of IL-4 (25 ng/mL), IL-21 (25 ng/mL), IL-6 (25 ng/mL), IL-13 (25 ng/mL), IL-1 (25 ng/mL), and CD40L (25 ng/mL) for 48 hours. CD79b protein in viable CLL cells was examined by immunoblot. (H) CLL samples were cocultured in the absence (MED) or presence of IL-4 for 48 hours. CD79b mRNA expression was examined by quantitative PCR. Lines represent mean ± SEM of 5 samples. Each spot represents an individual sample. (I) M-CLL samples were cocultured in the absence (MED) or presence of IL-4 (25 ng/mL), or a combination of IL-4 (25 ng/mL) and CD40L (25 ng/mL) for 48 hours. Results in (A,D-G,I), represent 1 of 5 independent experiments. Membranes were stripped and reprobed with anti-actin antibody as a loading control. ***P < .005; ****P < .001. NC, noncultured; NS, not significant.
It is possible that HS-5 cells in the coculture also affect CD79b protein expression or play a synergistic role in IL-4–induced CD79b protein expression. To test these, CD79b protein was examined by immunoblot in parent CLL samples and CLL samples cocultured with HS-5 cells, IL-4, HS-5 cells, and IL-4. We find that HS-5 cells neither change CD79b protein expression nor affect IL-4–restored CD79b protein expression in cocultured CLL samples (Figure 3D).
To examine the dose response and time course of IL-4–restored CD79b protein expression, CLL samples were first cocultured with different doses of IL-4 for 48 hours. IL-4 restores CD79b protein at as low as 5 ng/mL and CD79b protein expression reaches a plateau at 25 ng/mL (Figure 3E). Further, CLL samples were cocultured with IL-4 (25 ng/mL) for 0, 24, 48, and 72 hours. IL-4–restored CD79b protein reaches a plateau at 48 hours (Figure 3F). To test whether other environmental factors also induce CD79b protein expression, CLL samples were cocultured with IL-4 (25 ng/mL), IL-21 (25 ng/mL), IL-6 (25 ng/mL), IL-13 (25 ng/mL), IL-1 (25 ng/mL), and CD40L (25 ng/mL) for 48 hours, and CD79 protein was examined by immunoblot. We find that IL-4 is the unique cytokine restoring CD79b protein expression in CLL samples (Figure 3G). To explore the nature of IL-4–induced CD79b protein expression in CLL samples, CD79b mRNA expression was examined by quantitative PCR. IL-4 treatment increases CD79b mRNA expression by three- to fivefold (Figure 3H; increase ≈3.8 ± 1.1-fold [P = .0021]). Considering the IL-4–induced increase in CD79b protein is much greater than the increase in CD79b mRNA, IL-4 may induce CD79b protein expression through both transcriptional and posttranscriptional mechanisms. The latter is the dominant one.
IL-4 is insufficient to fully restore CD79b protein in M-CLL samples (Figure 3A-B). To test whether CD40L enhances IL-4–induced CD79b protein in M-CLL samples, M-CLL samples were cocultured with or without IL-4, or a combination of IL-4 and CD40L, and CD79b protein was examined by immunoblot. We find that CD40L does not affect IL-4–induced CD79b protein expression in M-CLL samples (Figure 3I).
IL-4 rescues sIgM expression in CLL cells
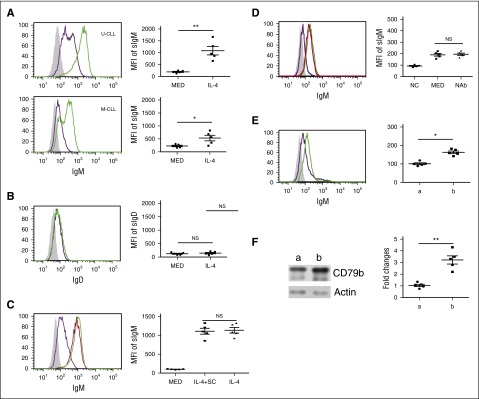
IL-4 rescues CD79 protein expression, suggesting IL-4 may rescue sIgM expression in CLL cells. To test this, CLL cells were cocultured with or without IL-4 for 48 hours. The sIgM on the CLL cells was examined by flow cytometric analysis. IL-4 significantly restores sIgM expression on CLL cells (Figure 4A). Interestingly, although IL-4 rescues sIgM expression, it does not affect sIgD expression (Figure 4B). As noted above, IL-4 rescues much more CD79b in U-CLL cells than it does in M-CLL cells. In line with this, IL-4 restores more sIgM in U-CLL cells than it does in M-CLL cells (Figure 4A; increase ≈5.2 ± 2.2-fold [P = .0032] and 2.8 ± 2.6-fold (P = .0254) in U-CLL and M-CLL cells, respectively).
Figure 4.
IL-4 rescues sIgM on CLL cells. (A) IL-4 significantly rescues sIgM expression in CLL cells. U-CLL cells (n = 5; top) and M-CLL cells (n = 5; bottom) were cocultured in the presence (green line) or absence (purple line) of IL-4 (25 ng/mL) for 48 hours. The cocultured CLL cells were stained with mouse anti-human IgM antibody or isotype control mouse antibody (shaded area) and analyzed by flow cytometric assay. (B) IL-4 does not significantly change sIgD expression. CLL cells (n = 5) were cocultured in the presence (green line) or absence (purple line) of IL-4 (25 ng/mL) for 48 hours. The cocultured CLL cells were stained with mouse anti-human IgD antibody or isotype control mouse antibody (shaded area) and analyzed by flow cytometric assay. (C) Stromal cells do not affect IL-4–restored sIgM protein expression. CLL cells (n = 5) were cocultured in the presence (green line) or absence (purple line) of IL-4 (25 ng/mL), or cultured with IL-4 (25 ng/mL) alone (red line) for 48 hours. Viable CLL cells were stained with mouse anti-human IgM antibody or isotype control mouse antibody (shaded area) and analyzed by flow cytometric analysis. (D) Coculture-induced sIgM is IL-4 independent. CLL samples were noncultured (purple line), or cocultured in the absence (MED; green line) or presence of neutralizing anti-human IL-4 antibody (10 μg/mL) (NAb; red line). Viable CLL cells were stained with mouse anti-human IgM antibody or isotype control mouse antibody (shaded area) and analyzed by flow cytometric assay. (E) CLL samples were stained with mouse anti-human CD19 antibody, CXCR4 antibody, CD5 antibody, or isotype control mouse antibody (shaded area). sIgM expression in CXCR4dimCD5hi subpopulation (green line; “b” in the right panel) and CXCR4dhiCD5dim subpopulation (purple line, “a” in the left panel) was examined by flow cytometric analysis. (F) CLL samples were stained with mouse anti-human CD19 antibody, CXCR4 antibody, and CD5 antibody. CXCR4dimCD5hi subpopulation (“b”) and CXCR4dhiCD5dim subpopulation (“a”) were sorted, and CD79b protein was examined by immunoblot. Membranes were stripped and reprobed with anti-actin antibody as a loading control. In (A-F), the left panels represent 1 of 5 independent experiments and the right panels are the summarized results. Lines represent mean ± SEM of 5 samples. Each spot represents 1 sample. **P < .01; *P < .05. MFI, mean fluorescence intensity; NAb, neutralizing antibody; NC, noncultured; NS, not significant; SC, stromal cells.
To test the possibility that HS-5 cells affect IL-4–restored sIgM expression, CLL samples were cultured with IL-4 alone or cocultured with or without IL-4 for 48 hours. We find that HS-5 cells do not change IL-4–rescued sIgM expression (Figure 4C). A previous study37 showed that culture in medium alone reverses sIgM expression and BCR signaling in CLL samples. To test whether this restoration is IL-4 dependent, CLL samples were uncultured or cocultured with or without neutralizing anti–IL-4 antibody for 48 hours. The sIgM was examined by flow cytometric analysis. We find that coculture slightly upregulates sIgM expression, and this increase is IL-4 independent (Figure 4D).
If sIgM and CD79b proteins in CLL cells are restored in the proliferation centers, it is possible that recently egressed CLL cells express higher sIgM and total CD79b protein than resting cells. To test this, CXCR4dimCD5hi cells and CXCR4hiCD5dim cells were sorted from individual CLL samples, and sIgM and CD79b protein were examined by flow cytometric analysis and immunoblot, respectively. We find that CXCR4dimCD5hi cells express more sIgM (increase ≈1.6 ± 0.2-fold [P = .032]) and CD79b (increase ≈3.2 ± 1.1-fold [P = .0092]) than CXCR4hiCD5dim cells (Figure 4E-F).
IL-4 promotes IgM maturation and IgM-BCR assembly in CLL cells
IL-4 can restore sIgM expression in CLL samples. One possible explanation is that IL-4 increases total IgM protein expression, which will proportionally elevate sIgM. To test this, CLL cells were cocultured with or without IL-4 for 48 hours. We find that no matter the presence or absence of IL-4, CLL cells express the same amount of total IgM (data not shown), suggesting that IL-4–restored sIgM expression is not through an increase in total IgM protein expression.
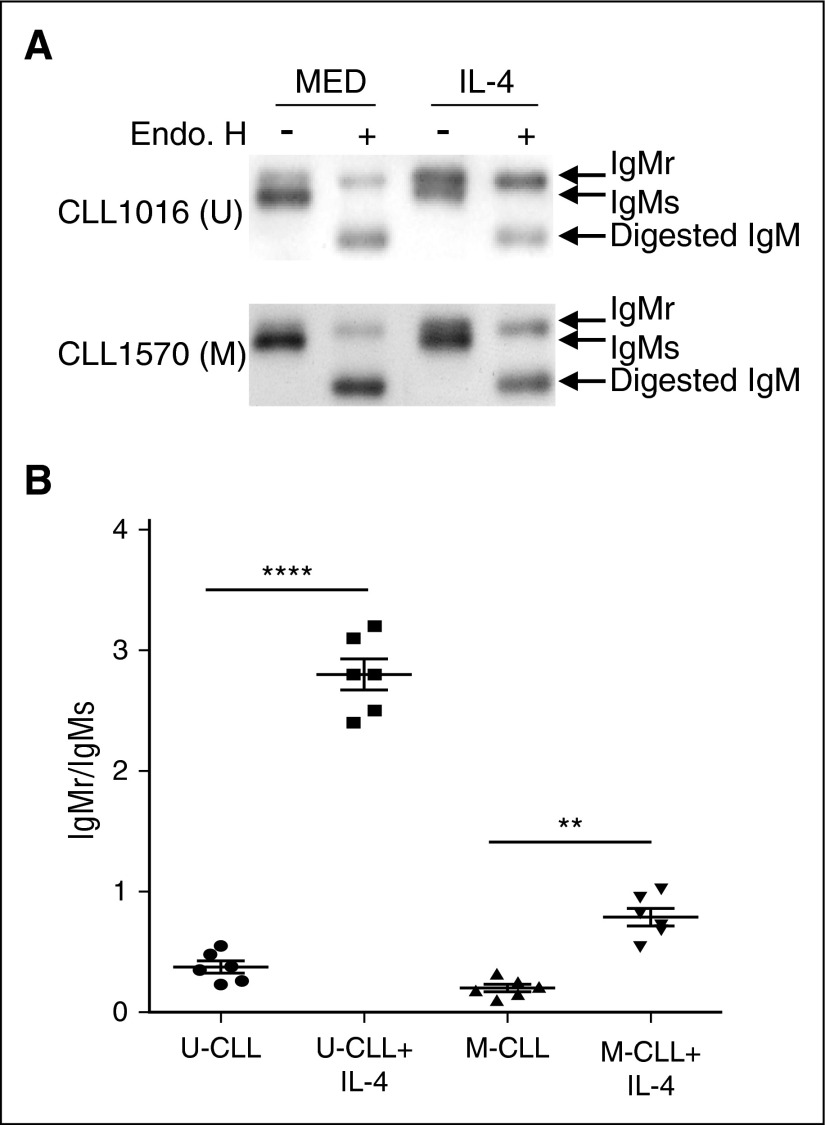
Another possibility is that IL-4 changes IgM maturation status. To test this, CLL cells were cocultured with or without IL-4 for 48 hours. The cell lysates were subjected to endo H digestion followed by immunoblot. After IL-4 treatment, more IgM protein became resistant to endo H digestion (Figure 5A), suggesting that IL-4 substantially promotes IgM maturation. Furthermore, we find that more immature IgM is converted into the mature form by IL-4 in U-CLL cells than M-CLL cells (Figure 5A-B; ratios of endo H-resistant IgM to endo H-sensitive IgM increases from 0.35 to 2.9 [P = .00094] in U-CLL samples and from 0.19 to 0.78 [P = .0086] in M-CLL samples).
Figure 5.
IL-4 promotes IgM maturation in CLL cells. (A-B) IL-4 promotes IgM maturation in CLL samples. U-CLL cells (A, top) and M-CLL cells (A, bottom) were cocultured in the presence (IL-4) or absence (MED) of IL-4 (25 ng/mL) for 48 hours. Viable CLL cells were sorted and lysed in NP-40 buffer. Cell lysates were subjected to endo H digestion followed by immunoblot using anti-IgM antibody to identify endo H-resistant (IgMr) and endo H-sensitive (IgMs) species. The summarized results in (B) represent ratios of IgMr to IgMs in U-CLL and M-CLL cell samples, with or without IL-4 (lines represent mean ± SEM of 6 samples). The results in (A) show 1 representative experiment with 1 U-CLL sample and 1 M-CLL sample. **P < .01; ****P < .001.
IL-4 restores total CD79b protein and sIgM, suggesting that IL-4 may recover the defective association of IgM-BCR. To test this, CLL cells were cocultured with or without IL-4 for 48 hours. Viable cells were lysed in digitonin buffer and total IgM was immunoprecipitated. IgM-associated CD79a and CD79b proteins were examined by immunoblot. We find that IL-4 markedly enhances the association of IgM with CD79a and CD79b in CLL samples (Figure 6A-C). Because IL-4 rescues more CD79b protein in U-CLL samples (Figure 3A-B), a greater association of IgM with CD79a and CD79b is observed in IL-4–experienced U-CLL samples than IL-4–experienced M-CLL samples (Figure 6A-C).
Figure 6.
IL-4 recovers assembly of IgM-BCR complexes. (A) U-CLL and M-CLL samples were cocultured in the presence (IL-4) or absence (MED) of IL-4 (25 ng/mL) for 48 hours. Viable CLL cells were sorted and extracted in digitonin buffer. IgM protein was immunoprecipitated, and IgM-associated CD79a and CD79b proteins were examined by immunoblot. Membranes were stripped and reprobed with anti-IgM antibody as a loading control. The summarized results in (B-C) represent the relative association of CD79a and CD79b to IgM in U-CLL and M-CLL cell samples with or without IL-4 (lines represent mean ± SEM of 6 samples), with total IgM-associated CD79a or CD79b protein in U-CLL samples set at “1” (lines represent mean ± SEM of 6 samples). The values of IgM-associated CD79a and CD79b are normalized to anti–IgM-precipitated IgM in all samples. The results in (A) show 1 representative experiment with 2 samples of each population. **P < .01; ***P < .005; ****P < .001. IB, immunoblot; IP, immunoprecipitated.
IL-4 enhances CLL cell activation initiated by BCR crosslinking
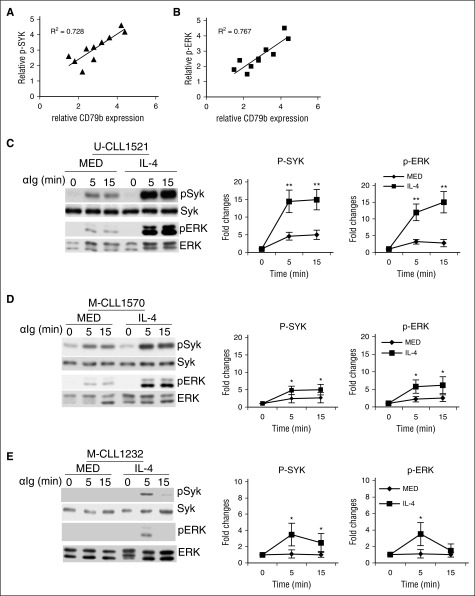
Generally, U-CLL samples are more responsive to BCR crosslinking than M-CLL, and U-CLL samples express more CD79b protein than M-CLL samples (Figure 1E), suggesting CD79b protein levels may determine the responsiveness of CLL samples to BCR crosslinking. To test this, CD79b protein and BCR-stimulated pSYK and pERK were examined by immunoblot. We find that, although CD79b expression is variable between individual CLL samples, BCR-stimulated pSYK (Figure 7A; R2 = 0.767) and pERK (Figure 7B; R2 = 0.728) are generally correlated with CD79b protein levels.
Figure 7.
IL-4 enhances CLL cell activation in response to BCR crosslinking. (A-B) Responsiveness of CLL samples to BCR crosslinking is correlated with pCD79 protein levels. CLL samples were stimulated with anti-IgM antibody (αIg; 10 μg/mL) for 0 and 5 minutes. The correlation of pSYK (A) and pERK (B) to CD79b protein expression is analyzed. (C-D) IL-4 enhances pSYK and pERK in CLL samples stimulated by BCR crosslinking. U-CLL samples (C) and M-CLL samples (D) were cocultured in the presence (IL-4) or absence (MED) of IL-4 (25 ng/mL) for 48 hours. Viable CLL cells were stimulated with anti-IgM antibody (αIg; 10 μg/mL) for 0, 5, and 15 minutes. Phosphorylation of SYK and ERK was examined by immunoblot. Membranes were stripped and reprobed with SYK- and ERK-specific antibodies as loading controls. (E) IL-4 breaks down CLL cell anergy. Anergic CLL samples were cocultured in the presence (IL-4) or absence (MED) of IL-4 (25 ng/mL) for 48 hours. Viable CLL cells were stimulated with anti-IgM antibody (αIg; 10 μg/mL) for 0, 5, and 15 minutes. Phosphorylation of SYK and ERK was examined by immunoblot. Membranes were stripped and reprobed with SYK- and ERK-specific antibodies as loading controls. Left panels in (C-E) show 1 representative experiment; right panels represent the summarized results. Lines represent mean ± SEM of 6 samples. *P < .05; **P < .01.
IL-4 rescues sIgM expression, suggesting that IL-4 may enhance CLL cell activation initiated by BCR crosslinking. To test this, both U-CLL and M-CLL cells were cocultured with or without IL-4 for 48 hours. Viable cells were stimulated with anti-IgM. BCR-crosslinking stimulates pSYK and pERK in CLL cells at relatively low levels. IL-4 substantially enhances BCR-stimulated pSYK and pERK (Figure 7C-D). Because IL-4 salvages more sIgM expression on U-CLL cells than M-CLL cells, IL-4 enhances more pSYK and pERK in U-CLL cells (Figure 7C; increase ≈3.12 ± 0.65-fold [P = .009] at 5 minutes and 2.95 ± 0.62-fold [P = .007] at 15 minutes for pSYK, and increase ≈3.75 ± 0.58-fold [P = .007] at 5 minutes and 5.31 ± 0.76-fold [P = .006] at 15 minutes for pERK) than M-CLL cells (Figure 7D; increase ≈1.82 ± 0.22-fold [P = .032] at 5 minutes and 2.05 ± 0.28-fold [P = .045] at 15 minutes for pSYK, and increase ≈2.63 ± 0.25-fold [P = .022] at 5 minutes and 2.02 ± 0.26-fold [P = .036] at 15 minutes for pERK) in response to anti-IgM stimulation.
Some CLL cells, in particular M-CLL cells, are anergic to BCR crosslinking. IL-4 restores sIgM expression and enhances BCR signaling, suggesting that IL-4 may break down CLL cell anergy. To examine this, CLL cells, which were anergic to BCR crosslinking, were cocultured with or without IL-4 for 48 hours. Viable cells were stimulated with anti-IgM. BCR-stimulated pSYK and pERK are undetectable in anergic M-CLL cells (Figure 7E). However, after culture with IL-4, these anergic CLL cells exhibit phosphorylation of SYK and ERK in response to BCR crosslinking (Figure 7E; increase ≈3.32 ± 0.85-fold [P = .038] at 5 minutes and 2.11 ± 0.46-fold [P = .045] at 15 minutes for pSYK, and increase ≈3.18 ± 0.35-fold [P = .042] at 5 minutes and 1.51 ± 0.18-fold [not significant] at 15 minutes for pERK), even though BCR-stimulated SYK and ERK phosphorylation are much lower in anergic CLL cells than other CLL cells after IL-4 treatment (Figure 7C,E).
Discussion
CLL B cells are anergic with low sIgM, presumably resulting from antigen engagement.38,39 Our results identify a specific lesion that is responsible for impaired BCR expression and responsiveness in CLL cells. We show that BCR component expression is severely unbalanced in CLL cells. Deficient CD79b expression leads to decoupling with CD79a and IgM, which blocks BCR assembly and reduces sIgM expression in CLL cells. Surplus CD79a and IgM remain immature in endoplasmic reticulum due to a lack of CD79b, which may explain glycosylation abnormalities previously noted for these molecules in CLL cells.31 We find that IL-4 induces CD79b protein, increases sIgM expression, and upregulates BCR responsiveness to antigen. Antigen binding leads to internalization of BCR complexes that is counterbalanced by IL-4. However, when CLL cells migrate into the blood stream where the IL-4 concentration is very low, CD79b protein expression is markedly downregulated by an unknown mechanism and BCR complexes cannot be properly assembled. As a consequence, recent nodal emigrants (CXCR4dimCD5hi) express more sIgM that is lost in resting CXCR4hiCD5dim CLL cells that had egressed at an earlier time point.
Evidence of BCR signaling and proliferation was observed in CLL cells located in nodal tissue.17 Our data suggest that reduced sIgM is restored in proliferation centers. Here, we find that IL-4, a key microenvironmental factor in the proliferation centers, significantly salvages sIgM in CLL samples. When CLL cells return to proliferation centers, CD79b protein and sIgM are restored by the resident IL-4. To our knowledge, IL-4 is the first microenvironmental factor that markedly restores sIgM in CLL cells. Although IL-4 increases CD79b mRNA expression in CLL cells, our results showed that IL-4–restored CD79b protein is predominantly regulated at posttranscriptional levels. Components of the BCR complex are heterogeneously expressed in CLL cells, suggesting that CD79b protein expression is inhibited in a gene-specific way. MicroRNAs (miRs) are small endogenous RNAs that direct posttranscriptional repression through specifically targeting the 3′ untranslated region of mRNA of genes. Aberrant expression of certain miRs has been observed in CLL and is implicated in CLL disease.40 CLL cells express high levels of miR-29a and miR-29b.40,41 M-CLL cells express more miR-29a and miR-29b than U-CLL cells. This expression pattern is negatively correlated with CD79b protein. A recent study showed that IL-4 could significantly downregulate miR-29a.42 Furthermore, computational analysis showed that the 3′ untranslated region of CD79b mRNA has binding sites for miR-29a, miR-29b, and miR-29c, suggesting that CD79b protein expression may be inhibited by these miRs. Thus, IL-4 may restore CD79b protein expression by inhibiting the expression of these miRs in CLL cells. However, the detailed mechanism remains to be determined.
A previous report37 showed that sIgM and BCR signaling are reversed in CLL cells when cultured in medium alone. We confirmed a small increase in sIgM expression during in vitro coculture (without IL-4). However, BCR signaling was not reversed unless IL-4 was added. The different observations between these 2 studies may be due to usage of different culture systems. In the previous study, CLL cells were cultured in medium alone. In such a culture system, CLL cells will undergo rapid apoptosis.43 Apoptotic CLL cells will release substances such as auto-antigens, cytokines, and chemokines44-50 that potentially change the nature of CLL cells and may result in less anergy. In contrast, our coculture system imitates the supportive microenvironment in proliferation centers, and keeps CLL cells alive and in good condition. The sIgM is only slightly restored and this increase is IL-4 independent. The anergy is not reversed until IL-4 is added, which strongly upregulates CD79b and sIgM. Furthermore, our results show that IL-4 acts differentially between U-CLL and M-CLL cells. In general, IL-4 rescues more sIgM on U-CLL cells than M-CLL cells, which leads to more pSYK and pERK upon BCR crosslinking in U-CLL cells than M-CLL cells. These findings are consistent with a recent report51 showing that the gene expression response to IL-4 in CLL cells depends on the zeta-chain–associated protein kinase 70 status, which is enriched in U-CLL samples.52
Collectively, we find in this study, that CLL cells express a low level of total CD79b protein, which results in defective assembly of the IgM-BCR complex and reduced sIgM. Furthermore, we find that IL-4 specifically rescues CD79b protein expression and sIgM in CLL, providing for BCR responsiveness. Importantly, we find that IL-4 exerts much more impact on U-CLL cells than it does on M-CLL cells by rescuing much more CD79b and sIgM, and enhancing stronger signaling in U-CLL cells than M-CLL cells. Thus, this study reveals the potential mechanism by which CLL cells poorly express sIg, but vigorously proliferate in lymph nodes. CLL produces substantial morbidity and mortality. Over the past decades, numerous treatments for CLL have been developed.1,19,53 Because the etiology remains unknown, most of these treatments are designed to destroy CLL cells for the purpose of alleviating morbidity rather than prevention of progression. Currently, a “watchful waiting” mode is applied to early stage CLL patients who will not be given treatment until they have evidence for progressive or symptomatic disease. More importantly, morbid complications with symptomatic CLL patients, along with side effects and increased resistance to current therapies, are major concerns. Our finding indicates a new pathogenic mechanism by which CLL cells expand. Therefore, this study suggests a new treatment strategy, particularly for early intervention or prevention of CLL disease, by opposing IL-4 signaling and/or CD79b upregulation in proliferation centers.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (NIH) United States Public Health Service (029690) (T.L.R.), NIH National Cancer Institute (CA081554) (N.C.), and NIH National Institute of Allergy and Infectious Diseases (AI115544) (B.G.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.G. and T.L.R. designed the study and wrote the manuscript; B.G. performed experiments and analyzed the data; and N.C. and L.Z. provided patient samples and assisted in writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Benchang Guo, The Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; e-mail: bguo@northwell.edu.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Ternynck T, Dighiero G, Follezou J, Binet JL. Comparison of normal and CLL lymphocyte surface Ig determinants using peroxidase-labeled antibodies. I. Detection and quantitation of light chain determinants. Blood. 1974;43(6):789–795. [PubMed] [Google Scholar]
- 3.Fais F, Ghiotto F, Hashimoto S, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102(8):1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 5.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 6.Guarini A, Chiaretti S, Tavolaro S, et al. BCR ligation induced by IgM stimulation results in gene expression and functional changes only in IgV H unmutated chronic lymphocytic leukemia (CLL) cells. Blood. 2008;112(3):782–792. doi: 10.1182/blood-2007-12-127688. [DOI] [PubMed] [Google Scholar]
- 7.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol. 2003;21:841–894. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 8.Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013;34(12):592–601. doi: 10.1016/j.it.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packham G, Stevenson F. The role of the B-cell receptor in the pathogenesis of chronic lymphocytic leukaemia. Semin Cancer Biol. 2010;20(6):391–399. doi: 10.1016/j.semcancer.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Messmer BT, Albesiano E, Efremov DG, et al. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200(4):519–525. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatopoulos K, Belessi C, Moreno C, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood. 2007;109(1):259–270. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 12.Tobin G, Thunberg U, Karlsson K, et al. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104(9):2879–2885. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
- 13.Agathangelidis A, Darzentas N, Hadzidimitriou A, et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012;119(19):4467–4475. doi: 10.1182/blood-2011-11-393694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messmer BT, Messmer D, Allen SL, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115(3):755–764. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calissano C, Damle RN, Marsilio S, et al. Intraclonal complexity in chronic lymphocytic leukemia: fractions enriched in recently born/divided and older/quiescent cells. Mol Med. 2011;17(11-12):1374–1382. doi: 10.2119/molmed.2011.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuthill K, Zhang Y, Buggins A, et al. In-vivo labeling studies in patients with chronic lymphocytic leukemia studies demonstrate the existence of apparently distinct subpopulations that differ in phenotype and proliferative capacity. Blood. 2015;126(23):615. [Google Scholar]
- 17.Herishanu Y, Pérez-Galán P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petlickovski A, Laurenti L, Li X, et al. Sustained signaling through the B-cell receptor induces Mcl-1 and promotes survival of chronic lymphocytic leukemia B cells. Blood. 2005;105(12):4820–4827. doi: 10.1182/blood-2004-07-2669. [DOI] [PubMed] [Google Scholar]
- 19.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lannutti BJ, Meadows SA, Herman SE, et al. CAL-101, a p110delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood. 2011;117(2):591–594. doi: 10.1182/blood-2010-03-275305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas RS, Capocasale RJ, Lamb RJ, Nowell PC, Moore JS. Chronic lymphocytic leukemia B cells are resistant to the apoptotic effects of transforming growth factor-beta. Blood. 1997;89(3):941–947. [PubMed] [Google Scholar]
- 22.Rossmann ED, Lewin N, Jeddi-Tehrani M, Osterborg A, Mellstedt H. Intracellular T cell cytokines in patients with B cell chronic lymphocytic leukaemia (B-CLL). Eur J Haematol. 2002;68(5):299–306. doi: 10.1034/j.1600-0609.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- 23.Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood. 2009;114(16):3367–3375. doi: 10.1182/blood-2009-06-225326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker KF, Lappin DF, Takahashi K, Hope J, Macdonald DG, Kinane DF. Cytokine expression in periapical granulation tissue as assessed by immunohistochemistry. Eur J Oral Sci. 2000;108(3):195–201. doi: 10.1034/j.1600-0722.2000.108003195.x. [DOI] [PubMed] [Google Scholar]
- 25.Fluckiger AC, Rossi JF, Bussel A, Bryon P, Banchereau J, Defrance T. Responsiveness of chronic lymphocytic leukemia B cells activated via surface Igs or CD40 to B-cell tropic factors. Blood. 1992;80(12):3173–3181. [PubMed] [Google Scholar]
- 26.Lundin J, Kimby E, Bergmann L, Karakas T, Mellstedt H, Osterborg A. Interleukin 4 therapy for patients with chronic lymphocytic leukaemia: a phase I/II study. Br J Haematol. 2001;112(1):155–160. doi: 10.1046/j.1365-2141.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 27.Rothman JE. Polypeptide chain binding proteins: catalysts of protein folding and related processes in cells. Cell. 1989;59(4):591–601. doi: 10.1016/0092-8674(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 28.Schamel WW, Reth M. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity. 2000;13(1):5–14. doi: 10.1016/s1074-7613(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 29.Venkitaraman AR, Williams GT, Dariavach P, Neuberger MS. The B-cell antigen receptor of the five immunoglobulin classes. Nature. 1991;352(6338):777–781. doi: 10.1038/352777a0. [DOI] [PubMed] [Google Scholar]
- 30.Matsuuchi L, Gold MR, Travis A, Grosschedl R, DeFranco AL, Kelly RB. The membrane IgM-associated proteins MB-1 and Ig-beta are sufficient to promote surface expression of a partially functional B-cell antigen receptor in a nonlymphoid cell line. Proc Natl Acad Sci USA. 1992;89(8):3404–3408. doi: 10.1073/pnas.89.8.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuillier F, Dumas G, Magnac C, et al. Lower levels of surface B-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the mu and CD79a chains. Blood. 2005;105(7):2933–2940. doi: 10.1182/blood-2004-09-3643. [DOI] [PubMed] [Google Scholar]
- 32.Guo B, Rothstein TL. IL-4 upregulates Igα and Igβ protein, resulting in augmented IgM maturation and B cell receptor-triggered B cell activation. J Immunol. 2013;191(2):670–677. doi: 10.4049/jimmunol.1203211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo B, Rothstein TL. B cell receptor (BCR) cross-talk: IL-4 creates an alternate pathway for BCR-induced ERK activation that is phosphatidylinositol 3-kinase independent. J Immunol. 2005;174(9):5375–5381. doi: 10.4049/jimmunol.174.9.5375. [DOI] [PubMed] [Google Scholar]
- 34.Guo B, Blair D, Chiles TC, Lowell CA, Rothstein TL. Cutting Edge: B cell receptor (BCR) cross-talk: the IL-4-induced alternate pathway for BCR signaling operates in parallel with the classical pathway, is sensitive to Rottlerin, and depends on Lyn. J Immunol. 2007;178(8):4726–4730. doi: 10.4049/jimmunol.178.8.4726. [DOI] [PubMed] [Google Scholar]
- 35.Rai KR, Han T. Prognostic factors and clinical staging in chronic lymphocytic leukemia. Hematol Oncol Clin North Am. 1990;4(2):447–456. [PubMed] [Google Scholar]
- 36.Zhang W, Trachootham D, Liu J, et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol. 2012;14(3):276–286. doi: 10.1038/ncb2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mockridge CI, Potter KN, Wheatley I, Neville LA, Packham G, Stevenson FK. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood. 2007;109(10):4424–4431. doi: 10.1182/blood-2006-11-056648. [DOI] [PubMed] [Google Scholar]
- 38.Vilen BJ, Nakamura T, Cambier JC. Antigen-stimulated dissociation of BCR mIg from Ig-alpha/Ig-beta: implications for receptor desensitization. Immunity. 1999;10(2):239–248. doi: 10.1016/s1074-7613(00)80024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caligaris-Cappio F. B-chronic lymphocytic leukemia: a malignancy of anti-self B cells. Blood. 1996;87(7):2615–2620. [PubMed] [Google Scholar]
- 40.Moussay E, Wang K, Cho JH, et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2011;108(16):6573–6578. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz-Lafuente N, Alcaraz-García MJ, Sebastián-Ruiz S, et al. IL-4 up-regulates miR-21 and the miRNAs hosted in the CLCN5 gene in chronic lymphocytic leukemia. PLoS One. 2015;10(4):e0124936. doi: 10.1371/journal.pone.0124936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurer B, Stanczyk J, Jüngel A, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62(6):1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 43.Munk Pedersen I, Reed J. Microenvironmental interactions and survival of CLL B-cells. Leuk Lymphoma. 2004;45(12):2365–2372. doi: 10.1080/10428190412331272703. [DOI] [PubMed] [Google Scholar]
- 44.Catera R, Silverman GJ, Hatzi K, et al. Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol Med. 2008;14(11-12):665–674. doi: 10.2119/2008-00102.Catera. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hervé M, Xu K, Ng YS, et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005;115(6):1636–1643. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 1998;91(7):2387–2396. [PubMed] [Google Scholar]
- 47.Kipps TJ, Carson DA. Autoantibodies in chronic lymphocytic leukemia and related systemic autoimmune diseases. Blood. 1993;81(10):2475–2487. [PubMed] [Google Scholar]
- 48.Chu CC, Catera R, Hatzi K, et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood. 2008;112(13):5122–5129. doi: 10.1182/blood-2008-06-162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui X, Zhang L, Magli AR, et al. Cytoplasmic myosin-exposed apoptotic cells appear with caspase-3 activation and enhance CLL cell viability. Leukemia. 2016;30(1):74–85. doi: 10.1038/leu.2015.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess M, Cheung C, Chambers L, et al. CCL2 and CXCL2 enhance survival of primary chronic lymphocytic leukemia cells in vitro. Leuk Lymphoma. 2012;53(10):1988–1998. doi: 10.3109/10428194.2012.672735. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Lafuente N, Alcaraz-García MJ, Sebastián-Ruiz S, et al. The gene expression response of chronic lymphocytic leukemia cells to IL-4 is specific, depends on ZAP-70 status and is differentially affected by an NFκB inhibitor. PLoS One. 2014;9(10):e109533. doi: 10.1371/journal.pone.0109533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194(11):1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]