Abstract
Increasing evidence points to a complex interplay between genes and the environment in autism spectrum disorder (ASD), including rare de novo mutations in chromatin genes such as methyl-CpG binding protein 2 (MECP2) in Rett syndrome. Epigenetic mechanisms such as DNA methylation act at this interface, reflecting the plasticity in metabolic and neurodevelopmentally regulated gene pathways. Genome-wide studies of gene sequences, gene pathways and DNA methylation are providing valuable mechanistic insights into ASD. The dynamic developmental landscape of DNA methylation is vulnerable to numerous genetic and environmental insults: therefore, understanding pathways that are central to this ‘perfect storm’ will be crucial to improving the diagnosis and treatment of ASD.
Autism spectrum disorder (ASD) is a collection of neurodevelopmental disorders that is characterized by restricted interests and repetitive behaviours, and impairments in social communication1. Clinical presentation can vary from mild to severe and is further complicated by the frequent occurrence of comorbidities, including seizures, gastrointestinal disturbances, auditory disorders and psychiatric disorders2. The prevalence of ASD is estimated at 1 in 68 children in the United States3. There is a marked sex bias, with estimates of 1 in 42 boys versus 1 in 189 girls3. The prevalence of ASD has rapidly increased since the first epidemiological studies in the late 1960s, when estimates were 1 in 2,500 (REF. 3). The factors contributing to the rapid increase in ASD are complex and potentially include changes in the diagnostic criteria, increased public awareness and access to resources, as well as a potential rise in the prevalence of the disorder itself.
Epidemiological studies suggest a crucial role for both genetic and environmental contributions to the aetiology of ASD4–6. Although ASD is highly heritable, the genetic architecture is complex: hundreds of gene variants and copy number variants (CNVs) are associated with ASD (BOX 1). None of the individual genes identified to date accounts for more than 1% of ASD cases7. Together, genetic analyses — including karyotyping, CNV analysis and exome sequencing — can currently only identify a potentially causative genetic abnormality in 25% of clinically diagnosed ASD cases7. The combination of multiple common genetic variants may help to explain a larger portion of the genetic risk, but identifying individual risk loci and potential multi-gene risk interactions has proven difficult8. Together, the genetic data explain only a portion of ASD risk, and from this we infer a potentially substantial role for an environmental contribution to ASD. In a recent large survey of over 14,000 children with ASD in Sweden, heritable factors were found to contribute about half of ASD risk, which suggests that the other half comes from undetected genetic factors, environmental effects and/or stochastic effects9. In fact, a growing body of literature indicates that a wide variety of environmental exposures are associated with ASD, including a lower ASD risk associated with maternal prenatal vitamin use (odds ratio (OR): 0.61–0.62)10–12, and increased ASD risk associated with infection during pregnancy (OR: 1.24–1.37)13,14, in utero exposure to pesticides (OR: 1.3–3.0)15,16 or air pollution (OR: 1.04–1.5)5. Although each of these factors has a small to moderate effect size, when they occur in the context of specific genetic variants11 the combination may result in a ‘perfect storm’ that leads to disruption of normal neurodevelopment. How multiple environmental factors interact with the underlying genetics to alter patterns of brain development in ASD is currently unknown. However, epigenetic mechanisms that allow for stable regulation of gene expression without alterations to the underlying DNA sequence are optimally positioned at this interface between the genome and environment. There are multiple epigenetic layers — including DNA methylation, histone modifications, chromatin structure and non-coding RNAs — that interact to form a complex epigenomic landscape that coordinates gene expression during brain development17. This Review focuses on the methylation of cytosine bases, which are thought to be one of the most stable and crucial forms of epigenetic regulation of the genome.
Box 1. The genetic architecture of ASD: many genes but shared pathways.
Monozygotic twins show a higher concordance rate for autism spectrum disorder (ASD) than do dizygotic twins, but individual studies vary in the degree of reported concordance, which ranges from 36% to 92%105.
The ASD risk for an individual child is also higher if an older sibling has ASD, especially if there are multiple older siblings with the disorder106.
There is a 4:1 male bias in ASD owing to a poorly understood female protective effect107.
Monogenic syndromic forms of ASD, such as fragile X syndrome (mutations in fragile X mental retardation 1 (FMR1)), tuberous sclerosis (mutations in the TSC genes), Rett syndrome (mutations in methyl-CpG binding protein 2 (MECP2)) and Angelman syndrome (mutations in ubiquitin protein ligase E3A (UBE3A)), have been identified and characterized but account for less than an estimated 10% of cases of ASD and intellectual disability6.
Large copy-number variations are more common in children with ASD when compared with unaffected siblings108 or ancestry-matched controls109, but copy-number variations account for only 4% of ASD cases in the Simons Simplex collection44.
An increased global burden of both common and rare chromosomal duplications increases the risk for ASD110.
Exome sequencing in ASD cases versus unaffected family members identifies de novo loss-of-function variants in 7% of ASD cases44.
De novo genetic variants in ASD are enriched for pathways involved in chromatin modification, transforming growth factor-β (TGFβ), WNT and Notch signalling, synaptic transmission, and transcriptional regulation in embryonic development44,111–113.
Genes with lower expression in the cortex of individuals with ASD were enriched for synaptic functions, whereas genes with higher expression showed enrichment for immune and inflammatory-response functions86,114.
DNA methylation has been shown to have regulatory roles in maintaining genomic stability, defining tissue and cell-type-specific gene expression, and regulating cellular function in response to the environment. There is evidence for various different functions of DNA methylation in transcriptional regulation, such as silencing of repetitive elements, altering transcription factor binding sites and chromatin accessibility, and guiding alternative promoter usage and splicing. Recent advances in high-throughput genomic sequencing approaches have expanded our understanding of DNA methylation beyond a simple gene-silencing mechanism to encompass a much more dynamic and global view. This exciting exploration of DNA methylation at different regulatory genomic elements, across tissues and cell types, and during different developmental stages has revealed an unexpected complexity in how DNA methylation is tied to gene expression and, ultimately, to cellular function; dissecting this complex relationship will be crucial for understanding the prognostic usefulness of DNA methylation in ASD.
Complexity of DNA methylation in brain
Basic biochemistry of DNA methylation types and enzymes
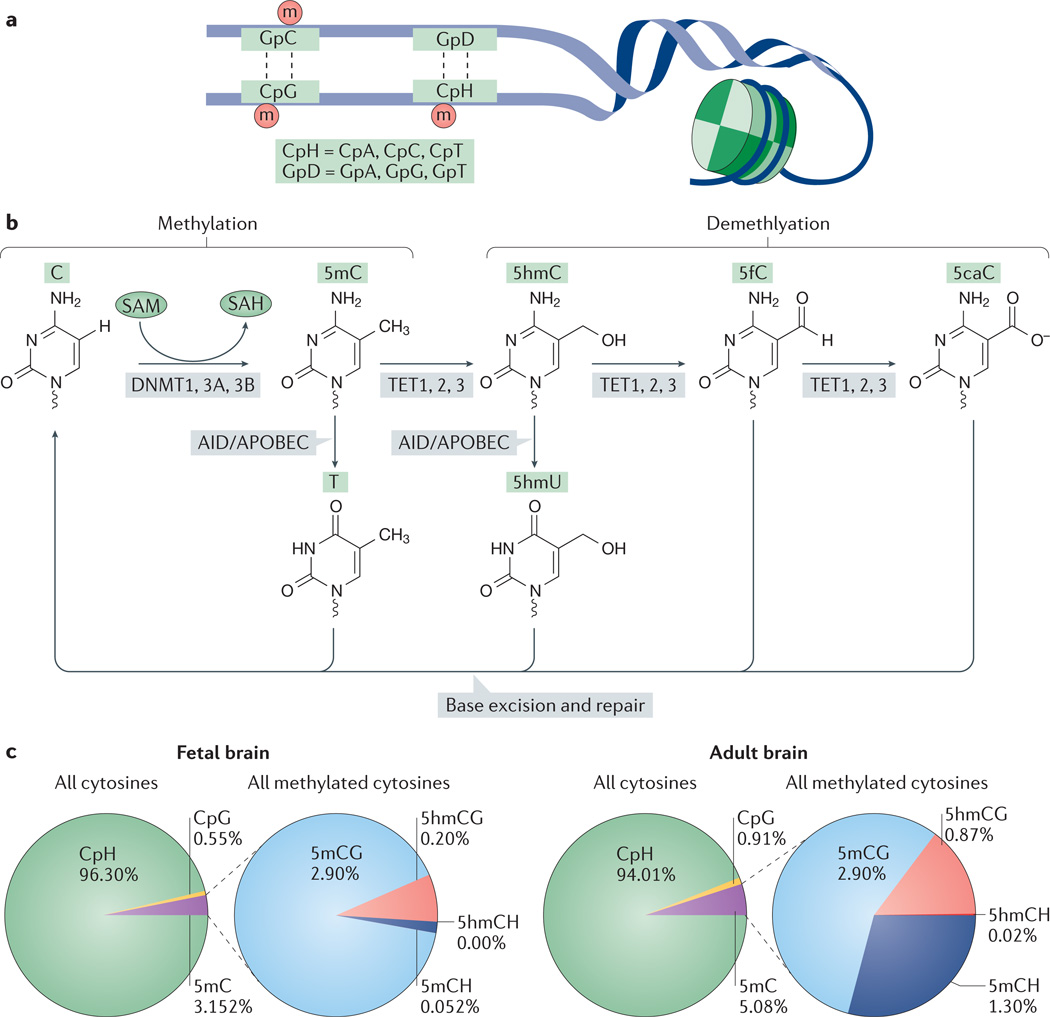
The conventional view of mammalian DNA methylation held that methyl groups were added to the fifth position of the pyrimidine ring of cytosine in the DNA context of cytosine followed by guanine (CpG). These methylated cytosines (5-methylcytosine (5mC)) were thought to be stable and heritable mechanisms of gene silencing and heterochromatin formation that were clustered at areas of high-density CpG (CpG islands). However, the first base-pair-resolution, genome-wide 5mC and 5-hydroxymethylcytosine (5hmC) (FIG. 1) maps in human cell lines and brain tissue revealed a strikingly more complex landscape of DNA methylation in the mammalian genome18,19. One of the most unexpected findings was that in human stem cells, 5mC occurred not only in the CpG context (methylated cytosine–guanine (5mCG)) but also in CpA, CpT and CpC (collectively termed CpH), the methylated forms of which are collectively referred to as 5mCH (FIG. 1a). Subsequent studies have examined 5mCG and 5mCH in tissues and at various developmental stages20–22, revealing that neurons acquire 5mCH postnatally19. 5mCH levels vary considerably with tissue type, with the highest levels occurring in embryonic stem cells, neuronal precursors and neurons21,22. In isolated neuronal populations from the mouse cortex, 5mCH was abundant in excitatory and inhibitory neurons (2–3% of total CpHs)23, which suggests a potential regulatory role for non-CpG methylation in neuronal development.
Figure 1. Types of DNA methylation in the mammalian brain.
a | DNA methylation in the CpG and CpH context. b | Proposed DNA methylation and demethylation pathways in neurons. DNA methyltransferase 1 (DNMT1), DNMT3A and DNMT3B catalyse the methylation of the fifth position of the pyrimidine ring of cytosine using S-adenosylmethionine (SAM) as a methyl donor. SAM is converted to S-adenosylhomocysteine (SAH). Members of the ten-eleven translocase (TET) family can catalyse the oxidation of the cytosine modification 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which can then be converted to different cytosine modifications (5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)). Activation-induced cytidine deaminase (AID) and members of the apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family are cytosine deaminases that are proposed to convert 5mC to thymine (T) or 5hmC to 5-hydroxymethyluracil (5hmU). The DNA base excision and repair pathway can recognize the cytosine modifications 5fC, 5caC, 5hmU and T and replace the mismatched base with an unmodified C. c | Percentage of methylated cytosines out of all cytosines in the genome of fetal and adult mouse brains. Methylated cytosines are further subdivided by type of methylation (5-hydroxymethylcytosine (5hmCG and 5hmCH) and 5-methylcytosine (5mCH and 5mCG)). Although CpH sites are much more abundant than CpG sites in the mammalian genome, CpG sites are much more likely to be methylated. However, in the adult brain, 5mCH and 5hmCG sites make up a larger fraction of the methylated cytosines than in the fetal brain. Data adapted from REF. 19.
DNA methylation is established and maintained in mammals by DNA methyltransferase 1 (DNMT1), DNMT3A and DNMT3B, which methylate DNA in the presence of S-adenosylmethionine (FIG. 1b). DNMT1 levels are highest before birth but decline during postnatal development19,24. In mice, homozygous deletion of Dnmt1 results in early embryonic lethality and threefold lower 5mCG levels in the developing embryo25, which is consistent with the role of DNMT1 as a replication-dependent maintenance methyltransferase. DNMT3B is expressed by neuronal progenitors within a narrow developmental window during early embryogenesis and is largely replaced by DNMT3A by mid-gestation26,27. Following birth, DNMT3A levels increase during the first 3 weeks of life and then decline to lower levels in adulthood19,27. However, both DNMT3A and DNMT3B are essential for life28. The continued expression of both DNMT1 and DNMT3A in the adult brain appears to regulate neuronal plasticity (BOX 2).
Box 2. Evidence for dynamic DNA methylation in adult neuronal plasticity.
Evidence is building for a role of dynamic DNA methylation changes in neuronal plasticity, as well as in learning and memory in the adult animal. Contextual fear conditioning regulates DNA methyltransferase 3a (Dnmt3a) and Dnmt3b levels in the adult rodent hippocampus, and pharmacologically inhibiting DNMTs in the hippocampus after learning prevents long-term memory formation115. Likewise, a learning event or induction of synaptic plasticity can induce both methylation and demethylation events regulating the expression of key plasticity related genes115,116. Neuronal activity resulted in dynamic 5mCG (5-methylcytosine-guanine) changes in 1.4% of sampled CpGs in the adult rodent hippocampal dentate gyrus, one-third of which remain stably modified for at least 24 hours117. DNMT1 and DNMT3A seem to mediate activity-dependent methylation in the adult brain118, whereas ten-eleven translocase 1 (TET1) and TET3 have crucial roles in activity-dependent demethylation119,121. Following these initial studies, DNA methylation has been shown to have a regulatory role in a variety of plasticity-related behaviours, including reward learning122, drug addiction123 and adaptations to stress124.
Several replication-independent DNA methylation removal pathways have been proposed to function in neurons (FIG. 1b). The ten-eleven translocase (TET) family of methylcytosine dioxygenases catalyses 5mC oxidation to form 5hmC, which can subsequently be further converted to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)29–31. In addition, activation-induced cytidine deaminase (AID) and members of the apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family may convert 5mC and 5hmC directly to thymine and 5-hydroxymethyluracil (5hmU), respectively32. The DNA base excision and repair pathway then recognizes the mismatched thymine, 5hmU, 5fC and 5caC and replaces them with an unmodified cytosine29–31. All three TETs (TET1, TET2 and TET3) are highly expressed in fetal mouse cortex, but their levels decrease after birth and their expression is maintained at a low level through adulthood. In humans, brain 5hmC levels spike during adolescence (12–25 years of age) and then return to a pre-adolescent baseline19. In both mice and humans species, TET3 is more abundant than TET2, and TET1 shows the lowest expression across multiple regions of the adult brain19,33. TET3 knockouts are embryonic lethal34. TET1-knockout mice35 and TET2-knockout mice36 are viable, but combined knockout shows depleted 5hmC, increased 5mCG, and a higher likelihood of perinatal lethality37. Together, these results suggest a crucial role for TET-mediated active demethylation in brain development and function.
5hmC and other oxidized forms of 5mC (5fC, 5acC and 5hmC) may represent intermediates in the demethylation pathway or stable modifications that serve unique regulatory roles. There is evidence for 5hmC having a functional role in regulating gene expression. 5hmC is enriched at intragenic regions and repetitive regions in the human cerebellum38, and levels of 5hmC increase during neuronal development19,38 from only 0.2% of total cytosines in fetal brain tissue to 0.9% in adult brain tissue19 (FIG. 1c). 5hmC levels in the gene body are often positively correlated with gene expression (FIG. 2): this is in contrast to promoter levels of 5mCG and 5mCH, which are higher at genes with lower expression. 5mC and 5hmC are both enriched at active gene bodies, the region between the transcription start and end sites of transcribed genes, but showed the opposite strand bias, with 5hmC showing a bias to the sense strand39. Not only does gene-body 5hmC not seem to impair transcriptional elongation, it might actually regulate exon inclusion. In the adult mouse and human prefrontal cortex, 5hmC was enriched over the 5′ splice site within internal exons, and the lower levels of methylation occurred at excluded exons39. In comparison with 5hmC, levels of 5fC and 5caC are low across mammalian tissues, including brain tissue31, and they may represent only a temporary demethylation transition state. Consistent with this idea, genome-wide profiling of 5fC in mouse embryonic stem cells revealed an enrichment of 5fC in regulatory regions that are poised for demethylation and subsequent developmental activation40. Future work will be needed to more fully characterize the localization and function of each of the oxidized DNA methylation derivatives.
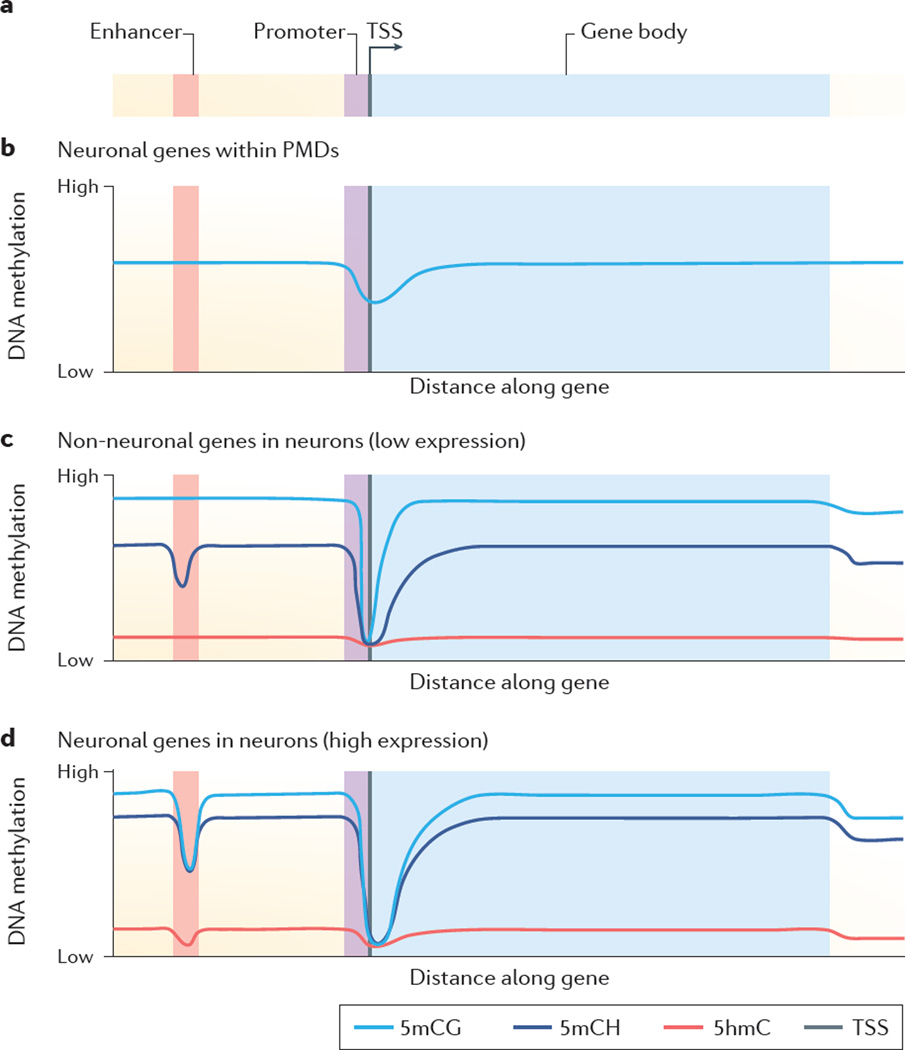
Figure 2. The landscape of DNA methylation in relation to gene expression.
a | DNA methylation over a neuronally active enhancer, promoter, transcription start site (TSS; denoted by arrow) and gene body (TSS to transcription end site)of representative neuronally and non-neuronally expressed genes. b | Neuronal genes in a partially methylated domain (PMD) in a PMD-containing tissue (such as a preimplantation embryo, placenta or tumour) are characterized by lower levels of 5-methylcytosine-guanine (5mCG) but relatively higher levels of promoter 5mCG compared to the promoters of active genes. c | Non-neuronal genes with low expression in neurons (such as glial-specific genes) are characterized by high 5mCG methylation except at the promoter, are enriched for 5-methylcytosine-(A, C or T) (5mCH), and are depleted of 5-hydroxymethylcytosine (5hmC). d | Neuronal genes with high expression in neurons are characterized by high levels of gene-body 5mCG and 5hmC but low levels of 5mCH, and low levels of enhancer and promoter 5mCG and 5hmC. Figures adapted from data in REFS 19,59. Methylation levels are shown as relative low and high values so that 5mCG, 5mCH and 5hmC can all be represented together.
Human genetic evidence for the importance of DNA methylation in neurodevelopment
Rare human genetic disorders on the autism spectrum have enlightened the field to the importance of DNA methylation and its recognition in neurodevelopment41,42. Heterozygous missense mutations in DNMT3A have been associated with overgrowth, intellectual disability43 and ASD44. The parentally imprinted disorders Angelman syndrome and Prader–Willi syndrome (PWS) result from loss of the maternal and paternal alleles, respectively, of 15q11.2–q13.3. Differential methylation in the gametes of an essential genetic locus called the imprinting control (IC) region is presumed to be responsible for the differences in chromatin and gene expression, as some rare cases of PWS lack the paternal IC (PWS-IC) and rare cases of Angelman syndrome are observed with two unmethylated PWS-IC copies45. Interestingly, one of the most common CNVs observed in ASD cases is maternal duplication of the same 15q11.2–q13.3 locus that is deleted in most Angelman syndrome and PWS cases46.
Rett syndrome (RTT), which is caused by mutations in the X-linked gene methyl-CpG binding protein 2 (MECP2), has revealed the crucial role for methyl-binding proteins in ASD47. Proteins containing a methyl-binding domain (MBD) can serve as ‘readers’ of DNA methylation and consequently link dynamic changes in DNA methylation to cellular function. Mutations in genes encoding other MBD family members, including MBD5 (REF. 44), MBD3 and MBD4 (REF. 48), have been found in ASD cases. MECP2 contains an MBD that serves as a reader of multiple forms of DNA methylation (5mCG, 5mCA, 5hmC and oxidized derivatives)49. MECP2 directly interacts with various transcription factors that are crucial for neuronal development50, and it can regulate higher chromatin structure47. The protein may serve as a critical link between environmentally driven perturbations in DNA methylation and cellular function. When exposure occurs in utero, chemicals such as flame retardants have been shown to interact with an MECP2-truncation mouse model, reducing both global DNA methylation and sociability in female offspring51. MECP2 therefore serves as an example for how altered patterns of DNA methylation could directly affect transcriptional regulation, leading to changes in brain development.
DNA methylation in specific genomic contexts
Understanding how DNA methylation regulates gene expression in the developing brain requires examination of cytosine methylation status across the entire genome. The pre-genomics era focused on DNA methylation at CpG islands — a specific class of regulatory elements that contain short (<1 kb) regions that are enriched for CpGs. The rest of the human genome is CpG-poor when compared with CpG islands, potentially owing to the conversion of 5mC to thymine by enzymatic deamination. CpG islands are thought to persist owing to their low or transient germline methylation status52, as most CpG islands near transcription start sites are unmethylated53,54. Methylated CpG-island promoters are found in developmentally or tissue specific silenced genes, as well as in imprinted or X-inactivated genes. For example, during neuronal differentiation, the stem cell marker zinc finger protein 42 (ZFP42) acquires promoter 5mCG and is silenced55. However, it is not clear whether the changes in promoter 5mCG are a cause or consequence of the rapid, cell-type-specific changes in gene expression. In at least a subset of the genes that acquire promoter 5mCG during differentiation, the increase in methylation occurs after the decrease in gene expression55.
DNA methylation outside CpG-island promoters has been shown to potentially be more dynamic, especially in terms of tissue and developmental stage specificity. In both mice and humans, cell-type-specific differential methylation in gene-body CpG islands overlaps potential alternative promoters that are embedded within genes, suggesting that at least some of the gene-body CpG islands might act as alternative promoters or regulate gene splicing54. For example, at the SH3 and multiple ankyrin repeat domains 3 (SHANK3) locus, two different CpG islands in the gene body show tissue and cell-type-specific methylation levels and produce unique alternative transcripts for SHANK3 (REF. 54). Furthermore, maternal-specific methylation at the small nuclear ribonucleoprotein N (SNRPN) 5′ CpG island is established by oocyte-specific use of upstream promoters, so that the canonical CpG island promoter becomes embedded within the gene body, and thus loses its usual protection from methylation56.
CpG sites outside CpG islands also undergo dynamic methylation changes during development and ageing, and in cell- and tissue-specific contexts. CpGs outside islands that showed age-dependent methylation changes in human cortex were more likely to become demethylated with increasing age, whereas the reverse trend was observed for CpG islands53,57. In a large survey of base-pair-resolution 5mC data from 30 different human cell lines and primary tissues, most tissue-specific methylation occurred outside CpG islands. The majority of tissue-specific differentially methylated regions (DMRs) overlapped with DNase hypersensitivity sites, enhancers and introns. Less than 12% of DMRs occurred in promoters, CpG islands, CpG-island shores or exons20. Similarly, among 18 different tissues, tissue-specific methylation sites were mostly found in distal regulatory regions. The strongest relationships between methylation and gene expression occurred in gene bodies21. The distribution of differential methylation across various genomic sites strongly suggests an important role for DNA methylation outside the classically studied CpG islands.
Genome-wide assessment of the DNA methylation landscape
Large genomic regions of partial methylation (defined as <70% 5mCG) have been identified in several cell lines18,58 and human tissues21,59. These domains tend to be over 100 kb in length and can cover as much as half of the human genome60. These partially methylated domains (PMDs) show histone marks that are indicative of a repressive chromatin environment18,21, where gene expression is largely stifled. In addition, genes in PMDs tend to be cell-type specific58,59, although they are also more variable in expression60. For example, placental PMDs contain neuronal genes that show lower gene-body methylation, higher promoter methylation and lower gene expression compared with genes in highly methylated domains (>70% 5mCG) (FIG. 2). Across eight different mammalian placentas, higher gene-body DNA methylation was a conserved feature of the epigenomic landscape, a property that is also seen in mammalian oocytes and preimplantation embryos61; this suggests that the placenta represents an early-life methylation pattern that is lost in most somatic tissues. As the placenta serves as a unique interface between the developing fetus and the environment62, future investigation into DNA-methylation changes in the placenta in families at high risk of autism may provide an epigenetic signature of in utero disruption.
Tissue-specific differences in PMDs mark distinct subsets of developmentally regulated genes and autism risk genes in the neuronal cell line SH-SY5Y58. Classically defined PMDs have not been identified in human brain19,58,59, and the adult brain tissues examined thus far show high levels of methylation across most CpG sites19,59. PMDs occur in fetal fibroblast and cancer cell lines18,58, as well as in human placenta tissue, oocytes and preimplantation embryos61. Thus, PMDs may serve as developmental intermediates during differentiation as cells transition from pluripotent embryonic cells to postmitotic neurons. Classically defined PMDs were not identified in mouse neuronal precursor cells (NPCs), but regions of low methylation (10–50% 5mCG) were identified. These regions occurred mostly at distal active enhancers adjacent to genes that are important for neuronal development, and they were enriched for neuronal transcription factor-binding motifs. They were also positively correlated with increased developmental expression of the nearest gene63. This suggests that domain-wide differences in DNA methylation around neuronal genes and their enhancers may regulate the transition to a permissive chromatin environment during neuronal differentiation.
DNA methylation across brain development
Global changes in DNA methylation during embryogenesis
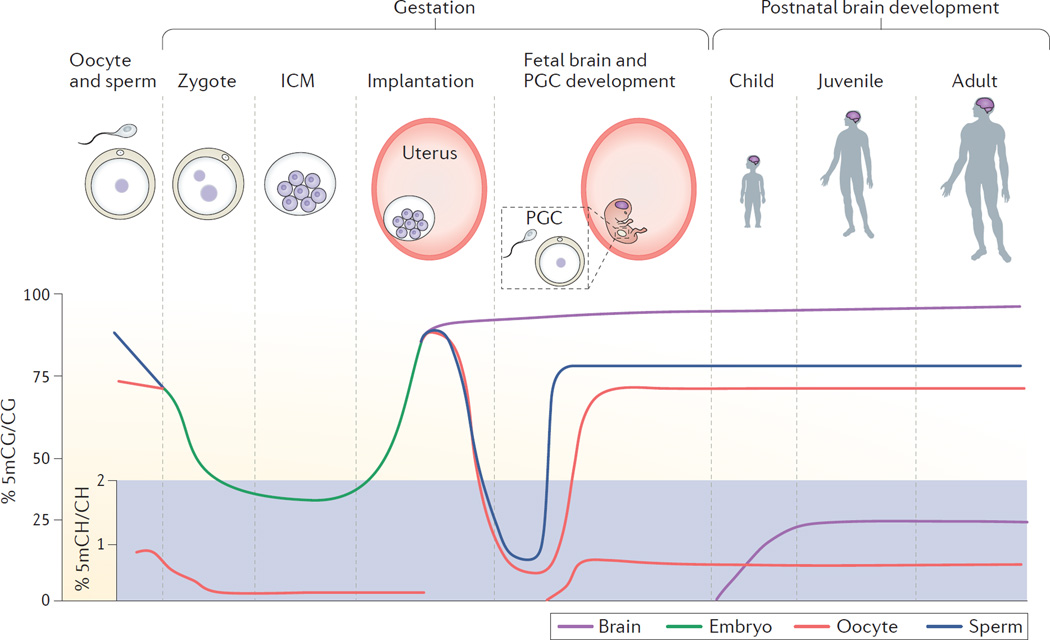
Dynamic changes to DNA-methylation patterns are crucial for re-establishing cellular identity in the early embryo (FIG. 3). In both mice and humans, sperm show high levels of 5mCG, whereas oocytes exhibit low to moderate methylation levels that increase with maturity64,65. After fertilization, sperm and oocyte methylomes are erased and re-established with new methylation patterns that are required for pluripotency in the inner cell mass (ICM) of the developing blastocyst. On fertilization in humans, zygotes and two-cell embryos show a dramatic decrease in 5mCG levels, and a further reduction occurs as the embryo progresses from the morula to the blastocyst stage64,65. In mice, levels of 5hmC increase in the paternal pronucleus as 5mCG levels decline, suggesting active demethylation by TET3 (REF. 34). The low methylation levels in the pluripotent ICM of the blastocyst are consistent with a state in which parental methylation is erased and replaced with new methylation over active gene bodies — a hallmark of a pluripotent epigenome. Following implantation, the ICM begins to show tissue-specific differentiation, and there is a sharp increase in 5mCG levels across the genome in both humans and mice64,65. These highly dynamic and global shifts in DNA methylation may represent a uniquely sensitive window in which environmental or genetic perturbations could epigenetically alter events during early embryogenesis.
Figure 3. DNA methylation landscape across development.
DNA methylation for 5-methylcytosine-guanine (5mCG) and 5-methylcytosine-(A, T or C) (5mCH) is shown on separate scales across human development. 5mCG levels in mature germ cells (sperm and oocyte) decrease after fertilization (zygote) and continue to decline in the inner cell mass (ICM) until implantation, when they rise rapidly. Primordial germ cells (PGCs) in the developing fetus then undergo a second global demethylation and subsequent re-methylation during maturation. Somatic cells, including brain cells, gradually gain 5mCG during the remainder of fetal and postnatal development. 5mCH levels are detectable in the oocyte (but not sperm), decline post-fertilization and return during subsequent PGC development. In the brain, 5mCH is detectable in glia and occurs at relatively high levels in neurons compared with other cell types. 5mCH levels in neurons rise after birth, peak in early childhood and are maintained into adulthood.
The second most dramatic erasure of DNA methylation in the mammalian genome occurs during the development of primordial germ cells (PGCs). As PGCs proliferate and migrate to the developing genital ridge, they lose nearly 95% of their DNA methylation and are nearly devoid of methylation by weeks 7–9 of human gestation. This is followed by a subsequent wave of global re-methylation in the developing germ cells. Global demethylation occurs across all genomic elements, with the exception of a subset of evolutionarily young repetitive elements66–68 and an interesting set of regions with overlapping genes that have few repeats and whose expression is significantly enriched in the brain. Expression of these genes is also enriched during neural development, and in PGCs that maintain partial methylation (20–30%)67. At least some of these regions are conserved in the mouse, and many show partial methylation in the ICM, indicating that they are ‘escapees’ of both rounds of epigenetic reprogramming and are therefore attractive candidate loci for transgenerational epigenetic inheritance67. However, whether these PGC escapee genes have true functional significance in neurodevelopment or are a phenomenon of other genomic features (such as increased length, number of promoters or composition of repeats) is an outstanding question of neuronal genes. Together, these two rounds of global methylation erasure and re-methylation suggest that early fetal development may be a uniquely sensitive time for epigenetic perturbation of brain development.
DNA methylation is highly dynamic across the lifespan
Methylation studies in human fetal and adult brains initially assayed predominantly CpG islands using various types of methylation arrays (BOX 3). Human prefrontal cortex samples collected from fetal brains (14–20 weeks gestational time), child brains (1–10 years old) and adult brains (>10 years old) showed distinct clustering of samples by age, with larger methylation differences found between the fetal and child brain samples than between the child and adult brain samples53. Similar findings were observed in a DNA methylation array study on 387 human subjects who ranged in age from 1–102 years old69. Although limited to a small percentage of the total CpGs in the human genome, this initial work suggested that the human brain methylome was highly dynamic across time.
Box 3. Genomic DNA methylation sequencing methods and features.
DNA methylation can be interrogated by methyl-sensitive restriction enzymes, methylation capture or bisulfite conversion. Most genome-wide methods for the quantification of DNA methylation adopted by studies in this Review involve bisulfite treatment of DNA, because methylated cytosines are protected from conversion and are therefore detected by hybridization or sequencing-based approaches. Illumina Infinium arrays are the most commonly used array-based platform and are available as a 27k platform primarily limited to gene promoters and CpG islands, or the 450k platform that expands the probe coverage to include gene bodies and intergenic regions. Another way to target dense regions of CpG sites is with reduced representative bisulfite sequencing (RRBS) using restriction enzymes or probe capture methods. The most comprehensive, least biased, but most expensive method is currently whole-genome bisulfite sequencing (WGBS or MethylC-seq). Although ×20–30 coverage of WGBS is required for accurate base pair resolution, lower coverage of ×2–3 is sufficient to detect methylation differences over 5–10 kb regions. With the increased read output and thereby decreased cost of sequencing on newer instruments (Illumina HiSeq 3000 and 4000) as well as the availability of multiplexing samples by barcodes in library construction, the use of WGBS approaches is expected to increase for epigenome-wide investigations in the future. One limitation to WGBS and Infinium arrays is that they cannot differentiate between 5-hydroxymethylcytosine (5hmC) and 5-methylcytosine (5mC) because both modifications are converted with bisulfite treatment. 5hmC and other oxidized derivatives (5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)) can be assayed by various different enzymatic or antibody pull-down sequencing techniques (MeDIP-seq) that currently must be conducted separately from 5mC analysis. For example, TET-assisted bisulfite sequencing (Tab-seq) uses β-glucosyltransferase to add a glucose to 5hmC (but not 5mC), and subsequent protection from oxidation by TET enzymes and bisulfite conversion allows direct, base-pair-resolution analysis of 5hmC125.
In mammals, dynamic changes in DNA methylation levels appear in the transition from fetal to early postnatal life. By comparing 35 fetal to 300 postnatal samples, Jaffe and colleagues70 identified widespread methylation changes associated in the transition from fetal to postnatal development. Differential methylation was observed at the individual CpG level (50.7% of CpGs on the array), DMR level (>6,000 DMRs) and large-scale level (896 multiple-kilobase regions), indicating global remodelling of the brain methylome70. These widespread changes in DNA methylation were largely attributed to shifts in cellular composition from progenitor-like cells before birth to terminally differentiated, lineage-specific cells in the postnatal brain. Differential methylation analysis between 73 young adult controls (aged 10–25 years) and 25 older controls (>25 years old) showed fewer CpG methylation differences than the postnatal–prenatal comparison70, indicating that early brain development has a particularly dynamic methylome.
Both 5hmC and 5mCH may also have crucial roles in neuronal development. Genome-wide 5hmC levels are low in the fetal mouse cortex (0.2% of all cytosines) and show a developmental increase in the transition from fetus to adult19,38,39. In mouse and human cerebellum, 5hmC enrichment was found mainly in CpG islands, shores, gene bodies and promoter regions that are associated with neuronal genes that increase their level of expression during development38,71. In further support for an association between 5hmC and neurodevelopmental gene expression, 5hmC DMRs in the human cerebellum were enriched for fragile X mental retardation protein (FMRP) target genes and ASD candidate genes, such as CACNA1C, SHANK3 and tuberous sclerosis 2 (TSC2)71. Similar to 5hmC, in both mice and humans 5mCH rapidly accumulates throughout postnatal and adolescent prefrontal cortex development from nearly undetectable levels to approximately 1.4%19. Because CpH sites are much more abundant than CpG sites, 5mCH represents a substantial and non-overlapping methylated portion of the adult neuronal methylome (FIG. 1c). The developmental increase in postnatal 5mCH and 5hmC suggests that DNA methylation may play a regulatory part in experience-dependent shaping of the neuronal transcriptome. For example, sensory input can dynamically regulate both 5mCG and 5hmC in the primary visual cortex of young but not adult mice. Inhibition of DNMTs blocked the negative functional impact of sensory deprivation, suggesting that dynamic DNA methylation changes modulate the experience-dependent transcription that is necessary for sensory input to correctly sculpt the developing visual system72. Additional work supports the role of active DNA methylation and demethylation in regulating experience-dependent plasticity and brain processes in adulthood (BOX 2).
Regions of differential methylation also seem to mark both cell-type- and age-specific regulatory DNA elements. Regions of CpG hypermethylation in neurons, when compared with similar regions in glia, showed a developmental gain in 5mCG from 1 to 6 weeks of age in mice and from fetal stages to 2 years of age in humans, after which levels stabilized. By contrast, DMRs that showed lower levels of 5mCG in neurons displayed the inverse pattern, with levels of 5mCG dropping during early development in both mice and humans19. Similarly, an even more specific analysis comparing three neuronal subtypes to fetal cortex and adult glia identified more than 250,000 5mCG DMRs, mostly in distal regulatory regions23. In the transition from fetus to adult, some of the fetal regulatory regions remained hypomethylated, whereas others gained methylation and lost active histone marks. These finely tuned patterns of enhancer and regulatory region usage are thought to coordinate gene expression during brain development73. In human prefrontal cortex, CpG sites with higher levels of 5hmC in the fetal compared with the adult brain were significantly enriched for enhancers, potentially marking a subset of regulatory regions that are poised for subsequent demethylation and activity in the adult brain39. Distal regulatory regions that are unique to the fetal frontal cortex gained 5mCG and 5mCH during later cortical development, suggesting that accumulation of 5mCG and 5mCH may mark the transition from active to inactive usage19. These neuron- and developmental-stage-specific methylation dynamics in regulatory regions suggest that DNA methylation might dictate enhancer usage during neuronal differentiation.
DNA methylation in ASD
DNA methylation is altered in ASD brains
Because most of the genomic views of the methylation landscape across development and cell type are recent high-resolution studies, interpreting the field of methylation differences in ASD is still in its infancy. Early work on DNA methylation in ASD focused on several candidate genes, including oxytocin receptor (OXTR), SHANK3, glutamic acid decarboxylase 65 (GAD65), reelin (RELN), ubiquitin protein ligase E3A (UBE3A) and MECP2 (REFS 74–78). Together, these targeted approaches provide tantalizing evidence that disruption of DNA methylation, at least at specific developmentally important loci, may play a part in the neuropathology underlying ASD.
To date, studies on genome-wide alterations in DNA methylation in ASD brain samples have been small in sample size and limited in CpG site coverage, but in spite of these inherent limitations they have shown promising preliminary evidence to support a role for DNA methylation in ASD. Nardone and colleagues examined both the prefrontal cortex (Brodmann area 10 (BA10); 12 cases and 12 controls) and anterior cingulate gyrus (BA24; 11 cases and 11 controls) in post-mortem human brains79. They identified 5,329 differentially methylated CpG sites between controls and ASD samples in BA10 and 10,745 significant CpGs in BA24. Differentially methylated sites occurred more often in areas of low CpG density, including gene bodies, and only rarely in CpG islands or at the transcription start site. CpGs that showed lower methylation in ASD BA10 compared with controls corresponded to several categories related to immune function and signalling. CpGs with higher methylation in ASD BA10 were enriched for functions involving synaptic transmission79. Ladd-Acosta and colleagues80 examined three brain regions: prefrontal cortex (6 cases and 5 controls), temporal cortex (6 cases and 10 controls) and cerebellum (7 cases and 6 controls) using the same type of Infinium 450k arrays. After adjusting for multiple comparisons, there were four significant DMRs (three in the temporal cortex and one in the cerebellum)80. Given the small sample size and the inherent heterogeneity of ASD cases, it is not surprising that few of the DMRs survived correction.
Both Nardone et al.79 and Ladd-Acosta et al.80 showed hypomethylation in the 3′ untranslated region of proline-rich transmembrane protein 1 (PRRT1) and the promoter regions of tetraspanin 32 (TSPAN32) and chromosome 11 open reading frame 21 (C11orf21) in ASD compared with control samples. PRRT1 was recently identified as an auxiliary protein for AMPA receptors that may influence the stability and electrophysiological properties of the receptor81. Interestingly, PRRT1 was recently found to have higher levels of gene-body 5mCG in the postnatal compared to fetal brain70, indicating an active gain in methylation with brain development that may fail to occur in ASD. TSPAN32 and C11orf21 are found in an imprinted region, but both escape imprinting and show biallelic expression82,83. TSPAN32 is expressed in very early embryonic development in haematopoietic stem cells and in adult haematopoietic progenitor cells in the adult bone marrow as well as in several types of immune cells82. A role for TSPAN32 in the brain has not been examined, but it does have a role in cellular immunity84, and other members of the tetraspanin family have been shown to be involved in brain function85. Little is known about C11orf21 function, but its expression is detected in various fetal tissues, including the brain83. Together PRRT1, TSPAN32 and C11orf21 represent novel sites of DNA methylation alterations in multiple regions of the ASD brain, but future studies will be needed to replicate and understand the functional implications.
The potential functional impact of DNA methylation alterations on gene expression in ASD is intriguing. To examine the relationship between DNA methylation and transcript levels in ASD, Nardone et al. examined genes with differential CpG methylation in BA10 compared with transcriptome data that were previously collected from BA9 in ASD cases and controls86. There was an overlap between genes showing hypomethylation and those that were overexpressed in BA9 ASD brains. Many of the genes showing altered CpG methylation79 and altered expression86 have immune-related functions, such as the genes that encode the complement cascade members C1Q, C3, integrin subunit beta 2 (ITGB2) and the cytokine tumour necrosis factor (TNF). Changes in immune-related gene expression and DNA methylation of immune-related genes may reflect alterations in the immune system in ASD. Prenatal infection is a risk factor for ASD, and children with ASD often display various immune-related abnormalities87. Given that immune-system-related genes are not highly prevalent among the genetic candidates for ASD risk, alterations in the expression and methylation of immune genes in ASD may occur in reaction to abnormal neurodevelopment and/or in response to environmental factors. Microglia are observed to be activated in the ASD brain88; however, differences in microglial-specific methylation patterns in ASD are currently unknown.
ASD brain samples seem to show a deficit in brain region specification for both DNA methylation and gene expression. In healthy controls, 52,000 differentially methylated CpG sites were identified between BA10 and BA24, indicating specialization of brain-region function79. Similar regional differences in DNA methylation have been observed among the frontal cortex, temporal cortex, pons and cerebellum of healthy controls across a wide range of ages69, and among the prefrontal, occipital and temporal cortex and the cerebellum in adults69,89. In comparison, only about 10,000 CpGs showed differential methylation between BA10 and BA24 in the ASD samples79. The lower regional specificity in DNA methylation in ASD brains is similar to what has previously been observed for transcription profiling. In control samples, 174 genes were differentially expressed between the superior temporal gyrus and prefrontal cortex, but no such regional expression differences were found for ASD samples86. Similarly, Ziats and Rennert90 found nearly 2,000 differentially expressed genes between the prefrontal cortex and cerebellum in controls and only 322 between brain regions in ASD samples. Regional differences in long non-coding RNA expression were also found between these two brain regions for controls but not for ASD samples90. However, not all brain regions show this lack of differentiation in regional expression in ASD, as gene expression differences between the occipital cortex (BA19) and the cerebellum seem to be intact in people with ASD91. Together, these data suggest that lack of brain-region-specific DNA methylation and gene expression may lead to a loss of cortical differentiation in at least some brain regions in ASD86.
Although these studies provide a promising basis for examining DNA methylation in ASD, target DMRs will need to be validated in larger cohorts of people with ASD and with methods that sample the entire genome and allow for examination of 5mCG, 5mCH and 5hmC. In addition, it is unclear how sex and age, as well as time of day, post-mortem interval and cause of death may influence methylation status in ASD samples compared to controls. Samples used by Ladd-Acosta et al.80 varied in age from 2 to 56 years of age at time of death, and 33% were female. Nardone et al.79 had age ranges from 16–51 and 13% were female. Given that both age-and sex-dependent changes in DNA methylation in the brain have been identified in mice and humans19, both factors need to be considered when interpreting DNA methylation and expression data in the context of ASD. All samples used by both studies had post-mortem intervals within 72 hours (median times: 21 hours80 and 23 hours79), a time window in which DNA methylation and integrity are relatively stable92. Furthermore, a study of 738 human post-mortem brains found significant differences in gene methylation by diurnal time of death93, so this may be an additional important variable for researchers to control for in brain methylation analyses.
Environment by genotype by epigenome
Prenatal exposure to a number of environmental chemicals has been associated with dysregulation of the fetal epigenome and potential consequences for brain function and developmental disorders. During embryogenesis, both the developing fetus and PGCs undergo massive changes in DNA methylation. Consequently, alterations in the levels of methyl donors or other perturbations of methylation pathways carry the potential for adverse effects on the fetal brain and germline development. There is growing epidemiological and laboratory evidence that prenatal exposure to a number of chemical compounds can negatively affect DNA methylation, gene expression and brain development. For example, in utero exposure to air-pollution-related polycyclic aromatic hydrocarbons, heavy metals, nicotine, alcohol and poor maternal nutrition have all been associated with alterations in DNA methylation and neurodevelopmental outcomes in the offspring94. Many of these environmental exposures can cross the placenta and fetal blood–brain barrier and alter a whole host of additional methylation-independent neurodevelopmental pathways. Future work is needed to dissect how DNA methylation alterations fit within the larger picture of brain dysregulation following in utero exposures. This work is crucial for understanding whether methylation alterations are a cause or consequence of altered gene expression in abnormal brain development and whether at least some of the methylation changes represent a form of molecular compensation for a perturbed epigenome.
The levels of methyl donors available to a developing fetus may be particularly important for early embryo-genesis. Folate serves as a major donor to the one carbon metabolism cycle that generates methyl donors that are crucial for DNA synthesis as well as methylation of DNA, proteins, phospholipids and neurotransmitters. Mammals are unable to synthesize the active form of folate and hence must rely on dietary sources. Prenatal vitamins typically contain several sources of methyl donors, including folic acid and vitamin B12. Mothers who increased their folic-acid intake with prenatal vitamins before conception and during the first month of pregnancy were at lower risk of having a child with ASD. This association was not observed for the second to ninth months of gestation, suggesting that early fetal development is particularly sensitive to levels of dietary methyl donors10,12,95. Consistent with this idea, mothers who took folic-acid-containing supplements before and/or during the first trimester had children with fewer behaviour problems96, whereas folate status during later pregnancy did not correlate with neurodevelopmental outcome97.
The increased sensitivity to levels of methyl donors during early pregnancy is consistent with the global levels of DNA de-methylation and subsequent re-methylation during embryo implantation in early gestation64,65. Mothers who took folic-acid supplementation for 4 weeks prior to and 8 weeks after conception had children with higher methylation of a known regulatory DMR for insulin-like growth factor 2 (IGF2) in whole blood samples. Higher IGF2 DMR methylation in the child was associated with higher blood levels of methyl donors in the mother and lower birth weight of the child98. Although methylation studies in whole blood samples are difficult to interpret owing to cell-composition heterogeneity (BOX 4), this study does suggest that maternal diet during early pregnancy can directly affect DNA methylation in the child. A recent, relatively large study (comprising 166 women in 24 villages in rural Gambia) showed that natural seasonal variation in maternal methyl-donor consumption correlated with changes in DNA methylation of metastable epialleles in the blood and hair of offspring99. Metastable epialleles, defined as epigenetic marks with precarious stability, are variably expressed owing to epigenetic modifications established during early embryonic development and, consequently, may be acutely sensitive to early-life environmental influences. In mice, high- and low-folic-acid diets correlated with alterations in DNA methylation in the cortex of early postnatal offspring. Specifically, both hyper and hypo levels of 5mCG and 5mCH were observed at a number of developmentally important sites, including several imprinted genes and ASD-risk candidates100. Together, these findings suggest that dietary manipulation of folate levels can directly affect DNA methylation status in the developing offspring. However, additional research is required to assess the long-term impact on the offspring’s health and brain function.
Box 4. The challenges of designing an ASD epigenome-wide association study.
Unlike genetic association studies, tissue-specific differences in DNA methylation and gene expression patterns complicate the choice of tissue for epigenome-wide association studies. Although post-mortem brain is the tissue of choice, both limited brain samples and incomplete clinical information about autism spectrum disorder (ASD) diagnosis are often problematic. Investigations are also underway of DNA methylation in various surrogate tissues that ideally would correlate with ASD symptoms and reflect misregulated pathways that are relevant to the brain. Whole blood is probably the most commonly used surrogate but is composed of various cell types, each with their own distinct DNA-methylation profile, and even in healthy controls DNA-methylation levels from whole blood reflect cell-type composition over individual differences126. Children with ASD have been reported to have alterations in immune cell composition in blood samples127, and consequently any DNA-methylation data from whole blood should be interpreted with caution. Buccal cells isolated from human saliva may be a better option because these cells and the brain both develop from the ectodermal layer. However, saliva is also contaminated with bacterial DNA, and the number of epithelial cells collected can vary substantially between individuals. In one study, DNA-methylation patterns from buccal cells did show a higher correlation with brain methylation than blood128. Sperm samples from fathers in a high-risk ASD cohort were used to correlate methylation changes with 12-month ASD assessments, identifying reduced methylation over a neuronally expressed small nucleolar RNA cluster in the Prader-Willi locus129. However, whether methylation differences in sperm could be predictive of an actual ASD diagnosis by 3 years of age is currently unknown. In addition, induced pluripotent stem cells (iPSCs) from patient samples may allow the study of individual ASD genetic mutations in iPSCs differentiated to neurons in vitro. However, iPSCs and iPSC-derived neurons show residual somatic DNA-methylation patterns130,131, aberrant novel methylation130 and aberrantly higher global methylation than the inner cell mass51. Abnormal DNA methylation in combination with the propensity for genetic abnormalities in iPSC cultures132 make the current iPSC methods problematic for the study of DNA-methylation alterations in ASD.
Dietary supplementation of methyl donors may help to offset genetic or environmental risk factors associated with ASD. The relationship between reduced risk of ASD and higher folic-acid intake was strongest when the mother and/or child had a genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR), a crucial enzyme in the one-carbon metabolism cycle10. The higher ASD risk variants of MTHFR (for example, 677T) are highly prevalent in the population (~60%) and show reduced enzyme activity, impaired folate metabolism and reduced methylation capabilities101. Polymorphisms in other components of the one-carbon metabolism cycle have also been identified in children with ASD95,102 and their mothers103. This suggests that folic-acid supplementation before and during early pregnancy may help to overcome some defects in the one-carbon metabolism cycle in individuals with increased genetic risk.
DNA methylation as a mark of past and future transcription
DNA methylation may serve as a marker or form of epigenetic ‘memory’ of past or future transcriptional events during brain development. Once established, DNA methylation marks can, in theory, be maintained into adulthood, suggesting that in utero environmental perturbations might alter transcriptional and methylation patterns for the lifetime of the organism. If unique and highly specific DNA methylation signatures were identified, they could serve as useful windows onto the transcriptional past and could potentially be used to predict future transcriptional regulation.
In rodents, at least some of the DNA methylation marks found in the adult brain show evidence of being laid down during fetal and early postnatal development. In a comparison between fetal and adult cells from the mouse prefrontal cortex, Mo et al. found that 51–67% of fetal DNA methylation valleys (DMVs) were conserved in adult neurons and glia and enriched for developmental transcription factors23. The largest gain in DMV methylation occurred at regions where gene expression decreased in the transition from the fetal to adult brain. These genes were enriched for neuron-subtype-specific transcription factors that are predicted to sculpt the function and relative abundance of neurons in the developing brain23. In addition, a small subset of active fetal enhancers (~9%) lose their active chromatin marks in the adult brain, but remain hypomethylated22. Hypomethylation combined with lack of active histone marks seems to serve as a unique form of epigenetic memory of past embryonic activity. 5hmC may also mark cell-type-specific regions in utero for subsequent regulation during development. For example, in the human brain, enhancers with higher levels of 5hmC in the fetal compared to adult prefrontal cortex may be poised for activation in development19,39.
Recent work in schizophrenia suggests that pre- to postnatal brain methylome changes may intersect with underlying genetic risk loci to potentially mark altered brain development. Using Illumina 450k arrays on post-mortem dorsolateral prefrontal cortex from 108 people with schizophrenia and 136 controls, Jaffe and colleagues70 found more than 2,100 differentially methylated CpGs, most of which showed lower methylation in people with the disorder. Ninety-four percent of these differentially methylated CpGs were specifically differentially methylated between fetal and postnatal life, and were also slightly enriched for schizophrenia genome-wide association study (GWAS) risk loci70. Similarly, DNA-methylation quantitative trait loci identified from DNA-methylation profiling and single --nucleotide polymorphism genotyping of 166 fetal human brain samples were also enriched (~fourfold) for schizophrenia GWAS-positive risk loci104. Together, these findings point to potential DNA methylation signatures of prenatal life in the adult brain methylome that may serve as useful markers of early life experience and adult disease risk.
Conclusions and perspective
Given that most cases of ASD result from a combination of genetic risk and environmental exposures, alterations in DNA methylation serve as an attractive mechanism to link environmental perturbations to lifelong changes in brain function and behaviour. DNA methylation may have various roles in brain development, including directly affecting gene expression by recruiting specific DNA methyl-binding proteins or serving as a mark of previous transcriptional events that are adaptive in poising genomic regions for future events. Examining DNA methylation in the context of developmental gene expression, other epigenomic markers of chromatin state and cell type can predict functional impact. How DNA methylation affects brain function is also inherently tied to genetic variation in transcription factors, methyl-binding proteins and other regulatory proteins and distal DNA regions. The combination of specific inherited genetic variants may make some individuals more susceptible to environmental risk factors such as chemical exposures, maternal infection or low folate levels. Individuals harbouring CNVs or single-nucleotide variants that affect chromatin and synaptic genes might also be especially sensitive to environmental perturbations. Given the widespread and highly dynamic changes in DNA methylation that occur in early fetal life, this time point in development may be particularly vulnerable to the cumulative impact of genetic and environmental risk factors.
Testing the causal relationship between DNA-methylation changes and altered brain development is difficult. DNA methylation is likely to be both a cause and consequence of the developmental regulation of gene expression and occurs in the context of other epigenomic mechanisms of regulation, such as histone modifications. Consequently, differences in methylation at any given point in development may or may not provide mechanistic information on the current state of neurodevelopment. For example, altered brain development caused by a genetic or environmental perturbation may indirectly affect DNA methylation as the brain attempts to compensate for suboptimal conditions. So far, there is only limited evidence of altered DNA-methylation patterns in ASD brain samples, but the levels of methyl donors available to the developing fetus during gestation are related to ASD risk. Although this is hardly causal evidence, it does suggest that supplements containing methyl donors taken before conception may provide some protection against ASD by mitigating the perfect storm of risk factors and aberrant DNA methylation in genetically susceptible individuals.
Acknowledgments
The authors thank D. Yasui, K. Dunaway and A. Nord for critical reading of the manuscript. Ongoing support for this research is supported by the US National Institute of Neurological Disorders and Stroke (grants R01NS081913 and R01NS076263), the US National Institute of Environmental Health Sciences (grant R01ES021707), the US Department of Defense (grant W81XWH-12-1-0491) and the US National Institute of Mental Health (grant T32MH073124-06).
Glossary
- CpG
Cytosine 5’ of guanine, where the ‘p’ stands for the phosphate
- 5-methylcytosine
(5mC). A cytosine base with a methyl group (CH3) in the 5’ position
- CpG islands
Short (<1 kb) regions in mammalian genomes with the highest density of CpG sites
- 5-hydroxymethylcytosine
(5hmC). A cytosine base with a hydroxymethyl (-CH2-OH) group in the 5’ position
- CpH
A cytosine followed by either adenine (CpA), cytosine (CpC) or thymine (CpT), where the ‘p’ stands for the phosphate
- Ten-eleven translocase
(TET). The three TET enzymes, TET1, TET2 and TET3, are methylcytosine dioxygenases that convert 5-methylcytosine to 5-hydroxymethylcytosine
- Gene body
The genomic region extending from the transcription start site to the transcription end site
- Methyl-binding domain
(MBD). A protein domain structure with a higher affinity to methylated DNA than to unmethylated DNA. The MBD family of proteins includes methyl-CpG binding protein 2
- Differentially methylated regions
(DMRs). Short clusters of CpG sites in regions showing significant differences in methylation between stages or samples, defined bioinformatically
- Partially methylated domains
(PMDs). Large genomic regions in the mammalian genome that are characterized by lower than 70% 5-methylcytosine-guanine (5mCG). PMDs are found in 30–50% of the genome in preimplantation embryos the placenta and most solid tumours. They are characterized by lower or more-variable transcript levels, repressive histone marks and nuclear lamina localization
- Inner cell mass
(ICM). The cells in the blastocyst of the preimplantation embryo that become fetal tissues
- Primordial germ cells
(PGCs). The embryonic lineage of cells that migrate to the genital ridge as precursors of the germline cells of the next generation (spermatagonia and oocytes)
- Methylenetetrahydrofolate reductase
(MTHFR). An enzyme in the one-carbon metabolism cycle that supplies methyl groups for DNA-methylation reactions. A common polymorphism in the MTHFR gene encodes a less-efficient enzyme and is a risk factor for neural tube defects and autism
- DNA methylation valleys
(DMVs). Regions of low methylation that are >5 kb in length and are found in most cell types. They can show differences in developmental methylation between fetal and adult tissues
- Induced pluripotent stem cells
(iPSCs). Adult somatic cells that are genetically reprogrammed in vitro to produce an embryonic stem cell-like state of pluripotency. iPSCs can then be differentiated into a number of different cell types
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Arlington: 2013. [Google Scholar]
- 2.Doshi-Velez F, Ge Y, Kohane I. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics. 2013;133:e54–e63. doi: 10.1542/peds.2013-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators & Centers for Disease Control and Prevention (CDC) Prevalence of autism spectrum disorder among children aged 8 years — autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill. Summ. 2014;63:1–21. [PubMed] [Google Scholar]
- 4.Hallmayer J, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalkbrenner AE, Schmidt RJ, Penlesky AC. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Curr. Probl. Pediatr. Adolesc. Health Care. 2014;44:277–318. doi: 10.1016/j.cppeds.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 8.Gaugler T, et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandin S, et al. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt RJ, et al. Maternal periconceptional folic acid intake and risk for developmental delay and autism spectrum disorder: a case-control study. Am. J. Epidemiol. 2012;175:S126. doi: 10.3945/ajcn.110.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt RJ. Maternal folic acid supplements associated with reduced autism risk in the child. Evid. Based. Med. 2013;18:e53. doi: 10.1136/eb-2013-101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suren P, et al. Association between maternal use of folic acid supplements and risk of autism spectrum disorders in children. J. Am. Med. Assoc. 2013;309:570–577. doi: 10.1001/jama.2012.155925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BK, et al. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behav. Immun. 2015;44:100–105. doi: 10.1016/j.bbi.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang SY, Wang S, Huang N, Yeh HH, Chen CY. Prenatal infection and autism spectrum disorders in childhood: a population-based case-control study in Taiwan. Paediatr. Perinat. Epidemiol. 2015;29:307–316. doi: 10.1111/ppe.12194. [DOI] [PubMed] [Google Scholar]
- 15.Keil AP, Daniels JL, Hertz-Picciotto I. Autism spectrum disorder, flea and tick medication, and adjustments for exposure misclassification: the CHARGE (CHildhood Autism Risks from Genetics and Environment) case-control study. Environ. Health. 2014;13:3. doi: 10.1186/1476-069X-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelton JF, et al. Neurodevelopmental disorders and prenatal residential proximity to agricultural pesticides: the CHARGE study. Environ. Health Perspect. 2014;122:1103–1109. doi: 10.1289/ehp.1307044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasalle J. Epigenetic layers and players underlying neurodevelopment. Trends Neurosci. 2013;36:460–470. doi: 10.1016/j.tins.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. This study was the first whole-genome bisulfite sequencing examination of DNA methylation in human cells to reveal mCH methylation
- 19. Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:629–643. doi: 10.1126/science.1237905. This was the first whole-genome bisulfite sequencing examination of DNA methylation dynamics in brain development in humans and mice
- 20.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mo A, et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. This study was the first to show DNA-methylation maps in specific neuronal subtypes from the mouse cortex, and it reveals cell-type-specific methylation signatures
- 24.Goto K, et al. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 25.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe D, Suetake I, Tada T, Tajima S. Stage-and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech. Dev. 2002;118:187–190. doi: 10.1016/s0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- 27.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 28.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 29.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y-F, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu T-P, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 35.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen L, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song C-X, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schanen NC. Epigenetics of autism spectrum disorders. Hum. Mol. Genet. 2006;15(Suppl. 2):R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 42.Lasalle JM. Epigenomic strategies at the interface of genetic and environmental risk factors for autism. 2013;58:396–401. doi: 10.1038/jhg.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatton-Brown K, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanders SJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. This is the most recent and comprehensive examination of de novo mutations in ASD
- 45.Leung KN, Chamberlain SJ, Lalande M, Lasalle JM. Neuronal chromatin dynamics of imprinting in development and disease. J. Cell. Biochem. 2011;112:365–373. doi: 10.1002/jcb.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaSalle JM, Reiter LT, Chamberlain SJ. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics. 2015;7:1213–1228. doi: 10.2217/epi.15.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaSalle JM, Yasui DH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics. 2009;1:119–130. doi: 10.2217/epi.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cukier HN, et al. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11:291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spruijt CG, et al. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat. Rev. Genet. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- 51. Woods R, et al. Long-lived epigenetic interactions between perinatal PBDE exposure and Mecp2308 mutation. Hum. Mol. Genet. 2012;21:2399–2411. doi: 10.1093/hmg/dds046. This paper presents an example of an in utero environmental exposure (to polybrominated diphenyl ether) and a genetic mutation (MECP2 truncation) interacting to alter DNA methylation in, and behaviour of, the offspring
- 52.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 53.Numata S, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim M, et al. Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Hum. Mol. Genet. 2014;23:657–667. doi: 10.1093/hmg/ddt453. [DOI] [PubMed] [Google Scholar]
- 56.Smith EY, Futtner CR, Chamberlain SJ, Johnstone Ka, Resnick JL. Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS Genet. 2011;7:e1002422. doi: 10.1371/journal.pgen.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christensen BC, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CPG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schroeder DI, Lott P, Korf I, LaSalle JM. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res. 2011;21:1583–1591. doi: 10.1101/gr.119131.110. PMDs mark distinct subsets of developmentally important genes in the neuronal cell line SH-SY5Y. These findings suggest that PMDs may be regions of DNA methylation that are relevant to the epigenetic regulation of ASD candidate genes acting at synapses
- 59.Schroeder D, et al. The human placenta methylome. Proc. Natl Acad. Sci. USA. 2013;110:6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeder DI, et al. Early developmental and evolutionary origins of gene body DNA methylation patterns in mammalian placentas. PLOS Genet. 2015;11:e1005442. doi: 10.1371/journal.pgen.1005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novakovic B, Saffery R. The ever growing complexity of placental epigenetics — role in adverse pregnancy outcomes and fetal programming. Placenta. 2012;33:959–970. doi: 10.1016/j.placenta.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2012;484:550–550. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 64.Guo H, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 65.Smith ZD, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gkountela S, et al. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang WWC, et al. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo F, et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 69.Hernandez DG, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jaffe AE, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2015;19:4–7. doi: 10.1038/nn.4181. This is a large 450k array study examining DNA methylation in human brain across fetal and postnatal development. The authors identify regions of differential methylation between fetal and postnatal ages that are enriched for schizophrenia GWAS risk loci, indicating a disruption of the fetal epigenome in this disorder
- 71.Wang T, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tognini P, et al. Experience-dependent DNA methylation regulates plasticity in the developing visual cortex. Nat. Neurosci. 2015;18:956–958. doi: 10.1038/nn.4026. [DOI] [PubMed] [Google Scholar]
- 73.Nord AS, Pattabiraman K, Visel A, Rubenstein JLR. Review genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gregory SG, et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu L, et al. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum. Mol. Genet. 2014;23:1563–1578. doi: 10.1093/hmg/ddt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhubi a, et al. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang YH, et al. A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am. J. Med. Genet. 2004;131:1–10. doi: 10.1002/ajmg.a.30297. [DOI] [PubMed] [Google Scholar]
- 78.Nagarajan RP, Hogard AR, Gwye Y, Martin MR, Lasalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:1–11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nardone S, et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry. 2014;4:e433. doi: 10.1038/tp.2014.70. This is one of the two 450k array studies to examine DNA methylation in ASD brain samples. In BA10, 5,000 differentially methylationed CpGs and in BA24 more than 10,000 differentially methylationed CpGs were identified between controls and ASD samples. In ASD brain samples, genes with lower levels of methylation in BA10 show an overlap with genes showing lower expression levels in BA9
- 80. Ladd-Acosta C, et al. Common DNA methylation alterations in multiple brain regions in autism. Mol. Psychiatry. 2014;19:862–871. doi: 10.1038/mp.2013.114. This is one of the two 450k array studies to examine DNA methylation in ASD brain samples from the prefrontal cortex, temporal cortex and cerebellum. Four significant ASD-associated DMRs were identified
- 81.Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 82.Nicholson RH, et al. Phemx, a novel mouse gene expressed in hematopoietic cells maps to the imprinted cluster on distal chromosome 7. Genomics. 2000;68:13–21. doi: 10.1006/geno.2000.6277. [DOI] [PubMed] [Google Scholar]
- 83.Zhu X, et al. C11orf21 a novel gene within the Beckwith-Wiedemann syndrome region in human chromosome 11p15.5. Gene. 2000;256:311–317. doi: 10.1016/s0378-1119(00)00377-2. [DOI] [PubMed] [Google Scholar]
- 84.Gartlan KH, et al. A complementary role for the tetraspanins CD37 and Tssc6 in cellular immunity. J. Immunol. 2010;185:3158–3166. doi: 10.4049/jimmunol.0902867. [DOI] [PubMed] [Google Scholar]
- 85.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 86. Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. This is the first transcriptome-wide analysis of gene expression in ASD compared with control brain samples. This study suggests that differences in gene expression between brain regions may be disrupted in ASD samples
- 87.Estes ML, Mcallister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat. Rev. Neurosci. 2015;16:469–486. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takano T. Role of microglia in autism: recent advances. Dev. Neurosci. 2015;37:195–202. doi: 10.1159/000398791. [DOI] [PubMed] [Google Scholar]
- 89.Xin Y, et al. Genome-wide divergence of DNA methylation marks in cerebral and cerebellar cortices. PLoS ONE. 2010;5:e11357. doi: 10.1371/journal.pone.0011357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. This large-scale transcriptome analysis of gene expression in ASD and control brain samples further supports a lack of regional expression differences in ASD for both coding and non-coding transcripts
- 91.Ginsberg MR, Rubin Ra, Natowicz MR. Patterning of regional gene expression in autism: new complexity. Sci. Rep. 2013;3:1831. doi: 10.1038/srep01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhein M, et al. DNA methylation results depend on DNA integrity — role of post mortem interval. Front. Genet. 2015;6:1–7. doi: 10.3389/fgene.2015.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim ASP, et al. 24-hour rhythms of DNA methylation and their relation with rhythms of RNA expression in the human dorsolateral prefrontal cortex. PLoS Genet. 2014;10:e1004792. doi: 10.1371/journal.pgen.1004792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Vaiserman A. Epidemiologic evidence for association between adverse environmental exposures in early life and epigenetic variation: a potential link to disease susceptibility? Clin. Epigenetics. 2015;7:96. doi: 10.1186/s13148-015-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schmidt R, et al. Prenatal vitamins, one-carbon metabolism gene variants, and risk for autism. Epidemiology. 2011;22:476–485. doi: 10.1097/EDE.0b013e31821d0e30. In this large, population-based case-control study, periconception and early prentatal vitamin intake are associated with lower risk for ASD. ASD risk was higher for children whose mothers failed to take prenatal vitamins and had specific genetic variants detrimental to one-carbon metabolism pathways
- 96.Julvez J, et al. Maternal use of folic acid supplements during pregnancy and four-year-old neurodevelopment in a population-based birth cohort. Paediatr. Perinat. Epidemiol. 2009;23:199–206. doi: 10.1111/j.1365-3016.2009.01032.x. [DOI] [PubMed] [Google Scholar]
- 97.Tamura T, et al. Folate status of mothers during pregnancy and mental and psychomotor development of their children at five years of age. Pediatrics. 2005;116:703–708. doi: 10.1542/peds.2004-2189. [DOI] [PubMed] [Google Scholar]
- 98.Steegers-Theunissen RP, et al. Periconceptional maternal folic acid use of 400 µg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS ONE. 2009;4:1–5. doi: 10.1371/journal.pone.0007845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dominguez-Salas P, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 2014;5:1–7. doi: 10.1038/ncomms4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barua S, et al. Single-base resolution of mouse offspring brain methylome reveals epigenome modifications caused by gestational folic acid. Epigenetics Chromatin. 2014;7:3. doi: 10.1186/1756-8935-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hustad S, et al. The methylenetetrahydrofolate reductase 677→T polymorphism as a modulator of a B vitamin network with major effects on homocysteine metabolism. Am. J. Hum. Genet. 2007;80:846–855. doi: 10.1086/513520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.James SJ, et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.James SJ, et al. A functional polymorphism in the reduced folate carrier gene and DNA hypomethylation in mothers of children with autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153:1209–1220. doi: 10.1002/ajmg.b.31094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hannon E, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156:255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- 106.Ozonoff S, et al. Recurrence risk for autism spectrum disorders: a baby siblings research consortium study. Pediatrics. 2011;128:e488–e495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Robinson EB, Lichtenstein P, Anckarsater H, Happe F, Ronald A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl Acad. Sci. USA. 2013;110:5258–5262. doi: 10.1073/pnas.1211070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 109.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Girirajan S, et al. Global increases in both common and rare copy number load associated with autism. Hum. Mol. Genet. 2013;22:2870–2880. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hormozdiari F, Penn O, Borenstein E, Eichler EE. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2015;25:142–154. doi: 10.1101/gr.178855.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gupta S, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun. 2014;5:5748. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miller Ca, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 116.Levenson J, Roth T, Lubin F. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 117.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaas Ga, et al. Hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rudenko A, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li X, et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc. Natl Acad. Sci. USA. 2014;111:7120–7125. doi: 10.1073/pnas.1318906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Day JJ, et al. DNA methylation regulates associative reward learning. Nat. Neurosci. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bali P, Im HI, Kenny PJ. Methylation, memory and addiction. Epigenetics. 2011;6:671–674. doi: 10.4161/epi.6.6.15905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zannas AS, West AE. Epigenetics and the regulation of stress vulnerability and resilience. NeuroScience. 2014;264:157–170. doi: 10.1016/j.neuroscience.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu M, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reinius LE, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ashwood P, et al. In search of cellular immunophenotypes in the blood of children with autism. PLoS ONE. 2011;6:e19299. doi: 10.1371/journal.pone.0019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smith AK, et al. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015;168:36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Feinberg JI, et al. Paternal sperm DNA methylation associated with early signs of autism risk in an autism-enriched cohort. Int. J. Epidemiol. 2015:1–12. doi: 10.1093/ije/dyv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;470:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ohi Y, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;470:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]