Abstract
Anopheles gambiae devotes over 2% (295) of its protein coding genes to structural cuticular proteins (CPs) that have been classified into 13 different families plus ten low complexity proteins not assigned to families. Small groups of genes code for identical proteins reducing the total number of unique cuticular proteins to 282. Is the large number because different structures utilize different CPs, or are all of the genes widely expressed? We used LC-MS/MS to learn how many products of these genes were found in five adult structures: Johnston's organs, the remainder of the male antennae, eye lenses, legs, and wings. Data were analyzed against both the entire proteome and a smaller database of just CPs. We recovered unique peptides for 97 CPs and shared peptides for another 35. Members of 11 of the 13 families were recovered as well as some unclassified. Only 11 CPs were present exclusively in only one structure while 43 CPs were recovered from all five structures. A quantitative analysis, using normalized spectral counts, revealed that only a few CPs were abundant in each structure. When the MS/MS data were run against the entire proteome, the majority of the top hits were to CPs, but peptides were recovered from an additional 467 proteins. CP peptides were frequently recovered from chitin-binding domains, confirming that protein-chitin interactions are not mediated by covalent bonds. Comparison with three other MS/MS analyses of cuticles or cuticle-rich structures augmented the current analysis. Our findings provide new insights into the composition of different mosquito structures and reveal the complexity of selection and utilization of genes coding for structural cuticular proteins.
Keywords: Cuticle, Crosslink, Mass spectrometry, R&R consensus, Sequence clusters
1. Introduction
Genes coding for putative structural cuticular proteins (CPs) have been identified in many species of Hexapoda, but the vast majority were named or had a function assigned based on resemblance to similar proteins in other species. It is only a tiny fraction of these proteins that actually were isolated from cuticle and sequenced or localized in cuticle with specific antibodies visualized with a colloidal gold-labeled secondary antibody. Only proteins identified by either of these two methods are authentic cuticular proteins; those identified solely by sequence comparisons are only putative cuticular proteins.
Early on in the annotation of the cuticular proteins of Anopheles gambiae we carried out a tandem MS/MS analysis of the proteins from larval head capsules and pupal cuticle left behind after a molt (He et al., 2007). We assumed these preparations would be primarily cuticle that had been stripped of adjacent cells in the process of molting. Data from the analysis was mapped back onto the databases of proteins available at that time. Thus there are two limitations of these data: annotation was at a preliminary stage and the cast cuticles might be devoid of many endocuticular proteins lost by digestion by molting fluid.
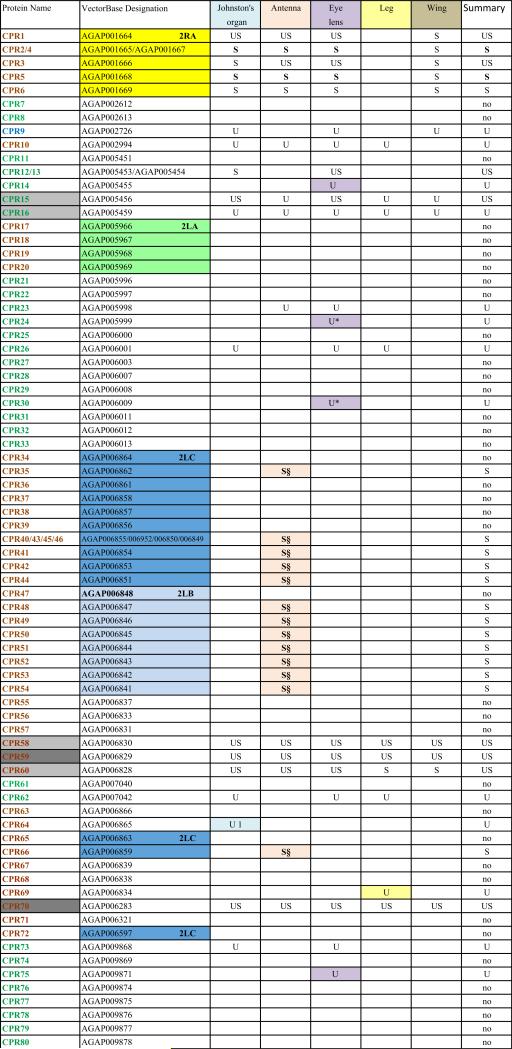
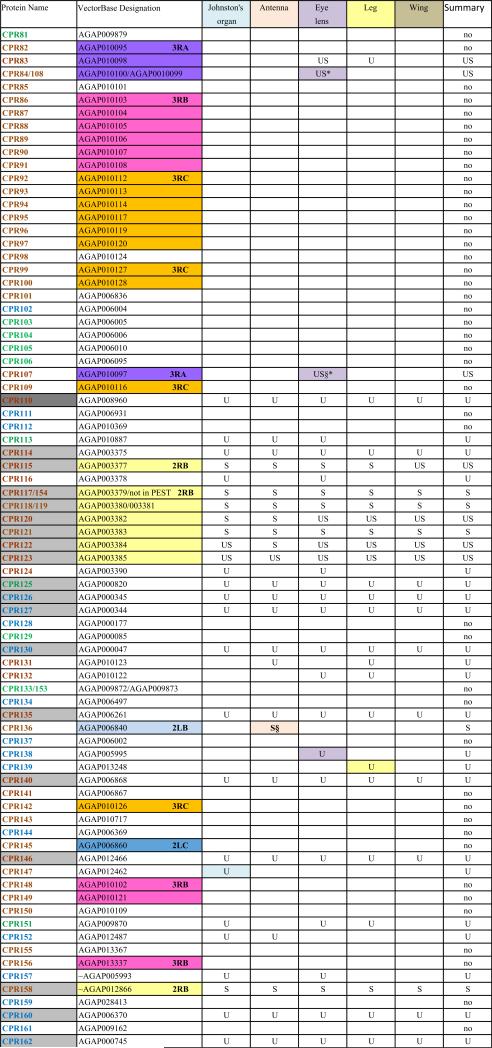
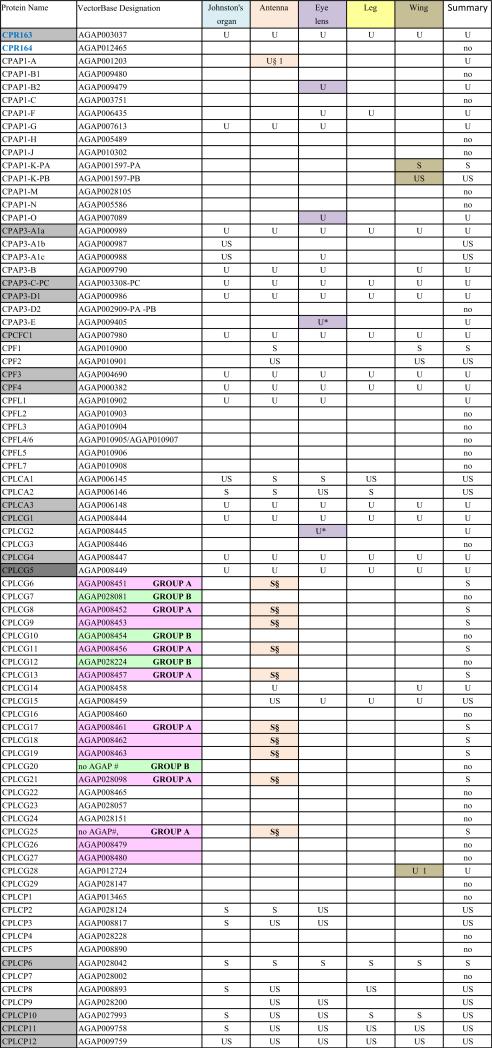
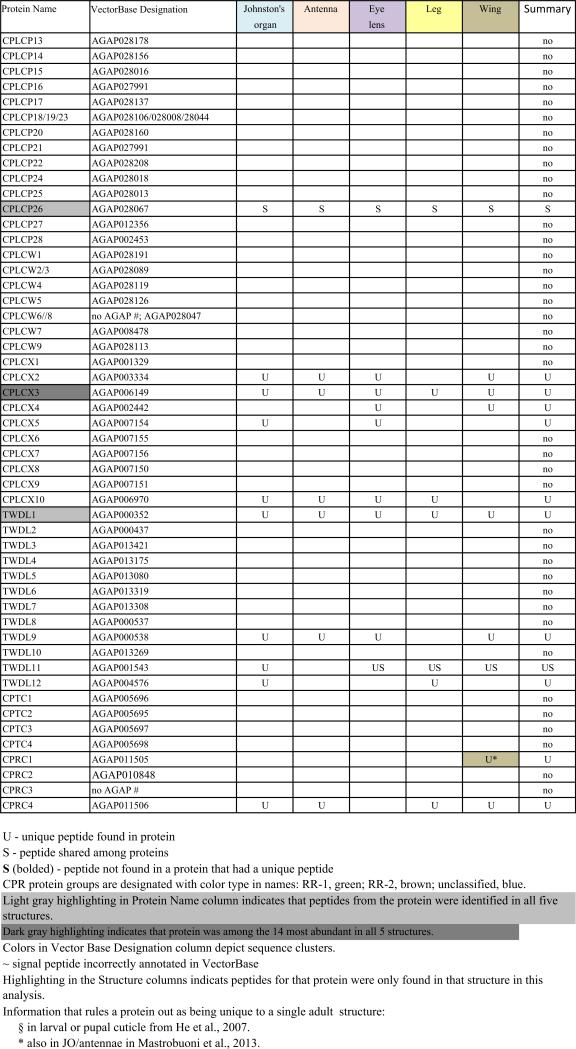
When He et al. (2007) was published, only a subset of An. gambiae putative cuticular proteins had been annotated. Despite its limitations, He et al. (2007) confirmed the authenticity of many of the CPs we had annotated and provided us with additional members of known CP families and several new families of CPs. Since then, three previously unrecognized families of CPs have been reported: CPAP1 and CPAP3 (Jasrapuria et al., 2010; Tetreau et al., 2015) and CPCFC (Vannini et al., 2015). Further annotation based on these results and a far more robust proteome (Vector Base AgamP4.2) resulted in a cuticulome of An. gambiae with 295 distinct genes. These genes code for only 282 distinct CPs, because the protein products of several genes are identical. The CP genes have been classified into 13 distinct families and some orphans (Willis, 2010; Willis et al., 2012; Ioannidou et al., 2014; Tetreau et al., 2015; Vannini et al., 2015). This information is summarized in Table 1 and all CP sequences where we identified peptides in this study are in Supplementary File 1.
Table 1.
Cuticular proteins identified in different structures.
A major limitation of our first study was that it was restricted to two preparations, neither of which contained adult components. Several recent studies have shown the importance of looking at specific organs for proteomes and transcriptomes since components specialized for a particular task might not be detected in analyses of whole organism because of the overwhelming contribution of those that are more widely used.
Furthermore, since our first study, we have carried out extensive in situ hybridization with probes based on An. gambiae CP sequences. From this we learned that a single structure such as the eyes, legs, or Johnston's organ could be the site of hybridization of numerous mRNAs for cuticular proteins. We have already reported on ten putative CP genes that had transcripts in the developing eye, most associated with cells that form the corneal lens and pseudocone (Vannini et al., 2014). We wondered if these transcripts were actually translated and exported into the cuticle. Was it really possible that so many different cuticular proteins would be used to build a single structure? Given the large number of structural cuticular proteins, one might expect that there would be proteins playing unique roles in specific structures. Indeed, recent analyses in Tribolium castaneum focused on just three proteins found, but not exclusively, in the elytra and demonstrated essential roles for each (Arakane et al., 2012; Noh et al., 2014, 2015).
Thus, we have repeated our proteomics analyses on adult structures: wings, legs, Johnston's organ (second antennal segment), the rest of the antennae, and the outer covering of the eye.
Data from this study have enabled us to expand the number of authentic CPs in An. gambiae, to confirm that individual structures are composed of multiple CPs and to demonstrate a provocative pattern of CP usage with almost none restricted to a single structure.
Many proteins annotated as structural cuticular proteins did not appear in our analyses. Could it be that the corresponding genes, based on annotation in PEST, are not present in the G3 strain? Fortunately, an extensive MS analysis with the G3 strain of several adult organs and parts as well as larvae (Chaerkady et al., 2011), helped to answer that possibility.
Two additional MS analyses, one analyzing antennae (Mastrobuoni et al., 2013) and the other analyzing both eyes and antennae (Champion et al., 2015) augmented our analysis and reduced the number of proteins that appeared to be unique for a single structure.
The two cast cuticles we had analyzed turned out to have numerous components in addition to the cuticular proteins we had annotated (He et al., 2007). Most of these had signal peptides and were not unexpected components of recently molted cuticles, i.e. molting fluid constituents, peptidases and chitinases. Others were enzymes associated with cuticular sclerotization, such as prophenyloxidases and laccases. There were also some muscle components.
In this study, we also identified peptides from proteins that obviously are not structural. More would be expected in this analysis because we were working with entire structures rather than cast cuticles.
2. Materials and methods
2.1. Tissue Collection
First instar larvae of An. gambiae (G3 strain), obtained from the breeding facility at the University of Georgia Entomology Department, were maintained in our laboratory in a 12/12 L/D photoperiod, at 27 °C and fed ground Koi food (Foster and Smith Aquatics). Adults had access to water and an 8% fructose solution and were kept in an insectary at 27 °C with a 16/8 L/D photoperiod. Most structures came from adults of both sexes that were frozen at −80 °C 5–6 days after eclosion. Johnston's organs and the rest of the antennae were only taken from males. Eyes were dissected to obtain the lens and the cuticular capsule that surrounds it. Johnston's organ is contained within a cuticular pedicle of the second antennal segment; hence it too has a cuticular capsule. All structures were immersed in PBS in 1.5 ml Kontes centrifuge tubes immediately after removal and frozen until enough were produced for further processing. Two batches of tissue were prepared except three for the eye lens. For each batch, Johnston's organs, antennae, eye lenses, wings and legs came from around 230, 230, 330, 100 and 80 individual adults, respectively.
2.2. Sample preparation
PBS was removed by centrifugation and structures were resus-pended in 80–300 μl of 1% SDS containing 50 mM dithiothreitol (DTT), homogenized with Kontes plastic pestles, and placed in a boiling water bath for 15 min. A rough approximation of the soluble protein concentration in the extracts was obtained with a Nano-Drop N-1000 (Thermo Scientific) using 2 μl for each of three replicates and the protein A280 method. SDS extracts were resolved in NuPAGE 4–12% Bis-Tris Gels (NP0323BOX, Invitrogen), with the same sample loaded onto 3 or occasionally 4 lanes. The gels were run at a constant voltage of 90–120 V for about 20 min for gels when we wanted only 1 slice and for 40 min to yield 4 slices. Gels were stained with Coomassie (Quick Coomassie Stain, Generon, Ltd, UK). Each gel slice was further cut into small pieces (~1 mm2). Gel pieces were destained by sequential rinses with water (once), 50% acetonitrile in water (twice), 25 mM NH4HCO3 (once) and twice with acetonitrile (ACN). Each time the volume used was sufficient to cover the pieces. Alkylation and reduction were carried out by incubating the gel pieces in 10 mM DTT/25 mM NH4HCO3 at 65 °C for 1 h, followed by 55 mM IDA (iodoacetamide)/25 mM NH4HCO3 in the dark for 1 h. The pieces were washed with 25 mM NH4HCO3, dehydrated with acetonitrile, then incubated with sequencing grade trypsin (Promega) (50:1 w/w protein/trypsin) overnight at 37 °C. The supernatant was transferred to a new tube; 50% acetonitrile/0.1% formic acid was added into the gel pieces to further extract the digested peptides. Pooled supernatants were dried in a Savant SpeedVac (Thermo Scientific) and re-dissolved in 20 μl of 5% ACN/0.1% formic acid solution before LC–MS/MS analysis. The data we present combines all the gel slices obtained from a single extract for we found nothing informative when we compared peptides from slow and fast moving proteins or the single slice with the four slices.
The pellets left after SDS extraction were washed 5 times with 25 mM ammonium bicarbonate, reduced with 10 mM DTT, alkylated with 55 mM IDA, digested with trypsin (50:1 w/w protein/ trypsin) and then filtered through a 30 kDA spin filter (EMB Millipore). Supernatants were dried and re-suspended in 5% ACN/0.1% formic acid.
2.3. LC-MS/MS analysis
Samples were subjected to LC-MS/MS analyses on different machines because of limitations on availability. Machines used for each sample are listed in Supplementary File 9. Most of the tryptic peptides from the gels of SDS extracts from eye lens, Johnston's organ, antenna, leg and wing were analyzed with the Finnigan LTQ linear ion trap mass spectrometer (Thermo-Fisher) and an 1100 Series Capillary LC system (Agilent Technologies) with an ESI source with spray tips built in-house. Samples were suspended in 15 μl of buffer A [0.1% formic acid (Sigma-Aldrich)/10 mM ammonium formate (Thermo-Fisher)] and 1 μl of buffer B (80% acetonitrile/0.1% formic acid/10 mM ammonium formate), and 8 ml of each sample were injected into the LC. Peptides were separated using a 200-μm × 150-mm HALO Peptide ES-C18 column packed with 5-μm diameter superficially porous particles (Advanced Materials Technology). The gradient used for each sample was 95–25% buffer A at a 2 μl/min flow rate for 120 min. The settings for the mass spectrometer included taking the 5 most intense ions from each full mass spectrum for fragmentation using collision-induced dissociation (CID) and the resulting MS/MS spectra were recorded.
One replicate of gel samples from antenna, Johnston's organ, leg and wing was run on an Orbitrap Elite (Thermo-Fisher) and a Dionex Ultimate 3000 Series LC System (Thermo-Fisher) with a nanospray ionization source. Samples were suspended in 15 μl of buffer A, and 2 μl of each sample were injected into the LC. Peptides were separated using a 75 μm × 150 mm Acclaim PepMap RSLC column packed with 2 μm diameter superficially porous particles (Thermo-Fisher). The gradient used for each sample was 96–35% buffer A over 94 min at a 0.3-μl/min flow rate. The settings for the mass spectrometer included taking the 5 most intense ions from each full mass spectrum for fragmentation using CID, and the resulting MS/MS spectra were recorded.
Peptides prepared from all final pellets (except the preliminary analysis, Batch 0, of the eye lens) were analyzed with an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Scientific). Digested material was acidified with 1% trifluoroacetic acid and desalting was performed using C18 spin columns (Silica C18, The Nest Group, Inc.). Peptides were dried down and resuspended with 39 μl of 0.1% formic acid and 1 μl of 80% acetonitrile/0.1% formic acid (buffer C). The samples were spun through a 0.2 μm filter (Nanosep, Pall Corp) before being loaded into an autosampler tube and racked into an Ultimate 3000 LC System (Thermo Scientific). LC-MS/MS analysis was performed utilizing a nanospray ionization source. Ten microliters of each sample were injected and separated via a gradient of 0–60% buffer C over 110 min with 0.1% formic acid as the diluent. We used a flow rate of approximately 200 nl/min. The settings for the mass spectrometer included collecting full mass spectra every 3 s and continuously fragmenting the most intense ions with CID and recording the resulting MS/MS spectra. Dynamic exclusion was utilized to exclude precursor ions from the selection process.
The final pellet from the first batch of eye lenses (batch 0) was analyzed somewhat differently. After spinning through the 0.2 μm Nanosep filter, the sample was offline bomb loaded on to a Picofrit emitter (New Objective) packed with 5 μm C18 silica and separated via a 160-min gradient of increasing buffer C. LC-MS/MS analysis was performed on an LTQ Orbitrap-XL (Thermo Scientific) utilizing a nanospray ionization source. An instrument method was used to collect a full mass spectrum with 30,000 resolution detected by the Orbitrap. The eight most intense parent ions were selected for MS/ MS fragmentation before taking a new full mass spectrum. Those parent ions were isolated and fragmented with 36% CID in the ion trap. Dynamic exclusion was utilized to exclude parent ions from the selection process for 15 s following a second selection within a 5 s window.
2.4. Database searching
Raw tandem mass spectra were converted using the Trans-Proteomic Pipeline (Seattle Proteome Center). MS/MS spectra of each sample were searched using Mascot (Matrix Scientific, Boston, MA, USA) against target and decoy protein databases. We searched against two databases, An. gambiae peptides (AgamP4.2) from VectorBase and a locally constructed database of 304 putative and previously verified structural cuticular proteins (see Introduction). Decoy databases were created by reversing the protein sequences of corresponding target databases.
We used the small CP database to facilitate our quantitative analysis of protein hits. Jagtap et al. (2013) have documented that the use of a second database constructed from expected proteins increased peptide detection. A total of 614 peptides attributed to CPs were identified. Of these 25 were found only using the P4.2 database and 77 using the CP database. This resulted in a small number of CPs identified with only one database. There were 11 with the CP database and 4 with the P4.2 database plus 14 peptides that belonged to the 2LB and 2LC sequences clusters and 10 peptides belonging to members of CPLCG group A. The peptides found exclusively in only one database are indicated in Supplementary File 2 and the proteins identified based on only one database are listed at the bottom of this file.
The following parameters were utilized in Mascot for all machines: a fragment tolerance of 0.6 Da, a maximum of two missed cleavages by trypsin, a fixed modification of carbamidomethylation of cysteine, and variable modifications of oxidation of methionine and deamidation of asparagine or glutamine. For the LTQ samples we used a peptide tolerance of 1000 ppm and an average mass search. For all Orbitrap samples we used a peptide tolerance of 100 ppm, and a monoisotopic mass search. Resulting Mascot files were analyzed using ProteoIQ (Premiere Biosoft; http://www.premierbiosoft.com/protein_quantification_software/index.html), where a 5% false discovery rate (FDR) was employed for confirmation of protein identifications.
2.5. Data handling and presentation
One of the challenges working with the CPs in An. gambiae and other mosquitoes is the presence of sequence clusters, groups of genes that code for proteins with considerable sequence identity (Cornman et al., 2008; Cornman and Willis, 2008, 2009). Nesvizhskii et al. (2003) address the problem of shared peptides in interpreting MS/MS data, but they were unaware of the magnitude of the problem in mosquito genomes. They suggested some solutions that appear to be similar to what we did manually. Hence we had to establish a method to handle such data so that we could recognize and report on all CP peptides and proteins that we recovered. Sequence clusters were found in the CPR, CPFL, CPLCP, CPLCG and CPLCW families. In 11 cases, we combined from 2 to 4 identical genes into single entries. Thus we ended with possible data for 282 proteins from 295 genes (Table 1 and Supplementary Files 1 and 3). Ultimately, for our own data and for each external study examined, it was necessary to determine whether peptide matches for each cuticular protein were unique (“U”) to that protein or potentially shared (“S”) with other proteins (Table 1, Supplementary Files 1–3). Nesvizhskii et al. (2003) also discuss the appropriateness of accepting as valid, proteins identified by only a single high-probability peptide.
2.6. Quantitative assessment
It is possible that a protein that is abundant in one structure may just be a minor component of another structure. Thus we looked at the quantitative data from Batch 1 and 2 analyses, reported as normalized spectral counts (NSAF) calculated by ProteoIQ. (http://www.premierbiosoft.com/protein_quantification_software/index.html). The calculation includes normalization for the length of the protein (including signal peptides). Hence short proteins may reach high values with fewer peptides than longer proteins. A case in point is AGAP012795 that codes for a protein similar to CPR117. We have not included it as a CP gene because it is a short fragment assigned to the unknown chromosome and appears to be missing its first two exons and the start of the third. It has 86 aa rather than the 162 found in CPR117. AGAP012795 was the top ranked protein in one leg and both wing samples. CPR117 had a normalized spectral count 59% of the shorter protein, similar to the 53% difference in length. We worried that other top hits might be due to a protein being small. In the data for spectral counts based on the entire An. gambiae database, the top hit for several structures was AGAP007549 (yellow-h). It has 507 aa, providing reassurance that size is not an overriding factor.
Other complications with normalized spectral counts were that they included the signal peptide in the length and that many CPs have long stretches with no sites that could be cut by trypsin to produce a peptide that would be recovered. Finally, when many proteins share the same peptide, its presence will be assigned to all of them, even though some may not be present in a particular structure. For our purposes, spectral counts provided useful information about protein abundance.
2.7. Comparisons to published data
We compared our data to that from an extensive analysis of proteins from larvae, pupae and several organs and residual tissues of adult An. gambiae of the G3 strain (Chaerkady et al., 2011). The data from Chaerkady et al. are on VectorBase, but we found it more effective to use the original data in Supplementary File 1 of that paper. All peptides (117,913 in total) were extracted from their Supplementary Materials using the PyPDF module for Python. The following operations were performed with a custom Python script: Extracted peptides at least 5 aa in length were condensed into a non-redundant list (51,676 peptides). All peptides in this list were checked against our full list of cuticular protein sequences, making sure that they could belong to a site cut by trypsin. Of the 573 peptides with matches to CP sequences, 364 were identical to peptides we had found in our analyses. Many of these revealed that proteins we had annotated based on the PEST genomic data were present in the G3 strain even though we had not detected them in our analyses, so strain differences do not appear to account for proteins we failed to detect (Supplementary File 3).
We also performed the same peptide analysis on two studies that have examined An. gambiae antennae with MS/MS (Mastrobuoni et al., 2013; Champion et al., 2015). Peptide data from Mastrobuoni et al. were provided by one of the authors (F. R. Dani) via e-mail, and data from Champion et al. were available on Peptide Atlas (PASS00300). Both studies had included JOin their antennal preparations (Duffield, personal communication; Dani, personal communication). Champion et al. separated data obtained from antennae, eyes, and total head appendages; we restricted our analysis to their eye and antenna data since we also had analyzed these structures. The An. gambiae analyzed by Mastrobuoni et al. were 2 day old adults of the GA-CAM-ST strain, M form and both sexes were analyzed. Those used by Champion et al. were females at least 4 days old of the Pimperena S form adults [MRA-861]. We also incorporated earlier data from our laboratory (He et al., 2007) into our analysis as these enabled us to learn whether proteins detected in adult structures were also present in larval and/or pupal cuticles.
3. Results and discussion
3.1. Overview of cuticular protein analyses
The cuticular proteins of An. gambiae present a challenge to tandem MS/MS analyses because of the sequence similarity among groups of proteins that we have described in detail (Cornman et al., 2008; Cornman and Willis, 2008, 2009). We have termed such sets of highly similar and generally linked genes, “sequence clusters.” Proteins from these clusters frequently shared peptides. Our data (Table 1) thus indicates where we recovered unique (U) and/or shared (S) peptides for each of the 282 distinct protein sequences from the annotated cuticular proteins. We have reported on all peptides from our data except for 38 copies of DGDVVK and 16 copies of KVPVYVEK that were removed as hits from 34 and 14 proteins, respectively. The former was retained in 4 others where it was present as a component of longer, unique peptides and the latter retained in 2 where other peptides were found (Supplementary File 2). We included all other cases where only a single peptide was present as unique or shared in Table 1. The complete data with peptides mapped to sequences are presented in Supplementary File 1 and all peptides recovered are in Supplementary File 2 along with the source (gel or final pellet) and the proteins to which they can be assigned. In many cases, multiple peptides were shared among a group of proteins. Sometimes one or more peptides were present in proteins that also had unique peptides. The shared peptides in such instances are given in brown type in Supplementary File 1. There were three instances when we found peptides that included all or part of the signal peptide. We cannot tell whether these were false positives or evidence of unprocessed protein, but in all three cases, other peptides were detected in those proteins.
We also included a summary of the data from He et al. (2007) that analyzed “cleaned” cuticles left behind after a molt, i.e. cast larval head capsules and cast pupal cuticles. That paper has lists of what were thought at the time to be non-CP proteins in its Supplementary Tables 2 and 5. A few of the peptides listed in these tables can now be assigned to CPs. They have been added to column C in Supplementary File 3 that summarizes findings from that paper.
These three other studies allowed us to rule out proteins that we thought, on the basis of our analysis, were unique for one or more structures, and the information was incorporated into Table 1 and will be discussed in detail in Section 3.2. The data from Champion et al. (2015) and Mastrobuoni et al. (2013) provided confirmation and augmentation of peptides we had assigned to the antennae and Johnston's organ since their antennal preparations included both structures. Champion et al. also had data on isolated eyes that include the eye lens (Supplementary File 3).
3.2. Analysis of adult structures
Each of the five adult structures that we analyzed, Johnston's organ, the rest of the male antenna, eye lens, leg, and wing had large numbers of CPs ranging from 61 to 95 distinct CPs (Table 1; see Table 2 for numerical summary). The 43 CPs found in all five structures consisted of 26 CPRs, 16 from 7 other families, and one in the miscellaneous group CPLCX (Tables 1 and 3). There were almost no proteins detected exclusively in one structure, with the eye lens having the most, with 9 exclusive proteins. Even eliminating the proteins identified solely by shared peptides and counting only those with unique peptides, we detected from 48 to 79 different proteins in a single structure (Table 2). What was especially surprising was the large number of structural CP found in the eye lens, 79 based on unique hits and 11 more identified with only shared peptides. The lenses had both the optical surface and a surrounding rim of sclerotized cuticle, and perhaps it is this combination of nonsclerotized (possibly soft) and sclerotized (hard) cuticle that accounts for the large number. That we had three replicates of the lens preparations may also have contributed to this larger number.
Table 2.
Cuticular proteins in different adult structures. Summary of findings in Table 1.
| Johnston's organ | Antenna | Eye lens | Leg | Wing | Summary | |
|---|---|---|---|---|---|---|
| Total unique/shared (US) | 12 | 15 | 22 | 11 | 12 | 30 |
| Total U | 60 | 55 | 79 | 51 | 48 | 97 |
| Total S | 31 | 55 | 33 | 21 | 27 | 65 |
| Total U only | 48 | 40 | 57 | 40 | 36 | 67 |
| Total S only | 19 | 40 | 11 | 10 | 15 | 35 |
| Total of all hits | 79 | 95 | 90 | 61 | 63 | 132 |
| Exclusive unique hits | 2 | 1 | 11 | 2 | 3 | |
| Exclusive based on shared | 24 | 1 | ||||
| Revised exclusivea | 2 | 0 | 5 | 2 | 2 | |
| Total with no hits out of 282 unique CP proteins | 150 |
Total U only is Total U minus Total US. Total S only is Total S minus Total US.
Other studies (Section 3.8 and Supplementary File 3) provided data revealing that some proteins were not restricted to a single structure.
Table 3.
Classification of CPs detected with peptides.
| Structure | CPAP1 | CPAP3 | CPCFC | CPF | CPFL | CLCA | CPLCG | CPLCP | CPLCW | CPLCX | TWDL | CPTC | CPRC | CPR RR-1 | CPR RR-2 | CPR RR unclassified |
RR-1/RR-2a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Johnston's organ | 1 | 6 | 1 | 2 | 1 | 3 | 3 | 8 | 0 | 4 | 4 | 0 | 1 | 9 | 27 | 9 | 0.78 |
| Antenna | 2 | 4 | 1 | 4 | 1 | 3 | 15 | 9 | 0 | 3 | 2 | 0 | 1 | 5 | 38 | 7 | 0.31 |
| Eye lens | 4 | 6 | 1 | 2 | 1 | 3 | 5 | 8 | 0 | 5 | 3 | 0 | 0 | 14 | 29 | 9 | 1.13 |
| Leg | 1 | 3 | 1 | 2 | 0 | 3 | 4 | 6 | 0 | 2 | 3 | 0 | 1 | 6 | 22 | 7 | 0.64 |
| Wing | 2 | 4 | 1 | 4 | 0 | 1 | 6 | 5 | 0 | 3 | 3 | 0 | 2 | 3 | 22 | 7 | 0.32 |
| present in all structures | 0 | 3 | 1 | 2 | 0 | 1 | 3 | 5 | 0 | 1 | 1 | 0 | 0 | 3 | 17 | 6 | |
| Total recovered based on all structures | 7 | 7 | 1 | 4 | 1 | 3 | 17 | 9 | 0 | 5 | 4 | 0 | 2 | 14 | 47 | 11 | |
| Total unique proteins in genome | 13 | 8 | 1 | 4 | 6 | 3 | 29 | 26 | 7 | 10 | 12 | 4 | 4 | 40 | 94 | 21 | 0.43 |
Values that appear substantially different are bolded.
Numbers of each group were normalized as a fraction of all members of that group.
We found peptides from all but two of the 13 CP families in An. gambiae (Table 3). There were also representatives of 5/10 proteins of a small group of low complexity CPs not assigned to families that we had named CPLCX# (Cornman and Willis, 2009). The two families with no detected peptides (CPLCW and CPTC) had yielded peptides in the study of He et al. (2007) and Chaerkady et al. (2011), but not the analyses of Mastrobuoni et al. (2013) and Champion et al. (2015). The genes are thus present in the G3 strain, hence the absence of the corresponding proteins from the five adult structures we analyzed is probably correct.
3.3. Distribution of CPR subfamilies, RR-1 and RR-2
Given the mounting evidence that RR-2 proteins are found predominantly in hard cuticle and RR-1 in soft, we asked whether the CPR proteins in the heavily sclerotized antennae and legs were classified as RR-2, and those in eye lens and wing as RR-1. So we calculated the ratio of RR-1/RR-2 for the 134 CPR proteins that could be classified into these two groups, adjusting for their relative abundance in the genome (Table 3). This speculation held for the eye lens where there was a disproportionate number of RR-1s, but not for the wing. With both the RR-2s and the CPLCGs in the antennae, the higher numbers were due to proteins identified based on shared peptides (Table 1).
An attempt to compare our data to a proteomic and transcriptomic analysis comparing rigid elytra with membranous hindwings of T. castaneum (Dittmer et al., 2012) was limited by the few CPs (19) identified by the MALDI/TOF analysis of spots on 2D gels. In their transcriptomic data from microarrays, 105 CPRs were identified with 87 having different transcript levels between the two wing types. RR-1s made up 14% of the CPRs in elytra and 54% in the hindwing, opposite in direction from our ratios of 64% and 32% in the hard leg and softer-appearing wing of An. gambiae (Table 3).
3.4. Proteins from sequence clusters
The presence of so many proteins with nearly identical sequences complicates our ability to verify that peptides from an individual protein were actually detected. Thus we have adopted the following principles. If a shared peptide was present in at least one protein for which there was at least one unique peptide then the amino acids of that peptide are in brown type in Supplementary Table 1, and designated in Table 1 with a simple S. Obviously, that peptide may have come solely from proteins that also had unique peptides. When peptides were shared among proteins for which no unique peptides were found, they are presented in other colors and marked with a bolded S, indicating that at least one of those similar proteins must have been present. In two cases where a single peptide (DGDVVK or KVPVYVEK) was present in multiple proteins, only a few of which had unique peptides, we required that at least one other peptide had to be present in order for the protein to be accepted. The proteins for which we did not use a peptide are shown in red in Column D of Supplementary File 2.
Proteins identified only by shared peptides account for the large number in the antennae (40/95 total in our data). Fourteen of these found solely in the antenna came from the 2LB and 2LC sequence clusters. We had previously suggested that the sequence clusters were a solution to having to synthesize and secrete CPs in the short pharate adult period found in mosquitoes (Cornman and Willis, 2008), but why this need would be present in antennae and not in other structures having morphologically similar hard cuticle, such as the legs, remains a mystery. Furthermore, all of the 2LB and 2LC proteins had been identified previously from cast larval head capsules (He et al., 2007). The additional 10 CPs (all CPLCGs) that were found exclusively in antennae based on shared peptides had also been detected in cast larval head capsules or cast pupal cuticle (He et al., 2007; Cornman and Willis, 2009).
The goal of this study, to learn about the distribution of CPs in different structures, was motivated in part by the large number of transcripts seen in a particular tissue following in situ hybridization. Vannini et al. (2014) listed 10 CPs where in situ hybridization had revealed transcripts in the eye. Probes for two, CPR10 and CPR132, had been found exclusively in the eye. In this study, we detected peptides for all 10 genes in the eye, but the only protein found exclusively in the eye was CPR75, whose transcripts had been detected in additional tissues. Furthermore, three CPs also discussed by Vannini et al. for which transcripts had not been found in the eye, had peptides in the eye.
3.5. Quantitative considerations
The conclusion from this analysis is that a single structure is composed of many CPs, but does the presence of a protein in the cuticle reflect that it is essential for the structure? It is possible that protein synthesis and secretion are not tightly regulated and some of our hits might be minor and non-essential components. We asked a similar question years ago when we discovered that mRNAs for two CPRs in the larvae of the Cecropia silkmoth (Hyalophora cecropia) that we thought should be present in either soft (abdominal surface) or hard (tubercle) epidermis, based on their assignment as RR-1 or RR-2, were present in both structures. A quantitative analysis based on dilution of cDNAs (RT-qPCR had not yet been invented), revealed that the unexpected mRNA was from 625- to 3125-fold less abundant (Gu and Willis, 2003). Thus it is likely that some of the low abundance proteins we detected in this study might not be playing an essential structural role or are present because of a lack of tight regulation of transcription/ translation.
Fortunately, we can use spectral counts to assess the relative contribution of individual proteins to a specific structure (See Section 2.6 for discussion of this method.).
Supplementary File 5 shows the top 14 hits for each structure based on normalized spectral counts. For each structure, the normalized spectral counts are given as a ratio to the top hit for that structure. The data revealed that relatively few proteins make up a substantial fraction of those detected in any structure. These 14 top hits accounted for from 74 to 98% of all the CPs detected in each structure. Most of these top hits were proteins found in abundance in all structures. For example, CPR59 and CPR70 were among the most abundant in all but wings where proteins in the 2RB sequence cluster predominated, but were then followed closely by CPR70 and CPR59.
In situ hybridization had revealed Johnston's organ to be the only structure with mRNA for CPR152 (unpublished observations), but the MS/MS analysis also found CPR152 in the rest of the antenna, but only in Johnston's organ was it a major protein. Several other proteins that appeared to be exclusive in a structure based on high rankings (indicated with pink highlighting) were ones with peptides that had been detected in all five structures. Examples are CPR146 and CPCFC1 in Johnston's organ, the RR-1 proteins CPR15 and CPR16 in the eye and CPLCG1 and CPAP3-C in the leg. These data indicate that the construction of an anatomical structure not only involves selection of proteins to incorporate, but also their relative abundance.
3.6. Chitin-binding domains
These data provide important information on the nature of chitin-binding. Identified peptides were frequently found for at least part of the extended R&R Consensus (pfam00379), the characteristic that defines the CPR family, and is necessary and sufficient for chitin-binding (Rebers and Willis, 2001). The Consensus usually begins with an aromatic triad (Y/F-X-Y/F-X-Y/F) and ends about 60 aa later with a G-F/Y. The Consensus is underlined where it occurs in Supplementary File 1. Specifically, of the 72 CPR proteins from which peptides were recovered, we found unique peptides in the R&R Consensus in 41 and shared peptides in an additional 27. Indeed only 4 CPR proteins that had matched peptides had none in the Consensus region.
There are two other major families of chitin-binding proteins, CPAP1 and CPAP3. Their chitin-binding domain/s consist of 6 cysteine residues. (Jasrapuria et al., 2010, 2012). These chitin-binding domains are known as the Chitin-binding Peritrophin-A domain (ChtBD2, CBM_14 or pfam01607). Distribution of their mRNAs and subsequent analyses with RNAi confirmed that spacing within and between these domains allows their classification as cuticular proteins rather than peritrophins where the domain was first identified (Jasrapuria et al., 2010, 2012). A recent paper (Tetreau et al., 2015) identified and named the members of these two families in An. gambiae. The CPAP1 and CPAP3 chitin-binding domains are underlined in Supplementary File 1 and peptides were recovered for both families in these regions. Of the 12 genes in the CPAP1 family, we recovered peptides for 6. Peptides for 4 of these included part of the chitin-binding domain. The situation with the CPAP3 family was more impressive. We recovered peptides in 14 of the 21 chitin-binding domains in the 7 genes in the CPAP3 family; all 14 regions had unique peptides.
Finally, a TWDL protein, Bombyx mori CPT1, has been shown experimentally to bind chitin (Tang et al., 2010). The specific domains have not been analyzed, but all TWDLs contain the domain: DUF243 superfamily (pfam03103). Unique peptides in that domain (underlined in Supplementary File 1) were found for the four TWDLs for which we had found matching peptides.
The presence of recovered peptides from mature adults from these three different chitin-binding domains adds data to the assumption that chitin-binding is not covalent.
3.7. Solubility of proteins
Proteins were extracted in a two-step process resulting in peptides from proteins that were SDS soluble and run out on acrylamide gels, and those that were only found in the final pellet. Thus we were in a position to learn which proteins in these structures were readily soluble. Peptides from only 8 proteins were found exclusively in the gel samples: 1 CPR, 5 CPAP1s and 2 CPLCGs. The majority (107) were found only in the final pellet, while 52 were found in both preparations. These data are in Supplementary File 4B, which also shows whether the CPRs had been classified as RR-1 or RR-2; both groups were in the final pellet and SDS soluble fractions. The enrichment of CPs in the final pellet is similar to the findings of He et al. (2007) that recovered peptides from cast larval head capsules and cast pupal cuticles.
We also examined whether there were differences in the solubility of proteins in the different structures. It appears that Johnston's organ and the antenna were enriched in peptides that were only recovered from proteins in the final pellet, while both leg and wing had relatively more gel peptides, still, almost all of the “gel” peptides from every structure were also recovered from final pellet proteins (Supplementary File 4A).
3.8. Comparison with published data
Data from He et al. (2007), summarized in Supplementary File 3 allowed us to eliminate 26 proteins we had designated as unique for one of the adult structures because they showed that the same proteins could be present in cast larval head capsules or cast pupal cuticle; such proteins were designated with a § in Table 1.
Champion et al. (2015) had analyzed the xentire eye and the entire antenna including Johnston's organ. For their eye, they had matches to 77 CPs, while we had found 95, but interestingly, their matches include 5 unique and 11 shared that we had not found. For the entire antennae, where we had 111 matched proteins, they had only 54, only 3 of which we had not also identified. This smaller number in the entire antennae no doubt reflects that although they had numerous samples, for they were looking for evidence for rhythmic behavior of antennal proteins, each sample had far fewer antennae than we had used.
Mastrobuoni et al. (2013) published a comprehensive shotgun MS/MS analysis of the antennae (including Johnston's organ) of An. gambiae (GA-CAM-ST strain). Proteins were extracted with 8M urea, and they had three biological replicates of 600 individuals of each sex and three technical replicates, for a total of 18 samples. We ran into similar problems with their CP data from shared peptides that we had discussed in Section 3.1. They had the peptide FEYGVK, which matched 28 proteins in our unique protein CP database. In 6 CPs, this peptide was accompanied by another peptide, but there were 22 instances when that peptide was the only one found in a protein and these hits are not included in Supplementary File 3.
Mastrobuoni et al. (2013) identified products from 151 CPs, 89 based on unique peptides. Their data also revealed that 7 of the 17 proteins we had designated as unique for a particular structure other than antennae or Johnston's organ could no longer be considered as unique for they were present in those two structures. These have been marked with a * in Table 1.
These two analyses provide additional, independent, evidence for the large number of proteins that contribute to individual adult structures. Combing all three analyses, the antennae, including Johnston's organ, contain up to 169 distinct CPs, 100 based on the presence of unique peptides (Supplementary File 3). But this large number must be tempered by the quantitative data discussed in Section 3.5 that revealed that most proteins are minor components. Nonetheless, these data reveal that An. gambiae does not build its different structures with unique proteins, rather each structure appears to use a group of cuticular proteins precisely selected from the almost 300 structural CPs available. This is more extensive and direct evidence than we had earlier based on electrophoretic patterns of cuticular proteins of the Cecropia silkmoth (Hyalophora cecropia), that CPs are utilized for their ability to contribute to physical properties of cuticle rather than being specific for a metamorphic stage (Willis, 1986).
These data now allow us to confirm that all but 52 of the 282 annotated CP proteins in An. gambiae are both transcribed and translated. Despite the more extensive analysis of stages and structures carried out by Chaerkady et al. (2011), our analyses plus those of Mastrobuoni et al. (2013) and Champion et al. (2015) found all but 16 of the proteins they had identified (Supplementary File 3, magenta letters in column K).
3.9. Proteins identied in the whole proteome database
While our quantitative analysis in Section 3.5 was based on the CP restricted database, we also ran the MS/MS data against the whole proteome (VectorBase version P4.2). Importantly, all peptides recovered using the CP database were found exclusively in CPs in the P4.2 database and not in any other proteins. The results, Supplementary File 6, show how enriched the preparations we used were for cuticular proteins. For every structure, the vast majority of the top 15 hits, using the criterion of normalized spectral counts, were CPs, indicating the relative contribution of CPs to these adult structures. A comparison of Supplementary File 5 with Supplementary File 6 shows how similar the top hits are irrespective of database used. The few proteins highlighted in gray in Supplementary File 6 show the major non-CPs we detected. These were primarily muscle proteins, with the provocative appearance of alpha crystallins in Johnston's organ, and the strange appearance of “alpha-tocopherol transfer protein-like protein” in antennae and wing. In addition to these, the gene for the yellow protein (AGAP007549) that is an ortholog of yellow-h in Drosophila melanogaster and T. castaneum was the most abundant protein in antennal preparations and close to the top in Johnston's organ (Supplementary File 6, orange highlighting). Its transcript is higher in elytra than hindwing in T. castaneum, but the protein was not detected by proteomic analysis using spots from 2D gels (Arakane et al., 2010). In neither of these species is the function known.
Using this comprehensive database, we recovered peptides for an additional 467 proteins; a quarter of them had signal peptides. It was not surprising to find so many cellular proteins given that we had processed whole tissues. Only 12% were identified in all structures and a quarter of these had signal peptides. A summary is given in Table 4 and details are in Supplementary File 7 that provides possible assignments to function based on several databases. A list of all the non-CP peptides and their source by structure and detection in gel slice or final pellet is in Supplementary File 8. A brief analysis of the data on these non-CPs follows.
Table 4.
Major non-cuticular proteins recovered from different structures.
| Protein family | Total | JO | Antenna | Eye | Leg | Wing |
|---|---|---|---|---|---|---|
| Dehydrogenase | 47 | 18 | 28 | 9 | 13 | 6 |
| Histone or histone related | 34 | 34 | 34 | 25 | 34 | 14 |
| Ribosomal protein | 25 | 6 | 21 | 0 | 2 | 5 |
| ATPase | 25 | 11 | 22 | 8 | 12 | 8 |
| Muscle-related (actin, myosin) | 17 | 8 | 9 | 8 | 13 | 9 |
| Mitochondrial-related | 13 | 10 | 8 | 3 | 5 | 2 |
| Actin not specified as muscle | 10 | 8 | 9 | 6 | 7 | 7 |
| Heat shock | 9 | 0 | 9 | 0 | 0 | 1 |
| Prophenoloxidase or PO inhibitor | 7 | 5 | 1 | 5 | 2 | 6 |
| Protease or protease inhibitor | 7 | 3 | 2 | 3 | 1 | 6 |
| Tubulin | 7 | 6 | 6 | 6 | 7 | 6 |
| Yellow protein | 5 | 4 | 4 | 4 | 2 | 4 |
| Chitin metabolism | 5 | 4 | 2 | 4 | 2 | 5 |
| Takeout | 5 | 4 | 4 | 5 | 2 | 3 |
| Odorant binding protein | 3 | 1 | 1 | 1 | 1 | 1 |
|
Summary | ||||||
| Total number of proteins per structure | 467 | 212 | 306 | 159 | 178 | 166 |
| Number of secreted proteins | 118 | 48 | 63 | 53 | 36 | 63 |
| Number of non-secreted proteins | 348 | 164 | 243 | 106 | 142 | 103 |
| Secreted/Non-secreted ratio | 0.34 | 0.29 | 0.26 | 0.50 | 0.25 | 0.61 |
| Number of unique proteins per structure | 43 | 114 | 20 | 22 | 27 | |
See Supplementary File 7 for complete listing of non-cuticular proteins.
Only 27 of the non-CPs (14 of which were histones) were present in every structure. The histones were the second-largest group of non-CP proteins identified; dehydrogenases were first. All of the histones were present in Johnston's organ, antennae, and legs, with fewer in the eye lens and in wings. This is consistent with the presence of fewer cellular components in wings compared to the other whole structures tested, as well as with our deliberate effort to remove non-cuticular components of the eye lens. This observation holds when comparing total numbers of secreted (i.e. signal peptide-possessing) and non-secreted non-cuticular proteins for each structure: the ratio of secreted to non-secreted proteins is substantially higher than average in the eye lens and leg, while all other structures are below average.
With the exception of a single α-tubulin, six tubulins were found in all structures. Other non-secreted protein groups found in all structures included ATPases and dehydrogenases, though members of these groups tended to have more structure-restricted expression than histones and tubulins. Strangely, 21 ribosomal proteins were found in the antenna, but no more than 6 were found in the other structures (none in the eye lens). Could it be that the antennae from these 5-day old adults were still active in protein synthesis, while the other structures are in a more maintenance mode?
Unsurprisingly, muscle proteins were found abundantly in all structures, with a single myosin heavy chain protein (AGAP010147, isoforms PA-PK) accounting for 9.4% of all identified non-CP peptides (Supplementary File 7). Eight of the nine heat shock proteins were found exclusively in the antenna; the one exception (AGAP002076) was also found in the wing. The brain of the flesh fly, Sarcophaga crassipalpis, exhibits a more sensitive heat shock response than other tissues, probably due to its greater susceptibility to damage by various stresses (Denlinger et al., 2001). Thus, exclusive expression in the antenna may be related to vulnerability of the structure and the importance of its proteins. Similarly to the results of He et al. (2007), yellow proteins were found in all structures, with two of five found in every structure and only one unique to a single structure.
There was a substantial disparity in number of proteins found uniquely in each structure. The higher number found in antennae and especially in Johnston's organ, given its small size relative to the other examined structures, is further evidence for the complexity of these structures. Eleven of the proteins unique to Johnston's organ were involved in electron transfer (e.g. NADH hydrogenase, cytochrome C oxidase). Two (AGAP007100 and AGAP007717) are likely, based on orthologs in D. melanogaster, to be involved in the sensory perception of sound, which is expected due to the organ's involvement in wing-beat frequency transduction.
3.10. Conclusions
These results indicate that individual structures with quite different morphology are constructed overwhelmingly with the same CPs. Only a tiny number of CPs (2–5) was unique for a single structure while all five structures shared 43 CPs. Quantitatively too, with a few exceptions, the most abundant CPs were similar in all five structures. Proteins were detected from 11 of the 13 CP families in An. gambiae, with most coming from the largest CPR family. A common notion that its subgroups, RR-1 and RR-2 would be associated with soft and hard cuticles, respectively, was not supported by these data. Four of the 13 families have verified chitin-binding domains, and we recovered peptides from all of these domains, supporting the notion that chitin-binding does not involve covalent bonds. When the MS/MS data were analyzed against the complete proteome, the major hits were primarily to CPs, indicating that they are the dominant proteins in these mature structures.
Supplementary Material
Acknowledgments
Giles Duffield, Matthew Champion and Francesca Romana Dani generously provided peptide sequences and information about their published analyses. We thank Mark R. Brown and Anne Elliot for maintaining the mosquito facility from which the animals were obtained and two anonymous reviewers for constructive suggestions. We are grateful to Lance Wells who provided access to the Orbitrap Fusion and Robert Bridger for his assistance in running samples with it. This research was funded by a grant from the U.S. National Institutes of Health R01AI055624.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http:// dx.doi.org/10.1016/j.ibmb.2016.05.001.
References
- Arakane Y, Dittmer NT, Tomoyasu Y, Kramer KJ, Muthukrishnan S, Beeman RW, Kanost MR. Identification, mRNA expression and functional analysis of several yellow family genes in Tribolium castaneum. Insect Biochem. Mol. Biol. 2010;40:259–266. doi: 10.1016/j.ibmb.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Lomakin J, Gehrke SH, Hiromasa Y, Tomich JM, Muthukrishnan S, Beeman RW, Kramer KJ, Kanost MR. Formation of rigid, non-flight forewings (elytra) of a beetle requires two major cuticular proteins. PLoS Genet. 2012;8:e1002682. doi: 10.1371/journal.pgen.1002682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaerkady R, Kelkar DS, Muthusamy B, Kandasamy K, Dwivedi SB, Sahasrabuddhe NA, Kim MS, Renuse S, Pinto SM, Sharma R, Pawar H, Sekhar NR, Mohanty AK, Getnet D, Yang Y, Zhong J, Dash AP, MacCallum RM, Delanghe B, Mlambo G, Kumar A, Keshava Prasad TS, Okulate M, Kumar N, Pandey A. A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res. 2011;21:1872–1881. doi: 10.1101/gr.127951.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MM, Sheppard AD, Rund SSC, Freed SA, O'Tousa JE, Duffield GE. Qualitative and quantitative proteomics methods for the analysis of the Anopheles gambiae mosquito proteome. In: Raman C, Goldsmith MR, Agunbiade TA, editors. Short Views on Insect Genomics and Proteomics: Insect Proteomics. Springer; 2015. pp. 37–62. [Google Scholar]
- Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC Genom. 2008;9:22. doi: 10.1186/1471-2164-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH. Extensive gene amplification and concerted evolution within the CPR family of cuticular proteins in mosquitoes. Insect Biochem. Mol. Biol. 2008;38:661–676. doi: 10.1016/j.ibmb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Mol. Biol. 2009;18:607–622. doi: 10.1111/j.1365-2583.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denlinger DL, Rinehart JP, Yocum GD. Stress proteins: a role in insect diapause? In: Denlinger DL, Giebultowicz JM, Saunders DS, editors. Insect Timing: Circadian Rhythmicity to Seasonalilty. Amsterdam. Elsevier; 2001. pp. 155–171. [Google Scholar]
- Dittmer NT, Hiromasa Y, Tomich JM, Lu N, Beeman RW, Kramer KJ, Kanost MR. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J. Proteome Res. 2012;11:269–278. doi: 10.1021/pr2009803. [DOI] [PubMed] [Google Scholar]
- Gu S, Willis JH. Distribution of cuticular protein mRNAs in silk moth integument and imaginal discs. Insect Biochem. Mol. Biol. 2003;33:1177–1188. doi: 10.1016/j.ibmb.2003.06.010. [DOI] [PubMed] [Google Scholar]
- He N, Botelho JM, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem. Mol. Biol. 2007;37:135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ. CutProtFam-Pred: detection and classification of putative structural cuticular proteins from sequence alone, based on profile hidden Markov models. Insect Biochem. Mol. Biol. 2014;52:51–59. doi: 10.1016/j.ibmb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap P, Goslinga J, Kooren JA, McGowan T, Wroblewski MS, Seymour SL, Griffin TJ. A two-step database search method improves sensitivity in peptide sequence matches for metaproteomics and proteogenomics studies. Proteomics. 2013;13:1352–1357. doi: 10.1002/pmic.201200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem. Mol. Biol. 2010;40:214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Jasrapuria S, Specht CA, Kramer KJ, Beeman RW, Muthukrishnan S. Gene families of cuticular proteins analogous to peritrophins (CPAPs) in Tribolium castaneum have diverse functions. PLoS One. 2012;7:e49844. doi: 10.1371/journal.pone.0049844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrobuoni G, Qiao H, Iovinella I, Sagona S, Niccolini A, Boscaro F, Caputo B, Orejuela MR, Della Torre A, Kempa S, Felicioli A, Pelosi P, Moneti G, Dani FR. A proteomic investigation of soluble olfactory proteins in Anopheles gambiae. PLoS One. 2013;8:e75162. doi: 10.1371/journal.pone.0075162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Noh MY, Kramer KJ, Muthukrishnan S, Kanost MR, Beeman RW, Arakane Y. Two major cuticular proteins are required for assembly of horizontal laminae and vertical pore canals in rigid cuticle of Tribolium castaneum. Insect Biochem. Mol. Biol. 2014;53C:22–29. doi: 10.1016/j.ibmb.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Noh MY, Muthukrishnan S, Kramer KJ, Arakane Y. Tribolium castaneum RR-1 cuticular protein TcCPR4 is required for formation of pore canals in rigid cuticle. PLoS Genet. 2015;11:e1004963. doi: 10.1371/journal.pgen.1004963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers JE, Willis JH. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem. Mol. Biol. 2001;31:1083–1093. doi: 10.1016/s0965-1748(01)00056-x. [DOI] [PubMed] [Google Scholar]
- Tang L, Liang J, Zhan Z, Xiang Z, He N. Identification of the chitin-binding proteins from the larval proteins of silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2010;40:228–234. doi: 10.1016/j.ibmb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Tetreau G, Dittmer NT, Cao X, Agrawal S, Chen YR, Muthukrishnan S, Haobo J, Blissard GW, Kanost MR, Wang P. Analysis of chitin-binding proteins from Manduca sexta provides new insights into evolution of peritrophin A-type chitin-binding domains in insects. Insect Biochem. Mol. Biol. 2015;62:127–141. doi: 10.1016/j.ibmb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Dunn WA, Reed TW, Willis JH. Changes in transcript abundance for cuticular proteins and other genes three hours after a blood meal in Anopheles gambiae. Insect Biochem. Mol. Biol. 2014;44:33–43. doi: 10.1016/j.ibmb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Bowen JH, Reed TW, Willis JH. The CPCFC cuticular protein family: anatomical and cuticular locations in Anopheles gambiae and distribution throughout Pancrustacea. Insect Biochem. Mol. Biol. 2015;65:57–67. doi: 10.1016/j.ibmb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH. The paradigm of stage-specific gene sets in insect metamorphosis time for revision. Archives Insect Biochem. Physiol. 1986;(Suppl. 1):47–58. [Google Scholar]
- Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ. Cuticular proteins. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic Press; London, Waltham & San Diego: 2012. pp. 134–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.