Abstract
The number of long noncoding RNAs (lncRNAs) has grown rapidly; however, our understanding of their function remains limited. Although cultured cells have facilitated investigations of lncRNA function at the molecular level, the use of animal models provides a rich context in which to investigate the phenotypic impact of these molecules. Promising initial studies using animal models demonstrated that lncRNAs influence a diverse number of phenotypes, ranging from subtle dysmorphia to viability. Here, we highlight the diversity of animal models and their unique advantages, discuss the use of animal models to profile lncRNA expression, evaluate experimental strategies to manipulate lncRNA function in vivo, and review the phenotypes attributable to lncRNAs. Despite a limited number of studies leveraging animal models, lncRNAs are already recognized as a notable class of molecules with important implications for health and disease.
Introduction
Approximately three-quarters of the mammalian genome is transcribed into RNA (1–3); however, only a fraction of this transcription produces mRNA, whose mature nucleotide sequence serves as a template for protein synthesis (3). The function of this nonprotein-coding RNA (or noncoding RNA) is mostly obscure, despite a larger number of noncoding genes than protein-coding genes. Given the broad functional repertoire derived from protein-coding RNA, it is perhaps not surprising that the relatively few known functions for noncoding RNAs also span diverse cellular processes, resulting in an array of noncoding RNA subclassifications (4). One particular subclass, long noncoding RNAs (lncRNAs), represents a large family of noncoding RNA molecules with potentially broad implications for basic science, health, and disease. In this Review, we focus on the use of animal models to discover novel lncRNAs and to investigate their significance in vivo.
lncRNA definitions and mechanisms
lncRNAs represent a burgeoning class of molecules broadly defined as RNA transcripts longer than 200 nucleotides, with no protein-coding potential. This length is somewhat arbitrary, but it serves to distinguish them from shorter, biologically distinct noncoding RNAs, such as microRNAs. In humans, high-throughput experimental approaches have led to the rapid identification of approximately 16,000 lncRNA genes thus far, rivaling the approximately 20,000 protein-coding genes (5–10) (Figure 1A). These lncRNAs are informatically predicted to lack protein-coding potential, although notable cases of presumed lncRNAs encoding micropeptides (11, 12) or acting as precursors to microRNAs (13, 14) exist. Therefore, current definitions attempt to corral this emerging class of molecules, whose validation and function in vivo remain to be fully elucidated (Figure 1B).
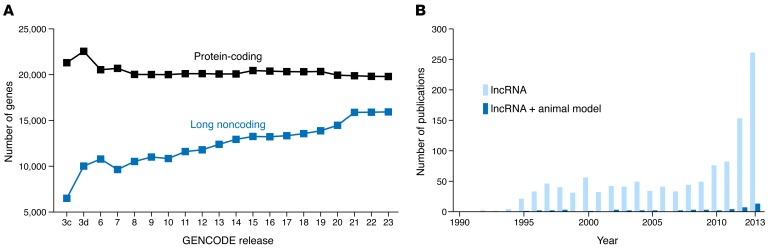
Figure 1. lncRNA biology is a burgeoning field.
(A) The number of genes designated as lncRNAs in humans has steadily increased over successive GENCODE releases (http://www.gencodegenes.org/releases/) to nearly equal the number of protein-coding genes. (B) The number of publications in PubMed returned by querying “lncRNAs” has rapidly increased in recent years. However, few publications have explored lncRNAs using animal models.
lncRNAs are found in both the nucleus and the cytoplasm. The majority of lncRNAs reside in the nucleus (10), where they can act proximally or distally to their site of transcription, functioning in cis or trans, respectively. For example, during X chromosome inactivation in mice, the lncRNA Xist functions in cis to initiate silencing of genes across the same X chromosome from which it was originally transcribed (15). Conversely, the mouse lncRNA Trp53cor1 (also known as lincRNA-p21) acts in trans to globally repress the expression of hundreds of genes distant from its site of transcription (16). Additionally, the transcription of a cis-acting lncRNA, per se, rather than its resulting RNA product, can also have a biological effect (17). As discussed below, this possibility raises important considerations when designing experiments to manipulate lncRNA expression and function.
The molecular mechanisms for most lncRNAs remain largely unknown. They can bind to DNA, RNA, or proteins, and many techniques have been developed to assay these interactions (Figure 2A). Techniques exploring lncRNA-DNA interactions, such as capture hybridization analysis of RNA targets (CHART) (18) and chromatin isolation by RNA purification (ChIRP) (19), utilize complementary oligonucleotides that hybridize to a lncRNA of interest and serve as an affinity handle to enrich for bound DNA. Similar hybridization approaches, such as radioimmunoassay sequencing (RIA-seq), can also be used to assay lncRNA-RNA interactions (20). Other techniques, such as RNA immunoprecipitation (RIP) or its variants, cross-linking immunoprecipitation (CLIP) (21, 22), and photoactivatable ribonucleoside–enhanced cross-linking immunoprecipitation (PAR-CLIP) (23), utilize antibodies to purify lncRNA-protein complexes. Additionally, high-throughput approaches to investigate RNA secondary structure, such as in vivo click selective 2′hydroxyl acylation and profiling experiment (icSHAPE) (24) or selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-seq) (25), have also been developed. Collectively, these techniques demonstrate that lncRNAs interact with diverse macromolecules, potentially impacting a wide range of biological functions.
Figure 2. Techniques to investigate lncRNA properties and tissue expression.
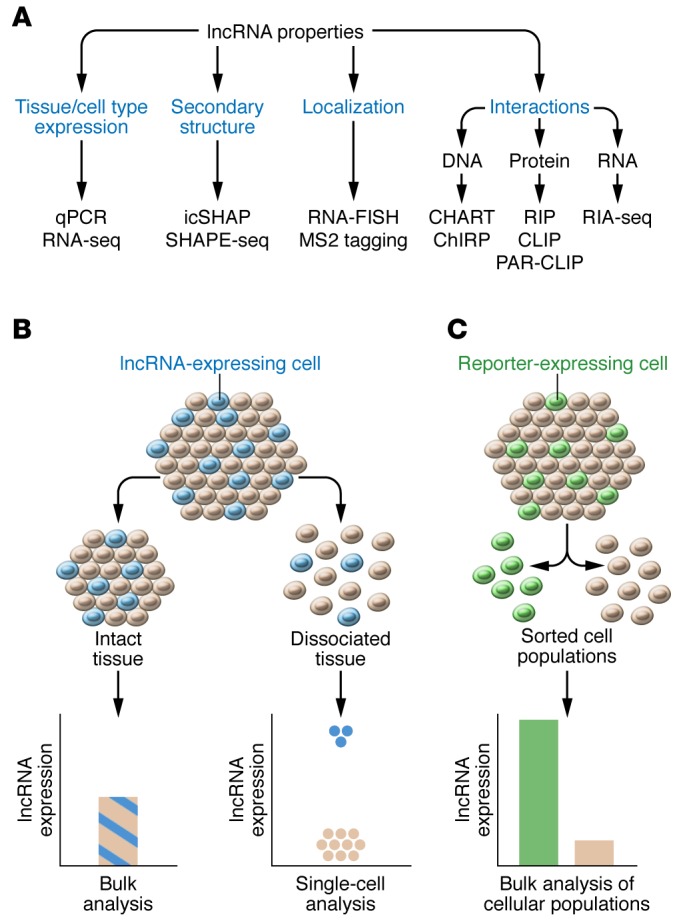
(A) These techniques include quantitative PCR (qPCR) and RNA-seq for transcript expression; RNA fluorescence ISH (FISH) or MS2 tagging (128) for localization; icSHAPE (24) or SHAPE-seq (25) for secondary structures; RIP, CLIP (21, 22), or PAR-CLIP (23) for protein interactions; RIA-seq (20) for RNA interactions; and CHART (18) or ChIRP (19) for DNA interactions. (B) Dissociation of tissue into single cells, followed by single-cell–sequencing analysis, can improve the sensitivity of detection for lncRNA expressed in a minority of cells within a tissue. (C) Fluorescently labeled cells within a tissue can be dissociated, sorted on the basis of fluorescence detection, and assayed for lncRNA expression to improve the sensitivity of detection.
lncRNAs and disease
Accumulating evidence suggests that lncRNA dysfunction promotes disease (26). Notably, approximately 40% of disease- or trait-associated SNPs are found within the noncoding regions flanking protein-coding genes, where a subclass of lncRNAs, termed long intergenic noncoding RNAs, reside (27). Although the functional consequences, if any, for many of these SNPs remain to be experimentally evaluated, they may promote disease by affecting lncRNA function and/or expression (28). For example, an SNP within the human lncRNA myocardial infarction–associated transcript (MIAT), which is associated with an increased risk for myocardial infarction, also increases the binding affinity of MIAT for nuclear proteins (29). This same SNP also increases expression levels of MIAT, although the precise molecular mechanisms remain unknown (29). Additionally, in a human cell line, the binding of lncRNAs to the protein complex Mediator promotes permissive chromatin states necessary for gene expression, and mutations in Mediator that result in intellectual disability have been shown to impair its interaction with lncRNAs (30). An alternative scenario, whereby an alteration of a lncRNA disrupts its interaction with Mediator, could conceivably produce similar, yet less pronounced, effects. These results contribute to a growing body of evidence implicating lncRNAs in disease.
Genetic linkage studies have uncovered new lncRNAs, providing additional evidence to suggest that lncRNA dysfunction promotes disease. For example, HELPP syndrome, which occurs in mothers during pregnancy and is characterized by hemolysis, elevated liver enzymes, and low platelet counts, is associated with a previously unknown placental lncRNA (31). Mutations within this lncRNA increase the in vitro proliferation of human placenta cells in a model of this syndrome (31). Additionally, brachydactyly type E can result from a chromosomal rearrangement that disrupts local interactions between the human lncRNA chondrogenesis-associated transcript (CISTR) and the spatially proximal protein-coding gene parathyroid hormone–like hormone (PTHLH), diminishing the expression of PTHLH mRNA (32). These findings highlight how lncRNA dysfunction impacts cellular processes with clinically relevant consequences.
Notably, lncRNA expression in human tumors is associated with clinical outcomes for a variety of cancers, and xenograft studies in mice have been instrumental when extending these findings in vivo. For example, more than 100 lncRNAs correlate with overall or progression-free survival for ovarian cancer, prostate cancer, glioblastomas, and lung squamous cell carcinomas (33). Nine of these lncRNAs consistently correlate across these cancer subtypes (33). Other examples include the lncRNA HOTAIR, which is overexpressed in primary breast tumors, and elevated expression of HOTAIR in these tumors correlates with an increased probability of metastasis and a decreased overall rate of survival (34). Additionally, xenografts overexpressing HOTAIR in a cell line derived from metastatic breast tissue have an increased propensity for metastasis in mice (34). Furthermore, approximately 1,900 lncRNAs are differentially expressed in T cell acute lymphoblastic leukemia (35). This finding is further supported in the same study by demonstrating in mice that one of these lncRNAs, LUNAR1, is necessary for xenograft tumor growth of a human T cell lymphoma cell line (35). Finally, xenograft studies can also support the in vivo significance of lncRNAs implicated in biological processes such as hypoxia that are known to promote cancer progression (36). For example, the lncRNAs NPTN-IT1 (also known as lncRNA-LET) (37), TP53COR1 (38), and LINC-ROR (39) all regulate hypoxia-induced signaling and affect xenograft growth. In summary, accumulating evidence implicates lncRNAs in cancer development, a finding supported by the use of laboratory animals for xenograft studies.
Diversity of animal models
Animal models vary in biological complexity, which can be leveraged depending on the area of investigation. Commonly used animal models more evolutionarily divergent from humans include the nematode (Caenorhabditis elegans), the fruit fly (Drosophila melanogaster), and the zebrafish (Danio rerio). The reduced complexity of these species can be advantageous for investigating the role of lncRNAs in highly conserved biological processes, whose dysfunction may contribute to human disease. For example, studies of protein-coding genes utilizing C. elegans and D. melanogaster have aided in understanding the molecular mechanisms of apoptosis, whose dysfunction contributes to cancer development and progression (40). Less evolutionarily divergent animal models include the rat (Rattus norvegicus), the mouse (Mus musculus), and the nonhuman primate. The greater biological complexity and physiological similarity of these organisms to humans may better model the complex biology of diseases, such as that underlying tumor growth and metastasis (41, 42). Additional considerations when choosing an appropriate animal model include financial cost, ethical considerations, potential throughput, and ease of experimental manipulation.
Evolutionary conservation of lncRNAs
In addition to humans, numerous lncRNAs are also found in the fruit fly (43–46), the nematode (47), the zebrafish (48, 49), and the mouse (1, 6). Cross-species comparisons have demonstrated that lncRNAs can contain short, conserved regions (9, 50) and also show evidence of purifying selection (51). In general, however, their primary sequence is weakly conserved across species (49, 52–56). For example, only 29 lncRNAs are conserved between zebrafish and humans (49). Strikingly, in the same study, the phenotype in zebrafish following functional inactivation of conserved lncRNAs can be rescued by an orthologous gene from either mice or humans, despite diverging primary sequences (49). These results suggest that the higher-order structure of a lncRNA, rather than its primary sequence, may be conserved. Given the approximately 500 computationally predicted, intergenic RNA secondary structures conserved across vertebrates (57), zebrafish may be a robust model for probing a conserved lncRNA function that is not readily apparent from the primary sequence.
Species-specific lncRNAs may broadly highlight the involvement of lncRNAs in certain conditions in which certain animal models have afforded advantages. For example, both the fruit fly and the nematode are preferentially used to model aging, as their life span is shorter than that of the mouse or the rat (58). Using a genetic model of aging in the nematode, it has been demonstrated that binding of the lncRNA tts-1 to the ribosome suppresses ribosomal levels and promotes longevity, implicating lncRNAs in this phenotype (59).
Other advantages of animal models more evolutionarily distant from humans include the ability to conduct genetic screens (60). For example, expanded nucleotide repeats within the human lncRNA ATXN8OS (also known as SCA8) result in the neurodegenerative disorder spinocerebellar ataxia (61). Using the retina of the fruit fly as a model system, the expression of ATXN8OS, either with or without expanded repeats, results in neurodegeneration (62). However, a genetic screen in this same study demonstrated that this phenotype is differentially modified by RNA-binding proteins (62), in agreement with accumulating evidence implicating aberrant RNA-protein interactions in neurodegenerative disorders (63). Therefore, the use of animal models more evolutionarily distant from humans to study lncRNA function may serve to uncover fundamental roles of lncRNAs and their dysfunction in disease.
Animal models and lncRNA profiling
Animal models generate a variety of tissue and cell types from which to profile lncRNA expression. This feature renders animal models well suited for the study of lncRNAs, given that lncRNA expression is more tissue and cell type specific than is protein-coding gene expression (5, 10, 64). When compared with cultured cells, material derived from animals is generally less abundant and more time-consuming to generate. Nonetheless, animal models provide direct, in vivo evidence of endogenous lncRNA expression and dramatically narrow the relevant search space when investigating phenotypes.
Modern techniques available for genomics research, such as RNA-seq, provide an unbiased, high-throughput approach to investigating endogenous lncRNA expression and to discovering and assembling novel lncRNAs de novo (64). lncRNA expression within grossly dissected tissue can reveal tissue specificity, although lncRNAs expressed in only a minority of cells may be undetectable. This limitation can be circumvented by assessing gene expression within individual cells (Figure 2B). This strategy may uncover rare or novel cells types contributing to diseases such as cancer, in which tumor relapses are thought to arise from a genetically transformed subpopulation of cells (65–67). Additionally, cellular populations labeled with a fluorescent reporter protein can be dissociated into individual cells, which are then sorted on the basis of fluorescence detection and separately analyzed (Figure 2C). This strategy can increase the sensitivity of lncRNA detection and has revealed novel lncRNAs necessary for the development of the cerebral cortex (64, 68). Profiling endogenous lncRNA expression may therefore highlight the diversity of genetically defined cell types and their potential to promote disease.
Additionally, environmental stimuli may affect lncRNA expression, making animal models an attractive resource for certain areas of research, such as neuroscience. Interestingly, lncRNAs are overrepresented in the human brain (10), where they may be fundamental to synaptic transmission (56). Additionally, both neuronal activation (69) and drugs of abuse (70, 71) promote lncRNA expression. The diversity of cell types within the brain, combined with myriad environmental stimuli, has the potential to reveal numerous unrecognized lncRNAs. In sum, animal models are a rich source of material from which lncRNA expression can be profiled across tissues or cell types, providing insight into their potential function in vivo.
Animal models and lncRNA function
The function of a gene is commonly inferred by attenuating or ablating its expression. Strategies to do so include physically interacting with its RNA (RNA-targeted approaches) or altering the underlying genetic locus (DNA-targeted approaches) (Figure 3, A and B). In animal models, these approaches may be applied at the zygotic stage, thereby modeling congenital conditions. They may also be applied to a specific cell population or during a defined time during development. Precise spatial and/or temporal control can refine the conclusions of a study or can circumvent deleterious developmental effects, such as embryonic lethality. Although these approaches have been successfully applied to protein-coding genes, their use requires caution. We discuss the advantages and limitations of these approaches below, highlighting examples from the literature to illustrate the diverse functions attributed to lncRNAs in vivo.
Figure 3. DNA- and RNA-targeted strategies and general workflow.
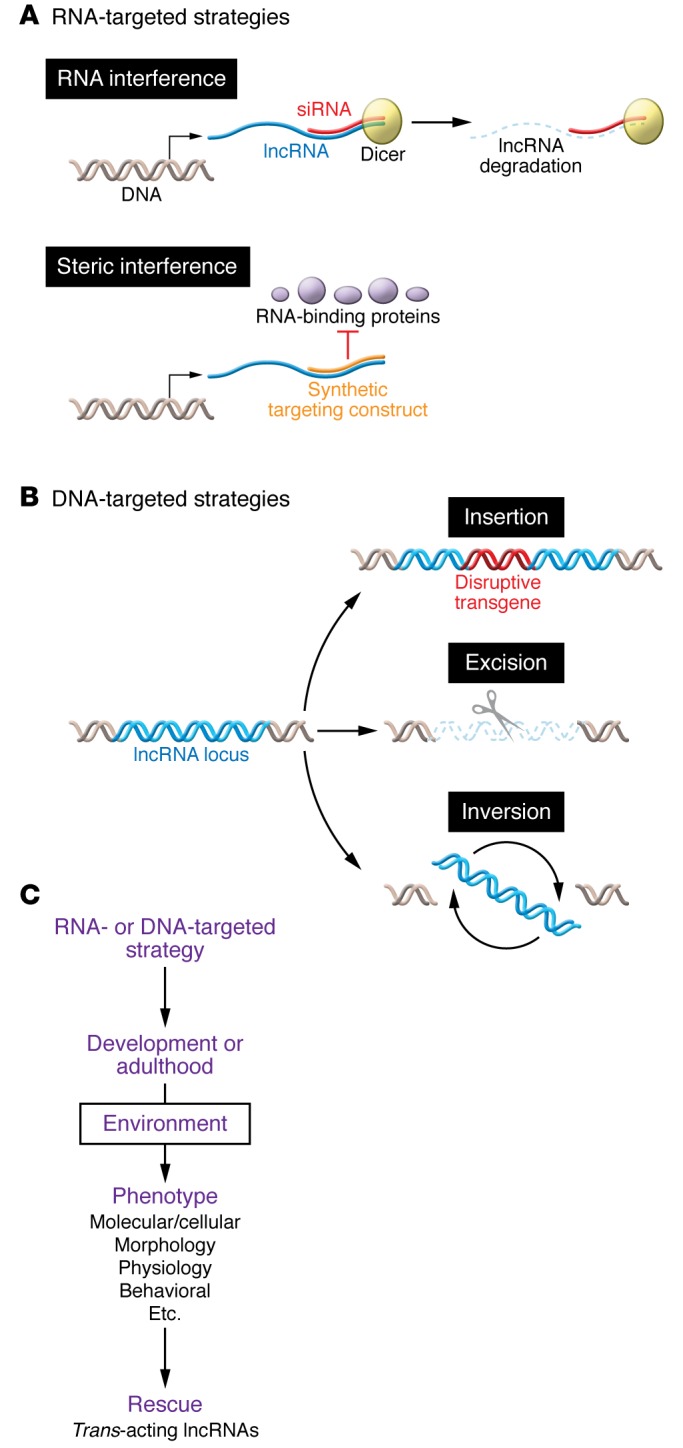
(A) RNA-targeted approaches to attenuate lncRNA expression include RNAi, which degrades the lncRNA complementary to the experimentally introduced RNA. Other RNA-targeted approaches sterically interfere with a lncRNA complementary to the experimentally introduced RNA. (B) DNA-targeted approaches to ablate lncRNA expression include introducing a disruptive transgene within the lncRNA loci, excising the lncRNA loci or its regulatory elements, or inverting the lncRNA loci. (C) DNA- or RNA-targeted approaches to interfere with lncRNA function can be used either during development or adulthood in animal models. Functional ablation of a lncRNA may be sufficient to produce a phenotype or may result from gene-environment interactions. For trans-acting lncRNA, rescue experiments that revert the observed phenotype control for possible nonspecific effects, such as off-target effects of morpholinos or manipulation of DNA regulatory elements.
RNA-targeted approaches and animal models.
RNA-targeted strategies introduce an exogenous RNA or an RNA analog that specifically binds to and functionally inactivates an endogenous lncRNA through complementary base pairing. Commonly used RNA-targeted approaches include RNAi and the use of morpholinos. These two approaches differ mechanistically. RNAi promotes lncRNA degradation, while morpholinos sterically hinder cellular processes, such as splicing, and prevent the formation or function of a mature lncRNA. Steric interference can also disrupt the binding of a lncRNA to other macromolecules, preventing the formation of functional complexes (49).
Zebrafish are an attractive animal model for investigating lncRNAs, because morpholinos can be readily microinjected at the embryonic, one-cell stage to attenuate lncRNA function throughout development and into adulthood (72). In these animals, the lncRNAs terminator, alien, and punisher are necessary for cardiovascular development, complementing similar inferences derived from cultured human and mouse cells (73). Additionally, attenuation of megamind (also known as tuna) in zebrafish impairs locomotion and disrupts brain development, while attenuation of cyrano in zebrafish results in neural tube defects and dysmorphic head and eyes (49). These results highlight the pronounced roles of lncRNAs in zebrafish development.
Mouse models are another valuable resource for understanding lncRNA function in vivo. Transgenic mice engineered to constitutively overexpress an antisense RNA targeted to Sfta3 (also known as Nanci) have abnormal epithelial morphogenesis in their developing lungs (74). A more localized, tissue-specific interference of lncRNAs using RNAi can also have phenotypic consequences. shRNA-mediated downregulation of Miat (also known as Gomafu) in the medial prefrontal cortex of the brain promotes anxiety-like behaviors (75), while local siRNA-mediated downregulation of Munc, which is specifically expressed in skeletal muscle, impairs myogenesis (76). Additionally, the use of an shRNA to revert the upregulation of Arid2-IR in the mouse kidney following renal inflammation also reverts the biochemical signatures of this condition (77). Future work using mutant- and virus-mediated strategies will likely reveal additional phenotypes directly attributable to lncRNAs.
Limitations of RNA-targeted approaches.
While informative, these RNA-targeted approaches have important caveats. RNAi is effective for RNA exported to the cytoplasm but is relatively inefficient for RNAs residing in the nucleus (78), where many lncRNAs are found (10). This inefficiency may be particularly apparent for cis-acting lncRNAs, which act near their site of origin and therefore may necessitate rapid binding for inactivation. This limitation may be circumvented by using RNA analogs, such as locked nucleic acids, which are better suited for nuclear targets because of their faster kinetics (79). However, sterically interfering RNA analogs, including both locked nucleic acids and morpholinos, cannot be genetically encoded, precluding the generation of transgenic animals. Furthermore, both strategies need sufficient expression levels to be effective, potentially inducing off-target effects and general toxicity within the cell. Conversely, insufficient levels for either strategy may only attenuate, rather than eliminate, lncRNA function and may not be sufficient to produce a phenotypic effect (80). Finally, binding of an exogenous RNA after transcription may not affect lncRNA function, because its transcription, per se, may have a biological effect. Despite these limitations, successfully applied RNA-targeted approaches have revealed significant phenotypes following lncRNA dysfunction.
Clinical application of RNA-targeted approaches.
Findings from animals models serve as a translational proof of concept for therapeutic interventions targeting lncRNAs, though significant hurdles exist (81). In mice, systemic administration of an antisense oligonucleotide can downregulate its complementary RNA in muscle tissue (82). For example, systemic administration of an antisense oligonucleotide complementary to the lncRNA Malat1 (also known as Neat2) can attenuate its expression by 80% (82). Although this approach was principally used to attenuate a pathological protein-coding RNA in a mouse model of myotonic dystrophy type 1, the attenuation of Malat1 suggests that lncRNAs are potential targets for this condition and possibly other muscle disorders. However, only RNAs retained in the nucleus are sensitive to this approach (82), a feature well suited for targeting the abundance of nuclear lncRNAs (10). This work not only highlights the value of animal models in advancing novel therapeutic approaches but also the feasibility of correcting pathological gene expression, including that of lncRNAs.
DNA-targeted approaches and animal models.
Strategies to manipulate the genetic locus encoding a lncRNA, many of which have recently been reviewed (83), represent an alternative to RNA-targeted approaches. Transcription of both cis- and trans-acting lncRNAs can be eliminated, in contrast to the dose-dependent, posttranscriptional effects of RNA-targeted approaches. Furthermore, if this ablation occurs in the germline, it is possible to generate transgenic animals whose offspring constitutively lack a given lncRNA. These aspects of DNA-targeted approaches confer several advantages compared with RNA-targeted approaches.
DNA-targeted approaches can facilitate expression profiling of a lncRNA if its locus is replaced with a reporter gene. This strategy was used in mice for 18 different lncRNA gene loci (84). For example, the lncRNA Peril was found to be expressed in discrete regions of the mouse brain and spinal cord, while Mdgt expression in mice appears restricted to the testes, brain, thymus, and colon (84). This methodology may also detect lncRNAs that are found only in a minority of cells. For example, the same study revealed that mouse Pantr2 (also known as linc-Brn1b) is expressed in select brain regions throughout cortical development and is selectively expressed in upper layers of the cortex in adulthood (84). This reporter strategy, therefore, provides sufficient resolution to assess lncRNA expression, not only between tissues, but also within a tissue.
A spectrum of phenotypes has been reported following the ablation of a lncRNA by DNA-targeted approaches. For example, mice lacking Peril or Mdgt die perinatally with varying degrees of penetrance, demonstrating an essential, life-supporting role for both genes (84). Surviving Peril- and Mdgt-deficient mice are developmentally stunted, having smaller body sizes and reduced body weight compared with WT animals (84). A similar developmental phenotype was observed in mice lacking the lncRNA Pint (84). lncRNA ablation can also result in more subtle phenotypes. For example, deletion of Sra1 in mice improves obesity-related measures when animals are fed a high-fat diet (85), and deletion of Hotair in mice results in skeletal abnormalities of the vertebrae and wrist (86). Thus, DNA-targeted approaches have demonstrated that lncRNAs may affect not only dramatic phenotypes but also more nuanced ones.
DNA-targeted approaches and genomic imprinting.
Animal models of lncRNA function using DNA-targeted approaches have been instrumental in investigating the phenotypic effects of genomic imprinting, whereby gene expression is derived from a parent-specific allele, while transcription from the other allele is epigenetically repressed. Both viability and body weight are influenced by imprinted lncRNA expression. For example, deletion of maternal Meg-3 (also known as Gtl2) in mice results in perinatal death, an effect not observed following paternal deletion (87). Similarly, ablation of maternal H19 in mice results in greater offspring body mass compared with that of offspring with paternal deletion (88, 89). Additionally, female mice lacking a single Tsix allele, a lncRNA implicated in X chromosome inactivation, produce fewer surviving offspring than do males lacking the same gene (90, 91). Finally, deletion of the X-linked Tsx in male mice results in smaller testes, reduced fear-related behaviors, and enhanced short-term memory (92). Collectively, these studies demonstrate diverse in vivo functions for imprinted lncRNAs.
Animal models of lncRNA function have also been instrumental when investigating the complementary molecular mechanisms underlying genomic imprinting (93). For example, deletion of paternal Kcnq1ot1 (also known as KvDMR1) in mice results in offspring with reduced body mass and de-repression of proximal genes, an effect not observed after maternal deletion (94). These results were further refined following a more targeted ablation strategy in mice, in which a premature termination sequence was inserted downstream of the Kcnq1ot1 promoter (95). This alteration also resulted in gene de-repression, demonstrating that Kcnq1ot1 transcription is necessary for gene silencing (95). Finally, complementary molecular studies have demonstrated that mouse Kcnq1ot1 interacts with chromatin and epigenetic modifiers with tissue specificity (96). This complementary use of animal models to investigate lncRNA function highlights the utility of animal models when exploring lncRNA function in vivo.
Limitations of DNA-targeted approaches.
DNA-targeted approaches also have important limitations. Oftentimes, a large region encompassing the majority, if not all, of a lncRNA gene is removed, although smaller domain-specific (97) and promoter-specific (90, 94, 95) deletions may be possible. Insertion of a premature termination sequence in the gene body may also ablate lncRNA function (98). These strategies differ from those used for protein-coding genes, whereby a single nucleotide deletion or insertion is often sufficient to abolish a protein product. These larger genomic alterations may introduce unintended and confounding consequences by removing regulatory elements within the deleted region to affect the expression of neighboring genes (83, 99). For example, during embryonic stem cell differentiation in mice, overexpression of the lncRNA Haunt diminishes the expression of the neighboring HOXA gene (100). Conversely, enhancer elements within the Haunt locus facilitate HOXA expression (100). These opposing influences within a locus may complicate efforts to alter a lncRNA locus without also perturbing regulatory elements embedded within that locus. Because of the extensive number of regulatory elements within both the human (101) and mouse (102) genomes, these concerns may be particularly acute and represent the norm rather than the exception. Rescue strategies in which the disrupted gene is experimentally reintroduced may control for these potentially confounding effects. However, this experimental design can only apply to trans-acting lncRNAs for which the integration site of the transgene is independent of its function. These limitations are important caveats to consider when interpreting data generated by DNA-targeted approaches.
Another important consideration is that different DNA-targeted approaches may result in different phenotypes. In mice, replacing the lncRNA gene Fendrr with a reporter gene results in lung defects and perinatal death (84), a phenotype in agreement with clinical studies demonstrating that deletions within this locus in humans results in abnormal lung development and neonatal death (103). In contrast, a second study demonstrated that insertion of a premature transcriptional termination sequence within the same mouse locus results in prenatal death, body wall abnormalities, and heart malfunction (80). These contrasting phenotypes exemplify how different DNA-targeting strategies may produce inconsistent phenotypes and highlight the fact that caution should be exercised in making premature functional conclusions based on a single approach. Notably, this second study (80) also reported an attempted RNA-targeted strategy, in which a constitutively expressed antisense oligonucleotide resulted in a 60% reduction of Fendrr expression levels in mice that lacked any abnormal phenotype. Thus, three different strategies to manipulate lncRNA expression produced three different phenotypes, highlighting the challenges encountered when designing and interpreting lncRNA studies.
Interpretation of an absent phenotype.
Loss of a lncRNA can result in no discernible phenotype. Despite being highly conserved throughout mammalian evolution, ablation of the brain-expressed Linc0046 (also known as Visc-2) in mice does not result in any overt anatomical or behavioral phenotype (104). Similarly, three independent mouse strains lacking Malat1, which is highly expressed in the brain and liver, appear to develop normally (105–107). Finally, the loss of Neat1 in mice does not result in any overt phenotype, except for the loss of mammalian-specific nuclear subregions termed paraspeckles, where Neat1 is usually localized (108). Because paraspeckles are thought to reflect higher-order compartmentalization within the nucleus that is necessary for intricate regulation of mammalian gene expression, the absence of any obvious phenotype following the loss of Neat1 prompted a critical reassessment of their function (109). Paraspeckles are induced following exposure to infectious diseases or cellular stressors, and it is possible that a phenotype in mice lacking Neat1 may only be apparent following an environmental manipulation, such as viral infection or exposure to microbes, that would normally induce paraspeckles (108).
This possibility extends to all studies using genetic ablation in animal models. Gene-environment interactions may unmask unrecognized phenotypes that are important in understanding complex diseases such as psychiatric disorders with known environmental risk factors (110, 111). The mechanism of risk conferral for many of these environmental factors, such as exposure to environmental pathogens, maternal stress during pregnancy, or maternal substance abuse during pregnancy, may be investigated by using animal models in a controlled laboratory setting to reveal latent phenotypes and resolve gene-environment interactions (Figure 3C) (112–116).
Moreover, highly exploratory research involving lncRNAs of unknown function and/or expression profile may constitute a “fishing expedition,” where only a fraction of the potential phenotypes are explored. A possible phenotype could span a large spectrum consisting of biochemical, anatomical, physiological, or behavioral effects, and thus the lack of a reported phenotype may simply represent a limited selection of unaffected phenotypes. Compensatory effects may further mitigate the appearance of a phenotype.
Alternative approaches.
Overexpression of a lncRNA is an alternative strategy to assess its phenotypic effects. This approach has been successfully applied to animal models using protein-coding genes to study neuropsychiatric disorders that may result from a failure to maintain homeostasis after a gain or loss of gene expression (117). When applied to lncRNAs, overexpression experiments may produce a phenotype opposite that occurring after lncRNA ablation, indicating bidirectional effects of lncRNA expression levels. Alternatively, they may induce a phenotype that is absent or seemingly unrelated to those observed following lncRNA ablation. However, this approach is only applicable for trans-acting lncRNAs, whose function is independent of their site of genomic integration.
Genome-editing technology.
Major technological advances in genome editing (118) open the possibility of altering the genomes not only of conventional model organisms, but also of less widely used animal models that have historically been less amenable to alterations. TALENs or clustered regularly interspaced palindromic repeat (CRISPR/Cas9) methods have been widely used to excise or invert regions of a lncRNA locus in zebrafish (119, 120), mice (121, 122), and rats (123). These technologies may be extended to other animal models, adopted for specialized areas of investigation, such as the pig used to model cystic fibrosis (124), the songbird used to model language acquisition (125), and the nonhuman primate used to model the neurodegenerative disorder Huntington’s disease (126, 127). However, caveats and concerns similar to those previously discussed for standard DNA-targeted approaches still apply, given the comparatively large segments of the genome that may need to be altered in order to affect lncRNA expression and function.
Conclusions
Animal models are promising tools for aiding in the discovery of novel lncRNAs and investigating the phenotypic significance of these molecules. However, lncRNAs inherently differ from their protein-coding counterparts, and hence their investigation requires overcoming a host of important new challenges and addressing new considerations with respect to experimental design and data interpretation. Despite these caveats, it has been possible to convincingly demonstrate that lncRNA dysfunction in animal models results in diverse phenotypes, ranging from lethality to subtle dysmorphia. So far, only a fraction of lncRNAs have been assessed, and this substantial gap in knowledge highlights a pressing need to progress beyond the initial cataloging of lncRNAs and to understand their impact in vivo. We anticipate that these initial findings represent a promising beginning to the diverse functions of lncRNAs in vivo and in understanding their relevance to disease.
Acknowledgments
We thank Andrew Holmes, Caroline Siebald, and members of the Goff laboratory for their helpful comments during the preparation of this manuscript.
Footnotes
Conflict of interest: L.A. Goff is a co-inventor on two patents, neither of which is directly related to the manuscript: (a) “High-throughput Methodology for Identifying RNA-Protein Interactions Transcriptome-wide,” B.D. Gregory, J.L. Rinn, F. Li, L.A. Goff, and C. Trapnell (U.S. Patent no. 9,097,708 B2); and (b) “Rational Probe Optimization for Microarray Detection of MicroRNAs,” R. Getts, R.P. Hart, and L.A. Goff (U.S. Patent Application no. PCT/US2005/038261).
Reference information:J Clin Invest. 2016;126(8):2783–2791. doi:10.1172/JCI84422.
References
- 1.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bono H, Kasukawa T, Furuno M, Hayashizaki Y, Okazaki Y. FANTOM DB: database of Functional Annotation of RIKEN Mouse cDNA Clones. Nucleic Acids Res. 2002;30(1):116–118. doi: 10.1093/nar/30.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson BR, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351(6270):271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28(5):491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froberg JE, Yang L, Lee JT. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 2013;425(19):3698–3706. doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338(6113):1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 18.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108(51):20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48(3):287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 22.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37(4):376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519(7544):486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci USA. 2011;108(27):11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015;43(Database issue):D181–D186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii N, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 30.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dijk M, et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J Clin Invest. 2012;122(11):4003–4011. doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maass PG, et al. A misplaced lncRNA causes brachydactyly in humans. J Clin Invest. 2012;122(11):3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trimarchi T, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158(3):593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 37.Yang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127(Pt 7):1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer. 2005;5(5):355–366. doi: 10.1038/nrc1611. [DOI] [PubMed] [Google Scholar]
- 41.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 42.Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26(3):513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- 43.Tupy JL, et al. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102(15):5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young RS, et al. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol. 2012;4(4):427–442. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JB, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512(7515):393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam J-W, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res. 2012;22(12):2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11(7):1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24(4):616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutter C, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8(7): doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babak T, Blencowe BJ, Hughes TR. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics. 2005;6: doi: 10.1186/1471-2164-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, et al. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431(7010) doi:10.1038/nature03016. [PubMed] [Google Scholar]
- 56.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 57.Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol. 2005;23(11):1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 58.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Essers PB, et al. A long noncoding rna on the ribosome is required for lifespan extension. Cell Rep. 2015;10(3):339–345. doi: 10.1016/j.celrep.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 60.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 61.Koob MD, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat Genet. 1999;21(4):379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 62.Mutsuddi M, Marshall CM, Benzow KA, Koob MD, Rebay I. The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr Biol. 2004;14(4):302–308. doi: 10.1016/j.cub.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 63.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154(4):727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molyneaux BJ, et al. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85(2):275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 66.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magrangeas F, et al. Minor clone provides a reservoir for relapse in multiple myeloma. Leukemia. 2013;27(2):473–481. doi: 10.1038/leu.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aprea J, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32(24):3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barry G, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19(4):486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 70.Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31(10):2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116(3):459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26(2):216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 73.Kurian L, et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131(14):1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herriges MJ, et al. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28(12):1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spadaro PA, et al. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol Psychiatry. 2015;78(12):848–859. doi: 10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mueller AC, et al. MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol Cell Biol. 2015;35(3):498–513. doi: 10.1128/MCB.01079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Q, Huang XR, Yu J, Yu X, Lan HY. Long noncoding RNA Arid2-IR is a novel therapeutic target for renal inflammation. Mol Ther. 2015;23(6):1034–1043. doi: 10.1038/mt.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8(7):855–860. doi: 10.1017/S1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107(51):22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 82.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24(10):594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2: doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu S, et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Biol Chem. 2014;289(19):13000–13009. doi: 10.1074/jbc.M114.564658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li L, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137(16):2643–2652. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 89.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 90.Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103(1):17–27. doi: 10.1016/S0092-8674(00)00101-X. [DOI] [PubMed] [Google Scholar]
- 91.Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128(8):1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 92.Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7(9): doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanduri C. Long noncoding RNAs: lessons from genomic imprinting. Biochim Biophys Acta. 2016;1859(1):102–111. doi: 10.1016/j.bbagrm.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 94.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32(3):426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 95.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20(10):1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 97.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137(15):2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 98.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 99.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3: doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin Y, et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015;16(5):504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 101.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Szafranski P, et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23(1):23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oliver PL, et al. Disruption of Visc-2, a brain-expressed conserved long noncoding RNA, does not elicit an overt anatomical or behavioral phenotype. Cereb Cortex. 2015;25(10):3572–3585. doi: 10.1093/cercor/bhu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eißmann M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakagawa S, Hirose T. Paraspeckle nuclear bodies — useful uselessness? Cell Mol Life Sci. 2012;69(18):3027–3036. doi: 10.1007/s00018-012-0973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rutter M. Environmentally mediated risks for psychopathology: research strategies and findings. J Am Acad Child Adolesc Psychiatry. 2005;44(1):3–18. doi: 10.1097/01.chi.0000145374.45992.c9. [DOI] [PubMed] [Google Scholar]
- 111.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7(7):583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 112.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13(10):1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cryan JF, Holmes A. The ascent of mouse: advances in modelling human depression and anxiety. Nat Rev Drug Discov. 2005;4(9):775–790. doi: 10.1038/nrd1825. [DOI] [PubMed] [Google Scholar]
- 114.Holmes A, Wellman CL. Stress-induced prefrontal reorganization and executive dysfunction in rodents. Neurosci Biobehav Rev. 2009;33(6):773–783. doi: 10.1016/j.neubiorev.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Adamec R, Holmes A, Blundell J. Vulnerability to lasting anxiogenic effects of brief exposure to predator stimuli: sex, serotonin and other factors-relevance to PTSD. Neurosci Biobehav Rev. 2008;32(7):1287–1292. doi: 10.1016/j.neubiorev.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Millstein RA, Holmes A. Effects of repeated maternal separation on anxiety- and depression-related phenotypes in different mouse strains. Neurosci Biobehav Rev. 2007;31(1):3–17. doi: 10.1016/j.neubiorev.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 117.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455(7215):912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu Y, Luo D, Zhao H, Zhu Z, Hu W, Cheng CH. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS ONE. 2013;8(10): doi: 10.1371/journal.pone.0076387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiao A, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41(14): doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Han J, et al. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014;11(7):829–835. doi: 10.4161/rna.29624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Welsh IC, et al. Chromatin architecture of the Pitx2 locus requires CTCF- and Pitx2-dependent asymmetry that mirrors embryonic gut laterality. Cell Rep. 2015;13(2):337–349. doi: 10.1016/j.celrep.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cheng X, et al. A CRISPR/Cas9 system based targeted disruption of a novel rat long non-coding RNA in a rat genetic model of hypertension. FASEB J. 2015;29(1 Supplement). http://www.fasebj.org/content/29/1_Supplement/814.1.
- 124.Rogers CS, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321(5897):1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Agate RJ, Scott BB, Haripal B, Lois C, Nottebohm F. Transgenic songbirds offer an opportunity to develop a genetic model for vocal learning. Proc Natl Acad Sci USA. 2009;106(42):17963–17967. doi: 10.1073/pnas.0909139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang SH, et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453(7197):921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sasaki E, et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459(7246):523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 128.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/S1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]