Abstract
Inflammatory myofibroblastic tumors (IMTs) are characterized by myofibroblast proliferation and an inflammatory cell infiltrate. Little is known about the molecular pathways that precipitate IMT formation. Here, we report the identification of somatic mutations in UPF1, a gene that encodes an essential component of the nonsense-mediated RNA decay (NMD) pathway, in 13 of 15 pulmonary IMT samples. The majority of mutations occurred in a specific region of UPF1 and triggered UPF1 alternative splicing. Several mRNA targets of the NMD pathway were upregulated in IMT samples, indicating that the UPF1 mutations led to reduced NMD magnitude. These upregulated NMD targets included NIK mRNA, which encodes a potent activator of NF-κB. In human lung cells, UPF1 depletion increased expression of chemokine-encoding genes in a NIK-dependent manner. Elevated chemokines and IgE class switching events were observed in IMT samples, consistent with NIK upregulation in these tumors. Together, these results support a model in which UPF1 mutations downregulate NMD, leading to NIK-dependent NF-κB induction, which contributes to the immune infiltration that is characteristic of IMTs. The molecular link between the NMD pathway and IMTs has implications for the diagnosis and treatment of these tumors.
Introduction
Inflammatory myofibroblastic tumors (IMTs) encompass a wide spectrum of irregular myofibroblastic proliferative growths accompanied by infiltrating immune cells that occur in almost all major organs of the human body (1). While IMTs are rare, constituting only 0.04% of general thoracic surgeries, they often occur in children and can grow to a large size and are often locally invasive (2, 3).
At present, the causes of IMTs are unknown. In this communication, we provide evidence that disruption of an RNA degradation pathway has a causal role in the majority of cases of IMT. This pathway — nonsense-mediated RNA decay (NMD) — degrades subsets of mRNAs and is responsible for regulating developmental events and other biological processes (4). We recently showed that the gene encoding an ATP-dependent RNA helicase essential for NMD, Up-frameshift 1 (UPF1; also called RENT1), is commonly mutated in pancreatic adenosquamous carcinomas (5), suggesting that mutations in UPF1 have a causal role in these malignant tumors. In the present study, we report that 86% of IMTs that we examined have UPF1 mutations. We provide evidence that NMD is strongly perturbed in these tumors, leading to aberrant activation of the NF-κB pathway and immune responses characteristic of these tumors. These findings afford opportunities to better diagnose and treat these tumors.
Results and Discussion
Somatic UPF1 mutations in IMTs elicit exon skipping.
Based on our previous identification of frequent UPF1 mutations in pancreatic tumors (5), we screened IMT lung samples to determine whether they also had UPF1 mutations. Sequence analysis of UPF1 revealed mutations in IMTs from 13 of 15 patients (Supplemental Table 1 and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI86508DS1). In total, 41 mutations were detected. All but one of these 41 mutations were clustered at 11 sites in exon 10 and intron 10 (Supplemental Figure 1B and Supplemental Table 1). Mutations at 8 of these 11 sites were found in more than one tumor (Supplemental Figure 1B). The mutations in the 13 IMTs were not detected in disease-free lung tissues and thus were somatic in origin (data not shown).
UPF1 exon 10 and intron 10 are unusually short (169 and 85 nt, respectively) and have a high GC content (59% and 73%, respectively), both of which are features that can weaken RNA splicing (6). This raised the possibility that intron 10 splicing depends on local splicing enhancers (7) and that these elements are disrupted by the mutations we identified in the UPF1 gene, leading to aberrant UPF1 splicing. To test this possibility, we constructed a UPF1 minigene containing the relevant region of UPF1 (Figure 1A). When transfected into HEK293 cells, this minigene was normally spliced, as shown by direct sequencing of the only band observed by RT-PCR analysis (Figure 1B). We then introduced the mutations found in IMTs (Figure 1A and Supplemental Figure 2A). This caused reduced expression of the normally spliced mRNA and the appearance of an alternatively spliced transcript lacking exons 10 and 11 (Figure 1B and Supplemental Figure 2B).
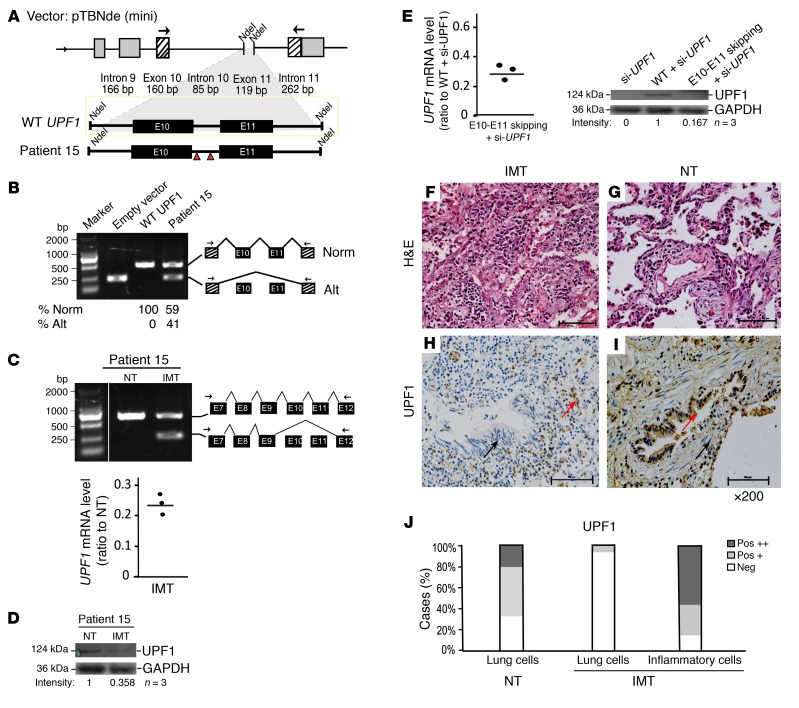
Figure 1. IMT-specific UPF1 mutations disrupt splicing of UPF1 mRNA and reduce UPF1 protein levels.
(A) Patient 15 mutations were inserted into the wild-type human UPF1 minigene as previously described (5). (B) RT-PCR analysis of HEK293 cells transfected with the constructs shown in A (primer locations are indicated by the arrows) (n = 5). Direct sequencing of the large (518 bp) and small (239 bp) bands indicated that they correspond to normally spliced (Norm) and exon-skipped transcripts (Alt), respectively. The numbers below the gel are the average values from 5 independent transfections. (C) RT-PCR (upper) and RT-qPCR (lower) analysis of endogenous UPF1 expression in IMT lung tissue and normal lung tissue (NT) from patient 15 (direct sequencing results correspond to the indicated schematics [n = 5]). (D) Western analysis of patient 15 (n = 3). Because of the large size difference, UPF1 and the loading control, GAPDH, were assayed on different percentage polyacrylamide gels (but with the same amount of sample loaded); the lanes shown were run on the same gel but were noncontiguous. (E) RT-qPCR (left) and Western (right) analysis of HEK cells transfected with the indicated UPF1 expression vectors (n = 3). GAPDH is the loading control (n = 3). Western analysis performed in parallel gels was done as in D. (F and G) H&E staining of patient 15 tissue. (H and I) IHC staining of patient 15 tissue (×200). Red arrows indicate strong staining; black arrows indicate weak or negative staining (n = 6). (J) Quantification of H and I. Pos +, positive staining of greater than 20% of cells; Pos ++, positive staining of greater than 50% of cells.
To examine endogenous UPF1 splicing, we analyzed the IMT tumor from patient 15, which was frozen, providing sufficiently high quality RNA for analysis. RT-PCR analysis revealed an alternatively spliced UPF1 transcript identical to that generated by the mutant minigene in IMT tissue (Figure 1C). This alternatively spliced UPF1 transcript was not present in normal lung tissue (NT) from the same patient. Consistent with the reduced level of UPF1 mRNA in IMT (Figure 1C), the UPF1 protein level was also reduced (Figure 1D). To investigate the mechanism, we generated a UPF1 cDNA construct lacking exons 10 and 11. Transfection of this mutant construct into HEK cells depleted of endogenous UPF1 revealed that it was expressed at a significantly lower steady-state RNA level than was the full-length UPF1 construct (Figure 1E), an effect that was mirrored at the protein level (Figure 1E). The simplest explanation for these findings is that the loss of exons 10 and 11 destabilizes UPF1 RNA.
As an independent means to analyze UPF1 protein expression levels, we examined IMT tissue sections, which unlike NT, had not only alveolar epithelial cells but also inflammatory cells (Figure 1, F and G, and Supplemental Figure 3), as is characteristic of IMTs (1, 3). Immunohistochemical (IHC) analysis with a highly specific UPF1 antibody showed that normal alveolar epithelial cells expressed high levels of UPF1, while most patient IMT alveolar epithelial cells exhibited little or no UPF1 expression (Figure 1, H–J, Supplemental Figure 3, and Supplemental Table 2). The only exceptions were the epithelial cells in the two IMTs lacking UPF1 mutations and the IMT with a missense UPF1 mutation (in exon 13), all of which expressed high levels of UPF1 (Supplemental Figure 3). In contrast with the alveolar epithelial cells, the inflammatory cells in IMTs had high UPF1 antibody reactivity regardless of UPF1 mutation status (Supplemental Figure 3 and Supplemental Table 2). These findings suggest that UPF1 mutations occur specifically in the alveolar epithelial cells in IMTs, leading to reduced UPF1 protein levels in these cells.
We next examined the functional consequences of the single UPF1 mutation we identified outside of the exon 10-11 region (c.1796A>T p.K599M from patient 13; Supplemental Table 1). The strong UPF1 IHC staining in this patient sample (Supplemental Table 2) suggested that this mutation does not destabilize UPF1 RNA, and raised the alternative possibility that it disrupts UPF1 activity. In support, we found that exogenous UPF1 harboring the missense mutation corresponding to patient 13 failed to rescue NMD activity in UPF1-depleted cells (as it did not significantly decrease the level of an NMD-sensitive NS39 β-globin mRNA), indicating that this mutant lacked detectable NMD function. Furthermore, this mutant conferred dominant-negative activity, as it reduced the activity of exogenous WT UPF1 (Supplemental Figure 4). Together with our analysis of the other UPF1 mutants described above, this suggested that two types of mutations are employed to reduce UPF1 activity in IMTs: those that generate a dominant-negative UPF1 or an unstable form of UPF1 mRNA.
IMTs have disrupted NMD and elevated expression of the proinflammatory molecule NIK.
The dramatically reduced level of UPF1 expression in most IMTs raised the possibility that NMD is depressed in IMTs, leading to upregulation of NMD target transcripts. In agreement with this hypothesis, we found that 11 endogenous NMD substrates were upregulated in the patient 15 IMT sample relative to NT from the same patient (Supplemental Table 3). We next considered that perturbed NMD would allow mRNAs encoding proinflammatory proteins to be stabilized, leading to the immune infiltration characteristic of IMTs. We identified mitogen-activated protein kinase kinase kinase 14 (MAP3K14 or NIK) as a good candidate, as it is an activator of the proinflammatory NF-κB signaling pathway (8). In support of its potential role, we found that NIK mRNA was upregulated in the patient 15 IMT relative to NT (Figure 2A) and in response to depletion of UPF1 in the normal lung epithelial cell line BEAS-2B (Figure 2B, upper). Upregulation of NIK in BEAS-2B cells was confirmed at the protein level (Figure 2B, lower) and in IMT tissue harboring UPF1 mutations, as detected by IHC analysis (Figure 2, C–E, and Supplemental Figure 5). In contrast, neither of the two IMT tumors lacking detectable UPF1 mutations had upregulated NIK protein expression (Supplemental Table 2). Upregulated NIK expression was specific to lung tumor cells, as the local infiltrating leukocytes had low/no NIK expression in all patients (Supplemental Figure 5 and Supplemental Table 2).
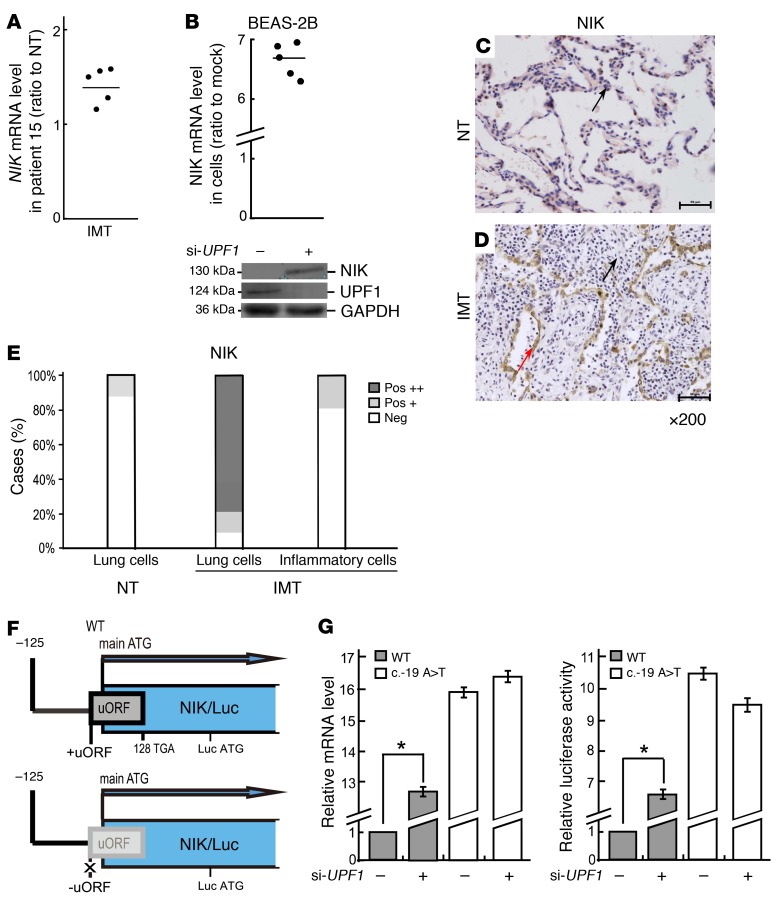
Figure 2. NIK expression is upregulated in IMTs harboring UPF1 mutations.
(A) RT-qPCR analysis of IMT lung tissue and normal lung tissue (NT) from patient 15 (n = 5). (B) RT-qPCR (top [n = 5]) and Western (bottom [n = 3]) analysis of BEAS-2B cells depleted of UPF1. GAPDH is the loading control. Western analysis performed in parallel gels was done as in Figure 1D. (C and D) IHC staining in patient 15 tissue (×200). Red arrow, strong staining; black arrows, weak or negative staining (n = 6). (E) Quantification of C and D. Pos +, positive staining in greater than 20% of cells; Pos ++, positive staining in greater than 50% of cells. (F) NIK 5′ UTR cloned upstream of a luciferase (Luc) reporter gene. The 147-nt uORF with or without a mutated start ATG is shown as a gray box. (G) Relative Luc mRNA levels (left [n = 5]) and Luc enzymatic activity (right [n = 5]) from transfected BEAS-2B cells. For G, the values are the average fold change (mean ± SEM) from 5 independent experiments (Student’s t test, *P < 0.05). 18S mRNA was used as the endogenous control.
NIK mRNA was previously suggested to be an NMD substrate (9, 10), and so we assessed the mechanism by which NIK mRNA is degraded by NMD. Stop codons terminating short ORFs upstream of the protein-coding ORF can elicit NMD (4). Thus, we considered the possibility that the upstream ORF (uORF) present in the 5′ UTR of NIK mRNA is responsible for it being degraded by NMD. In support of this, insertion of the NIK 5′ UTR in front of a luciferase (Luc) reporter (Figure 2F) led to increased reporter expression in UPF1-depleted cells, as measured at the RNA level or by Luc activity (Figure 2G). Mutation of the uORF start ATG by replacing the “A” with a “T” (–19A>T) (Figure 2F) completely abolished this reporter upregulation (Figure 2G). We conclude that NIK mRNA is an NMD substrate by virtue of its possessing a uORF.
Stabilized NIK elevated chemokine gene expression and IgE production.
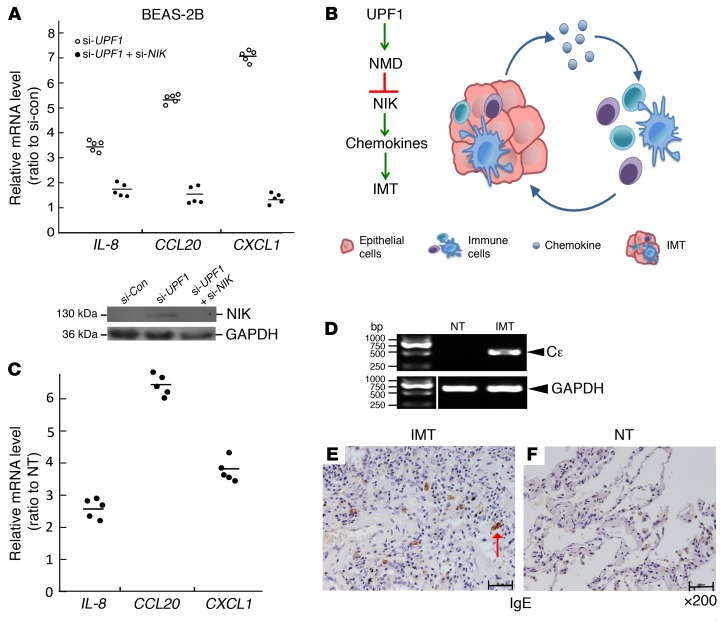
NIK is a serine/theonine protein kinase that stimulates NF-κB activity (8). Given that a hallmark of NF-κB activation is the generation of chemokines (11), this raised the possibility that the upregulation of NIK in IMTs has a causal role in these inflammatory tumors. In support of this possibility, we found that UPF1 depletion dramatically increased IL-8, CCL20, and CXCL1 mRNA expression in the BEAS-2B lung cell line (Figure 3A). This upregulated chemokine expression resulted, at least in part, from upregulated NIK expression, as it was largely abolished when we prevented NIK upregulation using RNAi (Figure 3A). Together, these data support a model in which NMD normally serves to suppress the chemokine response by destabilizing NIK mRNA (Figure 3B). When NMD is perturbed, this suppression is lifted, leading to high NIK expression and the generation of high levels of chemokines, thereby triggering immune cell infiltration, the hallmark of IMTs (1). As evidence that this model applies in vivo, RT-qPCR analysis revealed upregulated expression of the chemokine genes IL-8, CCL20, and CXCL1 in the patient 15 IMT relative to NT (Figure 3C).
Figure 3. Evidence for a UPF1-based circuit that elevates chemokine and IgE expression.
(A) RT-qPCR (top [n = 5]) and Western (bottom [n = 3]) analysis of BEAS-2B cells depleted of UPF1 and/or NIK. 18S mRNA and GAPDH are the endogenous controls. (B) Model depicting how NMD deficiency causes IMT formation. (C) RT-qPCR analysis of IMT lung tissue and normal lung tissue (NT) from patient 15 (n = 5). (D) RT-PCR analysis of patient 15 tissue (n = 5). Direct sequencing of the band indicated that it is a Cε germline transcript (exons 1 and 3; 487 bp). GAPDH is the loading control. The lanes were run on the same gel but were noncontiguous. (E and F) IHC analysis of patient 15 (×200). The red arrow, IgE-positive cell (n = 3).
Another property of NIK is its ability to promote B cell proliferation and induce B cells to differentiate into IgE-secreting plasma cells (12, 13). The commitment of a B cell to isotype class switch to an IgE-producing cell requires induction of Cε germline transcription (14, 15). We thus assessed whether IMTs have upregulated germline Cε transcripts. Using an RT-PCR assay, we observed a 0.5-kb band indicative of Cε transcription, which we confirmed was a germline Cε transcript by direct sequencing (Figure 3D). Consistent with activation of Cε transcription and rearrangement, IHC analysis showed that patient 15 had many IgE-positive cells (Figure 3E), whereas normal lung tissue from the same patient did not (Figure 3F). The IgE-positive cells in the IMT tissue had the morphology of plasma or mast cells.
Our data support a model in which NMD normally serves to confer tight regulation on NIK abundance in order to place cells below the threshold required for NF-κB pathway induction and inappropriate immune activation. By analogy, NMD has been shown to raise the threshold for triggering the unfolded protein response, thereby preventing programmed cell death in response to innocuous stimuli (16, 17). Interestingly, the vast majority of the mutations that we observed in IMTs were clustered in one of the same two regions of the UPF1 gene as in adenosquamous pancreatic cancer (the exon 10-intron 10-exon 11 region; Supplemental Figure 1B). Indeed, 7 of the pulmonary IMTs had UPF1 mutations at precisely the same site as in some ASC tumors (IVS10+32C>T and IVS10-25C>T). We speculate that the aberrations in NMD caused by genetic alterations in the UPF1 gene lead to different disease outcomes, depending on the cell type. In premalignant glandular pancreatic cells, this may lead to reprogramming towards a squamous, more malignant state (5). In lung epithelial cells, NMD perturbation may drive a proinflammatory response, as suggested by our results herein.
Our finding that UPF1 mutations are a common molecular alteration associated with IMTs suggest that UPF1 mutations and NMD activity may be useful diagnostic markers for this disease. New diagnostic markers are critical, as IMTs are often misdiagnosed as malignant tumors (1, 2). Further, our results described herein suggests that therapies directed at restoring the NMD pathway and NIK expression have the potential to ameliorate or cure the symptoms of IMT and related diseases, including IgG4-related disease (IgG4-RD), an autoimmune disorder that resembles IMTs (1, 3). New therapies are desired, as current methods to treat IMTs and IgG4-RD, including surgery and antiinflammatory agents, are invasive or have undesirable side effects (2, 3).
Methods
Subjects.
We evaluated a set of IMTs from 15 patients, provided by the First Affiliated Hospital of Bengbu Medical College. Among them, 14 specimens were formalin-fixed paraffin-embedded (FFPE) tissue sections, and one IMT of patient 15 was frozen immediately after collection. All patients provided written informed consent.
Statistics.
The program ESE Finder was used to locate several predicted ESEs and ISEs in the regions of the UPF1 gene. Pairwise comparisons were performed by 2-tailed Student’s t test using Excel software (Microsoft). Data are expressed as the mean ± SEM.
Study approval.
All aspects of the study were approved by the Bengbu Medical College and Tongji University School of Medicine.
For DNA/RNA sequencing and analysis, constructs, cell culture and transfections, and protein analysis, see Supplemental Methods.
Author contributions
CL, YW, GY, and TP performed UPF1 screening. JL and AH constructed wild-type and mutant mini-gene expression vectors. JL, FS, SL, and YW performed IHC, Western, and RNA analyses. FS, CS, YJ, and CY provided pathology samples, clinical data, and expertise on IMTs. JL and TP prepared figures and tables and wrote the manuscript. YL and MFW designed and supervised the project and assisted in writing the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the YangFan plan of Tongji University School of Medicine (for Y.J. Lu) and NIH grant GM111838 (for M.F. Wilkinson). T.D. Plank was supported by NIH grants T32 HD007203 and F32 GM113487.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(8):3058–3062. doi:10.1172/JCI86508.
Contributor Information
JingWei Lu, Email: 502506122@qq.com.
Fang Su, Email: sufang2899@163.com.
XiuJuan Shi, Email: xiujuansh@163.com.
Chen Liu, Email: 382943525@qq.com.
Yuan Ji, Email: newera_ji@yahoo.com.
ShuaiJun Li, Email: 1564920380@qq.com.
Andrew Huynh, Email: ahuynh9999@gmail.com.
Chao Shi, Email: wuweishichao@126.com.
Bo Zhu, Email: 35736791@qq.com.
Guang Yang, Email: naiveyangboy@163.com.
YanMing Wu, Email: yanmingsh@163.com.
YanJun Lu, Email: yanjunlu@hotmail.com.
References
- 1.Gleason BC, Hornick JL. Inflammatory myofibroblastic tumors: where are we now? J Clin Pathol. 2008;61(4):428–437. doi: 10.1136/jcp.2007.049387. [DOI] [PubMed] [Google Scholar]
- 2.Cerfolio RJ, Allen MS, Nascimento AG, Deschamps C. Inflammatory pseudotumors of the lung. Ann Thorac Surg. 1999;67(4):933–936. doi: 10.1016/S0003-4975(99)00155-1. [DOI] [PubMed] [Google Scholar]
- 3.Zen Y, et al. IgG4-positive plasma cells in inflammatory pseudotumor (plasma cell granuloma) of the lung. Hum Pathol. 2005;36(7):710–717. doi: 10.1016/j.humpath.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med. 2014;20(6):596–598. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amit M, et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012;1(5):543–556. doi: 10.1016/j.celrep.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Ke S, Chasin LA. Context-dependent splicing regulation: exon definition, co-occurring motif pairs and tissue specificity. RNA Biol. 2011;8(3):384–388. doi: 10.4161/rna.8.3.14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noort AR, et al. NF-κB-inducing kinase is a key regulator of inflammation-induced and tumour-associated angiogenesis. J Pathol. 2014;234(3):375–385. doi: 10.1002/path.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 10.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23(10):1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JM, Cho SJ, Oh YK, Jung HY, Kim YJ, Kim N. Nuclear factor-κB activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin Exp Immunol. 2002;130(1):59–66. doi: 10.1046/j.1365-2249.2002.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korenaga M, Akimaru Y, Shamsuzzaman SM, Hashiguchi Y. Impaired protective immunity and T helper 2 responses in alymphoplasia (aly) mutant mice infected with Trichinella spiralis. Immunology. 2001;102(2):218–224. doi: 10.1046/j.1365-2567.2001.01169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brady K, Fitzgerald S, Moynagh PN. Tumour-necrosis-factor-receptor-associated factor 6, NF-κB-inducing kinase and IkappaB kinases mediate IgE isotype switching in response to CD40. Biochem J. 2000;350(pt 3):735–740. [PMC free article] [PubMed] [Google Scholar]
- 14.Geha RS, Jabara HH, Brodeur SR. The regulation of immunoglobulin E class-switch recombination. Nat Rev Immunol. 2003;3(9):721–732. doi: 10.1038/nri1181. [DOI] [PubMed] [Google Scholar]
- 15.Delphin S, Stavnezer J. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline epsilon promoter: regulation by NF-IL-4, a C/EBP family member and NF-κB/p50. J Exp Med. 1995;181(1):181–192. doi: 10.1084/jem.181.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 2015;16(5):599–609. doi: 10.15252/embr.201439696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28(11):3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.