Abstract
The traditional view of genome organization has been upended in the last decade with the discovery of vast amounts of non–protein-coding transcription. After initial concerns that this “dark matter” of the genome was transcriptional noise, it is apparent that a subset of these noncoding RNAs are functional. Long noncoding RNA (lncRNA) genes resemble protein-coding genes in several key aspects, and they have myriad molecular functions across many cellular pathways and processes, including oncogenic signaling. The number of lncRNA genes has recently been greatly expanded by our group to triple the number of protein-coding genes; therefore, lncRNAs are likely to play a role in many biological processes. Based on their large number and expression specificity in a variety of cancers, lncRNAs are likely to serve as the basis for many clinical applications in oncology.
Introduction
Completion of sequencing of the human genome led to a surprising downward revision of the number of “genes” to approximately 25,000 (1), as prior estimates based on expressed sequence tag (EST) data ranged from 45,000 to 140,000 (2). However, disappointment was soon replaced by hopeful intrigue when the comprehensive expression analysis of the ENCyclopedia of DNA Elements (ENCODE) project found that 60%–70% of the bases in the human genome could be found in transcripts, while only approximately 1.5% of the genome codes for a protein. This seeming discrepancy posed a large looming question of the nature of this noncoding “dark matter” of the genome (3).
Initial concern that this noncoding RNA was simply leaky transcription noise, as had been shown in yeast (4, 5), gave way relatively quickly, as numerous noncoding RNAs were shown to have specific functions. While a number of classes of small noncoding RNA were already known, including microRNA and small nucleolar RNA (snoRNA), a new class of longer RNAs emerged that seemed to hold particular functional promise. Long noncoding RNAs (lncRNAs) were defined by length greater than 200 nt and similarities to protein-coding genes, including transcription mediated by RNA polymerase II, a 5′ cap, multiple exons, poly-adenylation, histone 3, lysine 4 tri-methylation (H3K4me3) around the transcription start site, and histone 3, lysine 36 tri-methylation (H3K36me3) throughout the transcribed gene body (6). Unbiased genome-wide searches were quickly able to identify thousands of lncRNAs (7–9), and more directed experiments are identifying lncRNAs involved in specific contexts, such as the effects of cigarette smoking (10).
In this Review, we will highlight the role of lncRNAs in cancer. As the number of lncRNAs has exploded, so too has the number of lncRNAs involved in cancer biology grown such that an exhaustive review is impossible. Therefore, we discuss lncRNA cancer biology themes as well as the translation of lncRNAs from bench research to clinical use as biomarkers and therapeutic targets. For several reasons that we discuss throughout, lncRNAs — the dark matter — now appear as a very promising class of genes for exploitation in the battle against cancer.
MiTranscriptome.
Early lncRNA search efforts, described above, were in the genomics era, but they also quickly fell victim to the extraordinary rapidity of technology development and their own success. The noncoding insights from ENCODE and the studies mentioned above were performed with microarray technology (3, 7–9). It quickly became apparent from these studies that transcription was much more complex and much less discrete than traditionally thought and therefore not assayed very accurately by microarray. Next-generation sequencing (NGS) technology, on the other hand, allows for global unbiased single-transcript interrogation. Based on this, our group undertook a pilot study to identify novel prostate cancer lncRNAs using early NGS technology to profile around 102 prostate tissues and cell lines, including 20 benign adjacent prostate, 47 localized prostate tumors, and 14 metastatic tumors. We identified 121 noncoding transcripts that were dysregulated in prostate cancer (termed prostate-associated cancer transcripts [PCATs]) and were not found in any gene annotation databases. We have gone on to show that several of the PCATs play important and disparate roles in prostate cancer biology. Importantly, one of the identified transcripts, which was renamed second chromosome locus–associated prostate-1 (SChLAP1), is a powerful prognostic biomarker of metastatic progression risk after prostatectomy (11–15).
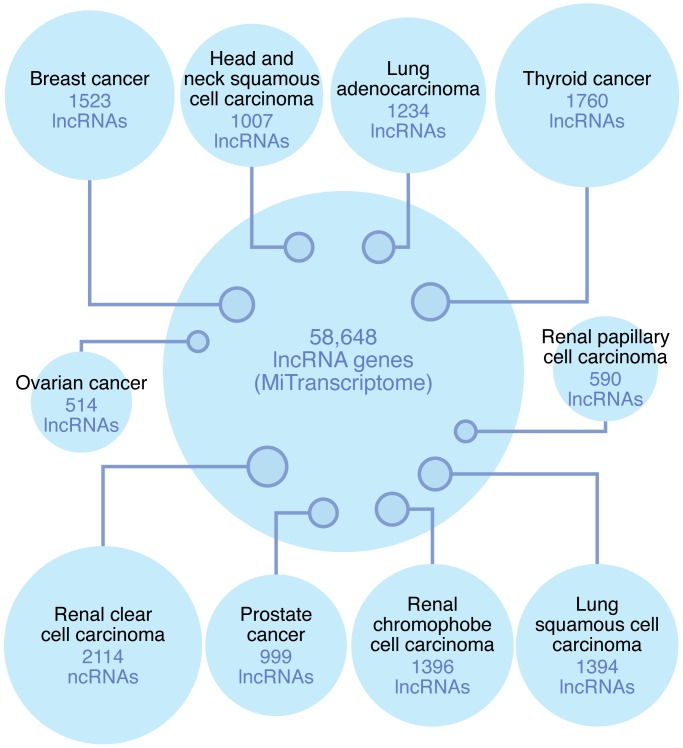
Based on the success of this pilot study, we undertook a much larger effort that took advantage of the vast RNA sequencing (RNA-seq) resource built by The Cancer Genome Atlas (TCGA, cancergenome.nih.gov) (16). We developed a robust bioinformatics pipeline to predict novel transcripts and genes with specific emphasis on lncRNAs that are often expressed at low levels. Rather than analyzing the sequencing data from each sample individually, which has limited sensitivity for lowly expressed transcripts, we used the sequencing information from all samples within a given cancer type coupled with a custom sequencing noise reduction algorithm. This pipeline was applied to over 7,000 sequencing samples across 18 organ systems from 25 independent studies, though 80% of these studies were from TCGA (16). This transcript discovery pipeline identified nearly three times as many lncRNA genes as protein-coding genes, and approximately 80% of these lncRNA genes did not exist in any annotated database (Figure 1). Interestingly, we found that 5.6% of lncRNAs contain conserved sequence and 597 novel intergenic lncRNAs harbor an ultra-conserved element (UCE) consisting of more than 200 nt with nearly perfect conservation across multiple organisms. Additionally, very few lncRNAs (<10%) with previously defined UCE-containing transcripts are altered to this level in cancer (17, 18). These findings suggest that conserved lncRNAs have an important role in higher organisms and are a class of lncRNAs with high potential for future study. Beyond general identification, we developed an algorithm to identify lncRNAs with cancer-type/lineage-specific expression or with cancer-specific expression in those organ systems with a sufficient number of normal samples for comparison. There were nearly 8,000 lncRNAs that were lineage and/or cancer specific, representing a vast tumor-specific resource for cancer biomarker and therapeutic target research (16). Much of the data on these 8,000 lncRNAs is available for public use at mitranscriptome.org, which will be maintained and updated in support of cancer lncRNA research efforts.
Figure 1. Scale of lncRNA genes.
The identification of lncRNA genes has progressed rapidly since their recognition. Early efforts identified several thousand lncRNA genes at a time, and there were initial indications that lncRNAs exhibited greater expression restriction than protein-coding genes. Through our large-scale MiTranscriptome bioinformatics effort, we greatly expanded the number of lncRNAs to nearly 60,000, while the number of protein-coding genes remained approximately 21,000. Additionally, our pipeline demonstrated that nearly 8,000 lncRNA genes were highly cancer and/or lineage specific.
lncRNAs in cancer biology
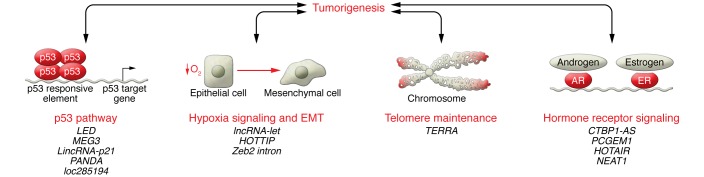
A precedent for involvement of lncRNAs in cancer biology had been set by the pregenomics era lncRNA H19 (19), and convincing evidence for the modern lncRNAs came relatively quickly with elucidation of the role of the HOX locus HOTAIR lncRNA in breast cancer. HOTAIR is overexpressed in breast tumor cells and causes genomic redistribution of the master epigenetic regulatory complex polycomb repressive complex 2 (PRC2). Consequent changes in the repressive epigenetic mark histone 3, lysine 27 methylation (H3K27me) mediate increased tumor cell metastatic potential (20). This example is illustrative of an early dominant theme in lncRNA function. Numerous lncRNAs have been demonstrated to interact with and modulate epigenetic regulatory complexes, most notably PRC2 (e.g., Xist [ref. 21], H19 [ref. 22], and the INK4/ARF tumor suppressor locus lncRNA ANRIL [refs. 22, 23]), but also PRC1 (24) as well as neuronal gene repressor H3K17 methylase CoREST (25), H3K4me3 demethylase SMCX (8), and histone methyltransferase MLL1 (26, 27). While these studies are quite intriguing, there remains some skepticism about the specificity of these interactions based on unbiased studies that detected thousands of lncRNAs (up to 24% of all lncRNAs known at the time) interacting with PRC2 (8, 28), CoREST, or SMCX (8). Beyond epigenetic mark regulators, our group showed that SChLAP1 interacts with the nucleosome positioning complex SWI/SNF through its component member SNF5 (also known as SMARCB1). This interaction results in genomic redistribution of SWI/SNF and elicits a global gene expression program that enhances metastatic potential in prostate cancer (14). Epigenetic regulation has been a dominant primary mechanism in lncRNA biology that affects many cellular processes, including oncogenic signaling. Through myriad molecular mechanisms, lncRNAs have been implicated in many classic cancer biology pathways. This list has grown so rapidly that it is impossible to describe exhaustively, and we highlight several illustrative examples in Figure 2.
Figure 2. Selected examples of lncRNAs in cancer biology.
The sheer number of lncRNA genes strongly suggests that lncRNAs are involved in every cellular and disease process, including tumorigenesis and cancer biology. Numerous lncRNA roles in cancer biology have already been described, so that it is impossible to list them exhaustively. Here, we have provided several select examples of classic cancer biology processes and listed several lncRNAs that have been implicated in each.
p53 pathway.
Several roles for lncRNAs have been found in the p53 pathway; interestingly, one of these novel RNA functions is found in p53 itself. TP53 mRNA can enhance p53 activation by directly binding to and inhibiting the E3 ubiquitin ligase MDM2, and this feed-forward loop for p53 activation can be mitigated by TP53 point mutations in the MDM2-binding region that have been found in patient tumor samples (29). Canonical p53 function also involves several lncRNA-mediated mechanisms. A recent global study identified 16 lncRNAs as p53 target genes that form a signaling network (30). Consistent with this, the lncRNA LED is a p53 target gene that activates strong enhancers including in cyclin-dependent kinase 1A (CDKN1A), thus providing a mechanism for p53-activated enhancers that do not have a p53-binding site (31). The lncRNA MEG3 selectively enhances transcriptional activation by p53 and downregulates MDM2, resulting in cell-cycle arrest and apoptosis in vitro as well as modulation of autophagy (32, 33). Consistent with this antineoplastic effect, MEG3 is downregulated in multiple cancer types (34–37). Several lncRNAs have been identified as p53 target genes. The lncRNA loc285194 is a p53 target gene that inhibits proliferation through binding of the proproliferation miR-211 (38). The CDKN1A tumor-suppressor locus lncRNA p21-associated ncRNA DNA damage activated (PANDA) dampens the apoptotic response following p53 activation by interacting with the transcription factor NF-YA to decrease induction of proapoptotic genes (39).
Linc-p21 is also a p53 target gene and provides a mechanism for the longstanding mystery of transcriptional repression in the p53 response. Linc-p21 directly interacts with and is required for genomic localization of heterogeneous nuclear ribonucleoprotein K (hnRNP-K), which mediates p53-associated transcriptional repression. RNAi-mediated knockdown of linc-p21 decreases apoptosis after doxorubicin treatment, suggesting a possible mechanism for tumor cells avoiding apoptosis after p53 activation (40). These examples demonstrate the importance of lncRNAs in the p53 response.
Hypoxia signaling and EMT.
Tumor cells often utilize hypoxia signaling to maintain a proliferative response in normoxia and escape growth arrest in hypoxia. lncRNA-LET normally represses hypoxia signaling by promoting degradation of nuclear factor 90 (NF90), which is required for hypoxia signaling. Under hypoxic conditions or in cancer cells, hypoxia-inducible histone deacetylase 3 (HDAC3) downregulates lncRNA-LET expression by promoter deacetylation, thus allowing hypoxia signaling to proceed (41). Moreover, hypoxia signaling often stimulates a cellular process known as the epithelial-mesenchymal transition (EMT), which is a critical mediator of metastasis. Several lncRNAs are known to affect EMT signaling in cancer cells. For example, HOTTIP is involved in EMT by activating Wnt/β-catenin signaling, which leads to E-cadherin (CDH1) downregulation (22). Additionally, the function of key EMT regulator Snail1 has recently been found to involve an antisense lncRNA produced from the region of the first intron of the Zeb2 gene and is upregulated by Snail1 expression. Expression of the lncRNA retains a Zeb2 5′-UTR intron, which contains an internal ribosomal entry site (IRES) that facilitates Zeb2 translation. ZEB2 protein then transcriptionally represses E-cadherin to facilitate EMT (42).
Telomere maintenance.
The lncRNA TERC has long been known to be involved in telomere maintenance (43). More recently, another ncRNA in this process has been discovered that provides insight into telomere maintenance in cancer cells. Telomeres produce a large heterogeneous ncRNA named TERRA that binds to both telomeres and TERT to inhibit TERT activity (44). Tumor cell lines and immortalized primary human cells that use TERT to maintain telomeres escape TERRA-mediated TERT repression through heavily methylated subtelomeric DNA to repress TERRA expression (45).
Hormone receptor signaling.
Signaling by the nuclear hormone receptors for androgens (AR) and estrogens (ER) is a fundamental aspect of prostate and breast cancer, and the role for the AR is expanding. A number of lncRNAs have been identified that participate in AR or ER signaling and may provide novel targeting strategies.
The lncRNA PCGEM1 is known to be overexpressed in prostate cancer patient samples, and its overexpression inhibited apoptosis in a prostate cancer cell line in an AR-dependent manner, confirming a functional interaction with AR (46). Subsequently, a detailed mechanism for PCGEM1 was uncovered that involves another lncRNA, PRNCR1, which was identified in our pilot study as PCAT-8 (11). PRNCR1 binds to the AR C-terminal which, in association with histone H3 methyltransferase DOT1L, facilitates recruitment of PCGEM1 to the AR N terminus. This recruitment enhances AR-mediated transcriptional activation by inducing looping of remote AR-bound enhancers to target gene promoters. Thus, overexpression of PRNCR1 and PCGEM1 contributes to castration-resistant AR signaling in prostate cancer (47). While this mechanism is intriguing, it was difficult to reproduce in our hands and further studies will be needed to verify this interaction (48). CTBP1-AS is an antisense lncRNA of the AR corepressor CTBP1. CTBP1-AS directly inhibits CTBP1 transcription via recruitment of the RNA-binding transcriptional repressor PSF and histone deacetylases. This mechanism was found to inhibit several tumor suppressor genes across the genome in an AR-dependent manner, resulting in enhanced cell-cycle progression (49).
A recent study demonstrated that approximately 25% of the genome is transcriptionally regulated by ER in breast cancer cells, including over 1,500 unannotated intergenic, antisense, and divergent transcripts; a substantial proportion of these transcripts are likely to be lncRNAs (50). More specifically, HOTAIR is transcriptionally upregulated by estrogen in a breast cancer cell line where it supports proliferation and suppresses apoptosis, a potential mechanism underlying its overexpression in primary breast tumors (51). In prostate cancer, transcriptional regulation by ERα upregulates the lncRNA NEAT1, whose expression was associated with poor outcome (52). These data demonstrate that lncRNAs are regulated by estrogen signaling and play a major role in estrogen-related cancers.
Competitive endogenous RNA.
lncRNA genes also have a functional interaction with another major class of noncoding RNAs, namely miRNA. Poliseno and colleagues hypothesized a regulatory role for long RNAs (such as lncRNAs) in the binding of miRNAs and uncovered a mechanism that has important implications in cancer biology (53). They found that the phosphatase and tensins homolog (PTEN) pseudogene PTENP1 acts as a molecular sponge for miRNAs that target PTEN mRNA for degradation. They named this class of lncRNAs competitive endogenous RNAs (ceRNAs). Interestingly, the PTENP1-sequestered miRNAs also target other tumor-suppressor genes, including E2F transcription factor 1 (E2F1), CDKN1A, and programmed cell death 4 (PDCD4). The authors were able to show that RNAi against PTENP1 resulted in downregulation of CDKN1A and increased proliferation in PTEN-null cells. Conversely, the PTENP1 locus was deleted in a cohort of colon tumor samples, which also exhibited decreased PTEN expression levels. As suspected, this mechanism is not specific to PTENP1. KRAS and its pseudogene KRAS1P share miRNA let-7–binding sites; their expression was positively correlated in a breast cancer cohort and the KRAS1P locus is narrowly amplified in several cancer types (53). These findings demonstrate that the ceRNA function of lncRNAs has a novel gene dysregulation function in cancer through modulation of miRNA function.
RNA processing.
Several lncRNA functions that appear to contribute to oncogenic phenotypes do not fall into classical pathways, and MALAT-1 provides one interesting example. MALAT-1 is a well-studied lncRNA that was initially identified by subtractive hybridization in early lung cancer tumor samples from patients who did or did not eventually develop metastasis; thus, MALAT-1 was associated with aggressive disease (54). Further research efforts bolstered this role in lung and prostate cancer by demonstrating that MALAT-1 promoted neoplastic behavior in multiple preclinical cancer models, including lung, colorectal, and prostate cancer (55–57). The molecular mechanisms underlying this role in cancer remain vague. MALAT-1 is involved in modulation of mRNA splicing and is found in nuclear paraspeckles, which are sites of RNA processing and editing (58, 59); however, there are conflicting reports over whether the mRNA-processing function of MALAT-1 is responsible for its neoplastic effects (60).
Difficulties in modeling lncRNA functions in mouse models.
Historically, transgenic mice have been arguably the most valuable preclinical model system for evaluating gene function and testing experimental therapeutic agents. However, lncRNA transgenic mice present inherent difficulties, few models have been developed, and the resulting data have been mixed. The primary inherent difficulty of lncRNA transgenic mouse models is that lncRNAs are conserved at much lower rates than protein-coding genes, so that many human lncRNAs do not exist as expressed genes in mice. Among the few that have been made, H19, MALAT-1, and NEAT-1 knockout mice are grossly normal (61), while deletion of Xist in hematopoietic progenitors led to hematologic neoplasms (62). Two knockout mice for HOTAIR have been generated with differing results, which is instructive. A mouse harboring deletion of most of the HOXC locus, which includes HOTAIR, was reported to have a minimal phenotype at the molecular or developmental level (63). Two more targeted HOTAIR knockout mice showed very similar mild but reproducible homeotic phenotypes that were consistent with the known function of HOTAIR (64, 65). These differences demonstrate the difficulty and importance of designing “clean” lncRNA mouse models. These difficulties may be addressed by advanced transgenic mouse models, wherein large human genome portions, including whole chromosomes, are added to or replace portions of the mouse genome (66).
lncRNA deregulation in cancer
There is intense interest in uncovering exactly how tumor cells co-opt lncRNA function to contribute to oncogenic phenotypes. As described above, the primary mode described to date is up- or -downregulation of lncRNA expression levels, but the molecular mechanisms underlying these transcriptional changes have yet to be studied in detail. A global study comparing proximal promoter sequences of protein-coding and noncoding genes found statistical differences in the presence of transcription factor–binding sites and chromatin states. This study also used machine learning to build a protein-coding versus noncoding promoter classifier; however, it is unclear whether these findings explain lineage or cancer specificity of lncRNA expression (67). Copy-number alteration (CNA) can alter lncRNA expression levels in a manner similar to that of protein-coding genes. The lncRNA FAL1 was identified through an unbiased global search for lncRNAs overexpressed by CNAs and for lncRNAs within recurrent CNAs from nearly 2,400 tumor samples. FAL1 expression is associated with outcome in ovarian cancer and interacts with PRC1 component BMI1 to repress numerous genes, including CDKN1A (68). Perhaps more strikingly, the lncRNA PVT1 is located on the 8q24.21 amplicon in MYC amplification and upregulation of PVT1 by coamplification is required for the oncogenic effects of MYC amplification (69, 70).
Oncogenic alterations of protein-coding genes, including point mutations, deletion, and gene fusion, are by and large easy to detect and predict based on the thorough understanding of protein-reading frames and domains. However, this level of understanding does not yet exist for lncRNAs, and so the effects of point mutations, deletion, or gene fusion are very difficult to predict. Indeed, methods for confident detection of lncRNA point mutation or deletion have not yet been developed, though several gene fusions involving lncRNAs have been identified. One of the lncRNAs identified in our PCAT pilot study, PCAT-14, can be found in a somatic gene fusion with the Ets transcription factor family member ETV1. This fusion retains the PCAT-14 promoter, which contains an AR-binding site, allowing for androgen regulation of ETV1 expression (11). In a B cell lymphoma patient, a GAS5-BCL6 fusion was found as a result of a t(1;3) translocation. Here again, the lncRNA GAS5 essentially supplies only its promoter; the entire coding sequence of BCL6 is retained, which is almost certainly the functional portion, given that other BCL6 fusions are common in this disease (71). It seems likely that oncogenic alterations of lncRNAs are occurring in cancer and that they will be uncovered with further refinement of bioinformatics and the sheer explosion in sequencing data.
lncRNAs in cancer risk and SNP studies
There have been numerous GWAS for germline SNPs that associate with a cancer predisposition. A troublesome aspect of these studies has been that most of the SNP loci discovered so far do not have a clear relationship with a known protein-coding gene. A recent survey of the GWAS catalogue identified 301 SNPs associated with increased cancer risk, of which only 12 (3.3%) had an effect on the amino acid sequence of a protein-coding gene (72). Using a catalogue of 11,194 disease-associated SNPs (not restricted to cancer) in our global MiTranscriptome study, we observed that our newly defined transcripts overlapped 2,181 intergenic SNPs (16). This finding provides strong support, though not proof, for the hypothesis that unassociated SNPs are actually associated with unidentified noncoding transcripts (73).
Several studies on individual cancer risk SNPs provide proof of principle for the mechanistic aspect of this hypothesis. The deletion allele of SNP rs10680577 correlates with increased hepatocellular carcinoma (HCC) risk and with upregulation of egl-9 family hypoxia-inducible factor 2 (EGLN2) and RERT-lncRNA, while the insertion allele alters the function of RERT-lncRNA (74). In papillary thyroid cancer, the 14q13.3 lncRNA PTCSC3 is repressed by the SNP rs944289 risk allele through reduction of C/EBP binding to the lncRNA locus and resultant derepression (75). Further analysis of other unexplained cancer risk regions may help identify particularly important lncRNAs for further study.
lncRNAs as diagnostic and prognostic biomarkers
Cancer molecular biomarkers have improved dramatically in the last 2 decades. Diagnostic, prognostic, and predictive biomarkers allow for confident identification of lung adenocarcinoma with transcription termination factor, RNA polymerase I (TTF-1) (76), poor prognosis of neuroblastoma with MYCN amplification (77), and anti-estrogen or -HER2 therapy in breast cancer with ER expression or HER2 amplification, respectively. TCGA results show marked molecular heterogeneity among most cancer types, suggesting that further tumor subclassification is needed; such analysis will almost by definition require molecular biomarkers. The primacy of specificity in diagnostic biomarkers points directly to lncRNAs, as there is mounting evidence that lncRNAs are expressed in a more tissue-specific manner than protein-coding genes (9). Consistent with this, the nearly 8,000 cancer- and/or lineage-specific lncRNAs from our MiTranscriptome study represent a rich resource for biomarker studies (16).
Several examples from our group and others highlight the potential of lncRNA biomarkers, including use in noninvasive body fluid tests. SChLAP1 was originally identified as an unannotated, noncoding transcript with outlier expression in metastases in the relatively small cohort for our PCAT pilot study (14). In nearly 1,100 patients undergoing prostatectomy followed by high-density microarray profiling of the prostatectomy sample and long-term follow-up, we performed an unbiased analysis wherein SChLAP1 was independently renominated and validated as a biomarker of metastatic progression risk (13). Based on these studies, our group has developed and validated an ISH test for SChLAP1 expression that could be used to guide therapy intensification (78). PCA3 is a prostate-specific lncRNA that is highly overexpressed in the majority of prostate cancers. A urine test developed by our group to detect PCA3 and TMPRSS2-ERG fusion transcripts for noninvasive prostate cancer diagnosis outperforms PSA or PCA3 tests alone, has been approved by the US FDA, and is showing promise in clinical settings (79, 80). The commercial prostate cancer prognostic signature Decipher includes several noncoding transcripts (81). Lung cancer lncRNA MALAT-1 as a plasma biomarker in non–small cell lung cancer had a sensitivity of 56% and a specificity of 96% (82). Additionally, several other lncRNAs have been detected in body fluids and may allow for noninvasive detection and monitoring of different cancers. For example, HULC can be found in HCC patient plasma (83), H19 in gastric cancer patients (84), and several lncRNAs in oral cavity squamous cell carcinoma patient saliva (85). These studies strongly indicate that lncRNAs will be developed as useful noninvasive biomarkers across a range of cancer types.
lncRNAs as therapeutic targets
lncRNA biology has already suggested many promising therapeutic targets. Few lncRNAs have been thoroughly validated as targets, but MALAT-1 provides an illustrative and promising example. MALAT-1 knockout mice are resistant to carcinogenesis in breast and lung cancer models and show a minimal phenotype, indicating that toxicity resulting from disruption of MALAT-1 would be unlikely (86–88). The most straightforward therapeutic targeting strategy is to directly target the RNA by sequence, which can be accomplished through several technologies. Antisense oligonucleotides (ASOs) have recently received FDA approval for two nonmalignant diseases, familial hypercholesterolemia (89) and transthyretin amyloidosis (90). Though ASOs have yet to be proven as an anticancer therapy, MALAT-1 ASOs have shown efficacy in a preclinical breast cancer model (91). siRNA-based therapeutics are in development and have entered clinical trials with therapeutics targeting a range of mRNAs in cancer and other diseases (92). Hammerhead ribozymes have self-contained nucleolytic activity and high-specificity sequence recognition, but have not been tested in humans (93). Synthetic RNAs or small RNAs could be used to redirect chromatin-modifying complexes for gene expression modulation, and antagoNATs could be used to target the natural antisense transcript (NAT) class of lncRNAs (94, 95). Sequence-specific therapies have many advantages, but require further development.
There is a host of other therapeutic strategies seeking to exploit other aspects of lncRNA biology. Efforts are underway to disrupt RNA-protein binding sites (96, 97). Another strategy takes advantage of the restricted expression of lncRNAs by using lncRNA regulatory elements. BC-819 is a plasmid containing the diphtheria toxin gene under H19 promoter control that has shown promising results as a cytoreduction agent in bladder, ovarian, and pancreatic tumors (98). An alternative strategy in myotonic dystrophy targets RNA structure with small molecules, in this case binding aberrant ncRNA structure caused by repeat expansion, thereby preventing the pathological binding to and inhibition of muscleblind-like splicing regulator 1 (MBNL1) (99). Based on their signal transduction relationship, mTOR inhibition can increase GAS5 levels in androgen-responsive prostate cancer cell lines and reduce proliferation (100). There does not appear to be a clear winner among these alternative technologies.
Conclusions
The role of lncRNAs in basic, translational, and clinical oncology is likely to equal and perhaps even surpass the role of protein-coding genes. We envision that lncRNA-based clinical tools will expand rapidly in the near future, including as diagnostic and prognostic biomarkers, and as therapeutic targets (Figure 3). While the door to this new world has been opened for us to see the vast potential of the dark matter of the genome, including several clinically available biomarker tests, we still await the first big steps that will benefit cancer patients.
Figure 3. Promise of lncRNAs in oncology.
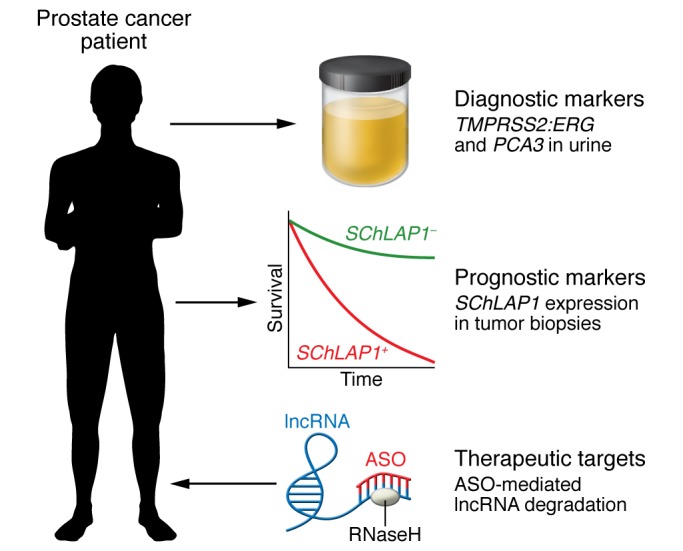
The vast number of lncRNAs and their cancer-specific expression portends great promise for taking advantage of lncRNAs in cancer patient management across diagnostic and prognostic biomarkers and as therapeutic targets. Here we have highlighted prostate cancer as an exemplar. The lncRNA PCA3 is expressed very specifically by prostate tumor cells, which led to its use in a commercial urine diagnostic test, along with the prostate tumor–specific gene fusion TMPRSS2-ERG RNA (79, 80). The lncRNA SChLAP1 has been shown to be a very powerful prognostic marker of metastatic progression risk after prostatectomy for high-risk prostate cancer and could be used to guide therapy intensification through an ISH test (78). lncRNAs are also very attractive therapeutic targets, and several targeting strategies have shown promise, including ASOs, which have been approved for 2 noncancer diseases (89, 90). We expect that lncRNA-based therapeutics will see a rapid clinical expansion in the near future.
Acknowledgments
We wish to thank John Prensner for his helpful discussions about the organization of the article, and Karen Giles for her diligence in managing correspondence. We thank RSNA (Research Resident Grant #RR1420 to JRE) and Prostate Cancer Foundation (Challenge Award to FYF and JRE) for funding lncRNA research.
Footnotes
Conflict of interest: F.Y. Feng and A.M. Chinnaiyan have filed a patent on the clinical uses of the PCATs (U.S. Patent Filing 13/299,000). A.M. Chinnaiyan holds patents on the urine PCA3/TMPRSS2-ERG prostate fusion (U.S. Patent 8,143,232 and U.S. Patent 8,759,301). A.M. Chinnaiyan is a cofounder and serves on the scientific advisory boards for OncoFusion Therapeutics, MedSyn Biopharma LLC, Armune BioScience, and Esanik Therapeutics. SChLAP1 has been licensed to GenomeDx Biosciences Inc. to be developed as a biomarker of prostate cancer (A.M. Chinnaiyan is named as an inventor). F.Y. Feng has received funds for travel, accommodations, or expenses from GenomeDx Biosciences Inc. None of these companies played a role in the design and conduct of this study, in the collection, analysis, or interpretation of the data, or in the preparation, review, or approval of the article.
Reference information:J Clin Invest. 2016;126(8):2775–2782. doi:10.1172/JCI84421.
References
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J. Gene index analysis of the human genome estimates approximately 120,000 genes. Nat Genet. 2000;25(2):239–240. doi: 10.1038/76126. [DOI] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 5.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14(2):103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 6.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prensner JR, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74(6):1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prensner JR, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik R, et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res. 2014;12(8):1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 18.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 19.Christofori G, Naik P, Hanahan D. Deregulation of both imprinted and expressed alleles of the insulin-like growth factor 2 gene during beta-cell tumorigenesis. Nat Genet. 1995;10(2):196–201. doi: 10.1038/ng0695-196. [DOI] [PubMed] [Google Scholar]
- 20.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 23.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Candeias MM, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10(9):1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- 30.Sánchez Y, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5: doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Léveillé N, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6: doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282(34):24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 33.Ying L, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9(3):407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 34.Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60(5):486–492. doi: 10.4149/neo_2013_063. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70(6):2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braconi C, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun M, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35(2):1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 38.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280(7):1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 39.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu W, Zhang Y, Liu D, Songyang Z, Wan M. Telomeres-structure, function, and regulation. Exp Cell Res. 2013;319(2):133–141. doi: 10.1016/j.yexcr.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38(17):5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37(4):1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25(3):135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prensner JR, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5(6):1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takayama K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32(12):1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425(19):3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chakravarty D, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5: doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt LH, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 56.Li D, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182(1):64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 57.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39(1):169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 58.Lin R, et al. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585(4):671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tripathi V, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3): doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 62.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7(5): doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lai KM, et al. Diverse Phenotypes and Specific Transcription Patterns in Twenty Mouse Lines with Ablated LincRNAs. PLoS ONE. 2015;10(4): doi: 10.1371/journal.pone.0125522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Devoy A, Bunton-Stasyshyn RK, Tybulewicz VL, Smith AJ, Fisher EM. Genomically humanized mice: technologies and promises. Nat Rev Genet. 2012;13(1):14–20. doi: 10.1038/nrg3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alam T, Medvedeva YA, Jia H, Brown JB, Lipovich L, Bajic VB. Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PLoS ONE. 2014;9(10): doi: 10.1371/journal.pone.0109443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang S, et al. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS ONE. 2014;9(8): doi: 10.1371/journal.pone.0105538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riquelme E, et al. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9(7):998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tseng YY, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakamura Y, et al. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet Cytogenet. 2008;182(2):144–149. doi: 10.1016/j.cancergencyto.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 72.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93(5):779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Z, et al. An insertion/deletion polymorphism within RERT-lncRNA modulates hepatocellular carcinoma risk. Cancer Res. 2012;72(23):6163–6172. doi: 10.1158/0008-5472.CAN-12-0010. [DOI] [PubMed] [Google Scholar]
- 75.Jendrzejewski J, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A. 2012;109(22):8646–8651. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srodon M, Westra WH. Immunohistochemical staining for thyroid transcription factor-1: a helpful aid in discerning primary site of tumor origin in patients with brain metastases. Hum Pathol. 2002;33(6):642–645. doi: 10.1053/hupa.2002.124910. [DOI] [PubMed] [Google Scholar]
- 77.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3(10): doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mehra R, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16(12):1121–1127. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tomlins SA, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3(94): doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laxman B, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68(3):645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Erho N, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE. 2013;8(6): doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weber DG, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6: doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013: doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arita T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33(8):3185–3193. [PubMed] [Google Scholar]
- 85.Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7(3):761–766. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 86.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eißmann M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McGowan MP, et al. Randomized, placebo-controlled trial of mipomersen in patients with severe hypercholesterolemia receiving maximally tolerated lipid-lowering therapy. PLoS ONE. 2012;7(11): doi: 10.1371/journal.pone.0049006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coelho T, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369(9):819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 91.Arun G, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv Drug Deliv Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tedeschi L, Lande C, Cecchettini A, Citti L. Hammerhead ribozymes in therapeutic target discovery and validation. Drug Discov Today. 2009;14(15–16):776–783. doi: 10.1016/j.drudis.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 94.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12(6):433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 96.Pedram Fatemi R, Salah-Uddin S, Modarresi F, Khoury N, Wahlestedt C, Faghihi MA. Screening for small-molecule modulators of long noncoding RNA-protein interactions using alphascreen. J Biomol Screen. 2015;20(9):1132–1141. doi: 10.1177/1087057115594187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Falk SP, Weisblum B. Aptamer displacement screen for flaviviral RNA methyltransferase inhibitors. J Biomol Screen. 2014;19(8):1147–1153. doi: 10.1177/1087057114533147. [DOI] [PubMed] [Google Scholar]
- 98.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12(5):607–616. [PubMed] [Google Scholar]
- 99.Lee MM, et al. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009;131(47):17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75(7):693–705. doi: 10.1002/pros.22952. [DOI] [PubMed] [Google Scholar]