Abstract
Chronic Helicobacter pylori infection triggers neoplastic transformation of the gastric mucosa in a small subset of patients, but the risk factors that induce progression to gastric metaplasia have not been identified. Prior to cancer development, the oxyntic gastric glands atrophy and are replaced by metaplastic cells in response to chronic gastritis. Previously, we identified schlafen 4 (Slfn4) as a GLI1 target gene and myeloid differentiation factor that correlates with spasmolytic polypeptide-expressing metaplasia (SPEM) in mice. Here, we tested the hypothesis that migration of SLFN4-expressing cells from the bone marrow to peripheral organs predicts preneoplastic changes in the gastric microenvironment. Lineage tracing in Helicobacter-infected Slfn4 reporter mice revealed that SLFN4+ cells migrated to the stomach, where they exhibited myeloid-derived suppressor cell (MDSC) markers and acquired the ability to inhibit T cell proliferation. SLFN4+ MDSCs were not observed in infected GLI1-deficient mice. Overexpression of sonic hedgehog ligand (SHH) in infected WT mice accelerated the appearance of SLFN4+ MDSCs in the gastric corpus. Similarly, in the stomachs of H. pylori–infected patients, the human SLFN4 ortholog SLFN12L colocalized to cells that expressed MDSC surface markers CD15+CD33+HLA-DRlo. Together, these results indicate that SLFN4 marks a GLI1-dependent population of MDSCs that predict a shift in the gastric mucosa to a metaplastic phenotype.
Introduction
Helicobacter pylori infection induces chronic inflammation in the stomach, which eventually leads to atrophy of the acid-producing glands, metaplasia, dysplasia, and then gastric cancer in some infected patients (1–3). The timeline for gastric cancer development occurs over several decades, raising the likelihood that Helicobacter infection is only one of several factors contributing to transformation of the normal gastric epithelium. Indeed, results from The Cancer Genome Atlas (TCGA) for gastric cancer reported only sporadic molecular signatures of Helicobacter in primary tumors (4). Therefore, infection with Helicobacter alone is not sufficient to predict who among the small subset of infected patients will develop gastric cancer. In most instances, the extent of the inflammatory response also contributes to cancer susceptibility (5, 6). Nevertheless, at least 50% of infected individuals develop chronic gastritis without symptoms or further progression of the mucosa to metaplasia and dysplasia (7). Both intestinal metaplasia and spasmolytic polypeptide-expressing metaplasia (SPEM) are histologic lesions strongly associated with neoplastic transformation (8–11). Identifying markers predictive of preneoplasia would allow clinicians to risk-stratify the subset of individuals at greater risk for progression to gastric cancer and who subsequently require more frequent monitoring, regardless of Helicobacter prevalence (12).
Hedgehog (HH) ligands expressed in the gastrointestinal epithelium activate GLI transcription factors in stromal and immune cells (13). Zavros and coworkers showed that the sonic hedgehog (SHH) ligand secreted from gastric parietal cells within 2 days after the infection is required for myeloid cell recruitment to the infected mouse stomach (14). After 2 months of Helicobacter infection, SHH expression in parietal cells gradually diminishes in response to chronic gastritis, despite adjacent mucous cells retaining ligand expression (15). Moreover, GLI1+ myeloid cells are recruited to the stomach within the initial months of the infection (16). By 6 months, most of the parietal and zymogenic chief cells have atrophied and are replaced by SPEM. However, deletion of one or both Gli1 alleles prevents Helicobacter-induced SPEM, indicating that targets of the HH pathway are required for SPEM development (16). Since HH signaling in the stomach is paracrine (17), blocking HH-dependent crosstalk between infiltrating immune cells and the gastric epithelium is sufficient to prevent histologic transition from chronic gastritis to metaplasia.
A myeloid cell differentiation factor and GLI1 target gene (18) called schlafen 4 (Slfn4) was identified in a microarray analysis of gastric tissue from Helicobacter-infected WT versus Gli1–/– mice (16). SLFN4+ myeloid cells express IL-1β and TNF-α (16), cytokines associated with myeloid-derived suppressor cell (MDSC) regulation (6, 19). MDSCs are a heterogeneous myeloid cell population that develops under conditions of infection, tissue injury, autoimmune disease, and cancer (20). Their ability to suppress T cell function dampens the immune response and creates a microenvironment favoring neoplastic transformation (21).
Here we tested the hypothesis that Slfn4 expression marks a GLI1-dependent population of myeloid cells with phenotypic characteristics of MDSCs. We found that SLFN4+ cells in the mouse stomach coincided with SPEM in the setting of chronic gastritis before the development of dysplasia. Since type I interferon induction of Slfn4 gene expression required GLI1, we concluded that HH signaling synergizes with regulatory cytokines to create a permissive environment for gastric metaplasia, a harbinger of possible neoplastic transformation.
Results
SPEM development requires Hedgehog signaling.
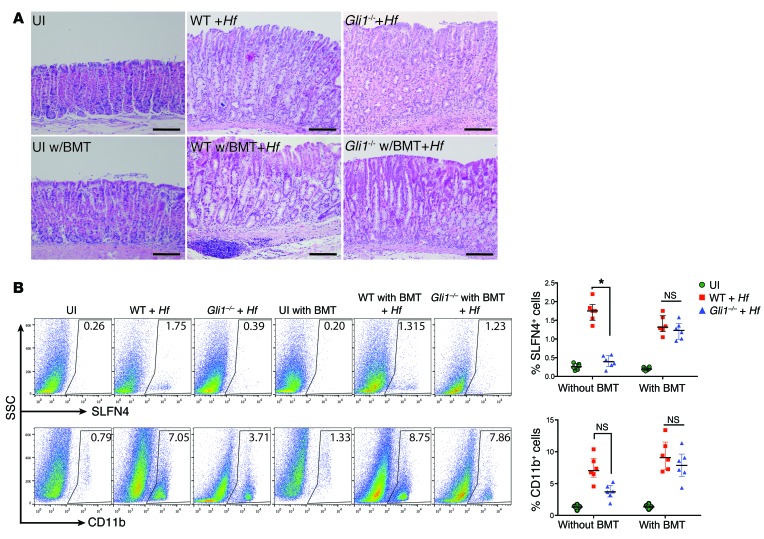
Helicobacter-induced SPEM in WT mice was observed at 6 months after infection. Infected Gli1nLacZ/nLacZ mice (referred to here as Gli1–/– mice) did not develop SPEM as previously reported, which implicates a significant role for canonical HH signaling in the emergence of this preneoplastic lesion (Figure 1A and ref. 16). In the stomach, stromal cells, specifically myofibroblasts and immune cells, express GLI1 (16, 17, 22). To determine whether the metaplastic change was due to bone marrow–derived cells (BMDCs), we transferred marrow from Slfn4-eGFP-CreERT2 Rosa26-LSL-tdTomato (Slfn4-tdT; see below) mice (that are Gli1+/+) into irradiated WT (WT w/bone marrow transplant [BMT]) or Gli1–/– mice (Gli1–/– w/BMT) prior to infection with H. felis for 6 months. Infected Gli1–/– w/BMT mice that received WT marrow developed SPEM, demonstrating that BMDCs were sufficient to transfer gastric susceptibility to Helicobacter-induced metaplasia (Figure 1A). SPEM is defined by the combined expression of chief cell (e.g., gastric intrinsic factor) and mucous neck cell (e.g., TFF2 and GSII-lectin) markers that represent reprogramming of the mature gastric chief cells in the setting of parietal cell atrophy (23–25). Overlap between these two cell-specific markers confirmed the presence of SPEM in the infected mice with WT marrow (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI82529DS1). WT mice developed metaplastic changes only if infected, and the SPEM correlated with an increase in CD11b+ and SLFN4+ cells (Figure 1B). However, the absence of SPEM in the Gli1–/– mice correlated with reduced numbers of SLFN4+, but not a significant reduction of CD11b+ cells (Figure 1B), suggesting a shift in the composition of the myeloid population rather than a defect in total myeloid cell recruitment.
Figure 1. Bone marrow–derived cells are sufficient to induce SPEM with H. felis.
(A) Representative H&E images (n = 6) for each group as indicated with and without bone marrow transplant (BMT) (from Slfn4-CreERT2 Rosa26-LSL-tdTomato [Slfn4-tdT] mice) with and without H. felis infection (Hf). UI, uninfected. Scale bars: 50 μm. (B) Flow analysis of SLFN4-tdT+ or CD11b+ cells versus side scatter (SSC). The percentage of SLFN4+ or CD11b+ cells per mouse is shown in the representative histograms and plotted in the adjacent scatter graphs for n = 6 mice per group over 3 experiments. Horizontal lines represent the median and interquartile range. Significance was determined using Kruskal-Wallis ANOVA with Dunn’s test of multiple comparisons. *P < 0.05.
SHH accelerates H. felis–induced gastric metaplasia.
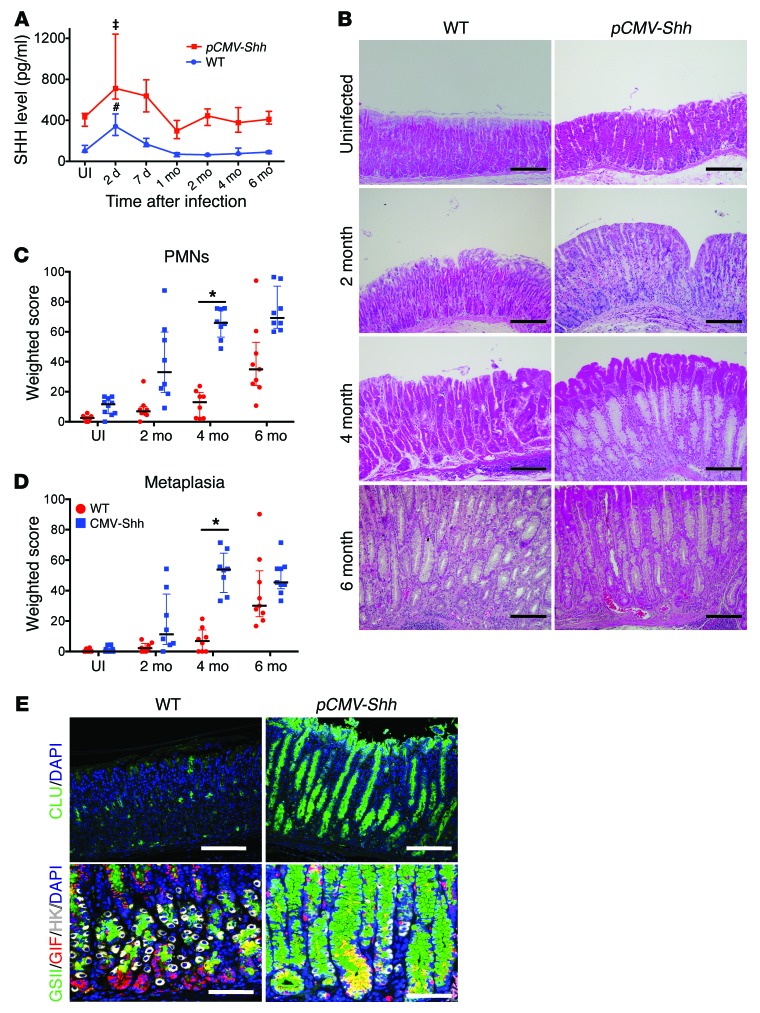
Although these results collectively indicated that HH signaling was necessary for SPEM development, the phenotype required several months to develop. To accelerate the timeline, we infected transgenic mice expressing the pCMV-Shh transgene (16) with H. felis over 6 months and compared them with infected nontransgenic mice. Serum SHH levels in uninfected pCMV-Shh transgenic mice were 4 times the levels observed in WT mice (Figure 2A). In addition, there were more immune cells observed in the pCMV-Shh transgenic mice in the absence of Helicobacter infection compared with WT mice (Supplemental Figure 2). With Helicobacter infection, there was a rapid increase in the blood levels of SHH, which peaked within days after the infection and then returned to baseline levels after 1 month (Figure 2A). The result was consistent with a prior report indicating that H. pylori infection increases circulating levels of SHH, which originates from gastric parietal cells (14). H. felis–infected pCMV-Shh mice exhibited greater inflammation in the gastric corpus at 2 months, and subsequently SPEM was observed at 4 months, which was 2 months earlier than observed in WT littermates (Figure 2B). The amount of polymorphonuclear (PMN) leukocytes infiltrating the mucosa and the extent of SPEM, as quantified by morphometry, were greater in the SHH-overexpressing mice (Figure 2, C and D). SPEM development in pCMV-Shh mice was also documented by de novo expression of clusterin, as well as coexpression of GSII lectin and GIF at the oxyntic gland base, with atrophy of H+-K+-ATPase–positive parietal cells (Figure 2E). Real-time quantitative PCR (RT-qPCR) analysis for H+-K+-ATPase α subunit (Atp4a), Gif, mucous neck cell and SPEM cell marker trefoil factor 2 (Tff2), and clusterin mRNAs were used to quantify the extent of the metaplastic epithelial response (Supplemental Figure 3). Unlike the corpus, the antrum exhibited no phenotypic changes (Supplemental Figure 4).
Figure 2. Elevated SHH levels accelerate H. felis–induced gastritis and SPEM development.
(A) Serum SHH levels in WT and pCMV-Shh mice infected with H. felis over 6 months. n = 5–10 mice per time point over 3 experiments. One-way ANOVA followed by Dunnett’s multiple comparisons test on log-transformed values was performed. P values are relative to UI; ‡P < 0.001, #P < 0.0001. (B) Representative H&E images of gastric mucosa from WT and pCMV-Shh mice at the indicated times. Scale bars: 100 μm. Histologic scoring of gastric (C) PMN infiltration and (D) metaplasia for n = 8–10 mice per time point over 3 experiments. Kruskal-Wallis ANOVA with Dunn’s test of multiple comparisons was performed. *P < 0.05; horizontal lines represent the median and interquartile range. (E) Images of 4-month-infected gastric corpus stained with GSII lectin (green), clusterin (CLU, green), GIF (red), and H+-K+-ATPase (HK, white) in WT and pCMV-Shh mice. Scale bars: 50 μm. n = 5 mice per group for B and E.
SHH accelerates the appearance of SLFN4.
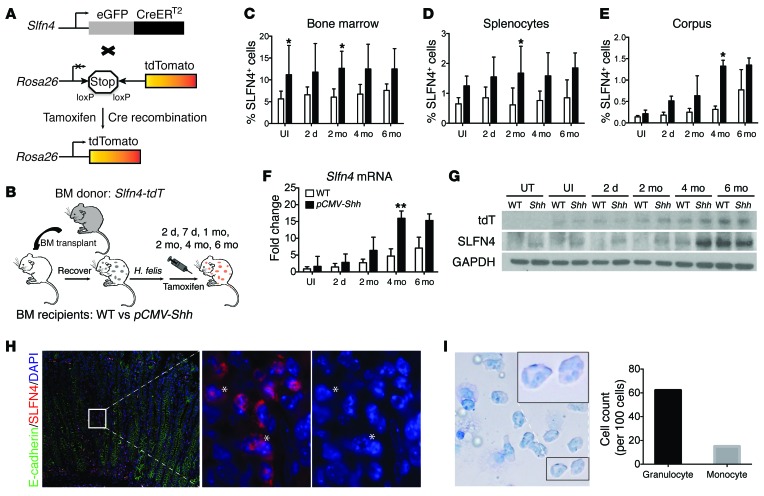
Since ectopic expression of SHH accelerated SPEM development, we queried whether the appearance of SLFN4+ cells also occurred earlier. A Slfn4-eGFP-CreERT2 BAC transgene was generated to trace SLFN4+ cells from the bone marrow by breeding this transgenic line to Rosa26-LSL-tdTomato mice to produce the transgenic line Slfn4-eGFP-CreERT2 Rosa26-LSL-tdTomato (referred to here as Slfn4-tdT mice), which served as bone marrow donors (Figure 3A). Bone marrow cells from the Slfn4-tdT mice were injected into 8-week-old irradiated WT and pCMV-Shh mice. After a 6-week engraftment period, the two groups were infected with H. felis, then pulsed with tamoxifen (Tx, 0.1 mg/g body weight/d) 2 weeks before euthanization at the indicated time points (Figure 3B). The percentage of SLFN4+ cells from the bone marrow, spleen, liver, intestine, and stomach corpus was analyzed. When we compared the uninfected pCMV-Shh mice with WT mice, there was a notable increase in the percentage of SLFN4+ cells in the bone marrow and spleen, with a pronounced trend toward significance (Supplemental Figure 5A), but no differences were observed in the corpus (stomach), liver, or intestine in the absence of the Helicobacter infection (Supplemental Figure 5, A, C, and D). After infection with H. felis, the number of SLFN4-expressing cells remained similar in the bone marrow and spleen of the pCMV-Shh mice compared with WT mice (Figure 3, C and D, and Supplemental Figure 5B). Moreover, there was no significant increase in the number of SLFN4+ cells in the liver and intestine with infection over 6 months (Supplemental Figure 5, C and D). However, in the gastric corpus, there was a time-dependent increase in SLFN4+ cells that correlated with the duration of Helicobacter infection (Figure 3E and Supplemental Figure 5B). Slfn4 mRNA and SLFN4 protein were detectable in total cell lysates from the corpus, and expression peaked at 4 and 6 months after H. felis infection with the appearance of SPEM in the pCMV-Shh and WT mice, respectively (Figure 3F and Figure 2B). Using gastric lysates, a Western blot analysis of the tdTomato (tdT) protein (red fluorescent protein) matched the pattern of endogenous SLFN4 expression, except in mice without BMT (untreated [UT]), which did not express tdT. Over the subsequent months after infection, both endogenous SLFN4 protein and the tdT protein were at low levels until 4 months after the infection, when Slfn4 expression peaked in the pCMV-Shh mice (Figure 3G). Epifluorescence revealed the presence of SLFN4+ cells in the lamina propria of the infected pCMV-Shh corpus 4 months after infection (Figure 3H). A magnified view of the tdT+ cells in the lamina propria showed the presence of granulocytic nuclei in the SLFN4-tdT+ cells. To further identify the nuclear morphology of the SLFN4+ cells from the stomach, FACS-sorted SLFN4-tdT+ cells were Giemsa stained and quantified, revealing that granulocytic cells constituted the majority of SLFN4-tdT+ cells in the stomach (Figure 3I).
Figure 3. SLFN4+ cells track to the stomach after H. felis infection.
(A) Schematic diagram of Slfn4-tdT (Slfn4-CreERT2 Rosa26-LSL-tdTomato) mice generated. (B) Protocol to lineage trace SLFN4-expressing cells from the bone marrow to peripheral organs. WT versus pCMV-Shh chimeric mice reconstituted with Slfn4-tdT bone marrow were treated with Tx 2 weeks prior to euthanization. Shown is the percentage of SLFN4+ cells in the (C) bone marrow, (D) spleen, and (E) corpus by flow cytometry, plotted as the median and interquartile range for n = 8–10 mice per group over 3 experiments. Kruskal-Wallis ANOVA with Dunn’s test of multiple comparisons was performed for C–E; *P < 0.05. The corpus was analyzed for (F) Slfn4 mRNA by RT-qPCR and (G) tdT, SLFN4, and GAPDH protein by Western blot for n = 8–10 mice. One-way ANOVA followed by Tukey’s multiple comparisons test on log-transformed RT-qPCR values was performed for F; **P < 0.01. UT, untreated. (H) Representative images from n = 5 pCMV-Shh mouse corpora infected for 4 months showing SLFN4-tdT red+ cells with E-cadherin (green) and DAPI. Left: original magnification, ×200. White box region enlarged in the middle (tdT and DAPI) and right (DAPI only) panels (magnification, ×1,000). Asterisks indicate granulocytic nuclear morphology. (I) Giemsa stain of SLFN4-tdT+ cells sorted from n = 3 infected pCMV-Shh mouse stomachs (original magnification, ×600) showing granulocytic and monocytic cells enumerated per 100 cells. Black box indicates enlarged region (original magnification, ×1,000).
SLFN4 marks MDSCs in the gastric mucosa.
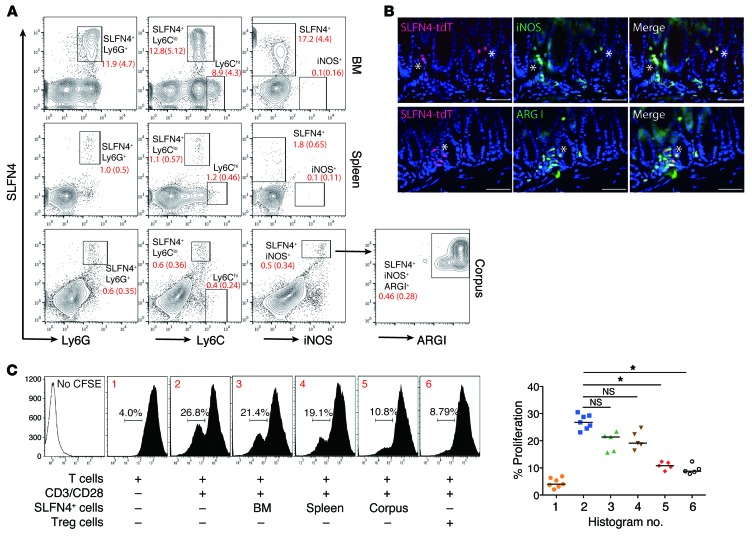
Ectopic expression of SHH increased the number of cells that were CD11b+Gr-1+SLFN4+ 4 months after infection compared with WT mice that acquired the same cell population at 6 months (Supplemental Figure 6). Mouse MDSC markers are defined as CD11b+Gr-1+ and subdivided into monocytic (Ly6G–Ly6Chi) and granulocytic (Ly6G+Ly6Clo) subtypes. Analysis for Gr-1 subunits confirmed that the SLFN4+ cells from bone marrow, spleen, and stomach were Ly6G+Ly6Clo, consistent with a granulocytic MDSC phenotype (Gr-MDSCs) (Figure 4A). Next, we queried whether the gastric SLFN4+ population acquired the ability to suppress T cells. MDSCs suppress T cells in part by depleting l-arginine from the microenvironment. l-Arginine depletion by MDSCs suppresses CD3-ζ chain biosynthesis, which subsequently blocks T cell proliferation during the G0–G1 phase of the cell cycle, ultimately reducing T cell cytokine production (20). MDSCs express high levels of inducible NOS (iNOS, or NOS2) and arginase I (ARG1), which consume l-arginine in the microenvironment to produce nitric oxide and ROS (26). We found that the SLFN4+ cells were positive for iNOS in the corpus but not in the bone marrow or spleen, suggesting that acquisition of the MDSC phenotype occurred in the stomach (Figure 4A). Gated SLFN4+iNOS+ cells from the corpus were further identified as ARG1+ cells. Indeed, further analysis of these cells in the stomach demonstrated colocalization of SLFN4-tdT with iNOS and ARG1 (Figure 4B). A CFSE-based T cell suppression assay was performed to detect the suppressive function of SLFN4+ cells. When the SLFN4+ cells from the stomach were sorted and cocultured with activated T cells, we observed a 52% suppression of T cell activation with the SLFN4+ cells isolated from the stomach, but only minimal suppression when the SLFN4+ cells were isolated from the bone marrow or spleen (Figure 4C). Tregs exhibited a 62% suppression of T cell activation. Therefore, we concluded that the appearance of SLFN4+ cells in the stomach was a three-step process requiring a pool of circulating SLFN4+ cells (which was expanded by SHH overexpression), accumulation of these cells in the corpus in response to the Helicobacter infection, and then acquisition of the ability to suppress T cell proliferation.
Figure 4. SLFN4+ cells from gastric corpus suppress T cell proliferation.
(A) SLFN4+ cells from bone marrow, spleen, and corpus were analyzed for Ly6G, Ly6C, and iNOS expression. Gated SLFN4+iNOS+ cells from the corpus were further analyzed for arginase I (ARG1). Median percentage (interquartile range) for each gated subpopulation shown in red for n = 4 mice per tissue. (B) Representative images from n = 4 mouse stomachs of SLFN4-tdT+ cells (red) colocalized with ARG1 (green) or iNOS (green). Scale bars: 20 μm. Asterisks indicate colocalization. (C) SLFN4+ cells were sorted from the indicated tissues of 3 pooled 4-month-infected infected pCMV-Shh mice and then cocultured with naive splenic T cells prestained with the CFSE dye. T cell proliferation was initiated with anti-CD3/CD28 microbeads cultured with or without SLFN4+ cells for 72 hours, and CFSE+ T cells were quantified by flow cytometry. The median percentage of proliferating T cells is shown in the representative histograms (nos. 1–6) plotted for n = 5 experiments in the scatter graph. Tregs (CD4+CD25+) from spleen were used as the positive control. Kruskal-Wallis ANOVA with Dunn’s test of multiple comparisons was performed. *P < 0.05.
Type I IFNs regulate SLFN4.
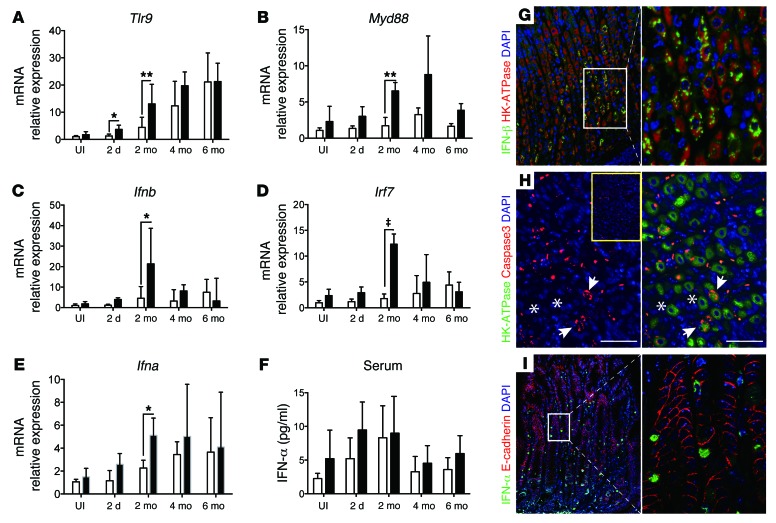
Van Zuylen et al. previously showed that type I interferons induce Slfn4 mRNA expression in bone marrow–derived macrophages through a MYD88-independent mechanism (18). Moreover IFN-α strongly induces all human schlafen family members (27). Since induction of Slfn4 expression correlated with the acquisition of MDSC function coincident with SPEM, we analyzed gastric lysates from infected WT and pCMV-Shh mice for type I interferon expression. Previous reports showed that macrophages, dendritic cells, and CD3+ cells increase gene expression of Toll-like expression receptor 9 (Tlr9) in mouse gastric tissue following H. pylori infection (28). TLR9 recognizes unmethylated bacterial CpG DNA and induces production of a variety of cytokines, including type I IFNs (IFN-α/β) (29). Moreover, damage-associated molecular patterns (DAMPs) from stressed, injured, or dying cells primarily activate TLR9 via MYD88, IRF3, and eventually IFN-β (30). Secreted IFN-β binds and activates the type I IFN receptor in an autocrine or paracrine manner, leading to IRF7 translocation to the nucleus and expression of IFN-α. Thus, we posited that Helicobacter infection activates TLR9 on gastric epithelial cells, triggering events that culminate in the local release of an immune modulator such as IFN-α. Indeed, Tlr9 expression gradually increased over 6 months in both groups of infected mice, whereas Myd88 expression was elevated in pCMV-Shh mouse stomachs, with peak expression at 2 and 4 months (Figure 5, A and B). By contrast, Ifnb1 and Irf7 expression in the pCMV-Shh mice peaked sharply at 2 months, then returned to near baseline by 4 months; while Ifna mRNA was highly expressed in the infected pCMV-Shh mouse stomachs and peaked at 2 months, then remained elevated in both groups (Figure 5, C–E). By comparison, serum IFN-α protein levels trended upward during the first 2 months of the infection, but there was no statistically significant difference in the levels between infected WT and pCMV-Shh mice (Figure 5F), suggesting that differential regulation of Ifna correlated with maximal Slfn4 expression and the histological changes only within infected gastric tissue rather than in response to circulating IFN-α (compare Figure 3F with Figure 5, E and F). Immunofluorescence demonstrated that IFN-β protein was expressed in H+-K+-ATPase+ parietal cells (Figure 5G). Coexpression of H+-K+-ATPase and cleaved caspase-3 suggested that the dying parietal cells were the likely source of DAMP signals (Figure 5H). IFN-α was expressed on both lamina propria cells and E-cadherin+ cells (Figure 5I). Therefore, the source of IFN-α was further analyzed by flow cytometry (Supplemental Figure 7), showing that gated IFN-α+ cells were a mixture of plasmacytoid dendritic cells with E-cadherin+PDCA-1+ surface markers (31, 32) and E-cadherin–CD8a+ cells, which are likely cytotoxic T or dendritic cells. Collectively, the results demonstrated a time-dependent increase in DAMP pathway components after Helicobacter infection, which correlated with Slfn4 expression and the emergence of MDSCs, whereas ectopic SHH production accentuated the extent of the induction.
Figure 5. Analysis of DAMP signaling components in H. felis–infected corpus.
Total RNA was extracted from H. felis–infected WT and pCMV-Shh corpus lysates and analyzed by RT-qPCR for (A) Tlr9, (B) Myd88, (C) Ifnb1, (D) Irf7, and (E) Ifna mRNA at the indicated time points, and then expressed as fold change relative to uninfected WT mice (UI). (F) Serum IFN-α levels determined by ELISA. Shown are the median and interquartile range for n = 8–10 mice per group over 3 experiments. One-way ANOVA followed by Tukey’s multiple comparisons test on log-transformed values was performed. *P < 0.05, **P < 0.01, ‡P < 0.001. Immunofluorescence images for (G) IFN-β1 and (H) cleaved caspase-3 with H+-K+-ATPase (HK-ATPase) in the gastric corpus of 2-month H. felis–infected pCMV-Shh mice. Inset: Image of uninfected pCMV-Shh gastric corpus stained for cleaved caspase-3. Asterisks and arrows indicate nonapoptotic and apoptotic cells, respectively. G: left, original magnification, ×200; G, right, and H: magnification, ×400); (I) Images for IFN-α with E-cadherin in 4-month-infected pCMV-Shh mice. I, left: original magnification, ×200; I, right: magnification, ×600. White boxes indicate enlarged regions. Images in G–I are representative of n = 3 mouse stomachs.
Type I interferon induction of Slfn4 and MDSC function is GLI1 dependent.
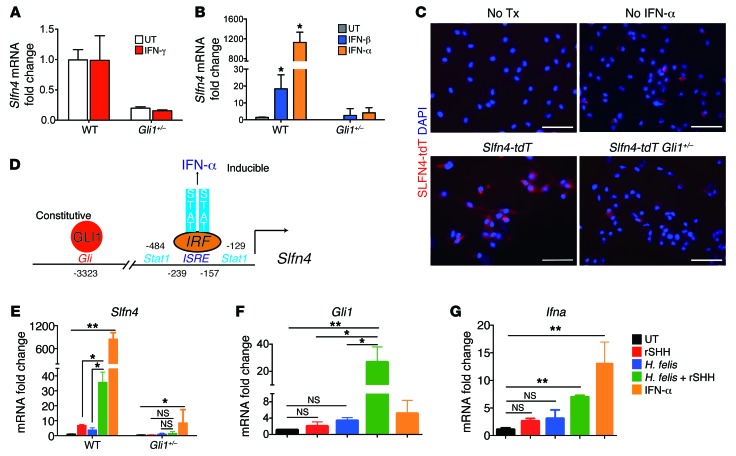
Since peak Slfn4 gene expression correlated with SPEM and the acquisition of MDSC function, we further analyzed interferon regulation of Slfn4 expression in vitro using primary myeloid cells elicited from the peritoneum. Helicobacter infection is typically associated with a Th1 immune response and elevated IFN-γ expression (33, 34). However, IFN-γ, a type 2 interferon, did not induce Slfn4 expression (Figure 6A). Prior studies reported that schlafens are regulated by type I interferons (18, 35). Comparing Slfn4 induction by either IFN-α or IFN-β revealed that IFN-α was a more potent inducer of Slfn4 gene expression than IFN-β and that both required GLI1 (Figure 6B). To directly confirm that the SLFN4+ cells responded directly to IFN-α, we generated primary myeloid cells from the Slfn4-tdT and Slfn4-eGFP-CreERT2 Rosa26-LSL-tdTomato Gli1+/nLacZ (referred to here as Slfn4-tdT Gli1+/–) mouse lines and then treated both populations of primary myeloid cultures with IFN-α. No epifluorescence was observed in the absence of Tx/with IFN-α or with Tx/without IFN-α (Figure 6C). However, after Tx and IFN-α treatment, epifluorescent SLFN4+ cells were observed in primary myeloid cells isolated from the Slfn4-tdT but not the Slfn4-tdT Gli1+/– mouse lines (Figure 6C).
Figure 6. SHH synergizes with H. felis to stimulate Gli1 and Slfn4 in primary myeloid cells.
(A) Peritoneal myeloid cells from WT or Gli1+/– mice were treated with IFN-γ (800 U/ml), and Slfn4 mRNA was analyzed by RT-qPCR. (B) Slfn4 mRNA expression determined after treating peritoneal cells with IFN-α (800 U/ml) or IFN-β (200 ng/ml) for 24 hours. (C) Representative image of SLFN4-tdT+ cells visualized after culturing primary myeloid cells from Slfn4-tdT mice treated ex vivo with IFN-α but no Tx, Tx but no IFN-α, or Tx with IFN-α; or Slfn4-tdT+ Gli1+/– cells treated with Tx and IFN-α. Scale bars: 20 μm. Replicated with 3 mice per genotype. (D) Schematic of Slfn4 promoter, with GLI1 and IRF binding sites indicated. Primary peritoneal myeloid cells collected from WT or Gli1–/– mice were treated with recombinant mouse SHH ligand (rSHH), 100 CFU live H. felis, or both. (E) Slfn4 mRNA in WT and Gli1–/– cells; (F) Gli1 mRNA or (G) Ifna mRNA in WT primary myeloid cells. Shown for A and B and E–G are the median and interquartile range for n = 3 experiments performed in triplicate per genotype. One-way ANOVA followed by Tukey’s multiple comparisons test on log-transformed values was performed. *P < 0.05, **P < 0.01.
The Slfn4 promoter consists of a Gli binding site at –3323 (16) and several STAT/ISRE elements at –239 near the start site of transcription (18) (Figure 6D). Slfn4 promoter activity required GLI1; however, elevated SHH levels did not appear sufficient to boost Slfn4 expression in the corpus (Figure 3F), whereas peak induction required IFN-α, suggesting synergy between the HH pathway and an inflammatory signal. To test this concept in the context of a Helicobacter infection, we treated peritoneal myeloid cells from WT and Gli1–/– mice with recombinant SHH (rSHH), H. felis, or both rSHH and H. felis. Although rSHH and H. felis treatment was sufficient to modestly induce Slfn4 mRNA, synergistic induction of Slfn4 was observed when the cells were treated with both HH ligand and H. felis (Figure 6E). In addition, IFN-α remained the more potent inducer of Slfn4 in WT cells, while 95% of the inducible expression was abolished when the Gli1–/– cells were treated, demonstrating that Gli1 was required for the Slfn4 promoter to be IFN-α responsive, consistent with synergy between the two pathways. Indeed synergistic induction of Gli1 mRNA was observed with rSHH and H. felis, but IFN-α did not strongly induce Gli1 (Figure 6F). By contrast, IFN-α treatment induced Ifna expression in myeloid cells, while rSHH and H. felis alone or combined were weak activators of Ifna gene expression (Figure 6G).
Finally, we examined whether IFN-α treatment of primary peripheral myeloid cells was sufficient to induce MDSC surface markers and function. IFN-α induced Slfn4 expression in CD11b+Gr-1+ to a greater extent than CD11b+Gr-1– populations (Supplemental Figure 8A). However, only Slfn4 mRNA induction in the double-positive CD11b+GR-1+ population was Gli1 dependent (Supplemental Figure 8, B and C). Thus, we concluded that IFN-α strongly induced Slfn4 expression in only a subset of CD11b+GR-1+ cells. Gli1-dependent iNos and Arg1 mRNA expression were highest in the SLFN4+ compared with SLFN4– cells after IFN-α treatment (Supplemental Figure 9, A and B). Moreover, the ability to suppress T cell function was greatest only in the CD11b+Gr-1+SLFN4+ cells compared with the SLFN4− population (Supplemental Figure 9C).
SLFNs associated with Gr-MDSCs and intestinal metaplasia in human stomach.
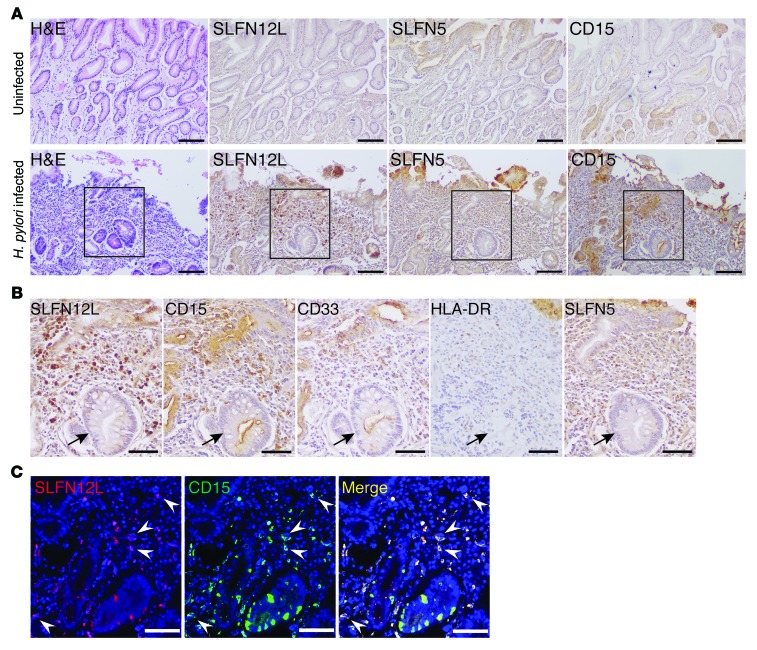
Since the mouse data suggested that SLFNs might function as potential biomarkers for gastric cancer precursor lesions such as intestinal metaplasia in human subjects, we examined whether SLFNs are expressed in H. pylori–infected individuals and related to MDSCs. Human MDSC markers are defined as CD33+HLA-DR−/lo, and subdivided into monocytic (CD14+) and granulocytic (CD15+) subtypes. Since SLFN4 does not exist in the human genome, we analyzed SLFN12L as the human ortholog to the mouse Slfn4 gene based on sequence homology (18) and SLFN5 as the predominant type I IFN–regulated human SLFN gene in human tissues (27, 36). Normal gastric mucosa without H. pylori infection was negative for both SLFN12L- and SLFN5-expressing immune cells, but SLFN+ cells were observed in H. pylori–infected gastric mucosa with intestinal metaplasia (Figure 7A). These immune cells were also positive for the Gr-MDSC marker CD15 and in each patient were near metaplastic glands in at least 2 fields. Further analysis showed that SLFN12L+ cells are CD15+CD33+HLA-DRlo, consistent with the Gr-MDSC designation (Figure 7, B and C). Therefore, human SLFN12L expression increases in H. pylori–infected individuals with intestinal metaplasia and marks a Gr-MDSC subpopulation.
Figure 7. SLFN12L and SLFN5 associated with Gr-MDSCs and intestinal metaplasia in human stomach.
(A) Representative images showing H&E and IHC for SLFN12L, SLFN5, and CD15 in stomach from n = 6 uninfected and n = 6 H. pylori–infected subjects with intestinal metaplasia. Scale bars: 100 μm. (B) Boxed region expanded in IHC for SLFN12L, CD15, CD33, HLA-DR, and SLFN5. Intestinal metaplastic glands are indicated (arrows). Scale bars: 50 μm. (C) Immunofluorescence images for SLFN12L (red) and CD15 (green). Arrowheads indicate colocalization. Scale bars: 50 μm.
Discussion
Myeloid cells home from the bone marrow to peripheral organs to fight invading organisms. Infection by Helicobacter induces acute and then chronic gastritis, initiating the development of gastric metaplasia in some individuals and cancer in about 1% of those infected (1). Yet it is not well understood what component of the chronic inflammatory milieu provides the impetus to propel the mucosa toward cellular atrophy and metaplasia, ultimately presaging the development of cancer. Petersen et al. recently reported that M2 macrophages promote the advancement of SPEM in the presence of inflammation induced by treatment of mice with DMP-777 and L635 (37). However, with chemical induction, oxyntic gland metaplasia develops very rapidly relative to bacterial pathogenesis and may not necessarily recapitulate the events occurring over a prolonged time course as observed in humans. Here we report that canonical HH signaling plays an essential role in the transition from chronic inflammation to gastric metaplasia (SPEM). Subtyping a population of immune cells responsible for the metaplasia, we found that the GLI1 target gene Slfn4 marks a subset of CD11b+Gr-1+Ly6G+ Ly6Glo cells, which acquire the ability to suppress T cell proliferation within the infected stomach. Examining the infected gastric mucosa in 2-month intervals over 6 months revealed that induction of mucosal IFN-α correlated with both SPEM development and the emergence of SLFN4+ cells. Recombinant SHH, live Helicobacter, or IFN-α treatment of peritoneal-derived myeloid cell cultures induced Slfn4 mRNA, providing an ex vivo approach to study the process of MDSC development. The data presented here support a novel pathway for metaplastic induction, where HH signaling synergizes with regulatory cytokines to create a permissive environment for preneoplastic transformation of the gastric mucosa.
The early phase of gastric cancer pathogenesis, especially with intestinal metaplasia, tends to express high levels of SHH ligand (38). However, the effect of SHH on gastric tumor initiation and growth has not been well documented. Acute Helicobacter infection in mice increased SHH blood levels (within 2 days), in agreement with a report by Schumacher et al. that secreted SHH diffuses from the stomach through the circulation and is interpreted by the immune system as a help signal to direct circulating immune cells to the infected stomach (14). Once specific myeloid cells express GLI1 in response to SHH ligand, they become primed to respond to accumulating DAMPs and TLR9 signal transduction, which ultimately stimulates the release of IFN-α into the microenvironment, perhaps with the initial intent to quench the inflammatory response initiated by Helicobacter (32).
Since oxyntic glands atrophy during chronic Helicobacter infection (39), stressed parietal cells would typically be the optimal candidates to release DAMP signals prior to undergoing apoptosis (30). We found that parietal cells express IFN-β, a proximal cytokine mediator of DAMP signaling that peaked 2 months after the H. felis infection. The time course for mucosal Ifna and Slfn4 mRNA expression was consistent with activation of this regulatory pathway, suggesting that induction of DAMPs and Tlr9 expression occurs 2 months after infection. We speculate that the lag period between chronic inflammation and metaplasia permits the accumulation of a sufficient amount of DAMPs from dying parietal cells that gradually increase Tlr9 and type I interferon expression. Indeed, initiation of DAMP signaling coincided with expression of cleaved caspase-3 by parietal cells, marking their entry into an apoptotic phase during this 2-month lag period prior to complete atrophy and their replacement by metaplastic mucous cells. Similarly, a recent study reported that H. pylori infection in mice gradually induces Tlr9 expression in the gastric mucosa over 6 months, enhances neutrophil recruitment, and dampens the proinflammatory cytokine response (28).
This cascade of events leads to induction of IFN-α coincident with the emergence of SLFN4-expressing MDSCs and SPEM. Thus, GLI1-dependent Slfn4 expression within myeloid cell subpopulations appears to be a reliable marker of the phenotypic transition, since it requires both the epithelial HH ligand and immune-modulatory cytokine IFN-α for peak expression. Type I interferons have not been frequently found to play a role in the proinflammatory response observed in a Helicobacter infection (40). Since myeloid cells normally encounter Helicobacter-mediated events only in the infected stomach, peak induction of Slfn4 appears to occur only when tissue IFN-α levels rise above a certain threshold, and could account for the time lag observed prior to the acquisition of the SLFN4+ MDSC phenotype. This transition period represents an important interventional window, since it is well accepted that once metaplasia segues into dysplasia, medical intervention is unlikely to reverse the histologic progression to frank cancer due to the genetic heterogeneity of many epithelial cancers.
Schlafens in immune cells were recently summarized as being a heterogeneous group of genes that potently suppress cell growth, are strongly induced by type I interferons, and primarily exhibit antiviral and antineoplastic effects (35, 41, 42). Yet little is understood regarding epithelial remodeling by infiltrating SLFN-expressing cells. Although we primarily used this GLI1 gene target to probe the role of HH signaling in preneoplasia, the current understanding of the function of schlafen family members suggests that they exhibit potent regulatory effects depending on the cell type (43, 44). Indeed, the analysis of natural mutations in both mouse and human schlafen loci suggests that they normally depress the cell cycle (45, 46). Specifically, the elektra phenotype in mice represents a missense mutation in Slfn2 that allows T cells to remain in a semiactivated state, suggesting a role in mediating T cell quiescence (46, 47). We report here that Slfn4 expression marks a GLI1-dependent population of myeloid cells with the ability to suppress T cell function and phenotypic characteristics of granulocytic MDSCs, consistent with one study showing that gastric cancer patients have elevated levels of MDSCs with a granulocyte-like phenotype (48). Moreover, we showed here that the human homologs SLFN12L and SLFN5 correlated with H. pylori infection and intestinal metaplasia, and coincided with Gr-MDSC markers in human gastric biopsy samples.
H. felis is not a human pathogen and does not express the virulence factors (i.e., VacA and the cag pathogenicity island) that presumably contribute to bacterial pathogenesis in human subjects (49, 50). Nevertheless, H. felis has routinely been used to accelerate the pathological changes in mice that are normally observed in humans (51, 52). Indeed, SLFN4+ MDSCs in our mouse model induced by infection with H. felis correlated with SLFN+ immune cells in H. pylori–infected patients with intestinal metaplasia, suggesting direct applicability to human cancer pathogenesis. Therefore, SLFNs might function as biomarkers for preneoplastic transformation of the gastric mucosa, a concept that we have begun to test in human samples (36). However, a recent study showed that gastric IL-22 expression correlated with CXCR2-dependent influx of monocytic MDSCs acutely into the stomach after H. pylori infection in mice (53). The difference between that report and our study might be related to the time points they studied (49 days after infection) compared with the extended time line reported here (180 days).
We conclude that Slfn4 is perhaps one of several HH-dependent immune genes expressed at low levels in circulating myeloid cells that becomes primed to respond to a threshold level of specific immune mediators after an acute increase in SHH ligand. In this way, SLFN+ MDSCs would remain quiescent until they engage regulatory signals (HH ligand and type I IFNs) at the appropriate time to dampen the proinflammatory response in the stomach. Presumably, sustained immune dysregulation creates a microenvironment capable of supporting cancer development.
Methods
Transgenic mice.
Generation of the pCMV-Shh transgenic mouse line was described previously, and it was genotyped using the following primer pairs: forward 5′-CTGGCTAACTAGAGAACC; reverse 5′-CTTTGGATTCATAGTAGACC (16). Gli1-nLacZ reporter mice were a gift from Alex Joyner (Memorial Sloan Kettering Cancer Center, New York, New York, USA). The mice contain a knock-in of the cDNA for nuclear β-galactosidase (nLacZ) into exon 2 of the mouse Gli1 gene, which disrupts expression, creating a null allele. The mice were maintained as heterozygous Gli1+/nLacZ and homozygous Gli1nLacZ/nLacZ and are herein referred to as Gli1+/– and Gli1–/–, respectively. Genotyping was achieved using the following primers for the WT Gli1 locus: forward 5′-TGATTTCCATGACCAGTTTCTG; reverse 5′-AGGTGAGACGACTGCCAAGT; and for the Gli1-nLacZ knock-in mice: reverse 5′-TCTGCCAGTTTGAGGGGACGAC. Rosa26-LoxSTOPLox-tdTomato reporter mice were purchased from The Jackson Laboratory (B6.Cg-Gt(ROSA)26Sortm27.1(CAG–COP4*H134R/tdTomato)Hze/J strain 012567). The mice were genotyped using two primer sets: one set for the native ROSA26 locus: ROSA26 locus forward 5′-AAGGGAGCTGCAGTGGAGTA, ROSA26 locus reverse 5′-GGCAAAATCTGTGGGAAGTC; and the second set to detect the Rosa-LSL-Cag-hChR2(H134R)-tdTomato-WPRE targeting vector (tdTomato, or tdT) inserted between exon 1 and exon 2: tdTomato reporter forward 5′-CTGTTCCTGTACGGCATGG, tdTomato reporter reverse 5′-GGCATTAAAGCAGCGTATCC. The eGFP-CreERT2 cassette was inserted into exon 5 of the mouse schlafen 4 (Slfn4) gene contained within BAC clone RPCI-23-211B8 (NCBI Clone DB, http://www.ncbi.nlm.nih.gov/clone/666411/) to generate the Slfn4-eGFP-CreERT2 plasmid (JrGang Cheng, University of North Carolina, Chapel Hill BAC Engineering Core, Chapel Hill, North Carolina, USA). The purified Slfn4 BAC plasmid was microinjected into the pronuclei of fertilized C57BL/6 × SJL mouse eggs (University of Michigan Transgenic Core). The presence of the Slfn4-EGFP-CreERT2 was confirmed using three PCR primer sets: (a) spanning the junction between exon 5 of the Slfn4 locus and the eGFP-CreERT2 insert cassette, forward 5′-CACCACACACCGTCCAAGTA; reverse 5′-CTTGTAGTTGCCGTCGTCCT using the Bio-Rad thermocycler settings 95°C (5 minutes), 10 cycles at 95°C (1 minute), 60°C (1 minute) (–1°C per cycle), 72°C (1 minute), 35 cycles 95°C (1 minute), 50°C (1 minute), 72°C (1 minute), 72°C (5 minute), 4°C (hold) to produce a 776-bp amplimer; (b) the eGFP sequence using forward 5′-GACGTAAACGGCCACAAGTT; reverse 5′-CCTCCTTGAAGTCGATGCCC primer set, and settings 95°C (5 minutes), 10 cycles 95°C (1 minute), 60°C (1 minute) (–1°C per cycle), 72°C (1 minute), 25 cycles 95°C (1 minute), 50°C (1 minute), 72°C (1 minute), 72°C (5 minutes), 4°C (hold) to produce a 337-bp product; (c) the Cre cDNA using forward 5′-CATGCTTCATCGTCGGTCC; reverse 5′-GATCATCAGCTACACCAGAG primer set and thermocycler settings 95°C (5 minutes), 35 cycles 95°C (30 seconds), 58°C (40 seconds), 72°C (40 seconds), 72°C (2 minutes), 4°C (hold) to generate a 412-bp PCR product (54). The PCR products were verified by DNA sequencing. To lineage trace the SLFN4+ cells, Slfn4 eGFP-CreERT2 transgenic mice were bred to the Rosa26-LSL-tdTomato reporter mice to generate Slfn4-CreERT2 Rosa26-tdTomato (Slfn4-tdT) mice. Slfn4-tdT mice and Gli1–/– mice were bred to generate Slfn4-CreERT2 Rosa26-tdTomato Gli1+/– (Slfn4-tdT Gli1+/–) mice. All mice were cohoused under the same specific pathogen–free conditions and fasted overnight prior to use, with free access to water. All mice were maintained on a C57BL/6 genetic background.
BMT.
The recipient mice (WT, pCMV-Shh, Gli1–/+, or Gl1–/–) were lethally irradiated using 2 doses of 5 Gy with a 1-hour interval between doses. Bone marrow cells (2 × 106) collected from hind leg long bones of 8- to 12-week-old donor mice (Slfn4-tdT mice) were injected into the tail vein of the irradiated recipient mice. After a 6-week engraftment period, chimeric mice were infected with H. felis. To determine BMT efficiency, 1 ng of DNA from the bone marrow of chimeric mice was analyzed by qPCR for the Rosa26-tdTomato (tdT) transgene. The amount of tdT DNA detected in the donor mice was set to 1. WT mice without BMT were negative for the tdT transgene, and the level was set to 0. The ratio of the chimeric mice tdT DNA versus donor mice tdT DNA is shown in Supplemental Figure 10, indicating approximately 70% engraftment efficiency. Samples with a score <0.5 were excluded from further analysis, since it indicated less than 50% engraftment efficiency.
H. felis culture and infection.
H. felis (CS1 strain) was cultured in sterile-filtered Brucella broth (BD) plus 10% horse serum (Atlanta Biologicals) using the GasPak EZ Campy Container System (BD Biosciences) at 37°C with shaking at 100 rpm. The cultures were pelleted at 1,700 g at room temperature and then resuspended in saline. The optical density of H. felis was measured, and the final concentration was adjusted to 109 organisms per ml of saline. Mice were gavaged 3 times over 5 days with 108 H. felis CFU per 100 μl. Control mice were gavaged with 100 μl saline alone.
Tissue preparation.
Mice were fasted overnight before euthanization. The squamous forestomach was removed prior to bisecting the stomach along the greater and lesser curvatures and then sectioning each half stomach into longitudinal strips. Tissue from the lesser and greater curvatures was fixed overnight in 4% paraformaldehyde and transferred to 70% ethanol for subsequent paraffin embedding. The remaining corpus tissue was minced for total RNA and protein extraction as well as tissue dissociation in EDTA for flow cytometry.
Flow cytometry.
Stomach or small intestine was minced, incubated in 20 ml Ca+2/Mg+2-free HEPES-buffered saline solution (CMF-HBSS) supplemented with 5% FBS (Thermo Fisher Scientific) and 2 mM EDTA (51201, Lonza) with orbital shaking at 250 rpm for 20 minutes at 37°C. The non-dissociated tissue was removed by passing the 20-ml cell suspension through a 100-μm cell strainer by gravity (Fisher Scientific). The tissue collected in the cell strainer was further digested a second time in 20 ml EDTA media (CMF-HBSS/5% FBS with 2 mM EDTA) at 250 rpm for 20 minutes at 37°C. After a second round of EDTA chelation, the cell suspension was passed through the cell strainer a third time, minced rapidly, and then transferred to 20 ml of collagenase solution (1.5 mg/ml type VIII collagenase dissolved in prewarmed CMF-HBSS/5% FBS with 40 μg/ml DNase I) for digestion at 200 rpm for 10–20 minutes at 37°C. After passing through the 100-μm cell strainer and centrifuging at 400 g for 5 minutes at 4°C, the cell pellet was resuspended in PBS containing 5% BSA. To collect bone marrow cells, the marrow from the femurs was directly flushed with PBS, and red blood cells were lysed in ACK lysis buffer (10-548E, Lonza). Cells were collected at 400 g and resuspended in 200 μl PBS containing 5% BSA (Roche). To collect splenocytes and hepatocytes, the spleen and liver were triturated with a 1-ml syringe and flushed through a 70-μm cell strainer into a 50-ml conical tube. The strainer was rinsed twice with HBSS to recover all cells. The cells were collected at 300 g for 10 minutes, and the pellet was resuspended in ACK lysis buffer with gentle swirling for 2 minutes to lyse red blood cells. The lysis was stopped by adding 12 ml of HBSS, centrifuging at 300 g for 10 minutes, and then resuspending the cells in PBS containing 5% BSA. Cells (106 in a 100-μl total volume) were blocked with purified rat anti–mouse CD16/CD32 Fc block (BD Biosciences) at 4°C for 5 minutes, prior to incubation with 0.25 μg CD11b-FITC (BioLegend), Gr-1–APC (BioLegend), Ly6G-APCCy7 (BioLegend), Ly6C–Pacific Blue (BioLegend), CD8a-APC (BioLegend), PDCA-1–PE-Cy7 (eBioscience), rabbit anti–E-cadherin (sc-7870, Santa Cruz Biotechnology Inc.), NOS2-FITC (Santa Cruz Biotechnology Inc.), rabbit anti–arginase I (sc-20150, Santa Cruz Biotechnology Inc.), IFN-α–FITC (Interferon Source). Prior to staining for intracellular proteins, cells were fixed and permeabilized with the Fix and Perm Kit (Thermo Fisher). Prior to initiating the multicolor analysis, we ran the fluorescence minus one (FMO) control to define the spectral overlap. A single-stained control for each fluorochrome was made for compensation for each experiment. A separate set of cells was stained with isotype control antibodies for the negative control. Isotype control antibodies used were PE isotype control (BioLegend, 400608), Pacific Blue isotype control (BioLegend, 400627), PE-Cy7 isotype control (BioLegend, 400618), FITC isotype control (eBioscience, 11-4614-80), APC isotype control (eBioscience, 11-4321-41), APC-Cy7 (BioLegend, 400623). Cell viability was determined with propidium iodide (Abcam, ab14083). Fluorescent-conjugated secondary antibodies goat anti-rabbit IgG APC conjugate (A-10931) and goat anti-rabbit IgG Alexa Fluor 555 conjugate (A27017) were purchased from Life Technologies. The cells were analyzed using an iCyt Synergy SY3200 flow sorter (Sony Biotechnology Inc.).
Histological analysis.
Five-micrometer sections were prepared, deparaffinized, and then rehydrated. Each slide contained sections of well-oriented gastric glandular mucosa, which extended along the greater curvature of the stomach. Slides were stained with H&E to evaluate the degree of PMN accumulation and metaplasia in the corpus. A veterinary pathologist blinded to the experimental conditions used a semiquantitative scale from 0 to 3 on ×200 histologic fields to score gastric lesions. The absence of inflammatory aggregates, atrophy, or metaplasia was given a score of 0. For PMNs, more than 3 occasional clusters (3 or more contiguous cells) was scored 1, widespread PMN aggregates at the base of glands were scored 2, and aggregates throughout the full mucosal thickness were scored 3. For mucous cell metaplasia (loss of parietal cells with replacement by mucous-type cells), the presence of “foamy” epithelial cells in 1–2 glands was scored 1, foamy cells in most fields but less than the entire field was scored 2, and the entire glandular epithelium replaced by foamy mucous cells was scored 3. The weighted score = [(number of fields with a score = 1) + (2× number of fields with a score = 2) + (3× number of fields with a score = 3)/(total number of fields ×3). For each slide, around 12–20 microscopic fields were scored.
Specific protein expression was identified by immunostaining. Antigen retrieval was performed using 10 mM citric acid buffer (pH 6). Slides were washed in 0.01% Triton X-100 in PBS twice, incubated with the serum of the animal in which the secondary antibody was raised for 1 hour. For immunofluorescence staining, slides were incubated with the following antibodies overnight: GSII-FITC (1:1,000, FL-1211, Vector Laboratories), arginase I (1:100, sc-20150, Santa Cruz Biotechnology Inc.), IFN-β (1:100, AP30401PU-N, Acris Antibodies), IFN-α–FITC (1:200, 22100-3, Interferon Source), H+-K+-ATPase-β (1:1,000, D032-3, Medical and Biological Laboratories), caspase-3 (1:400, 9661, Cell Signaling Technology), clusterin (1:2,000, sc-6420, Santa Cruz Biotechnology Inc.), E-cadherin (1:1,000, sc-7870, Santa Cruz Biotechnology Inc.), CD15 (1:7,500, 13-0159-1631, eBioscience), SLFN12L (1:100, NBP1-91060, Novus). Alexa Fluor–conjugated secondary antibodies (Molecular Probes, Invitrogen) were used to detect primary antibodies at a dilution of 1:500 and then mounted with ProLong Gold Antifade reagent with DAPI (Life Technologies). Species-matched secondary antibodies without primary antibodies were used as the negative controls. Immunofluorescence was visualized using a Nikon Eclipse E800 microscope or Nikon A-1 confocal microscope with NIS acquisition software. For light microscopy, the Vectastain ABC kit (Vector Laboratories) and DAB substrate kit (ab64238, Abcam) were used. Primary antibodies used for DAB staining were SLFN12L (1:100, NBP1-91060, Novus), CD15 (1:7,500, 13-0159-1631, eBioscience), HLA-DR (1:50, 340688, BD), CD33 (1:300, 303414, BioLegend), SLFN5 (1:500, ab121537, Abcam). Secondary antibodies used were goat anti-rabbit (1:1,000, 656140, Invitrogen) and donkey anti-mouse (1:500, BA9200, Vector Laboratories).
T cell suppression assay.
T cells from the mouse spleen were isolated using the EasySep Mouse T Cell Isolation Kit (StemCell Technologies) in which magnetically labeled cells to be excluded are retained by the magnetic field, while unlabeled T cells pass through and were collected for CFSE labeling using the CellTrace CFSE Cell Proliferation Kit (C34554; Molecular Probes). Purified T cells were cultured in RPMI supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 mM nonessential amino acids, 5 mM HEPES free acid, 10 ml of 0.05 μM β-mercaptoethanol, and 100 U/ml penicillin/streptomycin with 10% heat-inactivated FBS. To stimulate proliferation, T cells were cultured with anti-CD3/CD28–coated sulfate latex beads in the medium. Suppression of T cell proliferation was assayed after the addition of SLFN4+ (tdT, red) cells for 3 days at a T cell/SLFN4+ cell ratio of 10:1. SLFN4+cells were sorted from H. felis–infected mouse stomach, bone marrow, or spleen. To ensure that the EDTA-collagenase isolation procedures used to isolate the gastric SLFN4+ (tdT, red) cells did not differentially induce the gastric cell suppressor function relative to the SLFN4+ (tdT, red) cells, bone marrow cells and splenocytes were prepared in parallel with the gastric tissue. In brief, in order to isolate cells from the bone marrow and spleens to be used in the T cell suppression assay, the bone marrow was flushed from the long bones with the HBSS/FBS EDTA media (as described above), and spleens were triturated and passed the cell strainer into the EDTA media, with shaking for 20 minutes. The chelation step was repeated. The bone marrow and spleen cells were then incubated in the HBSS/FBS collagenase/DNAse solution for 20 minutes at 37°C with agitation as described above in detail for the minced gastric tissue. The CFSE content in T cells was analyzed by flow cytometry in which low CFSE labeling indicated more proliferation and high-intensity CFSE indicated less proliferation. T lymphocytes were cultured with (CD4+CD25+) splenic Tregs at the same 10:1 ratio were used as the positive control for T cell suppressor activity.
Thioglycollate-elicited peritoneal cells.
Mice were inoculated intraperitoneally with 1 ml of 3% thioglycollate 2 days prior to isolation of peritoneal myeloid cells. The harvested peritoneal cells were plated onto 6-well plates and allowed to adhere overnight to the plastic. The nonadherent cells were removed by aspiration. Flow cytometry was used to determine that most of the adherent peritoneal cells were positive for CD11b and therefore were designated primary myeloid cells. These cells were treated with 800 U/ml IFN-γ, 800 U/ml IFN-α, 200 ng/ml IFN-β, 200 ng/ml recombinant mouse SHH (rmSHH, R&D Systems), or 107 CFU of H. felis per well (~10 CFU:1 myeloid cell) for 12 hours prior to analysis. Primary myeloid cells prepared from the Slfn4-tdT mice were cultured in DMEM media with 10% FBS at 37°C and treated with 100 nM Tx (dissolved in DMSO) for 24 hours to induce Cre recombinase activity ex vivo.
Real-time quantitative PCR.
Total RNA from the corpus was extracted from freshly collected tissue in TRIzol (Invitrogen). The RNeasy Mini Kit (QIAGEN) was used for purification of total RNA. RNA (1 μg) was DNAse treated (Promega) and used for complementary DNA synthesis using iScript reverse transcriptase (Bio-Rad). RT-qPCR was performed using Platinum Taq DNA polymerase (Invitrogen) on a CFX96 real-time PCR detection system (Bio-Rad), using the following primer sequences (Tm = 65°C for all primers): arginase I forward 5′-AACACGGCAGTGGCTTTAACC, reverse 5′-GGTTTTCATGTGGCGCATTC; Gli1 forward 5′-TTGGGATGAAGAAGCAGTTG, reverse 5′-GGAGACAGCATGGCTCACTA; Hprt forward 5′-AGGACCTCTCGAAGTGTTGGATAC, reverse 5′-AACTTGCGCTCATCTTAGGCTTTG; Ifna forward 5′-CGCATCAAAGGACTCATCTGCTG, reverse 5′-CTGCTGCATCAGACAACCTTGC; Ifnb forward 5′-TTCCTGCTGTGCTTCTCCAC, reverse 5′-GATTCACTACCAGTCCCAGAGTC; Ifng forward 5′-TCAAGTGGCATAGATGTGGAAGAA, reverse 5′-TGGCTCTGCAGGATTTTCATG; iNos forward 5′-CGAAACGCTTCACTTCCAA, reverse 5′-TGAGCCTATATTGCTGTGGCT; Irf7 forward 5′-ACAGGGCGTTTTATCTTGCG, reverse 5′-TCCAAGCTCCCGGCTAAG; Myd88 forward 5′-CAAAAGTGGGGTGCCTTTGC, reverse 5′-AAATCCACAGTGCCCCCAGA; Slfn4 forward 5′-GCCCTCTGTTCAAGTCAAGTGTCC, reverse 5′-CCCAGATGAAATCCTTTCCACGA; clusterin forward 5′-CTGTCCACTCAAGGGAGTAGG; reverse 5′-GTGTCCTCCAGAGCATCCTC; Tff2 forward 5′-CTTGGCCCTGACCTGTATGT, reverse 5′-TAGGTTGCTCAGGTGTCACG; Gif forward: 5′-CCCTCTACCTCCTAAGTGTTCTC; reverse 5′-CTGAGTCAGTCACCGAGTTCT.
Western blot analysis.
Tissue from the corpus was homogenized in RIPA buffer (Pierce). Western blotting was performed with an SDS-PAGE Electrophoresis System (Invitrogen). Briefly, 30 μg protein was resuspended in sample buffer, and then electrophoresed on a Novex 4% to 20% Tris-Glycine gradient gel (Invitrogen) with Tris running buffer; blotted to PVDF membrane using the iBlot Dry Blotting System (Invitrogen) according to the manufacturer’s instructions. The membranes were incubated in nonfat dry milk for 1 hour at room temperature and then sequentially probed with primary antibodies against RFP (600901-379S, Rockland), SLFN4 (sc-8903, Santa Cruz Biotechnology Inc.), or GAPDH (MA5-15738, Thermo Fisher) proteins overnight at 4°C. Membranes were washed with TBS containing 0.1% Tween, incubated with IRDye 680–conjugated antibodies (LI-COR), and visualized on an Odyssey infrared imager (LI-COR).
Human gastric tissue analysis.
Archived, de-identified tissue sections of endoscopic biopsies from 6 urease-positive (Campylobacter-like organism [CLO] test) and 6 urease-negative subjects were analyzed for human MDSC markers CD15 (LewisX), CD33, and HLA-DR and schlafen proteins. Intestinal metaplasia was documented on the H&E-stained sections by Joel Greenson, Gastrointestinal Pathology, University of Michigan. Dual-positive CD15 and SLFN12L cells were identified in at least 2 fields with intestinal metaplasia at ×200.
Statistics.
For all ELISA and qPCR experiments, statistical analysis for significance was performed on the log-transformed values using 1-way ANOVA with Tukey’s post hoc test for multiple comparisons. Cell subpopulation computed as percentages and the weighted scores used in the morphometric analysis were analyzed using Kruskal-Wallis 1-way ANOVA with Dunn’s test for multiple comparisons (GraphPad Prism). All data were expressed as the median with the interquartile range. P values less than 0.05 were considered statistically significant. The number of mice per group and replicate experiments are indicated in the figure legends.
Study approval.
Animal use was approved by the University of Michigan Institutional Animal Care and Use Committee. The human tissue samples used in this study were deidentified archived specimens that do not require IRB regulation, as reviewed by the University of Michigan Medical School Institutional Review Board (IRBMED; ID: HUM00108090).
Author contributions
LD designed and conducted the experiments, analyzed the results, and drafted and edited the manuscript. MMH, KAE, QL, and ROR acquired data. AP performed immunohistochemical analysis. JLM designed the experiments, analyzed results, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank the University of Michigan Transgenic Rodent Core supported by the University of Michigan P30 DK Digestive Disease Center (5P30DK034933) and acknowledge support from PHS grant P01 DK62041 (to J.L. Merchant). The authors thank Jessy Deshane, Asma Nusrat, and Ben Allen for their careful review of the manuscript; JrGang Cheng for creating the Slfn4-eGFP-CreERT2 construct by BAC recombineering; and University of Michigan statisticians Kerby Shedden, Chris Andrews, and Joel Greenson (University of Michigan Gastrointestinal Pathology) for histopathologic analysis of archived human biopsies.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(8):2867–2880. doi:10.1172/JCI82529.
Contributor Information
Lin Ding, Email: dinglin@umich.edu.
Michael M. Hayes, Email: mmhayes@umich.edu.
Amanda Photenhauer, Email: alphote@med.umich.edu.
Kathryn A. Eaton, Email: kateaton@umich.edu.
Qian Li, Email: liqian0816@sina.com.
Ramon Ocadiz-Ruiz, Email: jocadizr@med.umich.edu.
References
- 1.Haenszel W, Correa P. Developments in the epidemiology of stomach cancer over the past decade. Cancer Res. 1975;35(11 pt 2):3452–3459. [PubMed] [Google Scholar]
- 2.Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211–217. doi: 10.1016/j.gtc.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007;117(1):60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Omar EM, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404(6776):398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 6.Tu S, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14(5):408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser MJ, Parsonnet J. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J Clin Invest. 1994;94(1):4–8. doi: 10.1172/JCI117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González CA, Sanz-Anquela JM, Gisbert JP, Correa P. Utility of subtyping intestinal metaplasia as marker of gastric cancer risk. A review of the evidence. Int J Cancer. 2013;133(5):1023–1032. doi: 10.1002/ijc.28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt PH, et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest. 1999;79(6):639–646. [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi H, Goldenring JR, Kaminishi M, Lee JR. Identification of spasmolytic polypeptide expressing metaplasia (SPEM) in remnant gastric cancer and surveillance postgastrectomy biopsies. Dig Dis Sci. 2002;47(3):573–578. doi: 10.1023/A:1017920220149. [DOI] [PubMed] [Google Scholar]
- 11.Goldenring JR, Nam KT, Wang TC, Mills JC, Wright NA. Spasmolytic polypeptide-expressing metaplasia and intestinal metaplasia: time for reevaluation of metaplasias and the origins of gastric cancer. Gastroenterology. 2010;138(7):2207–2210. doi: 10.1053/j.gastro.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeh JM, Hur C, Ward Z, Schrag D, Goldie SJ. Gastric adenocarcinoma screening and prevention in the era of new biomarker and endoscopic technologies: a cost-effectiveness analysis. Gut. 2016;65(4):563–574. doi: 10.1136/gutjnl-2014-308588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merchant JL. Hedgehog signalling in gut development, physiology and cancer. J Physiol. 2012;590(3):421–432. doi: 10.1113/jphysiol.2011.220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher MA, et al. Gastric Sonic Hedgehog acts as a macrophage chemoattractant during the immune response to Helicobacter pylori. Gastroenterology. 2012;142(5):1150–1159.e6. doi: 10.1053/j.gastro.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waghray M, et al. Interleukin-1β promotes gastric atrophy through suppression of Sonic Hedgehog. Gastroenterology. 2010;138(2):562–572. doi: 10.1053/j.gastro.2009.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Zaatari M, et al. Gli1 deletion prevents helicobacter-induced gastric metaplasia and expansion of myeloid cell subsets. PLoS One. 2013;8(3): doi: 10.1371/journal.pone.0058935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolterud A, et al. Paracrine Hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology. 2009;137(2):618–628. doi: 10.1053/j.gastro.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Zuylen WJ, et al. Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PLoS One. 2011;6(1): doi: 10.1371/journal.pone.0015723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sade-Feldman M, Kanterman J, Ish-Shalom E, Elnekave M, Horwitz E, Baniyash M. Tumor necrosis factor-α blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity. 2013;38(3):541–554. doi: 10.1016/j.immuni.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saqui-Salces M, et al. Inflammation and Gli2 suppress gastrin gene expression in a murine model of antral hyperplasia. PLoS One. 2012;7(10): doi: 10.1371/journal.pone.0048039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldenring JR, Nam KT, Mills JC. The origin of pre-neoplastic metaplasia in the stomach: chief cells emerge from the Mist. Exp Cell Res. 2011;317(19):2759–2764. doi: 10.1016/j.yexcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam KT, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139(6):2028–2037.e9. doi: 10.1053/j.gastro.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClelland JF. Perspective: photoacoustic spectroscopy: “Linearization and signal recovery in photoacoustic infrared spectroscopy” [Rev. Sci. Instrum. 78, 051301 (2007)] Rev Sci Instrum. 2007;78(5): doi: 10.1063/1.2736265. [DOI] [PubMed] [Google Scholar]
- 26.Köstlin N, et al. Granulocytic myeloid derived suppressor cells expand in human pregnancy and modulate T-cell responses. Eur J Immunol. 2014;44(9):2582–2591. doi: 10.1002/eji.201344200. [DOI] [PubMed] [Google Scholar]
- 27.Katsoulidis E, et al. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285(51):40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otani K, et al. Toll-like receptor 9 signaling has anti-inflammatory effects on the early phase of Helicobacter pylori-induced gastritis. Biochem Biophys Res Commun. 2012;426(3):342–349. doi: 10.1016/j.bbrc.2012.08.080. [DOI] [PubMed] [Google Scholar]
- 29.Radovic-Moreno AF, et al. Immunomodulatory spherical nucleic acids. Proc Natl Acad Sci U S A. 2015;112(13):3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou W, et al. Strange attractors: DAMPs and autophagy link tumor cell death and immunity. Cell Death Dis. 2013;4: doi: 10.1038/cddis.2013.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqui KR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32(4):557–567. doi: 10.1016/j.immuni.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119(7):1623–1633. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto T, Kita M, Ohno T, Iwakura Y, Sekikawa K, Imanishi J. Role of tumor necrosis factor-alpha and interferon-gamma in Helicobacter pylori infection. Microbiol Immunol. 2004;48(9):647–654. doi: 10.1111/j.1348-0421.2004.tb03474.x. [DOI] [PubMed] [Google Scholar]
- 34.Lindholm C, Quiding-Järbrink M, Lönroth H, Hamlet A, Svennerholm AM. Local cytokine response in Helicobacter pylori-infected subjects. Infect Immun. 1998;66(12):5964–5971. doi: 10.1128/iai.66.12.5964-5971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res. 2013;33(4):206–210. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Companioni Nápoles O, et al. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J Gastroenterol. doi: 10.1007/s00535-016-1202-4. http://dx.doi.org/10.1007/s00535-016-1202-4 [published online ahead of print March 31, 2016]. doi:10.1007/s00535-016-1202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen CP, Weis VG, Nam KT, Sousa JF, Fingleton B, Goldenring JR. Macrophages promote progression of spasmolytic polypeptide-expressing metaplasia after acute loss of parietal cells. Gastroenterology. 2014;146(7):1727–1738. doi: 10.1053/j.gastro.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SY, et al. Sonic hedgehog expression in gastric cancer and gastric adenoma. Oncol Rep. 2007;17(5):1051–1055. [PubMed] [Google Scholar]
- 39.Neu B, et al. TNF-alpha induces apoptosis of parietal cells. Biochem Pharmacol. 2003;65(10):1755–1760. doi: 10.1016/S0006-2952(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell P, et al. Chronic exposure to Helicobacter pylori impairs dendritic cell function and inhibits Th1 development. Infect Immun. 2007;75(2):810–819. doi: 10.1128/IAI.00228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9(5):657–668. doi: 10.1016/S1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- 42.Abdel-Mohsen M, et al. Select host restriction factors are associated with HIV persistence during antiretroviral therapy. AIDS. 2015;29(4):411–420. doi: 10.1097/QAD.0000000000000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuang CY, Yang TH, Zhang Y, Zhang L, Wu Q. Schlafen 1 inhibits the proliferation and tube formation of endothelial progenitor cells. PLoS One. 2014;9(10): doi: 10.1371/journal.pone.0109711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaturvedi L, Sun K, Walsh MF, Kuhn LA, Basson MD. The P-loop region of Schlafen 3 acts within the cytosol to induce differentiation of human Caco-2 intestinal epithelial cells. Biochim Biophys Acta. 2014;1843(12):3029–3037. doi: 10.1016/j.bbamcr.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Recher M, et al. Genetic variation in schlafen genes in a patient with a recapitulation of the murine Elektra phenotype. J Allergy Clin Immunol. 2014;133(5):1462–1465.e5. doi: 10.1016/j.jaci.2013.10.052. [DOI] [PubMed] [Google Scholar]
- 46.Berger M, et al. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol. 2010;11(4):335–343. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derbinski J, Schulte A, Kyewski B, Klein L. Pillars article: Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001. 2: 1032–1039. J Immunol. 2016;196(7):2915–2922. [PubMed] [Google Scholar]
- 48.Wang L, Chang EW, Wong SC, Ong SM, Chong DQ, Ling KL. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J Immunol. 2013;190(2):794–804. doi: 10.4049/jimmunol.1202088. [DOI] [PubMed] [Google Scholar]
- 49.Arnold IC, et al. Comparative whole genome sequence analysis of the carcinogenic bacterial model pathogen Helicobacter felis. Genome Biol Evol. 2011;3:302–308. doi: 10.1093/gbe/evr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang MY, Liu XF, Gao XZ. Helicobacter pylori virulence factors in development of gastric carcinoma. Future Microbiol. 2015;10(9):1505–1516. doi: 10.2217/fmb.15.72. [DOI] [PubMed] [Google Scholar]
- 51.Duckworth CA, Burkitt MD, Williams JM, Parsons BN, Tang JM, Pritchard DM. Murine models of Helicobacter (pylori or felis)-associated gastric cancer. Curr Protoc Pharmacol. 2015;69(14.34):1–35. doi: 10.1002/0471141755.ph1434s69. [DOI] [PubMed] [Google Scholar]
- 52.Sgouras DN, Trang TT, Yamaoka Y. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2015;20(suppl 1):8–16. doi: 10.1111/hel.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang Y, et al. A pro-inflammatory role for Th22 cells in Helicobacter pylori-associated gastritis. Gut. 2015;64(9):1368–1378. doi: 10.1136/gutjnl-2014-307020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grachtchouk M, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest. 2011;121(5):1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.