Abstract
Progressive tissue fibrosis is a major cause of the morbidity and mortality associated with repeated epithelial injuries and accumulation of myofibroblasts. Successful treatment options are limited by an incomplete understanding of the molecular mechanisms that regulate myofibroblast accumulation. Here, we employed in vivo lineage tracing and real-time gene expression transgenic reporting methods to analyze the early embryonic transcription factor T-box gene 4 (TBX4), and determined that TBX4-lineage mesenchymal progenitors are the predominant source of myofibroblasts in injured adult lung. In a murine model, ablation of TBX4-expressing cells or disruption of TBX4 signaling attenuated lung fibrosis after bleomycin-induced injury. Furthermore, TBX4 regulated hyaluronan synthase 2 production to enable fibroblast invasion of matrix both in murine models and in fibroblasts from patients with severe pulmonary fibrosis. These data identify TBX4 as a mesenchymal transcription factor that drives accumulation of myofibroblasts and the development of lung fibrosis. Targeting TBX4 and downstream factors that regulate fibroblast invasiveness could lead to therapeutic approaches in lung fibrosis.
Introduction
Progressive tissue fibrosis is a major cause of morbidity and mortality with limited therapeutic choices. Severe pulmonary fibrosis is the integrative result of repeated epithelial injuries and recruitment and activation of myofibroblasts, as well as matrix deposition. Idiopathic pulmonary fibrosis (IPF) is a terminal illness characterized by unremitting matrix deposition in the lung (1). The accumulation of myofibroblasts is one of the key features in tissue fibrosis.
The origins of myofibroblasts have been of great interest in understanding the pathogenesis of tissue fibrosis (2–5). Fibroblasts in IPF (6) and in mouse models (5) are extremely heterogeneous, suggesting that they could be derived from different cell types or represent different stages of activation, or that they may be influenced by the surrounding milieu (7). Identification of fibroblast-specific cell surface markers has been elusive (7). Markers such as α-smooth muscle actin (αSMA, encoded by the Acta2 gene), FSP1 (also known as S100A4), vimentin, desmin, and PDGFRβ are either not exclusively expressed by fibroblasts or not specific to all fibroblasts (5, 8). Recent reports suggest that several cellular sources contribute to myofibroblasts. Cells of extrapulmonary origin may be able to migrate to the fibrotic lesion and become myofibroblasts (9–11). Several studies have proposed that epithelial cells (12–14) or endothelial cells (4, 15, 16) transform into stromal cells in experimental fibrosis models. However, using genetic tracing approaches, recent studies have shown that lung epithelial cells such as adult Sftpc-lineage alveolar type II cells, as well as adult Scgb1a1-lineage club cells, do not give rise to fibroblasts (5). In addition, genetic lineage labeling studies have confirmed that pericytes trace-labeled with either NG2, FOXJ1, or FOXD1 (3, 5) contribute to αSMA-expressing myofibroblasts. Finally, GLI1-labeled perivascular cells with distinct characteristics of mesenchymal stem cells (MSCs) can differentiate into myofibroblasts in tissue fibrosis (17). These studies suggest that a number of cell types may contribute to myofibroblast accumulation, but key upstream regulatory molecules that may promote myofibroblast accumulation from a number of cellular origins have not been identified. In particular, we sought to determine whether there might be a transcription factor that promotes myofibroblast accumulation from different cells of origin.
T-box gene 4 (TBX4) is a mesenchymal-specific transcription factor that is expressed early during embryonic development and is essential for hindlimb development (18, 19) and airway branching (20). TBX4 expression is detected in lung buds at E9.25 (20), and TBX4 is expressed at E11.5 and later throughout the lung mesenchyme (20). GLI1 is detectable from E11.5, and FOXD1 from E13.5, both of which are later than TBX4 (3, 21). These reports raise the possibility that TBX4 might be upstream of other mesenchymal transcription factors that regulate the diversity of mesenchymal progenitor populations with different locations, patterns of migration, and recruitment mechanisms. Mice with both TBX4 and TBX5 deficiency exhibit severely reduced lung branching at mid-gestation, indicating that TBX4 is important in regulating the proliferation, migration, and differentiation of mesenchymal cells that support and help the generation of surrounding epithelium (20). We sought to determine whether TBX4 is playing a role in disease contexts such as fibrosis in the adult lung. In addition, we investigated the role of TBX4 in regulating key effector functions of fibroblasts such as proliferation, migration, and invasion as well as myofibroblast accumulation, activation, and extracellular matrix (ECM) gene expression in vitro and in vivo.
Through comprehensive analyses including tracking of mesenchymal markers in vivo, multicolor clonal cell labeling, cell-specific lineage ablation, and gene deletion in a cell type–specific fashion, we show here that Tbx4-derived cells give rise to a diversity of cell types including fibroblasts, smooth muscle cells, some pericytes, and a few endothelial cells in the adult lung. In addition, Tbx4-lineage cells expand, are proliferative, and form clonal-like patches within fibrotic foci. Furthermore, Tbx4-derived cells are major contributors to myofibroblast accumulation. Specific deletion of Tbx4-lineage cells or the Tbx4 gene in either collagen- or αSMA-expressing fibroblasts significantly attenuated lung fibrosis. Lastly, TBX4 binding sites in the hyaluronan synthase 2 (Has2) promoter regulate expression and enhance fibroblast invasiveness and hyaluronan (HA) production, revealing the importance of this pathway in fibrogenesis. Collectively, these data suggest that TBX4 may be an important regulator of fibrogenic potential of diverse cell types that contribute to myofibroblast accumulation.
Results
Tbx4-lineage cells give rise to fibroblasts, smooth muscle, and rare endothelial cells in the lung.
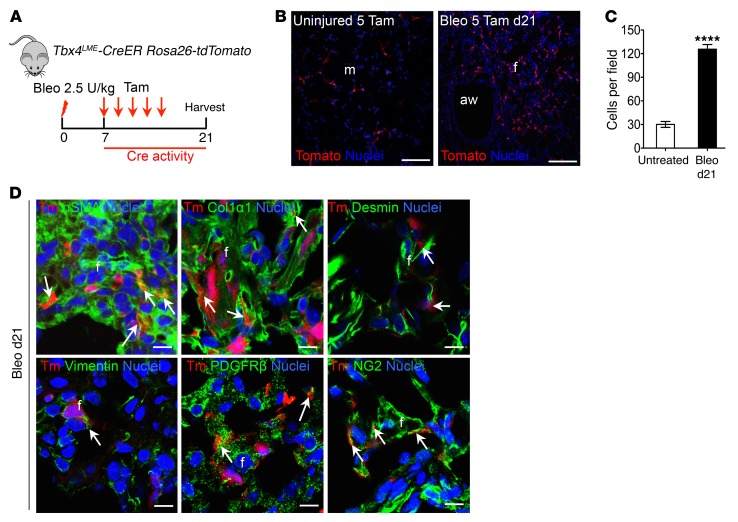
To permanently label TBX4-expressing cells and their descendants in the lung, Tbx4 lung enhancer-Cre transgenic mice (Tbx4LME-Cre, also known as Tbx4-CreTg) (22) or Tbx4-Cre knock-in mice (Tbx4-CreKi) (23, 24) were crossed with Rosa26-tdTomato (Tm) reporter mice (25). It has been shown that TBX4 is expressed as early as E9.25 in lung mesenchyme (20). We found that Tbx4 lineage–labeled stromal cells reside along blood vessels, underneath the bronchioles, and within the interstitial mesenchyme, of both embryonic (E15.5) and adult-stage (8W) lung tissue (Figure 1, A–C; and Supplemental Figure 1B; supplemental material available online with this article; doi:10.1172/JCI85328DS1). Tbx4-lineage cells include lipofibroblasts and a few endothelial cells (Figure 1, G and H). These results are consistent with recent data indicating that Tbx4-Cre labels undifferentiated mesenchyme, airway smooth muscle, vascular smooth muscle, and mesothelium (22, 26, 27). The locations of tdTomato-labeled (tdT-labeled) cells were identical in the lungs of both strains.
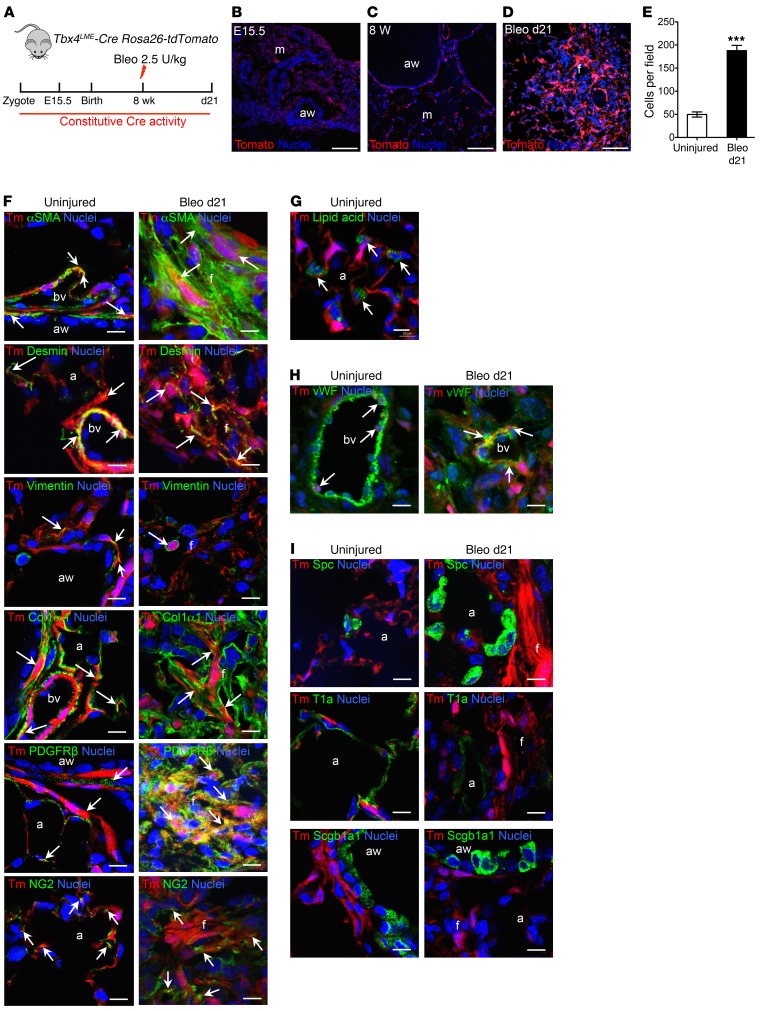
Figure 1. Tbx4-lineage cells include smooth muscle cells, fibroblasts, some pericytes, and endothelial cells in both normal and injured lung.
(A) Schematic depicting lineage analysis methodology. Tbx4LME-Cre Rosa26-tdTomato mice were used for all experiments in this figure. (B–D) Representative histological sections of Tbx4LME-Cre Rosa26-tdTomato mouse lungs at embryonic d15.5 (B) and uninjured 8-week-old (C) and bleomycin-treated (d21) (D) adult Tbx4LME-Cre Rosa26-tdTomato with Tbx4-lineage cells in red and nuclei in blue. (E) Quantification of Tbx4 lineage tracing, expressed as cells counted in C and D (n = 9 in each group of Tbx4LME-Cre Rosa26-tdTomato mice; ***P ≤ 0.001 by 2-tailed Student’s t test; mean ± SEM). (F–I) Representative immunofluorescent images of adult Tbx4LME-Cre Rosa26-tdTomato mouse lungs, uninjured or bleomycin-treated (d21) and stained with αSMA, desmin, vimentin, COL1α1, PDGFRβ, NG2 (F), lipid acid (G), vWF (H), SPC, T1a, and SCGB1A1 (I) antibody (n = 9 lungs examined). Arrows show cells with overlaps. Scale bars: 100 μm (B–D) and 10 μm (F–I). a, alveoli; aw, airway; bv, blood vessel; f, fibrotic foci; m, mesenchyme; Tomato and Tm, tdTomato.
To determine whether Tbx4-lineage cells expanded during injury, adult Tbx4LME-Cre Rosa26-tdTomato and Tbx4-CreKi Rosa26-tdTomato mice were administered 2.5 U/kg bleomycin intratracheally, and the lungs were examined 21 days after injury (Figure 1A and Supplemental Figure 1A). We found extensive expansion of Tbx4-derived cells on day 21 in comparison with saline-treated lung (Figure 1, D and E, and Supplemental Figure 1B).
We then costained with stromal markers and demonstrated overlap of Tbx4-lineage cells with several stromal cell markers, including SMA, desmin, vimentin, collagen type 1 α1 (COL1α1), and pericyte markers PDGFRβ and NG2 (Figure 1F) in the lungs of Tbx4LME-Cre Rosa26-tdTomato mice. Overlap of tdT labeling and αSMA staining can be easily seen in airway smooth muscle, vascular smooth muscle, and interstitial fibroblasts (Figure 1F). Overlap of tdT+ labeling and staining of fibroblast markers (COL1α1, desmin, and vimentin) can also be readily identified, especially in the lungs 21 days after bleomycin injury (Figure 1F). The antibody staining results were similar in the lungs of both Tbx4LME-Cre Rosa26-tdTomato and Tbx4-CreKi Rosa26-tdTomato mice.
Pericytes have been implicated in tissue fibrosis and are suggested to be a source of myofibroblasts (3, 28, 29). PDGFRβ and chondroitin sulfate proteoglycan 4 (CSPG4, or NG2), along with FOXD1 and FOXJ1, have each been used as pericyte markers (3, 5). In Ng2-YFP Tbx4-CreKi Rosa26-tdTomato triple-heterozygous mice, there were 0.40% and 0.88% NG2/tdT double-positive cells within total cells in uninjured and bleomycin-injured (day 21) lung single-cell homogenates, respectively (Supplemental Figure 2B). A few NG2+ cells are of Tbx4 lineage in both bleomycin-injured (22%) and uninjured (10%) Ng2-YFP Tbx4-CreKi Rosa26-tdTomato mouse lungs (Supplemental Figure 2C). Within tdT+ cells, 10% were NG2+ in both uninjured and bleomycin-injured lungs (Supplemental Figure 2C).
Epithelial cells defined by their expression of SCGB1A1, SFTPC, T1a (Figure 1I), or E-cadherin (not shown) were absent from the tdT-labeled fraction of lung cells. This is consistent with previous studies in embryonic lungs that also indicate that Tbx4-lineage cells do not include an epithelial subset (22, 24).
Tbx4-lineage cells were rarely observed in extrapulmonary tissues of either Tbx4LME-Cre Rosa26-tdTomato or Tbx4-CreKi Rosa26-tdTomato mice. In bone marrow, we found that only a very small fraction (less than 0.03%) of the total live cells expressing tdT in Tbx4-CreKi Rosa26-tdTomato mice, whereas no Tbx4-lineage cells were detected in Tbx4LME-Cre Rosa26-tdTomato bone marrow (Supplemental Figure 3). Other organs of 8-week-old Tbx4-CreKi Rosa26-tdTomato mice, including heart, liver, kidney, spleen, and intestine, were all negative for Tbx4-lineage cells (data not shown).
Next, we wanted to determine whether Tbx4-lineage cells give rise to MSCs. GLI1-labeled perivascular cells with distinct characteristics of MSCs can differentiate into myofibroblasts in tissue fibrosis (17). Our data suggest that Tbx4-lineage cells do not include cells expressing a classic MSC pattern (CD44+CD29+CD105+SCA1+ and CD31–CD34–CD45–), although about 70% of Tbx4-lineage cells express CD29 (integrin-β1) (Supplemental Figure 4).
We next compared the expression profiles of tdT+ versus tdT– cells, which were sorted from cultured lung fibroblasts of Tbx4LME-Cre Rosa26-tdTomato mice. Genes that were expressed at higher levels in tdT+ cells included fibroblast-related genes and ECM remodeling genes such as Acta2, Adam12, Pdgfra, Fgf10, and Fgf7, genes in the collagen family (Col1a2, Col3a1, Col4a1, Col4a2, Col4a5, Col4a6), Mmps (Mmp10, Mmp11, Mmp8), desmin, and Has2 (Supplemental Table 1). In addition, several transcription factors including SOX11, Smad genes (Smad1, Smad2, Smad3, Smad5, and Smad7), Snai1, Snai2, and T-box genes (Tbx2, Tbx3, Tbx5, Tbx18, and of course Tbx4 itself) were also upregulated in tdT+ cells (Supplemental Table 2). These data suggest that Tbx4-lineage cells express more fibroblast- and ECM remodeling–related genes indicative of a more activated state.
TGF-β signaling is a prominent driver in tissue fibrosis (30). It is has been reported that conditional deletion of TGF-β receptor II in COL1α2- and TBX4-expressing cells significantly reduces lung fibrosis (31, 32). We found that TBX4+ fibroblasts have an activated phenotype and express higher levels of Smad transcription factors (Supplemental Table 2). We wondered whether the activated phenotype in TBX4+ cells was dependent on TGF-β signaling. To our surprise, exogenous TGF-β did not increase Tbx4 gene expression in either mouse or human fibroblast cultures (Supplemental Figure 5, A–E). In our array data, both TGF-β1 and TGF-βR1 expression was decreased in TBX4+ fibroblasts compared with TBX4– fibroblasts. However, both TGF-β2 and TGF-βR2 expression was elevated in TBX4+ fibroblasts compared with TBX4– fibroblasts (Supplemental Table 3). Interestingly, we found that the TGF-β superfamily member BMP4 can induce the expression of Tbx4 mRNAs in cultured primary human lung fibroblasts. Similar results were found following WNT5A treatment (Supplemental Figure 5F). These data suggested a complex role for TBX4 in TGF-β signaling; further investigations are needed to elucidate the relationship.
Together, these results indicate that Tbx4-lineage cells in the lung represent a heterogeneous cell population that includes smooth muscle cells and different types of fibroblasts, as well as some pericytes and endothelial cells. Tbx4-lineage fibroblasts expand during pulmonary fibrosis and significantly contribute to αSMA+ myofibroblast accumulation.
The vast majority of myofibroblasts derive from Tbx4-lineage cells.
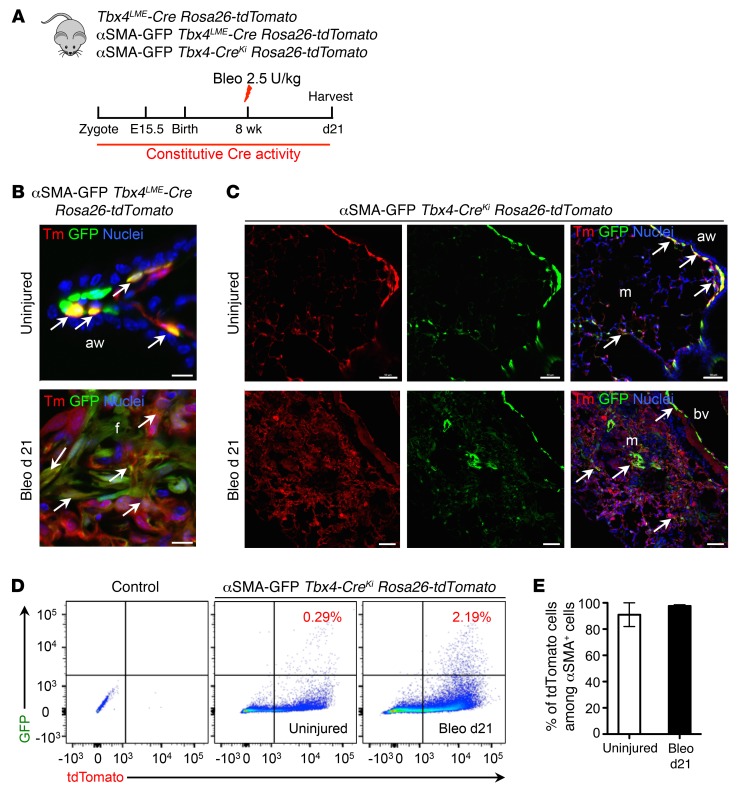
To further confirm the αSMA antibody staining results, we crossed the reporter mouse line Acta2-GFP (referred to here as αSMA-GFP mice) with Tbx4LME-Cre Rosa26-tdTomato and Tbx4-CreKi Rosa26-tdTomato mice. FACS analysis of lung single cells from uninjured αSMA-GFP Tbx4-CreKi Rosa26-tdTomato mice revealed that approximately 80% of the 7AAD–, EPCAM–, and CD31–CD45– cells are Tbx4-lineage cells (Supplemental Figure 6). The histology images of αSMA-GFP Tbx4LME-Cre Rosa26-tdTomato and αSMA-GFP Tbx4-CreKi Rosa26-tdTomato mouse frozen sections are consistent with the antibody staining results (Figure 2, B and C). The Acta2 promoter drives GFP expression well in the lung, as the expression pattern and the levels were comparable with antibody staining (Supplemental Figure 7). In uninjured αSMA-GFP Tbx4-CreKi Rosa26-tdTomato triple-heterozygous mice, 0.29% of total cells in lung single-cell homogenates were αSMA/tdT double-positive cells as determined by flow cytometric analysis (Figure 2D). The SMA/tdT double-positive cells increased more than 7-fold to 2.19% of total cells, in the lungs of mice 21 days after bleomycin exposure (Figure 2D). Among αSMA-GFP+ lung cells, 90.6% were tdT+ in uninjured mice, and 97.6% were tdT+ 21 days after bleomycin exposure (Figure 2E). These data confirmed that most myofibroblasts were derived from Tbx4 lineage–labeled resident lung stromal cells.
Figure 2. Tbx4-lineage cells in the lung are the major source of myofibroblasts.
(A) Schematic depicting lineage analysis methodology. Tbx4-CreLME Rosa26-tdTomato, αSMA-GFP Tbx4LME-Cre Rosa26-tdTomato, and αSMA-GFP Tbx4-CreKi Rosa26-tdTomato mice were used for all experiments in this figure. (B) Representative histology images of uninjured or bleomycin-treated (d21) adult αSMA-GFP Tbx4LME-Cre Rosa26-tdTomato mouse lung. (C) Representative histology images of uninjured or bleomycin-treated (d21) adult αSMA-GFP Tbx4-CreKi Rosa26-tdTomato mouse lung. (D) Representative FACS plots of mouse lung single cells isolated from uninjured or bleomycin-treated (d21) αSMA-GFP Tbx4-CreKi Rosa26-tdTomato mice. (E) Quantification of tdT+ cells among αSMA-GFP+ cells in D (n = 3 mice analyzed, mean ± SEM). Arrows show cells with overlaps. Scale bars: 10 μm (B) and 100 μm (C). aw, airway; bv, blood vessel; f, fibrotic foci; m, mesenchyme.
The majority of COL1α1+ cells are derived from Tbx4-lineage cells.
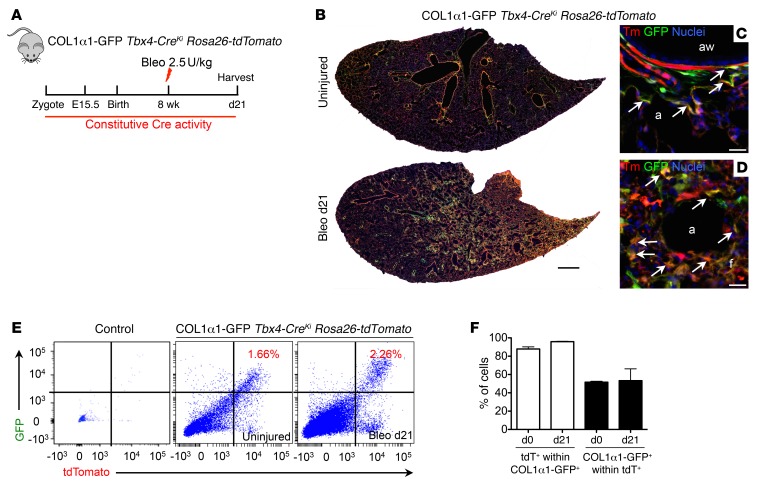
COL1α1-expressing cells are interstitial mesenchymal cells and fibrocytes in tissues (15, 33). The reporter mouse line Col1a1-GFP (referred to here as COL1α1-GFP mice) (33) was crossed with Tbx4-CreKi Rosa26-tdTomato mice to generate COL1α1-GFP Tbx4-CreKi Rosa26-tdTomato triple-heterozygous mice. FACS analysis of lung single cells from uninjured COL1α1-GFP Tbx4-CreKi Rosa26-tdTomato mice revealed that more than 80% of the 7AAD–, EPCAM–, and CD31–CD45– cells are Tbx4-lineage cells (Supplemental Figure 8). The mice were subjected to intratracheal bleomycin-induced lung injury (Figure 3A). Analysis of frozen lung section at day 21 after bleomycin showed that most of the COL1α1-GFP+ cells were labeled by mesenchymal-specific Tbx4-Cre (Figure 3, B–D), suggesting that COL1α1-expressing fibroblasts were predominantly derived from Tbx4-lineage mesenchymal cells. Following bleomycin-induced lung fibrosis, COL1α1-expressing and Tbx4-lineage fibroblast numbers were visibly increased. Flow cytometry analysis showed that 1.16% and 2.26% of the total lung single cells were COL1α1/tdT double-positive in uninjured and bleomycin-injured (day 21) lungs, respectively (Figure 3E). Moreover, 87% of all GFP+ cells were tdT+ in uninjured lungs, and 95% of total GFP cells were tdT+ in bleomycin-injured lungs. Among tdT+ cells, 50% of the cells are GFP+ in both uninjured and bleomycin-injured lungs (Figure 3F). Together, these results demonstrate that the majority of COL1α1-expressing fibroblasts are Tbx4 lineage–labeled cells and the Tbx4-derived population is a larger population than COL1α1+ cells.
Figure 3. Tbx4-lineage cells in the lung are the major source of COL1α1+ fibroblasts.
(A) Schematic depicting lineage analysis methodology. COL1α1-GFP Tbx4-CreKi Rosa26-tdTomato mice were used for all experiments in this figure. (B–D) Representative histology images of uninjured or bleomycin-treated (d21) adult COL1α1-GFP Tbx4-CreKi Rosa26-tdTomato mouse lung. (E) Representative FACS plots of mouse lung single cells isolated from uninjured or bleomycin-treated (d21) COL1α1-GFP Tbx4-CreKi Rosa26-tdTomato mice. (F) Quantification of tdT+ cells among COL1α1-GFP+ cells, and COL1α1-GFP+ cells within tdT+ cells in E (n = 3 mice analyzed, mean ± SEM). Arrows show cells with overlaps. Scale bars: 1 mm (B) and 10 μm (C and D). a, alveoli; aw, airway.
TBX4-expressing cells proliferate and expand during injury.
We next sought to determine the role of TBX4-expressing cells in regulating fibroblast accumulation in the adult mice. To address this question, we carried out tamoxifen-induced cell lineage tracing experiments to specifically label postnatal TBX4-expressing cell types. Tbx4LME-CreER transgenic mice (22) were bred with Rosa26-tdTomato mice, allowing tamoxifen-regulated tdT labeling of the TBX4-expressing cells (Figure 4A).
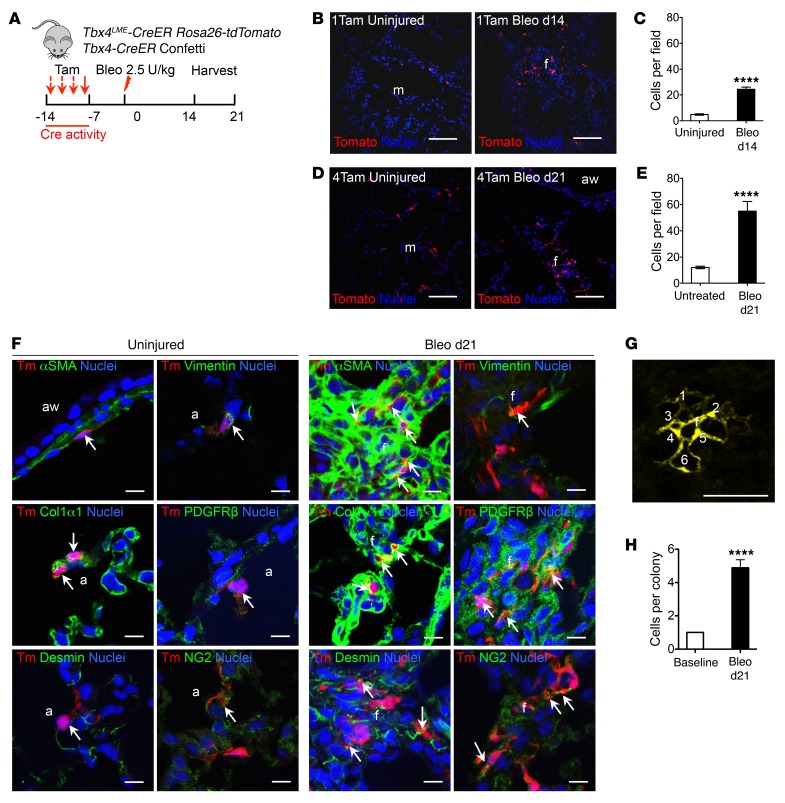
Figure 4. Tbx4-lineage cells are highly proliferative upon bleomycin injury in the lung.
(A) Experimental design for inducible TBX4 cell labeling using 1 or 4 doses of tamoxifen 1 week before bleomycin injury for Tbx4LME-CreER Rosa26-tdTomato mice. Lungs were harvested on d21. Tbx4-CreKi Rosa26-tdTomato (Tam, 20 mg/g/dose) and Tbx4LME-CreER Confetti (Tam, 10 mg/g/dose) were used for all experiments in this figure. (B and D) Representative lung sections from Tbx4LME-CreER Rosa26-tdTomato mice with 1 (B) or 4 (D) doses of tamoxifen injection showing Tbx4-lineage cells in red and nuclei in blue. (C and E) Quantification of Tbx4 cells counted in B and D (from 2 lobes per mouse, n = 9 mice; ****P ≤ 0.0001 by 2-tailed Student’s t test; mean ± SEM). (F) Representative immunofluorescent images of mice from B staining with αSMA, COL1α1, desmin, vimentin, PDGFRβ, and NG2. Arrows show cells with overlap in staining (n = 6 lungs examined). (G) TBX4 cells were marked in Tbx4LME-CreER Confetti mice using 4 doses of tamoxifen. Representative confocal images of a typical colony of TBX4 cells. Cells in clone are enumerated. (H) TBX4 colony size at baseline and d21 after bleomycin injury. n = 9 lungs examined. ****P ≤ 0.0001 by Student’s t test; mean ± SEM. Scale bars: 100 μm (B and D) and 10 μm (F and G). a, alveoli; aw, airway; f, fibrotic foci; m, mesenchyme.
Tamoxifen dose–dependent increases were observed in the percentage of tdT-labeled cells in adult Tbx4LME-CreER Rosa26-tdTomato mice (Supplemental Figure 9A). Mice were then given either 1 or 4 doses of tamoxifen 1 week before bleomycin injury. Lung tissue was harvested for histology at both 14 and 21 days after bleomycin administration (Figure 4A). We detected a significant increase in tdT+ cells in mouse lungs at both 14 and 21 days after bleomycin exposure compared with lungs from untreated control mice (Figure 4, A–E). These tdT+ cells costained with αSMA, COL1α1, desmin, vimentin, PDGFRβ, NG2, and lipid acid, as well as vWF antibodies (Figure 4F and Supplemental Figure 9, C and D). However, tdT+ cells did not costain with SPC, T1a, or SCGB1A1 (Supplemental Figure 9E). Additionally, BrdU labeling at 21 days indicated that TBX4-expressing cells lineage-labeled before injury were proliferating 21 days after bleomycin exposure of Tbx4LME-CreER Rosa26-tdTomato mice (Supplemental Figure 9, F and G).
Next, we crossed Tbx4LME-CreER mice with Confetti mice (34) to determine whether Tbx4-lineage cells are clonally expanded (Figure 4A). In light of the low frequency of lineage labeling within lung tissue, patches of adjacent lineage-labeled cells harboring a common fluorescent protein tag were considered to be clones and to reflect the clonal behavior of TBX4-expressing progenitors. Analysis of clone size as a function of time following bleomycin exposure indicated that Tbx4-lineage cells underwent progressive clonal expansion after bleomycin exposure (Figure 4, G and H, and Supplemental Figure 10). These data demonstrate that Tbx4-lineage fibroblastic progenitors undergo clonal expansion during lung injury and fibrosis. We therefore conclude that TBX4-expressing cells labeled before injury proliferated and expanded.
Tbx4-lineage cells are induced de novo by injury.
In order to test whether TBX4 expression is induced de novo by injury, we carried out experiments by labeling TBX4-expressing cells by tamoxifen after injury. We administered 5 doses of tamoxifen every other day beginning 1 week following bleomycin injection in Tbx4LME-CreER Rosa26-tdTomato mice, and the lungs were harvested on day 21 (Figure 5A). We observed a significant increase of tdT+ cells in bleomycin-treated mice compared with untreated control mice (Figure 5, B and C). The progeny of TBX4-expressing cells in bleomycin-treated mice costained with αSMA, COL1α1, desmin, vimentin, PDGFRβ, and NG2 (Figure 5D). Hence, these data suggest that TBX4 is induced de novo after injury and TBX4-expressing cells labeled after injury contributed to the expansion of Tbx4-lineage cells. TBX4 expression is high in the embryonic mouse lungs (24, 26) and is much lower in adult mouse lungs (22, 26). These data suggest that TBX4 signaling can be re-activated under pathological conditions such as tissue injury.
Figure 5. TBX4 cells are induced de novo upon bleomycin injury in the lung.
(A) Five doses of tamoxifen (20 mg/g/dose) were given to Tbx4LME-CreER Rosa26-tdTomato every other day starting from d7 after bleomycin treatment. Lungs were harvested for histology on d21 after bleomycin. (B) Representative lung sections from Tbx4LME-CreER Rosa26-tdTomato mice in A showing Tbx4-lineage cells in red and nuclei in blue. (C) Quantification of TBX4 cells in B (from 2 lobes per mouse, n = 9 mice; ****P ≤ 0.0001 by Student’s t test; mean ± SEM). (D) Representative images of αSMA, COL1α1, desmin, vimentin, PDGFRβ, and NG2 immunofluorescence staining. Arrows show the overlap cells. Scale bars: 100 μm (B) and 10 μm (D). aw, airway; f, fibrotic foci; m, mesenchyme.
Ablation of TBX4-expressing cells inhibits lung fibrosis.
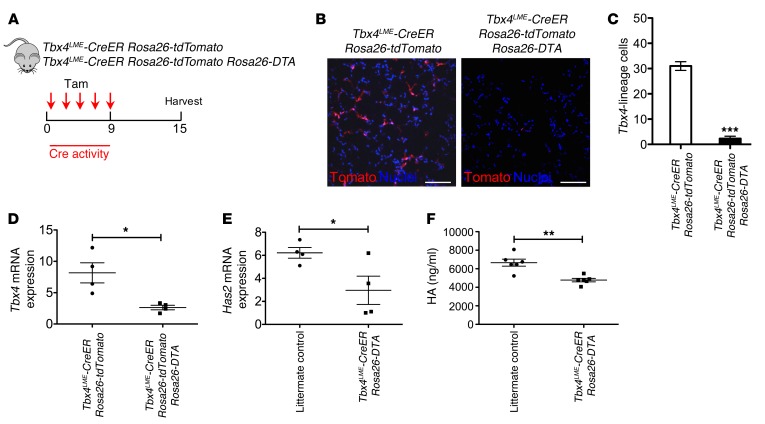
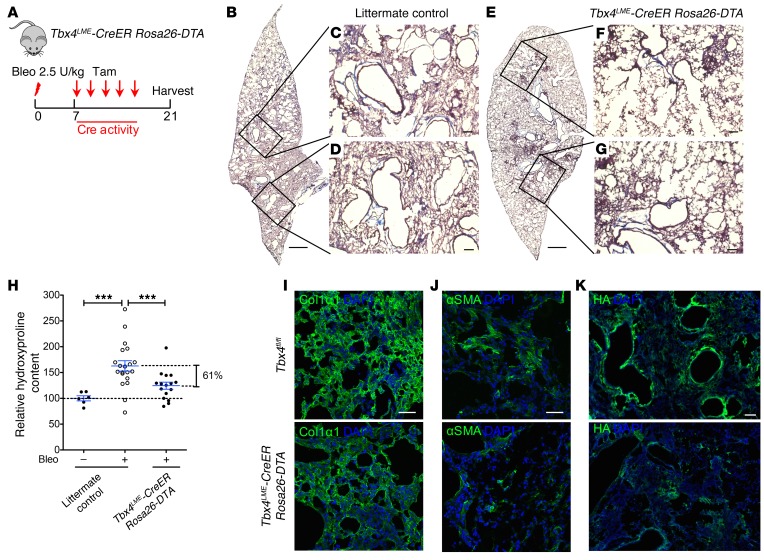
To directly demonstrate the functional role of TBX4-expressing fibroblasts and their cellular progeny in lung fibrosis, Tbx4LME-CreER mice were crossed with Rosa26-Stop-DTA or Rosa26-tdTomato Rosa26-Stop-DTA mice, in which the tamoxifen-inducible, Cre-mediated combination will result in deletion of TBX4-expressing cells. These mice were given 5 doses of tamoxifen for 1 week to induce the ablation of TBX4-expressing cells (Figure 6A). As expected, injection of tamoxifen induced a marked reduction of the progeny of TBX4 cells (tdT+ cells) (Figure 6, B and C), of Tbx4 and Has2 mRNA, and of HA production in lung fibroblasts (Figure 6, D–F). To determine whether there are any structural changes in Tbx4-lineage cell–ablated lungs at baseline, we performed H&E staining, and mean linear intercept calculation to assess for emphysema. Our data suggest that there are no significant structural changes in either uninjured or relatively normal areas of injured lung with short-term TBX4 cell deletion (Supplemental Figure 11, A and B). We detected a significant decrease in collagen content (~61%) in tamoxifen-injected Tbx4LME-CreER Rosa26-DTA mouse lungs 21 days after bleomycin by Masson’s trichrome staining (Figure 7, A–G) and hydroxyproline assay (Figure 7H). Immunofluorescence analysis also revealed decreasing of COL1α1, αSMA, and HA expression (Figure 7, I–K). These data indicate that Tbx4-lineage cells play a critical role in regulating fibrosis in the bleomycin-induced lung injury model. Furthermore, the loss of TBX4-expressing cells and their progeny attenuated interstitial lung fibrosis.
Figure 6. Ablation of TBX4 cells decreases HAS2 and HA expression.
(A) Verification of ablation of Tbx4-lineage cells using Tbx4LME-CreER Rosa26-tdTomato and Tbx4LME-CreER Rosa26-tdTomato Rosa26-DTA mice. Five doses of tamoxifen were injected, and lungs were harvested 1 week later. (B) Representative images were taken from Tbx4LME-CreER Rosa26-tdTomato and Tbx4LME-CreER Rosa26-tdTomato Rosa26-DTA mouse lung (n = 4 lungs examined). (C) Quantification of Tbx4 lineage tracing and ablation, expressed as cells counted in B (n = 4 in each group of mice). (D and E) Tbx4 mRNA (D) and Has2 mRNA (E) expression of lung fibroblasts from Tbx4LME-CreER Rosa26-tdTomato and Tbx4LME-CreER Rosa26-tdTomato Rosa26-DTA were analyzed by quantitative RT-PCR, and normalized by GAPDH (n = 4 mice per group examined). (F) HA contents in conditioned media of lung fibroblasts from Tbx4LME-CreER Rosa26-tdTomato and littermate controls were analyzed by HA ELISA. n = 6 mice per group examined. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by 2-tailed Student’s t test, mean ± SEM.
Figure 7. Ablation of TBX4 cells attenuates pulmonary fibrosis.
(A) Strategy for inducible ablation of TBX4 cells. Tbx4LME-CreER Rosa26-DTA mice were used in the subsequent experiment. The above-mentioned transgenic mice and their WT littermates (8–16 weeks old) were treated with bleomycin (2.5 U/kg), followed by 5 doses of tamoxifen (20 mg/g/injection) every other day starting on d7. The lungs were collected for hydroxyproline content determination and Masson’s trichrome staining on d21 after bleomycin. DTA, diphtheria toxin fragment A. (B–G) Representative Masson’s trichrome staining images of bleomycin-treated (d21) adult littermate control (B–D) or Tbx4LME-CreER Rosa26-DTA (E–G) mouse lung. (H) Targeting TBX4 cells reduced lung fibrosis (means ± SEM, ***P ≤ 0.001, 1-way ANOVA with Bonferroni test; uninjured littermate control, n = 6; bleomycin [Bleo] littermate control, n = 20; Tbx4LME-CreER Rosa26-DTA, n = 16). (I–K) Representative images of COL1α1 (I), αSMA (J), and HA (K) immunofluorescence staining of d21 Tbx4LME-CreER Rosa26-DTA mouse lung. n = 6 mice per group examined. Scale bars: 1 mm (B and E) and 100 μm (C, D, F, G, and I–K).
Conditional Tbx4 knockout in mesenchymal cells inhibits fibrosis.
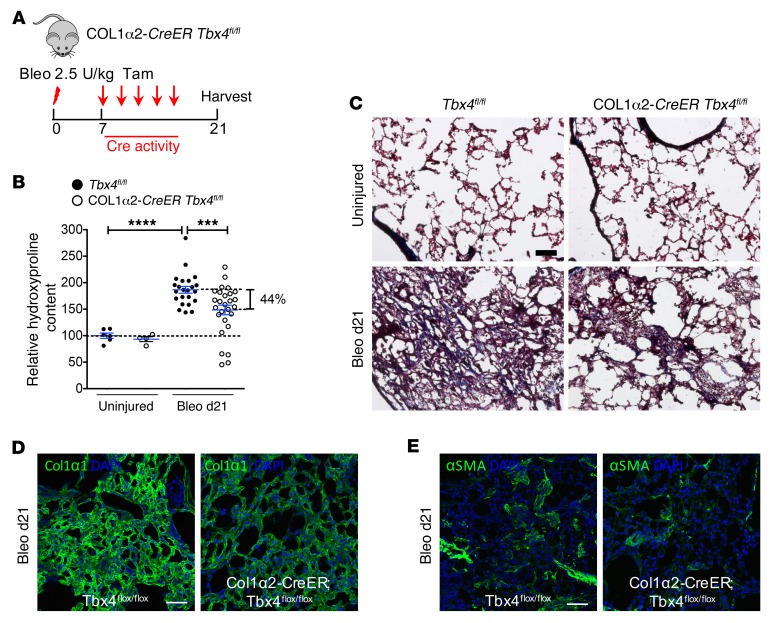
To directly demonstrate the role for TBX4 in lung fibrosis, we determined the impact of deletion of Tbx4 in either fibroblast or myofibroblast populations. Col1a2-CreER transgenic mice were crossed with Tbx4fl/fl mice (19), resulting in Tbx4 gene deletion in COL1α2-expressing cells (Figure 8A). Five doses of tamoxifen were administered 1 week after bleomycin. Hydroxyproline content in Col1a2-CreER Tbx4fl/fl mouse lungs was significantly decreased (~44%) in comparison with control mice 21 days after bleomycin administration (Figure 8B). Masson’s trichrome and immunofluorescence staining of the lung for collagen, COL1α1, and αSMA also showed similar results (Figure 8, C–E).
Figure 8. Loss of TBX4 expression in COL1α2-expressing cells attenuates pulmonary fibrosis.
(A) Strategy for inducible knockout of TBX4 expression in COL1α2+ cells. Col1a2-CreER Tbx4fl/fl mice were used in these experiments. The above-mentioned mice and their WT littermates (8–16 weeks old) were treated with bleomycin (2.5 U/kg), followed by 5 doses of tamoxifen (20 mg/g/injection) every other day starting on d7. The lungs were collected for hydroxyproline content determination on d21 after bleomycin. (B) Knockdown of Tbx4 in COL1α2-expressing cells decreased hydroxyproline content (means ± SEM, ***P ≤ 0.001, ****P ≤ 0.0001, n = 6 in uninjured Tbx4fl/fl group, n = 4 in uninjured Col1a2-CreER Tbx4fl/fl group, n = 23 in bleo Tbx4fl/fl group, n = 27 in bleo Col1a2-CreER Tbx4fl/fl group). (C) Representative Masson’s trichrome staining from lungs at d21 after bleomycin injection, showing decreased collagen deposition (blue) in Col1a2-CreER Tbx4fl/fl. (D and E) Representative images of COL1α1 and αSMA antibody staining for bleo d21 Col1a2-CreER Tbx4fl/fl mouse lung. n = 6 mice per group examined. Scale bars: 100 μm (C–E).
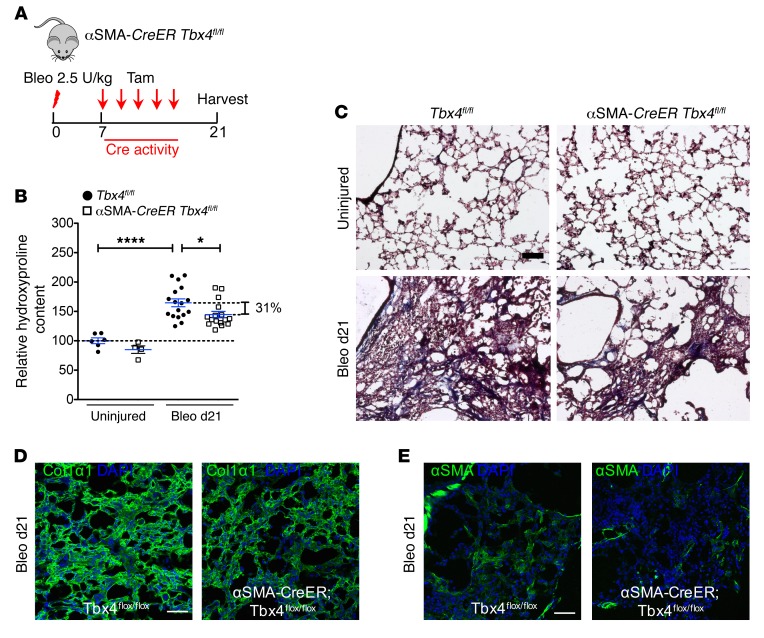
We then crossed Tbx4fl/fl mice with Acta2-CreER mice (referred to here as αSMA-CreER mice) in order to delete the Tbx4 gene in αSMA-expressing myofibroblasts upon tamoxifen administration (Figure 9A). Similar to the observation in COL1α2-expressing cells, αSMA-CreER Tbx4fl/fl mouse lungs had reduced collagen content and αSMA expression compared with control mice 21 days after bleomycin by hydroxyproline assay (~31%) and Masson’s trichrome staining and immunofluorescence (Figure 9, C–E).
Figure 9. Inhibition of Tbx4 in αSMA+ cells attenuates lung fibrosis.
(A) Strategy for inducible knockout of Tbx4 expression in αSMA+ cells. Acta2-CreER Tbx4fl/fl mice were used in these experiments. The above-mentioned mice and their WT littermates (8–16 weeks old) were treated with bleomycin (2.5 U/kg), followed by 5 doses of tamoxifen (20 mg/g/injection) every other day starting on d7. The lungs were collected for hydroxyproline content determination on d21 after bleomycin. (B) Knockdown of Tbx4 in αSMA-expressing cells decreased hydroxyproline content (means ± SEM, *P ≤ 0.05, ****P ≤ 0.0001, n = 6 in uninjured Tbx4fl/fl group, n = 4 in uninjured Acta2-CreER Tbx4fl/fl group, n = 17 in bleo Tbx4fl/fl group, n = 16 in bleo Acta2-CreER Tbx4fl/fl group). Hydroxyproline content values were expressed as percentage of control Tbx4fl/fl group, which is set to 100%. One-way ANOVA with Bonferroni test was used. (C) Representative Masson’s trichrome staining from lungs at d21 after bleomycin injection, showing decreased collagen deposition (blue) in Acta2-CreER Tbx4fl/fl mice. (D and E) Representative images of COL1α1 and αSMA antibody staining for bleo d21 Acta2-CreER Tbx4fl/fl mouse lung. n = 6 mice per group examined. Scale bars: 100 μm (C–E).
The lungs from the uninjured group of Col1a2-CreER Tbx4fl/fl and αSMA-CreER Tbx4fl/fl mice, which received tamoxifen injection to knock down the TBX4 expression, showed normal lung architecture (Supplemental Figure 12, A–G). The decreased expression of TBX4 in lung fibroblasts of the uninjured group of Col1a2-CreER Tbx4fl/fl and αSMA-CreER Tbx4fl/fl mice was verified (Supplemental Figure 12, H and K). Similar to TBX4 expression, the Has2 mRNA expression and HA production of the lung fibroblasts were reduced (Supplemental Figure 12, L and M). These results indicate that the short-term Tbx4 gene deficiency under the static conditions does not affect lung structure, although the impact of long-term Tbx4 gene deficiency needs to be determined.
In contrast, bleomycin treatment of tamoxifen-exposed Ng2-CreER Tbx4fl/fl mice revealed that deletion of TBX4 expression in NG2+ cells had no impact on fibrosis following bleomycin treatment (Supplemental Figure 13, A and B), consistent with the data showing that NG2/tdT double-positive cells represent a minor population of αSMA-expressing cells (Supplemental Figure 2, B and C).
In order to determine a TBX4 role in myofibroblast transformation, collagen production, and fibroblast viability and apoptosis, we performed additional in vitro experiments. In IPF lung fibroblasts, the knockdown of Tbx4 by transfection of Tbx4 siRNA decreased αSMA protein expression by Western blot analysis (Supplemental Figure 14A; see supplemental material for full uncut gels). The secreted COL1α1 production in IPF lung fibroblast supernatants was reduced in the presence and absence of TGF-β1 treatment following Tbx4 knockdown (Supplemental Figure 14B). In regard to TBX4-mediated fibroblast viability, we tested the cell viability upon Tbx4 knockdown by Calcein AM cell viability assay. The viability of Tbx4 knockdown cells was decreased as compared with control siRNA (Supplemental Figure 14C). In order to investigate whether loss of Tbx4 affects IPF lung fibroblast apoptosis, we performed a caspase-3 activity assay. We found that upon transfection of Tbx4 siRNA, the caspase-3 activity in IPF lung fibroblasts was elevated in comparison with control siRNA.
Collectively, these data strongly suggest that TBX4 plays an important role in lung fibroblast invasion, SMA expression, collagen production, proliferation, and inhibition of apoptosis. Furthermore, TBX4 signaling drives fibrogenesis in bleomycin-induced lung fibrosis in vivo.
TBX4 regulates fibroblast invasion through HAS2.
Our previous studies have shown that an invasive phenotype of fibroblasts was required for severe fibrosis. Fibroblasts isolated from lung tissue of IPF patients were more invasive than those from lung tissue of normal donors (35, 36). We next wanted to determine whether TBX4 plays a role in fibroblast invasion. Fibroblasts were isolated from Tbx4LME-Cre Rosa26-tdTomato mouse lungs and cultured in vitro for 3 passages, and the tdT+ and tdT– cells were separated by flow cytometry. Immunofluorescence staining of tdT+ cells showed costaining with αSMA, COL1α1, desmin, vimentin, PDGFRβ, and NG2 (Supplemental Figure 15). Microarray analysis showed that tdT+ cells had a more than 2-fold increase of Tbx4 mRNA expression (Figure 10A). Using the Matrigel invasion assay, we found that Tbx4-lineage cells were significantly more invasive than non–Tbx4-lineage cells (Figure 10B).
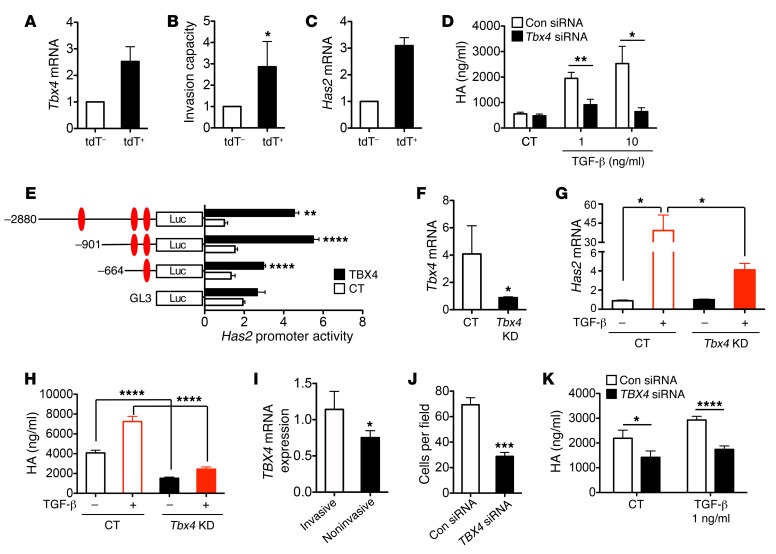
Figure 10. TBX4 regulates fibroblast invasion through regulation of Has2.
(A) Tbx4 mRNA expression was increased in sorted tdT+ cells from Tbx4-CreTg Rosa26-tdTomato mice. (B) tdT+ fibroblasts were more invasive. Invasion capacity values were expressed as relative fold change of control, which is set to 1. (C) Has2 mRNA level was elevated in tdT+ fibroblasts. (D) Tbx4 siRNA reduced TGF-β1–induced HA release by mouse fibroblast 3T3 cells. Cells were transfected with either Tbx4 siRNA or control siRNA (Con). Forty-eight hours later cells were treated with TGF-β1 at 1 ng/ml or 10 ng/ml concentration for 24 hours. HA was measured in conditioned media. CT, control. (E) TBX4 enhanced Has2 promoter activity. All Has2 promoter constructs showed increased activity over the promoterless vector pGL3 when cotransfected with a TBX4-expressing vector in HEK 293 cells. (F–H) MEF cells were isolated from E14 embryo of CMV-Cre Tbx4f/w and WT mice. MEF cells at passage 3 were used for experiments. (F) Tbx4 mRNA levels were compared between Tbx4-deficient and WT MEFs. (G) MEF cells were treated with or without TGF-β1 (5 ng/ml) overnight. Has2 mRNA expression was analyzed using RT-PCR. (H) HA in supernatants was measured with HA ELISA. (I) TBX4 gene expression was elevated in invasive human fibroblasts from IPF patients. (J) Invasion assay for IPF fibroblasts was performed 48 hours after transfection of control and TBX4 siRNA. Knocking down TBX4 with siRNA reduced the invasiveness of IPF fibroblasts. (K) Knocking down TBX4 with siRNA reduced HA production of primary fibroblasts from IPF patients with or without 1 ng/ml TGF-β1 treatment after 48 hours. n = 4 in each group were analyzed. All results are the mean of triplicate experiments ± SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, as analyzed by 2-tailed Student’s t test (B, F, I, and J) or 1-way ANOVA with Bonferroni test (D, E, G, H, and K).
We previously determined that HAS2 regulates fibroblast invasion. To determine whether the increased invasiveness of Tbx4-lineage fibroblasts was due to enhanced HAS2 expression, we compared the expression levels of HAS2 between Tbx4-lineage and non–Tbx4-lineage fibroblasts, and found that Has2 mRNA was increased in Tbx4-lineage fibroblasts compared with non–Tbx4-lineage cells (Figure 10C and Supplemental Table 1). Furthermore, knocking down Tbx4 with a specific siRNA decreased TGF-β–induced HA production in 3T3 mouse fibroblasts, suggesting that Has2 is regulated by TBX4 (Figure 10D) in agreement with results from the allantois (37). There are 3 putative T-box response elements in the Has2 promoter (37). Using a luciferase promoter assay, we showed that TBX4 could significantly induce the activity of vectors containing Has2 promoters (Figure 10E), suggesting that TBX4 regulates Has2 through transcriptional regulation.
In addition, we obtained MEF cells from CMV-Cre Tbx4f/w mice, designated Tbx4KD cells. Tbx4 mRNA expression in Tbx4KD MEF cells was significantly less than that in control MEF cells (Figure 10F). TGF-β–induced Has2 mRNA expression was decreased in Tbx4KD MEF cells (Figure 10G). HA production in Tbx4KD MEFs was dramatically reduced when treated with or without TGF-β (Figure 10H). Furthermore, we found that the TBX4 gene expression was elevated in invasive human fibroblasts from IPF patients when compared with noninvasive human fibroblasts (Figure 10I). In lung fibroblasts from IPF patients, TBX4 siRNA transfection inhibited both invasiveness and TGF-β–induced HA production (Figure 10, J and K). Taken together, these data show that TBX4 mediates fibroblast invasion through HAS2.
Discussion
This study identifies a TBX4-driven fibrogenic cell population as a heterogeneous mesenchymal population, containing interstitial fibroblasts, vascular and airway smooth muscle cells, some pericytes, and a few endothelial cells in the adult mouse lung in normal and disease states. TBX4-labeled cells did not give rise to any lung cells expressing epithelial cell markers. More importantly, Tbx4-lineage cells expanded, proliferated, and formed organized clonal patches during lung fibrosis in mice in vivo. Almost all of the αSMA+ myofibroblasts and COL1α1+ fibroblasts were Tbx4-derived cells, suggesting a critical role for this population of cells in lung fibrosis. This is further supported by data that lung fibrosis is significantly reduced by ablation of Tbx4-lineage cells. When Tbx4 was deleted in SMA- or COL1α2-expressing fibroblasts, lung fibrosis was also significantly reduced. Thus several lines of evidence support a nonredundant role for TBX4 in regulating the pathobiology of lung fibrosis. We further showed that TBX4 mediated fibroblast invasiveness and HA production by transcriptional regulation of HAS2.
A major finding in this study was that virtually all αSMA-expressing myofibroblasts as well as COL1α1-expressing fibroblasts were derived from TBX4-expressing cells. These data suggest that within the lung, a local lineage of interstitial resident fibroblasts promote fibrogenesis. The observation is consistent with recent studies in skin fibrosis (2, 38), lung fibrosis (3), and liver fibrosis (33). This appears to be in contrast to kidney fibrosis, where only 50% of myofibroblasts were suggested to arise from resident fibroblasts (4). The demonstration that Tbx4-lineage cells are devoid of all epithelial cell markers is in line with previous work (5) suggesting that cell lineage commitment does not cross between epithelial and stromal compartments in lung fibrosis. Epithelial-mesenchymal transition (EMT) has been suggested in the pathogenesis of cardiac fibrosis (39), lung fibrosis (13, 14, 40, 41), and kidney fibrosis (42, 43). Although our study did not directly examine the role of EMT in lung fibrosis, the data did not support a major role for EMT in lung fibrosis. In addition, extrapulmonary contribution of fibroblasts and myofibroblasts is not regulated by Tbx4-derived cells. Bone marrow–derived stromal progenitor cells such as fibrocytes (44) have been suggested in lung fibrosis (9, 45). A very small fraction of Tbx4-derived cells were observed in the bone marrow. Collectively, these data suggest that the majority of myofibroblasts and fibroblasts that accumulate during fibrosis are Tbx4 lineage–derived mesenchymal intrapulmonary cells regardless of the particular cell surface expression marker.
A critical goal for the field of tissue fibrosis is to define fibroblast origins under homeostatic and disease states. Current views on the origins of fibroblasts during development and in diseases have been established mostly based on antibody staining, rather than the use of transgenic reporter markers or systematic quantitative analysis of fibroblast lineages present in the lung. A few recent studies have attempted to genetically trace the origin of myofibroblasts during tissue fibrosis. For example, resident perivascular Adam12- and Gli1-lineage cells, and Foxd1-lineage pericytes, stained positive for αSMA antibody after tissue injury (2, 3, 17). Among them, Foxd1-lineage pericytes coexpress Col1a1 reporter GFP (3). Furthermore, Engrailed 1–derived cells are positive for collagen type I antibody staining in skin fibrosis (38). In the current study, we revealed that Tbx4-lineage cells constitutively labeled from the embryonic stage or inducibly labeled postnatally can differentiate into heterogeneous stromal cell types including αSMA+ and COL1α1+ cells by antibody staining. To confirm our antibody staining results, we used several transgenic reporter lines, including αSMA-GFP and COL1α1-GFP mice, to detect the colocalization of αSMA- and Col1α1-expressing cells within the Tbx4-lineage cells by means of confocal microscopy and FACS analysis. Our observations showed that αSMA-expressing cells are dramatically increased in the fibrotic lung. Almost all of the αSMA-expressing cells (over 90%) are Tbx4-lineage cells. We also detected a large portion of Tbx4-lineage-only (TBX4+αSMA–) cells, indicating that not only αSMA+ cells but other non-αSMA mesenchymal cells accumulate during fibrosis. This is in line with a previous study showing that multiple stromal populations contribute to pulmonary fibrosis, including αSMA, vimentin, desmin, PDGFRβ, and NG2, by using antibody staining (5). In addition, we showed that the vast majority of COL1α1+ cells are Tbx4-lineage cells. COL1α1+ cells are considered resident fibroblasts (3, 17, 33), while some data suggest that circulating fibrocytes can express COL1α1 (44, 46). The data presented in this study suggest that the majority of COL1α1+ cells are resident fibroblasts, although a small proportion of COL1α1 single-positive cells, the circulating fibrocytes, were observed.
Pericytes are defined as mesenchymal cells that share a common basement membrane with endothelial cells (47). They have been suggested as 1 of the sources of fibroblasts during tissue fibrosis (17, 28). Our lineage labeling data demonstrate that a maximum of 20% of NG2+ cells are Tbx4-lineage cells. Considering that the majority of αSMA+ cells are Tbx4-lineage cells, our results suggest that NG2+ cells represent a minor population of αSMA+ cells. This observation is consistent with a previous study using Ng2-CreER mice, which demonstrated that NG2+ pericytes do not express high levels of αSMA in fibrotic lungs (5). These also support recent reports that NG2 may not be a reliable marker for all pericytes (3, 4, 48). Antibody staining with another pericyte marker, PDGFRβ, showed much more colocalization with Tbx4-lineage cells than NG2 staining. This suggested that PDGFRβ may be a better marker for pericytes than NG2. Collectively, these data suggest that NG2+ pericytes are not the major contributor to myofibroblasts in lung fibrosis.
Tbx4-lineage progenitor cells not only contain αSMA+, COL1α1+, and NG2+ cells, as we have demonstrated with transgenic gene reporter mice, but also include a few endothelial cells by antibody staining. This population does not increase during fibrosis, consistent with a report that PECAM-1 costained with Tbx4-lineage cells in embryonic mouse lung (26). It has been reported that mesenchymal-endothelial transition contributes to neovascularization during heart injury (49). As TBX4 is expressed early in lung mesenchyme, the data suggest that mesenchymal-to-endothelial transition might occur during development. It would be interesting to use additional lineage tracing tools to determine when Tbx4-lineage cells start to express endothelial cell markers. Whether a portion of mesenchymal cells differentiate to endothelial cells or a small portion of endothelial cells require TBX4 signaling is unknown.
We used mesenchyme-specific clonal analysis (22) to examine expansion of TBX4-expressing cells during fibrosis. We found that TBX4-expressing cells expanded clonally. Within 3 weeks after bleomycin injury, clone size can be as many as 6 cells. Furthermore, these patches seemed to be monoclonal, indicating that regeneration foci are derived from a single Tbx4 progenitor. These data suggest that the mesenchymal progenitors during fibrogenesis are similar to the embryonic-stage mesenchymal cells, which are highly proliferative and progressively expand (22). The adult mesenchymal progenitors are unlike the alveolar stem cells, which undergo long-term self-renewal and diffuse proliferation (50, 51). These findings suggest that mesenchymal progenitors proliferate and self-renew during lung fibrogenesis. These data support the previously suggested concept that during lung fibrosis, embryonic gene programs are reactivated (52). However, more comprehensive clonal analysis of Tbx4 progenitor cells such as long-term tracing, single dose versus multiple doses of tamoxifen, before and after bleomycin injury, and coexpression with other mesenchymal/lineage markers is needed to further delineate the clonal nature of these cells.
As a mesenchymal transcription factor, TBX4 could control downstream gene expression and impact key fibroblast effector functions. In order to gain insights into possible target genes, we analyzed Tbx4-lineage and non–Tbx4-lineage cell gene expression, and identified mesenchymal genes that could be regulated by TBX4. Among them, Has2 is upregulated in TBX4-expressing cells. Has2 is 1 of the 3 HA synthase genes that have been identified. We have previously shown that fibroblasts from Acta2-Has2 transgenic mice (referred to here as αSMA-Has2 mice) are more invasive, while fibroblasts from Fsp1-Cre Has2fl/fl mice (also known as S100a4-Cre Has2fl/fl mice) had diminished invasive capacity (35). αSMA-Has2 mice showed increased fibrosis after bleomycin injury, and mice with conditional deletion of Has2 in mesenchymal cells exhibited less fibrosis (35). We then analyzed how TBX4 regulates Has2 and impacts the fibroblast invasive phenotype. We found that TBX4 regulates both fibroblast invasiveness and HA production. TBX4 binds to the Has2 promoter and triggers Has2 gene expression. These data establish TBX4 as a proximal regulator of Has2 expression, HA production, and fibroblast invasiveness. Interestingly, the array data suggest that TBX4 may regulate additional fibroblast and ECM remodeling genes, including Fgf, Pdgfr, Mmp, and Adam family genes. It has been shown that TBX4 and TBX5 are linked to the activity of FGF, BMP, and WNT signaling pathways that are required for limb outgrowth and patterning (53). TBX4 and TBX5 are critical for expression of mesenchymal FGF10 in embryonic mouse lung (54). It has been reported that TBX4 and TBX5 trigger limb formation by the induction of both Wnt and Fgf (55). There are no reports to date directly linking Tbx4 and Mmp gene expression, although inhibition of the T-box transcription factor Brachyury showed downregulation of Mmp2 and Mmp24 in the context of EMT in cancer (56). Moreover, T-BET, another T-box transcription factor, was found increased together with MMP3 when TGF-β was blocked in human gut (57). Further investigations into the interrelationships between TBX4 and matrix remodeling genes are warranted.
An intriguing possibility is raised of a hierarchical relationship of TBX4-lineage fibroblasts with other mesenchymal populations, such as FGF10+, GLI1+, ADAM12+, PDGFRα+, and FOXD1+ cells. TBX4 is a potential upstream regulator of these genes and is detected as early as E9.25 exclusively in lung mesenchyme without epithelial expression (18, 20, 22, 24, 58, 59). TBX4 induces mesenchymal genes and the pluripotent stem cell–related transcription factors (Gata2 and Gata3; Smad1, Smad2, Smad3, and Smad5; and Snail) in adult mice, suggesting that TBX4 can be a critical driver of fibroblast differentiation. Furthermore, the identification of TBX4 as a regulator of Has2 expression points to this pathway as a potential target for therapeutic intervention.
Collectively, these data demonstrate that Tbx4-lineage cells represent most of the resident mesenchymal progenitors, expand during fibrosis, and form clonal patches. TBX4 drives fibroblast matrix production and invasiveness through the regulation of Has2. Further understanding the origin, signaling partners, and functions of Tbx4-derived mesenchymal cells could provide new insights into the pathogenesis of progressive fibrotic diseases.
Methods
Mice.
Tbx4 lung enhancer-Cre transgenic mice (Tbx4LME-Cre or Tbx4-CreTg) and Tbx4LME-CreER transgenic mice were recently described (22). Tbx4-Cre knock-in (Tbx4-CreKi) mice were reported (24). Tbx4-Cre Rosa26-tdTomato mouse lines were obtained by crossing of Tbx4-Cre knock-in (Tbx4-CreKi) mice or transgenic mice with lineage reporter Rosa26-tdTomato mice (25) (Brigid L.M. Hogan, Duke University, Durham, NC). To obtain Tbx4LME-CreER Rosa26-tdTomato, Tbx4LME-CreER mice were crossed to Rosa26-tdTomato mice. Tbx4LME-CreER Confetti mice were obtained by crossing of Tbx4LME-CreER mice with Rosa26 Confetti mice (34) (Jackson Laboratory). Triple-heterozygous αSMA-GFP Tbx4-Cre Rosa26-tdTomato mice were created by crossing of Tbx4-Cre Rosa26-tdTomato mice with Acta2-GFP mice (referred to here as αSMA-GFP mice) (60) (Robert N. Fariss, National Eye Institute, Bethesda, MD). Triple-heterozygous Tbx4-Cre Rosa26-tdTomato mice were bred with Col1a1-GFP mice (referred to here as COL1α1-GFP mice) (33) (David A. Brenner, University of California, San Diego, La Jolla, CA) to generate COL1α1-GFP Tbx4-Cre Rosa26-tdTomato. Tbx4-Cre Rosa26-tdTomato mice were bred with Ng2-YFP mice (4) (Raghu Kalluri, University of Texas MD Anderson Cancer Center, Houston, TX) to create the Ng2-YFP Tbx4-Cre Rosa26-tdTomato triple-heterozygous mice. Tbx4LME-CreER mice were bred with Rosa26-DTA mice (61) (Jackson Laboratory) to generate Tbx4LME-CreER Rosa26-DTA mice.
Tbx4fl/fl mice were described previously (20). Acta2-CreER mice (referred to here as αSMA-CreER mice) (62) were crossed with Tbx4fl/fl mice to create the αSMA-CreER Tbx4fl/fl mice. Col1α2-CreER Tbx4fl/fl mice were generated by breeding of Col1a2-CreER mice (referred to here as COL1α2-CreER mice) (63) (Jackson Laboratory) with Tbx4fl/fl mice. Conditional NG2 cell–specific Tbx4 knockout mice (Ng2-CreER Tbx4fl/fl mice) were generated by breeding of Ng2-CreER (also known as Cspg4-CreER) mice (64) (Jackson Laboratory) with Tbx4fl/fl mice. CMV-Cre mice were from Jackson Laboratory. Tamoxifen (Sigma-Aldrich) dissolved in corn oil was injected i.p. at 1–5 doses of 10 μl/g or 20 μg/g body weight to induce Cre-mediated recombination. All mice were on a C57BL/6 background.
Murine lung fibrosis model.
Mice (both male and female), 8 to 16 weeks old, were subjected to bleomycin-induced lung injury (35, 36). Bleomycin at 2.5 U/kg was injected intratracheally. Mouse lungs were harvested on day 14 or day 21 for histology, hydroxyproline assay, and single-cell isolation.
Histology, immunofluorescence, and confocal imaging.
Immunofluorescent staining on frozen sections (5 μm or 12 μm) was performed using primary antibodies to αSMA (catalog A2547, clone 1A4, Sigma-Aldrich), desmin (catalog RB9014-P0, Thermo Scientific), NG2 (catalog AB5320, Millipore), vimentin (catalog sc-7557, Santa Cruz Biotechnology), Col1α1 (catalog NB600-408, Novus Biologicals), PDGFRβ (catalog 04-397, Millipore), vWF (catalog Ab6994, Abcam), lipid acid (Vybrant DiO Cell-Labeling Solution, catalog v22886, Life Technologies), SPC (catalog AB3786, Millipore), Scgb1a1 (a gift of the Barry Stripp laboratory, Cedars-Sinai Medical Center, Los Angeles, CA), T1 (clone 8.1.1-c, Developmental Studies Hybridoma Bank), BrdU (catalog OBT0030, Accurate Chemical and Scientific), biotinylated recombinant human aggrecan (staining for HA, custom order from R&D Systems Inc., lot PQO051505A), and associated Alexa Flour 488–conjugated secondary antibodies (Life Technologies) per manufacturer instructions. RFP was stained with paraffin lung sections (catalog 600-401-379, Rockland). Stained sections were imaged using a Zeiss 780 reverse Laser Scanning Confocal Microscope.
BrdU incorporation assay.
Cell proliferation was quantified by BrdU labeling. Briefly, BrdU (5 mg/ml, catalog B-9285, Sigma-Aldrich) was injected i.p. into bleomycin-treated Tbx4LME-CreER Rosa26-tdTomato mice at a dose of 10 μl/g body weight at day 21. After 3 hours, lungs were collected and fixed by 10% formaldehyde. Lungs were embedded with OCT compound (Sakura Finetek). Frozen sections underwent antigen retrieval by application of 1.5 N HCl to the section for 30–45 minutes. Sections were then processed for immunofluorescent staining with anti-BrdU antibody (1:250; catalog OBT0030, Accurate Chemical and Scientific), followed by Alexa Fluor 488–conjugated secondary antibody (1:500; Invitrogen).
Scale processing and imaging.
Tbx4LME-CreER Confetti mouse lungs were fixed with 10% formalin for 4–16 hours and washed with PBS overnight. Tissues were transferred to Scale A2 and kept on a rocker at 4°C for 1–2 weeks until maximal tissue clearing occurred. Whole-mount samples immersed in Scale A2 solution were imaged on the inverted Zeiss 780 confocal microscope in a 35-mm glass-bottom microwell dish with a ×20 objective. Z-stack and tiled images were acquired from large areas and volumes of tissue. Images were processed by ZEN software.
Hydroxyproline assay.
Collagen content in lung tissue was measured with the conventional hydroxyproline method (65). The ability of the assay to completely hydrolyze and recover hydroxyproline from collagen was confirmed using samples containing known amounts of purified collagen.
Fibroblast isolation and culture.
Primary fibroblasts were derived from mouse lungs as described previously (66). The mouse primary fibroblasts were used from 3 to 6 passages. Human lung fibroblasts were isolated from lung transplant explants obtained from patients with IPF as described previously (35, 66, 67). All cases fulfilled multidisciplinary diagnostic criteria described in the American Thoracic Society/European Respiratory Society consensus statement. In addition, pathological confirmation was obtained for every case. IPF was confirmed by the identification of a usual interstitial pneumonia under the light microscope. Patient demographics were as follows: 2 females and 2 males; ages between 51 and 78 with median age 67; percentage forced vital capacity (FVC%) 51–78, median 57; and percentage diffusing capacity factor of the lung for carbon monoxide (DLCO%) 18–42, median 30.75. Samples of whole lung tissue were obtained at the time during orthotopic lung transplantation surgery. Samples were immediately processed following removal from the body. The specimens were cut into small pieces (~1 mm in diameter), and cultured in DMEM supplemented with 15% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml gentamicin, and 0.25 μg/ml amphotericin B. The cells of passages 4–8 were used for invasion assay, siRNA interference assays, and COL1α1 and HA amount measurements. Four cell lines were analyzed per group.
Matrigel invasion assay.
The invasive behavior of Tbx4-lineage and non–Tbx4-lineage fibroblasts isolated from Tbx4-Cre Rosa26-tdTomato mouse lungs was analyzed essentially as described previously (35). Equal numbers of fibroblasts (5 × 104) in 500 μl of 10% FBS complete medium were plated onto the BioCoat Matrigel Invasion Chamber (BD Biosciences). Seven hundred fifty microliters of 10% FBS complete medium with 10 ng/ml PDGF (120-HD-001, R&D Systems Inc.) was added to the bottoms of wells. After 24 hours of incubation in a CO2 incubator, media were removed, and the polycarbonate filters with the invaded cells were washed once with PBS followed by fixing and staining with the Protocol Hema 3 stain set. Matrigel matrix and noninvading cells on the upper surface of the filter were removed by wiping with a cotton swab, and the filters were removed from the insert by a scalpel blade and were mounted onto glass slides. The invading cells of each sample were counted in 9 randomly selected fields of duplicate filters under a microscope at ×400 magnification. Tbx4 siRNA effects on 3T3 cell line and human IPF lung fibroblast invasion were assessed using 30 nM siRNA. The cells were transfected with siRNAs for 48 hours before the invasion assay was performed.
MEF isolation and culture.
Mouse embryonic fibroblast cells were derived from E14.5 embryos of CMV-Cre Tbx4f/w or control (CMV-Cre only or Tbx4f/w only) mice as described previously (68). The cells were used from 3 to 6 generations. MEFs were treated with 5 ng/ml TGF-β1 overnight. RNA was isolated for RT-PCR, and supernatant was collected for HA ELISA.
Flow cytometry.
Mouse lung or bone marrow was isolated from Tbx4LME-Cre Rosa26-tdTomato, αSMA-GFP Tbx4LME-Cre Rosa26-tdTomato, Col1α1-GFP Tbx4LME-Cre Rosa26-tdTomato, and NG2-YFP Tbx4LME-Cre Rosa26-tdTomato mice after perfusion. Single mouse lung cells were dissociated using a mouse lung dissociation kit and gentleMACS dissociator (Miltenyi Biotec). Single bone marrow cells from Tbx4LME-Cre Rosa26-tdTomato mice were dissociated with a pipette, followed by rbc lysis. Cells were analyzed with Fortessa (BD Biosciences). Fluorescence intensity is expressed in arbitrary units on a logarithmic scale or on a linear scale for forward scatter.
Cultured passage 3 fibroblasts from untreated Tbx4LME-Cre Rosa26-tdTomato mice were sorted for tdT+ and tdT– cells with FACS Aria 3 (BD Biosciences) to 95%–98% purity.
Quantitative real-time PCR.
Real-time RT-PCR was used to quantify the relative mRNA levels of Has2 in CMV-Cre Tbx4f/w or control MEF cells with or without human recombinant TGF-β1 treatment using gene-specific primers. In brief, total RNA was purified using RNAqueous-4PCR kit (Life Technologies) and was reversed to cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies) according to the manufacturer’s instructions. Has2 gene levels in the resultant cDNAs were examined using the ABI 7500 Fast Detection system (Applied Biosystems) with SYBR Green as fluorescent dye enabling real-time detection of PCR products according to the manufacturer’s protocol (Power SYBR Green PCR Master Mix, Applied Biosystems). The relative expression levels of the gene were determined against Gapdh levels in the samples. The following primers were used: mouse Has2 (GenBank accession no. NM_008216) forward, 5′-ACGACGACCTTTACATGATGGA-3′; mouse Has2 reverse, 5′-GATGTACGTGGCCGATTTGCT-3′; mouse Tbx4 (GenBank accession no. NM_011536.2) forward, 5′-TCACTGGATGCGGCAGTTGGTCTCT-3′; mouse Tbx4 reverse, 5′-CACGTGGGTGCAAAAGGCTGTGTTT-3′; mouse Gapdh forward, 5′-ATCATCTCCGCCCCTTCTG-3′; and mouse Gapdh reverse, 5′-GGTCATGAGCCCTTCCACAAC-3′.
Affymetrix cDNA microarray.
Cultured passage 3 fibroblasts from (untreated) Tbx4LME-Cre Rosa26-tdTomato mice were sorted for tdT+ and tdT– cells with FACS Aria 3 (BD Biosciences) to 95%–98% purity. Total RNA was assessed for quality with Agilent 2100 Bioanalyzer G2939A (Agilent Technologies) and Nanodrop 8000 spectrophotometer (Thermo Scientific/Nanodrop). Hybridization targets were prepared with the Ovation Pico WT Amp V2 kit (NuGen) and the Encore Biotin Module (NuGen) from total RNA, hybridized to GeneChip Mouse 430 2.0 arrays in Affymetrix GeneChip hybridization oven 645, washed in Affymetrix GeneChip Fluidics Station 450, and scanned with Affymetrix GeneChip Scanner 7G according to standard Affymetrix GeneChip Hybridization, Wash, and Stain protocols.
Data were background-subtracted and normalized within the array using the LOESS normalization by GeneSpring GX 11 software. The complete microarray data set is available at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE68427.
HA quantification.
The HA contents in cultured media of human lung fibroblasts, 3T3 cell lines, and MEF cells were quantified using the Hyaluronan DuoSet ELISA (DY3614, R&D Systems Inc.).
RNA interference.
Tbx4 siRNA effects on 3T3 cell line and human IPF lung fibroblast HA production were assessed with siRNA specific to Tbx4 (mTbx4 siRNA: 5′-rGrCrArCrUrGrCrCrArArGrArArArCrArUrGrGrArArArGGT-3′; hTbx4 siRNA: 5′-rUrGrCrArArUrUrArUrCrUrArArGrArArGrUrGrArCrUrUTG-3′; Universal scrambled negative control siRNA duplex, SR30004, OriGene Technologies). The cells were transfected with 30 nM siRNAs for 48 hours, and then treated with 5 mg/ml TGF-β1 overnight. The supernatants were collected thereafter for HA measurement. Invasion assays were done using cells transfected for 48 hours.
Promoter luciferase assay.
The 2,880-bp Has2 promoter luciferase reporter plasmid was provided by Chin Chiang (69) (Vanderbilt University Medical Center, Nashville, TN) and contains sequence from –2,326 to exon 1 of Has2 in pGL3 Basic plasmid (Promega). The 901-bp (chromosome 15, NC_000081.6: –436 to exon 1) and 664-bp (–112 to exon 1) Has2 promoter luciferase reporter plasmid was generated by PCR and cloned upstream of pGL3 Basic plasmid. Primers used were as follows: forward 1: 5′-CAAGCTCGAGGGAATCCTTGTAACG-3′; forward 2: 5′-CCGCCTCGAGTCCCGCCCAGTCCCT-3′; and reverse: 5′-TGCCAAGCTTCTTGTTCAGCTCCTGCTCATAGA-3′. CMV promoter–driven GFP-tagged TBX4 expression plasmid was purchased from GeneCopoeia. Luciferase assays were performed essentially as previously (69), using the Promega Dual luciferase Reporter assay system. To determine the effect of TBX4 on different lengths of the Has2 promoter activity, HEK 293 cells were cotransfected with Has2 promoter or pGL3 Basic plasmid and TBX4-expressing plasmid for 48 hours prior to the luciferase assay. All reporter assays were normalized using Renilla luciferase as an internal control. Each data point represents the mean of triplicate wells with error bar representing the SEM.
Calculation of the internal surface of a lung.
To ensure consistent and reproducible results, all (left and right) lungs were fixed by 10% formalin and with the same pressure, degassed, and embedded in OCT. We performed step sectioning on each block of lung. For the slides of each step section, more than 5 lung images were sampled randomly under Zeiss AXIO microscopy (×10 objective). Lung morphometric analysis was performed on the images of the lung parenchymal area for uninjured and injured lung without severe fibrotic lesion. Mean linear intercept, also known as mean chord length (70), was calculated by point and intersection counting using a grid line system (71).
Western blot.
To detect αSMA and β-actin expression, 25 μg of cell lysate was separated through 10% SDS-PAGE electrophoresis, blotted onto a nitrocellulose membrane (Life Technologies), and incubated with mAbs against αSMA (1:1,000; F3777, Sigma Chemical Co.) and β-actin (1:1,000; 12620, Cell Signaling Technology). The bands were visualized using the ECL system (Bio-Rad).
Collagen 1α1 ELISA.
The collagen 1α1 contents in cultured media of human lung fibroblasts were quantified using a collagen 1α1 direct ELISA developed in the laboratory. Standard curve for the assay was generated using collagen type I solution (C3867, Sigma-Aldrich) and collagen 1α1 antibody (ab21286, Abcam). The concentration of the collagen 1α1 in the cell supernatants was determined by extrapolation from the appropriate standard curve.
Caspase-3 activity assay.
IPF fibroblasts were cultured on 96-well plates. Caspase-3 activity assay was performed 48 hours after Tbx4 siRNA transfection. The amount of protein in cell lysates was determined using a BCA protein assay kit (Pierce Chemical Co.). Caspase3/CPP32 Fluorometric Assay Kit (BioVision) was used to examine the caspase-3 activity assay.
Calcein AM cell viability assay.
The cell viability of IPF fibroblasts 48 hours after Tbx4 siRNA transfection was analyzed by Calcein AM Cell Viability Assay Kit (Trevigen).
Statistics.
Data are expressed as the mean ± SEM. Differences in measured variables between experimental and control groups were assessed by Student’s t tests (2-tailed) or Wilcoxon rank-sum test with nonparametric data. One-way ANOVA with Bonferroni test was used for multiple comparisons. The survival curves were compared using the log-rank test. Results were considered statistically significant at P ≤ 0.05. GraphPad Prism software was used for statistical analysis.
Study approval.
All human lung fibroblast experiments were approved by the Cedars-Sinai Medical Center IRB and were in accordance with the guidelines outlined by the board. Informed consent was obtained from each subject (IRB: Pro00032727).
All mouse studies were approved by the IACUC at Cedars-Sinai Medical Center (protocols IACUC004722 and IACUC004751).
Author contributions
PWN, DJ, TX, and JL conceived and designed the study. TX performed most of the in vivo fate-mapping and in vitro experiments. JL, NL, and CH participated in in vivo experiments. JL, YZ, and WL participated in culture experiments. MK, DM, PC, VEP, BRS, RX, and JD provided critical reagents, intellectual discussions, and interpretation of data. TX, JL, DJ, and PWN analyzed data and wrote the paper. All authors discussed the data. DJ and PWN approved the submission of the manuscript.
Supplementary Material
Acknowledgments
We thank Brigid L.M. Hogan for Rosa26-tdTomato mice, Robert Fariss for αSMA-GFP mice, David Brenner for COL1α1-GFP mice, and Raghu Kalluri for NG2-YFP mice. We thank Mark Krasnow for suggestions during the initiation of the project. We thank Chin Chiang (Vanderbilt University Medical Center) for pHas2 luciferase plasmids. We thank Patricia Lin and Laura Dieu at the Cedars-Sinai Medical Center Flow Cytometry Core for cell sorting. We thank Laura-Leigh Rowlette and Zhengzheng Wei (Duke University) for microarray analysis. This work was supported by NIH grants P01-HL108793, R01-HL060539, and AI052201 (to P.W. Noble), R01-HL122068 (to D. Jiang), and R37H-D033082 (to V.E. Papaioannou), and a grant from California Institute for Regenerative Medicine (RB5-07302 to P.W. Noble).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(8):3063–3079. doi:10.1172/JCI85328.
Contributor Information
Ting Xie, Email: ting.xie@cshs.org.
Jiurong Liang, Email: Carol.Liang@cshs.org.
Ningshan Liu, Email: Ningshan.Liu@cshs.org.
Caijuan Huan, Email: hcj520@sina.com.
Yanli Zhang, Email: zhyanli@gmail.com.
Weijia Liu, Email: weijia902@126.com.
Maya Kumar, Email: mayak@stanford.edu.
Rui Xiao, Email: ruixiao85@gmail.com.
Daniel Metzger, Email: daniel.metzger@igbmc.fr.
Pierre Chambon, Email: pierre.chambon@igbmc.fr.
Barry R. Stripp, Email: barry.stripp@cshs.org.
Dianhua Jiang, Email: Dianhua.Jiang@cshs.org.
Paul W. Noble, Email: paul.noble@cshs.org.
References
- 1.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest. 2012;122(8):2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dulauroy S, Di Carlo SE, Langa F, Eberl G, Peduto L. Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat Med. 2012;18(8):1262–1270. doi: 10.1038/nm.2848. [DOI] [PubMed] [Google Scholar]
- 3.Hung C, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;188(7):820–830. doi: 10.1164/rccm.201212-2297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBleu VS, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med. 2013;19(8):1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108(52):E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordana M, et al. Heterogeneous proliferative characteristics of human adult lung fibroblast lines and clonally derived fibroblasts from control and fibrotic tissue. Am Rev Respir Dis. 1988;137(3):579–584. doi: 10.1164/ajrccm/137.3.579. [DOI] [PubMed] [Google Scholar]
- 7.Zeisberg M, Kalluri R. Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis. Am J Physiol Cell Physiol. 2013;304(3):C216–C225. doi: 10.1152/ajpcell.00328.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krenning G, Zeisberg EM, Kalluri R. The origin of fibroblasts and mechanism of cardiac fibrosis. J Cell Physiol. 2010;225(3):631–637. doi: 10.1002/jcp.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto N, Jin H, Liu T, Chensue SW, Phan SH. Bone marrow-derived progenitor cells in pulmonary fibrosis. J Clin Invest. 2004;113(2):243–252. doi: 10.1172/JCI18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson-Sjöland A, et al. Fibrocytes are a potential source of lung fibroblasts in idiopathic pulmonary fibrosis. Int J Biochem Cell Biol. 2008;40(10):2129–2140. doi: 10.1016/j.biocel.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Phillips RJ, et al. Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. J Clin Invest. 2004;114(3):438–446. doi: 10.1172/JCI20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degryse AL, et al. Repetitive intratracheal bleomycin models several features of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2010;299(4):L442–L452. doi: 10.1152/ajplung.00026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KK, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103(35):13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanjore H, et al. Contribution of epithelial-derived fibroblasts to bleomycin-induced lung fibrosis. Am J Respir Crit Care Med. 2009;180(7):657–665. doi: 10.1164/rccm.200903-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto N, et al. Endothelial-mesenchymal transition in bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol. 2010;43(2):161–172. doi: 10.1165/rcmb.2009-0031OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Jimenez SA. Protein kinase Cδ and c-Abl kinase are required for transforming growth factor β induction of endothelial-mesenchymal transition in vitro. Arthritis Rheum. 2011;63(8):2473–2483. doi: 10.1002/art.30317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramann R, et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell. 2015;16(1):51–66. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi JK, et al. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 1999;398(6730):810–814. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- 19.Naiche LA, Papaioannou VE. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development. 2007;134(1):93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- 20.Arora R, Metzger RJ, Papaioannou VE. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8(8): doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du J, et al. Different expression patterns of Gli1-3 in mouse embryonic maxillofacial development. Acta Histochem. 2012;114(6):620–625. doi: 10.1016/j.acthis.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Kumar ME, Bogard PE, Espinoza FH, Menke DB, Kingsley DM, Krasnow MA. Mesenchymal cells. Defining a mesenchymal progenitor niche at single-cell resolution. Science. 2014;346(6211): doi: 10.1126/science.1258810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luria V, Krawchuk D, Jessell TM, Laufer E, Kania A. Specification of motor axon trajectory by ephrin-B:EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron. 2008;60(6):1039–1053. doi: 10.1016/j.neuron.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Naiche LA, Arora R, Kania A, Lewandoski M, Papaioannou VE. Identity and fate of Tbx4-expressing cells reveal developmental cell fate decisions in the allantois, limb, and external genitalia. Dev Dyn. 2011;240(10):2290–2300. doi: 10.1002/dvdy.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, et al. Spatial-temporal targeting of lung-specific mesenchyme by a Tbx4 enhancer. BMC Biol. 2013;11: doi: 10.1186/1741-7007-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greif DM, et al. Radial construction of an arterial wall. Dev Cell. 2012;23(3):482–493. doi: 10.1016/j.devcel.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphreys BD, et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176(1):85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SL, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol. 2008;173(6):1617–1627. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331(19):1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 31.Hoyles RK, et al. An essential role for resident fibroblasts in experimental lung fibrosis is defined by lineage-specific deletion of high-affinity type II transforming growth factor β receptor. Am J Respir Crit Care Med. 2011;183(2):249–261. doi: 10.1164/rccm.201002-0279OC. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y, et al. A novel profibrotic mechanism mediated by TGFβ-stimulated collagen prolyl hydroxylase expression in fibrotic lung mesenchymal cells. J Pathol. 2015;236(3):384–394. doi: 10.1002/path.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwaisako K, et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. 2014;111(32):E3297–E3305. doi: 10.1073/pnas.1400062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, et al. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med. 2011;208(7):1459–1471. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovgren AK, et al. β-arrestin deficiency protects against pulmonary fibrosis in mice and prevents fibroblast invasion of extracellular matrix. Sci Transl Med. 2011;3(74): doi: 10.1126/scitranslmed.3001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora R, del Alcazar CM, Morrisey EE, Naiche LA, Papaioannou VE. Candidate gene approach identifies multiple genes and signaling pathways downstream of Tbx4 in the developing allantois. PLoS One. 2012;7(8): doi: 10.1371/journal.pone.0043581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinkevich Y, et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015;348(6232): doi: 10.1126/science.aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112(12):1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman HA. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol. 2011;73:413–435. doi: 10.1146/annurev-physiol-012110-142225. [DOI] [PubMed] [Google Scholar]
- 41.Willis BC, et al. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-β1: potential role in idiopathic pulmonary fibrosis. Am J Pathol. 2005;166(5):1321–1332. doi: 10.1016/S0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeisberg M, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9(7):964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 43.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110(3):341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bucala R, Spiegel LA, Chesney J, Hogan M, Cerami A. Circulating fibrocytes define a new leukocyte subpopulation that mediates tissue repair. Mol Med. 1994;1(1):71–81. [PMC free article] [PubMed] [Google Scholar]
- 45.Moore BB, et al. CCR2-mediated recruitment of fibrocytes to the alveolar space after fibrotic injury. Am J Pathol. 2005;166(3):675–684. doi: 10.1016/S0002-9440(10)62289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Deventer HW, Palmieri DA, Wu QP, McCook EC, Serody JS. Circulating fibrocytes prepare the lung for cancer metastasis by recruiting Ly-6C+ monocytes via CCL2. J Immunol. 2013;190(9):4861–4867. doi: 10.4049/jimmunol.1202857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro-oncology. 2005;7(4):452–464. doi: 10.1215/S1152851705000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duffield JS. Cellular and molecular mechanisms in kidney fibrosis. J Clin Invest. 2014;124(6):2299–2306. doi: 10.1172/JCI72267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ubil E, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514(7524):585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messier B, Leblond CP. Cell proliferation and migration as revealed by radioautography after injection of thymidine-H3 into male rats and mice. Am J Anat. 1960;106:247–285. doi: 10.1002/aja.1001060305. [DOI] [PubMed] [Google Scholar]
- 51.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med. 2008;5(3): doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua Belmonte JC. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 1999;398(6730):814–818. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- 54.Cebra-Thomas JA, Bromer J, Gardner R, Lam GK, Sheipe H, Gilbert SF. T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev Dyn. 2003;226(1):82–90. doi: 10.1002/dvdy.10208. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi JK, Koshiba-Takeuchi K, Suzuki T, Kamimura M, Ogura K, Ogura T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130(12):2729–2739. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- 56.Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120(2):533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Sabatino A, et al. Blockade of transforming growth factor beta upregulates T-box transcription factor T-bet, and increases T helper cell type 1 cytokine and matrix metalloproteinase-3 production in the human gut mucosa. Gut. 2008;57(5):605–612. doi: 10.1136/gut.2007.130922. [DOI] [PubMed] [Google Scholar]
- 58.Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130(12):2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- 59.Sakiyama J, Yamagishi A, Kuroiwa A. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development. 2003;130(7):1225–1234. doi: 10.1242/dev.00345. [DOI] [PubMed] [Google Scholar]
- 60.Condren AB, et al. Perivascular mural cells of the mouse choroid demonstrate morphological diversity that is correlated to vasoregulatory function. PLoS One. 2013;8(1): doi: 10.1371/journal.pone.0053386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voehringer D, Liang HE, Locksley RM. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol. 2008;180(7):4742–4753. doi: 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wendling O, Bornert JM, Chambon P, Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis. 2009;47(1):14–18. doi: 10.1002/dvg.20448. [DOI] [PubMed] [Google Scholar]
- 63.Zheng B, Zhang Z, Black CM, de Crombrugghe B, Denton CP. Ligand-dependent genetic recombination in fibroblasts: a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol. 2002;160(5):1609–1617. doi: 10.1016/S0002-9440(10)61108-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, Nishiyama A. Age-dependent fate and lineage restriction of single NG2 cells. Development. 2011;138(4):745–753. doi: 10.1242/dev.047951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adamson IY, Bowden DH. The pathogenesis of bleomycin-induced pulmonary fibrosis in mice. Am J Pathol. 1974;77(2):185–197. [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang D, et al. Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J Clin Invest. 2010;120(6):2049–2057. doi: 10.1172/JCI38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meltzer EB, et al. Bayesian probit regression model for the diagnosis of pulmonary fibrosis: proof-of-principle. BMC Med Genomics. 2011;4: doi: 10.1186/1755-8794-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jozefczuk J, Drews K, Adjaye J. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J Vis Exp. 2012;(64): doi: 10.3791/3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J, Li Q, Kuehn MR, Litingtung Y, Vokes SA, Chiang C. Sonic hedgehog signaling directly targets Hyaluronic Acid Synthase 2, an essential regulator of phalangeal joint patterning. Dev Biol. 2013;375(2):160–171. doi: 10.1016/j.ydbio.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campbell H, Tomkeieff SI. Calculation of the internal surface of a lung. Nature. 1952;170(4316):116–117. doi: 10.1038/170116a0. [DOI] [PubMed] [Google Scholar]
- 71.Foronjy RF, Mercer BA, Maxfield MW, Powell CA, D’Armiento J, Okada Y. Structural emphysema does not correlate with lung compliance: lessons from the mouse smoking model. Exp Lung Res. 2005;31(6):547–562. doi: 10.1080/019021490951522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.