ABSTRACT
The Neisseria gonorrhoeae ferric uptake regulator (Fur) protein controls expression of iron homeostasis genes in response to intracellular iron levels. In this study, using transcriptome sequencing (RNA-seq) analysis of an N. gonorrhoeae fur strain, we defined the gonococcal Fur and iron regulons and characterized Fur-controlled expression of an ArsR-like DNA binding protein. We observed that 158 genes (8% of the genome) showed differential expression in response to iron in an N. gonorrhoeae wild-type or fur strain, while 54 genes exhibited differential expression in response to Fur. The Fur regulon was extended to additional regulators, including NrrF and 13 other small RNAs (sRNAs), and two transcriptional factors. One transcriptional factor, coding for an ArsR-like regulator (ArsR), exhibited increased expression under iron-replete conditions in the wild-type strain but showed decreased expression across iron conditions in the fur strain, an effect that was reversed in a fur-complemented strain. Fur was shown to bind to the promoter region of the arsR gene downstream of a predicted σ70 promoter region. Electrophoretic mobility shift assay (EMSA) analysis confirmed binding of the ArsR protein to the norB promoter region, and sequence analysis identified two additional putative targets, NGO1411 and NGO1646. A gonococcal arsR strain demonstrated decreased survival in human endocervical epithelial cells compared to that of the wild-type and arsR-complemented strains, suggesting that the ArsR regulon includes genes required for survival in host cells. Collectively, these results demonstrate that the N. gonorrhoeae Fur functions as a global regulatory protein to repress or activate expression of a large repertoire of genes, including additional transcriptional regulatory proteins.
IMPORTANCE Gene regulation in bacteria in response to environmental stimuli, including iron, is of paramount importance to both bacterial replication and, in the case of pathogenic bacteria, successful infection. Bacterial DNA binding proteins are a common mechanism utilized by pathogens to control gene expression under various environmental conditions. Here, we show that the DNA binding protein Fur, expressed by the human pathogen Neisseria gonorrhoeae, controls the expression of a large repertoire of genes and extends this regulon by controlling expression of additional DNA binding proteins. One of these proteins, an ArsR-like regulator, was required for N. gonorrhoeae survival within host cells. These results show that the Fur regulon extends to additional regulatory proteins, which together contribute to gonococcal mechanisms of pathogenesis.
INTRODUCTION
Neisseria gonorrhoeae is an obligate human pathogen and is the etiological agent of the sexually transmitted infection gonorrhea. In 2013, there were 333,004 reported cases in the United States (http://www.cdc.gov/std/stats13/gonorrhea.htm), with the total number estimated to be much higher due to underreporting. Gonococcal infection of the male urethra results in inflammatory symptoms, characterized by a purulent discharge and a polymorphonuclear leukocyte (PMN) influx (1). In contrast, a large percentage of gonococcal infections of the female genital tract are asymptomatic (1). Like many other human pathogens, this organism must adapt to environments encountered during infection, including low pH and varying oxygen and iron levels (2–8). Tight control of gene expression in the gonococcus is mediated through several mechanisms, including phase variation (9), promoter mutations (10), small RNAs (sRNAs) (11, 12), and transcription factors that alter expression of target genes. In contrast to the case for Escherichia coli, in which there are ∼150 transcription factors, physiological studies have revealed that the gonococcus expresses relatively few (∼50) classical DNA binding proteins.
Within the family of DNA binding proteins encoded by N. gonorrhoeae is a homologue of the ferric uptake regulator (Fur) protein. Fur controls expression of iron homeostasis genes in response to iron and ensures a crucial balance between the requirement for iron and the avoidance of iron toxicity (13, 14). The gonococcal Fur protein has strong sequence conservation to the Fur proteins of Gram-positive and Gram-negative bacteria (15–18). The conserved Fur binding sequence of N. gonorrhoeae, an NATWATNATWAT repeat sequence, is also similar to those in other bacteria (19, 20). The classic mechanism of Fur-mediated regulation involves Fur binding directly to DNA sequences to inhibit transcription. In the absence of iron, Fur exists as an inactive monomer; however, increasing ferrous iron levels leads to dimerization and Fur binding to the promoter regions of target genes, subsequently blocking RNA polymerase binding and leading to decreased transcription (21). In Helicobacter pylori Fur has also been shown to bind DNA in the absence of iron in what is termed apo-Fur-mediated regulation (22), though this has not yet been definitely shown in N. gonorrhoeae. One of the best examples of iron-mediated direct repression by Fur involves transcriptional control of gonococcal genes that encode proteins acting to scavenge free iron from the host, including the transferrin binding proteins (TbpAB) and the ferric binding protein (FbpABC) (23). The importance of these proteins is shown by the avirulent nature of a tbp-deficient strain of N. gonorrhoeae in experimental infection of the male genital tract in humans (24). In addition, we have reported that fur, tbpB, and fbp are expressed during natural gonococcal infection, supporting a role for Fur-mediated control of gene expression during the pathogenic process (2, 3, 25).
Fur can also regulate genes indirectly by repressing a repressor such as a small RNA (sRNA). The targets of such repressors are then transcribed and translated at higher rates. This is the case for the Fur-repressed sRNA NrrF, which controls transcription of the gonococcal succinate dehydrogenase (sdhC and -A) genes (26). Fur has also been demonstrated to function as a direct activator via binding to promoter regions and facilitating RNA polymerase binding, resulting in increased transcription of target genes, including the norB gene (27, 28). Using in silico analysis of the gonococcal genome together with electrophoretic mobility shift assay (EMSA) analysis, we previously identified 16 additional putative Fur-activated genes (20). Further analysis utilizing a newly constructed gonococcal fur strain revealed >1.5-fold-lower expression levels of these 16 genes in the absence of Fur, suggesting that Fur may positively regulate their expression (20). However, this study was focused only on a subset of genes for which a Fur binding site was predicted.
In the current study, we compared the complete transcriptomes of N. gonorrhoeae wild-type (WT) and fur strains under iron-replete and -depleted conditions to define the repertoire of genes under both iron- and Fur-mediated control. This analysis revealed differential expression of a large number of genes via the presence of iron and Fur. We further characterized a DNA binding protein whose expression was activated by Fur, an ArsR-like regulator (NGO1562) here referred to as ArsR, and demonstrated that ArsR is required for gonococcal survival within human endocervical epithelial cells. These studies are the first to define the Fur and iron regulon of N. gonorrhoeae using a fur mutant strain and demonstrate how the Fur regulon extends to additional regulatory proteins.
MATERIALS AND METHODS
Construction of gonococcal arsR-complemented strains.
An N. gonorrhoeae F62 arsR (NGO1562) mutant strain was kindly provided by Virginia L. Clark (University of Rochester School of Medicine and Dentistry). The arsR gene was disrupted in N. gonorrhoeae F62 (29) by replacing a portion of the coding region with a kanamycin resistance gene cassette (27). An arsR strain was also constructed from FA1090 with all 11 opacity-coding (opa) genes inactivated (FA1090 opaA-K) (provided by Janne Cannon, University of North Carolina) (30). The kanamycin resistance cassette in the F62 arsR strain was amplified using primers listed in Table 1 and transformed into the FA1090 opaA-K strain. FA1090 opaA-K transformants were screened on plates supplemented with 100 μg/ml kanamycin and confirmed by DNA sequencing. To construct arsR-complemented strains from the F62 and FA1090 opaA-K arsR strains, the arsR gene with the 200-bp putative promoter region in F62 was amplified by PCR using the primers listed in Table 1, digested, and ligated into FstI-PacI doubly digested pGCC4 plasmid (31), generating the pGCC4/arsR plasmid (Table 2). This plasmid contains a kanamycin resistance gene to be used for selection in E. coli and acts to insert an erythromycin resistance gene as a marker into a nonfunctional intergenic region of N. gonorrhoeae between the aspC and lctP genes. The ligation product was transformed into E. coli TOP10 and screened using LB plates supplemented with 100 μg/ml kanamycin. Resistant colonies with the pGCC4/arsR plasmid were confirmed by DNA sequencing. The plasmid was then linearized by NotI digestion, purified using a gel purification kit (Qiagen, Hilden, Germany), and transformed into both the F62 arsR and the FA1090 opaA-K arsR strains by electroporation. Transformants containing the arsR coding region and promoter along with an erythromycin resistance gene were screened on GCB plates supplemented with 2 μg/ml erythromycin. Expression of the arsR gene in F62 (see Fig. S1 in the supplemental material) and FA1090 opaA-K arsR (data not shown) complemented strains was confirmed by reverse transcription-PCR (RT-PCR).
TABLE 1.
Primers used in this study
| Function and primer | Sequencea | PCR product size (bp) |
|---|---|---|
| Construction of arsR FA1090 opaA-K strain | ||
| arsR-Kan-Fw | TACCGCGATACCCCTGTAAC | 2,211 |
| arsR-Kan-Rv | ACACGTTGGCACAACTGGAC | |
| Construction of arsR-complemented strain | ||
| arsR-PacI-Fw | GGGGGGTTAATTAAGTAGGGGCGGCAGTTTTATG | 698 |
| arsR-FseI-Rv | TTTTTTGGCCGGCCGGAAATTTGTCCTGAT | |
| Construction of His-tagged protein | ||
| NdeI_arsR_Fw | CATATGAACACAATACCGCTCCAC | 291 |
| BamHI_arsR_Rv | GGATCCATTCTAACACTATGCCCG | |
| RT-PCR | ||
| arsR-RT-Fw | ATGAACACAATACCGCTCCA | 191 |
| arsR-RT-Rv | TAACGCGTAAAGTCCACCAG | |
| fbpA-RT-Fw | CTCCACCATCAACGAAACAC | 216 |
| fbpA-RT-Rv | GCGACAATCTGTTCCAAGAA | |
| Cloning of promoter regions | ||
| BamHI-arsR-p-Fw | CCCCCCGGATCCAGGGTAGGGGCGGCA | 271 |
| BamHI-arsR-p-Rv | CCCCCCGGATCCGGATCAGTATCGCCA | |
| BamHI-fur-p-Fw | CCCCCCGGATCCGGGTTTTGTTC | 256 |
| BamHI-fur-p-Rv | CCCCCCGGATCCCAATACCCCTGTA | |
| Competition assay | ||
| fur forward | CTGCTTGAATATTTTATAAAAGCGAACGATAATCATACGCTTAAGCGGT | 49 |
| fur complemented | ACCGCTTAAGCGTATGATTATCGTTCGCTTTTATAAAATATTCAAGCAG | |
| rmp forward | TAGTGGAGACTGAAATATCCGATTTGCCGCCATGTTTCTACAGCGGCCTG | 50 |
| rmp complemented | CAGGCCGCTGTAGAAACATGGCGGCAAATCGGATATTTCAGTCTCCACTA | |
| ArsR binding assay | ||
| norB-p forward | GAATAATGATAGTTATTATCATATATGTATTTGTTCTATATGTGAGTTTG | 50 |
| norB-p complementary | CAAACTCACATATAGAACAAATACATATATGATAATAACTATCATTATTC | |
| GC-rich forward | GCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGC | 50 |
| GC-rich complementary | GCTGGCTGGCTGGCTGGCTGGCTGGCTGGCTGGCTGGCTGGCTGGCTGGC |
Underlining indicates restriction enzyme sites.
TABLE 2.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| N. gonorrhoeae | ||
| F62 | ||
| Wild type | Isolated from a case of uncomplicated infection and is highly competent | 29 |
| fur mutant | fur gene was removed by replacing with a kanamycin resistance cassette | 36 |
| fur-complemented strain | A single copy of a fur gene allele was inserted into the chromosome of the fur mutant strain using plasmid pGCC4/fur | 36 |
| arsR mutant | A portion of the coding region of the arsR gene was removed via insertion of a kanamycin resistance cassette | 27 |
| arsR-complemented strain | A single copy of an arsR gene allele was inserted into the chromosome of the arsR mutant strain using plasmid pGCC4/arsR | This study |
| FA1090 opaA-K | ||
| Wild type | Isolated from a female patient with disseminated gonococcal infection | 30 |
| arsR mutant | A portion of the coding region of the arsR gene was removed via insertion of a kanamycin resistance cassette | This study |
| arsR-complemented strain | A single copy of an arsR gene allele was inserted into the chromosome of the arsR mutant strain using plasmid pGCC4/arsR | This study |
| E. coli | ||
| HBMV119 | A Δfur strain (QC1732) infected with λDE3 to construct an expression strain | 32 |
| BL21(DE3) | fhuA2 (lon) ompT gal (λDE3) (dcm) ΔhsdS λDE3 = λ sBamHIo ΔEcoRI-B int::(lacI::PlacUV5::T7 gene 1) i21 Δnin5 | NEB |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacΧ74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG λ− | Invitrogen |
| Plasmids | ||
| pET15b_NgFur | The gonococcal fur ORF of strain F62 was cloned into pET15b using NdeI and BamHI sites | 20 |
| pET15b_NgArsR | The gonococcal arsR ORF of strain F62 was cloned into pET15b using NdeI and BamHI sites | This study |
| pGCC4/arsR | The arsR (NGO1562) gene with the 200-bp putative promoter region upstream of the ATG start codon was cloned into pGCC4 using FstI and PaeI sites | This study |
| pGEMT/arsR-p | The 271-bp putative promoter region upstream of the ATG start codon was PCR amplified and cloned into a pGEMT-Easy vector | This study |
| pGEMT/fur-p | The 256-bp putative promoter region upstream of the ATG start codon was PCR amplified and cloned into a pGEMT-Easy vector | 20 |
Purification of gonococcal Fur and ArsR proteins.
The gonococcal Fur protein was overexpressed in E. coli HBMV119 (32) and purification performed as described previously (20). For purification of the ArsR protein, the gonococcal arsR gene was amplified from N. gonorrhoeae F62 genomic DNA using primers with the restriction sites NdeI and BamHI (Table 1). The PCR product was cloned into a pET15b-TEV vector (Novagen, San Diego, CA), and the protein was overexpressed in E. coli BL21(DE3) with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight and purified using Ni-charged resin according to the manufacturer's protocol (Novagen) (33, 34). The purified protein was dialyzed against buffer (50 mM Tris-Cl, 500 mM NaCl, 100 μM MnCl2, pH 7.9) and the 6×His tag cleaved using a 6×His-tagged tobacco etch virus (TEV) protease (35). The uncleaved protein and the cleaved 6×His tag were removed by passing through another Ni-charged resin.
qRT-PCR.
N. gonorrhoeae fur and fur-complemented strains were generated in a previous study (36). All strains were plated on GCB plates, grown overnight at 37°C with 5% CO2, and inoculated into chemically defined medium (CDM) (37) containing 0.042% Na2HCO3 with shaking for 2 h. Cultures were diluted into fresh CDM to an optical density at 600 nm (OD600) of 0.1 and incubated for an additional 2 h. Subsequently, 100 μM ferric nitrate (for iron-replete [+Fe] conditions) or 100 μM desferal (for iron-depleted [−Fe] conditions) was added to the culture. The bacterial pellets were collected 1 h after the addition of iron or desferal. For quantitative real-time PCR (qRT-PCR), RNA was purified from these pellets using an RNeasy kit (Qiagen, Hilden, Germany) and treated with DNase I (38). qRT-PCR was performed using the QuantiTect SYBR green RT-PCR kit (Qiagen) and analyzed using an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) with the primers listed in Table 1. Each reaction mixture contained 25 ng of purified total RNA. The relative mRNA levels were evaluated using the comparative cycle threshold (ΔΔCT) method as described by the manufacturer (39). The relative expression level of each gene was normalized to the endogenous 16S rRNA gene and represented as the ratio to that of the WT strain grown under iron-replete conditions. The results are presented as the mean ± standard deviation from three independent experiments.
RNA sequencing and data analysis.
For RNA sequencing, N. gonorrhoeae wild-type, fur, and fur-complemented strains were grown to an OD600 of 0.25 before the addition of iron or desferal (100 μM) to induce iron-replete and -depleted conditions, respectively. This was done to ensure that all strains would be in the same growth phase for transcriptome sequencing (RNA-seq) analysis. Strains were grown for 1 h, and RNA was purified from the resulting pellets using TRIzol according the manufacturer's instructions. Silica membranes were not used so as to retain the isolation of small RNA transcripts. Following RNA isolation, 2.5 to 3.0 μg of RNA was depleted of rRNA using the Agilent Microbe Express kit and prepared for sequencing using the BioChain High Yield Directional mRNA Sample Prep kit per the manufacturer's instructions. cDNA libraries were subject to electrophoresis on an 8% Tris-borate-EDTA (TBE) gel for 60 min at 140 V. cDNA fragments between 200 and 500 bp were extracted from the gel in Tris-EDTA (TE) buffer with agitation, and libraries were analyzed on an Agilent Technologies 2100 Bioanalyzer to confirm cDNA library size and quantity. Libraries were sequenced using 72-bp single-end reads on an Illumina GAIIX machine. Reads were aligned to the N. gonorrhoeae FA1090 genome (accession number AE004969) using the Rockhopper program (40). Alignment data are shown in Table S1 in the supplemental material. The output of Rockhopper consisted of each gene being assigned a reads per kilobase per million (RPKM) expression value normalized using the upper quartile of expressed genes. Genes showing a statistically significant difference (q value of <0.1) in expression values of at least 2.0-fold when comparing any two conditions were included in the analysis. To increase robustness of data, if a gene showed expression values equal to zero under one of the two conditions under analysis, that gene was not included in the analysis of transcriptome data sets. When describing complementation of genes putatively regulated by Fur, only those genes that showed less than a 2-fold change in expression, regardless of q value, when comparing the wild-type strain to the fur-complemented strain were classified as Fur regulated.
DNase I foot printing.
The PCR products of the promoter regions of target genes were cloned into a pGEMT-Easy vector (Qiagen). One primer, either T7 or Sp6 (Table 1), was labeled with fluorescent 6-carboxyfluorescein (FAM) (Invitrogen). The singly labeled probes of the promoter regions of each gene were PCR amplified with one labeled primer and one coupled unlabeled primer from the respective pGEMT-Easy vectors and purified by ethanol precipitation. DNase I footprinting assays were carried out in 50 μl buffer (10 mM Tris-Cl [pH 7.9], 100 mM KCl, 1 mM CaCl2, 200 μg/ml bovine serum albumin [BSA], 2 mM MgCl2, 0.5 mM β-mercaptoethanol [β-ME], 8 mM MnCl2, 10% glycerol), containing 1 μg poly(dIdC). After a 30-min incubation of the protein with 2,000 ng of the probes at room temperature, 1 μl of DNase I (Ambion, Applied Biosystems, 2 U/μl) was added and DNA digestion was carried out at 37°C for 4 min. Fifty microliters of stop solution (0.1 M EDTA [pH 8.0], 0.6 M Na acetate) was subsequently added to the reaction mixture to terminate DNA digestion (41). Digested DNA was purified using the QIAquick PCR purification kit (Qiagen) and eluted into 30 μl H2O. Ten microliters of the digested DNA was mixed with 9.5 μl HiDi formamide (Applied Biosystems, Foster City, CA) and 0.5 μl of GENEScan-600 LIZ size standards (Applied Biosystems). The samples were analyzed using fragment analysis and results visualized using the software Peakscanner (Applied Biosystems).
EMSA.
For Fur binding assays, the putative promoter regions (∼ 200 bp) of each open reading frame (ORF) were PCR amplified with the primers listed in Table 1. Probes (10 μM) were incubated with gonococcal Fur in the binding buffer [20 mM Tris-Cl (pH 7.9), 5 mM MgCl2, 40 mM KCl, 0.125 mM MnCl2, 2 mM dithiothreitol (DTT), 10% glycerol, 0.19 μg/μl poly(dIdC), 3.125 μg/μl BSA] at room temperature for 30 min. For ArsR binding assays, a 50-bp double-stranded DNA (dsDNA) sequence of the norB promoter region was created by a standard hybridization method: the forward strand and the complementary strand solutions were mixed at 20 μM, heated at 80°C for 10 min, and then slowly cooled to room temperature. One microliter of dsDNA solution was mixed with purified ArsR protein in the above-described binding buffer at room temperature for 30 min. For electrophoretic mobility shift assay (EMSA) with either protein, the reaction mixture was separated on a native 6% polyacrylamide gel (acrylamide/bisacrylamide ratio, 37.5:1 [wt/wt]) (Bio-Rad, Hercules, CA) and stained with SYBR green (Invitrogen) following the manufacturer's instructions. For Fur competition assays, 50-bp double-stranded DNA probes were incubated with the reaction mixtures (38). The probes containing a Fur box (fur positive) from the promoter region of the fur gene and the probes without a Fur box (rmp negative) from the promoter region of the rmp gene are listed in Table 1.
Infection of human epithelial cells.
Approximately 5 × 105 endocervical epithelial cells (End/E6E7) (42) were seeded into each well of six-well tissue culture plates and grown to confluence (106/well) in 2 ml of KSFM medium (Invitrogen). Concurrently, N. gonorrhoeae F62 and FA1090 opaA-K wild-type, arsR, and arsR-complemented strains were streaked from pilin-negative stocks on an overnight GCB plate and suspended in fresh KSFM medium. Bacterial cells were then incubated with epithelial cells at a target multiplicity of infection (MOI) of 100 at 37°C in an incubator containing 5% CO2. An aliquot of the initial inoculum was also plated to determine the exact MOI. At 2 h, 5 h, and 24 h postincubation, total cell-associated and intracellular bacteria were measured by counting CFU. Counting of total cell-associated bacteria and intracellular bacteria was carried out as described previously (36). The experiments were repeated four times with F62 strains and three times using N. gonorrhoeae FA1090 opaA-K wild-type, arsR, and arsR-complemented strains. The final results are represented as the percentage of the inoculum (mean ± standard error). The means were considered significantly different at a P value of <0.05 using the Student t test.
Accession number.
RNA sequencing data are available from the Gene Expression Omnibus (GEO) database with accession number GSE83138.
RESULTS
Gonococcal global transcriptional response to iron and the influence of Fur.
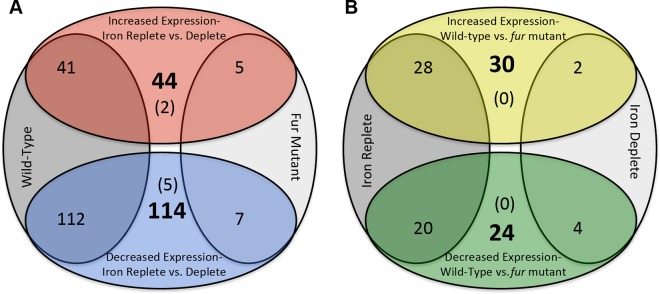
To define the gonococcal iron and Fur regulon, we performed RNA-seq analysis of N. gonorrhoeae wild-type (F62), fur, and fur-complemented strains grown under either iron-replete (+Fe) or -depleted (−Fe) conditions. There were no significant differences in the growth of each strain under either iron-replete or -depleted conditions (see Fig. S2 in the supplemental material). We identified 153 gonococcal protein-coding genes that showed changes in expression in the wild-type strain as a function of iron (Fig. 1; see Table S2 in the supplemental material). Of these, 41 were expressed 2.0-fold higher under iron-replete versus -depleted conditions (iron activated), and 112 were expressed 2.0-fold lower under iron-replete versus -depleted conditions (iron repressed). In contrast to the wild-type strain, only a few (12) genes showed iron-dependent changes in expression in a fur strain (Fig. 1; see Table S2 in the supplemental material).
FIG 1.
Iron- and Fur-mediated gene expression in N. gonorrhoeae, showing the number of genes in each category. (A) Iron-mediated expression is shown in the top and bottom ovals, and strains in which iron-mediated expression was observed are shown in the left and right ovals. Numbers in bold indicate the total number of genes showing alterations in expression, while numbers in parentheses indicate genes that were iron activated or repressed regardless of the strain under analysis. (B) Fur-mediated expression is shown in the top and bottom ovals, and iron conditions are shown in the left and right ovals. Numbers in bold indicate the total number of genes showing alterations in expression, while numbers in parentheses indicate genes that were Fur activated or repressed regardless of the iron condition under analysis.
There were large differences in the categorization of genes with altered expression in the presence of iron in the wild-type strain (Fig. 2A). Aside from genes encoding uncharacterized hypothetical proteins, genes that showed increased expression in the presence of iron were dominated by those encoding respiration proteins, such as NADH dehydrogenase protein complex (NGO1737 to NGO1750), a nitrate reductase (NGO1276), and a cytochrome c (NGO1769). In contrast, categorization of genes showing decreased expression in response to iron showed enrichments of transposase and phage-associated genes (Fig. 2A). Two genes involved in recombination (come2 and -e4) showed decreased expression in response to iron, while we did not observe any such genes whose expression was increased in response to iron. Several genes that encode iron transport proteins showed increased expression under iron-depleted conditions, including tbp (NGO1495 and NGO1496) and fbp (NGO0215 and NGO0217), in conjunction with their roles in maintaining iron homeostasis levels within the host cell.
FIG 2.
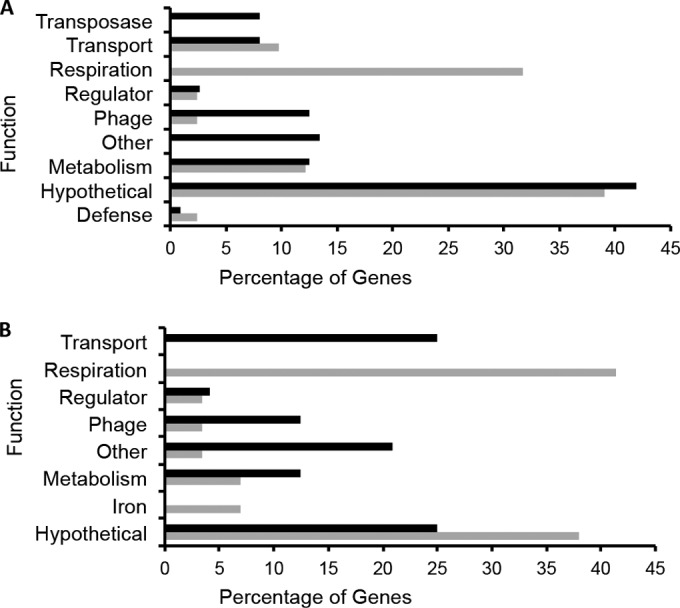
Categorization of genes responding to iron and Fur. (A) Categorization of genes showing decreased (black bars) or increased (gray bars) expression in response to iron in the wild-type strain. The percentage of genes in each category is shown on the x axis, and the functions of categories are shown on the y axis. (B) Categorization of genes showing decreased (black bars) or increased (gray bars) expression in response to the presence of Fur.
To determine the role of Fur in controlling gene expression, we compared the RNA-seq data for the wild-type and fur strains grown under either iron-replete or -depleted conditions. This revealed a total of 54 protein-coding genes that showed Fur-dependent changes in expression, including the previously described tbpAB and fbp genes (43–45) (Fig. 1; see Table S3 in the supplemental material). When iron was present, a greater number of genes exhibited increased expression in a wild-type strain than in a fur strain, suggesting that Fur functions to activate rather than to repress gene expression. As Fur has traditionally been understood to be a repressor, this raises the possibility that Fur-mediated activation of gene expression results from the use of additional regulators such as the sRNA NrrF or other protein-based regulators. In contrast, only six ORFs showed changes in expression in a wild-type compared to a fur strain under iron-depleted conditions (Fig. 1; see Table S3 in the supplemental material). There were no genes that showed changes in expression under both iron-replete and -depleted conditions when comparing the fur strain to the wild-type strain and only a few that showed changes in expression in a fur strain when comparing iron-replete to -depleted conditions. Categorization of genes with expression differences in response to Fur showed that genes encoding respiration proteins exclusively showed increased expression in response to the presence of Fur (Fig. 2B). This corroborates our findings above showing these genes to be induced by iron. As iron also activates Fur activity (46), it makes biological sense that these respiration genes would also be induced by Fur. Several transport genes exhibited increased expression in response to the absence of Fur, including the previously characterized Fur-regulated tbp and fbp iron transport genes (23). Finally, we observed significant overlap with our previous studies of Fur- and iron mediated regulation in N. gonorrhoeae and Neisseria meningitidis (20, 47).
Extension of the Fur regulon to additional regulators.
It is likely that Fur-mediated regulation extends via control of additional regulators such as sRNAs, including NrrF (12, 26), or other DNA binding proteins. To identify possible Fur-controlled regulators, we examined our data set to identify either sRNAs or DNA binding proteins that show differential expression in a fur strain compared to a wild-type strain. Under iron-replete conditions, our RNA-seq analysis identified seven putative sRNAs (defined as transcription originating either within an intergenic region or opposite an annotated ORF) that showed increased expression in the fur strain compared to the wild-type strain, including the previously identified sRNA NrrF, and four sRNAs that showed decreased expression in the fur strain compared to wild-type strain. (see Table S4 in the supplemental material). Under iron-depleted conditions, we identified three sRNAs that showed decreased expression in the fur strain compared to the wild-type strain (see Table S4 in the supplemental material).
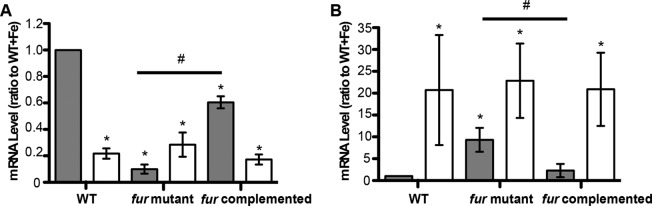
To determine if additional regulatory proteins were controlled by Fur, we queried these data sets to identify putative transcription factors that exhibited altered transcription when comparing the wild-type, fur, and fur-complemented strains grown under iron-replete or -depleted conditions. This analysis revealed one such gonococcal gene controlled via Fur under iron-replete conditions, mpeR (NGO0025), an AraC regulator, which was expressed at a level approximately 47-fold higher in the fur strain than in the wild-type strain. In addition, another regulator, arsR (NGO1562), was expressed at a level 4.2-fold lower in the fur strain than in the wild-type strain, though the q value associated with this regulation (0.107) was slightly above our imposed cutoff of 0.1. Neither of these ORFs exhibited changes in expression under iron-depleted conditions when comparing the wild-type and fur strains, though both showed iron regulation in the wild-type strain (see Tables S2 and S3 in the supplemental material). Because arsR encodes a regulatory protein that has previously been shown to be crucial to anaerobic growth of N. gonorrhoeae (27) and regulation was altered in response to iron and Fur, we selected arsR for further study. To confirm putative Fur-mediated changes in expression of arsR obtained by RNA-seq analysis, we utilized qRT-PCR to confirm arsR transcription levels in wild-type, fur, and fur-complemented strains. We observed that the arsR gene was expressed at a level approximately 5-fold higher in the N. gonorrhoeae wild-type strain under iron-replete conditions than under iron-depleted conditions (Fig. 3A). However, in the fur strain, such changes in expression via iron were absent and the arsR gene was expressed constitutively at low levels, a phenotype that was reversed in the fur-complemented strain (Fig. 3A). These results corroborate our RNA-seq analysis and suggest that Fur activates transcription of the arsR gene in an iron-dependent manner. The mRNA levels of fbpA, a well-established N. gonorrhoeae Fur-repressed gene, were also examined as a positive control. Transcription of fbpA was increased in the wild-type strain under iron-depleted conditions and was increased in the absence of the fur gene across iron conditions (Fig. 3B). Repression of fbpA transcription by Fur was reestablished in the fur-complemented strain, confirming the phenotypes of the strains used (Fig. 3B).
FIG 3.
RT-PCR analysis of arsR and fbpA in a fur strain (A) Transcriptional changes in arsR (NGO1562) expression were analyzed using quantitative real-time PCR. RNA samples were purified from cultures of wild-type (WT), fur, and fur-complemented strains grown under iron-replete (+Fe, gray bars) or -depleted (−Fe, white bars) conditions at 1 h after addition of 100 μM ferric nitrate or 100 μM desferal. (B) Transcriptional changes in expression of fbpA, a gene that is repressed by iron-bound Fur (43, 44), were used as a positive control for iron and Fur responses in N. gonorrhoeae. The mRNA levels observed for the five conditions (WT strain under −Fe conditions, fur strain under +Fe and −Fe conditions, and fur-complemented strain under +Fe and −Fe conditions) were compared to the value for the WT strain under +Fe conditions. *, significantly different compared to the mRNA level of the WT strain with Fe; #, significantly different between the mRNA levels of the fur strain with Fe and the fur-complemented strain with Fe. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
Fur binds to the promoter region of the arsR gene.
To determine if Fur bound to the promoter of the arsR gene, we performed an in vitro EMSA. At relatively low Fur levels (100 nM), we observed a complete shift of the arsR promoter (Fig. 4A). Binding of Fur to the arsR promoter region was specific as demonstrated through the loss of binding with the addition in excess of an unlabeled dsDNA probe known to contain a Fur box. In contrast, we did not observe displacement of Fur binding to the arsR promoter region using a dsDNA probe that contained the promoter region of the rmp gene, a negative control that does not contain a Fur box (Fig. 4B). These results indicate that Fur most likely regulates expression of arsR via direct binding to the promoter of arsR and not through the use of an additional regulator.
FIG 4.
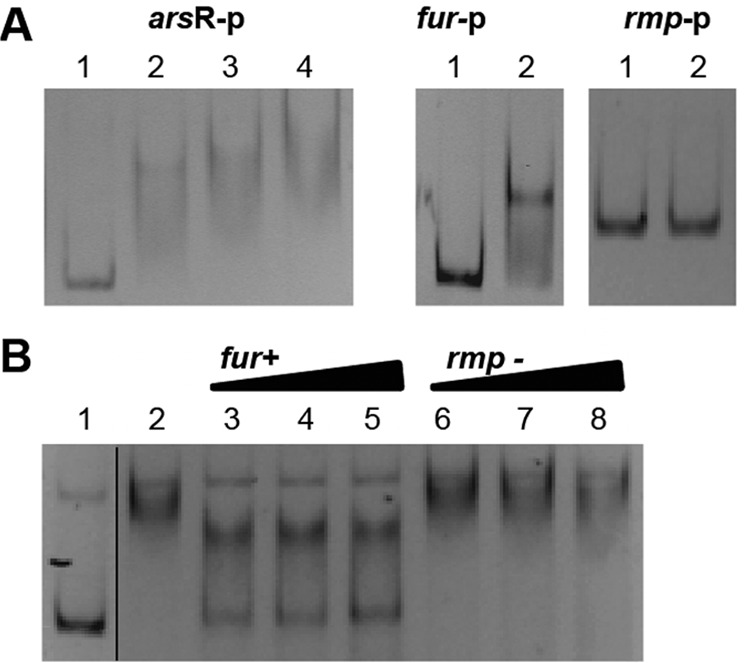
EMSA analysis of Fur binding to the putative arsR promoter region. (A) Left panel, the putative promoter region of arsR (271 bp) was amplified by PCR and incubated with Fur protein. Lane 1, free DNA; lanes 2 through 4, gonococcal Fur was added at 50 nM, 100 nM, and 200 nM, respectively. Middle panel, Fur protein binding to the fur promoter region (positive control). Lane 1, free DNA; lane 2, 200 nM Fur protein. Right panel, lack of Fur protein binding to the rmp promoter region (negative control). Lane 1, free DNA; lane 2, 500 nM Fur protein. (B) Competition of Fur binding to the putative arsR promoter region. Lane 1, free labeled DNA; lane 2, gonococcal Fur protein was added at 200 nM; lanes 3 through 5, 200 nM Fur protein and 50-bp unlabeled dsDNA probes of the fur promoter region (positive control) were added with excesses of 25-fold, 50-fold, and 100-fold, respectively (indicated by the triangles); lanes 6 through 8, 200 nM Fur protein and 50-bp unlabeled dsDNA probes of the rmp promoter region (negative control) were added with excesses of 25-fold, 50-fold, and 100-fold, respectively (indicated by the triangles). The line between lanes 1 and 2 denotes splicing of the gel to remove extraneous lanes.
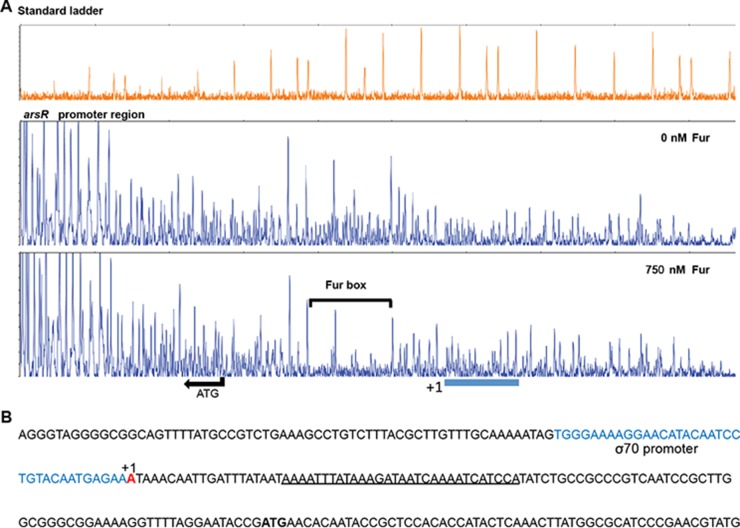
Mapping of the Fur binding site within the promoter region of the arsR gene.
The specific Fur binding site in the promoter region of arsR was next determined using DNase I foot printing. We observed that a 30-bp region, AAAATTTATAAAGATAATCAAAATCATCCA, was protected following incubation with Fur (750 nM) (Fig. 5A). The sequence of the Fur box in the arsR promoter region was similar to that of the Fur box in the gonococcal fur promoter region, with both containing NATWAT repeats (20). The Fur-protected region in the promoter region of the fur gene was used as a positive control for these experiments, and DNase I foot printing confirmed the Fur box sequence as previously determined (see Fig. S3 in the supplemental material) (3, 20). A predicted σ70 promoter region, TGGGAAAAGGAACATACAATCCTGTACAATGAGAA, and the transcription start site for the arsR gene had also been identified in a previous study (48) and were found to be upstream of the Fur binding site, as shown in Fig. 5B, which may function to block RNA polymerase.
FIG 5.
Analysis of the promoter region of arsR. (A) DNase I footprinting of the Fur binding site. DNA regions protected by Fur are indicated by a bracket. The arrow indicates the translation start site (ATG) and the transcriptional direction of the arsR gene. (B) Nucleotide sequence spanning the promoter region of the arsR gene. The predicted σ70 promoter region is indicated in blue, and the transcriptional start site is indicated in red. The Fur binding site in the predicted promoter region of arsR resulting from the DNase I footprinting is underlined.
ArsR contributes to N. gonorrhoeae survival within human endocervical cells.
To define the role of ArsR in gonococcal interactions with host cells, we examined the ability of arsR and arsR-complemented strains in the F62 background to survive in a human endocervical epithelial cell line (End/E6E7) (49, 50). No significant differences were observed in the growth of the three strains (wild-type, arsR, and arsR-complemented strains) grown in CDM (iron-replete or iron-depleted conditions). However, differences were seen in the growth rates of all 3 strains during growth under iron-replete versus -depleted conditions (see Fig. S4 in the supplemental material), with slower growth under iron-depleted conditions. We observed that the arsR strain showed a significant decrease in intracellular survival in the endocervical epithelial cell line compared to the wild-type and arsR-complemented strains at 2 and 5 h postincubation (Fig. 6A). Restored intracellular survival was observed with the arsR-complemented strain at 5 and 24 h postincubation compared to the arsR strain (Fig. 6A). In contrast, there was no obvious or significant trend when examining adherence to host cells when the arsR strain and the wild-type or arsR-complemented strain were compared (Fig. 6B). These results suggest that the arsR mutation alters the ability of N. gonorrhoeae to either invade or survive within the endocervical epithelial cell line. To confirm that these findings were due to the arsR mutation and not due to changes in Opa expression, the arsR and arsR-complemented strains were constructed from a gonococcal strain with all 11 opa genes inactivated (FA1090 opaA-K), and the experiments with adhesion and invasion of endocervical epithelial cells were repeated. As with experiments using F62, we observed that the arsR strain showed a significant decrease in intracellular survival in the endocervical epithelial cell line compared to the wild-type and the arsR-complemented strains at 2 and 5 h postincubation (see Fig. S5A in the supplemental material). However, while there was decreased intracellular survival of the arsR strain in the F62 background at 24 h postincubation, this was not observed in the FA1090 opaA-K background. These observations may be due to differences in the strain backgrounds (F62 and FA1090) or to general differences in growth at later time points. There was no obvious or significant trend when examining adherence to host cells when the arsR strain and the wild-type or arsR complemented strain were compared in the FA1090 opaA-K background (see Fig. S5B in the supplemental material).
FIG 6.
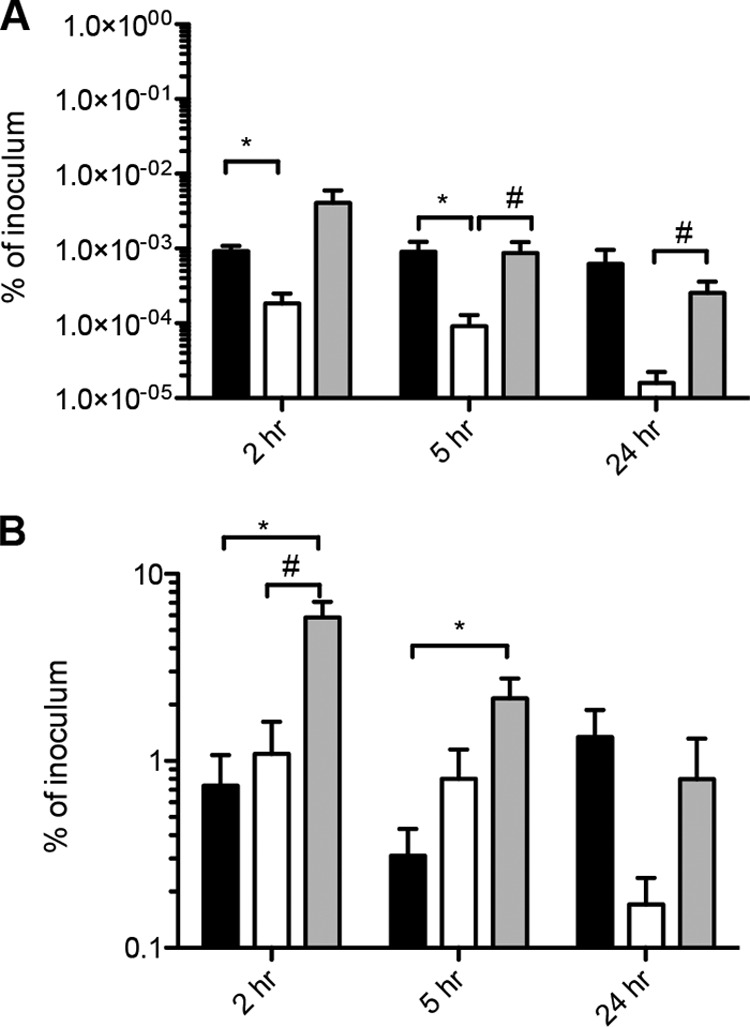
Reduced survival of the N. gonorrhoeae arsR strain in the human endocervical epithelial cell line End E6/E7 compared to the wild-type strain. (A) Intracellular bacteria were recovered after gentamicin treatment at 2, 5, and 24 h after addition of bacteria, and CFU were normalized to the inoculum of the same strain at time zero. The results are the means from four experiments, and error bars indicate standard errors. The means were considered significantly different at a P value of <0.05 using the Student t test. *, significantly different from the WT strain at each time point; #, significantly different from the complemented strain at each time point. Black bars, N. gonorrhoeae wild-type strain; white bars, arsR strain; gray bars, arsR-complemented strain. (B) Cell-associated bacteria (both intracellular and bacteria attached to endocervical epithelial cell surface) were determined at 2, 5, and 24 h after addition of bacteria, and CFU were normalized to the inoculum of the same strain at time zero.
Putative ArsR targets identified in the gonococcal genome.
The results obtained above suggest that genes within the ArsR regulon may have a role in gonococcal survival within epithelial cells. To begin to define genes under ArsR-mediated control, we examined the gonococcal genome for genes containing a putative ArsR binding sequence. We first confirmed that the ArsR binding sequence within the norB promoter gene was CATATAnnTATTTG, as was previously predicted (27), via EMSA (see Fig. S6 in the supplemental material). Once this sequence was confirmed, a genome-wide search for additional ArsR binding sites using the CATATATGTATTTG sequence revealed two additional homologous sequences in the intergenic regions of the gonococcal genome. One predicted binding site (CATATATTTATTTG) was localized upstream of the NGO1411 gene (encoding a hypothetical protein), and the other (CCTCTATGTATTTG) was localized upstream of the NGO1646 gene (encoding a phage-associated protein). As these genes have putative arsR binding sites in their promoters and since deletion of the arsR gene resulted in reduced intracellular survival, it is plausible that NGO1411 and NGO1646 encode proteins that contribute to survival of the gonococcus in human epithelial cells.
DISCUSSION
In this study, we have further defined the N. gonorrhoeae iron and Fur regulons and characterized a Fur-activated DNA binding protein, ArsR, which was required for gonococcal survival in human epithelial cells at early time points. We demonstrate that a significant percentage of the gonococcal genome responds to iron and that several of these genes exhibit altered expression in response to Fur. In total, 153 genes (8% of the gonococcal genome) showed changes in expression when comparing iron-replete to -depleted conditions in a wild-type strain. A total of 54 genes (2.6% of the genome) showed changes in expression when comparing a fur strain to a wild-type strain. There was significant overlap between the genes showing changes in expression via iron and via Fur, with 77% (42/54 genes) responding to Fur and iron. There were also few genes that showed changes in expression when comparing an iron-replete to -depleted conditions with the fur strain. Fur has been demonstrated to respond to iron; therefore, the observation that only a few genes showed iron-dependent changes in expression in the absence of Fur is biologically intuitive.
As iron is of paramount importance during infection of the human genital tract, it is possible that several of the genes that show changes in expression in response to iron and Fur are involved in pathogenesis of N. gonorrhoeae. Genes showing changes in response to iron included NGO1682, encoding an EmrB/QacA family multidrug transporter, as well as tonB (NGO1379) and a tonB-dependent receptor (NGO0553), which were previously shown to be crucial for intracellular survival (51). We also observed decreased expression in response to iron of the fitA/B locus (NGO0907-NGO0908), which is intimately involved in gonococcal trafficking across epithelial cell monolayers (52, 53). Genes showing changes in response to Fur included an opacity protein gene (NGO1277), showing increased expression in the presence of Fur, and the above-mentioned tonB, showing decreased expression in the presence of Fur. Several of the genes that we report here were also found in previous studies on Fur-mediated regulation in N. gonorrhoeae (20, 54) and N. meningitidis (47). The differences observed between the results obtained here and in our previous studies are most likely due to differences in how the bacteria were grown and the time points at which gene expression was examined. Regardless of these differences, we still found a significant overlap of genes between the studies when examining Fur-mediated changes in gene expression.
In addition to protein-coding genes, expression of other, noncoding transcripts, including 13 previously uncharacterized sRNA transcripts and the well-characterized sRNA NrrF, were altered in response to Fur (12, 26). NrrF was demonstrated here to be Fur repressed, in agreement with previous studies (12, 26), and an NrrF target, sdhB (NGO0920), exhibited a decrease in expression in the absence of fur, further demonstrating how Fur repression of a repressor (NrrF) results in the activation of a separate gene (sdhB). The expansion of transcription factor target genes through the use of RNA-based regulators may be more common in N. gonorrhoeae than in other Proteobacteria due to the fact that N. gonorrhoeae contains relatively few classical DNA binding proteins (∼50, compared to >200 in E. coli). We demonstrate here that among the genes showing changes in expression via Fur was a gene for an additional DNA binding protein, an ArsR-type regulator, arsR, whose expression was directly activated by Fur. We envision several possible mechanisms by which Fur can control the expression of arsR. Fur may compete with an additional DNA binding protein that functions as a repressor. Fur-mediated occlusion of this binding site may allow for subsequent binding of the RNA polymerase complex and induce expression of the arsR mRNA. This mechanism has been described for Fur and ArsR-mediated regulation of the norB gene (see below). It is also possible that Fur binding to the promoter region of the arsR gene stabilizes the interaction of the RNA polymerase complex to the promoter region, resulting in increased transcription of the arsR gene. We further propose that these two mechanisms are not mutually exclusive and may operate simultaneously under certain growth conditions.
Our results also demonstrate that ArsR functions in gonococcal survival in human endocervical epithelial cells and suggest that the targets of ArsR may be crucial for gonococcal survival at early time points. The known ArsR target norB encodes a nitric oxide reductase utilizing NO that is produced by gonococcal denitrification metabolism (55). A gonococcal norB mutant strain was demonstrated to be impaired in anaerobic growth (55). Since anaerobic conditions exist in vivo during gonococcal infection, it is possible that the decreased survival of the arsR mutant in human endocervical epithelial cells shown in this study is attributable to norB. Further studies are required to define the role of norB in the arsR mutant phenotype observed in the endocervical cells.
An examination of the gonococcal genome for the presence of an “ArsR box” within intergenic regions identified NGO1411 and NGO1646 as putative targets of ArsR. While the protein product of NGO1411 is classified as hypothetical in the N. gonorrhoeae FA1090 genome, in E. coli a homologue, NhaD (56), is classified as an Na+/H+ antiporter involved in arsenite metabolism. While there is little evidence that arsenite metabolism is crucial for intracellular survival of N. gonorrhoeae, NGO1411 may be involved in the transport of other metabolites required for growth within endocervical cells. Lack of expression of NGO1411 may deprive N. gonorrhoeae of a required metabolite or small molecule that is abundant in cell culture media but is limiting within endocervical cells, leading to reduced survival of the gonococcus. A second target, NGO1646, is predicted to encode a phage-associated protein in the N. gonorrhoeae FA1090 genome. While little is known regarding the role of phage-associated proteins in the regulatory pathways of N. gonorrhoeae, previous studies by our group have identified one such phage-associated protein, Npr, which is involved in gonococcal pathogenesis (36). Lack of Npr leads to increased adherence to and invasion of epithelial cells as well as enhanced colonization in a gonococcal mouse model of infection. It is possible that NGO1646 is also involved in pathogenic mechanisms and that lack of regulation of this protein by ArsR leads to the reduced survival phenotype seen here.
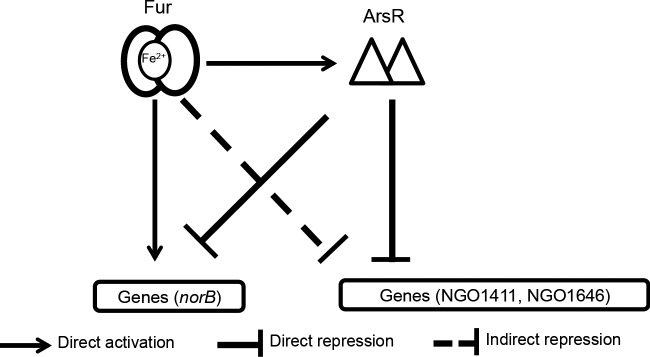
Our results support a model by which ArsR and Fur function together to control gene transcription in N. gonorrhoeae. The observation that the expression of arsR is both Fur and iron regulated suggests that targets of ArsR are also likely affected by the presence of iron. By virtue of the fact that expression of arsR is activated by Fur, Fur may then indirectly regulate the transcription of genes under ArsR-mediated control (Fig. 7). In this way, ArsR and Fur likely cooperate to regulate transcription of a subset of genes, including norB and possibly NGO1411 and NGO1646 (Fig. 7). Previous studies demonstrated that expression from the norB promoter was partially reduced in a fur strain compared to the wild-type strain and was restored in the arsR strain under anaerobic conditions, suggesting that both DNA binding proteins act together to regulate the norB gene (27). This dual mode of Fur-mediated regulation (direct control of a target and control of a regulatory protein of the same target) in the gonococcus likely allows for precise and temporally targeted gene expression patterns during infection. Though neither NGO1411 nor NGO1646 was found to be Fur regulated in the current study, changes in gene expression may be minimal, particularly since an additional regulator, ArsR, is positioned between Fur and these targets in the regulatory pathway. It is also possible that these genes, if they are Fur regulated, show changes in expression at time points later than the ones examined in this study. Here we examined an early time point (1 h after the addition of iron or desferal) that induced changes in the expression of Fur followed by changes in expression of ArsR. An additional step in regulation, changes in expression of NGO1411 and NGO1646, would likely not have occurred in the time frame examined.
FIG 7.
Schematic of cross talk between the Fur and ArsR proteins. Fur directly activates expression of ArsR, which results in the indirect control of ArsR-controlled genes. In addition, Fur and ArsR cooperate to regulate transcription of a subset of genes. As an example, Fur binds to the promoter region of norB to compete out the repressor ArsR, resulting in transcriptional activation of norB.
The ability of Fur to interact with other regulatory proteins has the potential to expand Fur-mediated control of gene expression in the gonococcus. Others have demonstrated that Fur controls transcription of the regulatory protein OxyR (45). The expression of downstream targets of these proteins is thus indirectly altered by Fur, enabling control of expression of a wide array of genes, including those involved in resistance to ROS (via OxyR) (57). Fur has been shown to repress expression of the MarR/FarR and MpeR regulatory proteins (58, 59). Targets of these proteins include genes involved in antimicrobial resistance (MtrR) (60), and thus these genes are indirectly regulated by Fur. Further characterization of gonococcal regulatory pathways will provide an avenue to design future antimicrobial therapies, a global health need that is likely to increase as N. gonorrhoeae continues to become resistant to known antibiotics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Virginia L. Clark for providing the gonococcal arsR strain. We thank Kenneth Barth for helpful discussions of the manuscript.
This work was supported by NIH/NIAID grant RO1AI048611 to C.A.G.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00166-16.
REFERENCES
- 1.Edwards JL, Apicella MA. 2004. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin Microbiol Rev 17:965–981. doi: 10.1128/CMR.17.4.965-981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal S, King CA, Klein EK, Soper DE, Rice PA, Wetzler LM, Genco CA. 2005. The gonococcal Fur-regulated tbpA and tbpB genes are expressed during natural mucosal gonococcal infection. Infect Immun 73:4281–4287. doi: 10.1128/IAI.73.7.4281-4287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal S, Sebastian S, Szmigielski B, Rice PA, Genco CA. 2008. Expression of the gonococcal global regulatory protein Fur and genes encompassing the Fur and iron regulon during in vitro and in vivo infection in women. J Bacteriol 190:3129–3139. doi: 10.1128/JB.01830-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark VL, Knapp JS, Thompson S, Klimpel KW. 1988. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb Pathog 5:381–390. doi: 10.1016/0882-4010(88)90038-1. [DOI] [PubMed] [Google Scholar]
- 5.Graver MA, Wade JJ. 2011. The role of acidification in the inhibition of Neisseria gonorrhoeae by vaginal lactobacilli during anaerobic growth. Ann Clin Microbiol Antimicrob 10:8. doi: 10.1186/1476-0711-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma B, Forney LJ, Ravel J. 2012. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol 66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newkirk GR. 1996. Pelvic inflammatory disease: a contemporary approach. Am Fam Physician 53:1127–1135. [PubMed] [Google Scholar]
- 8.Zheng HY, Alcorn TM, Cohen MS. 1994. Effects of H2O2-producing lactobacilli on Neisseria gonorrhoeae growth and catalase activity. J Infect Dis 170:1209–1215. doi: 10.1093/infdis/170.5.1209. [DOI] [PubMed] [Google Scholar]
- 9.Belland RJ. 1991. H-DNA formation by the coding repeat elements of neisserial opa genes. Mol Microbiol 5:2351–2360. doi: 10.1111/j.1365-2958.1991.tb02081.x. [DOI] [PubMed] [Google Scholar]
- 10.Sarkari J, Pandit N, Moxon ER, Achtman M. 1994. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol Microbiol 13:207–217. doi: 10.1111/j.1365-2958.1994.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 11.Cahoon LA, Seifert HS. 2013. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog 9:e1003074. doi: 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducey TF, Jackson L, Orvis J, Dyer DW. 2009. Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae. Microb Pathog 46:166–170. doi: 10.1016/j.micpath.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 15.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 16.de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169:2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills SA, Marletta MA. 2005. Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry 44:13553–13559. doi: 10.1021/bi0507579. [DOI] [PubMed] [Google Scholar]
- 18.Sheikh MA, Taylor GL. 2009. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol 72:1208–1220. doi: 10.1111/j.1365-2958.2009.06718.x. [DOI] [PubMed] [Google Scholar]
- 19.Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J Bacteriol 187:4865–4874. doi: 10.1128/JB.187.14.4865-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C, Genco CA. 2012. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J Bacteriol 194:1730–1742. doi: 10.1128/JB.06176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez CA, Daaboul GG, Ahn S, Reddington AP, Monroe MR, Zhang X, Irani RJ, Yu C, Genco CA, Cretich M, Chiari M, Goldberg BB, Connor JH, Unlu MS. 2011. Biomolecular detection employing the interferometric reflectance imaging sensor (IRIS). J Vis Exp doi: 10.3791/2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter BM, Gilbreath JJ, Pich OQ, McKelvey A, Maynard EL, Li ZZ, Merrell DS. 2013. Identification and characterization of novel Helicobacter pylori apo-Fur-regulated target genes. J Bacteriol 195:5526–5539. doi: 10.1128/JB.01026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray-Owen SD, Schryvers AB. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol 4:185–191. doi: 10.1016/0966-842X(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 24.Cornelissen CN, Kelley M, Hobbs MM, Anderson JE, Cannon JG, Cohen MS, Sparling PF. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol Microbiol 27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 25.McClure R, Nudel K, Massari P, Tjaden B, Su X, Rice PA, Genco CA. 2015. The gonococcal transcriptome during infection of the lower genital tract in women. PLoS One 10:e0133982. doi: 10.1371/journal.pone.0133982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. 2007. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J Bacteriol 189:3686–3694. doi: 10.1128/JB.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isabella V, Wright LF, Barth K, Spence JM, Grogan S, Genco CA, Clark VL. 2008. cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154:226–239. doi: 10.1099/mic.0.2007/010470-0. [DOI] [PubMed] [Google Scholar]
- 28.Gong Q, Wang J, Ahmad KM, Csordas AT, Zhou J, Nie J, Stewart R, Thomson JA, Rossi JJ, Soh HT. 2012. Selection strategy to generate aptamer pairs that bind to distinct sites on protein targets. Anal Chem 84:5365–5371. doi: 10.1021/ac300873p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider H, Griffiss JM, Williams GD, Pier GB. 1982. Immunological basis of serum resistance of Neisseria gonorrhoeae. J Gen Microbiol 128:13–22. [DOI] [PubMed] [Google Scholar]
- 30.Cohen MS, Cannon JG, Jerse AE, Charniga LM, Isbey SF, Whicker LG. 1994. Human experimentation with Neisseria gonorrhoeae: rationale, methods, and implications for the biology of infection and vaccine development. J Infect Dis 169:532–537. doi: 10.1093/infdis/169.3.532. [DOI] [PubMed] [Google Scholar]
- 31.Mehr IJ, Seifert HS. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol 30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 32.Baumler AJ, Tsolis RM, van der Velden AW, Stojiljkovic I, Anic S, Heffron F. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207–213. doi: 10.1016/S0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 33.Zheng S, Lan P, Liu X, Ye K. 2014. Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast. J Biol Chem 289:22692–22703. doi: 10.1074/jbc.M114.584490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng S, Wang J, Feng Y, Ye K. 2012. Solution structure of MSL2 CXC domain reveals an unusual Zn3Cys9 cluster and similarity to pre-SET domains of histone lysine methyltransferases. PLoS One 7:e45437. doi: 10.1371/journal.pone.0045437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S, Ye K. 2014. Purification, crystallization and preliminary X-ray diffraction analysis of Imp3 in complex with an Mpp10 peptide involved in yeast ribosome biogenesis. Acta Crystallogr F Struct Biol Commun 70:918–921. doi: 10.1107/S2053230X14010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daou N, Yu C, McClure R, Gudino C, Reed GW, Genco CA. 2013. Neisseria prophage repressor implicated in gonococcal pathogenesis. Infect Immun 81:3652–3661. doi: 10.1128/IAI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morse SA, Bartenstein L. 1980. Purine metabolism in Neisseria gonorrhoeae: the requirement for hypoxanthine. Can J Microbiol 26:13–20. doi: 10.1139/m80-003. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Gong Q, Maheshwari N, Eisenstein M, Arcila ML, Kosik KS, Soh HT. 2014. Particle display: a quantitative screening method for generating high-affinity aptamers. Angew Chem 53:4796–4801. doi: 10.1002/anie.201309334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Applied Biosystems. 1997. User bulletin no. 2. Relative quantification of gene expression. Applied Biosystems, Foster City, CA. [Google Scholar]
- 40.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Rudzinski JF, Gong Q, Soh HT, Atzberger PJ. 2012. Influence of target concentration and background binding on in vitro selection of affinity reagents. PLoS One 7:e43940. doi: 10.1371/journal.pone.0043940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fichorova RN, Desai PJ, Gibson FC III, Genco CA. 2001. Distinct proinflammatory host responses to Neisseria gonorrhoeae infection in immortalized human cervical and vaginal epithelial cells. Infect Immun 69:5840–5848. doi: 10.1128/IAI.69.9.5840-5848.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai PJ, Angerer A, Genco CA. 1996. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J Bacteriol 178:5020–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forng RY, Ekechukwu CR, Subbarao S, Morse SA, Genco CA. 1997. Promoter mapping and transcriptional regulation of the iron-regulated Neisseria gonorrhoeae fbpA gene. J Bacteriol 179:3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson LA, Ducey TF, Day MW, Zaitshik JB, Orvis J, Dyer DW. 2010. Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J Bacteriol 192:77–85. doi: 10.1128/JB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun 77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, Rappuoli R, Grandi G, Genco CA. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc Natl Acad Sci U S A 100:9542–9547. doi: 10.1073/pnas.1033001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remmele CW, Xian Y, Albrecht M, Faulstich M, Fraunholz M, Heinrichs E, Dittrich MT, Muller T, Reinhardt R, Rudel T. 2014. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res 42:10579–10595. doi: 10.1093/nar/gku762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fichorova RN, Rheinwald JG, Anderson DJ. 1997. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol Reprod 57:847–855. doi: 10.1095/biolreprod57.4.847. [DOI] [PubMed] [Google Scholar]
- 50.Follows SA, Murlidharan J, Massari P, Wetzler LM, Genco CA. 2009. Neisseria gonorrhoeae infection protects human endocervical epithelial cells from apoptosis via expression of host antiapoptotic proteins. Infect Immun 77:3602–3610. doi: 10.1128/IAI.01366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagen TA, Cornelissen CN. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol Microbiol 62:1144–1157. doi: 10.1111/j.1365-2958.2006.05429.x. [DOI] [PubMed] [Google Scholar]
- 52.Hopper S, Wilbur JS, Vasquez BL, Larson J, Clary S, Mehr IJ, Seifert HS, So M. 2000. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect Immun 68:896–905. doi: 10.1128/IAI.68.2.896-905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilbur JS, Chivers PT, Mattison K, Potter L, Brennan RG, So M. 2005. Neisseria gonorrhoeae FitA interacts with FitB to bind DNA through its ribbon-helix-helix motif. Biochemistry 44:12515–12524. doi: 10.1021/bi0511080. [DOI] [PubMed] [Google Scholar]
- 54.Yu C, Genco CA. 2012. Fur-mediated global regulatory circuits in pathogenic Neisseria species. J Bacteriol 194:6372–6381. doi: 10.1128/JB.00262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Householder TC, Fozo EM, Cardinale JA, Clark VL. 2000. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect Immun 68:5241–5246. doi: 10.1128/IAI.68.9.5241-5246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. 1995. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol 177:2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seib KL, Wu HJ, Srikhanta YN, Edwards JL, Falsetta ML, Hamilton AJ, Maguire TL, Grimmond SM, Apicella MA, McEwan AG, Jennings MP. 2007. Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol Microbiol 63:54–68. doi: 10.1111/j.1365-2958.2006.05478.x. [DOI] [PubMed] [Google Scholar]
- 58.Mercante AD, Jackson L, Johnson PJ, Stringer VA, Dyer DW, Shafer WM. 2012. MpeR regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism. Antimicrob Agents Chemother 56:1491–1501. doi: 10.1128/AAC.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee EH, Rouquette-Loughlin C, Folster JP, Shafer WM. 2003. FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism. J Bacteriol 185:7145–7152. doi: 10.1128/JB.185.24.7145-7152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warner DM, Shafer WM, Jerse AE. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol 70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.