ABSTRACT
The assembly of lipopolysaccharide (LPS) in the outer leaflet of the outer membrane (OM) requires the transenvelope Lpt (lipopolysaccharide transport) complex, made in Escherichia coli of seven essential proteins located in the inner membrane (IM) (LptBCFG), periplasm (LptA), and OM (LptDE). At the IM, LptBFG constitute an unusual ATP binding cassette (ABC) transporter, composed by the transmembrane LptFG proteins and the cytoplasmic LptB ATPase, which is thought to extract LPS from the IM and to provide the energy for its export across the periplasm to the cell surface. LptC is a small IM bitopic protein that binds to LptBFG and recruits LptA via its N- and C-terminal regions, and its role in LPS export is not completely understood. Here, we show that the expression level of lptB is a critical factor for suppressing lethality of deletions in the C-terminal region of LptC and the functioning of a hybrid Lpt machinery that carries Pa-LptC, the highly divergent LptC orthologue from Pseudomonas aeruginosa. We found that LptB overexpression stabilizes C-terminally truncated LptC mutant proteins, thereby allowing the formation of a sufficient amount of stable IM complexes to support growth. Moreover, the LptB level seems also critical for the assembly of IM complexes carrying Pa-LptC which is otherwise defective in interactions with the E. coli LptFG components. Overall, our data suggest that LptB and LptC functionally interact and support a model whereby LptB plays a key role in the assembly of the Lpt machinery.
IMPORTANCE The asymmetric outer membrane (OM) of Gram-negative bacteria contains in its outer leaflet an unusual glycolipid, the lipopolysaccharide (LPS). LPS largely contributes to the peculiar permeability barrier properties of the OM that prevent the entry of many antibiotics, thus making Gram-negative pathogens difficult to treat. In Escherichia coli the LPS transporter (the Lpt machine) is made of seven essential proteins (LptABCDEFG) that form a transenvelope complex. Here, we show that increased expression of the membrane-associated ABC protein LptB can suppress defects of LptC, which participates in the formation of the periplasmic bridge. This reveals functional interactions between these two components and supports a role of LptB in the assembly of the Lpt machine.
INTRODUCTION
Gram-negative bacteria are surrounded by two lipid bilayers, the inner membrane (IM) and outer membrane (OM), showing distinct composition, structural, and functional properties (1). The two membranes delimit an aqueous compartment, the periplasm, containing a thin peptidoglycan layer. The IM is a symmetric bilayer made of phospholipids, whereas the OM is an asymmetric membrane composed of glycerophospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet (1). LPS is a complex glycolipid assembled at the outer leaflet of the OM, where it forms a permeability barrier that prevents entry of many hydrophobic toxic compounds, including antibiotics (2). In Escherichia coli, LPS export to the cell surface is performed by a transenvelope complex of seven essential Lpt proteins (LptABCDEFG) spanning across the four-cell compartment from cytoplasm to OM (3–7). At the IM, LptB2FG constitute an ATP binding cassette (ABC) transporter that provides energy to the LPS transport system (8–10). LptC is a bitopic IM protein that associates with the IM LptB2FG transporter (8). The β-barrel LptD protein and the LptE lipoprotein constitute the OM translocon, characterized by a unique plug and barrel architecture, responsible for the final stages of LPS assembly at the cell surface (11–14). LptA is the key periplasmic component of the machinery that connects the IM and OM complexes (5, 15, 16).
The crystal structures of five components of the Lpt machinery, LptC from E. coli (17), LptA from E. coli and Pseudomonas aeruginosa (18, 19), the LptD-LptE complex from Shigella flexneri and Salmonella enterica serovar Typhimurium (12, 20), and the cytoplasmic ABC protein LptB from E. coli (10, 21), have been solved. Interestingly, LptA, the periplasmic region of LptC, and the periplasmic N-terminal region of LptD, despite a lack of sequence similarity, share a very similar β-jellyroll fold, made of the juxtaposition of a variable number of antiparallel β strands. Such a β-jellyroll fold (the Lpt fold) appears to be a key element in driving the assembly of the Lpt machinery. In fact, through these structurally homologous domains, the C terminus of LptC interacts with the N terminus of LptA, and the C terminus of LptA interacts with the N-terminal periplasmic domain of LptD, thus forming a protein bridge that connects the IM and the OM (12, 16, 20, 22).
The assembly of the transenvelope bridge appears to be finely regulated to prevent LPS mistargeting. Proper interaction of LptC with LptB2FG is necessary for LptA recruitment (22). Interaction of the N-terminal domain of LptD with LptA requires the correct maturation of the LptDE complex that in turns depends on nonconsecutive disulfide bond formation in LptD (16); LptE and the chaperone/protease BepA have been implicated in this process (23). Based on in vivo photo-cross-linking experiments, LPS molecules appear to cross the periplasm inside the β jellyroll of LptC and LptA (9). ATP hydrolysis by LptB, the cytoplasmic ATPase of the LptB2FG transporter, is required for LPS extraction from the IM and its transfer to LptA via LptC. Energy does not seem to be required for the assembly of the transenvelope bridge (9). Nevertheless, LptB also plays a role in the IM LptCFG subcomplex assembly, as shown by lptB mutants defective in assembly but proficient in the ATPase activity (10).
The molecular role of LptC in LPS transport is still elusive. LptC does not seem to be a functional component of the IM ABC transporter, as its association with the LptB2FG complex does not affect its ATPase activity (8); on the other hand, it appears relevant for proper IM complex assembly (22). Mutational analyses suggest that the interaction of LptC with the IM LptB2FG complex is mediated by the N-terminal region of the β jellyroll, whereas its transmembrane domain appears to be dispensable, as a periplasmic soluble version of LptC and a LptC chimera carrying a heterologous transmembrane segment is functional and proficient in Lpt complex assembly (22).
To gain further insights into the role of LptC in the LPS export pathway, we dissected E. coli LptC (Ec-LptC) functional domains by analyzing the phenotype of E. coli lptC mutants and the properties of LptC from P. aeruginosa (Pa-LptC) and Ec-/Pa-LptC chimeras in E. coli. We show that lptB ectopic expression suppresses the lethality of C-terminal deletion of LptC and allows the functioning of hybrid Lpt machinery that carries the highly divergent LptC orthologue from P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli bacterial strains used in this study are listed in Table 1. Left and right junctions of TnSS2 inserted in the ST-190 mutants were sequenced upon amplification of the two regions with oligonucleotide pairs AP23-FG1676 and FG690-AP179, respectively. P. aeruginosa PAO1 (25) was used as a source of P. aeruginosa DNA. Plasmids are listed in Table 2 with a brief outline of their construction by standard cloning techniques and, where indicated, three-step PCR (26). Chimeric lptC alleles and proteins are designated by three letters following the gene/protein name indicating sequentially the source, from either E. coli (C) or P. aeruginosa (P), of regions 1, 2, and 3, as defined in Fig. 1; -H indicates the His6 tag at the C terminus. Oligonucleotides used as primers for plasmid engineering and/or sequencing are listed in Table S1 in the supplemental material. All cloned PCR products were verified by DNA sequencing. Bacteria were grown in LD medium (27). When required, 0.2% (wt/vol) l-arabinose (as an inducer of the araBp promoter), 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 25 μg/ml chloramphenicol, and 100 μg/ml ampicillin were added. Solid media were prepared as described above with 1% (wt/vol) agar.
TABLE 1.
E. coli strains
| Strain | Descriptiona | Reference |
|---|---|---|
| AM604 | MC4100 Ara+ | 3 |
| AMM04 | AM604 lptD-SPA::kan | 22 |
| DH5α | Δ(argF-lac)169 ϕ80dlacZ58(M15) glnV44(AS) λ− rfbD1 gyrA96 recA1 endA1 spoT1 thi-1 hsdR17 | 40 |
| DH10B | araD139 Δ(ara-leu)7697 ΔlacX74 galU galK rpsL deoR ϕ80dlacZΔM15 endAI nupG recAl mcrA Δ(mrr hsdRMS mcrBC) | 41 |
| FL905 | AM604 Φ(kan araC araBp-lptC) | 5 |
| MC4100 | F− araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | 42 |
| MG1655 | K-12 F− λ− ilvG rfb-50 rph-1 | 43 |
| ST-190 | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB (Rifr) argE(Am) recA1 λR lptC::TnSS2 | 24 |
AS, amber suppressor.
TABLE 2.
Plasmids
| Plasmid | Parental plasmid/replicon | Relevant characteristic(s) | Construction/origina or reference |
|---|---|---|---|
| pGS100 | pGZ11EH | ptac-TIR cat oriVColD | 44 |
| pGS103 | pGS100 | ptac-lptC cat | 44 |
| pGS104 | pGS100 | ptac-lptCAB cat oriVColD | 44 |
| pGS103G153R | pGS100 | ptac-lptCG153R cat | 15 |
| pGS108 | pGS100 | ptac-lptC-H cat | 44 |
| pGS111 | pGS100 | ptac-Pa-lptC cat | Pa-lptC was PCR amplified with AP175 and AP176 primers from P. aeruginosa PAO1 genomic DNA and cloned into EcoRI-HindIII sites of pGS100 |
| pGS200 | pGS100 | ptac-Pa-lptC-H cat | Pa-lptC was PCR amplified with AP175 and AP237 primers from pGS111 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS201 | pGS100 | ptac-lptC-CPP cat | lptC-CPP was obtained by three step PCR with AP91, AP192, AP92, AP191 primers from pGS103 and pGS111 as templates and cloned into EcoRI-HindIII sites of pGS100 |
| pGS201H | pGS100 | ptac-lptC-CPP-H cat | lptC-CPP-H was PCR amplified with AP91 and AP237 primers from pGS201 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS202 | pGS100 | ptac-lptC-PCC cat | lptC-PCC was obtained by three-step PCR with AP91, AP195, AP92, AP196 primers from pGS103 and pGS111 as templates and cloned into EcoRI-HindIII sites of pGS100 |
| pGS202H | pGS100 | ptac-lptC-PCC-H cat | lptC-PCC-H was PCR amplified with AP91 and AP63 primers from pGS202 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS203 | pGS100 | ptac-lptC-CPC cat | lptC-CPC was obtained by three-step PCR with AP91, AP194, AP91, and AP193 primers from pGS201 and pGS103 as templates and cloned into EcoRI-HindIII sites of pGS100 |
| pGS203H | pGS100 | ptac-lptC-CPC-H cat | lptC-CPC-H was PCR amplified with AP91 and AP63 primers from pGS203 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS204 | pGS100 | ptac-lptC-PCP cat | lptC-PCP was obtained by three-step PCR with AP91, AP198, AP92, and AP197 primers from pGS202 and pGS111 as templates and cloned into EcoRI-HindIII sites of pGS100 |
| pGS204H | pGS100 | ptac-lptC-PCP-H cat | lptC-PCP-H was PCR amplified with AP91 and AP237 primers from pGS204 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS206 | pGS100 | ptac-lptC-CCP cat | lptC-CCP was obtained by three-step PCR with AP91, AP198, AP92, and AP197 primers from pGS103 and pGS111 as templates and cloned into EcoRI-HindIII sites of pGS100 |
| pGS206H | pGS100 | ptac-lptC-CCP-H cat | lptC-CCP-H was PCR amplified with AP91 and AP237 primers from pGS206 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS207 | pGS100 | ptac-lptC-PPC cat | lptC-PPC was obtained by three-step PCR with AP91, AP194, AP92, and AP193 primers from pGS111 and pGS103 as templates and cloned into EcoRI-HindIII sites of pGS100 |
| pGS207H | pGS100 | ptac-lptC-PPC-H cat | lptC-PPC-H was PCR amplified with AP91 and AP63 primers from pGS207 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS208 | pGS100 | ptac-lptC-CCΔ cat | lptC-PCP was PCR amplified with AP91 and AP63 primers from pGS103 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS208H | pGS100 | ptac-lptC- CCΔ-H cat | lptC-PCP-H was PCR amplified with AP91 and AP361 primers from pGS103 and cloned into EcoRI-HindIII sites of pGS100 |
| pGS321 | pGS100 | ptac-lptA cat | lptA was PCR amplified with AP55 and FG2723 primers from pGS104 and cloned into EcoRI-XbaI sites of pGS100 |
| pGS401 | pGS100 | ptac-SD1-EcoRI-XbaI-SD2-SalI-HindIII cat | 19 |
| pGS402 | pGS401 | ptac-lptC cat | 19 |
| pGS403 | pGS401 | ptac-Pa-lptC cat | Pa-lptC was PCR amplified with AP175 and AP176 primers from P. aeruginosa PAO1 DNA and cloned into EcoRI-XbaI sites of pGS401 downstream of SD1 |
| pGS404 | pGS402 | ptac-lptC-lptA cat | 19 |
| pGS407 | pGS403 | ptac-Pa-lptC-lptA cat | Ec-lptA was PCR amplified with FG2935 and FG2936 primers from MG1655 DNA and cloned into SalI-HindIII sites of pGS403 downstream of SD2 |
| pGS408 | pGS401 | ptac-lptC190N cat | lptC190N was PCR amplified with FG2978 and FG2979 primers from ST-190 genomic DNA and cloned into EcoRI-XbaI sites of pGS401 |
| pGS411 | pGS407 | ptac-lptC190N-lptA cat | lptC190N was PCR amplified with FG2978 and FG2979 primers from ST-190 genomic DNA and cloned into EcoRI-XbaI sites of pGS407 |
| pGS412 | pGS407 | ptac-lptCG153R-lptA cat | lptCG153R was obtained by EcoRI-XbaI digestion of pGS103G153R and cloned into EcoRI-XbaI sites of pGS407 |
| pGS413 | pGS412 | ptac-lptCG153R-lptAB cat | Ec-lptAB was PCR amplified with FG2935 and FG3058 primers from MG1655 DNA and cloned into SalI-HindIII sites of pGS412 |
| pGS414 | pGS408 | ptac-lptC190N-lptAB cat | Ec-lptAB was PCR amplified with FG2935 and FG3058 primers from MG1655 DNA and cloned into SalI-HindIII sites of pGS408 |
| pGS415 | pGS402 | ptac-lptC-lptAB cat | Ec-lptAB was PCR amplified with FG2935 and FG3058 primers from MG1655 DNA and cloned into SalI-HindIII sites of pGS402 |
| pGS416 | pGS401 | ptac-void-lptAB cat | 19 |
| pGS417 | pGS401 | ptac-lptCΔ139-191 cat | Ec-lptC1-138 was PCR amplified with FG2978 and FG3088 primers from pGS402 and cloned into EcoRI-XbaI sites of pGS401 |
| pGS418 | pGS407 | ptac-lptCΔ139-191-lptA cat | Ec-lptC1-138 was PCR amplified with FG2978 and FG3088 primers from pGS402 and cloned into EcoRI-XbaI sites of pGS407 |
| pGS419 | pGS416 | ptac-lptCΔ139-191-lptAB cat | Ec-lptC1-138 was PCR amplified with FG2978 and FG3088 primers from pGS402 and cloned into EcoRI-XbaI sites of pGS416 |
| pGS428 | pGS401 | ptac-void-lptB cat | 19 |
| pGS429 | pGS402 | ptac-lptC-lptB cat | Ec-lptB was obtained by SalI-HindIII digestion of pGS428 and cloned into SalI-HindIII sites of pGS402 |
| pGS429-LptBE163Q | pGS429 | ptac-lptC-lptBE163Q cat | E163Q substitution was obtained by site-directed mutagenesis with AP471 and AP472 primers from pGS429 as the template |
| pGS429-LptBF90A | pGS429 | ptac-lptC-lptBF90A cat | F90A substitution was obtained by site-directed mutagenesis with AP467 and AP468 primers from pGS429 as the template |
| pGS429-LptBF90Y | pGS429 | ptac-lptC-lptBF90Y cat | F90Y substitution was obtained by site-directed mutagenesis with AP469 and AP470 primers from pGS429 as the template |
| pGS430 | pGS408 | ptac-lptC190N-lptB cat | Ec-lptB was obtained by SalI-HindIII digestion of pGS428 and cloned into SalI-HindIII sites of pGS408 |
| pGS431 | pGS417 | ptac-lptCΔ139-191-lptB cat | Ec-lptB was obtained by SalI-HindIII digestion of pGS428 and cloned into SalI-HindIII sites of pGS417 |
| pGS431-LptBE163Q | pGS431 | ptac-lptCΔ139-191-lptBE163Q cat | E163Q substitution was obtained by site-directed mutagenesis with AP471 and AP472 primers from pGS431 as the template |
| pGS431-LptBF90A | pGS431 | ptac-lptCΔ139-191-lptBF90A cat | F90A substitution was obtained by site-directed mutagenesis with AP467 and AP468 primers from pGS431 as the template |
| pGS431-LptBF90Y | pGS431 | ptac-lptCΔ139-191-lptBF90Y cat | F90Y substitution was obtained by site-directed mutagenesis with AP469 and AP470 primers from pGS431 as the template |
| pGS434 | pGS412 | ptac-lptCG153R-lptB cat | Ec-lptB was obtained by SalI-HindIII digestion of pGS428 and cloned into SalI-HindIII sites of pGS412 |
| pGS439 | pGS401 | ptac-lptCG153R cat | lptCG153R was obtained by EcoRI-XbaI digestion of pGS412 and cloned into EcoRI-XbaI sites of pGS401 |
| pGS440 | pGS401 | ptac-void-lptA cat | lptA was obtained by SalI-HindIII digestion of pGS404 and cloned into SalI-HindIII sites of pGS401 |
| pGS448 | pGS416 | ptac-Pa-lptC-lptAB cat | Pa-lptC was obtained by EcoRI-XbaI digestion of pGS407 and cloned into EcoRI-XbaI sites of pGS416 |
| pGS456 | pGS428 | ptac-Pa-lptC-lptB cat | Pa-lptC was PCR amplified with AP175 and AP176 primers from P. aeruginosa PAO1 genomic DNA and cloned into EcoRI-XbaI sites of pGS428 |
Ec-lptC1-138, Ec-lptC encoding amino acids 1 to 138.
FIG 1.
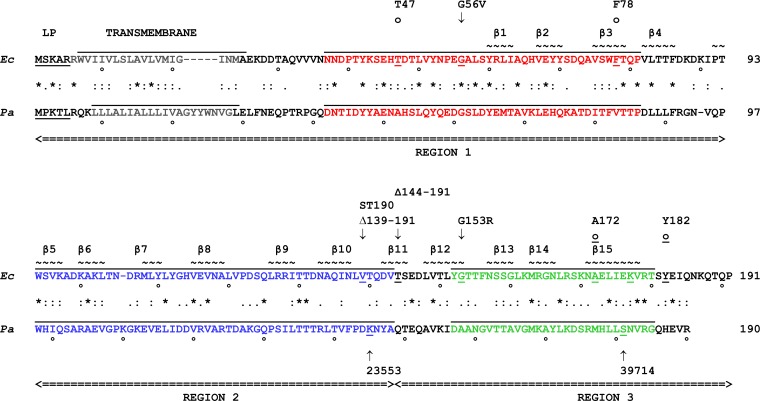
Comparison of E. coli and P. aeruginosa LptC amino acid sequences and structures. Amino acid sequence alignment of LptC from E. coli (Ec-LptC) and P. aeruginosa (Pa-LptC). Amino acid identity (asterisk) and similarity (colon and period) are labeled. Leader peptide (LP) and transmembrane regions are indicated. Regions 1, 2, and 3 swapped in chimera constructions are delimited by arrowheads at the end of double underlining. Sequences corresponding to the MEME motifs are overlined and color coded as follows: blue, motif 1 (residues 94 to 143); red, motif 2 (residues 37 to 81); green, motif 3 (residues 152 to 180). Relevant LptC mutations are indicated above and below the E. coli and P. aeruginosa sequences, respectively. Arrows indicate the mutated amino acid (for point mutations), the first deleted amino acid (for deletions), and the first amino acid at the right of the inserted transposon. ST-190, transposon insertion in ST-190 (24); 23553 and 39714 indicate two transposon insertions in Pa-lptC (http://ausubellab.mgh.harvard.edu). o and o indicate amino acids photo-cross-linked to LPS and LptA, respectively (9, 16). Tilde marks indicate amino acids in β strands, progressively numbered (17).
Identification of motifs in LptC.
The MM algorithm (28) from the MEME suite (29) (http://meme.nbcr.net/meme/tools/meme) was applied to analyze protein sequences for the occurrence of amino acid motifs. A motif is a sequence pattern that occurs repeatedly in a group of evolutionarily related proteins. The MM algorithm is capable of discovering different motifs with different number occurrences in a single data set. The algorithm estimates how many times each motif occurs in each sequence in the data set, ordering the motifs found with a statistical significance. Default settings have been applied to perform an ab initio motif discovery procedure to search for no more than 3 motifs on amino acid sequences of LptC homologues from a set of 13 representative taxa within Gammaproteobacteria (see Fig. S1 in the supplemental material). Motifs 1, 2, and 3 were numbered in an order based on statistical significance (Fig. 1; see also Fig. S1A). To correlate motif divergence and genetic distance among LptC homologues, a multialignment was performed with Multalin software, available online (http://multalin.toulouse.inra.fr/multalin/) (30), applying a modified blosum62 (blocks substitution) matrix for the amino acid substitution in proteins alignment (31), setting gap weight and gap length weight as 1 and 2, respectively. An LptC dendrogram was finally depicted, expressing the phylogenetic distance as a Dayhoff-Eck or point-accepted mutation (PAM), setting 20 PAMs as the minimal distance between sequences (see Fig. S1B). The overall phylogeny of the representative taxa selected among Gammaproteobacteria was compared to the LptC dendrogram, using the phylogenic tree proposed by Williams et al. (32) (see Fig. S1B).
The three-dimensional structures of Pa-LptC (PA4459) and Ec-LptCΔ139-191 (Ec-LptC with amino acids 139 to 191 deleted) were predicted using the online platform I-TASSER (iterative threading assembly refinement [http://zhanglab.ccmb.med.umich.edu/I-TASSER/]) (33). The computational models were obtained by threading, recognizing the E. coli LptC X-ray crystal structure (PDB no. 3MY2) (17) as the best template. Graphical modeling and superimposition were performed by Pymol (Pymol molecular graphics system, version 1.8; Schrödinger, LLC; www.pymol.org).
Complementation assay.
FL905 (araBp-lptC) carrying plasmids expressing different Ec-lptC or Pa-lptC alleles or chimeras, alone or in combination with Ec-lptA, lptB, or lptAB (see Table 2), were grown at 37°C in LD containing chloramphenicol (25 μg/ml) and arabinose (0.2%) for 18 h. Serial 10-fold dilutions in microtiter wells of the cultures were then replica plated on agar plates containing 25 μg/ml chloramphenicol, with or without 0.2% arabinose, and incubated overnight at 37°C.
Determination of LptA, LptC, LptB, and LptE levels.
LptA, LptC, LptB, and LptE levels were assessed in FL905 coexpressing wild-type or truncated LptC proteins with LptA or LptB or LptAB by Western blotting using polyclonal antibodies raised in mouse or rabbit against peptides (LptA and LptB) or whole proteins (LptE, LptC). Bacterial cultures grown up to an optical density at 600 nm (OD600) of 0.2 at 37°C in LD supplemented with 0.2% arabinose and 25 μg/ml of chloramphenicol were harvested by centrifugation, washed in LD, and diluted 500-fold in fresh media with or without 0.2% arabinose and with 25 μg/ml chloramphenicol. Growth was monitored by measuring the OD600. Samples for protein analysis were collected 240 min after the shift to nonpermissive conditions and centrifuged (16,000 × g, 5 min), and pellets were resuspended in a volume (in ml) of SDS sample buffer equal to 1/24 of the total OD of the sample. Samples were boiled for 10 min, and equal volumes (20 μl) were analyzed by 12.5% polyacrylamide-SDS gel electrophoresis. Proteins were transferred onto nitrocellulose membranes (GE Healthcare), and Western blot analysis was performed as previously described (4). Polyclonal sera raised against LptC and LptA (GenScript Corporation) were used as the primary antibody at a dilution of 1:500, whereas polyclonal sera against LptE and LptB (kindly provided by D. Khane and N. Ruiz, respectively) were used at a dilution of 1:5,000 and 1:1,000, respectively. Polyclonal serum raised against the S1 protein (kindly provided by F. Briani) was used at a dilution of 1:10,000. As secondary antibodies, anti-goat, anti-rabbit, and anti-mouse immunoglobulins (Li-Cor) were used at a dilution of 1:15,000.
Affinity purification of membrane Lpt complexes.
AMM04 cells harboring plasmids expressing His-tagged Ec-LptC, Pa-LptC, and LptC chimeras were subjected to affinity purification as previously described (7), with few modifications. The cells were lysed by a single cycle through a cell disrupter (One Shot Model; Constant Systems, Ltd) at a pressure of 22,000 lb/in2, and membranes were collected by ultracentrifugation of the supernatant at 100,000 × g for 1 h. The membranes were extracted at 4°C for 30 min with 5 ml of 50 mM Tris-HCl (pH 7.4), 10% glycerol, 1% dodecyl β-d-maltoside (Anatrace), and 5 mM MgCl2. The mixture was centrifuged again at 100,000 × g for 1 h, and insoluble material was discarded. The supernatant was incubated with 0.5 ml Talon resin suspension for 30 min at 4°C, and the mixture was then loaded onto a column. The column was washed with 10 ml of 50 mM Tris-HCl (pH 7.4), 10% glycerol, 0.05% dodecyl maltoside, and 5 mM imidazole, and eluted with 5 ml of 50 mM Tris-HCl (pH 7.4), 10% glycerol, 0.05% dodecyl maltoside, and 200 mM imidazole. The eluate was concentrated using an ultrafiltration device (Amicon Ultra, Millipore) by centrifugation at 5,000 × g to a final volume of 50 μl. Samples were mixed with 2× loading buffer, boiled, and separated on SDS-PAGE gel, electroblotted, and immunodetected using anti-His monoclonal antibodies (1:3,000) (Sigma-Aldrich) to detect wild-type Ec- and Pa-LptC-H and LptC-H chimeras and anti-LptA (1:2,000), LptB (1:1,000), LptD (1:500), and LptF (1:2,000), as previously described (22).
RESULTS
Complementation of E. coli lptC conditional mutant by lptC from P. aeruginosa.
A BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search using the amino acid sequence of LptC from Escherichia coli K-12 (Uniprot accession no. P0ADV9) as a query against the translated P. aeruginosa strain PAO1 genomic sequence (accession no. NC_002516.2) (http://www.pseudomonas.com/) identified a conserved putative protein of 190 amino acids with ∼20% sequence identity (Fig. 1) encoded by PA4459. This putative gene is located upstream of lptH (PA4460) and experimentally is demonstrated as the Pseudomonas homologue of lptA (19). Despite the low sequence similarity, the Pfam algorithm (34) predicts for the PA4459-encoded protein the same β-jellyroll domain found in Ec-LptC (17) (see Fig. S2 in the supplemental material). Overall, these data strongly suggest that PA4459 is the homologue of Ec-lptC and is herewith designated Pa-lptC.
We then tested whether Pa-lptC could complement the E. coli lptC conditional expression mutant under nonpermissive conditions. The reference lptC complementation test was performed in FL905, an E. coli araBp-lptC arabinose-dependent mutant (5), in which the complementing lptC allele is ectopically expressed at the basal level from the ptac promoter of the pGS100 vector. It should be noted that the level of lptC expressed from the plasmid is higher than that expressed from a native chromosomal promoter(s) (15). Moreover, under nonpermissive conditions (no chromosomal lptC expression in the absence of arabinose), expression of the downstream lptAB genes is driven by the minor promoters lptAp1 and lptAp2 (σE- and σD-dependent, respectively) located within the lptC coding region (35). FL905 was thus transformed with plasmid pGS200, which expresses Pa-lptC from the ptac promoter. As shown in Fig. 2, Pa-lptC, unlike the Ec-lptC control, did not complement FL905 for growth in the nonpermissive (no arabinose) conditions.
FIG 2.
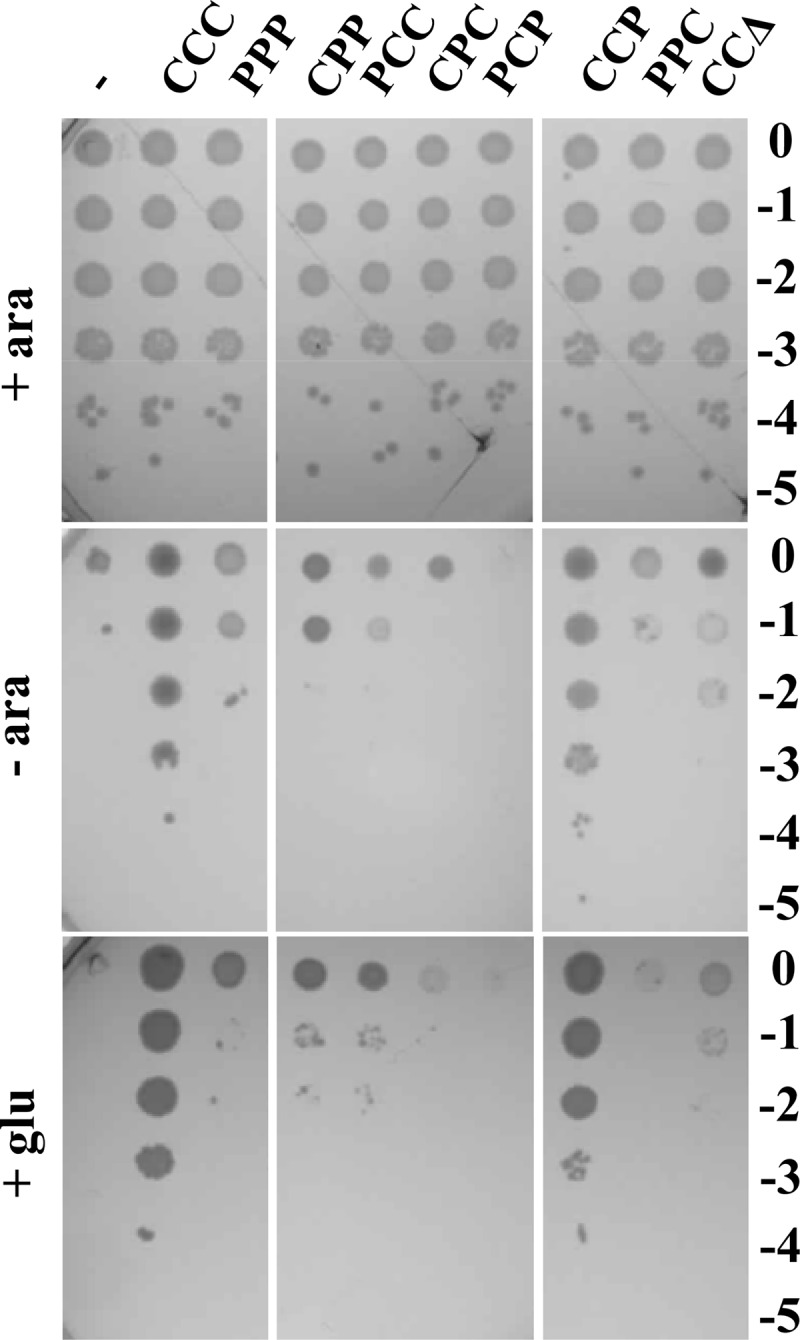
Complementation test of LptC depletion mutants with different E. coli and P. aeruginosa wild-type or chimeric LptC constructs. Cultures of FL905 (araBp-lptC) strains freshly transformed with pGS100 derivatives expressing Ec-LptC (pGS103, CCC), Pa-LptC (pGS111, PPP), LptC chimeras (pGS201, CPP; pGS202, PCC; pGS203, CPC; pGS204, PCP; pGS206, CCP; pGS207, PPC), or a truncated Ec-LptC protein missing region 3 (pGS208, CCΔ) and grown in LD-chloramphenicol-arabinose were serially diluted 1:10 in microtiter wells and replica plated in agar plates with (+ ara) or without (− ara) arabinose or with glucose (+ glu) to fully repress the araBp promoter. The log of the serial dilutions is indicated on the right of the panel.
Construction and expression of E. coli-P. aeruginosa LptC chimeras.
Sequence alignments among LptC homologues in a subset of representative taxa within Gammaproteobacteria using the MEME suite software (29) were performed to identify conserved sequence motifs in the LptC family of proteins. LptC multiple alignments recognized three sequence motifs in all representative LptC homologues taken into consideration, with the exception of those from P. aeruginosa and Legionella pneumophila (see Fig. S1A in the supplemental material). This observation is in agreement with the phylogenetic distances of such taxa (32) (see Fig. S1B). According to MEME analysis, motif 3 (which spans amino acids 152 to 180) is the least conserved across species, whereas motif 1 (amino acids 94 to 143), which is the most conserved in other bacteria, was recognized in neither L. pneumophila LptC nor P. aeruginosa LptC. As shown in Fig. 1 and in Fig. S1A in the supplemental material, motif 2 is located in the N-terminal region of LptC (region 1) and the most conserved motif, motif 1, lies in the central portion of the protein (region 2), whereas motif 3 is located at the C-terminal end of LptC (region 3).
To assess whether the lack of complementation by Pa-LptC could be imputed to any of the three regions, we swapped these three different regions of Ec- and Pa-LptC in all possible combinations, cloned the entire set of chimeric constructs in the pGS100 plasmid vector under the control of the ptac promoter, and tested the lptC chimeras by the reference lptC complementation assay. Only the chimeric protein that carries E. coli regions 1 and 2 and P. aeruginosa region 3 (CCP chimera) was able to support the growth of the araBp-lptC conditional mutant (Fig. 2).
Lpt complex assembly by LptC chimeras.
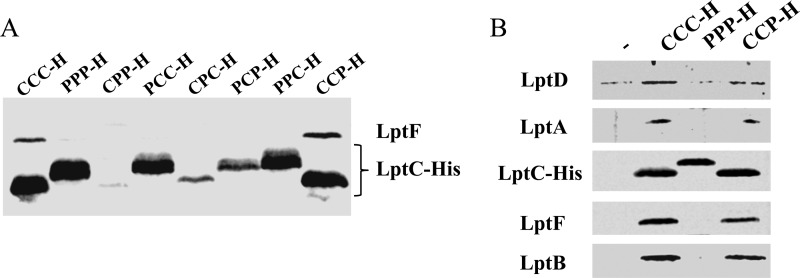
LptC is a component of the Lpt machinery and interacts with both the IM protein complex LptBFG and the periplasmic component LptA (8, 15, 22). The assembly of the Lpt machinery is a highly coordinated multistep process, as interaction of LptC with the IM complex LptBFG is required to recruit LptA and the OM complex LptDE (22). We therefore investigated by affinity purification experiments on the purified membrane fraction, as previously described (7, 22), whether the LptC chimeras are able to properly interact with the IM Lpt components and then to assemble the Lpt export machinery. C-terminal His-tagged Ec-LptC, Pa-LptC, and LptC chimeras expressed from the pGS100 vector in AM604 strain were used as baits in pulldown experiments. All His-tagged constructs behaved as the untagged counterparts for the ability, or lack thereof, to complement the araBp-lptC conditional mutant for growth (see Fig. S3 in the supplemental material). Total membranes collected from cells expressing C-terminally His-tagged Ec-, Pa-, or chimeric LptC proteins were solubilized, and the Lpt complexes with the different LptC baits were affinity purified. Samples were then processed by immunoblotting with a panel of specific antibodies. First, we assessed the ability of Pa-LptC and the chimeric proteins to recruit the IM complex LptBFG by revealing the presence of LptF with anti-LptF antibodies. As judged by the copurification profile shown in Fig. 3A it appears that only the complementing CCP chimera binds to the inner membrane component LptF at a level comparable to that of Ec-LptC. On the contrary, Pa-LptC and the noncomplementing chimeras fail to assemble the IM complex, as they are all unable to interact with LptF (Fig. 3A). Indeed, based on the copurification profile of LptA, LptD, and LptE shown in Fig. 3B, the CCP chimera but not Pa-LptC is able to recruit LptA and the LptDE complex and thus to assemble the whole Lpt machine.
FIG 3.
Assembly of the Lpt complex by Pa-LptC and LptC chimeras. Dodecyl β-d-maltoside-solubilized total membranes from AMM04 strains harboring pGS100 derivatives expressing the His-tagged proteins of interest were affinity purified using a Talon metal affinity resin, as described in Materials and Methods. Proteins were then fractionated by SDS-PAGE and immunoblotted with suitable antibodies to detect the corresponding proteins in the fraction. (A) Assembly of the Lpt IM complex. Samples were prepared from AMM04 harboring the pGS100 derivatives pGS108 (expressing Ec-LptC-H, CCC), pGS200 (expressing Pa-LptC-H, PPP), or those expressing His-tagged LptC chimeras, namely, pGS201H (CPP-H), pGS202H (PCC-H), pGS203H (CPC-H), pGS204H (PCP-H), pGS206H (CCP-H), and pGS207H (PPC-H). Immunoblotting was performed with anti-LptF and anti-His antibodies to detect LptF and the different His-tagged LptC forms, respectively, which display different electrophoretic mobility on SDS-PAGE. (B) Assembly of the transenvelope Lpt complex. Samples were prepared from AM604 harboring pGS108 (Ec-LptC-H, CCC), pGS200 (Pa-LptC-H, PPP), the His-tagged LptC CCP chimera (pGS206H), or pGS100 expressing the His tag (−) as a negative control. Immunoblotting was performed with the antibodies anti-His (to detect the different LptC forms) and anti-LptA, anti-LptB, anti-LptF, and anti-LptD.
These results implicate E. coli LptC regions 1 and 2 in the interaction with the LptB2FG complex. Moreover, the fact that a xenogeneic region 3 allows the recruitment of Ec-LptA and therefore the assembly of the Lpt machine suggests that either LptC region 3 is not relevant for LptC functionality or that the Lpt fold, rather than a specific amino acid sequence, is required to fulfill LptC function(s).
LptC region 3 may be dispensable.
Previous data (15, 16, 22) implicated the C-terminal region of Ec-LptC (which encompasses region 3) (Fig. 1) in binding with LptA. In particular, the Ec-LptC residues A172 and Y182 are thought to interact with LptA (16), whereas the G153R amino acid change in Ec-LptC (encoded by Ec-lptCG153R) not only is lethal but also impairs LptC-LptA copurification (15, 22). However, the results presented here suggest that structural features rather than specific amino acids residues are relevant to establish LptA-LptC interaction.
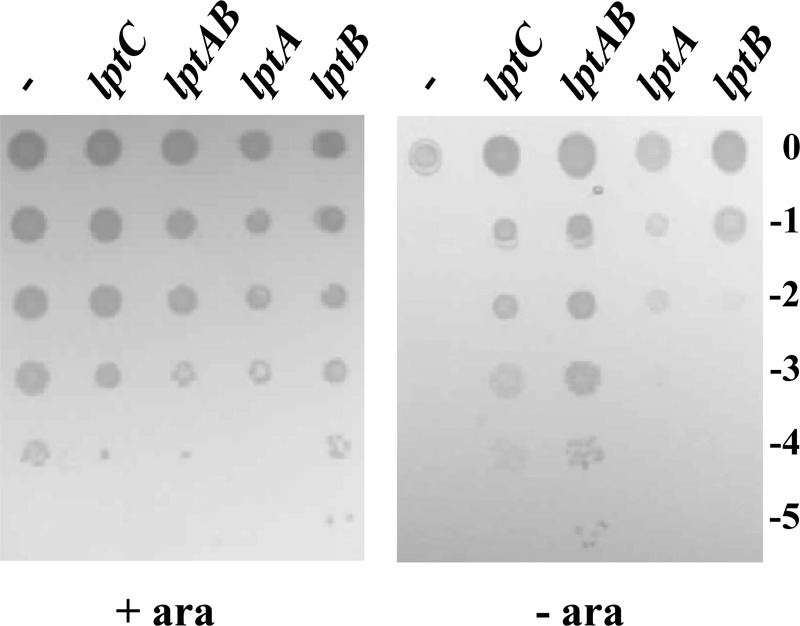
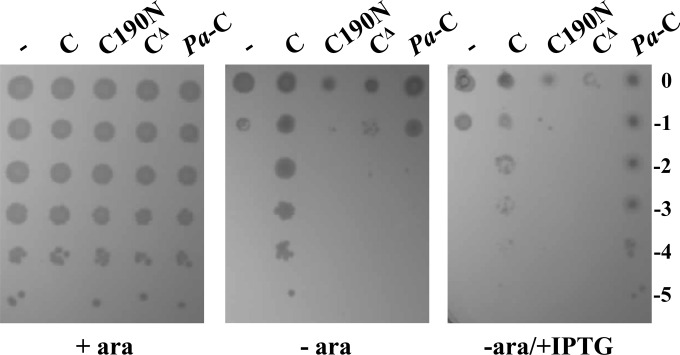
To gain further insights into this issue, we revisited the previously described conditional expression mutant ST-190 (24), in which a minitransposon (TnSS2) with the arabinose-inducible araBp promoter oriented toward lptAB is inserted after nucleotide 413 of lptC, as assessed by sequencing of this region. We resequenced the right junction of the TnSS2 insertion and, while we confirmed our previous data (24), we also found that the minitransposon insertion had created an 8-nucleotide direct repeat of the target site after nucleotide 413 of lptC (within codon 138). In the resulting LptC mutant protein, the L138 residue was preserved and amino acids 139 to 191 were substituted by the transposon-encoded CLLIRSGHLGRIPGDPVID sequence (Fig. 1). Thus, this mutant expresses, from the major promoter of the yrbG operon, an lptC substitution allele (henceforth named lptC190N) which encodes the 1 to 138 N-terminal peptide of LptC fused to a 19-amino-acid-long C terminus encoded by the left end of the inserted minitransposon (Fig. 1). Therefore, the LptC truncation in ST-190 largely overlaps region 3 of the protein. The minitransposon insertion occurs immediately upstream of the minor promoters lptAp1p2 located within lptC (35). In nonpermissive (no arabinose) conditions, expression of lptAB might be driven by these minor promoters only, whereas, in permissive conditions, the strong promoter araBp is also active. The viability of ST-190 in the presence of arabinose (24) indicates that, under such conditions, LptC190N is functional, whereas the nonviability of the mutant in nonpermissive condition suggests that lptAB expression is either lacking or insufficient. Here, we show (Fig. 4) that ST-190 viability in nonpermissive condition can be rescued not only by ectopic expression of lptC but also by a plasmid harboring lptAB. This indicates that, in this mutant strain (i) lptAB genes are indeed expressed from the minor promoters and their expression level is sufficient for viability in the presence of a wild-type LptC; (ii) the truncated lptC190N allele is defective when lptAB is expressed only from the ancillary promoters lptAp1p2; and (iii) a higher expression level of lptAB (from araBp in permissive conditions and/or from the complementing plasmid in the absence of arabinose) suppresses the lptC190N growth defect. It thus appears that the lptAB expression level from the ancillary lptAp1p2 promoters is limiting for the truncated lptC190N allele and that the C-terminal 53 residues of LptC are dispensable at least under conditions of higher (nonlimiting) lptAB expression levels.
FIG 4.
Growth of the ST-190 conditional expression mutant with different levels of LptA and/or LptB expression. Cultures of ST-190 transformed with pGS100 derivatives expressing the genes indicated on the top of each lane (lptC carried on pGS103; lptAB carried on pGS416; lptA carried on pGS321; lptB carried on pGS428) and grown in LD-chloramphenicol-arabinose were serially diluted 1:10 in microtiter wells and replica plated on agar plates supplemented (+ ara) or not supplemented (− ara) with arabinose. The log of the serial dilutions is indicated on the right of the panel.
To rule out that the observed phenotype depends on the ST-190 genetic background or the 19-amino-acid C-terminal substitution of the lptC190N allele, we tested whether plasmids expressing lptC190N or the truncated lptCΔ139-191 allele either alone or coexpressed with lptA, lptB, or both could complement the conditional FL905 mutant. As shown in Table 3, neither lptC190N nor lptCΔ139-191 complemented FL905 in the standard lptC complementation assay, whereas both mutants supported the growth of FL905 under nonpermissive conditions (no arabinose) when coexpressed with lptAB. Interestingly, whereas neither lptC mutant could complement when coexpressed with lptA only, coexpression of either lptC190N or lptCΔ139-191 with lptB was able to rescue FL905 growth under nonpermissive conditions (Table 3). Thus, expression of lptB over the basal lptAp1p2 level is required to suppress the lptC190N and lptCΔ139-191 defects, whereas the expression level of lptA does not seem to be a limiting factor. As in ST-190 (carrying the lptC190N allele), growth under nonpermissive conditions requires both lptA and lptB overexpression; we speculate that, in this mutant strain, the minitransposon insertion in close proximity to lptAp1p2 and/or the different genetic background might negatively affect the expression of the downstream lptAB operon. On the contrary, the defective lptCG153R mutation was not suppressed by increased lptB and/or lptAB expression (Table 3).
TABLE 3.
Complementation assays in LptC-depleted cells with additional ectopic expressiona of lptA-lptB
| Suppressing gene | Efficiency of plating with complementing lptC allele: |
|||||
|---|---|---|---|---|---|---|
| None | Ec-lptC | Ec-lptC190N | Ec-lptCΔ139-191 | Ec-lptCG153R | Pa-lptC | |
| None | − | + | − | − | − | − |
| lptAB | − | + | + | + | − | ± |
| lptA | − | + | − | − | − | − |
| lptB | − | + | + | + | − | + |
FL905 strains harboring pGS401 derivatives expressing different combinations of lptC complementing alleles and lptA-lptB suppressor genes were grown in LD with 0.2% arabinose and 25 μg/ml chloramphenicol. Serial dilutions in microtiter plates were replica plated on the same medium or without arabinose. Efficiency of plating in the nonpermissive condition (no arabinose) is expressed as follows: +, about 1; ±, between 10−2 and 10−3; −, <10−3.
LptB is a limiting factor for stabilization of LptCΔ139-191.
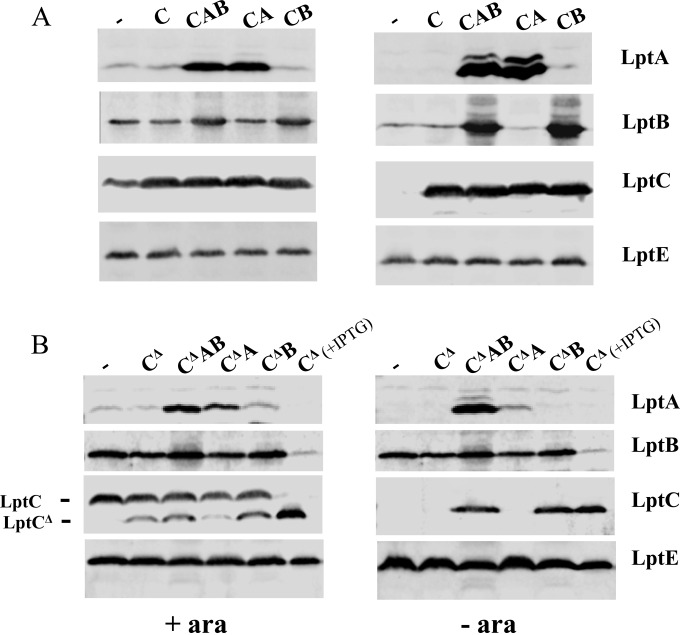
Suppression of LptC defects by increased expression of LptB is intriguing, as it reveals functional interactions between the cytoplasmic LptB and the bitopic IM LptC protein. LptB has been implicated not only in providing energy through ATP hydrolysis but also in the assembly of the IM complex (10). It could be hypothesized that the assembly of LptC lacking the C-terminal domain is partially defective, thus leading to LptC instability, and that a level of LptB higher than that provided by the ancillary promoters lptAp1 and lptAp2 is necessary for C-terminally truncated LptC assembly and stability. We therefore examined the levels of C-terminally truncated LptC with lptB expressed from a plasmid. FL905 cells coexpressing wild-type lptC or lptCΔ139-191 with lptA, lptB, or lptAB were grown up to the exponential phase and shifted into a medium lacking arabinose to deplete the chromosomally encoded wild-type LptC while allowing expression of the alleles from the complementing plasmids. Samples were then taken from cultures grown in the presence or in the absence of arabinose 240 min after the shift to nonpermissive conditions and analyzed by Western blotting using anti-LptA, anti-LptC, and anti-LptB antibodies. The level of the LptE protein was used as a sample loading control. As shown in Fig. 5, the expression level of LptCΔ139-191 in depleted FL905 cells was detectable when lptAB or lptB alone was also ectopically expressed, whereas it was not when the lptAB operon was expressed from the minor chromosomal promoters only. Interestingly, in the lptCΔ139-191 mutant background, the LptA level also increased when lptA was coexpressed with lptB (Fig. 5B, compare lanes LptCΔ139-191 LptAB and LptCΔ139-191 LptA), in keeping with our previous observations (15) that the increased amount of wild-type or truncated LptC helps in stabilizing LptA.
FIG 5.
Expression levels of LptA, LptB, and LptC or LptCΔ139-191 proteins in FL905 upon depletion of the chromosomally encoded LptC. FL905 cells transformed with pGS100 (−) or pGS100 derivatives harboring wild-type LptC (C, carried by pGS103) or LptCΔ139-191 (CΔ, carried by pGS417) as indicated on the left of panels A and B, respectively, coexpressed with LptA (A, carried by pGS404 and pGS418, respectively), LptB (B, carried by pGS429 and pGS431, respectively), or LptAB (AB, carried by pGS415 and pGS419, respectively), as indicated on the top of the lanes, were grown in LD in the presence (+ ara) or absence (− ara) of arabinose and chloramphenicol, as described in Materials and Methods. Samples collected 4 h after a shift to the nonpermissive condition were analyzed by Western blotting using anti-LptA, anti-LptB, anti-LptC, and anti-LptE antibodies. An equal amount of cells (OD600, 0.6) was loaded onto each lane. The migrations of full-length LptC and LptCΔ are indicated on the left side of panel B. The last lane of each image in panel B (CΔ + IPTG) was loaded with 10 μl of cell extract of FL905 harboring pGS417 (LptCΔ139-191) arabinose depleted for 3 h and further incubated 1 h with 0.1 mM IPTG to induce expression of the truncated LptC protein. LptE level was used as a sample loading control.
The above data suggest that a higher level of lptB expression mediates stabilization of LptCΔ139-191, thus suppressing the lethal phenotype of FL905 cells carrying the truncated lptC allele. However, neither LptCΔ139-191 nor LptC190N overexpressed from an IPTG-inducible promoter were able to rescue growth of FL905 cells under nonpermissive conditions (Fig. 6), thus confirming that an increased level of LptB is able to stabilize the otherwise unstable LptC truncated proteins. In line with these data is the finding that His-tagged truncated Ec-LptC protein missing region 3 (CCΔ) is not detectable when lptB is not overexpressed (see panel E in Fig. S4 in the supplemental material); moreover, this truncated form of LptC appears to be highly toxic when expressed from an IPTG-inducible promoter (see panel D in Fig. S4).
FIG 6.
Rescue of FL905 growth by overexpression of truncated Ec-lptC or Pa-lptC alleles. FL905 cells transformed with pGS100 (−) or pGS100 derivatives harboring wild-type LptC (pGS103), LptCΔ139-191 (pGS417), LptC190N (pGS408), or Pa-LptC (pGS111), as indicated on top of the lanes, were grown in LD-chloramphenicol-arabinose, serially diluted 1:10 in microtiter wells, and replica plated on LD-agar plates with chloramphenicol and with (+ ara) or without (− ara) arabinose and with IPTG (+ IPTG) to induce the lptC allele on the plasmids, as described in Materials and Methods. The log of the serial dilutions is indicated on the right.
Overexpression of lptB rescues LptC-depleted cells complemented by Pa-lptC.
We showed (Fig. 2) that expression of the highly divergent Pa-lptC gene from a plasmid did not complement FL905 for growth in the absence of arabinose. To assess whether lptB expression could be a limiting factor for complementation by the xenogeneic Pa-LptC, we tested whether increased expression of lptB could rescue growth of Ec-LptC-depleted cells. FL905 cells were transformed with plasmids coexpressing Pa-lptC with lptA, lptB, or lptAB from E. coli. As shown in Table 3, ectopic expression of lptB rescued growth of FL905 complemented by Pa-lptC. However, overexpression of Pa-lptC with (see Fig. S2 in the supplemental material) and without (Fig. 6) the His tag from an IPTG-inducible promoter also rescued FL905 growth, thus supporting the view that Pa-LptC might have a lower affinity for the E. coli IM Lpt complex. Although His-tagged Pa-LptC is able to complement the FL905 growth defect, the level of His-tagged Pa-LptC is very low and not detectable with anti-His antibodies (see Fig. S4). Overall, these data confirm that P. aeruginosa open reading frame PA4459 encodes the functional homologue of E. coli LptC.
LptB mutants in either ATPase or assembly function do not suppress the growth defect of C-terminally deleted LptC.
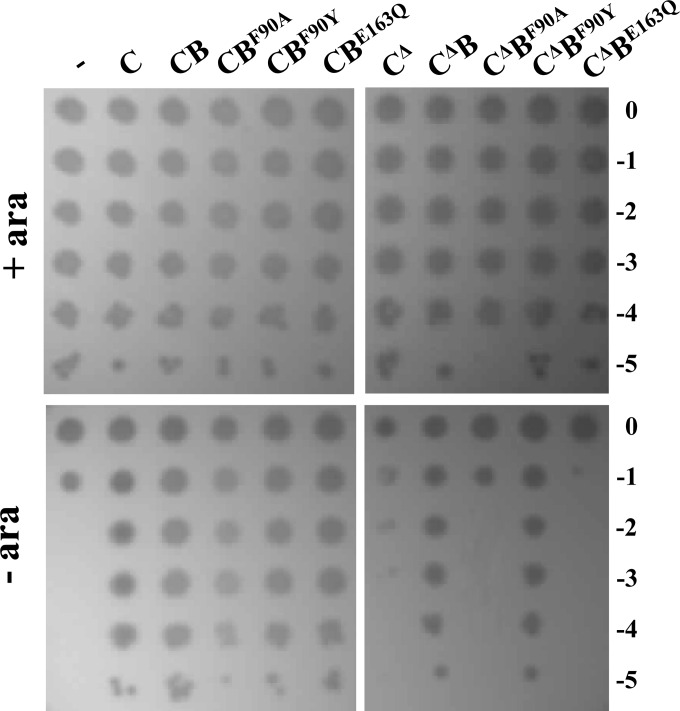
The ATPase activity of LptB can be genetically separated from its ability to assemble the IM LptBCFG complex (10). Indeed, LptBE163Q mutant is catalytically inactive but retains the ability to interact with the transmembrane regions of LptFG. On the contrary, LptB F90Y and F90A amino acid substitutions do not affect LptB ATPase activity, although they impair to different extents the LptB assembly properties (10). We therefore tested whether ectopic expression of lptB mutants E163Q, F90Y, and F90A suppresses lethality of the truncated lptCΔ139-191 allele. It should be remembered that, in our experimental setting, a wild-type copy of lptB is expressed from the ancillary lptAp1p2 chromosomal promoters. None of the coexpressed lptB mutants affected the growth of FL905 complemented by wild-type lptC (Fig. 7). On the contrary, ectopic expression of LptBE163Q and LptBF90A did not rescue the growth of FL905 complemented by LptCΔ139-191, whereas the partial loss of function mutant LptBF90Y was able to suppress the LptCΔ139-191 growth defect (Fig. 7).
FIG 7.
Suppression of LptC defects by lptB mutants. FL905 cells transformed with pGS100 (−) or pGS100 derivatives harboring wild-type LptC (C, carried by pGS103) or LptCΔ139-191 (CΔ, pGS417) coexpressed with LptB (B, carried by pGS429 and pGS431, respectively), LptBF90A (BF90A carried by pGS429-LptBF90A and pGS431-LptBF90A, respectively), LptBF90Y (BF90Y, carried by pGS429-LptBF90Y and pGS431-LptBF90Y, respectively), or LptBE163Q (BE163Q, carried by pGS429-LptBE163Q and pGS431-LptBE163Q, respectively), as indicated on top of the lanes, were grown in LD-chloramphenicol-arabinose, serially diluted 1:10 in microtiter wells, and replica plated on LD-agar plates with chloramphenicol in the presence (+ ara) or in the absence (− ara) of arabinose, as described in Materials and Methods. The log of the serial dilutions is indicated on the right.
DISCUSSION
LptC is an unusual component of the IM LptB2FG complex, an atypical ABC exporter (36) that energizes LPS transport. LptC role in LPS transport is not completely understood. Its transmembrane N-terminal domain is dispensable, whereas the soluble periplasmic domain interacts with both LptA and the IM LptB2FG complex via the C- and N-terminal regions, respectively (16, 22). LptC is thought to bind LPS and to transfer it to LptA in an energy-dependent way (9, 17), although its association with LptB2FG does not affect LptB ATPase activity (8). Here, we have further dissected the LptC functional domains by analyzing the properties of LptC C-terminal deletions and of Pa-LptC and Ec-/Pa-LptC chimeras in E. coli.
The LptC C-terminal region can be replaced by a xenogeneic divergent sequence.
Among the seven components of the Lpt machinery, Pa-LptC is the most divergent from the E. coli homologue (19% amino acid sequence identity) and does not seem to bear the most conserved of three motifs (encompassing amino acids 94 to 143) identified in LptC on the basis of sequence alignments of a panel of homologues (Fig. 1; see also Fig. S1 in the supplemental material).
Ectopic expression of Pa-LptC, unlike Ec-LptC expression, does not complement Ec-LptC-depleted E. coli mutants in the reference lptC complementation test. We exploited these noncomplementing conditions to dissect Pa-LptC and implicate specific regions in this phenotype. The Pa-LptC protein appears to be defective in the assembly with the E. coli components of the Lpt multiprotein machinery, as it does not interact with the Ec-LptB2FG IM subcomplex (Fig. 3); as previously shown, such interaction is necessary in vivo for LptA recruitment and for the whole Lpt complex assembly (22). On the other hand, overexpression of Pa-LptC by IPTG induction complements FL905 for growth in nonpermissive conditions, thus suggesting that a viable amount of hybrid Lpt machine can be assembled and that the inefficient interactions of Pa-LptC with the Ec-Lpt proteins may be overcome by Pa-LptC overexpression (Fig. 6).
Analysis of Ec-/Pa-LptC chimeras, in which the three LptC regions from the two organisms were swapped in all possible combinations, indicates that the E. coli LptC protein tolerates the substitution of the C-terminal region with the corresponding P. aeruginosa homologous region. Indeed, the LptC CCP chimera complements LptC-depleted E. coli in the lptC complementation test (Fig. 2) and assembles the LptC complex (Fig. 3B), whereas the noncomplementing Ec-/Pa-LptC chimeras, like Pa-LptC, are unable to bind the IM LptB2FG subcomplex in vivo. Based on these data, it appears that regions 1 and 2 are relevant for LptC function and Lpt complex assembly, as their substitutions with xenogeneic divergent sequences is not tolerated, whereas the C-terminal region 3 may have an ancillary role. As discussed below, it is possible that the presence of a structured C-terminal domain from P. aeruginosa helps to stabilize the CCP chimera.
Functional interaction between LptB and LptC.
The C-terminal region of LptC has been previously implicated in binding to LptA. Indeed, the unviable LptCG153R mutant fails to interact with LptA and to assemble the transenvelope complex, although it associates with the IM LptB2FG subcomplex (15, 22). Moreover, by in vivo photo-cross-linking, the C-terminal LptC residues A172 and Y182 have been implicated in binding LptA (16). These three residues, therefore, define the LptA-LptC interaction interface.
In contrast, we show here not only that the substitution of the C-terminal region with the highly divergent P. aeruginosa homologous region is viable but also that lptC alleles that are either C-terminally truncated or substituted by an unrelated sequence (lptCΔ139-191 and lptC190N, respectively) and the highly divergent Pa-lptC homologue, although unable to complement in the reference lptC complementation test, are viable under conditions of nonlimiting expression of lptB. Thus, a higher expression of lptB suppresses the growth defect of these lptC alleles. This suggests that, upon repression of chromosomal araBp-lptC, lptA expression from lptAp1p2 promoters is sufficient for growth, whereas lptB expression is the limiting factor for viability of lptCΔ139-191, lptC190N, or Pa-lptC-complemented E. coli.
LptCΔ139-191 stabilization appears crucial for FL905 viability upon LptC depletion. Indeed, the LptCΔ139-191 steady-state level increases when lptB but not lptA is overexpressed (Fig. 5). Interestingly, LptCΔ139-191 stabilization by increased expression of lptB seems also to positively affect LptA abundance (Fig. 5), thus implicating LptB in the control of LptC-LptA interactions. The tight genetic association of lptCAB, which belong to the same yrbG operon (24), and the presence of two ancillary promoters within lptC for the nested lptAB operon (4, 35) may be instrumental in maintaining balanced expression levels of the three encoded proteins.
The finding that ectopic expression of lptB allows functioning of mutant LptC or P. aeruginosa-E. coli hybrid Lpt machineries raises questions on the functional interactions between LptC and LptB. LptB is the IM-associated ATPase that interacts with the IM complex and provides the energy required for LPS transport (8, 9). LptB plays also a role in the assembly of the IM LptB2FGC subcomplex and, interestingly, these two functions can be genetically separated (10). In fact, the LptBE163Q mutant is catalytically inactive but retains the ability to interact with the transmembrane regions of LptFG. On the contrary, the F90A and F90Y amino acid substitutions do not affect LptB ATPase activity but impair, the former completely, the latter only partially, LptB assembly properties. We found that ectopic expression of both the catalytically inactive LptBE163Q and assembly-defective LptBF90A mutants (10) does not rescue the lethal phenotype of FL905 cells complemented by lptCΔ139-191, whereas LptBF90Y does. It should be remembered that, although partially defective in the LptCFG complex assembly, LptBF90Y is able to complement a lptB-deficient mutant (10); therefore, our results show that a nonlimiting expression of LptB endowed with both ATPase and assembly functions is required for the suppression of the lethal phenotype of C-terminally truncated LptC.
The role of LptB in the assembly of the Lpt machine (10) may explain why the level of LptB expression may be a limiting factor in the presence of the C-terminally truncated LptC. In fact, such LptC mutant, although fully proficient for interaction with LptB2FG, is highly unstable and may cause a rapid functional inactivation of the IM Lpt subcomplex, therefore affecting the recruitment of LptA and of the OM LptDE subcomplex. In LptC-depleted cells complemented by wild-type LptC, the expression level of lptB from the ancillary promoters may be sufficient for the assembly of an adequate number of stable Lpt IM subcomplexes. On the contrary, the rapid inactivation of IM subcomplexes containing C-terminally truncated LptC would result in a higher turnover of such Lpt IM subcomplexes, and thus, an increased amount of LptB would be required to assemble new functional IM subcomplexes sufficient for cell growth.
Interestingly, growth of LptC-depleted E. coli cells complemented by Pa-LptC can be rescued by both ectopic lptB expression and overexpression of Pa-lptC from an inducible IPTG promoter, whereas overexpression of E. coli C-terminally truncated or substituted lptC alleles does not. These data suggest that, in this case, the limiting factor, rather than Pa-LptC stability, is a lower affinity of the P. aeruginosa component for E. coli LptFG (Fig. 3A) and that the amount of IM subcomplexes sufficient to support growth could be achieved by increasing the level of either Pa-LptC or Ec-LptB. These findings are in agreement with a previous work showing that LptH (the P. aeruginosa LptA homologue) can replace Ec-LptA (19) when a functional IM LptCB2FG complex is in place to recruit LptA/LptH and, therefore, the OM LptDE subcomplex. Overall, the results here reported reinforce the view that the assembly of the IM complex is a control step for the formation of a functional transenvelope bridge.
On the other hand, the defective lptCG153R mutation was not suppressed by increased lptB expression. LptCG153R is a loss-of-function mutant protein able to assemble the IM LptB2FG complex but unable to interact with LptA (15, 22). As judged by its crystal structure, the LptCG153R protein maintains the overall β-jellyroll fold, and we proposed that the protein can be defective either in a conformational change required for LptA interaction or in LPS binding (22); likely, none of these defects can be suppressed by increased LptB levels.
What is the role of LptC in LPS transport?
The current model for LPS export postulates that LPS in the outer leaflet of the IM is extracted by the LptCFG complex with energy provided by the associated LptB ATPase component (9, 10). LptC cannot extract LPS on its own, but LPS likely is delivered to LptC by LptF or LptG or by both. LptC, which is functional even without its transmembrane domain (22), binds LPS at the N-terminal region (via residue T47) and transfers it to LptA, and both steps require ATP hydrolysis (9). LptA is thought to interact with the N-terminal region of LptD and to deliver the LPS to the LptDE translocon for assembly at the OM (12, 16, 20). The Lpt fold that is shared by LptA, LptC, and LptD (12, 17, 18, 20) and predicted in the periplasmic region of LptF and LptG (15, 22) appears to be a key element in driving the assembly of the transenvelope bridge. It has been suggested that these proteins may form a hydrophobic groove that accommodates the lipid moiety of LPS for its transport from the inner membrane to the outer membrane (9, 16).
Here, we show that LPS transport can be accomplished by Lpt machines carrying either C-terminally truncated LptC proteins or the highly divergent Pa-LptC (under specific conditions, i.e., increased lptB level and/or Pa-lptC overexpression) or the chimeric CCP LptC protein in which the C terminus is derived from P. aeruginosa. Since the CCP chimera appears fully functional, we speculate that the C-terminal region of Pa-LptC can fully substitute the corresponding region in Ec-LptC in recruiting Ec-LptA and building the hydrophobic groove for LPS export. Similarly, we propose that Pa-LptC is not defective in LptA recruitment and that its inability to assemble a functional Lpt machine relies on its low affinity to the IM LptB2FG complex. Such a defect is suppressed either by an increased level of LptB or by Pa-LptC overexpression, which helps stabilize the IM Pa-LptC/Ec-LptB2FG hybrid subcomplex and, as a consequence, allows the recruitment of Ec-LptA. The functional interaction between LptC and LptB is in line with a regulatory role of LptC in modulating the Lpt protein bridge formation (22) and/or the activity of the IM LptB2FG complex (37) It is not clear at the structural level how a C-terminally truncated LptC may function in recruiting LptA and assembling a functional Lpt machine. Based on structure prediction (see Fig. S2 in the supplemental material), the C-terminally truncated LptC protein seems to maintain the β-jellyroll structure; therefore, the binding between the C-terminally truncated LptC and LptA might occur through the edge of their respective β jellyroll, thus allowing the assembly of a functional Lpt machine.
Interestingly, deletions of the C-terminal regions of LptC seem to be tolerated also by species other than E. coli, as witnessed by the isolation of transposon insertion mutants in the C-terminal region of LptC in P. aeruginosa (http://ausubellab.mgh.harvard.edu) (Fig. 1) and Ralstonia solanacearum (38) and of an lptC frameshift mutation after codon 134 in Salmonella enterica conferring bile resistance (39).
It will be crucial to understand which are the structural bases for the functioning of Lpt machineries carrying truncated LptC proteins. Despite extensive studies on the Lpt machinery, we still lack structural data on the LptFG IM components and on LptC-LptA and LptC-LptFG interactions, which are fundamental to understand at the molecular level the functioning of LPS export machinery.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Khane and Natividad Ruiz for providing antibodies against LptE and LptB, respectively.
This research was supported in part by Fondazione Fibrosi Cistica, grant FFC13/2010 (Pseudomonas aeruginosa lipopolysaccharide cell surface transport is a target process for developing new antimicrobials), by MIUR Regione Lombardia, project 30190679 (Nuovi antibiotici mediante rational design), by MIUR PRIN 2012WJSX8K (Host-microbe interaction models in mucosal infections: development of novel therapeutic strategies), and by Fondazione Cariplo grant 2010.0653 (Outer membrane biogenesis in Gram-negative bacteria as a target for innovative antibacterial drugs).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00329-16.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Dehò G, Polissi A. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol 189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Dehò G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chng SS, Gronenberg LS, Kahne D. 2010. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita S, Tokuda H. 2009. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett 583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 9.Okuda S, Freinkman E, Kahne D. 2012. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherman DJ, Lazarus MB, Murphy L, Liu C, Walker S, Ruiz N, Kahne D. 2014. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc Natl Acad Sci U S A 111:4982–4987. doi: 10.1073/pnas.1323516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freinkman E, Chng SS, Kahne D. 2011. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci U S A 108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. 2014. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 13.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A 107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chimalakonda G, Ruiz N, Chng SS, Garner RA, Kahne D, Silhavy TJ. 2011. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sperandeo P, Villa R, Martorana AM, Samalikova M, Grandori R, Dehò G, Polissi A. 2011. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J Bacteriol 193:1042–1053. doi: 10.1128/JB.01037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freinkman E, Okuda S, Ruiz N, Kahne D. 2012. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51:4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran AX, Dong C, Whitfield C. 2010. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem 285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suits MD, Sperandeo P, Dehò G, Polissi A, Jia Z. 2008. Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J Mol Biol 380:476–488. doi: 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 19.Bollati M, Villa R, Gourlay LJ, Benedet M, Dehò G, Polissi A, Barbiroli A, Martorana AM, Sperandeo P, Bolognesi M, Nardini M. 2015. Crystal structure of LptH, the periplasmic component of the lipopolysaccharide transport machinery from Pseudomonas aeruginosa. FEBS J 282:1980–1997. doi: 10.1111/febs.13254. [DOI] [PubMed] [Google Scholar]
- 20.Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C. 2014. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511:52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Xiang Q, Zhu X, Dong H, He C, Wang H, Zhang Y, Wang W, Dong C. 2014. Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport. Biochem Biophys Res Commun 452:443–449. doi: 10.1016/j.bbrc.2014.08.094. [DOI] [PubMed] [Google Scholar]
- 22.Villa R, Martorana AM, Okuda S, Gourlay LJ, Nardini M, Sperandeo P, Dehò G, Bolognesi M, Kahne D, Polissi A. 2013. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J Bacteriol 195:1100–1108. doi: 10.1128/JB.02057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narita S, Masui C, Suzuki T, Dohmae N, Akiyama Y. 2013. Protease homolog BepA (YfgC) promotes assembly and degradation of beta-barrel membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 110:E3612–E3621. doi: 10.1073/pnas.1312012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serina S, Nozza F, Nicastro G, Faggioni F, Mottl H, Dehò G, Polissi A. 2004. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res Microbiol 155:692–701. doi: 10.1016/j.resmic.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 26.Li XM, Shapiro LJ. 1993. Three-step PCR mutagenesis for linker scanning. Nucleic Acids Res 21:3745–3748. doi: 10.1093/nar/21.16.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghisotti D, Chiaramonte R, Forti F, Zangrossi S, Sironi G, Dehò G. 1992. Genetic analysis of the immunity region of phage-plasmid P4. Mol Microbiol 6:3405–3413. doi: 10.1111/j.1365-2958.1992.tb02208.x. [DOI] [PubMed] [Google Scholar]
- 28.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 29.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A 89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. 2010. Phylogeny of Gammaproteobacteria. J Bacteriol 192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punta M, Simon I, Dosztanyi Z. 2015. Prediction and analysis of intrinsically disordered proteins. Methods Mol Biol 1261:35–59. doi: 10.1007/978-1-4939-2230-7_3. [DOI] [PubMed] [Google Scholar]
- 35.Martorana AM, Sperandeo P, Polissi A, Dehò G. 2011. Complex transcriptional organization regulates an Escherichia coli locus implicated in lipopolysaccharide biogenesis. Res Microbiol 162:470–482. doi: 10.1016/j.resmic.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang WC, Lin YM, Cheng YS, Cheng CP. 2013. Ralstonia solanacearum RSc0411 (lptC) is a determinant for full virulence and has a strain-specific novel function in the T3SS activity. Microbiology 159:1136–1148. doi: 10.1099/mic.0.064915-0. [DOI] [PubMed] [Google Scholar]
- 39.Hernández SB, Cota I, Ducret A, Aussel L, Casadesus J. 2012. Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet 8:e1002459. doi: 10.1371/journal.pgen.1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 41.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J Mol Biol 104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 43.Bachmann BJ. 1987. Derivatives and genotypes of some mutant derivatives of Escherichia coli K-12, p 1191–1219. In Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella Typhimurium cellular and molecular biology, vol 2 ASM Press, Washington, DC. [Google Scholar]
- 44.Sperandeo P, Pozzi C, Dehò G, Polissi A. 2006. Nonessential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol 157:547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.