ABSTRACT
FimV is a Pseudomonas aeruginosa inner membrane protein that regulates intracellular cyclic AMP (cAMP) levels—and thus type IV pilus (T4P)-mediated twitching motility and type II secretion (T2S)—by activating the adenylate cyclase CyaB. Its cytoplasmic domain contains three predicted tetratricopeptide repeat (TPR) motifs separated by an unstructured region: two proximal to the inner membrane and one within the “FimV C-terminal domain,” which is highly conserved across diverse homologs. Here, we present the crystal structure of the FimV C terminus, FimV861–919, containing a TPR motif decorated with solvent-exposed, charged side chains, plus a C-terminal capping helix. FimV689, a truncated form lacking this C-terminal motif, did not restore wild-type levels of twitching or surface piliation compared to the full-length protein. FimV689 failed to restore wild-type levels of the T4P motor ATPase PilU or T2S, suggesting that it was unable to activate cAMP synthesis. Bacterial two-hybrid analysis showed that TPR3 interacts directly with the CyaB activator, FimL. However, FimV689 failed to restore wild-type motility in a fimV mutant expressing a constitutively active CyaB (fimV cyaB-R456L), suggesting that the C-terminal motif is also involved in cAMP-independent functions of FimV. The data show that the highly conserved TPR-containing C-terminal domain of FimV is critical for its cAMP-dependent and -independent functions.
IMPORTANCE FimV is important for twitching motility and cAMP-dependent virulence gene expression in P. aeruginosa. FimV homologs have been identified in several human pathogens, and their functions are not limited to T4P expression. The C terminus of FimV is remarkably conserved among otherwise very diverse family members, but its role is unknown. We provide here biological evidence for the importance of the C-terminal domain in both cAMP-dependent (through FimL) and -independent functions of FimV. We present X-ray crystal structures of the conserved C-terminal domain and identify a consensus sequence for the C-terminal TPR within the conserved domain. Our data extend our knowledge of FimV's functionally important domains, and the structures and consensus sequences provide a foundation for studies of FimV and its homologs.
INTRODUCTION
Type IV pili (T4P) are filamentous surface appendages produced by a wide range of bacteria and archaea (1, 2), where they assist in DNA uptake, surface attachment, and twitching motility (3–5). There are two major subfamilies of T4P: T4aP and T4bP. T4aP are typically associated with twitching (2), a process in which pili undergo repeated cycles of extension, adhesion, and retraction, thus acting as molecular grappling hooks to pull cells along a surface.
The T4aP machinery is composed of four structural subcomplexes (6). In the model bacterium Pseudomonas aeruginosa, an inner membrane motor subcomplex consisting of the platform protein PilC and three hexameric ATPases—PilB, PilT, and PilU—provide energy for T4aP extension and retraction (7–9). A second inner membrane alignment subcomplex composed of PilMNOP connects the motor subcomplex with an outer membrane secretin composed of multimeric PilQ (2, 10–13). Finally, the pilus fiber is composed of the major pilus subunit PilA and a set of minor pilins that prime pilus assembly and export the adhesin PilY1 to the cell surface (14). These proteins comprise the T4aP machine, but its synthesis and function are further regulated by a number of components. A chemotaxis-like system called Chp controls T4aP function in part by regulating activity of the major adenylate cyclase, CyaB, and thus the levels of the second messenger molecule cyclic AMP (cAMP) (15). The transcriptional activator Vfr (virulence factor regulator) binds cAMP and controls the expression of more than 200 gene products—many involved in pathogenicity, including members of the T4aP alignment subcomplex—and the minor pilins (16).
Also implicated in regulation of T4aP assembly and function is the inner membrane protein, FimV (17, 18). FimV is involved in a number of virulence mechanisms, including twitching motility, type II secretion (T2S) of lipases and proteases, and regulation of cAMP production, and thus Vfr function (15, 17, 19). FimV is a 919-residue protein consisting of a large periplasmic domain with a peptidoglycan-binding lysin motif (LysM) (20), a single transmembrane segment, and a highly acidic cytoplasmic domain that is predicted to contain three discontinuous tetratricopeptide repeat (TPR) protein-protein interaction motifs (21, 22). TPR motifs are typically composed of 3 to 16 tandem repeats of an ∼34-residue α-helical hairpin. Individual pairs of helices associate to form a superhelical structure that mediates protein-protein interactions (23). TPR protein sequences are highly variable, although a core consensus sequence, W4-L7-G8-Y11-A20-F24-A27-P32, was identified two decades ago (24). None of the positions of the consensus are invariable, although hydrophobic residues are commonly observed at those positions (24–26). Of the three predicted TPR motifs in P. aeruginosa FimV, two are organized in tandem, proximal to the inner membrane (TPR1 and TPR2, residues 544 to 611), while the third (TPR3, residues 873 to 906) is at the C terminus, separated from TPR1 and TPR2 by a long, highly acidic region predicted to lack a regular secondary structure (17).
In addition to controlling cAMP production and related phenotypes, FimV functions in a cAMP-independent manner, promoting multimerization of the PilQ secretin (18). Based on recent work in other species, FimV may be a hub protein, similar to HubP of Vibrio cholerae and Shewanella putrefaciens, that coordinates interactions of a number of landmark proteins involved in motility and chromosome segregation at the poles of rod-shaped bacteria (27, 28). These large proteins have very limited sequence identity aside from their LysM motifs, single transmembrane segment, and a highly conserved “FimV C-terminal sequence” of ∼50 residues (NCBI Conserved Domain Database designation TIGR03504) (18) predicted to include a single TPR motif. FimV's domain architecture and C-terminal sequence are shown in Fig. 1.
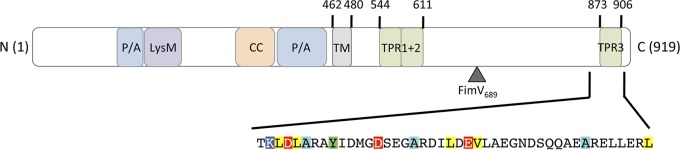
FIG 1.
Schematic of FimV domain organization. FimV has a peptidoglycan-binding LysM motif, two proline/alanine rich regions (P/A), and a coiled-coil (CC) motif in its periplasmic domain (residues 1 to 462), followed by a single transmembrane segment (TM) (18). The cytoplasmic domain (residues 480 to 919) has three predicted TPR motifs (22): two in tandem (residues 544 to 611) and a third at the C terminus (residues 873 to 906). The “FimV C-terminal domain” (TIGR03504) is highly conserved among orthologs. The amino acid sequence of the conserved C-terminal domain is shown. The domains are drawn to scale. The boundary of the truncated FimV689 is shown with an arrowhead.
The way in which FimV regulates cAMP production by the major P. aeruginosa adenylate cyclase CyaB is unclear. While other regulators of cAMP levels have been identified, including the Chp chemotaxis-like system, the CyaB activator FimL, and the phosphodiesterase CpdA (15, 29, 30), only FimL interacts with FimV (31). All the regulators identified so far are cytoplasmic, suggesting that interactions with FimV are likely to occur with its cytoplasmic domain. Here, we present the X-ray crystal structure of the highly conserved TPR3-containing C-terminal domain at 2.05 Å and provide evidence that it plays a key functional role in the regulation of motility and cAMP production and therefore in the production of many P. aeruginosa virulence factors.
MATERIALS AND METHODS
Bacterial growth conditions.
Bacterial strains and plasmids are listed in Table 1. Unless otherwise stated, P. aeruginosa strains were grown on Luria-Bertani (LB) agar at 37°C on medium containing 30 μg ml−1 gentamicin. Escherichia coli BL21(DE3) and B834(DE3) strains were grown in medium containing 100 μg ml−1 ampicillin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAK | Strain PAK | 15 |
| PAK pilA | PAK with a deletion of pilA | 60 |
| PAK pilU | PAK with a deletion of pilU | Matthew C. Wolgang |
| PAK cyaAB | PAK with deletions of cyaA and cyaB | 15 |
| PAK cyaB-R456L | PAK encoding an arginine-to-leucine change in CyaB at amino acid position 456 | This study |
| PAK fimV | PAK with a deletion of fimV | 15 |
| PAK fimV cyaB-R456L | PAK carrying a deletion of fimV and encoding an arginine-to-leucine change in CyaB at amino acid position 456 | This study |
| PAK fimV1194 | PAK with an FRT insertion at nucleotide position 1194 of fimV | This study |
| E. coli | ||
| BL21(DE3) | Protein overexpression strain | Invitrogen |
| B834(DE3) | Methionine auxotrophic protein expression strain | Invitrogen |
| BTH101 | Bacterial two-hybrid system reporter strain | Euromedex |
| Plasmids | ||
| pRK793 | IPTG-inducible expression vector encoding TEV protease | 41 |
| pBADGr | Arabinose-inducible protein expression vector | 61 |
| pBADGr::fimV | Arabinose-inducible vector encoding full-length FimV | This study |
| pBADGr::fimV689 | Arabinose-inducible vector encoding FimV residues 1 to 689 | This study |
| pET151/D-TOPO::fimV862–919 | IPTG-inducible protein overexpression vector encoding FimV residues 862 to 919 | This study |
| pKNT25 | IPTG-inducible expression vector encoding T25 fragment of B. pertussis CyaB | Euromedex |
| pKNT25::fimV488–618 | IPTG-inducible expression vector encoding T25 fragment of B. pertussis CyaB fused to the C terminus of FimV TPR1+2 | This study |
| pKNT25::fimV811–919 | IPTG-inducible expression vector encoding T25 fragment of B. pertussis CyaB fused to the C terminus of FimV TPR3 | This study |
| pKNT25::fimL | IPTG-inducible expression vector encoding T25 fragment of B. pertussis CyaB fused to the C terminus of FimL | This study |
| pKNT25::pilG | IPTG-inducible expression vector encoding T25 fragment of B. pertussis CyaB fused to the C terminus of PilG | This study |
| pUT18 | IPTG-inducible expression vector encoding T18 fragment of B. pertussis CyaB | Euromedex |
| pUT18::fimV488–618 | IPTG-inducible expression vector encoding T18 fragment of B. pertussis CyaB fused to the C terminus of FimV TPR1+2 | This study |
| pUT18::fimV811–919 | IPTG-inducible expression vector encoding T18 fragment of B. pertussis CyaB fused to the C terminus of FimV TPR3 | This study |
| pUT18::fimL | IPTG-inducible expression vector encoding T18 fragment of B. pertussis CyaB fused to the C terminus of FimL | This study |
Sequence analysis.
The CDvist server, which uses an array of protein sequence analysis tools and protein feature databases (32), was used to delineate complete domain architectures of FimV and orthologous sequences that were downloaded from the EggNOG database (33). A multiple-sequence alignment was constructed using MAFFT-LINSI with the legacygappenalty option (34). A maximum-likelihood tree was built using MEGA with pairwise deletion and the JTT substitution (35).
Twitching motility assay.
Twitching motility was tested as previously described (36) with some modifications. In brief, cells from an overnight culture were stab inoculated to the interface between LB medium–1% agar and a plasma-treated polystyrene petri dish (Thermo Fisher) and then incubated at 37°C for 16 h. The medium was supplemented with 0.1% (wt/vol) arabinose to induce the expression of FimV. Twitching zones were visualized by removing the agar and staining cells on the petri dish with 1% (wt/vol) crystal violet and washing with water to remove unbound dye. Twitching zones were measured by analyzing the diameter of each twitching zone in pixels using ImageJ software (National Institutes of Health [NIH]). Twitching zones were normalized to the twitching diameter of wild-type P. aeruginosa strain PAK in each individual experiment. Data are representative of three independent experiments.
Sheared surface protein preparation.
Surface pili were analyzed as previously described (36). In brief, strains of interest were streaked in a grid-like pattern onto LB medium–1.5% agar supplemented with 0.1% (wt/vol) arabinose and grown overnight at 37°C. Cells were gently scraped from the plates using a sterile coverslip and resuspended in 4.5 ml of phosphate-buffered saline (PBS; pH 7.4). Surface appendages were sheared by vortexing the cells for 30 s. The optical density at 600 nm (OD600) for each strain was measured, and an amount of cells equivalent to 4.5 ml of the sample with the lowest OD600 was pelleted by centrifugation at 16,100 × g for 5 min. When necessary, PBS was added to samples to a final volume of 4.5 ml prior to centrifugation. Supernatants were removed and centrifuged again at 16,100 × g for 20 min to remove the remaining cells. Supernatants were collected, mixed with 5 M NaCl and 30% (wt/vol) polyethylene glycol (Sigma; molecular weight, ∼8,000) to a final concentration of 0.5 M NaCl and 3% (wt/vol) polyethylene glycol, and incubated on ice for 30 min. Precipitated surface proteins were collected by centrifugation at 16,100 × g for 30 min. The supernatants were discarded, and samples were centrifuged again at 16,100 × g for 2 min. Pellets were resuspended in 150 μl of 1× SDS-PAGE sample buffer (80 mM Tris [pH 6.8], 5.3% [vol/vol] 2-mercaptoethanol, 10% [vol/vol] glycerol, 0.02% [wt/vol] bromophenol blue, 2% [wt/vol] SDS). Samples were boiled for 10 min and resolved by 15% SDS-PAGE. Bands were visualized by staining with Coomassie brilliant blue (Sigma). Data are representative of three independent experiments.
Western blotting.
Western blotting of whole-cell lysates was performed as previously described with some modifications (37). In brief, whole-cell lysates prepared from strains grown overnight on LB medium–1.5% agar supplemented with 0.1% (wt/vol) arabinose. Cell growth was resuspended in 1× PBS and normalized to an OD600 of 0.6. Cells were pelleted by centrifugation at 2,300 × g for 5 min. The pellets were then resuspended in 175 μl of 1× SDS-PAGE loading dye. Cell lysates were resolved on 15% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked in 5% skim milk dissolved in PBS (pH 7.4) for 1 h, washed in PBS, and incubated with antiserum raised against the FimV periplasmic domain (FimVperi) (18) or PilU (38), diluted in PBS (anti-FimVperi, 1:1,000; anti-PilU, 1:5,000) for 1 h, washed, incubated with alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (1:3,000; Bio-Rad) for 1 h, and washed. Blots were developed using BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium. The data are representative of three independent experiments.
Densitometric analysis.
The pixel density of the band corresponding to PilU and a nonspecific loading control band were determined using ImageJ software (NIH) (39). The pixel density of the PilU band was normalized to the pixel density of the loading control band, expressed as a percentage of the normalized PilU levels in wild-type PAK. The data are representative of the average of three independent experiments. A pairwise Student t test was performed using GraphPad Prism 5 software (GraphPad Software) to determine statistical significance. The results were considered significantly different if P < 0.05.
Type II secretion assay.
Overnight cultures grown on LB medium–1.5% agar were resuspended in PBS and diluted to an OD600 of 0.6. A 2-μl portion of the normalized suspension was spot inoculated onto Trypticase soy agar supplemented with 2% (wt/vol) skim milk and 0.1% arabinose (wt/vol) and incubated for 40 h at 30°C. Secretion of the type II secretion substrate elastase was visualized as a zone of clearance (degradation of casein) around the colony. Secretion was quantified by calculating the difference between the diameter of the zone of clearance and the diameter of the bacterial growth and was expressed as a percentage of the wild-type PAK. Data are representative of three independent experiments.
Bacterial two-hybrid assay.
E. coli BTH101 cells were cotransformed with TPR1 and 2 (FimV488–618), TPR3 (FimV811–919), TPR1-3, including the linker region (FimV488–919), or FimL, fused to either the T18 or T25 fragments of Bordetella pertussis CyaB (40) by heat shock. Single colonies were resuspended in 5 ml of LB medium supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin and grown overnight. A 5-ml subculture using 100 μl of the overnight culture was grown to an OD600 of 0.6. Then, 5 μl of each strain was spot plated onto MacConkey agar supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin, 1% (wt/vol) maltose, and 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and LB medium–1.5% agar (wt/vol) supplemented with 100 μl of 20 mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) spread and allowed to dry onto the surface of the plate, followed by incubation at 30°C for 24 h. Positive interactions were visualized by pink bacterial growth representing the fermentation of maltose on MacConkey agar or by blue growth on LB medium–1.5% agar supplemented with X-Gal representing X-Gal cleavage.
Protein overexpression and purification.
Tobacco etch virus (TEV) protease was expressed from plasmid pRK793 (Addgene plasmid 8827) (41). E. coli BL21 transformed with pRK793 was grown overnight at 37°C in 10 ml of LB supplemented with ampicillin. The overnight culture was subcultured in 1 liter of fresh medium and grown at 37°C until reaching an OD600 of 0.6. Expression was induced with 1 mM IPTG, and the cells grown at 30°C for 4 h. The cells were harvested, and the pellet was resuspended in 20 mM Tris (pH 8), 500 mM NaCl, 10% glycerol, and 25 mM imidazole for lysis by sonication and centrifugation as described above. TEV protease was purified by nickel affinity chromatography on an AKTA fast-performance liquid chromatograph (FPLC) as described above, where the protein was eluted in 220 mM imidazole after washes with 25, 40, 50, and 65 mM imidazole. The elution fraction was diluted up to 50 ml with 50 mM Tris (pH 8), 1 mM EDTA, 5 mM dithiothreitol, and 10% glycerol. TEV protease was further purified by cation exchange chromatography using a 5-ml HiTrap SP HP column (GE Healthcare) on an AKTA FPLC. The protein was eluted a gradient of 150 mM–500 mM KCl, the fractions were analyzed by SDS-PAGE, and those with TEV protease were pooled. TEV protease was concentrated to 1 mg/ml, flash frozen with liquid nitrogen, and stored at −80°C until used.
The DNA fragment encoding FimV862–919 was amplified using forward 5′-CACCGATGACTTCGACTTCCTCTCCGGTGC-3′ and reverse 5′-TCAGGCCAGGCGCTCCAGCAACTC-3′ primers (Table 2) and cloned into pET151/D-TOPO for expression with a cleavable His6-V5 tag. E. coli BL21(DE3) transformed with pET151::fimV862–919 was grown in LB medium supplemented with ampicillin for 6 h at 37°C and subcultured into Terrific Broth with ampicillin for 72 h at 20°C. The cells were harvested, and the pellet resuspended in 20 mM Tris (pH 7.5)–500 mM NaCl for lysis by sonication on ice for 2 min in 10-s pulses with 10 s of cooling in between using a Sonicator 3000 (Misonix) at power level 3. The cell lysate was centrifuged at 8,891 × g for 1 h at 4°C, and DNase I was added to the supernatant (1 μg ml−1) before purification. His6-V5-FimV862–919 was purified with Ni-nitriolotriacetic acid-agarose (Qiagen), where the protein was collected in 20, 40, and 80 mM imidazole elution fractions. These fractions were pooled and diluted with 20 mM Tris (pH 7.5) to lower the salt concentration to 100 mM NaCl for anion-exchange chromatography where a gradient of 100 mM–500 mM KCl was used to elute His6-V5-FimV862–919. The protein eluted at ∼250 mM KCl and dialyzed into 20 mM Tris (pH 7.5)–100 mM NaCl overnight, and the tags were removed by TEV protease cleavage. A second nickel affinity purification step was performed to purify FimV862–919 without the His6-V5 tag. The flowthrough fraction containing purified FimV862–919 in 20 mM Tris (pH 7.5)–100 mM NaCl was concentrated to 2 mg ml−1 using a Vivaspin 20 concentrator.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| cyaB-R456L F1 | GCGGGTACCTTCACCGCACTGGTCATCG |
| cyaB-R456L R1 | GCGTCTAGAAAGTCAACGGAGTCGTCACC |
| cyaB-R456L SDM F | TGCAACAGGCCGCCGAGCTCCTGCGCGACAAGGTC |
| cyaB-R456L SDM R | GACCTTGTCGCGCAGGAGCTCGGCGGCCTGTTGCA |
| fimV862–919 F | CACCGATGACTTCGACTTCCTCTCCGGTGC |
| fimV862–919 R | TCAGGCCAGGCGCTCCAGCAACTC |
| FimV F | TTAGTACTAAGGGATTACACTATGGTTCGGCT |
| FimV R | CAGTTCTAGATCAATGGTGATGGTGATGATGGGCCAGGCGCTCCAGCAACTC |
| FimV689 F | TAAGAATTCCAAGGGATTACACTATGGTTCGGCTT |
| FimV689 R | AATAAGCTTTCAGAGGTCGGCCTGCACGTCG |
| B2H FimV TPR1F | GCGTCTAGAGATGAATGCGCAGAAAGAGAAGG |
| B2H FimV TPR1R | GCGGGTACCGCCTTGAGCTGCTCGAC |
| B2H FimV TPR3F | GCGTCTAGAGATGGAGAAGGGCGAGGACAG |
| B2H FimV TPR3R | GCGGGTACCGCGGCCAGGCGCTCCAG |
| B2H FimL-F | GCG TCT AGA GAT GGT CAC AGG AGC CAC GTC CC |
| B2H FimL-R | GCG GGT ACC GCG GCG GCC ACC GG |
| B2H PilG-F | GCG TCT AGA AAT GGA ACA GCA ATC CGA CGG |
| B2H PilG-R | TAA GGT ACC CGG GAA ACG GCG TCC ACC |
For SAD phasing, selenomethionine (SeMet) labeled proteins were expressed from E. coli B834(DE3) in M9 SeMet high-yield growth medium (Shanghai Medicilon) supplemented with ampicillin (100 μg ml−1). A 10-ml starter culture was grown for 7.5 h at 37°C, subcultured into a 100 ml of fresh medium, and grown overnight at 37°C. The overnight culture was subcultured into 1 liter of fresh medium and grown at 37°C until reaching an OD600 of 0.6. SeMet-labeled His6-V5-FimV862–919 expression was induced with 1 mM IPTG, and the cells were grown overnight at 20°C. Purification was performed as described above except for nickel affinity chromatography; a 1-ml HiTrap chelating HP column (GE Healthcare) precharged with 100 mM NiCl2 was used in an AKTA FPLC system where the protein eluted in 300 mM imidazole. An additional buffer exchange step into 20 mM Tris (pH 7.5)–100 mM NaCl was also added following the second round of nickel affinity purification. SeMet-labeled proteins were concentrated to 2.87 mg ml−1 using a Vivaspin 20 concentrator.
Crystallization.
Broad crystallization screening was performed using the hanging-drop method with commercially available crystallization screens. Native FimV862–919 crystals grew in a 1:1 ratio of protein (2 mg/ml) and precipitant solution (1 M NH4H2PO4, 100 mM trisodium citrate [pH 5.6]) and equilibrated over 1.5 M ammonium sulfate at 18°C. SeMet-FimV862–919 (2.87 mg ml−1) was mixed in a 1:1 ratio with buffer containing 1.6 M NaH2PO4, 0.4 M KH2PO4, and 0.1 M phosphate citrate (pH 4.2) and equilibrated over 1.5 M ammonium sulfate at 18°C overnight. Crystallization was seeded with native FimV862–919 microcrystals grown in the same buffer. Small SeMet-FimV862–919 crystals grew within 10 to 25 days. Additional growth was achieved by further incubation at 4°C for 7 to 10 days.
Structure determination.
Diffraction data were collected at beamline X29 α β of the National Synchrotron Light Source (NSLS; Brookhaven, NY) at wavelengths of 1.075 Å for native FimV and 0.9791 Å for SeMet-FimV. Data sets were processed using the HKL-2000 program suite (42). SAD phasing, density modification, auto model building, and refinement were performed using the PHENIX suite of programs (43, 44). Manual model building and refinement were performed using Coot (45) until the R and Rfree values could no longer be improved. The refined FimV862–919-SeMet structure (PDB 4MAL) was used as an initial search model for structure determination of native FimV862–919 by molecular replacement in PHENIX (43, 44). Model refinement was performed using PHENIX (43, 44). The stereochemical quality of the models was verified using MolProbity (46). The data collection and model refinement statistics are listed in Table 3.
TABLE 3.
Data collection and refinement statisticsa
| Parameter | Value(s) for: |
|
|---|---|---|
| SeMet-FimV | Native FimV | |
| Data collection statistics | ||
| Beamline | NSLS X29 | NSLS X29 |
| Wavelength (Å) | 0.9791 | 1.075 |
| Space group | P41212 | P212121 |
| Unit cell dimensions | ||
| a, b, c (Å) | 42.33, 42.33, 139.48 | 34.0, 58.5, 136.7 |
| α, β, γ (˚) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Molecules per ASU | 2 | 6 |
| Resolution range (Å) | 50.0–2.05 (2.09–2.05) | 50.0–2.00 (2.07–2.00) |
| Total reflections | 122,397 | 246,901 |
| Unique reflections | 8,648 (412) | 18,054 (1,394) |
| Redundancy | 14.2 (11.1) | 13.7 (10.6) |
| Completeness (%) | 99.9 (99.3) | 94.6 (76.2) |
| Mean I/σI | 33.9 (6.2) | 20.8 (2.5) |
| Rmerge (%)b | 7.2 (47.8) | 15.1 (78.1) |
| Refinement statistics | ||
| Rwork/Rfree (%)c | 21.5/24.2 | 20.8/25.8 |
| Resolution range (Å) | 40.50–2.05 | 44.45–2.01 |
| No. of reflectionsd | 8,314 | 17,404 |
| No. of atoms | 984 | 2,244 |
| Protein | 949 | 2,159 |
| Water | 35 | 85 |
| RMSD | ||
| Bond length (Å) | 0.013 | 0.002 |
| Bond angle (˚) | 1.042 | 0.541 |
| Avg B (Å2)e | 38.8 | 39.8 |
| Protein | 38.8 | 39.7 |
| Water | 40.4 | 42.2 |
| Coordinate error (Å)f | 0.23 | 0.25 |
| Ramachandran statistics (%)g | ||
| Most favored regions | 95.97 | 99.31 |
| Allowed regions | 4.03 | 0.69 |
| Disallowed regions | 0.00 | 0.00 |
| PDB ID | 4MAL | 4MBQ |
Values in parentheses correspond to the highest-resolution shell.
Rmerge = Σhkl Σi | Ii (hkl) − [I(hkl)] |/Σhkl Σi Ii(hkl).
Rwork = Σ | |Fobs| − k|Fcalc| |/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (9.92 and 10.08% for SeMet-FimV and native FimV, respectively) excluded from all stages of the refinement.
Only reflections with Fobs/σ(Fobs) of ≥1.34 were used for SeMet-FimV model refinement. All unique reflections in the resolution range of 44.45 to 2.01 Å were included without further cutoff criteria for refinement of native FimV.
Maximum-likelihood-based coordinate error, as determined by PHENIX (62).
As calculated using MolProbity (63).
Protein structure accession numbers.
The structures of native and SeMet-derivatized FimV TPR3 can be accessed through the RCSB Protein Data Bank (PDB) using accession codes 4MBQ and 4MAL, respectively.
RESULTS AND DISCUSSION
The C-terminal TPR sequence of FimV is highly conserved among diverse homologs.
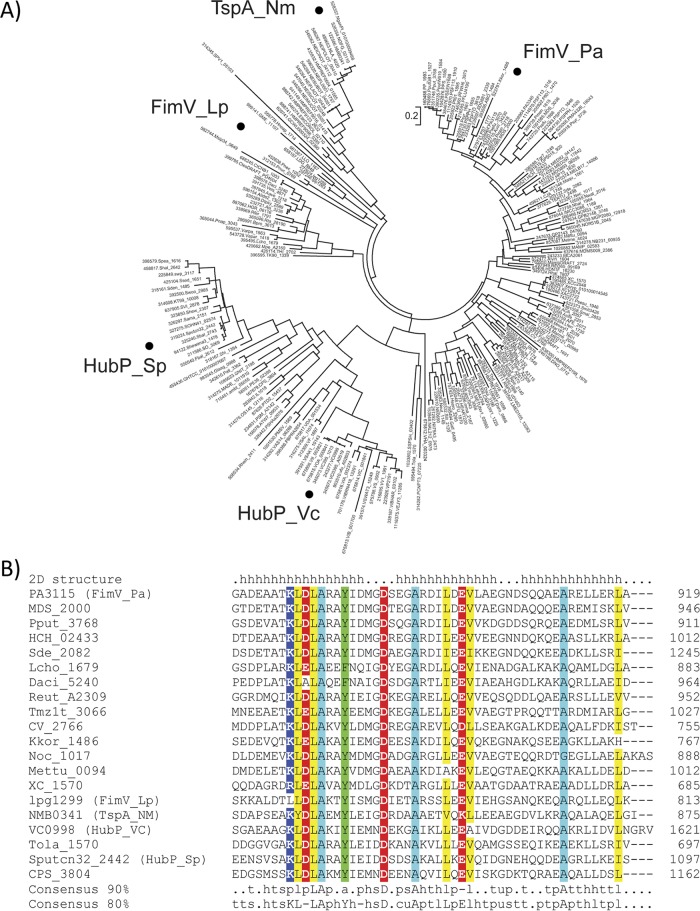
We downloaded all FimV ortholog sequences from the EggNOG database, which generates orthologous groups of proteins from complete genomes (33). The set of FimV ortholog sequences (COG3170) contained 235 sequences, including the previously characterized proteins FimV from Legionella pneumophila (47), TspA from Neisseria meningitidis (48), and HubP from V. cholerae and S. putrefaciens (27, 28). A multiple-sequence alignment was constructed based on the full-length sequence of FimV orthologs and the resulting phylogenetic tree revealed three major clades, exemplified by FimV from P. aeruginosa, HubP, and TspA, respectively (Fig. 2A). The functions of FimV orthologs in the context of T4P vary. Although they are in the same clade, deletion of N. meningitidis tspA had no effect on twitching motility or surface piliation (48), whereas L. pneumophila fimV mutants have reduced motility (47). Representative sequences from all three major clades (selected from smaller subgroups to avoid bias) were subjected to in-depth domain architecture analysis using the CDvist tool (32), revealing that only two domains are strongly conserved: the periplasmic LysM domain and the C-terminal TPR-containing domain, designated TIGR03504 in the TIGRFAM database of protein families. Multiple-sequence alignment revealed a dozen highly conserved residues within the C-terminal TPR-containing domain (Fig. 2B) that were further analyzed in the context of the corresponding structure (below).
FIG 2.
The C-terminal domain is highly conserved across FimV homologs. (A) Maximum-likelihood phylogenetic tree based on full-length FimV homolog sequences (COG3170 from the EggNOG database). Experimentally studied proteins are indicated as follows: Pa, P. aeruginosa (17); Vc, V. cholerae (27); Sp, S. putrefaciens (28); Lp, L. pneumophila (47); Nm, N. meningitidis (48). (B) Conservation of the C-terminal TPR3-containing domain in representative FimV proteins. Locus tags are shown as sequence identifiers. Conserved positions are highlighted: positively charged (blue), negatively charged (red), aromatic (green), aliphatic (yellow), and alanine (cyan). Consensus was calculated for the complete set of homologs shown in panel A. Secondary structure elements (h, alpha-helix) corresponding to the TPR motif (first two alpha-helices) and the C-terminal capping helix are shown above the alignment.
The C terminus of FimV is required for function.
The high level of conservation of FimV's C terminus implies a key functional role, supported by early observations (17) that transposon insertion ∼230 nucleotides from the 3′ end of fimV markedly decreased twitching motility. To determine whether this region was required for FimV's functions, we generated an arabinose-inducible expression construct (FimV689) encoding FimV 1-689 (of 919), lacking the region encompassing predicted TPR3. The boundaries selected for the FimV689 construct were based on the phenotype of a previously characterized transposon mutant, fimV2091, which expresses a stable fragment of FimV lacking the C-terminal region encompassing TPR3 (18, 49). The truncated construct or full-length fimV was used to complement a fimV deletion mutant. To examine cAMP-independent phenotypes controlled by FimV, a fimV mutant encoding a constitutively active point mutant of the adenylate cyclase CyaB (fimV cyaB-R456L) (50) was used to restore cAMP biogenesis in the fimV background. Both full-length and truncated proteins were stable and expressed to levels comparable to those in the wild-type control, as determined by Western blot with FimV-specific antisera (Fig. 3A). It is worth noting that FimV exhibits anomalous migration on SDS-PAGE (18), so it is not possible to accurately gauge its mass using that method.
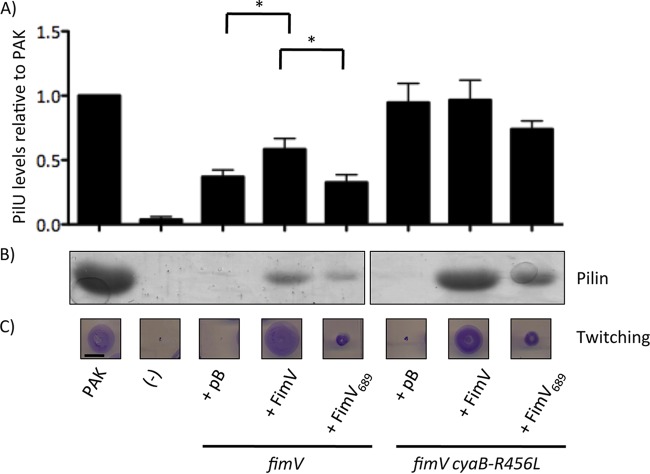
FIG 3.
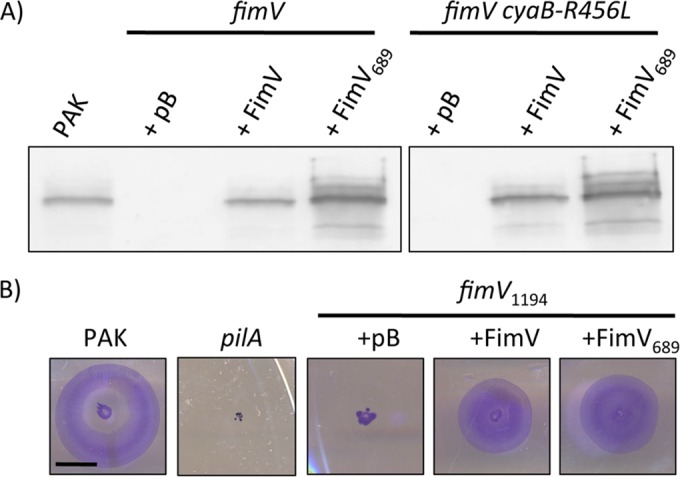
FimV689 is a stable and functional fragment. (A) fimV and fimV cyaB-R456L strains were transformed with arabinose-inducible expression vectors encoding full-length FimV (FimV) or a truncated form lacking TPR3 (FimV689). As a control, strains were also transformed with empty vector (pB). Cell lysates prepared as described in Materials and Methods from cells grown on LB medium–1.5% agar supplemented with 0.1% (wt/vol) arabinose were resolved on 10% SDS-PAGE and transferred to a nitrocellulose membrane. FimV was visualized by immunoblotting with anti-FimV antiserum. The previously reported anomalous migration pattern of FimV on SDS-PAGE (17, 18) means that the full-length and truncated versions appear similar in mass despite their differences in length. Data are representative of three experiments. (B). A fimV strain that expresses only the cytoplasmic region of the protein (fimV1194 strain) was transformed with arabinose-inducible expression constructs encoding either full-length FimV or a truncation mutant lacking TPR3 (FimV689). As a control, the fimV1194 strain was also transformed with empty vector (pB). Twitching motility was determined by stab inoculation of the strains into LB medium–1% agar supplemented with 0.1% (wt/vol) arabinose. Strains were incubated for 16 h, and twitching zones were visualized by removing the agar layer and staining the bacteria with 1% (wt/vol) crystal violet. Motility was measured as the diameter of the stained twitching zone. Scale bar, 1 cm.
To confirm that FimV689 was functional, we used it to complement a fimV mutant disrupted at nucleotide 1194, fimV1194 (18). This mutant expresses the cytoplasmic portion of FimV and can be complemented in trans with an N-terminal FimV construct expressing residues 1 to 507, FimV507 (18). Expression of FimV689 in fimV1194 restored twitching motility to the same degree as full-length FimV (Fig. 3B), suggesting that it is correctly folded and localized to the inner membrane.
As a proxy to assess relative levels of intracellular cAMP, we examined levels of the T4P motor ATPase PilU (Fig. 4A), which are positively correlated with levels of cAMP (see Fig. S1 in the supplemental material) (16). The fimV mutant complemented with empty vector had low levels of PilU, suggesting low levels of cAMP, while the fimV cyaB-R456L mutant had high levels of PilU, confirming that CyaB R456L was active in the absence of fimV. Complementation of fimV in trans with the full-length fimV gene in trans restored PilU levels to ∼66% of the wild type, but expression of FimV689 failed to increase PilU levels relative to the empty vector control, suggesting that FimV689 was incapable of activating CyaB.
FIG 4.
FimV TPR3 is critical for FimV function. fimV and fimV cyaB-R456L strains were transformed with arabinose-inducible expression constructs encoding full-length FimV (FimV) or a truncation mutant lacking TPR3 (FimV689). As controls, strains were also transformed with empty vector (pB). (A) The levels of PilU were determined by preparing cell lysates as described in Materials and Methods of cells grown overnight on LB medium–1.5% agar supplemented with 0.1% (wt/vol) arabinose. Samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. PilU was visualized by immunoblotting with anti-PilU antiserum. Densitometric analysis (average, n = 3) was carried out using ImageJ, normalizing band intensity of the PilU band to a nonspecific control band, and the results are shown as percentages of the normalized band intensity of the wild type ± the standard error. Samples were considered significantly different if a pairwise Student t test gave P values of <0.05 (*). (B) The levels of surface pilins were assessed by sheared surface protein preparation of strains grown on LB medium–1.5% agar supplemented with 0.1% (wt/vol) arabinose. The surface protein fractions were prepared as described in Materials and Methods and were resolved by SDS-PAGE and visualized by Coomassie blue staining. (C) Twitching motility was determined by stab inoculation of the indicated strains into LB medium–1% agar supplemented with 0.1% (wt/vol) arabinose. Strains were incubated for 16 h, and twitching zones were visualized by removing the agar layer and staining the bacteria with 1% (wt/vol) crystal violet. Motility was measured as the diameter of the stained twitching zone. Scale bar, 1 cm. A “−” represents the negative control for each experiment, pilU strains for the anti-PilU immunoblots or a nonpiliated pilA strain for surface piliation and twitching motility. The images are representative of three experiments.
Activation of CyaB via FimV is required for transcription of structural components of the T4P machinery, including the PilMNOP alignment subcomplex (16). The periplasmic domain of FimV promotes multimerization of the PilQ secretin (18), and thus surface piliation, in a cAMP-independent manner. We characterized levels of surface piliation in fimV and fimV cyaB-R456L mutants upon complementation with FimV689 (Fig. 4B). fimV complemented with empty vector had no recoverable surface pili, but piliation was restored with full-length fimV in trans. In contrast, FimV689 restored ∼65% of surface pilin levels relative to full-length FimV. The fimV cyaB-R456L mutant with empty vector had few recoverable surface pili, but piliation was restored upon complementation with full-length FimV. Complementation of fimV cyaB-R456L with FimV689 restored ∼50% of piliation relative to complementation with full-length FimV. Since FimV689 complemented inefficiently compared to full-length protein in either fimV or fimV cyaB-R456L, we conclude that the FimV C-terminal domain containing TPR3 is required for both cAMP-dependent and -independent functions of FimV.
We also examined twitching motility by subsurface stab assay (Fig. 4C). Consistent with previous reports, mutants lacking FimV—either the fimV or the fimV cyaB-R456L mutant—were unable to twitch (17, 18). Twitching was restored in both strains upon complementation with full-length FimV. However, the FimV689 construct only restored ∼31% of twitching relative to full-length FimV in the fimV background and ∼40% in fimV cyaB-R456L strains. The discrepancy between surface pilin levels land twitching likely reflects differences in the balance between pilus extension and retraction in these strains. For example, cyaAB mutants have almost no recoverable surface pili, but are motile (15), whereas pilT mutants are hyperpiliated but nonmotile. Given that the fimV cyaB-R456L mutant had high levels of PilU, and thus cAMP (Fig. 4A), these data suggest that failure of FimV689 to restore twitching is independent of its inability to activate CyaB.
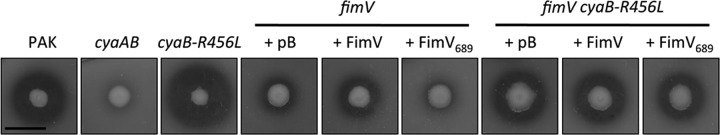
We next examined T2S in fimV and fimV cyaB-R456L strains complemented with FimV689 (Fig. 5). Transcription of T2S genes is cAMP dependent (16), and FimV regulates T2S at least in part through the cAMP-binding protein, Vfr (19). We tested whether FimV689 could complement defects in T2S by assessing casein hydrolysis using skim milk agar. fimV was defective for protease secretion, an observation consistent with previous findings (19). Complementation with full-length FimV restored ∼48% of T2S relative to the wild type, whereas FimV689 failed to restore secretion. The fimV cyaB-R456L mutant had ∼70% of wild-type T2S, suggesting that T2S is a mostly cAMP-dependent phenotype and thus likely to be indirectly regulated by FimV. Expression of FimV or FimV689 in fimV cyaB-R456L did not increase secretion relative to the vector control, confirming that the involvement of FimV in T2S is cAMP dependent.
FIG 5.
FimV TPR3 regulates type II secretion through modulation of cAMP levels. fimV and fimV cyaB-R456L strains were transformed with arabinose-inducible expression vectors encoding full-length FimV (FimV) or a truncation mutant lacking TPR3 (FimV689). As controls, the strains were also transformed with empty vector (pB), and representative high cAMP (cyaB-R456L) and low cAMP (cyaAB) strains were included. Cells were grown overnight and resuspended in PBS at an OD600 of 0.6. The, 2 μl of resuspended cells was spotted onto Trypticase soy broth–1.5% agar supplemented with 0.1% (wt/vol) arabinose and 2% (wt/vol) skim milk, followed by incubation for 40 h at 30°C. Elastase secretion was visualized as a zone of clearance around the bacterial colony. Secretion for each strain was measured as the difference between the diameter of the zone of clearance and the diameter of the colony (average for n = 3). The images are representative of three experiments. Scale bar, 1 cm.
FimV TPR3 interacts with FimL.
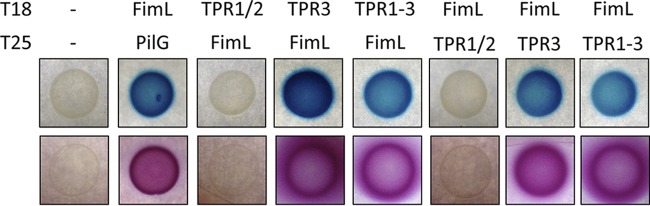
Inclan et al. recently reported that the CyaB-activating component FimL interacts with FimV (31) but did not define the specific interaction domain. Since FimL is a cytoplasmic protein, we tested whether the TPR segments of FimV, TPR1 and -2 (FimV488–618), or TPR3 (FimV811–919) were sufficient to interact with FimL using a bacterial two-hybrid (BTH) assay. T18 or T25 fragments of Bordetella pertussis adenylate cyclase CyaB were fused to the C termini of the entire cytoplasmic domain of FimV (FimV488–919), FimV488–618, FimV811–919, and FimL, and the constructs were expressed pairwise in E. coli BTH101 (Fig. 6). As a positive control, we coexpressed FimL-T18 and PilG-T25, whose interaction was also reported by Inclan et al. (31). FimL interacted with FimV488–919 and FimV811–919 but not with FimV488–618, suggesting that TPR3 alone was sufficient. These data provide additional evidence that the C-terminal domain of FimV plays an important role in regulation of CyaB, via an interaction between TPR3 and FimL.
FIG 6.
TPR3 interacts with FimL. Fusions of FimV488–618, FimV811–919, FimV488–919, or FimL with B. pertussis CyaB T18 or T25 fragments were expressed in E. coli BTH101 in a pairwise manner. As a positive control, we used FimL-T18 and PilG-T25 (31). Single colonies from the transformations were resuspended in 5 ml of LB medium and incubated at 37°C overnight. A 5-ml subculture using 100 μl of each overnight culture was made the following morning and grown to an OD600 of 0.6. Next, 5 μl was spotted onto MacConkey agar supplemented with 1% maltose and 0.5 mM IPTG, LB medium–1.5% agar with 100 μl of 20 mg/ml X-Gal was spread onto the surface, and the sample was dried. The plates were incubated at 30°C for 24 h. Protein-protein interaction was visualized by pink growth on MacConkey agar or by blue growth on LB medium plus X-Gal, indicating protein-protein interaction and reconstitution of B. pertussis CyaB function.
Structure of the conserved C-terminal TPR3-containing domain.
To gain further insight into FimV function, we initially attempted to solve the structure of the entire cytoplasmic domain; however, its large predicted unstructured region made the construct highly susceptible to degradation, and smaller fragments—while soluble even at high concentrations—failed to crystallize. Because of its pronounced conservation (and mutant phenotypes that suggested TPR3 plays a key role in FimV's virulence-related functions), we focused on solving the crystal structure of the C-terminal region. Using the predicted boundaries of the TPR3 region, we generated a construct spanning residues 862 to 919 (FimV862–919) for structure determination.
The structure of FimV862–919 was solved by SAD phasing with SeMet-labeled protein. SeMet-FimV862–919 crystallized as a dimer in the space group P41212. The final model was refined to 2.05 Å (PDB 4MAL). Native FimV862–919 crystallized in space group P212121 and contained six copies of the protein in the asymmetric unit, arranged as two trimers. The final model for native FimV862–919 was refined to 2.01 Å (PDB 4MBQ). Statistics for data collection and model refinement are presented in Table 3.
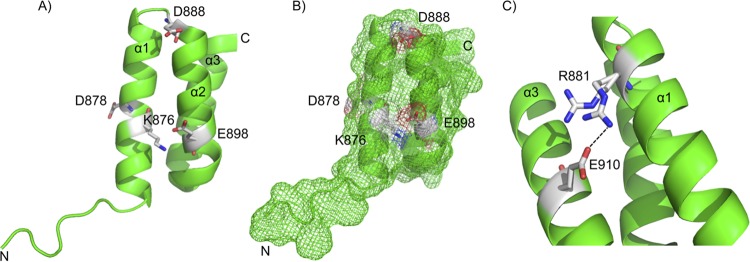
FimV862–919 includes the conserved FimV C-terminal domain (TIGR03504), which consists of two antiparallel α-helices forming the predicted TPR motif, followed by a C-terminal capping helix, which may play a role in enhancing solubility (26) (Fig. 7A). The SeMet labeled structure (PDB 4MAL) contained two biological assembly monomers (chain A is shown in Fig. 7). Each monomer contained three α-helices, with hydrophobic residues directed between the three helices and hydrophilic residues extending outwards. Surface representation suggests that conserved charged residues (Fig. 2B) are located primarily on one face of TPR3 (Fig. 7B). The side chain of R881 was found in two possible conformations (Fig. 7C), one of which formed a salt bridge with E910 on the capping helix in two-thirds of monomers.
FIG 7.
X-ray crystal structure of the FimV C-terminal domain. The X-ray crystal structure of SeMet-FimV TPR3 (residues 862 to 919) was solved to 2.05 Å (PDB 4MAL). (A) Chain A is shown, with identified conserved residues shown as sticks. Conserved charged residues are labeled directly. The two α-helices making up the TPR and the capping helix have been labeled as α1, α2, and α3. (B) Surface mesh representation of 4MAL chain A from the same view as panel A. Conserved charged residues are shown as white patches. (C) Residue R881 was observed to occupy two different states. Two-thirds of the time, R881 was in close proximity to E910, likely forming a salt bridge between α1 and α3.
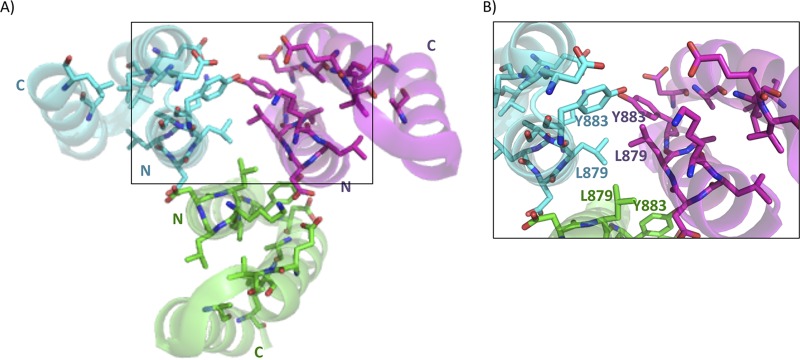
FimV native crystals contained two similar trimeric assemblies within the asymmetric unit (PDB 4MBQ) (Fig. 8). Subunits within each trimer associated primarily through helix 1 of neighboring monomers, and were arranged with helix 1 of each TPR at the center with the capping helix facing outward (Fig. 8, right panel). Conserved hydrophobic residues of each TPR motif were arranged in the center of a trimer, with hydrophilic residues from helix 2 and the capping helix facing outward. Structural comparison of the individual protomers from the SeMet and native structures showed that despite differences in crystal and monomer packing, they were very similar, with root mean square deviation (RMSD) values of ≤0.64 Å (see Table S1 in the supplemental material). The closest structural hit to 4MAL and 4MBQ as predicted by DaliLite (51, 52) (RMSD, 1.51 Å over 49 residues) was the type III secretion system chaperone, IpgC (PDB 3GZ1), a small, negatively charged all α-helical protein that escorts partly unfolded Shigella flexneri effectors IpaB and IpaC to the base of the secretion apparatus (53).
FIG 8.
X-ray crystal structure of FimV C-terminal domain homotrimer (PDB 4MBQ). The X-ray crystal structure of native FimV TPR3 was solved to 2.01 Å (PDB 4MBQ). Individual monomers are colored in cyan, green, or magenta. Highly conserved residues are shown as sticks with side chains colored by monomer. Residues and N and C termini are labeled in cyan, green, or magenta based on the associated monomer. Residues are numbered based on their position in full-length FimV. The boxed area is enlarged in panel B.
Our data suggest that FimV plays key roles in both cAMP-dependent and -independent regulation of virulence factor expression and function and that its conserved TPR3 region is critical for both roles. Previously identified cytoplasmic regulators of CyaB—FimL and PilG (15, 29)—require FimV for function (31), connecting FimV to surface mechanosensing through the Chp system. Loss of an indirect FimV-PilG interaction through loss of FimL binding may explain the inability of FimV689 to complement either cAMP-dependent and -independent phenotypes, since PilG functions in both pathways (15).
FimL failed to interact with TPR1 and 2 (Fig. 6), but other regulatory components may bind these motifs. Since TPRs typically form superhelical structures composed of three or more repeats, it is possible that transient interactions of the region between TPRs 1 and 2 and TPR3 with specific regulatory components lead to its restructuring, bringing the three into proximity to form a new interface for protein binding. Studies of the p67phox subunit of phagocyte NADPH suggested that the linker region between TPR motifs can bind target proteins (54). The linker regions in N. meningitidis TspA, V. cholerae HubP, and L. pneumophila FimV each contain different sets of multiple tandem repeats (27, 47, 55), which are absent or degenerate in Pseudomonas, and alteration of repeat numbers influenced pigmentation, cell length, and twitching in Legionella (47) and abolished interactions with some partners of Vibrio HubP (27). These differences may reflect the individual repertoires of interaction partners in each species.
The conservation of TPR3 among divergent species and homologs suggests that it might interact with a component that is widely conserved. Here we showed interactions of TPR3 and FimL, while the C terminus of V. cholerae HubP (which is 45% identical to P. aeruginosa TPR3 over 42 residues) interacts with FlhG, a protein responsible for controlling flagellar polar localization (27). This finding raises the possibility that FimV has a broader role in subcellular spatial organization in P. aeruginosa. Whereas both P. aeruginosa and L. pneumophila encode FlhG orthologs called FleN (56, 57), Neisseria lacks flagella and thus FlhG/FleN orthologs, arguing against the evolutionarily conserved partner hypothesis. In V. cholerae, deletion of HubP's C terminus failed to mislocalize another of its interaction partners, ParA1, suggesting that like FimV, it has multiple cytoplasmic protein-protein interaction domains specific for different partners.
Self-interactions mediated by hydrophobic residues were observed in TPR3 crystals (Fig. 8, right panel), but it is not yet clear whether those are physiologically relevant. The remarkably well-conserved TPR3 unit could also potentially contribute to intermolecular interactions, promoting functional homo-oligomerization of FimV monomers. TPR motif-dependent homo-oligomerization has been reported in yeast (58) and bacteria (59). In E. coli, trimerization of YbgF, a part of the Tol system, is mediated by a single C-terminal TPR motif (59). Although involvement in self-interactions might explain the high sequence similarity of this region in very diverse FimV/HubP family members, we saw no TPR3-TPR3 interactions in a BTH assay (data not shown), arguing against homo-oligomerization as a driver of sequence conservation.
Wehbi et al. (18) showed that the LysM motif of FimV was required for optimal PilQ multimerization and that the periplasmic and cytoplasmic domains of FimV could reconstitute FimV function when expressed individually. However, our prediction that the periplasmic and cytoplasmic domains had separate structural and regulatory roles was not supported by the phenotype of the FimV689 complemented strains. TPR3 is required to promote cAMP synthesis by CyaB, likely through its FimL interaction; however, FimV689 only partly restored piliation and motility in the cAMP-replete fimV cyaB-R456L background, suggesting that TPR3 is important for cAMP-independent function(s) of FimV as well. Efforts to define the full repertoire of protein interaction partners of P. aeruginosa FimV are ongoing.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Canadian Institutes of Health Research grant MOP 93585 to L.L.B. and P.L.H. and MOP 89903 to M.S.J. and by NIH grant R01GM072285 to I.B.Z. All X-ray diffraction data used in this study were collected at beamline X29 of the NSLS. NSLS funding came primarily from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and from the National Center for Research Resources (grant P41RR012408) and the National Institute of General Medical Sciences (grant P41GM103473) of the NIH.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00322-16.
REFERENCES
- 1.Albers SV, Pohlschroder M. 2009. Diversity of archaeal type IV pilin-like structures. Extremophiles 13:403–410. doi: 10.1007/s00792-009-0241-7. [DOI] [PubMed] [Google Scholar]
- 2.Pelicic V. 2008. Type IV pili: e pluribus unum? Mol Microbiol 68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- 3.Averhoff B, Friedrich A. 2003. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch Microbiol 180:385–393. doi: 10.1007/s00203-003-0616-6. [DOI] [PubMed] [Google Scholar]
- 4.Bradley DE. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol 26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 5.Burrows LL. 2005. Weapons of mass retraction. Mol Microbiol 57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- 6.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 7.Whitchurch CB, Mattick JS. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol Microbiol 13:1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 8.Lauer P, Albertson NH, Koomey M. 1993. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol Microbiol 8:357–368. doi: 10.1111/j.1365-2958.1993.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 9.Nunn D, Bergman S, Lory S. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol 172:2911–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampaleanu LM, Bonanno JB, Ayers M, Koo J, Tammam S, Burley SK, Almo SC, Burrows LL, Howell PL. 2009. Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. J Mol Biol 394:143–159. doi: 10.1016/j.jmb.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 11.Tammam S, Sampaleanu LM, Koo J, Sundaram P, Ayers M, Chong PA, Forman-Kay JD, Burrows LL, Howell PL. 2011. Characterization of the PilN, PilO, and PilP type IVa pilus subcomplex. Mol Microbiol 82:1496–1514. doi: 10.1111/j.1365-2958.2011.07903.x. [DOI] [PubMed] [Google Scholar]
- 12.Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J 19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCallum M, Tammam S, Little DJ, Robinson H, Koo J, Shah M, Calmettes C, Moraes TF, Burrows LL, Howell PL. 2016. PilN binding modulates the structure and binding partners of the Pseudomonas aeruginosa type IVa pilus protein PilM. J Biol Chem 291:11003–11015. doi: 10.1074/jbc.M116.718353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen Y, Sugiman-Marangos S, Harvey H, Bell SD, Charlton CL, Junop MS, Burrows LL. 2015. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem 290:601–611. doi: 10.1074/jbc.M114.616904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/S1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 17.Semmler AB, Whitchurch CB, Leech AJ, Mattick JS. 2000. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146(Pt 6):1321–1332. doi: 10.1099/00221287-146-6-1321. [DOI] [PubMed] [Google Scholar]
- 18.Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL. 2011. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol 193:540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michel GP, Aguzzi A, Ball G, Soscia C, Bleves S, Voulhoux R. 2011. Role of fimV in type II secretion system-dependent protein secretion of Pseudomonas aeruginosa on solid medium. Microbiology 157:1945–1954. doi: 10.1099/mic.0.045849-0. [DOI] [PubMed] [Google Scholar]
- 20.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 21.Blatch GL, Lassle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939. doi:. [DOI] [PubMed] [Google Scholar]
- 22.Karpenahalli MR, Lupas AN, Soding J. 2007. TPRpred: a tool for prediction of TPR-, PPR-, and SEL1-like repeats from protein sequences. BMC Bioinformatics 8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das AK, Cohen PW, Barford D. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J 17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb JR, Tugendreich S, Hieter P. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci 20:257–259. doi: 10.1016/S0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 25.Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, Staud F, Stulik J. 2013. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun 81:629–635. doi: 10.1128/IAI.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. 2012. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossmann F, Brenzinger S, Knauer C, Dorrich AK, Bubendorfer S, Ruppert U, Bange G, Thormann KM. 2015. The role of FlhF and HubP as polar landmark proteins in Shewanella putrefaciens CN-32. Mol Microbiol 98:727–742. doi: 10.1111/mmi.13152. [DOI] [PubMed] [Google Scholar]
- 29.Inclan YF, Huseby MJ, Engel JN. 2011. FimL regulates cAMP synthesis in Pseudomonas aeruginosa. PLoS One 6:e15867. doi: 10.1371/journal.pone.0015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs EL, Brutinel ED, Klem ER, Fehr AR, Yahr TL, Wolfgang MC. 2010. In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J Bacteriol 192:2779–2790. doi: 10.1128/JB.00168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inclan YF, Persat A, Greninger A, Von Dollen J, Johnson J, Krogan N, Gitai Z, Engel JN. 2016. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol Microbiol doi: 10.1111/mmi.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adebali O, Ortega DR, Zhulin IB. 2015. CDvist: a webserver for identification and visualization of conserved domains in protein sequences. Bioinformatics 31:1475–1477. doi: 10.1093/bioinformatics/btu836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kus JV, Tullis E, Cvitkovitch DG, Burrows LL. 2004. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150:1315–1326. doi: 10.1099/mic.0.26822-0. [DOI] [PubMed] [Google Scholar]
- 37.Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL, Burrows LL. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol 394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 38.Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. 2013. The platform protein is essential for type IV pilus biogenesis. J Biol Chem 288:9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and Nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coil DA, Anne J. 2010. The role of fimV and the importance of its tandem repeat copy number in twitching motility, pigment production, and morphology in Legionella pneumophila. Arch Microbiol 192:625–631. doi: 10.1007/s00203-010-0590-8. [DOI] [PubMed] [Google Scholar]
- 48.Oldfield NJ, Bland SJ, Taraktsoglou M, Dos Ramos FJ, Robinson K, Wooldridge KG, Ala'Aldeen DA. 2007. T-cell stimulating protein A (TspA) of Neisseria meningitidis is required for optimal adhesion to human cells. Cell Microbiol 9:463–478. doi: 10.1111/j.1462-5822.2006.00803.x. [DOI] [PubMed] [Google Scholar]
- 49.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, Hancock RE. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res 15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Topal H, Fulcher NB, Bitterman J, Salazar E, Buck J, Levin LR, Cann MJ, Wolfgang MC, Steegborn C. 2012. Crystal structure and regulation mechanisms of the CyaB adenylyl cyclase from the human pathogen Pseudomonas aeruginosa. J Mol Biol 416:271–286. doi: 10.1016/j.jmb.2011.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasegawa H, Holm L. 2009. Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol 19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. 2009. IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci U S A 106:9661–9666. doi: 10.1073/pnas.0812900106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lapouge K, Smith SJ, Walker PA, Gamblin SJ, Smerdon SJ, Rittinger K. 2000. Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol Cell 6:899–907. doi: 10.1016/S1097-2765(05)00091-2. [DOI] [PubMed] [Google Scholar]
- 55.Coil DA, Vandersmissen L, Ginevra C, Jarraud S, Lammertyn E, Anne J. 2008. Intragenic tandem repeat variation between Legionella pneumophila strains. BMC Microbiol 8:218. doi: 10.1186/1471-2180-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ensminger AW, Yassin Y, Miron A, Isberg RR. 2012. Experimental evolution of Legionella pneumophila in mouse macrophages leads to strains with altered determinants of environmental survival. PLoS Pathog 8:e1002731. doi: 10.1371/journal.ppat.1002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bansal PK, Nourse A, Abdulle R, Kitagawa K. 2009. Sgt1 dimerization is required for yeast kinetochore assembly. J Biol Chem 284:3586–3592. doi: 10.1074/jbc.M806281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krachler AM, Sharma A, Kleanthous C. 2010. Self-association of TPR domains: lessons learned from a designed, consensus-based TPR oligomer. Proteins 78:2131–2143. doi: 10.1002/prot.22726. [DOI] [PubMed] [Google Scholar]
- 60.Watson AA, Mattick JS, Alm RA. 1996. Functional expression of heterologous type 4 fimbriae in Pseudomonas aeruginosa. Gene 175:143–150. doi: 10.1016/0378-1119(96)00140-0. [DOI] [PubMed] [Google Scholar]
- 61.Asikyan ML, Kus JV, Burrows LL. 2008. Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J Bacteriol 190:7022–7034. doi: 10.1128/JB.00938-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.