ABSTRACT
Using the squid-vibrio association, we aimed to characterize the mechanism through which Vibrio fischeri cells signal morphogenesis of the symbiotic light-emitting organ. The symbiont releases two cell envelope molecules, peptidoglycan (PG) and lipopolysaccharide (LPS) that, within 12 h of light organ colonization, act in synergy to trigger normal tissue development. Recent work has shown that outer membrane vesicles (OMVs) produced by V. fischeri are sufficient to induce PG-dependent morphogenesis; however, the mechanism(s) of OMV release by these bacteria has not been described. Like several genera of both beneficial and pathogenic bacteria, V. fischeri cells elaborate polar flagella that are enclosed by an extension of the outer membrane, whose function remains unclear. Here, we present evidence that along with the well-recognized phenomenon of blebbing from the cell's surface, rotation of this sheathed flagellum also results in the release of OMVs. In addition, we demonstrate that most of the development-inducing LPS is associated with these OMVs and that the presence of the outer membrane protein OmpU but not the LPS O antigen on these OMVs is important in triggering normal host development. These results also present insights into a possible new mechanism of LPS release by pathogens with sheathed flagella.
IMPORTANCE Determining the function(s) of sheathed flagella in bacteria has been challenging, because no known mutation results only in the loss of this outer membrane-derived casing. Nevertheless, the presence of a sheathed flagellum in such host-associated genera as Vibrio, Helicobacter, and Brucella has led to several proposed functions, including physical protection of the flagella and masking of their immunogenic flagellins. Using the squid-vibrio light organ symbiosis, we demonstrate another role, that of V. fischeri cells require rotating flagella to induce apoptotic cell death within surface epithelium, which is a normal step in the organ's development. Further, we present evidence that this rotation releases apoptosis-triggering lipopolysaccharide in the form of outer membrane vesicles. Such release may also occur by pathogens but with different outcomes for the host.
INTRODUCTION
In symbiotic associations involving environmentally acquired microorganisms, mechanisms to deliver signals between a bacterium and its potential host are essential for the proper establishment of symbiosis. The association within the light-emitting organ of the Hawaiian bobtail squid, Euprymna scolopes, by its specific bioluminescent symbiont, Vibrio fischeri, provides a simple binary model with which to study the molecular communication that develops between this bacterium and its host (1, 2).
In the squid-vibrio model, symbiosis begins with the harvesting of planktonic V. fischeri cells from the ambient seawater by each generation of newly hatched hosts. The juvenile squid's nascent light organ exposes two pairs of prominent ciliated epithelial appendages whose activity facilitates the recruitment of symbionts. During the first few hours after the juveniles hatch into seawater containing V. fischeri, five to 15 cells attach to and aggregate on these cilia; they then move to and through surface pores at the base of the appendages. These bacteria travel down ducts, eventually reaching internal crypts where the inoculating symbionts multiply, filling the crypt space (3, 4). Once the host has been successfully colonized, the light organ undergoes an irreversible morphogenesis that leads to the loss of its ciliated appendages and differentiation into the mature functional state. While complete regression of the appendages takes more than 4 days (5, 6), indicators of its developmental induction can be identified within hours of inoculation. For example, the migration of macrophage-like hemocytes into the sinus of the appendages reaches a peak after 18 h, and programmed cell death (i.e., late-stage apoptosis) of the epithelial fields of the appendages can be detected after 24 h (5, 7). In the absence of V. fischeri cells, morphogenesis can be experimentally triggered simply by adding peptidoglycan (PG) derivatives, like the PG monomer, together with lipopolysaccharide (LPS), to seawater containing newly hatched animals (8).
PG derivatives and LPS are common microbe-associated molecular patterns (MAMPs), released by both beneficial and pathogenic bacteria, that induce responses in their host's tissues (9). Thus, the context in which those MAMPs are delivered determines the nature of the response and, ultimately, the outcome of the association. In a previous study, we reported that the addition of purified bacterial outer membrane vesicles (OMVs) to the seawater surrounding juvenile squid was sufficient to trigger PG-inducible phenotypes associated with the light organ's irreversible morphogenesis (8, 10). The release of OMVs, which frequently contain PG derivatives and display on their surface not only the primary components of LPS (i.e., lipid A and O antigen) but also outer membrane proteins (OMPs), has been hypothesized to facilitate signaling among bacteria and between bacteria and host tissues (11). Nevertheless, the dynamics underlying the release of OMVs remain poorly understood.
Certain bacteria, including V. fischeri, assemble flagella that are surrounded by an outer membrane-derived sheath (12). Transmission electron micrographs (TEM) of V. fischeri revealed vesicle-like structures on their sheathed polar flagella (13), and a recent study demonstrated that the rotation of sheathed flagella by at least two species of bacteria promotes the release of LPS (14). Given these findings, we aimed to determine whether the flagellar sheath plays a role in host morphogenesis by releasing OMVs. Using the squid-vibrio model, we showed that (i) almost all of the LPS released by this bacterium is associated with OMVs, and OMVs are responsible for LPS-triggered morphogenesis in the host; (ii) the amount of OMVs produced correlates with the number of sheathed flagella and is promoted by their rotation; and (iii) both the release and average size of a cell's OMVs are influenced by the presence of sheathed but not unsheathed bacterial flagella.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. V. fischeri strain ES114 (15) is the wild-type parent of all mutant derivatives. For overnight cultures, strains of Vibrio fischeri and Vibrio parahaemolyticus were grown in Luria-Bertani salt (LBS) medium (10 g of Bacto tryptone, 5 g of yeast extract, 20 g of NaCl, and 50 ml of 1 M Tris-HCl buffer [pH 7.5] per liter of deionized water). For motility assessments and squid inoculations, the strains were grown in a seawater tryptone (SWT) medium(5 g of Bacto tryptone, 3 g of yeast extract, 3 ml of glycerol, 700 ml of Instant Ocean [IO; Aquarium Systems, Inc., Mentor, OH] at a salinity between 33 and 35 ppt, and 300 ml of deionized water per liter). Vibrio cholerae and Escherichia coli strains were grown in Luria-Bertani medium (10 g of Bacto tryptone, 5 g of yeast extract, and 10 g of NaCl per liter of deionized water). When appropriate, erythromycin (Erm) was added to a final concentration of 5 μg/ml.
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| Vibrio fischeri | ||
| ES114 | Sequenced wild-type E. scolopes light organ isolate | 15 |
| ES114 HS (DM73) | ES114 spontaneous hyperflagellated swarmer mutant | 20 |
| ES114 cheA | VF_1831::TnErm; chemotaxis histidine autokinase | 21 |
| ES114 flrA | VF_1856::TnErm; sigma-54-dependent regulator | 21 |
| ES114 motB1 | VF_0715::TnErm; flagellar motor protein | 21 |
| ES114 ompC1 | ΔVF_1795; outer membrane porin | This study |
| ES114 ompC2 | ΔVF_1010; outer membrane porin | This study |
| ES114 ompU | VF_0475::TnErm; outer membrane porin | 39 |
| ES114 waaL | VF_0151::TnErm; O-antigen ligase | 18 |
| Vibrio parahaemolyticus KNH1 | Environmental isolate from the coastal waters of Oahu, HI | 40 |
| Vibrio cholerae | ||
| O395N1 | Wild type | 48 |
| O395N1 flrA | ΔVC0395 mutant | 48 |
| Escherichia coli | ||
| RP437 | Wild type | 49 |
| RO437 flrA | ΔLJ08_4617 mutant | 49 |
Construction of OMP mutants.
V. fischeri deletion mutants were constructed using a previously established allelic exchange method (16). Briefly, flanking regions of the target gene were amplified by PCR, ligated, and then inserted into the mobilizable suicide vector pKV363. The primers used to generate the ompC1 (VF_1795) and ompC2 (VF_1010) mutants were VF1795_salI_F1 (ATCTAGGTCGACCCAAAGACCTGTGCTTCTTTACCATC), VF1795_nheI_R1 (ATCTAGGCTAGCCTTTTTCATGTTCTATACCCTATATATTTAAATTTTATATTGGTTTGAT), VF1795_nheI_F2 (ATCTAGGCTAGCGAATTCTAATCTGACCCGATAATATTGATACAACTGTTAATCTATAAG), VF1795_sphI_R2 (ATCTAGGCATGCGATATGACAAATGCTCAACGCTTAATCCTTTC), VF1010_salI_F1 (ATCTAGGTCGACCTAACCGATTTGTTTAATGACTCTAACCACG), VF1010_nheI_R1 (ATCTAGGCTAGCACAACACATCTAATCGTGTGATTAATCAGTACTTC), VF1010_nheI_F2 (ATCTAGGCTAGCTACTTCTAATAGTATTACATAAAGTCAAGTAATTATTAAGCCAGTC), and VF1010_sphI_R2 (ATCTAGGCATGCCATAATAAGTCCACTGCCTTTTCTTAGAAAGAT). An E. coli strain carrying the allelic exchange vector was conjugated into V. fischeri ES114. Stable transconjugants were then cultured without antibiotic selection, plated on LBS agar, and assayed for antibiotic sensitivity. Antibiotic-sensitive clones were verified as deletion mutants by PCR, using the appropriate F1 and R2 primers, followed by sequencing.
OMV preparation.
OMVs were isolated from culture supernatants using a modified version of a previously described procedure (17). Briefly, all strains were grown to an optical density at 600 nm (OD600) of 2.0. The cells were removed by centrifugation at 4,500 × g for 15 min, and the resulting supernatant was successively filtered through 0.45-μm- and 0.22-μm-pore-size polyvinylidene difluoride (PVDF) membrane filters (Millipore Corp., Billerica, MA). OMVs were separated from other extracellular products by ultracentrifugation at 173,000 × g for 2 h at 4°C in a 90 Ti rotor (Beckman Coulter, Inc., Brea, CA). The resulting pellet was washed and resuspended in Dulbecco's phosphate-buffered saline (dPBS) (0.2 g of KCl, 0.2 g of KH2PO4, 11.7 g of NaCl, 1.1 g of Na2HPO4, 0.1 g of MgCl2(H2O)6, and 0.1 g of CaCl2 per liter of deionized water), supplemented with an additional 11.7 g/liter NaCl, and filter sterilized. The protein concentration of OMVs was determined using a Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY), according to the manufacturer's protocol. OMV preparations were stored at −20°C until use. For OMV characterization experiments, OMVs were further separated from flagella and cell debris using a discontinuous 20% to 55% sucrose gradient and centrifuged either at 100,000 × g for 12 h in a SW40 Ti rotor or at 90,000 × g for 15 h in an MLA-50 rotor (Beckman Coulter, Indianapolis, IN). The resulting fractions were washed and resuspended in dPBS. OMVs from cells suspended in Percoll were isolated by diluting the suspensions to <5% Percoll before the centrifugation steps. This procedure allowed a quantitative recovery of OMVs, as indicated by measurement of the amount of lipid in the resuspended pellet compared to that in the cell-free supernatant before and after pelleting by ultracentrifugation (see below).
Quantification of OMVs by total-lipid and LPS assays.
A second method to determine the relative amounts of OMVs in a preparation was by quantification of total lipid and LPS. The total lipid amount was determined by staining with a lipid-binding fluorophore and measuring fluorescence intensity, as described previously (10). Briefly, cell-free supernatants or OMV preparations were incubated with FM4-64 (Life Technologies/Molecular Probes) in dPBS for 10 min at 37°C. After excitation at 535 nm, the emission at 670 nm was measured using a Tecan Genios Pro plate reader fluorometer (Tecan Group, Männedorf, Switzerland) on three replicate samples. The controls included OMVs alone and the FM4-64 probe alone. The wild-type V. fischeri parent strain ES114 was used as the reference to compare relative OMV production by V. fischeri mutants and other species. Analysis of the different-size OMVs produced by the wild type and several mutants indicated that the ratio of protein to lipid remained within a factor of 2 of the wild type. This result was consistent with the finding that in V. fischeri OMVs, >90% of the OMV protein is membrane associated (i.e., <10% is cargo).
To quantify LPS, cell-free supernatants or OMV preparations were serially diluted in pyrogen-free water, and reactogenic LPS was detected using the ToxinSensor chromogenic LAL endotoxin assay kit (GenScript, Piscataway, NJ), according to the manufacturer's instructions but halving the suggested volumes. Reactogenic LPS units for each sample were normalized to the OD600 of the culture. All data are presented as the mean and standard error of the mean (SEM) for at least three sets of biological replicates.
Determination of OMV size and flagellar structures.
To visualize V. fischeri OMVs and flagellar structures, either cells grown in SWT to an approximate OD600 of 0.3 or purified OMVs were applied to Pioloform-coated copper grids (Ted Pella Co., Tustin, CA) for 1 min, washed twice with sterile water for 30 s, and negatively stained for 1 min with NanoW (Nanoprobes, Yaphank, NY). Grids were dried for 2 min and immediately examined using a Philips CM120 transmission electron microscope (University of Wisconsin Medical School Electron Microscope Facility, Madison, WI).
Comparison of OMV protein composition.
To compare the protein composition of OMVs from different strains, OMVs were prepared and purified with a discontinuous sucrose gradient, as described above. The resulting preparation was then boiled at 70°C for 10 min in NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Novex, Carlsbad, CA). Isolated proteins were then run in NuPAGE 4% to 12% Bis-Tris gels (Invitrogen, Carlsbad, CA) in NuPAGE morpholineethanesulfonic acid (MES) SDS running buffer for 35 min at 200 V. Gels were stained with ProtoBlue Safe (National Diagnostics, Atlanta, GA), according to the manufacturer's suggestions, and imaged on a ChemiDoc-It2 imager (UVP, LLC, Upland, CA).
Assessment of phenotypes associated with light organ maturation.
Newly hatched juvenile squids in filter-sterilized IO seawater were exposed to either 10,000 CFU/ml V. fischeri cells or OMVs (50 μg/ml OMV protein). To assess the onset of late-stage apoptosis, juveniles were fixed overnight in mPBS (50 mM sodium phosphate buffer containing 0.45 M NaCl [pH 7.4]) containing 4% paraformaldehyde, and washed 3 times for 10 min each in mPBS. The light organs were then dissected and rinsed in mPBS prior to staining. To label the actin cytoskeleton, the samples were incubated overnight in mPBS containing 1% Triton X-100 and 0.19 mM rhodamine phalloidin (Life Technologies) and then washed 3 times for 10 min each in mPBS. To label apoptotic nuclei, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining (which indicates DNA fragmentation) was performed using the DEADEnd fluorometric assay kit (Promega, Madison, WI) according to the manufacturer's instructions. To label host cell nuclei, light organs were washed 3 times for 10 min each in mPBS and incubated for 20 min in mPBS containing 1 μM TOTO-3. After incubation, the samples were washed 3 times for 10 min each in mPBS, and fluorescently labeled samples were analyzed using a Zeiss 510 laser scanning confocal microscope with a 40× objective.
To quantify hemocyte trafficking into the epithelial sinuses of the appendages, juvenile squids were anesthetized in fluorosilicone oil (FSIO) containing 2% ethanol and fixed with 4% paraformaldehyde in mPBS for 18 h at 4°C. The light organ was then exposed by dissection and permeabilized for 18 h with a solution of mPBS containing 1% Triton X-100 at 4°C. To label hemocytes and filamentous actin, respectively, the light organs were incubated with 0.64 mM Alexa Fluor 488-conjugated DNase I and 0.19 mM tetramethyl rhodamine isocyanate (TRITC)-rhodamine phalloidin (Invitrogen, Carlsbad, CA) for 48 h at 4°C. The samples were washed 4 times for 15 min each with mPBS. The light organs were mounted on a glass depression slide using Vectashield medium (Vector Laboratories, Inc., Burlingame, CA) to reduce photobleaching and examined by confocal laser scanning microscopy, using a 40× objective.
All experiments were performed with three biological replicates. Twenty squids were used per condition, except where stated otherwise. Statistics were performed using the GraphPad Prism software (GraphPad, La Jolla, CA). Asterisks in the figures indicate groups of statistically different means, determined with a one-way analysis of variance (ANOVA) of differences, a post hoc Bonferroni correction, and a Tukey's test, when appropriate.
To assess the extent of regression of the ciliated appendages, 10 animals were either inoculated with 10,000 CFU of V. fischeri per ml of IO for 24 h or exposed to OMVs (50 μg of protein per ml of IO) for 12 h each day. The IO was changed every 12 h. After 4 days, animals were fixed for 14 h in mPBS containing 4% paraformaldehyde. After 5 min, the mantle was opened to expose the light organ and ensure sufficient fixation of tissues. Then, the squids were washed 2 times in mPBS and dehydrated through an ethanol series. The samples were dried for scanning electron microscopy (SEM), using a Tousimis Samdri 780 critical point drier, and sputter coated with a SeeVac Auto Conductavac IV. Samples were mounted on stubs and examined with a Hitachi S-570 LaB6 scanning electron microscope.
RESULTS
OMVs are sufficient to induce light organ maturation.
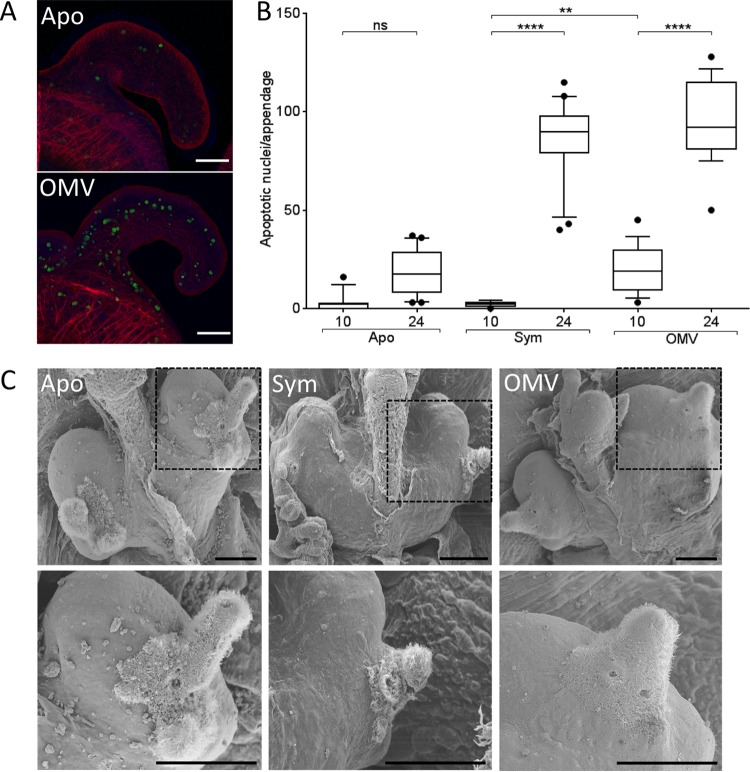
To address whether OMVs produced by V. fischeri present morphogenesis-inducing MAMPs to the host, we first asked whether the OMVs trigger two morphogenic changes known to be induced by the synergy of PG fragments and LPS: late apoptosis in and regression of the light organ's surface epithelial fields (6). We found that on its own, OMV addition induces pronounced levels of late-stage apoptosis after 24 h (Fig. 1A). We quantified the mean number of apoptotic nuclei present in the appendages after 10 h or 24 h of exposure either to the symbiont or to OMVs. We found that while a maximum level of late apoptosis was induced at 24 h by either treatment (Fig. 1B), after only 10 h, the addition of OMV had already triggered a significantly higher level of apoptosis than symbiotic cells alone, relative to uncolonized (aposymbiotic) animals. This accelerated response is similar to that reported for the experimental addition of purified MAMPs (8) and is consistent with the idea that both OMVs and soluble MAMPs can diffuse rapidly into the crypts. In contrast, the inoculating cells in symbiont-colonized animals must first migrate into the crypts and then proliferate before they can induce a response (10). Moreover, by 4 days postcolonization, we observed comparable regression of the ciliated appendages in both symbiont-exposed and OMV-treated light organs relative to aposymbiotic light organs (Fig. 1C). Thus, OMVs not only are sufficient to induce morphogenic changes characteristic of normal symbiotic light organ maturation but also trigger development more rapidly than bacterial colonization.
FIG 1.
OMVs induce phenotypes associated with LPS-linked light organ morphogenesis. (A) Representative Z-stack confocal images of TUNEL-stained aposymbiotic juvenile light organs, with (OMV) or without (Apo) the addition of OMVs. Filamentous actin was stained with rhodamine-633 (red), nuclei were stained with TOTO-3 (blue), and apoptotic nuclei were TUNEL stained (green). Scale bars = 50 μm. (B) Induction of apoptosis in the superficial epithelium. Animals were untreated (Apo), exposed to 10,000 CFU V. fischeri/ml (Sym), or exposed to 50 μg of OMV protein/ml (OMV) for either 10 or 24 h. Apoptotic nuclei were counted in one appendage per light organ. Error bars indicate one-way ANOVA (F = 184); here and elsewhere, the horizontal lines in the boxes indicate the median values and the boxes indicate the limits of the first and third quartiles. ****, P < 0.0001; **, P < 0.01; ns, not significant. (C) Two levels of scanning electron microscopy magnification (each image on the bottom is a magnification of the image in the dashed box above it) of the ventral light organ surfaces of representative 4-day-old animals, revealing the degree of ciliated epithelial field regression. Scale bars = 100 μm. Aposymbiotic animals (Apo) show an intact ciliated ridge, and both the anterior and posterior appendages are present and functional. Animals treated with 50 μg OMV protein/ml (OMV) show a level of regression similar to that of symbiotic animals (Sym).
OmpU, but not the O antigen, potentiates OMV-induced responses.
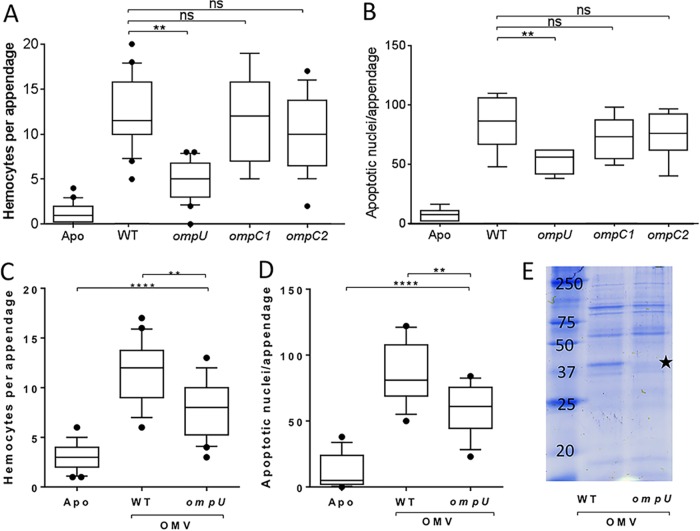
The two major classes of molecules exposed on a Gram-negative bacterium's cell envelope are outer membrane proteins (OMPs) and the O antigen of LPS (17). OMVs are derived from this envelope; thus, we asked whether OMPs and O antigen were involved in the OMV signaling perceived by the host's tissues. First, juvenile squids were exposed to OMVs isolated from one of three OMP mutants lacking an individual OMP: ompU (VF_0475), VF_1010, or VF_1795 (the latter two each encode a homolog of Escherichia coli OmpC, which we have termed OmpC1 and OmpC2, respectively). We determined the capacity of these mutants to induce two phenotypes linked to light organ maturation (i.e., hemocyte trafficking and late-stage apoptosis) by quantifying (i) the mean number of hemocytes present in the appendages after 18 h and (ii) the number of apoptotic nuclei appearing at 24 h after inoculation. While the response to colonization by the two OmpC mutants was indistinguishable from that of the wild type, colonization by the ompU mutant resulted in a significant reduction in the levels of both phenotypes (Fig. 2A and B), even though the ompU mutant produces significantly more OMVs than the wild type (see Fig. S1 in the supplemental material). Similarly, this differential response was also found after exposure to the OMVs isolated from wild-type and ompU mutant cells (Fig. 2C and D). Finally, to confirm that the decreased induction of both phenotypes was coincident with the absence of OmpU in the OMVs, we compared the general protein profiles of OMVs produced by the wild-type strain and the ompU mutant. The only obvious difference between the profiles was the expected absence of OmpU on the mutant OMVs (Fig. 2E). Collectively, these data support the hypothesis that OmpU is required for OMVs to mediate normal MAMP signaling.
FIG 2.
OmpU is required for induction of two symbiotic phenotypes. (A and C) Levels of hemocyte trafficking in the anterior appendages of animals after 18 h. (B and D) Counts of apoptotic nuclei in the anterior appendages of animals after 24 h. (A and B) Animals were exposed to no V. fischeri cells (Apo), wild-type V. fischeri (WT), or one of three OMP mutants: the ompU mutant, the ompC1 mutant, or the ompC2 mutant. (C and D) Animals were exposed to OMVs (50 μg of OMV protein/ml) released by either the wild-type (WT) or the ompU mutant. One-way ANOVA was carried out (F = 31, 54, 39, and 15 for panels A, B, C, and D, respectively). ****, P < 0.0001; **, P < 0.01; ns, not significant. (E) Coomassie-stained SDS-PAGE gel of the proteins of OMVs released by either the wild-type (WT) or the ompU mutant (5 μg of OMV protein was loaded in each well). The star indicates the migration position of OmpU.
V. fischeri has been reported to express an unusual O antigen on its LPS, so we next asked whether this major envelope surface element plays a role in OMV recognition. Specifically, we determined how host development was affected by symbionts carrying a mutation in the waaL gene, which eliminates the bacterium's ability to ligate the O antigen to the core LPS (18). In contrast to the ompU mutant, neither colonization by the waaL mutant nor treatment with waaL mutant-derived OMV resulted in a detectable difference in phenotypic responses from that of wild-type cells (see Fig. S2 in the supplemental material), even though the waaL mutant colonizes at a lower level (18). These results suggest that OMV recognition is not dependent on O-antigen recognition.
OMV production is primarily responsible for LPS release from V. fischeri.
LPS is a common inducer of the innate immune system in both vertebrate and invertebrate animals (19). In the squid-vibrio symbiosis, complete apoptosis of the light organ's ciliated epithelium is induced by the lipid A component of LPS (7) in synergy with PG (8). LPS release has been directly tied to the rotation of the sheathed flagella of V. fischeri (14); however, the form in which it is released was unknown. To determine how much of the cell-derived LPS is soluble or, instead, associated with OMVs, both the total lipids and the LPS present in cell-free culture supernatants of exponentially growing V. fischeri cells were quantified before and after removing the OMVs by ultracentrifugation. We found that essentially, all the LPS released by the bacteria was associated with OMVs (Fig. 3). These data support the hypothesis that OMV production is the major mechanism by which LPS, and perhaps other MAMPs, are released from symbiont cells.
FIG 3.
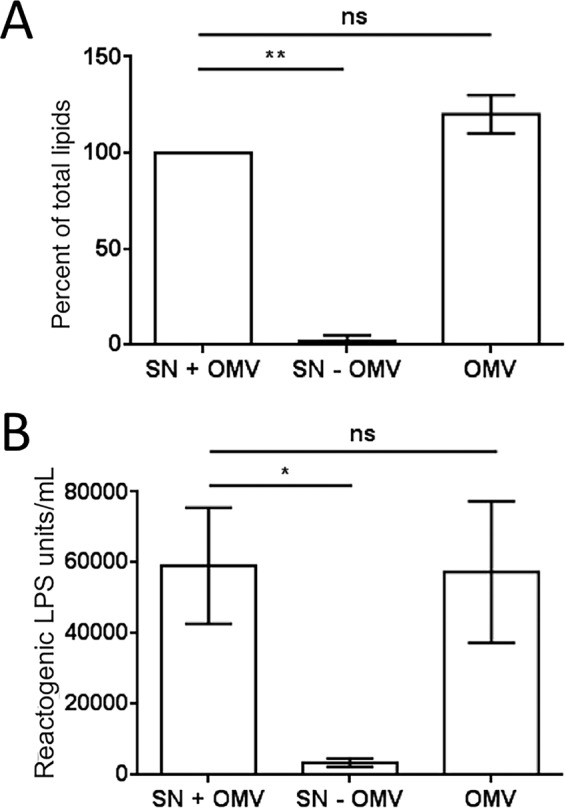
Lipid and LPS are released almost entirely as OMVs. OMVs were prepared from cell-free cultures of V. fischeri. Both total lipids and LPS were quantified in three fractions: culture supernatant (SN + OMV), the supernatant after OMV removal (SN − OMV), and the pelleted OMVs (OMV). (A) FM4-64-based total-lipid assay. (B) Limulus amoebocyte lysate (LAL) assay for LPS. A one-way repeated-measures ANOVA with a post hoc Bonferroni correction was performed. **, P < 0.01; *, P < 0.1; ns, not significant.
Rotation of flagella promotes OMV production.
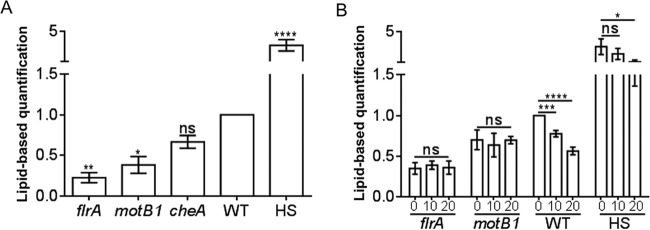
To further characterize the mechanism by which OMVs might be released, we investigated the contribution of flagellar rotation to OMV production. We observed that both a motB1 mutant, which lacks a functional motor to drive flagellar rotation, and an aflagellate flrA mutant release significantly fewer OMVs than wild-type cells. In contrast, a hyperflagellated swarmer (HS) derivative of the wild type that expresses 3 to 4 times more flagella (20) releases significantly more OMVs (Fig. 4A). In contrast, a chemotaxis mutant (cheA) releases about the same amount of OMVs as the wild type, suggesting that motility, but not chemotaxis, contributes to the production of OMVs. To confirm that motB1 and flrA mutants release fewer OMVs because of their lack of a rotating flagellum, we quantified the OMV production of cells in medium containing Percoll, an inert viscosity-increasing additive containing colloidal silica particles. At the higher viscosity, wild-type and HS cells were observed to have decreased motility, as well as a reduced production of OMVs (Fig. 4B). In contrast, the addition of Percoll had little effect on OMV release by motB1 and flrA mutants, consistent with the hypothesis that flagellar rotation is an important potentiator of OMV production. Finally, the major motor protein of the Vibrio flagellar machinery, MotB1, is a sodium pump (21). Treatment with phenamil, an amiloride analog, irreversibly blocks MotB1 activity, reducing flagellar rotation (22). A notable side effect of such inhibitors is that other sodium-dependent activities can also be blocked, thereby affecting regulation and metabolism, if not growth rate (23, 24). Perhaps due to a confounding effect of these sorts of off-target perturbations, we observed no decrease in lipid (i.e., OMV) accumulation in the culture supernatant upon the addition of phenamil to either wild-type or motB1 mutant cells; nevertheless, the inhibitor did significantly lower the amount of lipid released by the HS mutant, consistent with a decreased level of flagellar rotation (see Fig. S3 in the supplemental material). Collectively, these genetic, physical, and chemical approaches support the hypothesis that flagellar rotation increases OMV release.
FIG 4.
Flagellar rotation and motility promote OMV production. (A) Fold difference, relative to the wild type (WT), in the level of OMV production by four motility mutants: the flrA mutant (no flagellum), the motB1 mutant (nonrotating flagellum), the cheA mutant (chemotaxis defect), and a hyperflagellated swarmer mutant (HS). (B) Influence of medium viscosity (i.e., addition of 0, 10, or 20% Percoll) on relative levels of OMV production. Values are means ± standard errors of the means (SEM; n = 3). ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.1; ns, not significant.
OMV release is correlated with the presence of a sheathed flagellum.
We next asked whether the link between flagellar rotation and OMV production might be confined to bacteria with an outer membrane-derived sheath surrounding their flagella (14), and whether the amount of OMVs released was proportional to the number of sheathed flagella per cell. A survey of three Vibrio species with sheathed polar flagella (V. fischeri, V. cholerae, and V. parahaemolyticus), as well as E. coli, which expresses only unsheathed flagella, found that OMV production is correlated with the number of sheathed flagella. Specifically, V. cholerae has only a single polar sheathed flagellum and releases fewer OMVs than V. parahaemolyticus, which like V. fischeri, has several polar sheathed flagella (in addition to its multiple peritrichous but unsheathed flagella). (Fig. 5). We also found that, as with V. fischeri, an aflagellate mutant of V. cholerae releases fewer OMVs, whereas an aflagellate E. coli releases the same amount of OMVs as the parent (see Fig. S4 in the supplemental material). Thus, the presence of sheathed flagella increases the potential for OMV release in members of the genus Vibrio, and perhaps in other genera.
FIG 5.
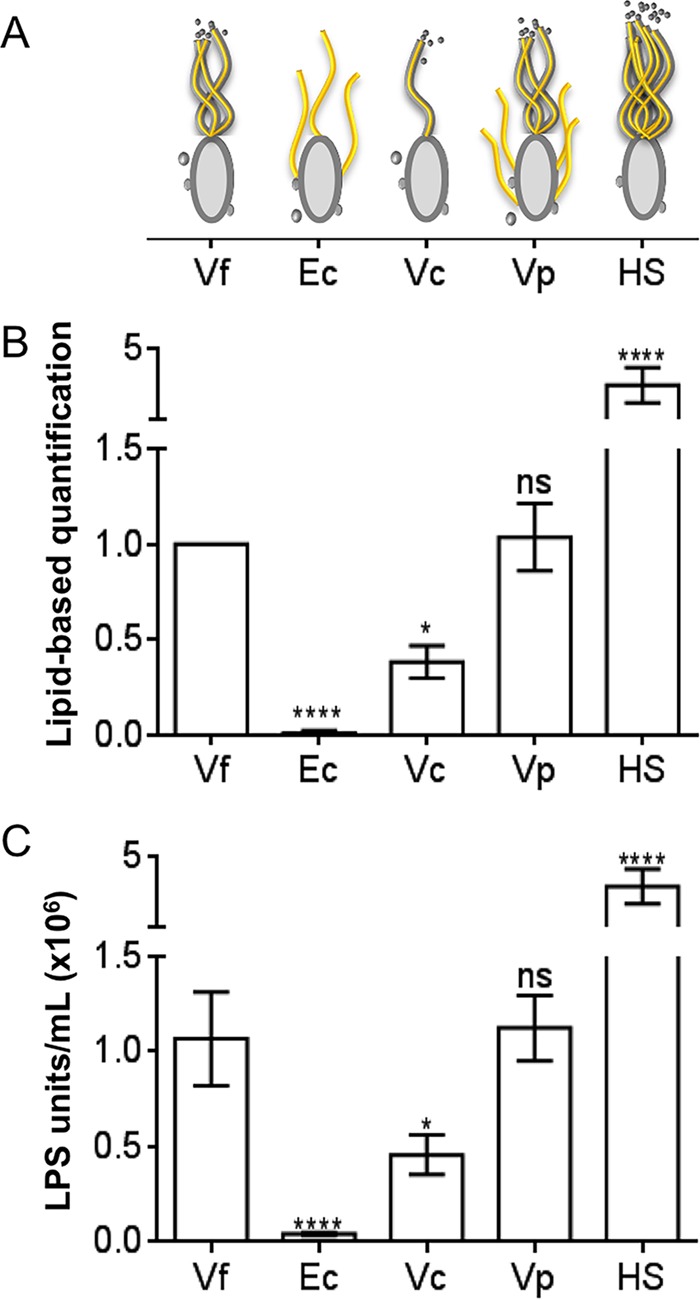
Presence of sheathed flagella correlates with OMV release. (A) Cartoon comparing different bacterial strains, their flagella (yellow), and the presence of a flagellar sheath (gray). Vf, V. fischeri; Ec, E. coli; Vc, V. cholerae; Vp, V. parahaemolyticus; HS, a hyperflagellated swarmer V. fischeri mutant. (B and C) Fold difference, relative to V. fischeri, in OMV production. (B) Total-lipid-based assay for OMVs. (C) LPS-based assay for OMVs. A one-way repeated-measures ANOVA with a post hoc Bonferroni correction was performed. ****, P < 0.0001; *, P < 0.1; ns, not significant.
Rotation of sheathed flagella influences the mean and range of OMV size.
Previous studies have reported that V. cholerae OMVs have a mean size of 72.8 nm, with a range between 20 and 200 nm, while OMVs produced by V. fischeri are generally smaller, averaging 29.8 nm (25, 26). This smaller size raised the question of whether the additional OMVs released during the rotation of the multiple sheathed flagella of V. fischeri might somehow skew the range of OMV sizes released compared to those of V. cholerae. That is, there might be a difference between the size of OMVs released from the cell body and those generated during the rotation of sheathed flagella (Fig. 5A and 6A). When the mean diameter of purified OMVs from three different strains was determined, we found that while fewer OMVs were produced by the immotile motB1 mutant strain (Fig. 4A), those that were released tended to be larger than those generated by wild-type cells (Fig. 6B). In contrast, the HS mutant, which is apparently enriched in OMVs released by rotating flagella (Fig. 4A), produces a higher proportion of smaller OMVs. These results suggest that the presence and functionality of a bacterium's sheathed flagella influence the size distribution of the OMVs it releases (Fig. 6B and C).
FIG 6.
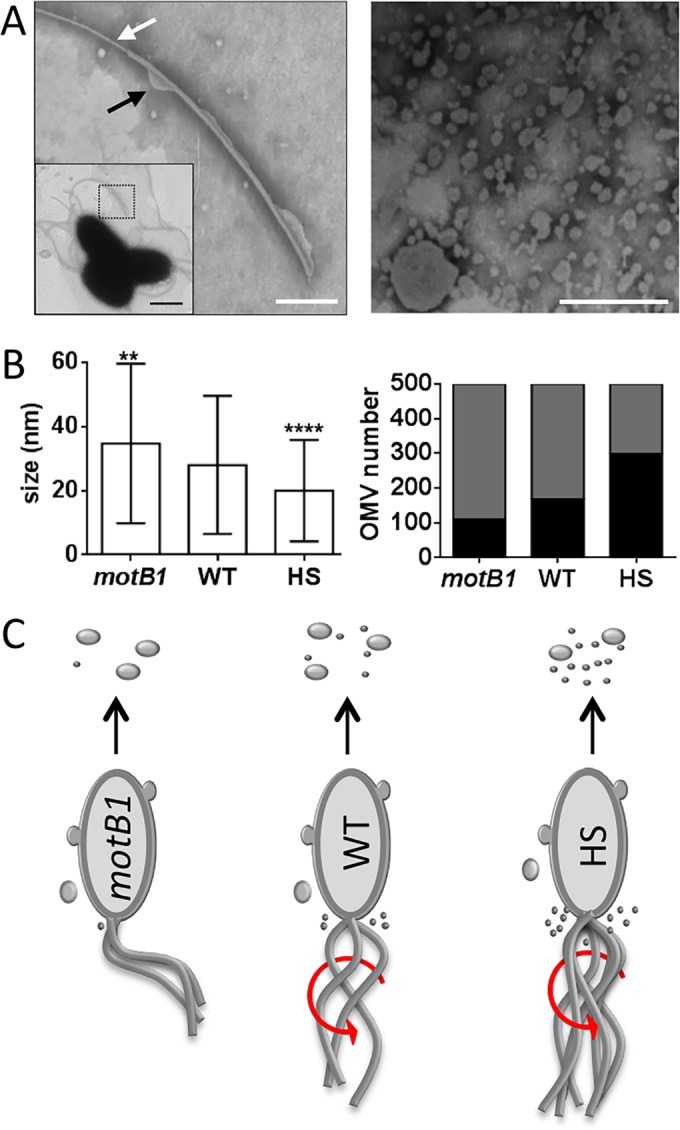
Flagellar activity is associated with reduced OMV size. (A) Left, negative-stained transmission electron micrograph (TEM) enlargement of a V. fischeri sheathed flagellum; white arrow, area exposing flagellar filament; black arrow, bleb forming on sheath; scale bar = 250 nm. Inset, TEM of three cells, with the flagellum in the area of enlargement (dotted box); scale bar = 1 μm. Right, TEM image of an OMV preparation; scale bar = 200 nm. (B) Left, mean size of OMVs produced by wild-type cells (WT) or by immotile mutant (motB1) or hyperflagellated swarmer (HS) derivatives. Right, proportion of OMVs whose size is less than (black bar) or greater than (gray bar) 20 nm. Error bars in the left panel indicate SEM (n = 500). ****, P < 0.0001; **, P < 0.01 (t test). (C) Cartoon depicting OMV production by different V. fischeri strains; the presence of sheathed flagella and their ability to rotate (red arrows), as well as the relative proportion of OMV sizes released, are illustrated.
OMVs produced by either sheathed flagella or the cell body induce hemocyte trafficking.
The induction of late-stage apoptosis is apparently independent of the size or specific LPS structure of bacterial OMVs, because this phenotype can be triggered by OMVs/LPS produced by a variety of Gram-negative species (7, 27). Here, we compared the protein profiles of OMVs produced by different strains used in this study and found that while the overall composition is similar, the relative abundances differ (Fig. 2E; see also Fig. S5A in the supplemental material). Specifically, we found that the protein composition of OMVs produced by the motility mutants appears to be similar to that of the wild type, although the relative proportions may be different, as is seen with the HS strain (see Fig. S5A). The appearance of other bands becoming dominant may indicate a different protein composition in the smaller flagellum rotation-derived OMVs (Fig. 6; see also Fig. S5B). As for nonprotein cargo, the smaller OMVs released during flagellar rotation not only have a smaller volume but also might have a different content, especially of PG fragments (10), the class of MAMPs that induces hemocyte trafficking (8).
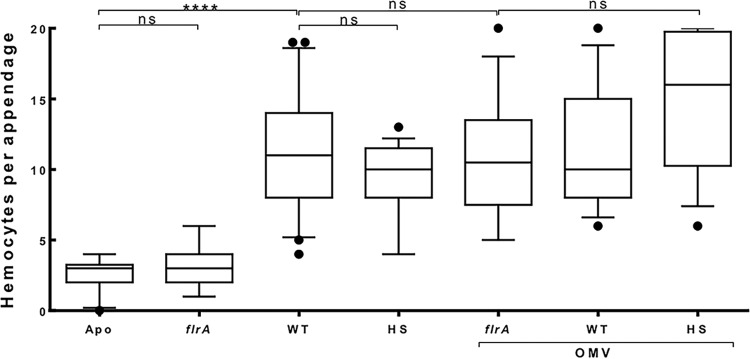
To address whether OMVs produced by either the V. fischeri cell body or its sheathed flagella present such inducers to the host, thereby triggering morphogenesis, we asked whether preparations enriched in OMVs originating from the cell body (flrA mutant) or driven by flagellar rotation (HS mutant) induce similar levels of hemocyte trafficking. We found that while the noncolonizing flrA mutant bacterium does not itself induce hemocyte trafficking (10), OMVs produced by either this strain or the HS mutant both induce a level of hemocyte trafficking similar to that of the wild type (Fig. 7). These data indicate that neither the size nor the site of origin of OMVs influence their capacity to induce key steps in light organ morphogenesis.
FIG 7.
Induction of hemocyte trafficking by OMVs released by motility mutants. Levels of hemocyte trafficking in the anterior appendages of animals were determined 18 h after exposure to cells or OMVs. Treatment was with no V. fischeri cells (Apo), wild-type V. fischeri (WT), or motility mutants lacking either flagella (flrA) or hyperflagella (HS). Animals were also exposed to OMVs (50 μg of OMV protein/ml) released by the three strains. One-way ANOVA, F = 22. ****, P < 0.0001; ns, not significant.
DISCUSSION
Arriving at an understanding of the function(s) of sheathed flagella in bacteria has been particularly challenging, because there are no known mutations that result in the loss of the sheath alone. Nevertheless, beyond the motility function typical of flagella, the presence of an encasing membrane in such host-associated genera as Vibrio, Helicobacter, and Brucella has led to several proposed functions, including physical protection of the flagella and masking of its immunogenic flagellins (28, 29). Using the squid-vibrio model, we demonstrate another role: in a successful colonization of the squid light organ, V. fischeri cells require functional flagella, not only to migrate through the ducts into the deep crypts (21) and release PG (10) but also to release LPS that induces apoptotic cell death within the light organ's surface epithelium (14). Here, we present evidence that this LPS is released in the form of OMVs and that rotation of the cell's sheathed flagellum increases the amount of OMVs produced by the bacterium. Finally, we showed that the cellular location of OMV production does not affect their development-triggering impact on host tissue (Fig. 7).
Several phenotypes associated with light organ morphogenesis are induced by the synergic activity of PG fragments and LPS (8, 30). These two classes of MAMPs have different effects in symbiont-induced morphogenesis: the phosphorylated lipid A of LPS triggers early stage apoptosis, while PG fragments, like tracheal cytotoxin (TCT), activate both hemocyte trafficking and late-stage apoptosis (8). Exposure to OMVs, which contain PG derivatives, has recently been shown to be sufficient to induce hemocyte trafficking in juvenile squid (10). In the current study, we demonstrate that OMVs also present morphogenic LPS, inducing full apoptotic regression of the light organ's ciliated fields (Fig. 1). To provide insight into what other envelope elements associated with OMVs might be involved in inducing host responses, we investigated the role of two major constituents of the cell envelope: outer membrane proteins (OMP) and the O antigen of LPS. OMV production is increased in a Helicobacter tolB mutant with an altered interaction with OMPs (31), indicating a connection between these membrane-stabilizing proteins and the rate of OMV generation (32, 33). Similarly, OmpU, the major OMP of Vibrio spp., plays roles in both pathogenesis and symbiosis within this genus (6, 34). For instance, in pathogenic relationships, OmpU facilitates colonization (35), allowing binding to host tissue and inducing specific immune responses, including cell death (36–38). Conversely, in the mutualistic V. fischeri, although an ompU mutant has no growth defect in culture, it is only 60% as effective in the colonization process (39) and is compromised in the ability to avoid phagocytosis by the host's hemocytes (40), indicating a role for this OMP in the normal establishment and persistence of symbiosis. This susceptibility to phagocytosis is relieved in the presence of wild-type cells, suggesting that the absence of OmpU results in an inappropriate interaction with these host cells rather than an inherent incapacity to colonize the tissue. In this study, we showed that even though an ompU mutation actually increases the rate of OMV generation (see Fig. S1 in the supplemental material), the host fails to respond normally to the mutant or its OMVs (Fig. 2). We hypothesize that OmpU may promote either OMV binding to a host surface receptor and/or their translocation to a specific host cell compartment (38). The binding and internalization of V. fischeri OMV by squid hemocytes and epithelial cells have been recently demonstrated (10), and future work will aim to determine the mechanism underlying the connection between the OmpU of OMVs and these host responses.
In several bacterial genera, such as Klebsiella and Porphyromonas, mutations that modify the O antigen have been reported to affect OMVs by altering the protein content and reducing OMV production, respectively (41, 42). Similarly, while Pseudomonas O-antigen mutants have no effect on the rate of OMV release, they do affect both their contents and size (43). The LPS of V. fischeri cells includes an unusual O antigen that plays a critical role in both flagellar motility and efficient colonization (18), but there is little understanding of this relationship or its relationship to broader changes in either the cell's surface or its OMVs. Exposure to LPS from any of several bacterial sources induces normal apoptosis in the ciliated fields of the light organ (7), suggesting that this host response is not specific to LPS expressed by V. fischeri or its unusual O antigen (27). Thus, it is not surprising that, as in Pseudomonas, the absence of an O antigen does not affect OMV release by V. fischeri, nor does it have a significant effect on hemocyte trafficking or apoptosis in the light organ epithelia (see Fig. S2 in the supplemental material), reinforcing the notion that lipid A is the critical driver of these LPS-induced host developmental responses (7). The squid encodes one LPS-binding protein (LBP) and three of the structurally related bactericidal/permeability-increasing proteins (BPIs) (44). Determining whether any of these host pattern recognition receptors recognize the LPS/lipid A of symbiont OMV during colonization will help identify how this surface MAMP triggers host morphogenesis.
In conclusion, our studies using V. fischeri have revealed a new role for sheathed flagella. Rotation of this motility apparatus was previously shown to increase LPS shedding (14), but the form in which the LPS is released was undetermined. While free LPS is sufficient to experimentally induce light organ apoptosis (7), here, we show that the LPS normally released by the symbionts is almost entirely associated with OMVs (Fig. 3) and that the rotating flagella of V. fischeri are a major source of these OMVs. It remains a mystery how the turning of the flagellum results in the release of OMVs; membrane blebs associated with the tip (13) or the shaft (35) (Fig. 6A) have been reported. We also hypothesize that because the cell's PG-linked outer membrane is contiguous with the rapidly rotating membrane surrounding the flagellar filament, it may be that OMVs are produced at this membrane-membrane interface when the bacteria are motile. If this model is correct, one might predict that by differing with cell surface-produced OMVs in both the mechanism and the location of their generation, flagellar OMVs would be distinct not only in their size (Fig. 6 and 7) but also in their contents. In addition, cryotomographic imaging has revealed unusual structural features of the V. fischeri flagellar basal body that likely play a role in stabilizing the cell's outer membrane at its interface with the sheath (45, 46), providing a new window into understanding flagellum-dependent OMV generation. Because pathogenic species in the genera Brucella, Helicobacter, and Vibrio also express sheathed flagella (31, 47), it will be important to determine whether these structures play a similar role in OMVs/LPS release (10) but with a critically different outcome for the host.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Ruby and McFall-Ngai laboratories for their insight into this research. In particular, Mirjam Knop provided valuable technical help. We thank Rodney Welch (University of Wisconsin—Madison) for sharing laboratory space and Ben August (University of Wisconsin—Madison) and Tina Carvalho (University of Hawaii—Manoa) for electron microscopy training.
Funding was provided by NIH grants from NIGMS (GM099507), NIAID (AI050661), and ORIP (RR012294/OD011024) and an NIAID training grant (T32AI055397).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00101-16.
REFERENCES
- 1.Ruby EG. 2008. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, Heath-Heckman EAC, Gillette AA, Peyer SM, Harvie EA. 2012. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koropatnick TA, Kimbell JR, McFall-Ngai MJ. 2007. Responses of host hemocytes during the initiation of the squid-Vibrio symbiosis. Biol Bull 212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- 4.Kremer N, Philipp EER, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–1729. [DOI] [PubMed] [Google Scholar]
- 6.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster JS, Apicella MA, McFall-Ngai MJ. 2000. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol 226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- 8.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 9.Pirofski L, Casadevall A. 2012. Q and A: what is a pathogen? A question that begs the point. BMC Biol 10:6. doi: 10.1186/1741-7007-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aschtgen M-S, Wetzel K, Goldman W, McFall-Ngai M, Ruby E. 2015. Vibrio fischeri-derived outer membrane vesicles trigger host development. Cell Microbiol 18:488–499. doi: 10.1111/cmi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferooz J, Letesson J-J. 2010. Morphological analysis of the sheathed flagellum of Brucella melitensis. BMC Res Notes 3:333. doi: 10.1186/1756-0500-3-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millikan DS, Ruby EG. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol 186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CA, Hunt JR, Kremer N, Krasity BC, Apicella MA, McFall-Ngai MJ, Ruby EG. 2014. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. eLife 3:e01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/S0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 17.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Post DMB, Yu L, Krasity BC, Choudhury B, Mandel MJ, Brennan CA, Ruby EG, McFall-Ngai MJ, Gibson BW, Apicella MA. 2012. O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: composition and analysis of their role in Euprymna scolopes light organ colonization. J Biol Chem 287:8515–8530. doi: 10.1074/jbc.M111.324012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi KB, Wong F, Harlan JM, Chaudhary PM, Hood L, Karsan A. 1998. Lipopolysaccharide mediates endothelial apoptosis by a FADD-dependent pathway. J Biol Chem 273:20185–20188. doi: 10.1074/jbc.273.32.20185. [DOI] [PubMed] [Google Scholar]
- 20.Millikan DS, Ruby EG. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol 68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. Microbiologyopen 2:576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaques S, Kim YK, McCarter LL. 1999. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc Natl Acad Sci U S A 96:5740–5745. doi: 10.1073/pnas.96.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen L, White EL, Pathak A, Ayala JC, Wang H, Wu J-H, Benitez JA, Silva AJ. 2011. A high-throughput screening assay for inhibitors of bacterial motility identifies a novel inhibitor of the Na+-driven flagellar motor and virulence gene expression in Vibrio cholerae. Antimicrob Agents Chemother 55:4134–4143. doi: 10.1128/AAC.00482-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atsumi T, Sugiyama S, Cragoe EJ Jr, Imae Y. 1990. Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J Bacteriol 172:1634–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata S, Visick KL. 2012. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol 194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee D, Chaudhuri K. 2011. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett 585:1357–1362. doi: 10.1016/j.febslet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 27.Krasity BC, Troll JV, Lehnert EM, Hackett KT, Dillard JP, Apicella MA, Goldman WE, Weiss JP, McFall-Ngai MJ. 2015. Structural and functional features of a developmentally regulated lipopolysaccharide-binding protein. mBio 6(5):e01193-15. doi: 10.1128/mBio.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gewirtz AT. 2006. Flag in the crossroads: flagellin modulates innate and adaptive immunity. Curr Opin Gastroenterol 22:8–12. doi: 10.1097/01.mog.0000194791.59337.28. [DOI] [PubMed] [Google Scholar]
- 29.Yoon SS, Mekalanos JJ. 2008. Decreased potency of the Vibrio cholerae sheathed flagellum to trigger host innate immunity. Infect Immun 76:1282–1288. doi: 10.1128/IAI.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFall-Ngai M, Nyholm SV, Castillo MG. 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes–Vibrio fischeri symbiosis. Semin Immunol 22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner L, Praszkier J, Hutton ML, Steer D, Ramm G, Kaparakis-Liaskos M, Ferrero RL. 2015. Increased outer membrane vesicle formation in a Helicobacter pylori tolB mutant. Helicobacter 20:269–283. doi: 10.1111/hel.12196. [DOI] [PubMed] [Google Scholar]
- 32.Baker JL, Chen L, Rosenthal JA, Putnam D, DeLisa MP. 2014. Microbial biosynthesis of designer outer membrane vesicles. Curr Opin Biotechnol 29:76–84. doi: 10.1016/j.copbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwechheimer C, Sullivan CJ, Kuehn MJ. 2013. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry (Mosc) 52:3031–3040. doi: 10.1021/bi400164t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathur J, Davis BM, Waldor MK. 2007. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol 63:848–858. [DOI] [PubMed] [Google Scholar]
- 35.Vanhove AS, Duperthuy M, Charrière GM, Le Roux F, Goudenège D, Gourbal B, Kieffer-Jaquinod S, Couté Y, Wai SN, Destoumieux-Garzón D. 2015. Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ Microbiol 17:1152–1165. doi: 10.1111/1462-2920.12535. [DOI] [PubMed] [Google Scholar]
- 36.Duperthuy M, Schmitt P, Garzón E, Caro A, Rosa RD, Le Roux F, Lautrédou-Audouy N, Got P, Romestand B, de Lorgeril J, Kieffer-Jaquinod S, Bachère E, Destoumieux-Garzón D. 2011. Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc Natl Acad Sci U S A 108:2993–2998. doi: 10.1073/pnas.1015326108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Chen J, Liu R, Jia J. 2011. Identification and evaluation of an outer membrane protein OmpU from a pathogenic Vibrio harveyi isolate as vaccine candidate in turbot (Scophthalmus maximus). Lett Appl Microbiol 53:22–29. doi: 10.1111/j.1472-765X.2011.03062.x. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, Prasad GVRK, Mukhopadhaya A. 2015. Vibrio cholerae porin OmpU induces caspase-independent programmed cell death upon translocation to the host cell mitochondria. J Biol Chem 290:31051–31068. doi: 10.1074/jbc.M115.670182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aeckersberg F, Lupp C, Feliciano B, Ruby EG. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J Bacteriol 183:6590–6597. doi: 10.1128/JB.183.22.6590-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. 2009. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol 11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cahill BK, Seeley KW, Gutel D, Ellis TN. 2015. Klebsiella pneumoniae O antigen loss alters the outer membrane protein composition and the selective packaging of proteins into secreted outer membrane vesicles. Microbiol Res 180:1–10. doi: 10.1016/j.micres.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Nakao R, Hasegawa H, Ochiai K, Takashiba S, Ainai A, Ohnishi M, Watanabe H, Senpuku H. 2011. Outer membrane vesicles of Porphyromonas gingivalis elicit a mucosal immune response. PLoS One 6:e26163. doi: 10.1371/journal.pone.0026163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy K, Park AJ, Hao Y, Brewer D, Lam JS, Khursigara CM. 2014. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J Bacteriol 196:1306–1317. doi: 10.1128/JB.01463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ. 2011. LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans 39:1039–1044. doi: 10.1042/BST0391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Müller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:E1917–E1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fretin D, Fauconnier A, Köhler S, Halling S, Léonard S, Nijskens C, Ferooz J, Lestrate P, Delrue R-M, Danese I, Vandenhaute J, Tibor A, DeBolle X, Letesson J-J. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol 7:687–698. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- 48.Gosink KK, Häse CC. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J Bacteriol 182:4234–4240. doi: 10.1128/JB.182.15.4234-4240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol 180:2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.