ABSTRACT
Corynebacterium glutamicum metabolizes sialic acid (Neu5Ac) to fructose-6-phosphate (fructose-6P) via the consecutive activity of the sialic acid importer SiaEFGI, N-acetylneuraminic acid lyase (NanA), N-acetylmannosamine kinase (NanK), N-acetylmannosamine-6P epimerase (NanE), N-acetylglucosamine-6P deacetylase (NagA), and glucosamine-6P deaminase (NagB). Within the cluster of the three operons nagAB, nanAKE, and siaEFGI for Neu5Ac utilization a fourth operon is present, which comprises cg2936, encoding a GntR-type transcriptional regulator, here named NanR. Microarray studies and reporter gene assays showed that nagAB, nanAKE, siaEFGI, and nanR are repressed in wild-type (WT) C. glutamicum but highly induced in a ΔnanR C. glutamicum mutant. Purified NanR was found to specifically bind to the nucleotide motifs A[AC]G[CT][AC]TGATGTC[AT][TG]ATGT[AC]TA located within the nagA-nanA and nanR-sialA intergenic regions. Binding of NanR to promoter regions was abolished in the presence of the Neu5Ac metabolism intermediates GlcNAc-6P and N-acetylmannosamine-6-phosphate (ManNAc-6P). We observed consecutive utilization of glucose and Neu5Ac as well as fructose and Neu5Ac by WT C. glutamicum, whereas the deletion mutant C. glutamicum ΔnanR simultaneously consumed these sugars. Increased reporter gene activities for nagAB, nanAKE, and nanR were observed in cultivations of WT C. glutamicum with Neu5Ac as the sole substrate compared to cultivations when fructose was present. Taken together, our findings show that Neu5Ac metabolism in C. glutamicum is subject to catabolite repression, which involves control by the repressor NanR.
IMPORTANCE Neu5Ac utilization is currently regarded as a common trait of both pathogenic and commensal bacteria. Interestingly, the nonpathogenic soil bacterium C. glutamicum efficiently utilizes Neu5Ac as a substrate for growth. Expression of genes for Neu5Ac utilization in C. glutamicum is here shown to depend on the transcriptional regulator NanR, which is the first GntR-type regulator of Neu5Ac metabolism not to use Neu5Ac as effector but relies instead on the inducers GlcNAc-6P and ManNAc-6P. The identification of conserved NanR-binding sites in intergenic regions within the operons for Neu5Ac utilization in pathogenic Corynebacterium species indicates that the mechanism for the control of Neu5Ac catabolism in C. glutamicum by NanR as described in this work is probably conserved within this genus.
INTRODUCTION
The Gram-positive Corynebacterium glutamicum is mostly known for its application in the industrial production of amino acids, mainly l-glutamate and l-lysine (1, 2), and has become a versatile cell factory for the production of various commodity products (3–5). C. glutamicum utilizes a large variety of sugars and organic acids as sources of carbon and energy (6, 7) and additionally has been genetically engineered for the utilization of alternative feedstocks such as starch, glycerol, xylose, glucuronic acid, and N-acetylglucosamine (8–12). In contrast to many other bacterial species, C. glutamicum prefers to use multiple carbon sources simultaneously. It simultaneously metabolizes glucose together with other carbon sources such as sucrose, fructose, maltose, gluconate, ribose, pyruvate, and acetate and thereby exhibits monophasic growth (13–18). Diauxic growth of C. glutamicum and consecutive utilization of the provided carbon sources have been observed so far for media that contain glutamate or ethanol in addition to glucose (6, 19–21). Recently, uptake and utilization of the sialic acid N-acetylneuramic acid (Neu5Ac) were reported to be inhibited in glucose-cultivated C. glutamicum cells (22). This finding suggests the presence of a third example of carbon catabolite repression (CCR) in C. glutamicum, although neither the transcription of genes for Neu5Ac utilization has been analyzed nor have the underlying mechanisms been identified and characterized.
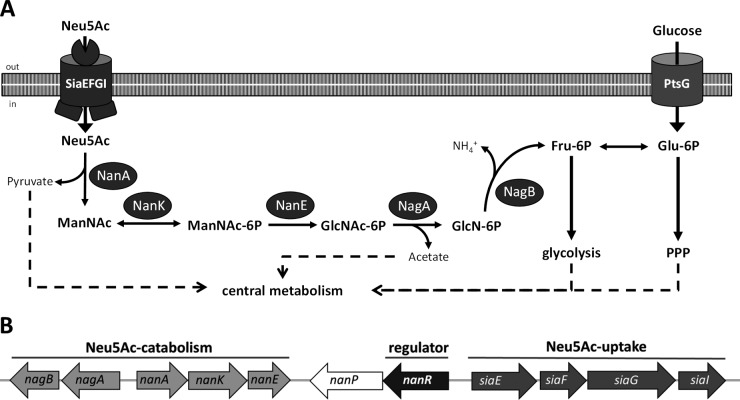
C. glutamicum harbors a full set of genes for Neu5Ac catabolism, which are clustered within the genome (23). Uptake of Neu5Ac is brought about in C. glutamicum by the ABC transporter here named SiaEFGI, encoded by cg2937 (siaE), cg2938 (siaF), cg2939 (siaG), and cg2940 (siaI) (22). As depicted in Fig. 1, Neu5Ac is then metabolized by the consecutive action of the N-acetylneuraminic acid lyase NanA (encoded by cg2931 [nanA]), the N-acetylmannosamine kinase NanK (encoded by cg2932 [nanK]), the N-acetylmannosamine-6-phosphate epimerase NanE (encoded by cg2935 [nanE]), the N-acetylglucosamine-6-phosphate deacetylase NagA (encoded by cg2929 [nagA]), and the glucosamine-6-phosphate deaminase NagB (encoded by cg2928 [nagB]) to pyruvate, acetate, ammonia, and fructose-6-phosphate (fructose-6P) (22). The enzyme NagB is also required for utilization of glucosamine as a source of carbon and nitrogen, which is taken up via the glucose specific permease EIIGlc of the phosphoenolpyruvate sugar phosphotransferase system (PTS) (24). For the efficient utilization of glucosamine by C. glutamicum increased expression of nagB is required, which is brought about either by ectopic expression or by a point mutation within the promoter region of the nagAB operon in the spontaneous mutant C. glutamicum M4 (24). The genes for Neu5Ac utilization are organized in C. glutamicum in three clustered operons, namely, nagAB, nanAKE, and siaEFGI. Within this cluster of operons a fourth operon is present, which comprises the open reading frame (ORF) cg2936 for a putative GntR-type transcriptional regulator and ORF cg2935 (nanP) for a putative sialidase (22).
FIG 1.
(A) Schematic diagram of the pathway for Neu5Ac metabolism in C. glutamicum. SiaEFGI, ABC transporter for Neu5Ac; NanA, N-acetylneuraminic acid lyase; NanK, N-acetylmannosamine kinase; NanE, N-acetylmannosamine-6P epimerase; NagA, N-acetylglucosamine-6P deacetylase; NagB, glucosamine-6P deaminase; PtsG, glucose-specific EII permease of the PTS. (B) Genetic organization of the genes for Neu5Ac utilization in C. glutamicum.
The sialic acid Neu5Ac is the terminal moiety of glycan molecules present on the surface of eukaryotic cells (25–28). Besides serving as attachment and recognition point for various pathogens, Neu5Ac is also an important source of carbon and energy available in various host niches such as the oral cavity and the respiratory, intestinal, and urogenital tracts (29–32). Therefore, Neu5Ac metabolism and its control have been studied in detail for various, often pathogenic, bacteria such as Escherichia coli, Vibrio vulnificus, Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, Clostridium perfringens, and the probiotic Bifidobacterium breve (33–38). In H. influenzae, transcription of the nan and the siaPT operons for Neu5Ac utilization is repressed by SiaR and is activated in the presence of glucosamine 6-phosphate (see Table S1 in the supplemental material), which is an intermediate of Neu5Ac catabolism and acts as a coactivator of the RpiR-type regulator SiaR (37, 39–41). The RpiR-type transcriptional repressor NanR of V. vulnificus controls nanTPSIAR (for the triapartite ATP-independent transporter, Neu5Ac aldolase, and the nan gene repressor), nanEK, and the nagA operons (42, 43). Binding of the Neu5Ac catabolism intermediate N-acetylmannosamine-6-phosphate (ManNAc-6P) to V. vulnificus NanR mediates relocation of residues in the ligand binding site, thus alleviating the interaction between the NanR dimer and DNA and subsequently relieving the repression by NanR and inducing transcription of the nan operons (42) (see Table S1). Different from the RpiR-type regulators of Neu5Ac metabolism of H. influenzae and V. vulnificus, the RpiR-type regulator NanR of S. pneumoniae acts as a transcriptional activator of Neu5Ac catabolism genes (34). In E. coli and B. breve, transcriptional control of genes for Neu5Ac utilization is brought about by GntR-type transcriptional repressors each also named NanR, which both depend on Neu5Ac as the sole inducer (44–46) (see Table S1). Mechanisms for the control of Neu5Ac catabolism have hitherto not been studied for C. glutamicum.
In this communication, we show that in C. glutamicum the cg2936-encoded GntR-type transcriptional regulator, here named NanR, represses transcription of its own gene and of all operons for Neu5Ac catabolism, namely, nagAB, nanAKE, and siaEFGI, by binding within the respective promoter regions. Repression of Neu5Ac catabolism operons in C. glutamicum is shown to be responsible for the preferential utilization of glucose or fructose compared to NeuAc, and the role of NanR for this new example of CCR in C. glutamicum is analyzed. As opposed to the GntR-type repressors of Neu5Ac catabolism of E. coli and B. breve, Neu5Ac did not interfere with C. glutamicum NanR as the inducer. Instead, we show that the presence of ManNAc-6P as well as N-acetylglucosamine-6P (GlcNAc-6P) alleviates binding of C. glutamicum NanR to DNA fragments carrying NanR-binding sites. Finally, we discuss the relevance of these findings for the control of Neu5Ac metabolism in the related pathogenic species Corynebacterium ulcerans, Corynebacterium diphtheriae, and Corynebacterium pseudotuberculosis.
MATERIALS AND METHODS
Microorganisms, plasmids, and cultivation conditions.
Strains and plasmids used in this study are listed in Table 1. The CGXII minimal medium used for cultivation of C. glutamicum has been described previously (47) and contained glucose, fructose, and/or Neu5Ac at concentrations indicated in Results. LB medium (48) was used as a complex medium for C. glutamicum and E. coli. When appropriate, kanamycin (50 μg ml−1) and/or isopropyl-β-d-thiogalactopyranoside (IPTG; 100 μM) was added to the medium. C. glutamicum was grown aerobically at 30°C as 10-ml cultures in 125-ml baffled Erlenmeyer flasks, and E. coli was grown at 37°C as 50-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm. Growth of E. coli and of C. glutamicum strains in liquid cultures was followed by measuring the optical density at 600 nm (OD600).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 phoA supE44 hsdR1 recA1 endA1 gyrA96 thi-1 relA1 | 89 |
| BL21(DE3) | ompT hsdSB(rB− mB−) gal dcm (DE3) | 90 |
| C. glutamicum strains | ||
| ATCC 13032 | Wild type | American Type Culture Collection |
| ΔnanR mutant | WT C. glutamicum with deletion of nanR (cg2936) | This work |
| Plasmids | ||
| pEKEx2 | Expression vector; ptac lacIq Kmr | 91 |
| pEKEx2-nanR | pEKEx2 carrying the nanR (cg2936) gene | This work |
| pEKEx2-siaEFGI | pEKEx2 carrying the genes siaE (cg2937), siaF (cg2938), siaG (cg2939), and siaI (cg2940) | This work |
| pK19mobsacB | Kmr; mobilizable E. coli vector for the construction of insertion and deletion mutations in C. glutamicum (oriV sacB lacZα) | 51 |
| pK19mobsacBΔnanR | Kmr; pK19mobsacB with the deletion construct for the gene nanR (cg2936) | This work |
| pASK_IBA3 | Expression vector, tetA promoter, C-terminal Strep-tag II; Ampr | IBA GmbH |
| pASK_IBA3_nanR | pASK_IBA3 carrying the nanR gene | This work |
| pEPRI | Promoter probe vector carrying the promoterless gfp gene; Kmr | 52 |
| pEPRI_PRnagAB_WT | pEPRI containing the nagA (cg2929) promoter fragment from C. glutamicum ATCC 13032 | 24 |
| pEPRI_PRnagAB_M4 | pEPRI containing the nagA (cg2929) promoter fragment from C. glutamicum M4 | 24 |
| pEPRI-PRnanA | pEPRI containing the nanA (cg2931) promoter fragment | This work |
| pEPRI-PRsiaE | pEPRI containing the siaE (cg2937) promoter fragment | This work |
| pEPRI-PRnanH | pEPRI containing the nanH (cg1756) promoter fragment | This work |
| pEPRI-PRnanR | pEPRI containing the nanR (cg2936) promoter fragment | This work |
Quantification of glucose, fructose, and Neu5Ac.
For quantification of glucose, fructose, and Neu5Ac in the culture broth, aliquots of the culture were withdrawn, cells were removed by centrifugation (5 min at 13,000 × g and 4°C), and the supernatant was used for the determination of sugars with a Hitachi high-performance liquid chromatography (HPLC) system equipped with a refractive index detector (L2490; used for glucose and fructose quantification) and a UV detector (L2400; used for Neu5Ac quantification) and a Nucleo Sugar 810H column (Macherey & Nagel). The mobile phase was 0.01 M H2SO4 with a flow rate of 0.5 ml min−1, the column temperature was 40°C, and fluorescence was recorded at 210 nm.
DNA preparation, manipulation, and transformation.
Standard procedures were employed for chromosomal as well as plasmid DNA isolation and for molecular cloning and transformation of E. coli, as well as for electrophoresis (48). Isolation of plasmids and chromosomal DNA of C. glutamicum was performed as described previously (49). Transformation of C. glutamicum was performed by electroporation as described previously (50). PCR experiments were performed in a Flexcycler (Analytik Jena) with Phusion DNA polymerase (New England BioLabs) with oligonucleotides obtained from Eurofins MWG Operon and listed in Table S1 in the supplemental material. All restriction enzymes, T4 DNA ligase, and shrimp alkaline phosphatase were obtained from New England BioLabs and used according to the manufacturer's instructions.
Construction of plasmids and strains.
Inactivation of the chromosomal nanR gene in C. glutamicum was performed using crossover PCR and the suicide vector pK19mobsacB. Flanking regions of the gene of roughly 560 bp were amplified by using the primer pairs D_nanR_P1 and D_nanR_P2 and D_nanR_P3 and D_nanR_P4. The PCR products were fused via the complementary artificial overhangs provided in the primers and in a PCR using both of the PCR products together with the primers D_nanR_P1 and D_nanR_P4. Using the PCR-generated 5′ XmaI and 3′ PstI restriction sites of the 1,146-bp fusion PCR product, the construct was ligated into pK19mobsacB and transformed into E. coli. The recombinant plasmid pK19mobsacBΔnanR was isolated from E. coli and electroporated into WT C. glutamicum. By application of the method described in reference 51, the complete chromosomal nanR gene was deleted via homologous recombination (double crossover). Deletion of nanR was verified by PCR using the primer check_D_nanR_fwd and check_D_nanR_rev, resulting in an 848-bp fragment for the WT and a 197-bp fragment for the deletion mutant.
For IPTG-inducible overexpression in C. glutamicum strains, vector pEKEx2 was used. For construction of pEKEx2-nanR, the gene was amplified via PCR from genomic DNA of C. glutamicum ATCC 13032 using the oligonucleotide primer OE_nanR_fwd and OE_nanR_rev (see Table S2 in the supplemental material); the resulting 801-bp PCR product was then cut using BamHI and SbfI and cloned into the PstI/BamHI-cut plasmid pEKEx2. For overexpression of siaEFGI, the genes were PCR amplified with the primers siaEFGI_fwd and siaEFGI_rev. The 5,619-bp PCR product was cloned using the PCR-generated 5′ SbfI site and the 3′ blunt end into the PstI- and EcoIRI-cut pEKEx2, leading to pEKEx2-siaEFGI. The recombinant plasmids were isolated from E. coli DH5α, controlled by sequencing (GATC Biotech AG, Constance, Germany), and transformed into C. glutamicum strains.
Construction of gfp reporter gene fusions.
In order to monitor the activity of promoters from C. glutamicum, transcriptional fusions with the promoterless gfp gene were used based on the corynebacterial promoter-probe vector pEPR1 (52). The siaEFGI promoter region (−207 to +45 in relation to the adenosine residue of the siaE ATG start codon) was amplified by PCR from genomic DNA of WT C. glutamicum by using primers PsiaE_for and PsiaE_rev, resulting in a 272-bp PCR product. The nanAKE promoter region promoter region (−179 to +29 in relation to the adenosine residue of the nanA ATG start codon) was amplified by PCR using primers PnanA_for and PnanA_rev, resulting in a 229-bp PCR product. The nanR promoter region (−278 to +59 in relation to the adenosine residue of the nanR ATG start codon) was amplified by PCR using primers PnanR_for and PnanR_rev, resulting in a 359-bp PCR product. The nanH promoter region (−136 to +179 in relation to the adenosine residue of the ATG nanH start codon) was amplified using primers PnanH_fwd and PnanH_rev, resulting in a 336-bp PCR product. The PCR products were digested at primer-generated restriction sites (see Table S2) and ligated into the multiple-cloning site of pEPRI in front of the gfp gene, resulting in the plasmids pEPRI-PsiaE, pEPRI-PnanA, pEPRI-PnanR, and pEPRI-PnanH. Promoter activities of C. glutamicum strains carrying promoter test vectors were measured by determining the gfp fluorescence in relation to the OD600 using a Tecan M200 Infinite plate reader (Tecan, Crailsheim, Germany).
[14C]Neu5Ac and [14C]glucose uptake studies.
C. glutamicum cells were grown to early exponential phase (3 h), harvested by centrifugation, washed twice with ice-cold CGXII medium, suspended to an OD600 of 2 with CGXII medium, and stored on ice until the measurement. Before the transport assay, cells were incubated for 3 min at 30°C; the reaction was started by addition of 1 μM to 500 μM [14C]N-acetylneuraminic acid (specific activity, 55 mCi mmol−1; American Radiolabeled Chemicals). At given time intervals (15, 30, 45, 60, and 90 s), 200-μl samples were filtered through fiberglass filters (type F; Millipore, Eschborn, Germany) and washed twice with 2.5 ml of 100 mM LiCl. The radioactivity of the samples was determined using scintillation fluid (Rotiszinth; Roth, Germany) and a scintillation counter (LS 6500; Beckmann, Krefeld, Germany). Kinetic parameters as well as standard errors were derived from nonlinear regressions according to the Michaelis-Menten equation by using Sigma Plot software. Uptake of [14C]glucose was analyzed at a concentration of 100 μM as described previously (53).
NagA and NagB activity assays.
Exponentially growing cells of C. glutamicum strains were harvested by centrifugation (10 min at 3,200 × g and 4°C) and washed twice with 50 mM Tris-HCl, pH 8.0. Cells were disrupted by ultrasonic treatment (UP 200S; Dr. Hielscher GmbH, Teltow, Germany) with an amplitude of 50% and a duty circle of 0.5 for 7 min. The cell suspension was centrifuged for 1 h at 4°C and 16,000 rpm. Glucosamine 6-phosphate deaminase and N-acetylglucosamine-6P deacetylase activities of the supernatant were determined according to the assays described by Uhde et al. (24) and Matano et al. (10), respectively.
Purification of NanR.
For heterologous expression, the nanR gene was amplified from genomic DNA of C. glutamicum ATCC 13032 by PCR using the primers IBA_nanR_fwd and IBA_nanR_rev, and the resulting 792-bp PCR product was then cloned into the vector pASK-IBA3 (IBA GmbH, Göttingen, Germany) according to the supplier's manual. Expression of the nanR-strep gene from plasmid pASK_IBA3_nanR was induced by the addition of anhydrotetracycline (2 μg ml−1) at an OD600 of 1, and cells were harvested 6 h later by centrifugation at 5,000 × g for 5 min. The cells were washed twice with wash buffer (100 mM Tris-HCl, pH 8.0, and 150 mm NaCl), resuspended in wash buffer, and disintegrated by ultrasonic treatment with a Branson 250 sonifier at an output control of 2.5 and a duty cycle of 25% for 1.5 min. After centrifugation at 15,000 × g for 20 min, the supernatant was subjected to Strep-tag purification (IBA GmbH) according to the manufacturer's protocol. For storage, glycerol was added to the purified NanR protein (final amount 10% [vol/vol]), and aliquots were then stored at −20°C until further use. Protein concentrations were determined using the Roti-Nanoquant kit (Roth) with bovine serum albumin as the standard. SDS-PAGE was performed according to the method of Laemmli (54). Loading buffer (4×) contained 8% (wt/vol) SDS, 20% (vol/vol) glycerol, 10 mM EDTA, 100 mM Tris-HCl (pH 6.8), 2% (vol/vol) β-mercaptoethanol, and 1 mg/ml of bromphenol blue. Western blot experiments for detection of the streptavidin-tagged NanR protein by using antibodies raised against Strep-tag II (IBA GmbH) were performed as described for the uptake carrier BetP (55).
EMSA.
Electophoretic mobility shift assays (EMSAs) with NanR were performed as described previously (56). Briefly, various concentrations (0 to 0.3 μg) of purified NanR were mixed with 15 ng of DNA probes generated by PCR in DNA binding buffer (40 mM Tris-HCl, 10% [vol/vol] glycerol, 0.2 M KCl, 4 mM dithiothreitol [DTT] [pH 7.5]) in a total volume of 10 μl. The DNA fragments were obtained by PCR with the primers listed in Table S2 in the supplemental material using genomic DNA from C. glutamicum ATCC 13032 or C. glutamicum M4 as the template and primer combinations mentioned in the Results section and purified by gel extraction using the NucleoSpin PCR cleanup and gel extraction kit (Macherey & Nagel) according to the manufacturer′s instructions. After incubation for 20 min at 30°C, the samples were separated on 9% native polyacrylamide gels at a constant electric current of 30 mA at 4°C, stained, and photographed as described previously (56). To test for possible effectors, the protein was incubated with glucosamine-6-phosphate (50 μg), glucosamine (50 μg), Neu5Ac (up to 100 μg), GlcNAc (50 μg), GlcNAc-6P (up to 50 μg), glucose (50 μg), glucose-6-phosphate (50 μg), and ManNAc-6P (up to 50 μg) in the binding buffer for 5 min before addition of DNA fragment PnagA and incubation for another 30 min. With the exception of ManNAc-6P and GlcNAc-6P (from Carbosynth Limited), the sugars and sugar phosphates tested as effector molecules in EMSAs were purchased from Sigma-Aldrich.
Gene expression analysis.
For the comparison of transcriptomes of C. glutamicum strains, cells growing exponentially in LB medium were harvested at an OD600 of 3.5. RNA purification, transcription to cDNA, fluorescent labeling, hybridization, and data analysis were performed as described previously (57–59). Only hybridization signals exceeding background noise by at least a factor of 3 were considered (GENEPIX 3.0). Normalized ratios of medians were taken to reflect relative mRNA levels. Slot blot experiments were performed as described previously (60). For hybridization, digoxigenin (DIG)-11-dUTP-labeled gene-specific antisense RNA probes against ptsG mRNA and the 16S rRNA were prepared from PCR products (generated with oligonucleotides listed in Table S2) carrying the T7 promoter by in vitro transcription (1 h at 37°C) using T7 RNA polymerase (MBI Fermentas) as described previously (61).
Computational analysis.
Databank searches were carried out by using BLAST (62), protein sequences were analyzed using CLUSTAL W (63), and protein domain assignments were performed using SUPERFAMILY (64). The following NCBI-GI accession numbers for protein sequences were retrieved from the KEGG database (65): 476417323 for B. breve NanR, 49176329 for E. coli NanR, 62391489 for C. glutamicum NanR, 375292402 for C. diphtheriae NanR, 300859481 for C. pseudotuberculosis NanR, and 384516670 for C. ulcerans NanR. Discovery of motives in sets of sequences was performed using GLAM2 (66), and FIMO (67) was used for motif scanning in the C. glutamicum ATCC 13032 (GenBank accession number NC_006958), C. diphtheriae INCA402 (NC_016783), C. ulcerans 809 (NC_017317), and C. pseudotuberculosis FRC41 (NC_014329) genome sequences.
RESULTS
Utilization and uptake of the Neu5Ac are inhibited in glucose-cultivated as well as fructose-cultivated C. glutamicum cells.
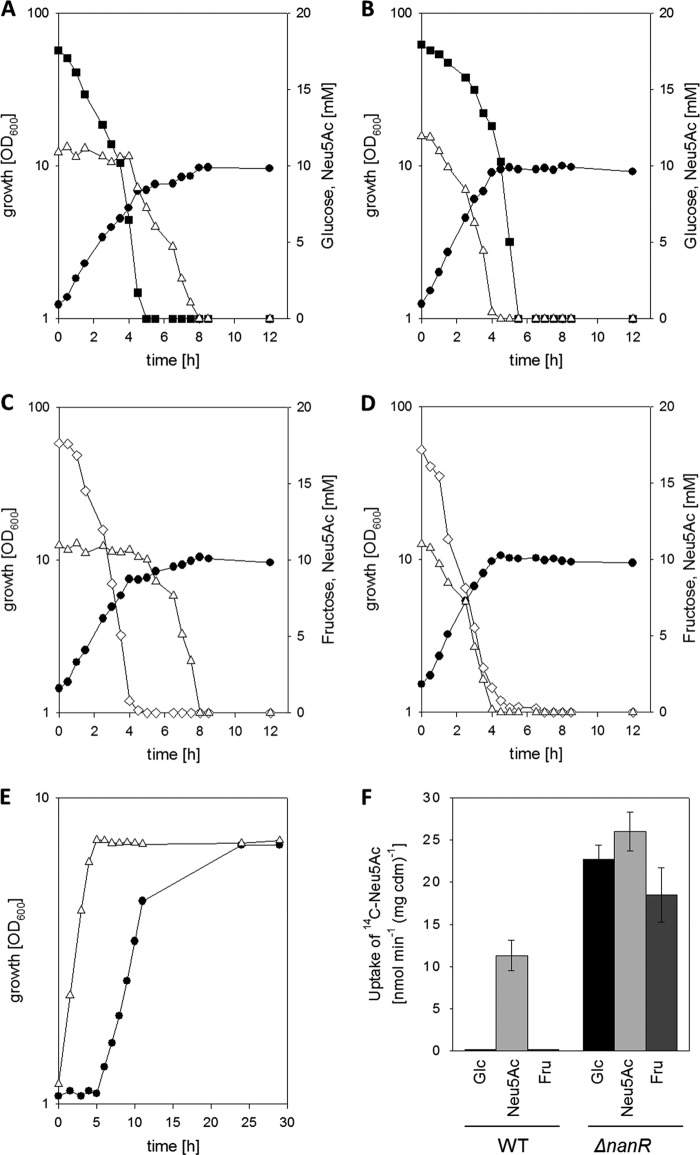
Uptake and utilization of Neu5Ac were recently reported to be inhibited in glucose-cultivated C. glutamicum cells (22); however, utilization of both carbon sources at the same time was hitherto not analyzed. In growth experiments in minimal medium with a mixture of 0.3% (wt/vol) glucose and 0.2% (wt/vol) Neu5Ac, consecutive utilization of the two substrates was observed for WT C. glutamicum. As depicted in Fig. 2A, utilization of Neu5Ac only started after 5 h of cultivation, after the initially provided glucose was completely consumed. The growth rate decreased from initially 0.35 ± 0.03 h−1 in the course of glucose utilization to 0.09 ± 0.02 h−1 when Neu5Ac was utilized. In addition, coutilization of Neu5Ac and fructose was tested. As shown in Fig. 2C, when 0.3% fructose and 0.2% Neu5Ac were used as substrates, WT C. glutamicum also showed consecutive utilization of these two sugars and utilization of Neu5Ac only began after 4 h of cultivation, after fructose was completely consumed. The growth rate decreased from initially 0.43 ± 0.04 h−1 in the course of fructose utilization to 0.09 ± 0.02 h−1 during Neu5Ac utilization. To analyze Neu5Ac uptake in C. glutamicum cells from different cultivations, transport assays with 14C-labeled Neu5Ac were established. Neu5Ac uptake in Neu5Ac-cultivated cells showed simple saturation kinetics with a Km of 10 ± 2 μM and a maximum uptake rate (Vmax) of 9 ± 1 nmol min−1 mg of cells (dry mass)−1 (see Fig. S1 in the supplemental material). To analyze the influence of precultivation on different carbon sources, transport measurements were performed at a Neu5Ac concentration of 100 μM. No uptake of 14C-labeled Neu5Ac was detected in cells of WT C. glutamicum cultivated on glucose and fructose, whereas uptake of 14C-labeled Neu5Ac proceeded at a rate of 11 ± 2 nmol min−1 mg of cells (dry mass)−1 in WT C. glutamicum cells cultivated on Neu5Ac (Fig. 2F). To test for carbon source-dependent transcriptional regulation of the siaEFGI operon for the Neu5Ac transporter, a transcriptional fusion between the siaE promoter region and the promoterless gfp gene was constructed in the promoter probe vector pEPPRI and the resulting plasmid, pEPRI-PRsiaE, was transformed into WT C. glutamicum. Whereas high green fluorescent protein (GFP) fluorescence was observed for WT C. glutamicum(pEPRI-PRsiaE) cultivated on Neu5Ac, only residual GFP fluorescence was detected when this strain was cultivated on glucose or fructose (Fig. 3A). Taken together, these results indicate the presence of a CCR mechanism in C. glutamicum for the inhibition of Neu5Ac utilization in the presence of glucose and fructose.
FIG 2.
(A to D) Growth and substrate consumption in minimal medium with 0.3% (wt/vol) glucose plus 0.2% (wt/vol) Neu5Ac (A and B) or with 0.3% (wt/vol) fructose plus 0.2% (wt/vol) Neu5Ac (C and D) of WT C. glutamicum (A and C) and C. glutamicum ΔnanR (B and D). Solid circles, growth; open triangles, Neu5Ac; solid squares, glucose; open diamonds, fructose. (E) Growth of WT C. glutamicum (solid circles) and C. glutamicum ΔnanR (open triangles) in minimal medium with 0.2% (wt/vol) Neu5Ac. Three independent cultivations were performed; data from one representative experiment are shown. Results of all of the cultivations were comparable. (F) Rates of [14C]Neu5Ac uptake of WT and ΔnanR C. glutamicum cells cultivated in minimal medium with different substrates. Data represent mean values and standard deviations of three independent measurements from two independent cultivations.
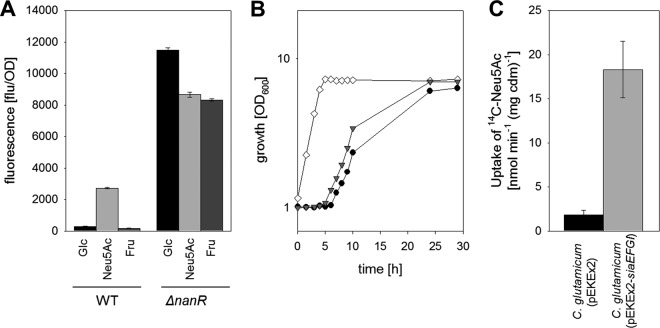
FIG 3.
(A) Analyses of siaE promoter activity, measured as relative fluorescence of the GFP reporter upon expression of the gfp gene under the control of the siaE promoter in C. glutamicum(pEPRI-PRsiaE) and C. glutamicum ΔnanR(pEPRI-PRsiaE) cells cultivated for 8 h in minimal medium plus glucose, Neu5Ac, or fructose. Data represent mean values and standard deviations of two independent measurements each from three independent cultivations. (B) Growth of C. glutamicum ΔnanR (open diamonds), C. glutamicum(pEKEx2) (solid circles), and C. glutamicum (pEKEx2-siaEFGI) (gray triangles) in minimal medium with 0.2% (wt/vol) Neu5Ac. (C) Rates of [14C]Neu5Ac uptake of C. glutamicum(pEKEx2) and C. glutamicum(pEKEx2-siaEFGI) cells cultivated in minimal medium with 0.3% (wt/vol) fructose. Data represent mean values and standard deviations of three independent measurements from two independent cultivations.
Deletion of cg2936 enables Neu5Ac and glucose as well as Neu5Ac and fructose coutilization in C. glutamicum.
The genes for Neu5Ac utilization in C. glutamicum comprise four clustered operons, namely, nagAB, nanAKE, and siaEFGI (22, 23) and a fourth operon with cg2936 encoding a putative GntR-type transcriptional regulator (Fig. 1B). Upon deletion of cg2936, the resulting mutant strain was able to grow without any lag phase in minimal medium with Neu5Ac as the sole source of carbon and energy (Fig. 2E). As expected (22), a lag phase of about 5 to 6 h was observed for the parental strain (Fig. 2E). Furthermore, transport assays with 14C-labeled Neu5Ac showed high transport activities, 23 ± 2 nmol min−1 mg of cells (dry mass)−1, 19 ± 3 nmol min−1 mg of cells (dry mass)−1, and 26 ± 2 nmol min−1 mg of cells (dry mass)−1, for cells of the cg2936-deficient strain cultivated on glucose, fructose, and Neu5Ac, respectively (Fig. 2F). Subsequently, the siaE promoter activity was assayed in WT C. glutamicum and in the cg2936 deletion mutant using the promoter test plasmid pEPRI-PRsiaE. Independent of the carbon source used for cultivation, higher GFP fluorescence was observed for the cg2936 deletion mutant than for WT C. glutamicum(pEPRI-PRsiaE) (Fig. 3A). Taken together, these data indicate that the cg2936 gene encodes a transcriptional regulator for the genes for Neu5Ac transport, and we therefore refer to it as nanR. Accordingly, the cg2936 mutant was designated C. glutamicum ΔnanR.
When C. glutamicum ΔnanR was cultivated in minimal medium with glucose plus Neu5Ac as sources of carbon and energy, growth proceeded at a rate of 0.45 ± 0.02 h−1 and the two substrates were consumed in parallel (Fig. 2B). However, in these cultivations the glucose consumption rate of the ΔnanR mutant was lowered to 3.6 ± 0.2 mmol of C g of cells (dry mass)−1 · h−1. For the parental strain, WT C. glutamicum, a glucose consumption rate of 10.3 ± 0.6 mmol of C g of cells (dry mass)−1 · h−1 had been determined when the strain was cultivated on glucose as the sole substrate (15) as well as on glucose plus Neu5Ac (Fig. 2A). In addition, growth of the ΔnanR mutant was also slowed down in cultivations with glucose as the sole carbon source compared to that of WT C. glutamicum (growth rates of 0.11 ± 0.02 h−1 and 0.35 ± 0.03 h−1 were determined, respectively [see Fig. S2A in the supplemental material]). These results indicate the presence of a limitation in glucose catabolism in C. glutamicum ΔnanR. Upon plasmid-encoded expression of nanR in C. glutamicum ΔnanR(pEKEX2-nanR), growth with glucose as the sole substrate was restored to a rate of 0.27 ± 0.03 h−1 (see Fig. S2A). Growth of the empty-vector control strain C. glutamicum ΔnanR(pEKEX2) on glucose remained slow. Deletion of nanR did not negatively affect growth on the glycolytic substrates maltose, fructose, and sucrose (data not shown). The metabolic pathways for glucose, maltose, and fructose in C. glutamicum differ only in their initial steps until glycolytic intermediates are formed (7). For glucose utilization, just the first two steps, namely, uptake and phosphorylation to glucose-6-phosphate, are specific, which both are brought about by the ptsG-encoded EIIGlc of the PTS (68). However, Northern blot analyses and uptake experiments with 14C-labeled glucose showed that neither ptsG transcript levels (see Fig. S2C) nor activity of the glucose uptake system (see Fig. S2B) is reduced in C. glutamicum ΔnanR compared to that in WT C. glutamicum. Thus, the reason for the poor growth of C. glutamicum ΔnanR in minimal medium with glucose and the decreased glucose consumption rate in cultivations of this strain with glucose plus Neu5Ac remains elusive. Although glucose utilization was slightly slower in C. glutamicum ΔnanR, we conclude that deletion of nanR enables coutilization of glucose and Neu5Ac, and NanR probably acts as a transcriptional repressor of the siaEFGI operon coding for the Neu5Ac transporter.
When the NanR-deficient strain C. glutamicum ΔnanR was cultivated in minimal medium with fructose plus Neu5Ac, growth proceeded at a rate of 0.50 ± 0.04 h−1 and the two carbon sources were coutilized. A fructose consumption rate of 10 ± 1 mmol of C g of cells (dry mass)−1 · h−1 was determined for C. glutamicum ΔnanR in cultivations with fructose plus Neu5Ac. Nearly identical fructose consumption rates of 11 ± 1 mmol of C g of cells (dry mass)−1 · h−1 and 11 ± 1 mmol of C g of cells (dry mass)−1 · h−1 were determined for the C. glutamicum ΔnanR mutant and WT in cultivations on fructose, respectively. Thus, it can be concluded that deletion of nanR enables the efficient coutilization of fructose and Neu5Ac in C. glutamicum and does not, in contrast to the situation with glucose, negatively affect fructose metabolism.
Identification of the C. glutamicum NanR regulon.
Transcription of the siaEFGI genes for Neu5Ac uptake is repressed by NanR in the presence of both glucose and fructose. Plasmid-encoded overexpression of siaEFGI indeed caused significantly increased Neu5Ac transport in fructose-cultivated cells of the strain C. glutamicum(pEKEx2-siaEFGI), whereas only residual Neu5Ac transport activity was measured for the empty-vector control strain C. glutamicum(pEKEx2) when cultivated on fructose (Fig. 3C). Despite the presence of high Neu5Ac transport activity, growth of C. glutamicum(pEKEx2-siaEFGI) on Neu5Ac after precultivation on fructose started only after a prolonged lag phase, as was also observed for C. glutamicum(pEKEx2) (Fig. 3B). In contrast, no lag phase was observed for the NanR-deficient mutant strain C. glutamicum ΔnanR under these conditions (Fig. 3B). This observation suggested that regulation by NanR might pertain to further genes for Neu5Ac utilization besides the Neu5Ac transporter siaEFGI operon. To identify the complete NanR regulon, the transcriptome of the mutant C. glutamicum ΔnanR was compared to that of the C. glutamicum WT by DNA microarray analyses during exponential growth in LB medium. Table S3 in the supplemental material lists genes that showed statistically significant (P < 0.05) expression changes by at least a factor of 4. It is noteworthy that deletion of nanR substantially increased expression of a small group of genes localized in proximity to nanR itself, i.e., 10-fold to several hundredfold. These genes, nagA, nagB, nanA, nanK, nanE, and, as expected, siaE, siaF, siaG, and siaI, are involved in Neu5Ac utilization and are derepressed in the absence of NanR. Moreover, expression of the propionate utilization operons prpDBC and prpD2B2C2, located elsewhere on the chromosome, increased up to 14-fold.
To further analyze transcriptional control of the nanAKE and nagAB operons as well as nanR itself, reporter plasmids which carry transcriptional fusions between the promoter regions and the promoterless gfp gene in the test vector pEPRI were constructed and transformed into WT and ΔnanR C. glutamicum. Analyses of the GFP fluorescence in samples of the resulting strains from cultivations on fructose showed a higher activity of all tested promoters in the NanR-deficient strain than in the WT (Fig. 4). These data confirm that NanR acts as a transcriptional repressor of the siaEFGI, nanAKE, and nagAB operons and additionally of its own gene, nanR.
FIG 4.
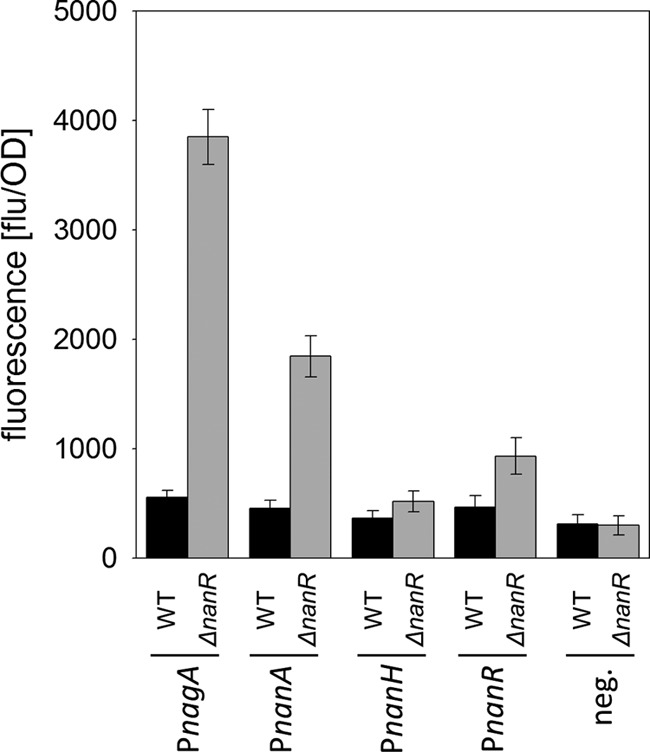
Analyses of nanA, nagA, nanH, and nanR promoter activities in WT and ΔnanR C. glutamicum strains cultivated for 8 h in minimal medium with 0.3% (wt/vol) fructose, expressed as relative fluorescence of the GFP reporter upon expression of the gfp gene under the control of the nanA, nagA, nanH, or nanR promoter present in the plasmid pEPRI-PRnanA, pEPRI-PRnagAB, pEPRI-PRnanH, or pEPRI-PRnanH, respectively. As a control, relative fluorescence was measured for cells of C. glutamicum(pEPRI) and C. glutamicum ΔnanR(pEPRI) carrying the empty vector pEPRI. Data represent mean values and standard deviations of two independent measurements each from three independent cultivations.
Deletion of nanR enables efficient utilization of glucosamine.
The increased expression levels of nagA and nagB in the absence of NanR are reflected by the specific activities of the encoded enzymes (see Table S4 in the supplemental material). The specific activities of NagA and NagB were about 20-fold and 40-fold higher, respectively, in C. glutamicum ΔnanR than in WT C. glutamicum. As increased expression of nagB was shown to be required for efficient utilization of glucosamine as the substrate (24), growth of C. glutamicum ΔnanR in minimal medium with glucosamine as the sole carbon source was tested. As depicted in Fig. S4A in the supplemental material, indeed good growth on glucosamine was observed for C. glutamicum ΔnanR (growth rate of 0.17 ± 0.03 h−1), whereas, as expected, only slow growth, with a rate of 0.08 ± 0.02 h−1, was observed for WT C. glutamicum under the same conditions. Ectopic expression of nanR in C. glutamicum ΔnanR using the plasmid pEKEx2-nanR resulted in slow growth of C. glutamicum ΔnanR(pEKEX2-nanR) on glucosamine, whereas the empty-vector control strain C. glutamicum ΔnanR(pEKEX2) grew well in the same medium (see Fig. S4B). Taken together, these results confirm that NanR also represses transcription of the nagAB operon and thus of glucosamine utilization. Moreover, the good growth of C. glutamicum ΔnanR on glucosamine also shows that the ptsG-encoded EIIGlc is present and active in NanR-deficient C. glutamicum strains, as uptake and phosphorylation of glucosamine are exclusively brought about by EIIGlc (24).
Binding of purified Strep-tagged NanR protein to promoter/operator regions and identification of NanR-binding sites.
To test for the binding of NanR to the siaEFGI, nagAB, and nanAKE promoter regions, we assayed the binding of purified NanR to promoter regions in vitro. For this purpose, NanR was synthesized as a Strep-tagged fusion protein in E. coli BL21(DE3) and purified to apparent homogeneity by affinity chromatography as described in Materials and Methods. For EMSAs, different amounts of purified NanR protein were incubated with DNA fragments and separated on 10% native polyacrylamide gels.
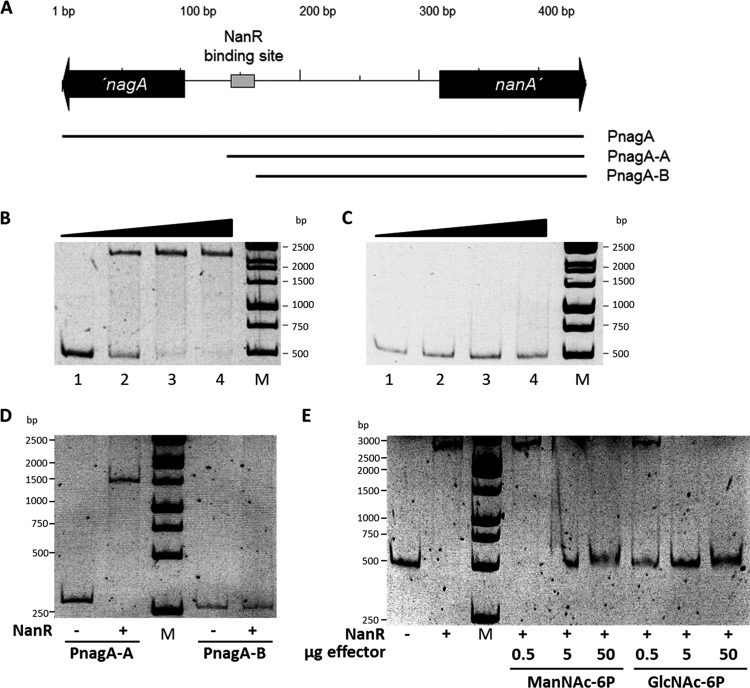
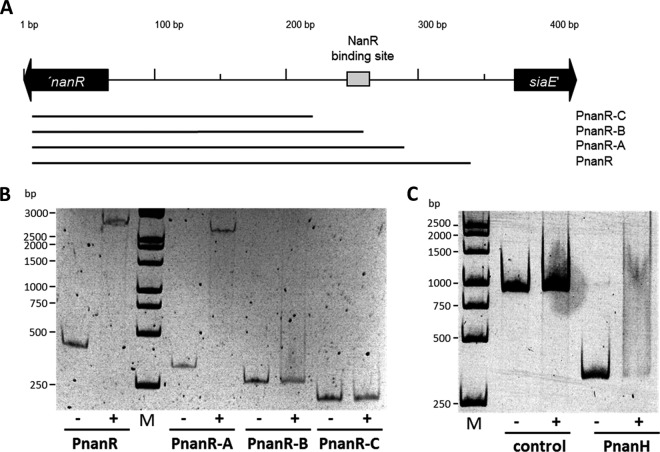
First, binding of NanR to the nagA and nanA intergenic region was tested using the 433-bp probe PnagA covering the region 121 bp downstream of the nanA ATG start codon and 98 bp downstream of the nagA ATG start codon, which was generated by PCR using the primers nagA-for and nagA-rev and WT C. glutamicum DNA as the template (Fig. 5A; see also Fig. S5 in the supplemental material). As shown in Fig. 5B, NanR indeed bound to the probe PnagA, which covers the nagA and nanA intergenic region. A complete mobility shift was observed when 0.2 μg of purified NanR protein was added to the probe (Fig. 5B, lane 3), which corresponds to a 120-fold molar excess. In C. glutamicum M4, a T-to-C point mutation within the nagA promoter region leads to high, constitutive expression of the nagAB operon (24). The probe PnagA-M4 was generated using the primers nagA-for and nagA-rev and DNA isolated from C. glutamicum M4 as the template and, thus, carries the T-to-C point mutation. No shift of the probe PnagA-M4 was observed in EMSAs using 0.1 to 0.3 μg of purified NanR (Fig. 5C). Also, when concentrations up to 2 μg of purified NanR were used (corresponding to a 1,200-fold molar excess), no shift upon adding the probe PnagA-M4 was observed. These data show that the exchanged nucleotide residue within the nanA-nagR intergenic region is required for binding of the repressor NanR. In addition, binding of purified NanR to the probes PnagA-A and Pnag-B, which carry truncated versions of the nagA-nanA intergenic region, was tested (Fig. 5A). In EMSAs, binding of NanR to the probe PnagA-A was observed, whereas no binding of NanR to the probe PnagA-B lacking the region 58 bp upstream of the nagA start codon was detected (Fig. 5D).
FIG 5.
Electrophoretic mobility shift assays with NanR and probes covering the nanA-nagA intergenic region. DNA probes containing the nagA-nanA intergenic region were incubated with various concentrations of NanR. (A) Schematic illustration of the intergenic region of the C. glutamicum nagA and nanA genes, probes used for EMSAs, and localization of NanR-binding site. (B and C) Representative EMSAs using NanR (0, 0.1, 0.2, and 0.3 μg) with 10 ng of the probes PnagA (B) and PnagA (C). (D) EMSAs using 0.3 μg of NanR (indicated by plus signs) with 10 ng of the probes PnagA-A or PnagA-B. (E) Identification of GlcNAc-6P and ManNAc-6P as NanR effector molecules. Shown are results of EMSAs using NanR (0.2 μg; 0.7 μM) with the Probe PnagA (10 ng; 3.7 nM) and 0.5, 5, or 50 μg of GlcNAc-6P (0.17 mM, 1.66 mM, and 16.6 mM) or 0.5, 5, or 50 μg of ManNAc-6P (0.14 mM, 1.44 mM, and 14.49 mM). Lanes M, molecular size markers.
Since NanR also represses transcription of siaEFGI and nanR in addition to nagAB and nanAKE, binding of purified NanR to the siaE-nanR intergenic region was analyzed in EMSAs using the 338-bp probe PnanR, which covers the region 30 bp upstream of the siaE ATG start codon and 56 bp downstream of the nanR ATG start codon (Fig. 6A; see also Fig. S6 in the supplemental material). A complete shift of the probe PnanR was observed when 0.2 μg of purified NanR was added (Fig. 6B), which corresponds to a 100-fold molar excess. To identify the NanR-binding site, the nucleotide sequences of the intergenic regions of nagA-nanA and siaE-nanR were compared using the program GLAM2. By this means the nucleotide sequences ACGTATGATGTCTTATGTCTACGGA within the nagA-nanA intergenic region and ACGTCTGATGTCTGATGTATATTGA within the siaE-nanR were found, which are identical in 20 of 25 positions. Actually, the T-to-C mutation in the nagA promoter of C. glutamicum M4 is located within this 25-bp nucleotide sequence, and moreover, the nucleotide sequence is not present in the truncated probe PnagA-B, which was not bound by NanR. When binding of NanR to truncated versions of the siaE-nanR intergenic regions was tested, no shifts were observed for the probes PnanR-B and PnanR-C (Fig. 6B), which lack the ACGTCTGATGTCTGATGTATATTGA nucleotide sequence.
FIG 6.
Electrophoretic mobility shift assays with NanR and probes covering the nanR-siaE intergenic region. DNA probes containing the nanR-siaE intergenic region were incubated with various concentrations of NanR. (A) Schematic illustration of the intergenic region of the C. glutamicum nanA and siaE genes, probes used for EMSAs, and localization of the NanR-binding site. (B) Representative EMSAs using 0.2 μg of NanR with 10 ng of each of the probes PnanR, PnanR-A, PnanR-B, and PnanR-C. (C) Representative EMSA using 0.2 μg NanR with 10 ng of the probe PnanH, which covers the nanH promoter. As a negative control, 10 ng of a PCR product of the nanR gene generated with the primers OE_nanR_fwd and OE_nanR_rev was used.
Based on the results from the EMSAs, the preliminary consensus motif ACGT[AC]TGATGTCT[TG]ATGT[AC]TA[CT][TG]GA for binding of NanR to the promoters of siaE, nanR, nagA, and nanA was determined. This consensus motif was then used to search within the C. glutamicum genome for further putative NanR-binding sites using the software FIMO. By this means, a further nucleotide sequence with a P value below 1 × 10−6 was identified upstream of cg1756, which is annotated as nanH and putatively encodes a secreted sialidase (23). The identified nucleotide sequence AAGCATGATGTCAGATGTCTAATTG overlaps the ATG start codon of cg1756 (indicated in bold). In EMSAs with purified NanR, a shift of the probe PnanH covering the promoter region of nanH was observed, whereas no shift of the control probe Pcont was detected at identical NanR concentrations (Fig. 6C).
To test for NanR-dependent transcriptional control of nanH, the promoter probe plasmid pEPRI-PnanH, which carries a transcriptional fusion between the nanH promoter region and the promoterless gfp gene, was constructed. Only residual GFP fluorescence was detected for WT C. glutamicum(pEPRI-PnanH) cultivated on fructose. GFP fluorescence was only slightly higher, not significantly increased, in the corresponding NanR-deficient strain C. glutamicum ΔnanR(pEPRI-PnanH) (Fig. 4). These results of the reporter gene experiments show that nanH possesses a weak promoter whose activity is only slightly affected by NanR. The low transcription might explain why no change of nanH transcript amounts was detected in microarray analyses of differential transcription in WT and ΔnanR C. glutamicum. These results demonstrate that NanR controls transcription of siaEFGI, nagAB, nanAKE, and nanR by binding within the promoter regions and at least binds also to the nanH promoter. Analyses of the promoter regions of aforementioned operons using GLAM2 led to the determination of the 21-bp consensus motif A[AC]G[CT][AC]TGATGTC[AT][TG]ATGT[AC]TA for NanR binding to its target promoters.
GlcNAc-6P and ManNac-6P inhibit binding of NanR to its cognate promoter regions.
Analyses of the NanR amino acid sequence using the program SUPERFAMILY suggested that this protein belongs to the GntR family of DNA binding proteins and possesses an N-terminal winged-helix DNA binding domain and a C-terminal GntR ligand-binding domain. Based on this C-terminal domain, NanR can be placed within the FadR branch of the large GntR family of transcriptional regulators (69). Moreover, NanR from C. glutamicum is highly similar to the well-characterized GntR-type (FadR subfamily) transcriptional regulators of sialic acid metabolism from B. breve (33% identity; E value, 5e−33) and E. coli (25% identity; E value, 5e−9), which both depend on Neu5Ac as the inducer (45, 46). Considering the observed induction of the genes for Neu5Ac utilization upon cultivation with Neu5Ac, the similar metabolic pathways present in C. glutamicum, B. breve, and E. coli, and the alignment of amino acid sequences of the NanR proteins from these three species, which demonstrates well-conserved C termini (see Fig. S7 in the supplemental material), we hypothesized that also C. glutamicum NanR repression might be relieved by binding of Neu5Ac. To test this hypothesis, in EMSAs with 0.3 μg of NanR, which ensures a complete shift of the probe PnagA, increasing concentrations of Neu5Ac (up to 30 mM) were added to the reaction mixture. Analysis of the gels demonstrated that NanR binding to the probe PnagA was not altered in the presence Neu5Ac (see Fig. S8C). Besides Neu5Ac, effects on NanR binding to PnagA of the metabolites GlcNAc, glucosamine, glucose-6-phosphate, glucose, GlcNAc-6P, glucosamine-6-phosphate, and ManNAc-6P were also tested in EMSAs. Only the presence of GlcNAc-6P and ManNAc-6P inhibited binding of NanR to the probe (Fig. 5E). Specifically, addition of 0.6 mM GlcNAc-6P or 2.1 mM ManNAc-6P to the reaction mixture inhibited the shift of the probe PnagA by NanR in EMSAs (see Fig. S8). These results suggest that conditions leading to high concentrations of either one of the two intermediates of Neu5Ac degradation, GlcNAc-6P and ManNAc-6P, in C. glutamicum will induce sialic acid catabolism by inhibiting binding of the GntR-type transcriptional regulator NanR to promoters of its target genes.
Transcription of siaEFGI, nagAB, nanAKE, and nanR is subject to catabolite repression.
In addition to the consecutive utilization of two carbon sources, a further characteristic of CCR is the repressed transcription of genes for the utilization of the nonpreferred substrate when both substrates are present in the culture broth. Analyses of kinetics of siaE promoter activities in WT C. glutamicum(pEPRI-PRsiaE) showed a delayed increase of GFP fluorescence in cultivations with Neu5Ac plus fructose compared to cultivations with Neu5Ac as the sole substrate (Fig. 7A and B). In cultivations of WT C. glutamicum(pEPRI-PRsiaE) with 0.2% (wt/vol) Neu5Ac plus 0.25% (wt/vol) fructose, GFP fluorescence started to increase after 6 h of cultivation, when the initially provided fructose was completely consumed (Fig. 7B). Analyses of siaE, nagA, nanA, and nanR promoter activities after 5 h of cultivation in minimal medium with 0.2% (wt/vol) Neu5Ac plus 0.25% (wt/vol) fructose or in minimal medium with 0.2% (wt/vol) Neu5Ac showed that transcription of all tested promoters was low when fructose was present in the medium (Fig. 7C). Furthermore, when glucose was added to cells of WT C. glutamicum(pEPRI-PRnagAB_WT) precultivated with Neu5Ac as the sole substrate, GFP fluorescence transiently deceased (see Fig. S9 in the supplemental material). This finding indicates that transcription from the nagA promoter stopped after glucose addition, which fits within the concept of CCR by presence of fructose or glucose. To analyze the role of NanR for CCR of Neu5Ac catabolism genes by fructose, siaE, nagA, nanA, and nanR promoter activities were analyzed in the NanR-deficient strain C. glutamicum ΔnanR after 5 h of cultivation in minimal medium with 0.2% (wt/vol) Neu5Ac plus 0.4% (wt/vol) fructose or in minimal medium with 0.2% (wt/vol) Neu5Ac. Whereas no significant reduction of siaE promoter activity by the additional presence of fructose in the culture broth was observed, nagA, nanA, and nanR promoter activities were significantly lower in cultivations of C. glutamicum ΔnanR in medium with Neu5Ac plus fructose than in cultivations with Neu5Ac as the sole substrate (Fig. 7C). These results show that despite the lack of NanR, the presence of fructose still leads to a partial repression of the transcription of the operons nagAB, nanAKE, and nanR.
FIG 7.
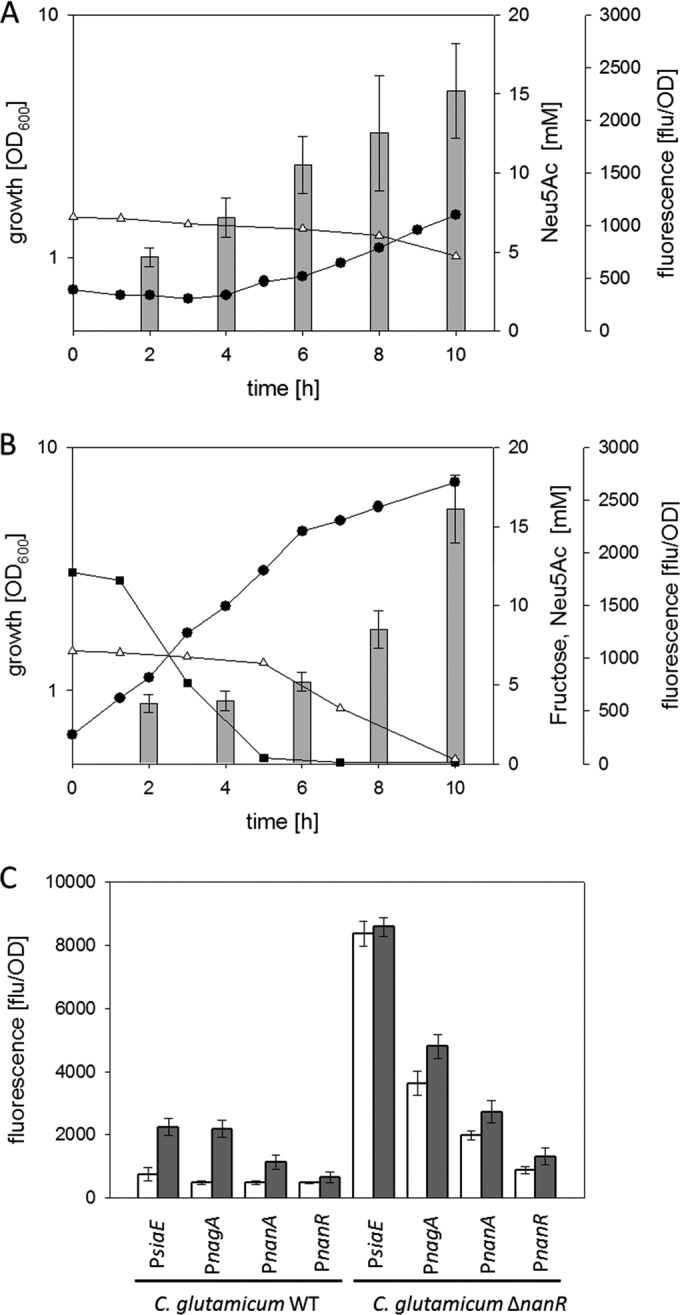
(A and B) Growth (black circles), substrate concentrations (white triangles, Neu5Ac; black squares, fructose), and relative fluorescence of the GFP reporter (gray bars) in cultivations of C. glutamicum WT (pEPRI-PRsiaE) on minimal medium with 0.2% (wt/vol) Neu5Ac (A) or 0.2% (wt/vol) Neu5Ac plus 0.25% (wt/vol) fructose (B) as substrates. (C) Analyses of siaE, nanA, nagA, and nanR promoter activities in WT C. glutamicum and C. glutamicum ΔnanR strains after 5 h of cultivation in minimal medium with 0.2% (wt/vol) Neu5Ac plus 0.4% (wt/vol) fructose (white boxes) or in minimal medium with 0.2% (wt/vol) Neu5Ac (gray boxes). Three independent cultivations were performed. Growth data from one representative experiment are shown; results of all of the cultivations were comparable. GFP fluorescence data represent mean values and standard deviations of two independent measurements each from three independent cultivations.
The presence of fructose and glucose, two sugars taken up via the PTS in C. glutamicum (7), prohibited induction of the genes for Neu5Ac utilization. In E. coli, activity of the PTS inhibits activity of several other transporters for sugar uptake, e.g., the lactose permease LacY and the ABC transporter for maltose uptake MalEFGK2, which leads to inducer exclusion (70, 71). To analyze the effects of the presence of glucose and fructose on Neu5Ac transport by SiaEFGI, uptake of 14C-labeled Neu5Ac was analyzed in cells of C. glutamicum(pEKEx2-siaEFGI) incubated for 3 min at 30°C with 500 μM either glucose or fructose before addition of the 14C-labeled Neu5Ac (cells were cultivated in LB medium). Surprisingly, uptake of 14C-labeled Neu5Ac was even faster in the presence of glucose (27.9 ± 3.4 nmol min−1 mg of cells [dry mass]−1) or fructose (26.4 ± 2.8 nmol min−1 mg of cells [dry mass]−1) than 14C-labeled Neu5Ac uptake in cells preincubated for 3 min in the absence of additional sugars (18.2 ± 3.2 nmol min−1 mg of cells [dry mass]−1). These results clearly show that inducer exclusion by glucose and fructose is not involved in the CCR of Neu5Ac catabolism genes in C. glutamicum.
Taken together, these findings indicate that besides NanR a second, hitherto-unidentified, NanR-independent regulatory mechanism is involved in the observed CCR of the genes for Neu5Ac catabolism in the presence of glucose and fructose.
DISCUSSION
In this study, we have shown that in C. glutamicum, the cg2939-encoded regulator protein NanR controls as a repressor transcription of the operons siaEFGI, nagAB, and nanAKE required for uptake and metabolization of Neu5Ac. In contrast to the GntR-type regulators of genes for Neu5Ac utilization from E. coli and B. breve, which both depend on the presence of Neu5Ac (45, 46) as an effector, binding of C. glutamicum NanR to its target promoters was relieved in the presence of 0.6 mM GlcNAc-6P and at slightly higher concentrations of ManNAc-6P. Control by intermediates of Neu5Ac degradation has also been reported for the RipR-type regulators of Neu5Ac metabolism from V. vulnificus, S. pneumoniae, S. aureus, and H. influenzae, which are activated in the presence of ManNAc-6P, ManNAc, and GlcN-6P, respectively (39, 42, 72, 73) (see Table S1 in the supplemental material). Thus, NanR of C. glutamicum is the first transcriptional regulator of sialic acid metabolism depending on the inducer GlcNAc-6P and the first of the GntR-type regulators independent of Neu5Ac (see Table S1). The use of GlcNAc-6P as a signal molecule for the control of Neu5Ac degradation in C. glutamicum is plausible, as this organism cannot utilize GlcNAc as the substrate for growth (10). In E. coli, for which the pathway for sialic acid utilization is regarded as an addition to the pathway for the utilization of the amino sugar GlcNAc (74), distinct regulatory circuits exist for differential control of the genes for Neu5Ac utilization and genes for GlcNAc utilization. Transcription of the genes nagE, nagA, and nagB for GlcNAc utilization is controlled in E. coli by the ROK-type transcriptional regulator NagC, which is displaced from its DNA targets by interacting with GlcNAc-6P (75). When Neu5Ac is metabolized in E. coli, GlcNAc-6P formed by the consecutive action of NanA, NanK, and NanE induces nagAB transcription via NagC and, thus, synthesis of the enzymes NagA and NagB, required for the last two steps of Neu5Ac degradation (74, 76). Similarly, GlcNAc-6P derived from Neu5Ac induces the NanR regulon in C. glutamicum. In the related C. glycinophilum, GlcNAc uptake and phosphorylation are accomplished by the nagE-encoded EII permease of the PTS (10). Alongside heterologous expression of nagE from C. glycinophilum, plasmid-encoded overexpression of endogenous nagA and nagB was shown to be necessary for efficient GlcNAc utilization in C. glutamicum (10). Moreover, plasmid-encoded overexpression of nagE from C. glycinophilum in C. glutamicum M4 did not lead to efficient growth on GlcNAc (10), despite the fact that this strain possesses high levels of NagA and NagB due to a point mutation within the promoter of nagAB and is thus able to efficiently utilize GlcN (24). This result indicates that besides the GlcNAc-6P-dependent transcriptional regulation of the operons for Neu5Ac metabolism mediated by NanR, further, hitherto-unidentified regulatory mechanisms for the control of amino sugar metabolism in C. glutamicum have to exist.
To accomplish the observed sequential utilization of glucose or fructose as preferred substrates together with Neu5Ac as the less-favorable substrate, a regulatory mechanism for CCR is required to inhibit induction of the Neu5Ac utilization genes. As transcription of Neu5Ac genes was still slightly repressed by the presence of fructose and glucose in C. glutamicum ΔnanR compared to cultivations with Neu5Ac as the sole substrate, CCR is probably controlled independently of NanR. In several bacteria, complex regulatory networks for the control of consecutive utilization of carbon sources have been described, and often the PTS is involved in the underlying signal transduction pathways (70, 71). Mechanisms underlying CCR by PTS substrates often include inducer exclusion (70, 71). When Neu5Ac uptake by the ABC transporter SiaEFGI was analyzed in C. glutamicum(pEKEx2-siaEFGI), preincubation with a 50 μM concentration of either of the preferred substrates glucose and fructose did not lead to a diminished rate for Neu5Ac uptake, as would have been expected if inducer exclusion occurs. Apart from inducer exclusion, also several other regulatory mechanisms for CCR are also controlled by PTS activity. In the Gram-positive S. pneumoniae, transcription of nanA is repressed by CcpA (34), the global regulator responsible for CCR in this organism (77). The presence of preferred substrates results in HPrK-catalyzed phosphorylation of the PTS component HPr at Ser-46, which, in turn, binds to and activates CcpA, and the resulting CcpA and P-Ser46-HPr complex then binds to DNA, which leads to repression of target promoters (71, 78). Although a serine residue is indeed present at position 46 in C. glutamicum HPr, it is unlikely that a CcpA-like mechanism underlies the observed repression of Neu5Ac catabolism in C. glutamicum in the presence of glucose or fructose, as neither a gene for nor activity of HPrK was detected in C. glutamicum (79, 80). In the Gram-negative bacteria V. vulnificus and H. influenzae, CCR is brought about by the cyclic AMP (cAMP) receptor protein (CRP) (37, 39, 43). The CRP orthologue GlxR has been identified in C. glutamicum as one of the central regulators of its metabolism, which controls more than 200 genes (81–83). However, no GlxR-binding sites have been identified within the nagA-nanA and nanR-siaE intergenic regions, nor has binding of GlxR to DNA fragments of this region been observed (81, 82, 84). Thus, it is unlikely that CCR of Neu5Ac metabolism in C. glutamicum is mediated by GlxR. Involvement of the PTS in the control of Neu5Ac metabolism remains to be investigated; however, the growth defect of the NanR-deficient strain on the PTS substrate glucose and the positive effects on Neu5Ac uptake indicate the presence of possible regulatory interactions.
Utilization of Neu5Ac as a substrate for growth is a common trait of a large variety of microorganisms inhabiting various niches within eukaryotic hosts and has therefore been associated with pathogenicity or at least commensal lifestyles (85). Therefore, the presence of a functional pathway for Neu5Ac utilization and distinct mechanisms for its control seem rather odd in a nonpathogenic soil bacterium such as C. glutamicum. Consequently, this trait of C. glutamicum was regarded as a remnant, possibly inherited from related pathogenic species such as Corynebacterium diphtheriae, Corynebacterium ulcerans, and Corynebacterium pseudotuberculosis (22). These three species indeed harbor gene clusters for Neu5Ac transport and utilization highly similar to the cluster described for C. glutamicum (22). Moreover, genes for putative GntR-type regulators are present within these clusters, and the gene products in fact are highly similar to NanR from C. glutamicum (see Fig. S10 in the supplemental material). Similar to the case with C. glutamicum, the genes encoding the putative Neu5Ac uptake systems are located in the genomes of C. diphtheriae (strain INCA 402), C. ulcerans (strain 809), and C. pseudotuberculosis (strain FRC41) adjacent to the gene for the corresponding transcriptional regulator, separated by a short intergenic region (86–88). In analyses of the genome sequences using FIMO and the consensus motif for C. glutamicum NanR-binding sites, we were able to identify highly conserved 21-bp motifs within the intergenic regions of these related species (see Fig. S11). The conserved genomic organization and the conserved regulator binding motifs indicate that the control of Neu5Ac metabolism by NanR in C. glutamicum may be rather similar to that of the related pathogenic species C. diphtheriae, C. ulcerans, and C. pseudotuberculosis and might thus be regarded as a model for the control of Neu5Ac utilization within the genus Corynebacterium.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ute Meyer and Eva Glees (Institute of Biochemistry, University of Cologne, Germany) for excellent technical assistance.
Funding Statement
This work, including the efforts of Volker F. Wendisch, Reinhard Krämer, and Gerd M. Seibold, was funded by Bundesministerium für Bildung und Forschung (BMBF) (0315589F, 0315589G, 03160717C, and 031A302D).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00820-15.
REFERENCES
- 1.Wendisch VF. 2007. Amino acid biosynthesis-pathways, regulation and metabolic engineering. Springer Verlag, Berlin, Germany. [Google Scholar]
- 2.Becker J, Wittmann C. 2012. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Heider SA, Wendisch VF. 2015. Engineering microbial cell factories: metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products. Biotechnol J 10:1170–1184. doi: 10.1002/biot.201400590. [DOI] [PubMed] [Google Scholar]
- 4.Wendisch VF. 2014. Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol 30C:51–58. [DOI] [PubMed] [Google Scholar]
- 5.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. 2013. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol 6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arndt A, Eikmanns BJ. 2008. Regulation of carbon metabolism in Corynebacterium glutamicum, p 155–182. In Burkovski A. (ed), Corynebacteria: genomics and molecuar biology. Caister Acadeic Press, Norfolk, United Kingdom. [Google Scholar]
- 7.Blombach B, Seibold GM. 2010. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of L-lysine production strains. Appl Microbiol Biotechnol 86:1313–1322. doi: 10.1007/s00253-010-2537-z. [DOI] [PubMed] [Google Scholar]
- 8.Hadiati A, Krahn I, Lindner SN, Wendisch VF. 2015. Engineering of Corynebacterium glutamicum for growth and production of L-ornithine, L-lysine, and lycopene from hexuronic acids. Bioresources Bioprocessing 1:25. [Google Scholar]
- 9.Kawaguchi H, Vertes AA, Okino S, Inui M, Yukawa H. 2006. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72:3418–3428. doi: 10.1128/AEM.72.5.3418-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matano C, Uhde A, Youn JW, Maeda T, Clermont L, Marin K, Krämer R, Wendisch VF, Seibold GM. 2014. Engineering of Corynebacterium glutamicum for growth and L-lysine and lycopene production from N-acetyl-glucosamine. Appl Microbiol Biotechnol 98:5633–5643. doi: 10.1007/s00253-014-5676-9. [DOI] [PubMed] [Google Scholar]
- 11.Rittmann D, Lindner SN, Wendisch VF. 2008. Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol 74:6216–6222. doi: 10.1128/AEM.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ. 2006. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol 124:381–391. doi: 10.1016/j.jbiotec.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 13.Cocaign M, Monnet C, Lindley ND. 1993. Batch kinetics of Corynebacterium glutamicum during growth on various carbon substrates—use of substrate mixtures to localize metabolic bottlenecks. Appl Microbiol Biotechnol 40:526–530. [Google Scholar]
- 14.Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J Bacteriol 182:3088–3096. doi: 10.1128/JB.182.11.3088-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause FS, Henrich A, Blombach B, Krämer R, Eikmanns BJ, Seibold GM. 2010. Increased glucose utilization in Corynebacterium glutamicum by use of maltose, and its application for the improvement of l-valine productivity. Appl Environ Microbiol 76:370–374. doi: 10.1128/AEM.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frunzke J, Engels V, Hasenbein S, Gatgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol 67:305–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nentwich SS, Brinkrolf K, Gaigalat L, Huser AT, Rey DA, Mohrbach T, Marin K, Pühler A, Tauch A, Kalinowski J. 2009. Characterization of the LacI-type transcriptional repressor RbsR controlling ribose transport in Corynebacterium glutamicum ATCC 13032. Microbiology 155:150–164. doi: 10.1099/mic.0.020388-0. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez H, CocaignBousquet M, Lindley ND. 1997. Simultaneous consumption of glucose and fructose from sugar mixtures during botch growth of Corynebacterium glutamicum. Appl Microbiol Biotechnol 47:600–603. doi: 10.1007/s002530050980. [DOI] [Google Scholar]
- 19.Arndt A, Eikmanns BJ. 2007. The alcohol dehydrogenase gene adhA in Corynebacterium glutamicum is subject to carbon catabolite repression. J Bacteriol 189:7408–7416. doi: 10.1128/JB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krämer R, Lambert C, Hoischen C, Ebbighausen H. 1990. Uptake of glutamate in Corynebacterium glutamicum. 1. Kinetic properties and regulation by internal pH and potassium. Eur J Biochem 194:929–935. [DOI] [PubMed] [Google Scholar]
- 21.Kronemeyer W, Peekhaus N, Krämer R, Sahm H, Eggeling L. 1995. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J Bacteriol 177:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruteser N, Marin K, Krämer R, Thomas GH. 2012. Sialic acid utilization by the soil bacterium Corynebacterium glutamicum. FEMS Microbiol Lett 336:131–138. doi: 10.1111/j.1574-6968.2012.02663.x. [DOI] [PubMed] [Google Scholar]
- 23.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Kramer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Puhler A, Rey DA, Ruckert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 24.Uhde A, Youn JW, Maeda T, Clermont L, Matano C, Krämer R, Wendisch VF, Seibold GM, Marin K. 2013. Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl Microbiol Biotechnol 97:1679–1687. doi: 10.1007/s00253-012-4313-8. [DOI] [PubMed] [Google Scholar]
- 25.Varki A. 2007. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 26.Varki A. 2008. Sialic acids in human health and disease. Trends Mol Med 14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varki A, Schauer R. 2009. Chapter 14. Sialic acids. In Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi C, Hart G, Etzler M (ed), Essentials of glycobiology, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 28.Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R. 2002. Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A 99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haines-Menges BL, Whitaker WB, Lubin JB, Boyd EF. 2015. Host sialic acids: a delicacy for the pathogen with discerning taste. Microbiol Spectrum 3(4):MBP-0005-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YL, Chassard C, Hausmann M, von Itzstein M, Hennet T. 2015. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun 6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vimr ER. 2013. Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol 2013:816713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Therit B, Cheung JK, Rood JI, Melville SB. 2015. NanR, a transcriptional regulator that binds to the promoters of genes involved in sialic acid metabolism in the anaerobic pathogen Clostridium perfringens. PLoS One 10:e0133217. doi: 10.1371/journal.pone.0133217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afzal M, Shafeeq S, Ahmed H, Kuipers OP. 2015. Sialic acid-mediated gene expression in Streptococcus pneumoniae and the role of NanR as a transcriptional activator of the nan gene cluster. Appl Environ Microbiol 81:3121–3131. doi: 10.1128/AEM.00499-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas GH, Boyd EF. 2011. On sialic acid transport and utilization by Vibrio cholerae. Microbiology 157:3253–3254; discussion, 3254–3255. doi: 10.1099/mic.0.054692-0. [DOI] [PubMed] [Google Scholar]
- 36.Mulligan C, Leech AP, Kelly DJ, Thomas GH. 2012. The membrane proteins SiaQ and SiaM form an essential stoichiometric complex in the sialic acid tripartite ATP-independent periplasmic (TRAP) transporter SiaPQM (VC1777-1779) from Vibrio cholerae. J Biol Chem 287:3598–3608. doi: 10.1074/jbc.M111.281030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, Gibson BW, Apicella MA, Munson RS Jr. 2007. Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol Microbiol 66:26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- 38.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston JW, Shamsulddin H, Miller AF, Apicella MA. 2010. Sialic acid transport and catabolism are cooperatively regulated by SiaR and CRP in nontypeable Haemophilus influenzae. BMC Microbiol 10:240. doi: 10.1186/1471-2180-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston JW, Apicella MA. 2008. Sialic acid metabolism and regulation by Haemophilus influenzae: potential novel antimicrobial therapies. Curr Infect Dis Rep 10:83–84. doi: 10.1007/s11908-008-0014-y. [DOI] [PubMed] [Google Scholar]
- 41.Jenkins GA, Figueira M, Kumar GA, Sweetman WA, Makepeace K, Pelton SI, Moxon R, Hood DW. 2010. Sialic acid mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence. BMC Microbiol 10:48. doi: 10.1186/1471-2180-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang J, Kim BS, Jang SY, Lim JG, You DJ, Jung HS, Oh TK, Lee JO, Choi SH, Kim MH. 2013. Structural insights into the regulation of sialic acid catabolism by the Vibrio vulnificus transcriptional repressor NanR. Proc Natl Acad Sci U S A 110:E2829–E2837. doi: 10.1073/pnas.1302859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim BS, Hwang J, Kim MH, Choi SH. 2011. Cooperative regulation of the Vibrio vulnificus nan gene cluster by NanR protein, cAMP receptor protein, and N-acetylmannosamine 6-phosphate. J Biol Chem 286:40889–40899. doi: 10.1074/jbc.M111.300988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalivoda KA, Steenbergen SM, Vimr ER, Plumbridge J. 2003. Regulation of sialic acid catabolism by the DNA binding protein NanR in Escherichia coli. J Bacteriol 185:4806–4815. doi: 10.1128/JB.185.16.4806-4815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalivoda KA, Steenbergen SM, Vimr ER. 2013. Control of the Escherichia coli sialoregulon by transcriptional repressor NanR. J Bacteriol 195:4689–4701. doi: 10.1128/JB.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egan M, O'Connell Motherway M, van Sinderen D. 2015. A GntR-type transcriptional repressor controls sialic acid utilization in Bifidobacterium breve UCC2003. FEMS Microbiol Lett 362:1–9. doi: 10.1093/femsle/fnu056. [DOI] [PubMed] [Google Scholar]
- 47.Eggeling L, Reyes O. 2005. Experiments, p 3535–3566. In Eggeling L, Bott M (ed), Handbook of Corynebacterium glutamicum. CRC, Boca Raton, FL. [Google Scholar]
- 48.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Eikmanns BJ, Thum-Schmitz N, Eggeling L, Ludtke KU, Sahm H. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140(Part 8):1817–1828. [DOI] [PubMed] [Google Scholar]
- 50.Tauch A, Kirchner O, Loffler B, Gotker S, Pühler A, Kalinowski J. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol 45:362–367. doi: 10.1007/s00284-002-3728-3. [DOI] [PubMed] [Google Scholar]
- 51.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 52.Knoppová M, Phensaijai M, Vesely M, Zemanova M, Nesvera J, Patek M. 2007. Plasmid vectors for testing in vivo promoter activities in Corynebacterium glutamicum and Rhodococcus erythropolis. Curr Microbiol 55:234–239. doi: 10.1007/s00284-007-0106-1. [DOI] [PubMed] [Google Scholar]
- 53.Lindner SN, Petrov DP, Hagmann CT, Henrich A, Krämer R, Eikmanns BJ, Wendisch VF, Seibold GM. 2013. Phosphotransferase system-mediated glucose uptake is repressed in phosphoglucoisomerase-deficient Corynebacterium glutamicum strains. Appl Environ Microbiol 79:2588–2595. doi: 10.1128/AEM.03231-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 55.Rübenhagen R, Rönsch H, Jung H, Krämer R, Morbach S. 2000. Osmosensor and osmoregulator properties of the betaine carrier BetP from Corynebacterium glutamicum in proteoliposomes. J Biol Chem 275:735–741. doi: 10.1074/jbc.275.2.735. [DOI] [PubMed] [Google Scholar]
- 56.Wennerhold J, Krug A, Bott M. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem 280:40500–40508. doi: 10.1074/jbc.M508693200. [DOI] [PubMed] [Google Scholar]
- 57.Polen T, Schluesener D, Poetsch A, Bott M, Wendisch VF. 2007. Characterization of citrate utilization in Corynebacterium glutamicum by transcriptome and proteome analysis. FEMS Microbiol Lett 273:109–119. doi: 10.1111/j.1574-6968.2007.00793.x. [DOI] [PubMed] [Google Scholar]
- 58.Wendisch VF. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J Biotechnol 104:273–285. doi: 10.1016/S0168-1656(03)00147-0. [DOI] [PubMed] [Google Scholar]
- 59.Hüser AT, Becker A, Brune I, Dondrup M, Kalinowski J, Plassmeier J, Pühler A, Wiegrabe I, Tauch A. 2003. Development of a Corynebacterium glutamicum DNA microarray and validation by genome-wide expression profiling during growth with propionate as carbon source. J Biotechnol 106:269–286. doi: 10.1016/j.jbiotec.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Möker N, Brocker M, Schaffer S, Krämer R, Morbach S, Bott M. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol Microbiol 54:420–438. doi: 10.1111/j.1365-2958.2004.04249.x. [DOI] [PubMed] [Google Scholar]
- 61.Henrich A, Kuhlmann N, Eck AW, Krämer R, Seibold GM. 2013. Maltose uptake by the novel ABC transport system MusEFGK2I causes increased expression of ptsG in Corynebacterium glutamicum. J Bacteriol 195:2573–2584. doi: 10.1128/JB.01629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 63.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. 2009. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res 37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frith MC, Saunders NF, Kobe B, Bailey TL. 2008. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol 4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon MW, Kim HJ, Oh TK, Shin CS, Lee JS, Kim SJ, Lee JK. 2005. Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol Lett 244:259–266. doi: 10.1016/j.femsle.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 69.Hoskisson PA, Rigali S. 2009. Chapter 1: Variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv Appl Microbiol 69:1–22. doi: 10.1016/S0065-2164(09)69001-8. [DOI] [PubMed] [Google Scholar]
- 70.Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol 11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 71.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 72.Olson ME, King JM, Yahr TL, Horswill AR. 2013. Sialic acid catabolism in Staphylococcus aureus. J Bacteriol 195:1779–1788. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gualdi L, Hayre JK, Gerlini A, Bidossi A, Colomba L, Trappetti C, Pozzi G, Docquier JD, Andrew P, Ricci S, Oggioni MR. 2012. Regulation of neuraminidase expression in Streptococcus pneumoniae. BMC Microbiol 12:200. doi: 10.1186/1471-2180-12-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plumbridge J, Vimr E. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol 181:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pennetier C, Dominguez-Ramirez L, Plumbridge J. 2008. Different regions of Mlc and NagC, homologous transcriptional repressors controlling expression of the glucose and N-acetylglucosamine phosphotransferase systems in Escherichia coli, are required for inducer signal recognition. Mol Microbiol 67:364–377. [DOI] [PubMed] [Google Scholar]
- 76.Chu D, Roobol J, Blomfield IC. 2008. A theoretical interpretation of the transient sialic acid toxicity of a nanR mutant of Escherichia coli. J Mol Biol 375:875–889. doi: 10.1016/j.jmb.2007.10.073. [DOI] [PubMed] [Google Scholar]
- 77.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. 2011. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One 6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fleming E, Lazinski DW, Camilli A. 2015. Carbon catabolite repression by seryl phosphorylated HPr is essential to Streptococcus pneumoniae in carbohydrate-rich environments. Mol Microbiol 97:360–380. doi: 10.1111/mmi.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parche S, Burkovski A, Sprenger GA, Weil B, Kramer R, Titgemeyer F. 2001. Corynebacterium glutamicum: a dissection of the PTS. J Mol Microbiol Biotechnol 3:423–428. [PubMed] [Google Scholar]
- 80.Kuhlmann N, Petrov DP, Henrich AW, Lindner SN, Wendisch VF, Seibold GM. 2015. Transcription of malP is subject to PTS-dependent regulation in Corynebacterium glutamicum. Microbiology 161:1830–1843. doi: 10.1099/mic.0.000134. [DOI] [PubMed] [Google Scholar]
- 81.Toyoda K, Teramoto H, Inui M, Yukawa H. 2011. Genome-wide identification of in vivo binding sites of GlxR, a cyclic AMP receptor protein-type regulator in Corynebacterium glutamicum. J Bacteriol 193:4123–4133. doi: 10.1128/JB.00384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kohl TA, Baumbach J, Jungwirth B, Pühler A, Tauch A. 2008. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J Biotechnol 135:340–350. doi: 10.1016/j.jbiotec.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 83.Kim HJ, Kim TH, Kim Y, Lee HS. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J Bacteriol 186:3453–3460. doi: 10.1128/JB.186.11.3453-3460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jungwirth B, Sala C, Kohl TA, Uplekar S, Baumbach J, Cole ST, Puhler A, Tauch A. 2013. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR in Corynebacterium glutamicum. Microbiology 159:12–22. doi: 10.1099/mic.0.062059-0. [DOI] [PubMed] [Google Scholar]
- 85.Almagro-Moreno S, Boyd EF. 2009. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol 9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trost E, Ott L, Schneider J, Schroder J, Jaenicke S, Goesmann A, Husemann P, Stoye J, Dorella FA, Rocha FS, Soares Sde C, D'Afonseca V, Miyoshi A, Ruiz J, Silva A, Azevedo V, Burkovski A, Guiso N, Join-Lambert OF, Kayal S, Tauch A. 2010. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics 11:728. doi: 10.1186/1471-2164-11-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trost E, Al-Dilaimi A, Papavasiliou P, Schneider J, Viehoever P, Burkovski A, Soares SC, Almeida SS, Dorella FA, Miyoshi A, Azevedo V, Schneider MP, Silva A, Santos CS, Santos LS, Sabbadini P, Dias AA, Hirata R Jr, Mattos-Guaraldi AL, Tauch A. 2011. Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genomics 12:383. doi: 10.1186/1471-2164-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trost E, Blom J, Soares Sde C, Huang IH, Al-Dilaimi A, Schroder J, Jaenicke S, Dorella FA, Rocha FS, Miyoshi A, Azevedo V, Schneider MP, Silva A, Camello TC, Sabbadini PS, Santos CS, Santos LS, Hirata R Jr, Mattos-Guaraldi AL, Efstratiou A, Schmitt MP, Ton-That H, Tauch A. 2012. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J Bacteriol 194:3199–3215. doi: 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 90.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 91.Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93–98. doi: 10.1016/0378-1119(91)90545-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.