STRUCTURED ABSTRACT
Purpose of review
The purpose of this review is to discuss recent advances and unsolved questions in our understanding of actin filament organization and dynamics in the red blood cell (RBC) membrane skeleton, a two-dimensional quasi-hexagonal network consisting of (α1β1)2-spectrin tetramers interconnecting short actin filament-based junctional complexes.
Recent findings
In contrast to the long-held view that RBC actin filaments are static structures that do not exchange subunits with the cytosol, RBC actin filaments are dynamic structures that undergo subunit exchange and turnover, as evidenced by monomer incorporation experiments with rhodamine-actin and filament disruption experiments with actin-targeting drugs. The malaria-causing parasite, Plasmodium falciparum, co-opts RBC actin dynamics to construct aberrantly branched actin filament networks. Even though RBC actin filaments are dynamic, RBC actin filament lengths are highly uniform (~37 nm). RBC actin filament lengths are thought to be stabilized by the capping proteins, tropomodulin-1 and αβ-adducin, as well as the side-binding protein tropomyosin, present in an equimolar combination of two isoforms, TM5b (Tpm1.9) and TM5NM1 (Tpm3.1).
Summary
New evidence indicates that RBC actin filaments are not simply passive cytolinkers, but rather dynamic structures whose assembly and disassembly play important roles in RBC membrane function.
Keywords: erythrocyte, actin polymerization, spectrin, tropomodulin-1, tropomyosin
INTRODUCTION
The mammalian red blood cell (RBC) is highly specialized, exhibiting characteristic biconcave shape and containing ~360 mg/ml hemoglobin for O2 delivery and CO2 clearance [1]. RBCs are highly deformable yet mechanically stable, withstanding large shear stresses in central arteries and traversing capillaries smaller than their diameter in peripheral tissues. RBCs lack intracellular organelles and a transcellular cytoskeleton; instead, the robust mechanical properties of RBCs are imparted by the spectrin-actin membrane skeleton, a quasi-hexagonal cytoskeletal network of long and flexible (α1β1)2-spectrin tetramers interconnecting short actin filament (F-actin) junctional complexes, or “nodes,” forming a two-dimensional lattice beneath the bilayer [2,3]. Recent data have challenged the long-standing view of RBC actin filaments as static “spot-welds” passively crosslinking adjacent (α1β1)2-spectrin tetramers. Here, we explore how the RBC membrane skeleton has served as a model system to understand spectrin-actin network organization and actin dynamics in diverse cell types. We then discuss recent advances in understanding RBC actin dynamics and length regulation in the membrane skeleton.
THE RBC MEMBRANE SKELETON: KEY CONCEPTS
Beneath the RBC plasma membrane, a quasi-hexagonal network of (α1β1)2-spectrin tetramers interconnects short F-actin nodes, establishing a two-dimensional lattice of “horizontal interactions” [2]. This network is tethered to the bilayer via “vertical interactions” mediated by ankyrin-B, an adaptor protein linking (α1β1)2-spectrin to Band3 [3], an abundant integral transmembrane protein that forms heteromultimeric complexes with other transmembrane glycoproteins (e.g, glycophorin-C, Rh, Duffy, Kell, XK, Glut1) [4,5]. Although purified RBC (α1β1)2-spectrin tetramers can extend into linear structures ~190 nm in length in vitro [6], the observed end-to-end distance of an (α1β1)2-spectrin tetramer in situ is considerably lower (~35–100 nm) [7–10], forming a folded configuration amenable to extension during RBC deformation (Fig. 1A). End-to-end lengths of (α1β1)2-spectrin tetramers vary widely due to conformational variability in the α1- and β1-spectrin polypeptides [6,11,12], but the F-actin nodes are relatively rigid, with highly uniform lengths of ~37 nm [2], orders of magnitude less than F-actin’s persistence length (~17–18 µm) [13,14]. Uniformity of RBC actin filament length enables ~6 (α1β1)2-spectrin attachments/filament, essential for quasi-hexagonal symmetry [2] (Fig. 1B), but five or seven attachments/filament are occasionally observed in negatively-stained spread membrane skeletons [15,16], suggesting (α1β1)2-spectrin attachment/detachment events [17]. Unexpectedly, branched (α1β1)2-spectrin strands, indicative of higher-order oligomers (i.e., hexamers and octamers), associated with bent actin filaments, have also been observed [10], further contributing to the heterogeneity and complexity of the membrane skeleton. Clearly, numerous questions regarding (α1β1)2-spectrin structure and dynamics in the RBC membrane skeleton still remain [18], but here we focus on the RBC actin filament nodes.
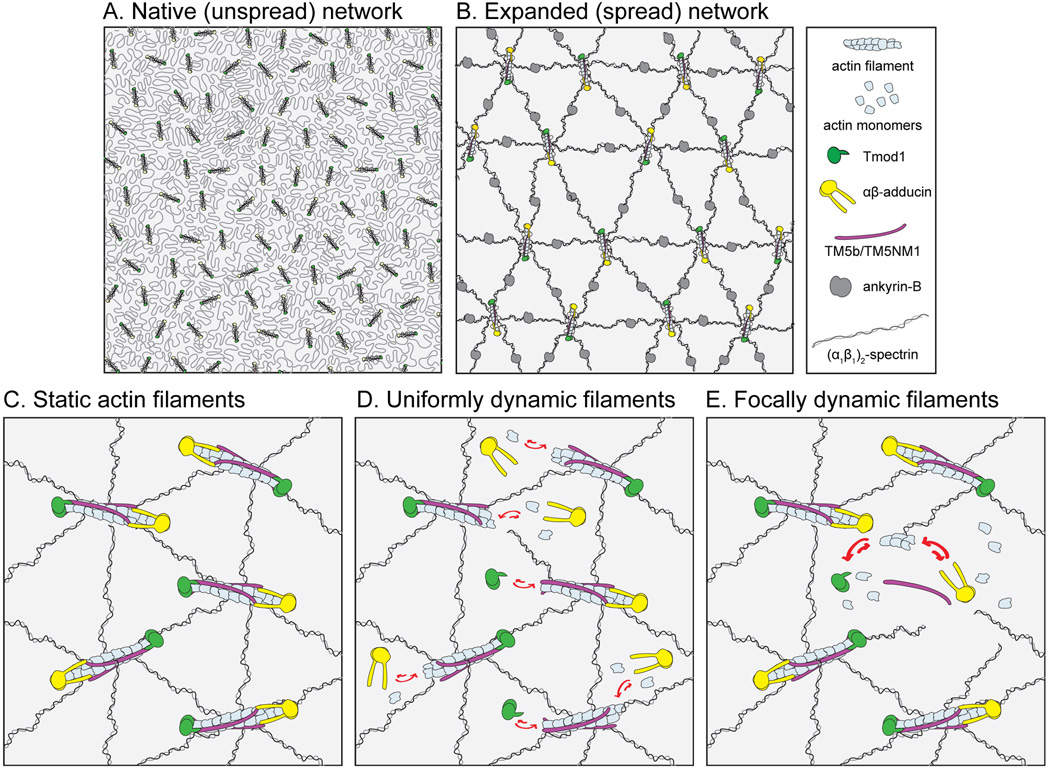
Figure 1. Membrane skeleton organization and models of actin dynamics in RBCs.
(A) In the native (unspread) RBC membrane skeleton, folded (α1β1)2-spectrin tetramers form a two-dimensional network interconnecting short actin filament nodes, which are capped by Tmod1 and αβ-adducin and stabilized along their sides by TM5b and TM5NM1. (B) When the (α1β1)2-spectrin network is expanded (spread), its quasi-hexagonal symmetry is revealed. Ankyrin-B molecules tether the (α1β1)2-spectrin network to the bilayer between actin filament nodes. (C) The now-obsolete “static network” model presumes that the actin subunits comprising the actin filament nodes do not exchange with cytosolic G-actin. (D) The “uniformly dynamic network” model presumes that most or all of the filaments can partially assemble and disassemble actin subunits and exchange them with cytosolic G-actin. (E) The “focally dynamic network” model presumes that a subset of filaments can completely assemble and disassemble, while the others remain static. (Panels A and B are adapted from reference [2].)
Uniformity of RBC actin filament lengths arises from regulation of actin polymerization and depolymerization by actin-binding proteins. In addition to binding (α1β1)2-spectrin, RBC actin filaments are stabilized along their lengths by two tropomyosin isoforms, TM5b (Tpm1.9) and TM5NM1 (Tpm3.1), and capped at their pointed and barbed ends by two tropomodulin-1 (Tmod1) molecules and an αβ-adducin heterodimer, respectively [2]. Additional RBC actin-binding proteins include protein 4.1R, which enhances β1-spectrin-F-actin binding, and dematin (protein 4.9), which bundles F-actin and/or enhances (α1β1)2-spectrin-F-actin binding [2]. Notably, RBC actin filaments provide additional sites of spectrin-actin network tethering to the bilayer [2,5]. For example, in addition to capping barbed ends, αβ-adducin binds Band3 [19], while protein 4.1R binds glycophorin-C [20,21] and Band3 [22]. Thus, RBC actin filaments can simultaneously form “vertical interactions” with the membrane via αβ-adducin-Band3, 4.1R-Band3, and 4.1R-glycophorin-C binding in conjunction with “horizontal interactions” via (α1β1)2-spectrin-F-actin binding. Additional tethering interactions have also been proposed [5].
The architecture of the spectrin-actin membrane skeleton was deciphered in RBCs, but spectrin-actin networks are actually versatile “building blocks” controlling membrane curvature, mechanics, and microdomain formation in diverse cells. For example, Tmod3- and αβ-adducin-capped actin filaments are essential for normal membrane skeleton organization and morphology of polarized epithelial cells [23,24]. A recent study indicates that dynamic rearrangements enable the membrane skeleton to actively “patrol” lateral epithelial membrane domains, inhibiting endocytosis and preventing membrane internalization and loss [25]. F-actin stability in the membrane skeleton is also essential for membrane morphology and physiological function in the hexagonally packed fiber cells of the ocular lens [26], sarcoplasmic reticulum of skeletal muscle [27,28], and demarcation membrane system of megakaryocytes [29,30]. Of note, recent advances in super-resolution fluorescence imaging have enabled identification of an unusual spectrin-actin membrane skeleton in axons of hippocampal neurons [31,32]. In this structure, adducin-capped actin filaments are organized into periodic rings encircling the axonal circumference, with (α2β2)2-spectrin tetramers connecting successive F-actin rings [31]. Axonal F-actin rings exhibit periodicity of ~180–190 nm [31], almost identical to the length of a fully extended spectrin tetramer [6], suggesting that spectrin tetramers may serve as molecular rulers governing spacing of successive rings. Even with this unconventional layout, the axonal membrane skeleton requires normal F-actin stability to establish its architecture [32], as in RBCs and other aforementioned cell types [23,24,26,27,30,33].
DYNAMIC CHARACTERISTICS OF RBC ACTIN FILAMENTS
A long-standing assumption is that RBC actin filament nodes are static structures that do not undergo subunit exchange with free monomers (Fig. 1C). However, recent data have suggested that this is not the case, i.e., that dynamic actin subunit exchange occurs between the cytosol and actin filaments in the RBC membrane skeleton (Fig. 1D,E). Indeed, actin has been visualized in the RBC cytosol via immunogold labeling and electron microscopy of ultra-thin cryosections of intact RBCs [34]. Early estimates of the cytosolic actin concentration in RBCs yielded ~0.24 µM, based on the ability of actin monomers (G-actin) to inhibit DNAse-I [35]. A more recent estimate of the cytosolic actin concentration in human RBCs yielded ~0.36 µM—50% higher than the previous estimate—based on western blotting of actin in Triton-X-100-extracted membrane skeletons vs. cytosolic fractions [36]. Even with this seemingly high cytosolic actin concentration, the overwhelming majority of human RBC actin (~96.3%) is F-actin in RBC membrane skeleton, while only a small minority (~3.7%) of RBC actin is cytosolic G-actin [36].
The presence of G-actin in the RBC cytosol is necessary, but insufficient, for dynamic actin subunit exchange. The first direct evidence for polymerization and depolymerization of RBC actin filaments came from human RBCs infected with the malaria-causing parasite, Plasmodium falciparum, wherein RBC actin filaments are completely disassembled and reassembled into an aberrant dendritic cytosolic network to facilitate export of virulence factors [34,37]. It is unknown whether P. falciparum depolymerizes F-actin throughout the membrane skeleton (Fig. 1D) by exporting parasite-derived F-actin-disassembly factors (e.g., cofilin/ADF [38,39] and/or gelsolin), or by activating RBC-endogenous disassembly factors. The latter scenario seems likelier, as peptides corresponding to F-actin-disassembly factors have been detected by mass spectrometry and proteomic analysis of both cultured human erythroblast-derived reticulocytes [40] and mature human RBCs [41,42]. Interestingly, hemolysates from RBCs with the mutant hemoglobins, HbSC (sickle cell trait hemoglobin) or HbCC constrain actin filament length in vitro and inhibit P. falciparum-induced RBC actin filament remodeling in vivo [34], potentially contributing to these hemoglobins’ ability to protect against malaria [43].
Nucleation of dendritic F-actin networks requires Arp2/3 complex [44,45], but, as with the F-actin-disassembly factors described above, it remains unclear whether P. falciparum exports Arp2/3 complex into the RBC or co-opts RBC-endogenous Arp2/3 complex [41,42]. The function of RBC-endogenous Arp2/3 complex in normal RBC homeostasis has yet to be demonstrated, but it is likely related to the function of Hem-1, a hematopoietic-cell-specific member of the large pentameric WAVE (Wiskott-Aldrich syndrome verprolin-homologous protein) heterocomplex and Arp2/3 activator, which is present in mature RBC lysates along with other WAVE complex components (WAVE1, WAVE2, and Abi2) [46]. Hem-1-null mice have hemolytic anemia with abnormal RBC shapes, reduced RBC lifespan, and aberrant F-actin and membrane skeleton protein composition, suggesting that Hem-1 is required for RBC membrane skeleton assembly and/or stability [46,47]. Notably, WAVE complex activation of Arp2/3-mediated F-actin assembly depends on WAVE activation by Rac GTPases [48], and Rac1/Rac2 GTPase-deficient mouse RBCs exhibit abnormal shapes, reduced deformability, and membrane skeleton disorganization [49].
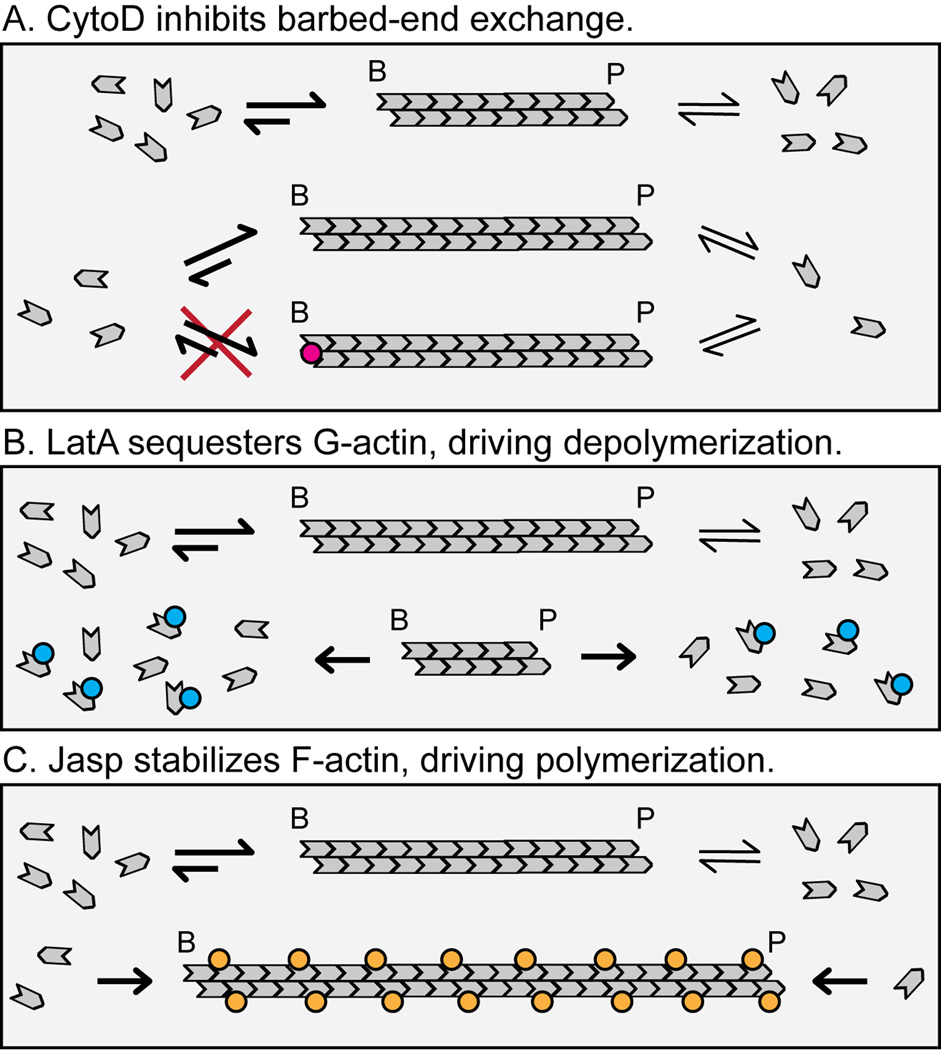
Recently, we directly observed RBC actin subunit dynamics by visualization of rhodamine-actin (rho-actin) incorporation into resealed human RBC ghosts. Rho-actin localizes to discrete puncta across the ghost membrane within 30 minutes, consistent with dynamic incorporation of rho-actin subunits into foci or “hotspots” in the membrane skeleton (Fig. 1E) [36]. These foci may represent permanent specialized microdomains within the membrane skeleton or transient structures reflecting a particular structural state of F-actin leading to localized filament assembly/disassembly events. Rho-actin incorporation can be blocked by treatment with 0.5 µM cytochalasin-D (CytoD), an inhibitor of barbed-end subunit exchange (Fig. 2A), indicating that barbed-end exchange mediates rho-actin incorporation into the RBC membrane skeleton [36]. A barbed-end-mediated mechanism for subunit exchange is consistent with a soluble G-actin concentration of ~0.36 µM [36]—halfway between the barbed-end and pointed-end critical concentrations of 0.1 µM and 0.6 µM, respectively [53]. Barbed-end assembly of rho-actin subunits may occur coincidently with pointed-end disassembly of endogenous subunits, resulting in treadmilling [54], or exogenous rho-actin subunits may exchange with endogenous subunits at barbed ends. The numbers of rho-actin foci assembling within 30 minutes were not determined, but visual inspection indicates at least an order of magnitude fewer foci than the 30,000–40,000 actin filaments per RBC [2,36,55]. Thus, only few filaments are dynamic at any instant, or, alternatively, all filaments may be dynamic but only infrequently. Future work will examine appearance, disappearance, kinetics and trajectories of these rho-actin foci by time-lapse imaging and computational analysis of intact RBCs, using techniques analogous to those developed for fluorescent speckle microscopy of actin filaments in migrating cells [56,57].
Figure 2. Effects of actin-disrupting drugs on actin filament assembly.
(A) CytoD inhibits barbed-end assembly of actin monomers. (B) LatA sequesters actin monomers, driving the F:G-actin balance toward the G-actin state [50]. LatA function requires that the actin filament be dynamic (i.e., capable of exchanging subunits with the G-actin pool). (C) Jasp stabilizes actin filaments, driving the F:G actin balance toward the F-actin state. The depicted distribution of Jasp along the actin filament is speculative and does not reflect known distributions or stoichiometries of Jasp required for filament stabilization in vitro or in vivo. Jasp competes with phalloidin for F-actin binding in vitro [51], and one phalloidin molecule can bind each subunit within F-actin under saturating conditions [52]. Thus, Jasp may recognize the same F-actin binding site as phalloidin with a similar stoichiometry, but this has not been proven.
In another approach, we labeled F-actin in intact human RBCs with a fluorescent jasplakinolide derivative (SiR-Jasp) [58] and investigated F-actin mobility by fluorescence recovery after photobleaching (FRAP). RBC F-actin has a mobile fraction of ~30% and fluorescence recovery half-time of ~2.5 minutes [36]; however, in this context, “mobility” likely involves both subunit assembly/disassembly (filament-level phenomena) and lateral movements of filaments (network-level phenomena). Inhibiting barbed-end assembly by CytoD treatment reduces the mobile fraction of F-actin from ~30% to ~25% [36], indicating that either ~1/6th of F-actin mobility can be explained by barbed-end assembly/disassembly, and/or that barbed-end assembly influences lateral movements of whole filaments. The mechanism for this crosstalk is unclear, but one possibility is that barbed-end assembly may influence (α1β1)2-spectrin-F-actin binding, such that barbed-end inhibition by CytoD treatment may alter the architecture of the (α1β1)2-spectrin-F-actin lattice. Indeed, barbed-end assembly is expected to require transient dissociation of the barbed-end cap, αβ-adducin, which would then weaken (α1β1)2-spectrin-F-actin interactions, since αβ-adducin recruits (α1β1)2-spectrin to F-actin [59,60].
Barbed-end regulation is critical for RBC physiology. Mice lacking α- or β-adducin have compensated hemolytic anemia with osmotically fragile, insufficiently deformable, and spherocytic RBCs, similar to hereditary spherocytosis [61–63], but neither RBC actin dynamics nor filament lengths have been studied. However, another barbed-end capping protein (CP, termed EcapZ) translocates from the RBC cytosol to the membrane skeleton in these mice [62,63], where it presumably caps adducin-deficient filament barbed ends [64], suggesting possible mechanisms for α- and β-adducin-null RBC phenotypes. For example, aberrant EcapZ-capped RBC actin filaments may have different dynamic properties than normal αβ-adducin-capped actin filaments [36], and EcapZ likely lacks αβ-adducin’s ability to bind Band3 [19] or recruit (α1β1)2-spectrin to F-actin [59,60]. A role for αβ-adducin in F-actin regulation has also been suggested by analysis of Rac1/Rac2-GTPase-null RBCs, which exhibit increased αβ-adducin phosphorylation, altered (α1β1)2-spectrin:F-actin ratios, and membrane skeleton disruption [49]. This agrees with in vitro observations that phosphorylation of αβ-adducin reduces its ability to cap barbed ends [65] and recruit (α1β1)2-spectrin to F-actin [65,66].
Dynamic F-actin assembly also contributes to RBC physiology. Treatment of human RBCs with latrunculin-A (LatA), a drug that drives actin depolymerization by sequestering G-actin (Fig. 2B), results in ~2-fold-increased soluble actin, whereas treatment with jasplakinolide (Jasp), a drug that drives actin polymerization by stabilizing F-actin (Fig. 2C), results in ~60%-decreased soluble actin [36]. These effects are consistent with dynamic filaments capable of polymerization and depolymerization. Importantly, disruption of RBC actin filaments with LatA or Jasp significantly alters the RBC membrane mechanics, with both LatA and Jasp treatment increasing the membrane deformability of human RBCs, determined by shorter RBC passage times through microfluidic channels [36]. Surprisingly, LatA and Jasp have differential effects on spontaneous membrane fluctuations (“flickering”), with LatA treatment increasing the variance of flickering amplitudes and Jasp treatment decreasing the variance [36]. Hence, RBC actin filament polymerization and depolymerization have differential effects on different aspects of RBC membrane mechanics. It is unclear whether the effects of LatA and Jasp on RBC cell mechanics are solely attributable to altered actin filament polymerization and depolymerization, or whether creation or elimination of (α1β1)2-spectrin attachments secondary to actin filament assembly/disassembly also plays a role.
How might actin dynamics drive RBC mechanical behaviors? Because such a small minority (~3.7%) of RBC actin is cytosolic [36], a ~2-fold increase in soluble actin induced by LatA or a ~60% decrease induced by Jasp does not imply wholesale restructuring of the RBC actin filament network. Rather, LatA- and Jasp-induced polymerization and depolymerization events most likely localize to foci capable of rho-actin incorporation [36], as described above. The classic “Brownian ratchet” theory postulates that actin polymerization near a lipid bilayer can rectify Brownian motion of G-actin and produce directed force against the bilayer [67,68]. Since RBC actin filament subunits exchange via barbed-end dynamics [36], Brownian ratchet forces may be generated where barbed ends within dynamic hotspots face toward the RBC membrane. However, analyses of actin filament orientation within the RBC membrane skeleton [69,70] have not assessed whether barbed or pointed ends are preferentially oriented towards the membrane. In the future, differential fluorescent labeling of barbed vs. pointed filament ends [71] followed by three-dimensional super-resolution imaging [72] of intact cells will enable such an analysis.
LENGTH REGULATION OF RBC ACTIN FILAMENTS
It seems paradoxical that RBC actin filaments can undergo dynamic subunit exchange while maintaining a uniform length of ~37 nm throughout the membrane skeleton. Length is not an intrinsic property of F-actin; when purified actin is polymerized to steady state in vitro, actin filaments assume an exponential distribution of lengths, with an abundance of short filaments and fewer long filaments [73,74]. Thus, RBC actin filaments must achieve their uniform in vivo lengths through concerted actions of actin-binding proteins. The barbed and pointed ends of RBC actin filaments are capped by αβ-adducin and Tmod1, respectively, which inhibit association and dissociation of actin subunits from their respective filament ends [75,76]. The mechanisms by which actin-capping proteins control filament lengths have primarily been elucidated in striated muscle cells, where actin (thin) filaments form antiparallel arrays within a sarcomere, with barbed ends anchored to Z-lines at the sarcomere periphery and pointed ends demarcating the H-zone at the sarcomere center. Like RBC actin filaments, sarcomeric thin filaments are long-lived cytoskeletal assemblies with precisely regulated and highly uniform lengths and actin-capping proteins at both ends [55]. However, sarcomeric thin filaments are much longer than RBC actin filaments (~1000 nm vs. ~37 nm, respectively [55]), and have barbed and pointed ends in spatial register. Thus, localizations of the barbed and pointed ends of sarcomeric thin filaments, and thin filament lengths, can be visualized and distinguished with conventional fluorescence microscopy.
In rho-actin-injected cardiac myocytes, subunits incorporate at both barbed and pointed thin filament ends [77]. CytoD treatment has no effect on thin filament length [77], indicating that barbed-end stability is dispensable for length regulation. However, overexpression of GFP-Tmod1 shortens thin filaments [77], while antibody inhibition of Tmod1’s pointed-end capping activity elongates thin filaments [78], indicating that pointed-end stability is essential for length regulation, with the extent of pointed-end capping by Tmod1 inversely related to lengths [79]. The extent to which these principles of length regulation extend to RBC actin filaments remains unclear. However, FRAP analyses of sarcomeric thin filaments in cultured muscle cells have identified an F-actin mobile fraction of ~25% and fluorescence recovery occurring within minutes [77,80,81], similar to RBCs [36], indicating similar kinetics of actin mobility despite markedly different cytoskeletal architectures.
To study the role of pointed-end stability in RBC actin filament length regulation, we examined Tmod1-null mice, which exhibit mild spherocytic elliptocytosis with osmotically fragile and inadequately deformable RBCs [33]. Tmod1-null RBCs exhibit compensatory appearance of Tmod3 [33], a Tmod isoform that is normally absent in mature RBCs and, instead, has critical functions during terminal erythroblast differentiation and enucleation [82]. In Tmod1-null RBCs, Tmod3 is present at ~20% of wild-type Tmod1 levels, with no changes in actin or other membrane skeleton protein levels [33]. Since in vitro experiments with purified Tmods demonstrate that Tmod3’s pointed-end capping activity is equivalent to that of Tmod1 [83], Tmod1-null RBCs represent a model of ~80% depletion of pointed-end capping activity, rather than complete ablation of pointed-end capping activity. Negative-staining electron microscopy reveals that average RBC actin filament lengths are unchanged in Tmod1-null RBCs, but the uniformity of lengths is disturbed, with emergence of both shorter and longer filament populations, leading to enlarged and variably sized membrane skeleton fenestrations (pores), likely due to improper assembly and/or instability of (α1β1)2-spectrin attachments [33]. This is distinct from Tmod1 perturbation studies in striated muscle, which reveal uniform actin filament lengthening upon Tmod1 depletion [78,79,84,85], with the caveat that the light microscopic methods applied to striated muscle can only detect changes in average thin filament length within a sarcomere, and not changes in the length of any individual filament. Additional studies are required to determine whether Tmod1-deficient RBCs actin filaments have altered dynamics, i.e., efficiency of rho-actin incorporation and responses to CytoD, LatA, or Jasp.
Another regulator of actin filament pointed-end stability is tropomyosin, which binds along actin filament sides and inhibits pointed-end depolymerization [86]. Ektacytometry experiments have demonstrated that Mg2+-free (tropomyosin-extracted) ghosts exhibit more rapid time-dependent decay in deformability than control Mg2+ (tropomyosin-containing) ghosts when subjected to constant shear flow [87], indicating that tropomyosin can regulate RBC membrane stability via its influence on RBC actin dynamics [36]. This effect is tropomyosin isoform-specific, as reconstitution of tropomyosin-extracted RBC actin filaments with purified RBC tropomyosin but not skeletal muscle tropomyosin restores normal membrane stability [87]. The two tropomyosin isoforms in RBCs, TM5b and TM5NM1, are present in an equimolar ratio (V.M. Fowler, unpublished data), and each rod-like tropomyosin molecule extends along most of the length of an RBC actin filament, possibly acting as a molecular ruler dictating filament length [2,55]. The failure of skeletal muscle tropomyosin to functionally substitute for RBC tropomyosin [87] supports this molecular ruler model, as skeletal muscle tropomyosins are “long” high-molecular-weight tropomyosins [88] that would extend beyond the ends of RBC actin filaments, unlike TM5b and TM5NM1, which are “short” low-molecular-weight tropomyosins [88]. Moreover, RBC tropomyosins interact with Tmod1 at the actin filament pointed end [89–91], enhancing Tmod1’s pointed-end capping activity [83] and defining structural scaffolding for RBC actin filaments. Whether RBC tropomyosin also interacts with αβ-adducin at the actin filament barbed end remains unknown, but the observation that mice lacking α- or β-adducin exhibit reduced tropomyosin levels in RBCs [62,63] suggests such an interaction, although it may be indirect.
The distribution and functional significance of TM5b and TM5NM1 in RBCs are unclear. Two dimeric tropomyosin molecules are associated with each RBC actin filament [2], but it remains unknown whether TM5b and TM5NM1 form homo- or heterodimers. If they form homodimers, it remains unknown whether the homodimers segregate into exclusively TM5b- or TM5NM1-containing filaments, exist in hybrid TM5b/TM5NM1-containing filaments, or a combination thereof. TM5b binds F-actin more strongly than TM5NM1 in vitro [83] and protects F-actin more effectively against depolymerization [92], suggesting that TM5b may outcompete TM5NM1 early in RBC membrane skeleton assembly and form exclusively TM5b-containing actin filaments, leaving compositionally distinct TM5NM1-containing filaments to assemble at later time-points. However, this is speculative, since the diversity of actin-binding proteins associated with RBC actin filaments in vivo may alter the isoform-specific affinities of RBC tropomyosins for F-actin. Microscopic analyses with isoform-specific tropomyosin antibodies are required to assess the spatial distribution of RBC tropomyosins in vivo.
A broader puzzle is how RBC actin filament length uniformity is specified during membrane skeleton assembly [93]. RBC actin filaments may be nucleated and assembled into short 37-nm-long filaments de novo, or, alternatively, RBC actin filaments may first be assembled into overlong filaments that are then pruned into proper-length filaments. The latter hypothesis appears likelier, given that tropomyosin promotes elongation in vitro [86], and levels of membrane skeleton-associated RBC tropomyosin decrease during reticulocyte maturation, with no accompanying changes in Tmod1 [94]. Moving forward, detailed microscopic analysis of F-actin and actin-binding protein localization and dynamics during erythroblast differentiation and reticulocyte maturation, in wild-type and gene-targeted mice, will help elucidate the origins and regulation of uniform RBC actin filament lengths.
CONCLUSION
The studies discussed here demonstrate that the RBC field should abandon its long-held assumption that RBC actin filament nodes are static cytolinkers. Rather, RBC actin filaments exhibit sophisticated and finely tuned dynamic properties, which enable RBC actin filaments to exchange actin subunits with the RBC cytosol while maintaining uniform lengths and mechanically fortifying the RBC membrane. Thanks to recent advances in F-actin drugs, detection, and imaging, we can now interrogate the composition, architecture, dynamics, and physiology of RBC actin filaments in unprecedented detail, ensuring that the RBC membrane skeleton will continue its productive tenure as the paradigmatic membrane skeleton.
KEY POINTS.
The red blood cell (RBC) membrane skeleton consists of (α1β1)2-spectrin tetramers interconnecting short actin filaments in a two-dimensional quasi-hexagonal network beneath the lipid bilayer.
RBC actin filaments dynamically exchange subunits with the cytosol during normal RBC homeostasis and are remodeled into an aberrantly branched network in malaria parasite-infected RBCs.
Uniform RBC actin filament lengths (~37 nm) and stability are coordinately regulated by actin-capping proteins, tropomodulin-1 and αβ-adducin, and two tropomyosin isoforms, TM5b (Tpm1.9) and TM5NM1 (Tpm3.1).
Acknowledgments
We are grateful to Roberta Nowak for assistance with preparing the figures.
FINANCIAL SUPPORT
This work was supported by National Institutes of Health grants K99-AR066534 (to D.S.G.) and R01-HL083464 (to V.M.F.).
Footnotes
CONFLICTS OF INTEREST
None.
REFERENCES
- 1.Kaushansky K, Lichtman MA, Beutler E, et al. Williams Hematology. 8th. New York: McGraw-Hill Medical; 2010. [Google Scholar]
- 2.Fowler VM. The human erythrocyte plasma membrane: a Rosetta Stone for decoding membrane-cytoskeleton structure. Curr Top Membr. 2013;72:39–88. doi: 10.1016/B978-0-12-417027-8.00002-7. [DOI] [PubMed] [Google Scholar]
- 3.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 4.van den Akker E, Satchwell TJ, Williamson RC, Toye AM. Band 3 multiprotein complexes in the red cell membrane; of mice and men. Blood Cells Mol Dis. 2010;45:1–8. doi: 10.1016/j.bcmd.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Mohandas N, Gallagher PG. Red cell membrane: past, present, and future. Blood. 2008;112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shotton DM, Burke BE, Branton D. The molecular structure of human erythrocyte spectrin. Biophysical and electron microscopic studies. J Mol Biol. 1979;131:303–329. doi: 10.1016/0022-2836(79)90078-0. [DOI] [PubMed] [Google Scholar]
- 7.Swihart AH, Mikrut JM, Ketterson JB, Macdonald RC. Atomic force microscopy of the erythrocyte membrane skeleton. J Microsc. 2001;204:212–225. doi: 10.1046/j.1365-2818.2001.00960.x. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi M, Miyamoto H, Sako Y, et al. Structure of the erythrocyte membrane skeleton as observed by atomic force microscopy. Biophys J. 1998;74:2171–2183. doi: 10.1016/S0006-3495(98)77926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGough AM, Josephs R. On the structure of erythrocyte spectrin in partially expanded membrane skeletons. Proc Natl Acad Sci U S A. 1990;87:5208–5212. doi: 10.1073/pnas.87.13.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nans A, Mohandas N, Stokes DL. Native ultrastructure of the red cell cytoskeleton by cryo-electron tomography. Biophys J. 2011;101:2341–2350. doi: 10.1016/j.bpj.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fung LW, Lu HZ, Hjelm RP, Jr, Johnson ME. Quantitative detection of rapid motions in spectrin by NMR. Life Sci. 1989;44:735–740. doi: 10.1016/0024-3205(89)90385-8. [DOI] [PubMed] [Google Scholar]
- 12.Mikkelsen A, Elgsaeter A. Human spectrin. V. A comparative electro-optic study of heterotetramers and heterodimers. Biochim Biophys Acta. 1981;668:74–80. doi: 10.1016/0005-2795(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 13.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ott A, Magnasco M, Simon A, Libchaber A. Measurement of the persistence length of polymerized actin using fluorescence microscopy. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1993;48:R1642–R1645. doi: 10.1103/physreve.48.r1642. [DOI] [PubMed] [Google Scholar]
- 15.Liu SC, Derick LH, Palek J. Visualization of the hexagonal lattice in the erythrocyte membrane skeleton. J Cell Biol. 1987;104:527–536. doi: 10.1083/jcb.104.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen BW, Josephs R, Steck TH. Ultrastructure of the intact skeleton of the human erythrocyte. J. Cell Biol. 1986;102:997–1006. doi: 10.1083/jcb.102.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Discher DE, Winardi R, Schischmanoff PO, et al. Mechanochemistry of protein 4.1's spectrin-actin-binding domain: ternary complex interactions, membrane binding, network integration, structural strengthening. J Cell Biol. 1995;130:897–907. doi: 10.1083/jcb.130.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lux SE. Anatomy of the red cell membrane skeleton: unanswered questions. Blood. 2016 doi: 10.1182/blood-2014-12-512772. in press. [DOI] [PubMed] [Google Scholar]
- 19.Anong WA, Franco T, Chu H, et al. Adducin forms a bridge between the erythrocyte membrane and its cytoskeleton and regulates membrane cohesion. Blood. 2009;114:1904–1912. doi: 10.1182/blood-2009-02-203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marfatia SM, Lue RA, Branton D, Chishti AH. In vitro binding studies suggest a membrane-associated complex between erythroid p55, protein 4.1, and glycophorin C. J Biol Chem. 1994;269:8631–8634. [PubMed] [Google Scholar]
- 21.Anderson RA, Lovrien RE. Glycophorin is linked by band 4.1 protein to the human erythrocyte membrane skeleton. Nature. 1984;307:655–658. doi: 10.1038/307655a0. [DOI] [PubMed] [Google Scholar]
- 22.Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. An alternative binding site for an element of the membrane skeleton. J Biol Chem. 1985;260:3676–3683. [PubMed] [Google Scholar]
- 23.Weber KL, Fischer RS, Fowler VM. Tmod3 regulates polarized epithelial cell morphology. J Cell Sci. 2007;120:3625–3632. doi: 10.1242/jcs.011445. [DOI] [PubMed] [Google Scholar]
- 24.Abdi KM, Bennett V. Adducin promotes micrometer-scale organization of beta2-spectrin in lateral membranes of bronchial epithelial cells. Mol Biol Cell. 2008;19:536–545. doi: 10.1091/mbc.E07-08-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jenkins PM, He M, Bennett V. Dynamic spectrin/ankyrin-G microdomains promote lateral membrane assembly by opposing endocytosis. Sci Adv. 2015;1:e1500301. doi: 10.1126/sciadv.1500301. This study proposes a novel mechanism of membrane assembly and maintenance in epithelial cells, whereby the membrane skeleton actively patrols over the membrane via dynamic rearrangements, thereby preventing endocytosis and membrane depletion. This mechanism may be relevant to other cell types, including RBCs.
- 26.Nowak RB, Fischer RS, Zoltoski RK, et al. Tropomodulin1 is required for membrane skeleton organization and hexagonal geometry of fiber cells in the mouse lens. J Cell Biol. 2009;186:915–928. doi: 10.1083/jcb.200905065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gokhin DS, Fowler VM. Cytoplasmic gamma-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. J Cell Biol. 2011;194:105–120. doi: 10.1083/jcb.201011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlahovich N, Kee AJ, Van der Poel C, et al. Cytoskeletal tropomyosin Tm5NM1 is required for normal excitation-contraction coupling in skeletal muscle. Mol Biol Cell. 2009;20:400–409. doi: 10.1091/mbc.E08-06-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel-Hett S, Wang H, Begonja AJ, et al. The spectrin-based membrane skeleton stabilizes mouse megakaryocyte membrane systems and is essential for proplatelet and platelet formation. Blood. 2011;118:1641–1652. doi: 10.1182/blood-2011-01-330688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sui Z, Nowak RB, Sanada C, et al. Regulation of actin polymerization by tropomodulin-3 controls megakaryocyte actin organization and platelet biogenesis. Blood. 2015;126:520–530. doi: 10.1182/blood-2014-09-601484. This study showed that actin stability, maintained by Tmod3, is important for the morphogenesis of the megakaryocyte demarcation membrane system, distribution of intracellular organelles, and sizing of nascent platelets. This is an excellent example of how actin dynamics, elucidated in RBCs but also in nonerythroid cell types, defines general principles that can be broadly applied to diverse membrane skeleton architectures.
- 31.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong G, He J, Zhou R, et al. Developmental mechanism of the periodic membrane skeleton in axons. Elife. 2014;3 doi: 10.7554/eLife.04581. This exciting study followed-up on the initial identification of the axonal spectrin-actin membrane skeleton and its hallmark F-actin rings. With particular relevance to RBCs, this study found that periodic spectrin tetramer assembly along the axon relies on normal actin dynamics, as demonstrated by disorganized spectrin organization in CytoD- and LatA-treated hippocampal neurons.
- 33.Moyer JD, Nowak RB, Kim NE, et al. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010;116:2590–2599. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cyrklaff M, Sanchez CP, Kilian N, et al. Hemoglobins S and C interfere with actin remodeling in Plasmodium falciparum-infected erythrocytes. Science. 2011;334:1283–1286. doi: 10.1126/science.1213775. [DOI] [PubMed] [Google Scholar]
- 35.Pinder JC, Gratzer WB. Structural and dynamic states of actin in the erythrocyte. J Cell Biol. 1983;96:768–775. doi: 10.1083/jcb.96.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gokhin DS, Nowak RB, Khoory JA, et al. Dynamic actin filaments control the mechanical behavior of the human red blood cell membrane. Mol Biol Cell. 2015;26:1699–1710. doi: 10.1091/mbc.E14-12-1583. This study was the first to directly probe actin subunit dynamics in RBCs by direct visualization of rho-actin monomer assembly into the RBC membrane skeleton. A critical finding is that CytoD blocks rho-actin incorporation, indicating that RBC actin filaments assemble via barbed-end dynamics. Moreover, disrupting RBC actin filaments with LatA or Jasp alters the RBC membrane deformability and spontaneous membrane oscillations (“flickering”), demonstrating a direct link between actin filament assembly/disassembly and RBC membrane mechanical properties.
- 37. Rug M, Cyrklaff M, Mikkonen A, et al. Export of virulence proteins by malaria-infected erythrocytes involves remodeling of host actin cytoskeleton. Blood. 2014;124:3459–3468. doi: 10.1182/blood-2014-06-583054. This study followed-up on the initial discovery of actin remodeling in P. falciparum-infected erythrocytes by demonstrating that the major. P. falciparum erythrocyte membrane protein 1 (PfEMP1)-trafficking protein 1 (PfPTP1) is required for Maurer’s cleft architecture in the cytosol of infected RBCs, where it directly links Maurer’s clefts to the organization and lengths of RBC actin filaments. PfPTP1 is present in a large complex with PfEMP1 and other parasite proteins, and is required for PfEMP1 trafficking to the P. falciparum-infected RBC surface, where PfEMP1 confers pathological RBC cytoadherence to the endothelium. This study provides direct evidence that cytosolic RBC actin filaments in infected RBCs are critical for this PfEMP1 trafficking pathway.
- 38.Wong W, Skau CT, Marapana DS, et al. Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1. Proc Natl Acad Sci U S A. 2011;108:9869–9874. doi: 10.1073/pnas.1018927108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong W, Webb AI, Olshina MA, et al. A mechanism for actin filament severing by malaria parasite actin depolymerizing factor 1 via a low affinity binding interface. J Biol Chem. 2014;289:4043–4054. doi: 10.1074/jbc.M113.523365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell AJ, Satchwell TJ, Heesom KJ, et al. Protein distribution during human erythroblast enucleation in vitro. PLoS One. 2013;8:e60300. doi: 10.1371/journal.pone.0060300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Alessandro A, Righetti PG, Zolla L. The red blood cell proteome and interactome: an update. J Proteome Res. 2010;9:144–163. doi: 10.1021/pr900831f. [DOI] [PubMed] [Google Scholar]
- 42.Kakhniashvili DG, Bulla LA, Jr, Goodman SR. The human erythrocyte proteome: analysis by ion trap mass spectrometry. Mol Cell Proteomics. 2004;3:501–509. doi: 10.1074/mcp.M300132-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 45.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 46.Chan MM, Wooden JM, Tsang M, et al. Hematopoietic protein-1 regulates the actin membrane skeleton and membrane stability in murine erythrocytes. PLoS One. 2013;8:e54902. doi: 10.1371/journal.pone.0054902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park H, Chan MM, Iritani BM. Hem-1: putting the "WAVE" into actin polymerization during an immune response. FEBS Lett. 2010;584:4923–4932. doi: 10.1016/j.febslet.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 49.Kalfa TA, Pushkaran S, Mohandas N, et al. Rac GTPases regulate the morphology and deformability of the erythrocyte cytoskeleton. Blood. 2006;108:3637–3645. doi: 10.1182/blood-2006-03-005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 51.Bubb MR, Senderowicz AM, Sausville EA, et al. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J Biol Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- 52.De La Cruz EM, Pollard TD. Transient kinetic analysis of rhodamine phalloidin binding to actin filaments. Biochemistry. 1994;33:14387–14392. doi: 10.1021/bi00252a003. [DOI] [PubMed] [Google Scholar]
- 53.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 54.Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–470. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 55.Fowler VM. Regulation of actin filament length in erythrocytes and striated muscle. Curr Opin Cell Biol. 1996;8:86–96. doi: 10.1016/s0955-0674(96)80052-4. [DOI] [PubMed] [Google Scholar]
- 56.Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- 57.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 58. Lukinavicius G, Reymond L, D'Este E, et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 2014;11:731–733. doi: 10.1038/nmeth.2972. In this study, the authors identify and characterize SiR-Jasp (also known as SiR-actin), a fluorescent derivative of jasplakinolide that binds specifically to F-actin and faithfully reports F-actin localization in live RBCs and other cell types. This probe has broad utility in studies on RBC actin dynamics.
- 59.Gardner K, Bennett V. Modulation of spectrin-actin assembly by erythrocyte adducin. Nature. 1987;328:359–362. doi: 10.1038/328359a0. [DOI] [PubMed] [Google Scholar]
- 60.Mische SM, Mooseker MS, Morrow JS. Erythrocyte adducin: a calmodulin-regulated actin-bundling protein that stimulates spectrin-actin binding. J Cell Biol. 1987;105:2837–2845. doi: 10.1083/jcb.105.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muro AF, Marro ML, Gajovic S, et al. Mild spherocytic hereditary elliptocytosis and altered levels of alpha- and gamma-adducins in beta-adducin-deficient mice. Blood. 2000;95:3978–3985. [PubMed] [Google Scholar]
- 62.Robledo RF, Ciciotte SL, Gwynn B, et al. Targeted deletion of alpha-adducin results in absent beta- and gamma-adducin, compensated hemolytic anemia, and lethal hydrocephalus in mice. Blood. 2008;112:4298–4307. doi: 10.1182/blood-2008-05-156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Porro F, Costessi L, Marro ML, et al. The erythrocyte skeletons of beta-adducin deficient mice have altered levels of tropomyosin, tropomodulin and EcapZ. FEBS Lett. 2004;576:36–40. doi: 10.1016/j.febslet.2004.08.057. [DOI] [PubMed] [Google Scholar]
- 64.Kuhlman PA, Fowler VM. Purification and characterization of an alpha 1 beta 2 isoform of CapZ from human erythrocytes: cytosolic location and inability to bind to Mg2+ ghosts suggest that erythrocyte actin filaments are capped by adducin. Biochemistry. 1997;36:13461–13472. doi: 10.1021/bi970601b. [DOI] [PubMed] [Google Scholar]
- 65.Matsuoka Y, Li X, Bennett V. Adducin is an in vivo substrate for protein kinase C: phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin-actin complexes and occurs in many cells, including dendritic spines of neurons. J Cell Biol. 1998;142:485–497. doi: 10.1083/jcb.142.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuoka Y, Hughes CA, Bennett V. Adducin regulation. Definition of the calmodulin-binding domain and sites of phosphorylation by protein kinases A and C. J Biol Chem. 1996;271:25157–25166. doi: 10.1074/jbc.271.41.25157. [DOI] [PubMed] [Google Scholar]
- 67.Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picart C, Dalhaimer P, Discher DE. Actin protofilament orientation in deformation of the erythrocyte membrane skeleton. Biophys J. 2000;79:2987–3000. doi: 10.1016/S0006-3495(00)76535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Picart C, Discher DE. Actin protofilament orientation at the erythrocyte membrane. Biophys J. 1999;77:865–878. doi: 10.1016/S0006-3495(99)76938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ofer N, Abu Shah E, Keren K. Differential mapping of the free barbed and pointed ends of actin filaments in cells. Cytoskeleton (Hoboken) 2014;71:341–350. doi: 10.1002/cm.21176. This study described a simple, straightforward technique to localize the barbed and pointed ends of actin filaments by permeabilization, introduction of fluorescently labeled actin monomers, brief fixation, and introduction of additional fluorescently labeled actin monomers in the presence of capping protein.
- 72.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawamura M, Maruyama K. Electron microscopic particle length of F-actin polymerized in vitro. J Biochem. 1970;67:437–457. doi: 10.1093/oxfordjournals.jbchem.a129267. [DOI] [PubMed] [Google Scholar]
- 74.Littlefield R, Fowler VM. Defining actin filament length in striated muscle: rulers and caps or dynamic stability? Annu Rev Cell Dev Biol. 1998;14:487–525. doi: 10.1146/annurev.cellbio.14.1.487. [DOI] [PubMed] [Google Scholar]
- 75.Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J. Cell Biol. 1994:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuhlman PA, Hughes CA, Bennett V, Fowler VM. A new function for adducin. Calcium/calmodulin-regulated capping of the barbed ends of actin filaments. J Biol Chem. 1996;271:7986–7991. doi: 10.1074/jbc.271.14.7986. [DOI] [PubMed] [Google Scholar]
- 77.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 78.Gregorio CC, Weber A, Bondad M, et al. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- 79.Gokhin DS, Fowler VM. Tropomodulin capping of actin filaments in striated muscle development and physiology. J Biomed Biotechnol. 2011;2011:103069. doi: 10.1155/2011/103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pappas CT, Krieg PA, Gregorio CC. Nebulin regulates actin filament lengths by a stabilization mechanism. J Cell Biol. 2010;189:859–870. doi: 10.1083/jcb.201001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- 82. Sui Z, Nowak RB, Bacconi A, et al. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood. 2014;123:758–767. doi: 10.1182/blood-2013-03-492710. This study demonstrated a critical role for Tmod3, a Tmod isoform not normally present in mature RBCs, in terminal erythroblast differentiation and enucleation. This study was the first to demonstrate that actin filament pointed-end stability is essential for RBC biogenesis.
- 83.Yamashiro S, Gokhin DS, Sui Z, et al. Differential actin-regulatory activities of Tropomodulin1 and Tropomodulin3 with diverse tropomyosin and actin isoforms. J Biol Chem. 2014;289:11616–11629. doi: 10.1074/jbc.M114.555128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gokhin DS, Tierney MT, Sui Z, et al. Calpain-mediated proteolysis of tropomodulin isoforms leads to thin filament elongation in dystrophic skeletal muscle. Mol Biol Cell. 2014;25:852–865. doi: 10.1091/mbc.E13-10-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gokhin DS, Ochala J, Domenighetti AA, Fowler VM. Tropomodulin1 directly controls thin filament length in both wild-type and tropomodulin4-deficient skeletal muscle. Development. 2016 doi: 10.1242/dev.129171. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Broschat KO, Weber A, Burgess DR. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28:8501–8506. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]
- 87.An X, Salomao M, Guo X, et al. Tropomyosin modulates erythrocyte membrane stability. Blood. 2007;109:1284–1288. doi: 10.1182/blood-2006-07-036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 89.Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. J Biol Chem. 1987;262:12792–12800. [PubMed] [Google Scholar]
- 90.Fowler VM. Tropomodulin: a cytoskeletal protein that binds to the end of erythrocyte tropomyosin and inhibits tropomyosin binding to actin. J Cell Biol. 1990;111:471–481. doi: 10.1083/jcb.111.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ursitti JA, Fowler VM. Immunolocalization of tropomodulin, tropomyosin and actin in spread human erythrocyte skeletons. J Cell Sci. 1994;107(Pt 6):1633–1639. doi: 10.1242/jcs.107.6.1633. [DOI] [PubMed] [Google Scholar]
- 92.Lewis RA, Yamashiro S, Gokhin DS, Fowler VM. Functional effects of mutations in the tropomyosin-binding sites of tropomodulin1 and tropomodulin3. Cytoskeleton (Hoboken) 2014;71:395–411. doi: 10.1002/cm.21179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hanspal M, Palek J. Synthesis and assembly of membrane skeletal proteins in mammalian red cell precursors. J Cell Biol. 1987;105:1417–1424. doi: 10.1083/jcb.105.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu J, Guo X, Mohandas N, et al. Membrane remodeling during reticulocyte maturation. Blood. 2010;115:2021–2027. doi: 10.1182/blood-2009-08-241182. [DOI] [PMC free article] [PubMed] [Google Scholar]