Abstract
Single positioned nucleosomes have been extensively employed as simple model experimental systems for analysis of various intranuclear processes. Here we describe an experimental system containing positioned mononucleosomes allowing transcription by various RNA polymerases. Each DNA template contains a pair of fluorescent labels (Cy3 and Cy5) allowing measuring relative distances between the neighboring coils of nucleosomal DNA using Forster resonance energy transfer (FRET). The single-particle FRET (spFRET) approach for analysis of DNA uncoiling from the histone octamer during transcription through chromatin is described in detail.
Keywords: Nucleosomes, chromatin, transcription, assembly, methods, spFRET
1. Introduction
All vital biological intranuclear processes (e.g. DNA replication, repair, recombination and transcription) occur on DNA organized in chromatin. At the first level of chromatin folding, 147-bp DNA regions are organized in 1 3/4 superhelical coils on the surface of the histone octamer forming nucleosome cores (1) . Short DNA fragments (150–350 bp) containing single nucleosomes have been used recently for analysis of transcription of nucleosomal templates (2–5) and many other intranuclear processes, including DNA repair in chromatin (6, 7), ATP-dependent chromatin remodeling (8, 9) and analysis of nucleosome structure and thermodynamics (10–13). In many cases the model templates faithfully recapitulate important aspects of these processes (2, 3, 6, 9, 14, 15). At the same time, these simple experimental systems can be analyzed with a high resolution. Positioning and histone composition of mononucleosomes that are often changed during various processes can be easily monitored by analysis in a native gel (16–19). Thus analysis of protein binding to nucleosomes and protein-induced changes in the conformation and/or structure of the complexes are relatively straightforward using mononucleosomal templates.
In the majority of recent studies uniquely positioned nucleosomes were assembled on a DNA fragment containing one of the strong nucleosome-positioning DNA sequences (20, 21). We routinely use two methods of nucleosome assembly: by dialysis of purified DNA and core histonesfrom 2M NaCl ( 22)or by transfer of the histone octamer from “donor” chromatin onto DNA (23). To avoid any traces of the donor chromatin in the resulting nucleosome preparation, we include an additional step of purification of the templates on Ni-NTA beads (4). In both protocols DNA or donor chromatin is present in an excess, allowing formation of nucleosomes only on the high-affinity DNA sequences.
Advanced insight into nucleosome structure and dynamics during various intranuclear processes can be achieved with modern experimental approaches based on single particle Forster resonance energy transfer (spFRET) analysis (24, 25). Thus spFRET analysis is informative for analysis of alterations in nucleosome structure induced by modification (methylation, acetylation) of DNA or histones (24, 26–28). spFRET records data from thousands nucleosomes, and each nucleosome is characterized individually. This allows study of structurally different subpopulations of nucleosomes that are often present during various processes of chromatin metabolism.
Here we describe the application of spFRET to analysis of DNA uncoiling from the histone octamer during transcription through chromatin. spFRETis a method of choice because of a large variety and high heterogeneity of chromatin states formed during transcription. We briefly describe basic principles of spFRET and present protocols for preparation of fluorescently labeled nucleosomes and stalled RNAP complexes for spFRET analysis.
2. Materials
2.1. Miscellaneous Items
-
2
Low adhesion tubes (USA Scientific).
-
3
Gele xtraction kit (Omega Bio-tek).
-
4
SpinX centrifuge tube filter(Amicon) .
-
5
Dialysis membranes (Spectra/Por; molecular weight cut-off of 8000 and 12,000–14,000).
-
6
Ni-NTA agarose (Qiagen).
-
7
8-well Lab-Tek chambered cover glasses( Thermo Scientific).
2.2. Enzymes
Restriction enzymes (New England Biolabs, NEB).
T4 DNA ligase (NEB).
Taq DNA polymerase (NEB).
RNA polymerase from Escherichia coli.
2.3. Buffers and Solutions
TAE buffer: 0.04M Tris–acetate, pH 8.0, and 1 mM EDTA.
TE buffer: 10 mM Tris-HCl, pH 8.0, 1 mMEDTA .
TB (transcription buffer): 20 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 2 mM 2-mercaptoethanol, and indicated concentration of KCl, mM.
EDTA stock solution: 0.5 M Na2–EDTA, pH 8.0.
RLB (RNA loading buffer): 95% formamide, 10 mM EDTA, 0.1% SDS, and 0.01% of each bromophenol blue and xylene cyanol dyes.
2x agarose loading buffer with 8 M urea: 45 mM Tris base, 45 mM Boric Acid, 8M Urea, 0.01% bromophenol blue, 0.01% xylene cyanol, 1 mM EDTA.
Annealing buffer: 10 mM TrisHCl, 100 mM NaCl, 1 mM EDTA pH7,5.
TB-PEG buffer for spFRET measurements: TB buffer supplemented with 0.1% poly(ethylene glycol) (MW 380–420 Da, Aldrich).
CRB 1 to 4 (Core Reconstitution Buffers): All four buffers contain HE, 5mM β-mercaptoethanol, 0.1% NP-40 and NaCl at the following concentrations: Buffer 1–1 M; 2 - 0.8 M; 3 - 0.6 M; 4 - 0.01 M.
3. Methods
3.1. Selectingposition sof fluorophore so n the nucleosomalDNA
FRET arises between two specially selected fluorophores, donor and acceptor, when the distance between them is ca. 10 nm or less. Donor excitation induces a donor fluorescence emission when the donor is far from an acceptor, or an acceptor emission when the donor is close to the acceptor (Fig. 1A,B). The efficiency of FRET (E) is related to the distance r between donor and acceptor according to the formula:
| (1) |
where R0 is the Forster radius. For example, R0 for fluorophores Cy3 (donor) and Cy5 (acceptor) (labels that are often used in spFRET) is of ca. 60 Å (29), and parameter E is a reliable reporterof changes in the distance between Cy3 and Cy5 in the rangeof 3–10 nm.
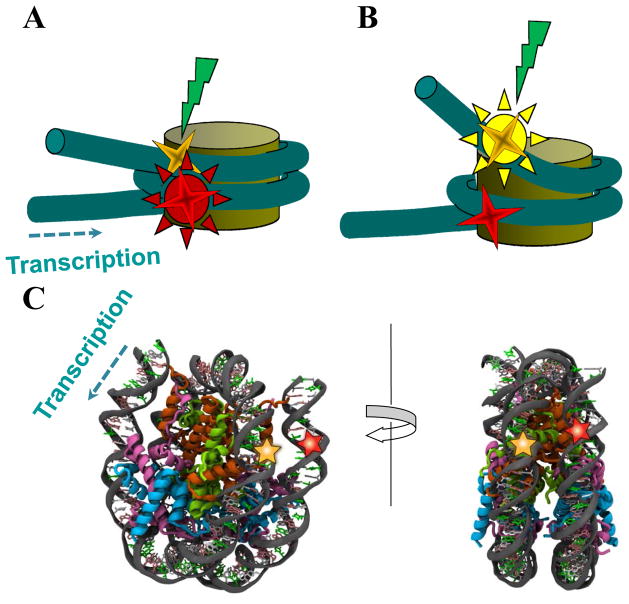
Figure 1. Experimental system for analysis of DNA uncoiling from the histone octamer.
Fluorescent donor (yellow star) and acceptor (red star) (Cy3 and Cy5 fluorophores, respectively) in a mononucleosome are shown. The direction of transcription is shown by dashed arrow. (A) A donor is close to an acceptor, and donor excitation results in acceptor emission (high FRET efficiency). (B) A donor is far from an acceptor, and donor excitation results in donor emission (no FRET). (C) Thymidines in positions +40 and +135 from the promoter-proximal boundary of the 603s nucleosome can be labeled with Cy3 and Cy5, respectively, to have inter-label distance of approximately 1.9 nm.
For spFRET analysis fluorescent labels (one donor and one acceptor) are introduced in the neighboring superhelical coils of DNA on a nucleosome. A distance between labels should be ≤ R0. In a case of the 603 DNA template used in our study the labels were incorporated in the positions +40 and +135 bp from the promoter-proximal boundary of the nucleosome (Fig. 1C).
Choice of these positions for DNA labeling is based on the following considerations. According to the structure of the nucleosome assembled on a high-affinity DNA sequence (13), both probes are incorporated into nucleosomal DNA in positions where DNA helix faces solution and the probes are in a close proximity to each other (Fig. 1C). This positioning minimizes the probability of interaction between the fluorescent probes and histones, and maximizes the FRET signal characteristic for the intact nucleosome. In the example described here the probes were incorporated to monitor uncoiling of the promoter-distal region of nucleosomal DNA; however they can be incorporated into different positions on nucleosomal DNA to monitor uncoiling of other DNA regions from the histone octamer.
3.2. Assembly of mononucleosomal templates containing Cy5/Cy3 probes for in vitro transcription
In the protocol described below nucleosomal DNA is obtained after annealing and ligation of overlapping DNA oligonucleotides. Alternatively, nucleosomal DNA can be obtained by PCR using shorter DNA oligonucleotides. DNA fragment containing the T7A1 promoter is amplified separately using a pair of primers. After digestion of both nucleosomal and promoter DNA with TspRI, the promoter fragment is ligated to the 147-bp 603 template through complementary TspRI sticky ends (30, 31), and the ligated template is gel-purified.
3.2.1. Purification of oligonucleotides with Cy5/Cy3 distal probes
-
Add 45 ul of water and 75 ul of 2x agarose loading buffer with 8 M urea to 30 ul of 100 uM solutions of the following oligonucleotides( Note 1):
6035AFP
5’-CCCGGTTCGCGCGCCCGCCTTCCGTGTGTTGTCTCCTCTCGG
3DW_Cy3_603-5A2
5’-GCGTCTAAGTACGCTTAGCGCACGGTAGAGCGCAATCCAAGGCTAACCACCGTGCATCGATGTTGAAAGAGGCCCTCCGTCCTTATTACTTCAAGTCCCTGGGGT
5DW_Cy5_603-5A2
5’-ACCCCAGGGACTTGAAGTAATAAGGACGGAGGGCCTCTTTCAACATCGATGCACGGTGGTTAGCCTTGGATTGCGCTCTACCGTGCGCTAAGCGTA
6035ARP
5’-CTTAGACGCCCGAGTGACGACTTCACTCGGCAGGCGGGCGCGCGAACCGGGCCCAGTGCC
Heats amples at 95ºC for 5 min and separate DNA fragments by denaturing PAGE (2).
Cut pieces of the gel, containing primers, and transfer them to 500 ul tubes with a hole in the bottom.
Insert tubes into the 1,5 ml tubes and spin on Eppendorf microcentrifuge at 13000 rpm for 10 sec.
Add 5 volumes of TE buffer to the samples after centrifugation and incubate overnight at 4ºC.
Collect supernatant by filtering through SpinX centrifuge tube filter by centrifugation at 13000 rpm on Eppendorf microcentrifuge for 10 min.
Precipitate DNA by isopropanol, wash with 70% ethanol and dissolve pellets in 100 ul of TE buffer .
Estimate DNA concentration by measuring absorption (A) at the wavelength 2 60 nm.
3.2.2. Assembly of DNA template containing Cy5/Cy3 probes
Mix 2 primers: 3DW_Cy3_603-5A2 and 5DW_Cy5_603-5A2 in equimolar amounts in the annealing buffer, heat at 95ºC for 3 min and slowly cool down from 95ºC to 16ºC during 2 hrs.
Extract the annealed product by phenol, then precipitate with ethanol and wash with 70% ethanol.
Dissolve pellet in the annealing buffer and then add a mixture of 6035AFP and 6035ARP primers in equimolar amounts.
Heat the mixture of four primers to 45ºC and slowly cool down from 45ºC to 16ºC during 1 hour.
Extract the annealed product by phenol, precipitate with ethanol and wash with 70% ethanol.
Dissolve pellet in 15 ul of 1X T4 ligase buffer (NEB), add 4 ul of T4 ligase and incubate overnight at 16ºC.
Cut the promoter and the nucleosomal Cy5/Cy3-labeled fragments by TspRI enzyme in NEB4 buffer (New England Biolabs) for 3 hrs at 65°C.
Resolve the obtained fragments in a 1.5% (w/v) agarose gel containing 4 M urea, 0.5 mg/ml ethidium bromide and 0.5 x TBE buffer at 4–6 V/cm for 1.5–2 hrs.
Excise the ~140-bp promoter fragment and the ~150-bp nucleosomal fragment avoiding UV illumination (see Note 1).
Purify both fragments by gel extraction kit (Omega Bio-tek) according to the manufacturer’s instructions.
Ligate 1–2 ug of the purified promoter DNA fragment (2) and the nucleosomal Cy5/Cy3-labeled fragments at molar ratio 1.15:1 in T4 ligase buffer for 1.5–2 hrs at room temperature.
Resolve the ligated from non-ligated contaminant fragment in a 1.8% (w/v) agarose gel containing 0.5 mg/ml ethidium bromide and 0.5 x TBE buffer at 4–6 V/cm for 2–2.5 hrs depending on the resolution required for clear band separation.
Excise the desired ligated~300 -bp fragment avoiding UV illumination(see Note 1).
Purify the ligated ~300-bp fragment by Gel Extraction kit (Omega Bio-tek) according to the manufacturer’s instructions.
Measure DNA concentration on A260 wavelength, using A260=20 for 1 mg/ml DNA.
3.2.3. Nucleosome assembly
Nucleosomes are assembled on the template described above by dialysis against decreasing concentrations of NaCl and using long chromatin without histone H1 as a donor of the histone octamer( Fig. 2A).
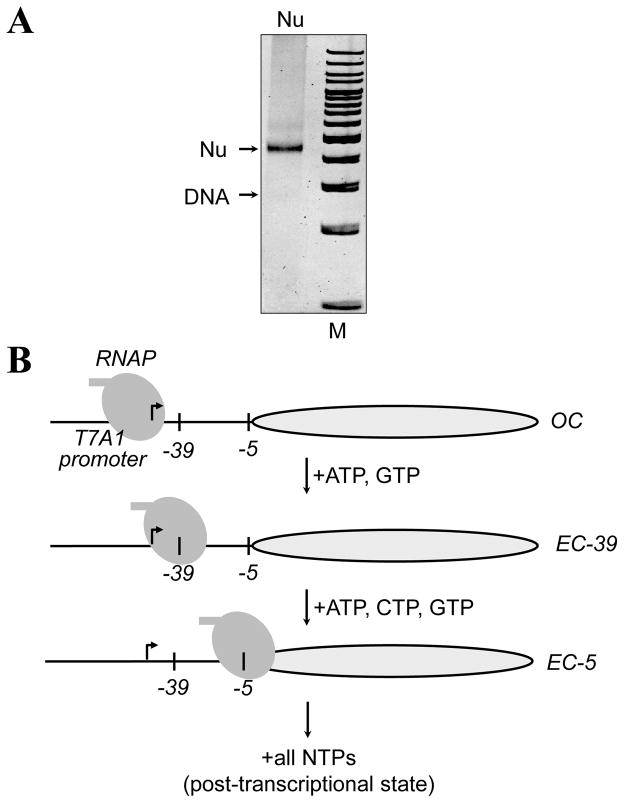
Figure 2. The experimental approach for stalling RNAP at unique positions on the 603 nucleosome.
A. The sequence of the 603 template allows formation of the open initiation complex (OC) and stalling of RNAP ECs at the_-5 or -39 positions (relative to the promoter-proximal nucleosome boundary) upon addition of different partial combinations of NTPs. Corresponding complexes are characterized and analyzed using DNase I footprinting or spFRET. B. Analysis of 603 mononucleosomal template containing Cy5/Cy3 fluorophores by native PAGE. Arrows indicate positions of DNA and the nucleosome (Nu). M – 100-b.p. DNA ladder (Fermentas).The gel was stained with ethidium bromide.
Cool 500 ml each of CRB1 to CRB4 buffers to 4ºC.
Mix one to three micrograms of the DNA fragment with long-H1/H5 donor chromatin at a ratio of 1:5 (w:w) in 0.04–0.1 ml of CRB1 buffer.
Dialyze successively against CRB2 and CRB3, each for 2 hours at 4ºC. Then dialyze the sample against CRB4 for 3 hours or overnight.
Transfer the reconstitute to a siliconized Eppendorf tube and store at 4ºC (do not freeze).
Check the samples by analysis by native PAGE (see below).
Utilize the prepared mononucleosomal templates for the transcription experiments in vitro described below .
3.3. Preparation of stalled nucleosome complexes with RNAP: open complex and elongation complexes stalled at the positions -39 and -5
The “minimal” 603 nucleosomal template for transcription by the RNAP contains a strong T7A1 promoter region with the start site localized 50 bp upstream of promoter-proximal boundary of one of the strong 147-bp 603 nucleosome-positioning sequences (30). Nucleosome positions on these templates are unique and were mapped with high resolution (32). The sequence of the template is designed to allow stalling of RNAP at different locations along the templates (30). The 603 template described here as an example; it allows stalling at the positions -39 and -5 (the numbers indicate positions of the active center of RNAP relative to promoter-proximal nucleosome boundary) using different combinations of NTPs (Fig. 2B (31, 33)).
All protein and DNA-protein complexes (E. coli RNAP and -H1 donor chromatin) were purified as described (2).
To form active initiation complexes, incubate 200 ng of nucleosomal templates or histone-free DNA with five-fold molar excess of the RNAP in 20 ul of the TB buffer containing 40 mM KCl (TB40) at 37°C for 10 min.
Take a small aliquot (2–3 ul) for FRET measurements (it contains the open complex, OC).
Add ApUpC RNA primer to 20 uM, and ATP and GTP to 1 uM to allow formation of the 11-nt labeled RNA transcript (EC-39 complex where the active center of the enzyme is positioned 39 bp upstream of promoter-proximal nucleosomal boundary) at 37°C for 10 min.
Add 20 ul of Ni2+-NTA agarose beads (50% suspension in alcohol) that was previously washed three times with 0.5 ml of TB40, incubated in the presence of 0.5 mg/ml of acetylated BSA (Sigma-Aldrich) for 10 min, and washed two times with 0.5 ml TB40.
Incubate obtained EC-39 with Ni2+-NTA agarose beads at room temperature for 5 min with gentle pipetting every 40–50 sec.
Spin the samples at 1600 rpm for 1 min at room temperature. Carefully take the supernatant.
Wash beads with the immobilized EC-39 complexes once with TB40, twice with TB300 and twice with TB40. Centrifuge after every wash on Eppendorf microcentrifugeat 1600 rpm for 1 min at room temperature.
Incubate the washed beads in 30 ul of TB150 buffer with 100 mM imidazole at room temperature for 5 min with gentle pipetting every 30 -40 sec.
Centrifuge on Eppendorf microcentrifuge at 1600 rpm for 1 min at room temperature. Carefully collect the supernatant.
Take a small aliquot (3–4 ul) for FRET measurements; it contains the elongation complex stalled at -39 position (EC-39).
Adjust volume to 20 ul. Incubate EC-39 with 1 uM CTP, 20 mg/ml rifampicin, 0.5 mg/ml acetylated BSA, 150 mM KCl at 37°C for 10 min to advance EC-39 to form EC-5 at the end of the -T DNA track on the upper DNA strand.
Take a small aliqoute (3–4 ul) for FRET measurements; it contains the elongation complex stalled at -5 position (EC-5).
Add 4mM rNTPs to 100 mM final concentration to start the chase reaction, incubate at room temperature for 10 min. The resulting reaction mixture contains post chase sample for FRET measurements(see Note 2).
3.4. spFRET measurements
spFRET analysis is performed in a diluted solution of nucleosomes or stalled elongation complexes using an experimental installation with key modules and elements shown in Fig. 4. Besides home-build experimental installations (24, 34, 35) any commercially available spectrometer for fluorescence correlation spectroscopy (FCS) or confocal microscope with the module for FCS can be used for spFRET measurements. FCS modules are usually equipped with avalanche-photodiodes (APDs), which surpass conventional photomultipliers installed in commercial confocal microscopes in sensitivity and suit ideally for spFRET measurements.
Figure 4. Experimental setup for the spFRET study of freely diffusing nucleosomes and their complexes.
Laser beam is directed to specimen through the objective using a wavelength selective mirror, which reflects light with the laser emission wavelength and transmit other wavelengths. Fluorescence of a specimen collected with the objective is filtered with the confocal diaphragm to reject signals coming from specimen layers situated above and below the focal point. As a result, the confocal diaphragm transmits a specimen signal coming from an ellipsoidal focal volume with main axes of ca. 0.3 and 1.5 μm (see insert at the top right showing the principle of single particle detection). Freely diffusing single nucleosomes labeled with donor and acceptor fluorophores pass through the focused laser beam (shown by green color), where their fluorescence is excited (shown by yellow star). Spatially filtered fluorescence is separated into two spectral parts (corresponding to donor and acceptor fluorescence) with the dichroic beam splitter, additionally spectrally filtrated with the longpass barrier filters (or band-pass filters) and registered with avalanche-photodiodes (APD). Insert at the bottom right: an example of fluorescence intensity dependences on time measured with two APDs. Green and red traces describe bursts of donor and acceptor fluorescence intensities, respectively, which arise when single nucleosomes diffuse through the focal volume. Intensities of these bursts are used to calculate FRET efficiencies (E) for each measured nucleosome.
We utilize the Confocor-3 module of the LSM710-Confocor3 laser scanning confocal microscope (Zeiss, Germany) to measure freely diffusing single nucleosomes and their complexes with RNAP. Measurements are performed using the C-Apochromat water-immersion 40× objective with the 1.2 numerical aperture (see Note 3). Fluorescence is excited with Ar+-ion laser using the 514.5 nm wavelength, allowing efficient absorption by Cy3. Intense absorption by Cy3 at the excitation wavelength allows reducing laser power to 2 μW (see Note 4). In our measurements a confocal diaphragm aperture is equal to the diameter of Airy disk in the projection of fluorescence emitting point object on the diaphragm. We utilize the 635 nm dichroic beam splitter and the 530 and 580 nm longpass barrier filters (Fig. 4) from a set of standard filters installed in Confocor3. Accordingly, fluorescent signals of Cy3 and Cy5 are collected within the 530–635 and 635–800nm wavelength ranges, respectively.
To study freely diffusing nucleosomes and their complexes with RNAP using spFRET technique, specimens are diluted with TB-PEG buffer to 0.1- 0.3 nM in order to have concentration of 0.1–0.3 particles per the focal volume (ca. 1 fL)( see Note 5).
Initiate measurement of Cy3 and Cy5 fluorescence intensities as a function of time (Fig. 4)( see Note 6).
Save the result of a measurement into a file.
Discriminate signals associated with nucleosomes, which are observed as a set of sharp spikes against a low intensity background in the measured traces (Fig. 4). Sharp fluctuations of signal intensity are assigned to fluorescence of single nucleosomes when they meet to the following threshold criterion: either Cy3- or Cy5- signal intensity is 5–10 times higher than the corresponding background level. Very intense signals should be rejected also, since they correspond to several nucleosomes which accidentally diffuse through the focal volume simultaneously( see Note 7).
Create a new file, where each identified nucleosome is characterized by the registered fluorescence intensities of Cy3 and Cy5.
-
Recalculate the set of Cy3 and Cy5 signals assigned to single nucleosomes into a set of FRET efficiencies (E) with a formula
(2) where IAa is intensity of Cy5 fluorescence in the acceptor channel, IDd is intensity of donor fluorescence in the donor channel (both corrected for background contribution),
α is the contribution of Cy3 fluorescence in the channel for Cy5 detection (spectral cross-talk) calculated as(3) where IDa is intensity of Cy3 fluorescence in the Cy5 detection channel corrected for background contribution (see Note 8).
Present the sets of FRET efficiencies calculated for the measured nucleosome samplings graphically as frequency distribution histograms and fit them with a sum of bands having a Gaussian shape( Fig. 5)( see Note 9).
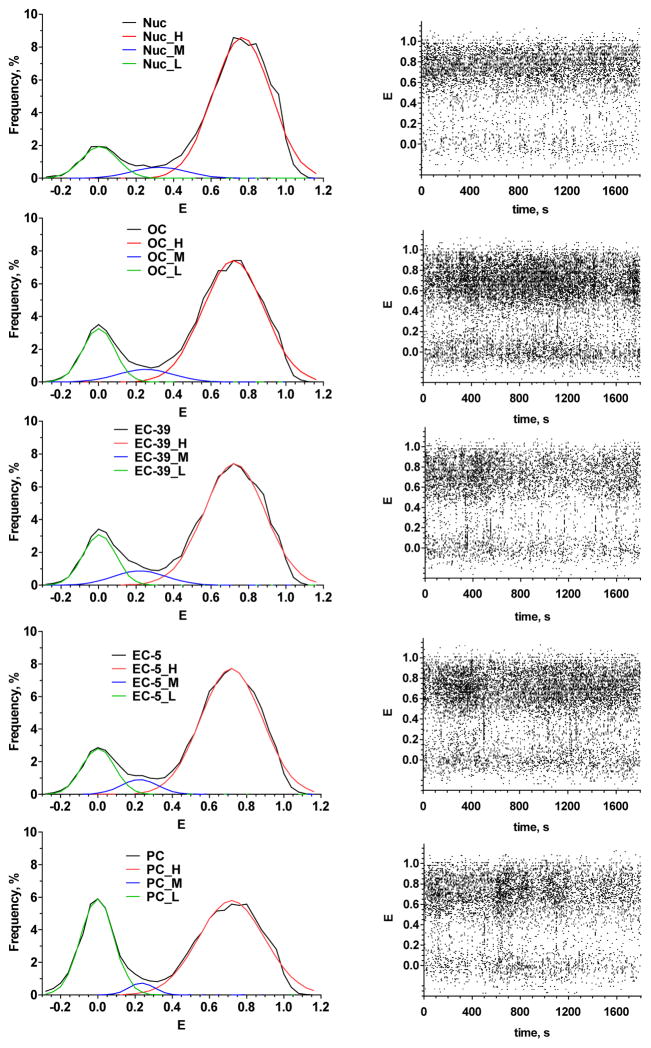
Figure 5. spFRET analysis of nucleosomes and their complexes with stalled RNAP.
Left column - frequency histograms of FRET efficiency (E) for nucleosomes (Nuc), open complex (OC), elongation complexes -39 (EC-39), -5 (EC-5) and post chase state (PC). Colored plots are the result of histogram deconvolution into three states: high FRET (H), middle FRET (M) and low FRET (L). Right column - time courses of FRET for nucleosomes and stalled complexes shown on the left. Dots represent E values measured for each single nucleosome (stalled complex) diffused through the focal volume in different moments of an observation period. High structural stability is an obvious property of the nucleosomes and stalled complexes.
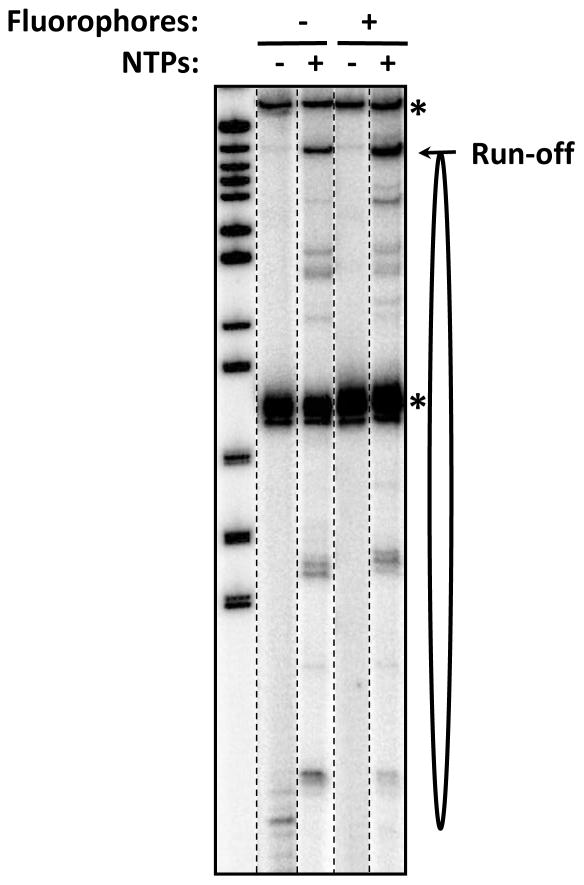
Figure 3. Analysis of transcription through 603 nucleosome containing intact and fluorophore-labeled DNA.
Transcription by the RNAP was conducted in the presence of NTPs for 30 seconds at 150 mM KCl. The locations of the 603 nucleosome (oval), labeled DNA (*) and the run-off transcript are shown. No additional pausing was detected on fluorophore-labeled DNA, suggesting that fluorophores do not interfere with progression of the enzyme.
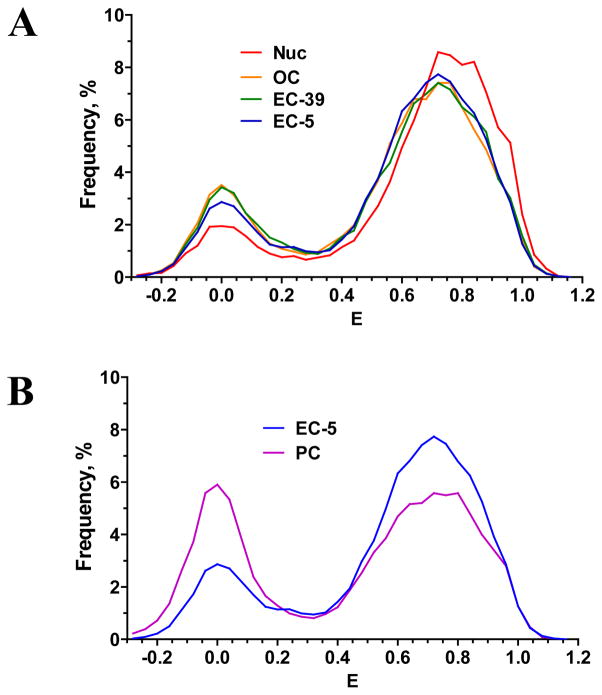
Figure 6. Comparison of frequency histograms of FRET efficiency (E).
for (A) nucleosomes (Nuc), open complex (OC), elongation complexes EC-39 and EC-5; (B) elongation complex EC-5 and post chasestate (PC).
Table 1.
Efficiencies of FRET and relative fractions (in parentheses) for three subpopulations (low-FRET, middle-FRET and high-FRET) observed in nucleosomes and their complexes with RNAP
| Low-FRET | Middle-FRET | High-FRET | |
|---|---|---|---|
| Nuc# | 0.00* (12%) | 0.32* (6%) | 0.77* (82%) |
| OC | 0.00 (18%) | 0.25 (6%) | 0.72 (76%) |
| EC-39 | 0.00 (17%) | 0.22 (7%) | 0.72 (76%) |
| EC-5 | 0.00 (14%) | 0.22 (6%) | 0.72 (80%) |
| PC | 0.00 (33%) | 0.23 (3%) | 0.72 (64%) |
Free nucleosomes.
The reported E values were calculated by fitting the histograms (Fig. 6) to a sum of three Gaussian distributions and finding their maxima
Acknowledgments
This work was supported by the NIH RO1 grant GM58650 to V.M.S.
Footnotes
All operations with fluorescently labeled oligonucleotides should be accompanied by minimal exposure to visible and UV light because the fluorophores are highly sensitive to light. The oligonuicleotides 3DW_Cy3_603-5A2 and 5DW_Cy5_603-5A2 were labeled by Cy3 and Cy5 at the positions 15 and 13, respectively.
The labeled RNA present in the sample should be characterized before and after transcription by denaturing PAGE (Fig. 3) to make sure that transcription is completed on the majority of the transcribed templates.
High numerical aperture of the objective is required to reduce the focal volume from which the fluorescent signal is recorded. Smaller focal volume enables higher concentration of nucleosomes to be used for spFRET analysis, which in turn promotes nucleosome structure stability. Signal intensity collected from a single nucleosome is proportional to the fourth power of the objective numerical aperture.
The lower laser power is important to diminish interference of Rayleigh scattering with Cy3 fluorescence and reduce probability of Cy5 photobleaching. High laser power can distort the E profile in frequency diagrams. This distortion is observed as a laser power-dependent shift of a high FRET value in an E-histogram to lower values.
Dilution should be performed directly in the well of a chambered cover glass to minimize the loss of nucleosomes because of their adsorption to the walls of the reaction tube. At the recommended concentration nucleosomes diffuse through the focal volume one by one, and signals from single nucleosomes coming to APDs are well separated in time (Fig. 1). Higher concentrations of nucleosomes result in an increase of the background signal and thus affect accuracy of FRET calculations. At lower concentrations the fraction of nucleosomes that are adsorbed on well walls and does not participate in the measurement is considerably increased .
It is recommended to perform recording of the signal as a function of time with 3–5 ms dwell time. Dwell time is selected to be roughly equal to an average time of nucleosome diffusion through the focal volume. Total measurement time is restricted by nucleosome structure stability and Cy5 photo stability. Under the described conditions measurements can be performed during at least 30 min (Fig. 5). Measurements during 10 min are sufficient to provide statistically reliable sampling (4000–8000 particles).
In our case, fluorescence spikes are assigned to single nucleosomes when either Cy3 -or Cy5- dependent signal intensities are higher than 10 or 5 kHz, respectively, and lower than 80 kHz. Signals having intensity higher than 80 kHz originate from two or sometimes several nucleosomes diffusing through the focal volume simultaneously. Spikes with intensities lower 10 and 5 kHz are enriched with Rayleigh scattering spikes and/or noise. Typical intensities of the background signals are ca. 0.9–1.1 and 0.4–0.6 kHz for Cy3 and Cy5 channels, respectively.
Fluorescence spectra of Cy3 and Cy5 overlap in the 635–700 nm region. Therefore fluorescence of Cy3 contributes to the measured signal of Cy5, and this contribution should be taken into account. Under our experimental conditions the spectral cross-talk α is equal to 0.19. Spectral cross-talk for Cy5 fluorescence in the 530–635 nm region as well as direct Cy5 excitation with the 514.5 nm wavelength are negligible.
Analysis of E histograms shows that both nucleosomes and stalled complexes OC, EC-39 and EC-5 contain three subpopulations of particles which differ in E (Fig. 5 and Table 1). These subpopulations can be described by E-values that correspond to the maxima of Gaussian bands utilized to decompose the histograms into particular states. The main subpopulation of particles (76–82%) is characterized by high FRET (high average E value): 0.77 for nucleosomes and 0.72 for OC, EC-39 and EC-5 (Table 1). These particles can be assigned to intact nucleosomes with the promoter-distal DNA (containing Cy3 and Cy5 labels) fully coiled on the surface of the histone octamer (Fig. 1C). The high values of FRET are in agreement with the expected short average distance between Cy3 (position +56) and Cy5 (position +135) in a fully assembled nucleosome (ca. 19 Å). The second subpopulation of particles has low FRET value (E=0.00), is observed in each specimen and contains 12–18% of particles (Table 1). This subpopulation most likely consists of nucleosomes containing DNA that is partially uncoiled from the octamer and histone-free DNA. Minor subpopulation of particles (6–7%) is characterized by intermediate average FRET efficiencies: 0.32 for nucleosomes and 0.22–0.25 for OC, EC-39 and EC-5 (Table 1). It has been suggested that these FRET efficiencies are characteristic for nucleosomes formed after dissociation of an H2A/H2B histone dimer (36). These are so called hexasomes (37); they have partially unwrapped DNA end and, as a result, are characterized by the increased distance between Cy3 and Cy5.
A remarkable feature of the studied nucleosomes, OC, EC-39 and EC-5 is their stability. Even at a high ionic strength (i.e. in the buffer containing 150 mM KCl) the equilibrium between high, middle and low FRET states is preserved during at least 30 min of measurements (Fig. 5). Thus we do not observe the instability detected for nucleosomes formed on similar 601 DNA sequences in single-molecule experiments at NaCl concentrations higher 100 mM because of rapid nucleosome dissociation (36). Both intact nucleosomal templates and their complexes with RNAP have comparable high stability.
The high FRET states of OC, EC-39 and EC-5 are very similar, but they are slightly different from the high FRET state of free nucleosomes (Table 1, Fig. 6A). Thus RNAP binding to the templates outside of the nucleosome core affects DNA packing in the region of Cy3 and Cy5 location. Bulk RNAP molecule likely sterically interferes with the binding of distal DNA end to the octamer and induces slight DNA unwrapping, thus increasing the distance and decreasing the efficiency of FRET between Cy3 and Cy5. The stalled complexes are also characterized by a small increase in the low FRET fraction and proportional decrease in the high FRET subpopulation (Table 1, Fig. 6A)
EC-5 complexes subjected to transcription are converted by RNAP into post chase (PC) state which is characterized by the presence of three similar subpopulations: low (E=0.00), middle (E=0.23) and high (E=0.72) FRET (Fig. 4, Table 1). According to biochemical data more than 95% of EC-5 complexes are transcribed (not shown). spFRET analysis reveals that ca. 16% of nucleosomes are converted from high to low FRET state after transcription (Fig. 6B). The fraction of middle FRET nucleosomes is also decreased (Table 1). The remaining 64% of nucleosomes preserve compact DNA packing which is similar to that of the EC-5. The data suggest that, consistent with the biochemical data (31), a large fraction of nucleosomes survives during transcription.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Gaykalova DA, Kulaeva OI, Pestov NA, Hsieh FK, Studitsky VM. Experimental analysis of the mechanism of chromatin remodeling by RNA polymerase II. Methods Enzymol. 2012;512:293–314. doi: 10.1016/B978-0-12-391940-3.00013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaykalova DA, Kulaeva OI, Bondarenko VA, Studitsky VM. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol Biol. 2009;523:109–123. doi: 10.1007/978-1-59745-190-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter W, Studitsky VM. Construction analysis, and transcription of model nucleosomal templates. Methods. 2004;33:18–24. doi: 10.1016/j.ymeth.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Walter W, Kashlev M, Studitsky VM. Transcription through the nucleosome by mRNA-producing RNA polymerases. Methods Enzymol. 2004;377:445–460. doi: 10.1016/S0076-6879(03)77029-3. [DOI] [PubMed] [Google Scholar]
- 6.Beard BC, Smerdon MJ. Analysis of DNA repair on nucleosome templates. Methods Enzymol. 2004;377:499–507. doi: 10.1016/S0076-6879(03)77032-3. [DOI] [PubMed] [Google Scholar]
- 7.Teng Y, Yu S, Reed SH, Waters R. Lux ex tenebris: nucleotide resolution DNA repair and nucleosome mapping. Methods. 2009;48:23–34. doi: 10.1016/j.ymeth.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Rowe CE, Narlikar GJ. The ATP-dependent remodeler RSC transfers histone dimers and octamers through the rapid formation of an unstable encounter intermediate. Biochemistry. 2010;49:9882–9890. doi: 10.1021/bi101491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hota SK, Bartholomew B. Approaches for studying nucleosome movement by ATP-dependent chromatin remodeling complexes. Methods Mol Biol. 2012;809:367–380. doi: 10.1007/978-1-61779-376-9_25. [DOI] [PubMed] [Google Scholar]
- 10.Andrews AJ, Luger K. A coupled equilibrium approach to study nucleosome thermodynamics. Methods Enzymol. 2011;488:265–285. doi: 10.1016/B978-0-12-381268-1.00011-2. [DOI] [PubMed] [Google Scholar]
- 11.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 12.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 13.Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the '601' strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 14.Mizuguchi G, Wu WH, Alami S, Luk E. Biochemical assay for histone H2A.Z replacement by the yeast SWR1 chromatin remodeling complex. Methods Enzymol. 2012;512:275–291. doi: 10.1016/B978-0-12-391940-3.00012-3. [DOI] [PubMed] [Google Scholar]
- 15.Cirillo LA, Zaret KS. Preparation of defined mononucleosomes dinucleosomes, and nucleosome arrays in vitro and analysis of transcription factor binding. Methods Enzymol. 2004;375:131–158. doi: 10.1016/s0076-6879(03)75009-5. [DOI] [PubMed] [Google Scholar]
- 16.Pennings S, Meersseman G, Bradbury EM. Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 17.Meersseman G, Pennings S, Bradbury EM. Mobile nucleosomes--a general behavior. EMBO J. 1992;11:2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studitsky VM, Clark DJ, Felsenfeld G. Structural Biology: the state of art. Adenine Press; 1994. Mechanism of nucleosome displacement by a transcribing polymerase; pp. 125–131. [Google Scholar]
- 19.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II. Loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 20.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 21.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 22.Simon RH, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owen-Hughes T, Utley RT, Steger DJ, West JM, John S, Cote J, Havas KM, Workman JL. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol Biol. 1999;119:319–331. doi: 10.1385/1-59259-681-9:319. [DOI] [PubMed] [Google Scholar]
- 24.Buning R, van Noort J. Single-pair FRET experiments on nucleosome conformational dynamics. Biochimie. 2010;92:1729–1740. doi: 10.1016/j.biochi.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Choy JS, Lee TH. Structural dynamics of nucleosomes at single-molecule resolution. Trends Biochem Sci. 2012;37:425–435. doi: 10.1016/j.tibs.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JY, Lee TH. Effects of histone acetylation and CpG methylation on the structure of nucleosomes. Biochim Biophys Acta. 2012;1824:974–982. doi: 10.1016/j.bbapap.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JY, Lee TH. Effects of DNA methylation on the structure of nucleosomes. J Am Chem Soc. 2012;134:173–175. doi: 10.1021/ja210273w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozov AV, Fortney K, Gaykalova DA, Studitsky VM, Widom J, Siggia ED. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37:4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh FK, Fisher M, Ujvari A, Studitsky VM, Luse DS. Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep. 2010;11:705–710. doi: 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S, Lee J, Hohng S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS One. 2010;5:e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widengren J, Kudryavtsev V, Antonik M, Berger S, Gerken M, Seidel CA. Single-molecule detection and identification of multiple species by multiparameter fluorescence detection. Anal Chem. 2006;78:2039–2050. doi: 10.1021/ac0522759. [DOI] [PubMed] [Google Scholar]
- 36.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Toth K, Langowski J, Seidel CA. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc Natl Acad Sci U S A. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]