Abstract
MicroRNAs are consistently reported to regulate gene expression in all cancer cell types by modulating a wide range of biological processes, including cell proliferation, differentiation, and apoptosis, which are associated with tumor development and progression. Previous studies have revealed that miR-490-3p regulates cell proliferation and apoptosis in cancers, such as hepatocellular carcinoma, lung cancer, bladder cancer, and ovarian carcinoma. In this study, we explored the hitherto unrevealed role of miR-490-3p in breast cancer. We tested miR-490-3p expression in breast cancer tissue and paracarcinoma tissue using reverse transcription–polymerase chain reaction. We also transfected the human breast cancer cell lines MCF-7 and T47D with miR-490-3p; subsequently, we determined the cell phenotype and the expression of Ras homolog gene family member A (RhoA), Bcl-xL, matrix metalloproteinase-9, and P70S6K (P70S6 kinase). Dual-luciferase reporter assay and a xenograft mouse model were used to reveal the roles of miR-490-3p and its target gene RHOA. We found that the levels of miR-490-3p were lower in the breast cancer tissue than in the paracarcinoma tissues. The overexpression of miR-490-3p suppressed breast cancer cell proliferation and promoted early stage apoptosis. Western blotting results revealed that the miR-490-3p overexpression reduced RhoA, Bcl-XL, matrix metalloproteinase-9, and P70S6K protein expression. The dual-luciferase reporter assay confirmed that RhoA is a target of miR-490-3p. The xenograft mouse model confirmed that miR-490-3p overexpression suppressed tumor growth and reduced RhoA expression. Our results indicate that miR-490-3p acts as oncosuppressive microRNA to inhibit breast cancer tumorigenesis and progression by targeting RhoA directly. It may contribute to breast cancer diagnosis and treatment.
Keywords: breast cancer, miR-490-3p, RhoA, tumorigenesis and progression
Introduction
Breast cancer is one of the most prevalent cancers and is ranked the second leading cause of cancer-related mortality in females worldwide.1,2 In recent years, the incidence of breast cancer has increased, accounting for nearly 22.9% of all female cancers worldwide.3,4 Diagnosis and treatment in the early stage are of great importance. Despite the impressive achievements in understanding breast cancer development and progression in the past decades, the underlying molecular mechanism remains elusive.
Ras homolog gene family member A (RhoA) is a small (~22 kDa) G protein/guanosine triphosphatase that affects tumor invasion and migration in various cancer cells.5–8 Overexpression of RhoA has been detected in some human tumors in which RhoA levels correlate with breast cancer stage.9 Studying the regulatory mechanism of RhoA and developing treatments targeting RhoA may be effective for treating breast cancer tumorigenesis and progression.
MicroRNAs are small, noncoding, single-stranded RNAs that inhibit gene expression in all cell types by directly binding to the 3′ untranslated region (3′ UTR) of target mRNAs and modulating mRNA degradation or inhibiting translation.10,11 The predicted seed region in the 3′ UTR of RhoA showed that RhoA is a direct target of miR-490-3p. Studies have suggested that miR-490-3p is downregulated in certain cancers and acts as a tumor suppressor. MiR-490-3p is a regulator in hepatocellular carcinoma cells, inhibiting cell proliferation, migration, and invasion.12 Epigenetic silencing of miR-490-3p promoted Helicobacter pylori-induced gastric carcinogenesis;13 miR-490-3p overexpression inhibits the proliferation of A549 lung cancer cells.14 However, the influence of miR-490-3p on breast cancer and the relationship between RhoA and miR-490-3p in breast cancer development and progression remain unknown. Our study reveals this in detail and is the first time such findings have been reported.
Materials and methods
Cell culture and transfection
The breast cancer cell lines MCF-7 and T47D were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 100 µL fetal bovine serum (FBS), 10 µL penicillin, and 10 µL streptomycin per mL medium at 37°C with 5% CO2. The medium was changed every 2 days.
The breast cancer cells were transiently transfected with miR-490-3p mimic (sense strand sequence: 5′-CAACCUGGAGGACUCCAGCG-3′) or mock control (sense strand sequence: 5′-ACUACUGAGUGACAGUAGA-3′) using Lipofectamine 2000 according to the manufacturer’s instructions.
Cell viability assay
We used the tetrazolium (MTT [3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide]) assay to measure cell viability spectrophotometrically. Cells (3×103) were seeded in a 96-well plate, 0, 24, 48, and 72 hours after adherence, 20 µL MTT solution was added to each well, and the cells were incubated at 37°C in 5% CO2. After 4 hours, 150 µL dimethyl sulfoxide was added to the wells and the absorbance measured at 490 nm spectrophotometrically within an hour. The optical density value obtained was related to the level of cell viability, that is, the number of live cells.
Cell cycle analysis
The cells were cultured at 37°C in 5% CO2 for 48 hours, digested by trypsinization, washed in phosphate-buffered saline (PBS) twice, and fixed in 4 mL 75% cold ethanol for over 2 hours. Then, the cells were washed twice in PBS and stained with 500 µL propidium iodide (PI; BD Biosciences, Franklin Lakes, NJ, USA) in the dark at room temperature, and then the PI signal was determined using flow cytometry.
Apoptosis assay
The cells were stained with PI and fluorescein isothiocyanatelabeled annexin V (KeyGen, Nanjing, People’s Republic of China), and then phosphatidylserine externalization, an indicator of early apoptosis, was determined according to the manufacturer’s instructions. Briefly, the cells were cultured at 37°C in 5% CO2 for 48 hours, washed twice in cold PBS, and resuspended in 200 µL 1× binding buffer. Then, 300 µL binding buffer, 5 µL PI, and 5 µL annexin V–fluorescein isothiocyanate were added to each tube, and the samples were assayed by flow cytometry in an hour.
Wound healing assay
We transfected the cells with miR-490-3p mimic or mock, seeded 1×106 cells in six-well plates, and cultured them for 24 hours with mitomycin. The cell monolayer was scratched using a 200 µL pipette tip, the wells were washed with PBS, and the cells were cultured in FBS-free medium. Photographs of the wounds were captured at 0, 24, and 48 hours; the wound healing rate was measured using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The wound healing rate = (area of original wound − area of actual wound at stated intervals)/area of original wound ×100%.
Cell invasion assay
Matrigel (40 µL from 8 mg/mL stock solution; Becton Dickinson Labware, Bedford, MA, USA) was used to coat the top chamber of 6.5 mm Transwell chambers (8 µm pore size; BD Biosciences, San Jose, CA, USA) and incubated at 37°C in 10% CO2 for 4 hours. Then, DMEM containing 10% FBS was added to the bottom chamber while FBS-free DMEM was added to the top chamber, in which 5×104 cells per well had been seeded. After 48 hours, cells that had not passed through the Matrigel were removed using cotton swabs. Cells that had passed through the Matrigel to the underside of the filters were fixed with methanol and stained with 0.1% crystal violet. The number of cells was observed and counted using an Olympus fluorescence microscope (Olympus Corporation, Tokyo, Japan) and was used to determine the invasive ability of the tumor cells. Five separate fields were counted for every filter.
Real-time reverse transcription–polymerase chain reaction
Total RNA was extracted from the breast cancer tissues or cell lines. Real-time reverse transcription–polymerase chain reaction was carried out using 2 µg RNA with AMV reverse transcriptase and random primers. MiR-490-3p primer (forward: 5′-CAACCTGGAGGACTCCATGC-3′; Takara, Kyoto, Japan) was certified by GenBank. Amplification of complementary DNA was carried out using a SYBR Premix Ex Taq II kit, and 18S rRNA (forward: 5′-ACGGACAGGATTGACAGATT-3′) was used as the internal control according to the manufacturer’s instructions. The expression level of the related gene was measured using the comparative threshold cycle method (2-ΔΔCt).
Western blot analysis
Western blotting was carried out using the Bradford method via a Bio-Rad protein assay kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis on 10% acrylamide gels was used to separate the denatured proteins, which were then transferred to Hybond membranes (Amersham, Freiburg, Germany). The membranes were blocked overnight with 5% skimmed milk in Tris-buffered saline with Tween 20 (10 mM Tris–HCl, 150 mM NaCl, 0.1% Tween 20). For immunoblotting, the membranes were cultured at 4°C overnight with the primary antibodies RhoA, P70s6K, Bcl-xL, and matrix metalloproteinase-9 (MMP-9) (1:500, ProteinTech, Chicago, IL, USA), rinsed with Tris-buffered saline with Tween 20, and cultured with antirabbit or antimouse immunoglobulin G antibodies conjugated to horseradish peroxidase (Dako, Carpinteria, CA, USA) at 1:5,000 dilution for 2 hours at room temperature. After applying electrochemiluminescence-Plus detection reagents (Santa Cruz Biotechnology Inc., Dallas, TX, USA), the protein bands were observed using X-ray film. Glyceraldehyde-3-phosphate (1:2,000; Proteintech) was used as the internal control.
Dual-luciferase reporter assay
HEK293 cells were seeded in 24-well plates in DMEM containing 10% FBS and cultured for 24 hours. The RhoA 3′ UTR containing the putative miR-490-3p binding site was designed and inserted downstream of the firefly luciferase reporter (Promega Corporation, Fitchburg, WI, USA) according to the human RHOA mRNA sequence in GenBank. The cultures were transiently cotransfected with 50 nM miR-490-3p mimic or mock and 600 ng dual-luciferase vectors (containing either wild type or mutant 3′ UTR). After 48 hours, the luciferase activity was determined using a dual-luciferase reporter assay system.
In vivo xenografts
Female BALB/c nude mice aged 4–5 weeks and weighing 20 g were purchased from Shanghai SLAC Laboratory Animal, Co., Ltd. (Shanghai, People’s Republic of China) and kept in a pathogen-free environment. Subcutaneous tumor xenografting was performed by subcutaneously injecting the mice with 200 µL PBS containing 1×107 exponential-growth MCF-7 cells15 that had been transfected with mock or miR-490-3p mimic. All animal procedures were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the China Medical University Animal Care and Use Committee.
Patients and tissues
A total of 137 cases of surgical resected breast cancer samples were collected from the Tumor Hospital of Liaoning Province (Shenyang, People’s Republic of China) between January 2007 and December 2013. All of the carcinomas had been histologically confirmed as invasive breast cancer according to the criteria of the World Health Organization and the molecular subtypes of breast carcinoma were identified. Paracarcinoma tissues were taken >2 cm away from the carcinomas according to Li et al.16 Written informed consent was provided by each participant. The study protocols conformed to the standards set by the Declaration of Helsinki and were approved by China Medical University Ethics Committee.
Immunohistochemistry
Consecutive tissue sections were deparaffinized using xylene, rehydrated with alcohol, and subjected to antigen retrieval by heating in target retrieval solution (Dako) for 15 minutes in a microwave oven (Oriental RotorLmt. Co., Tokyo, Japan). We used 3% hydrogen peroxide to quench the sections for 20 minutes to block endogenous peroxidase activity, and 5% bovine serum albumin was added to block nonspecific binding for 5 minutes. The sections were incubated with RhoA antibodies at 4°C overnight, and then incubated with horseradish peroxidase-conjugated antirabbit antibodies for 2 hours. Finally, the slides were washed with Tris-buffered saline with Tween 20 three times and the binding sites were stained using 3,3′-diaminobenzidine. After counterstaining with Mayer’s hematoxylin, the sections were dehydrated, cleared, and mounted.
Statistical analysis
Statistical evaluation was carried out using Spearman’s rank correlation coefficient to analyze ranked data, and the Mann–Whitney U-test was used to differentiate the mean of different groups. A P-value of <0.05 was considered statistically significant. SPSS v 17.0 (SPSS Inc., Chicago, IL, USA) was used to analyze the data.
Results
Overexpression of miR-490-3p inhibits breast cancer carcinogenesis
MiR-490-3p expression was significantly higher in para-carcinoma tissues than in the breast cancer tissues (Figure 1, P<0.05), proving that miR-490-3p suppresses breast cancer pathogenesis. Details can be found in Table S1.
Figure 1.
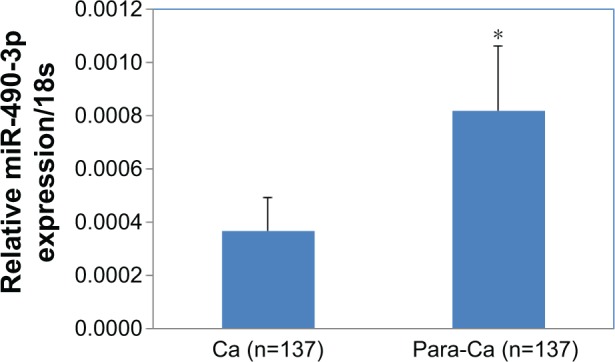
Overexpression of MIR4903P inhibits breast cancer carcinogenesis.
Notes: MiR-490-3p expression was significantly lower in breast cancer tissue than in the paracarcinoma tissue. *P<0.05.
MiR-490-3p inhibits breast cancer cell proliferation and migration in vitro
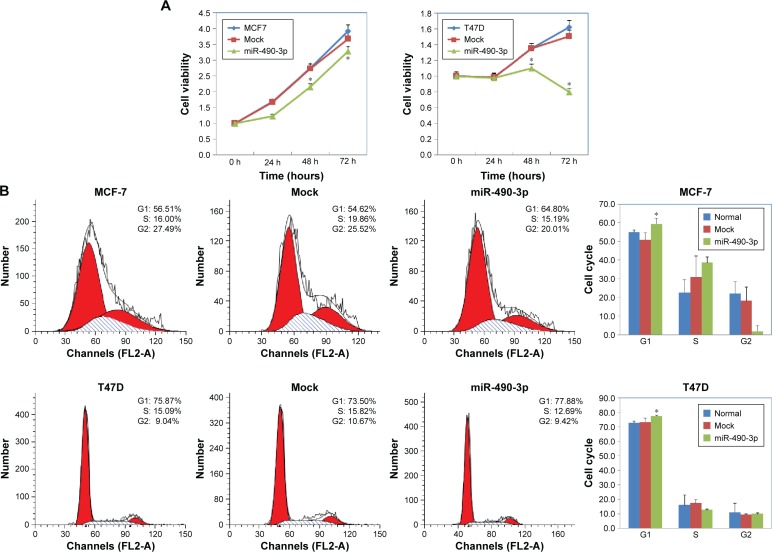
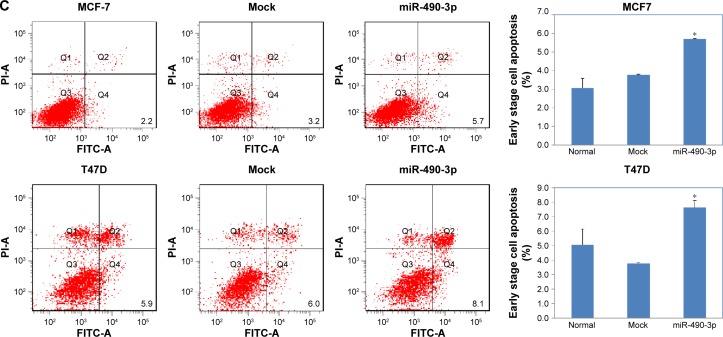
In order to check whether miR-490-3p influenced breast cancer cell proliferation and migration, we transfected MCF-7 and T47D cell lines with miR-490-3p mimics. Compared with the negative control and mock-transfected cells, the miR-490-3p-transfected cells grew markedly slower (Figure 2A, P<0.05). PI staining revealed that the miR-490-3p transfection induced G1 arrest (Figure 2B, P<0.05). Flow cytometry revealed that miR-490-3p transfection induced early stage apoptosis (Figure 2C, P<0.05). The wound healing assay (Figure 3A, P<0.05) and cell invasion assay (Figure 3B, P<0.05) showed that miR-490-3p transfection inhibited the migratory and invasive capability of the cancer cells.
Figure 2.
MiR-490-3p transfection in breast cancer cells inhibits cell proliferation and induces G1 arrest and apoptosis.
Notes: T47D and MCF-7 cells transfected with miR-490-3p (A) grew significantly slower, and (B) G1 arrest and (C) early stage apoptosis were induced as compared with the negative control and mock-transfected cells. Each result represents the mean of three independent experiments. *P<0.05.
Figure 3.
MiR-490-3p transfection in breast cancer cells inhibits cell migration and invasion ability.
Notes: MCF-7 and T47D cells transfected with miR-490-3p had decreased (A) migration ability and (B) invasion ability as compared with normal and mock-transfected cells. Each result represents the mean of three independent experiments. *P<0.05. Magnification: (A) ×4; (B) ×10.
MiR-490-3p targets RhoA and influences RhoA downstream factors directly in vitro
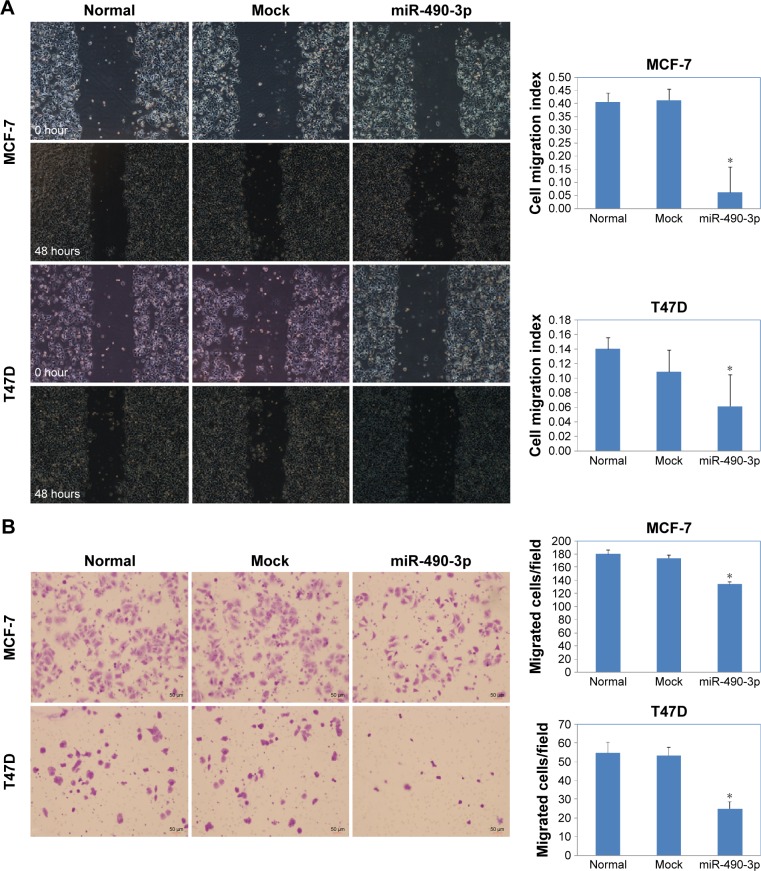
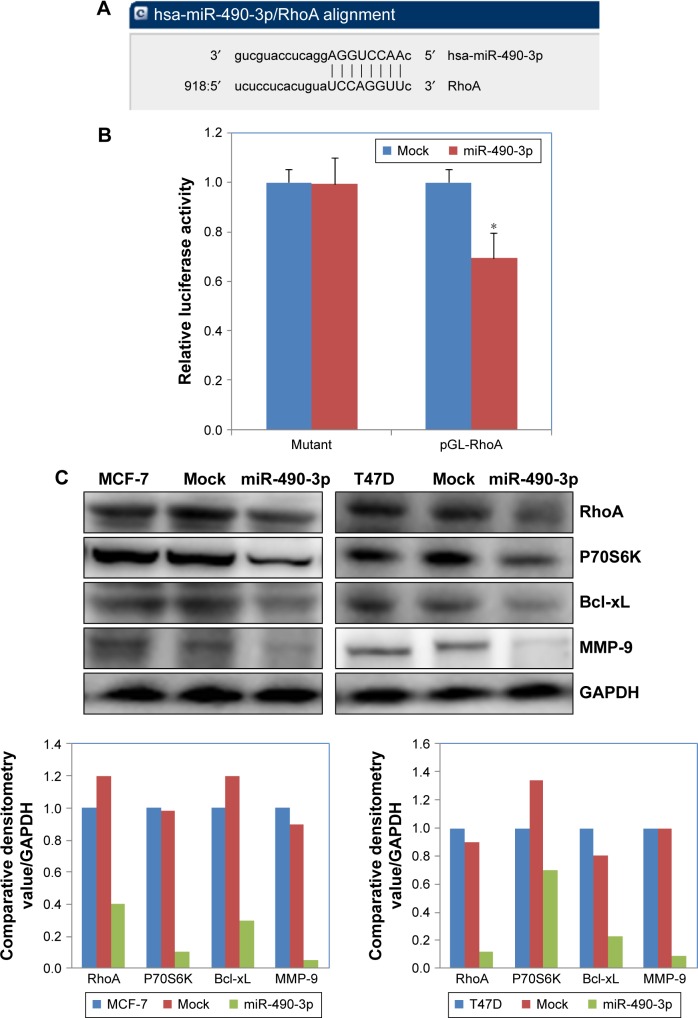
We searched microRNA.org (http://www.microrna.org) and found that the predicted binding site of miR-490-3p is the RhoA 3′ UTR (Figure 4A). The dual-luciferase reporter assay proved this prediction (Figure 4B, P<0.05). Western blotting results showed that miR-490-3p overexpression reduced RhoA, Bcl-xL, MMP-9, and P70S6 kinase (P70S6K) protein expression (Figure 4C).
Figure 4.
MiR-490-3p targets RhoA and influences RhoA downstream factors directly in vitro.
Notes: (A) The predicted seed region in the 3′ UTR of RhoA proved that RhoA is a direct target of miR-490-3p. (B) Dual-luciferase reporter assay showing that miR-490-3p targeted RhoA directly by binding its 3′ UTR. (C) MiR-490-3p overexpression reduced RhoA, Bcl-xL, MMP-9, and P70S6K protein expression. Each result represents the mean of three independent experiments. *P<0.05.
Abbreviations: MMP-9, matrix metalloproteinase-9; RhoA, Ras homolog gene family member A; UTR, untranslated region.
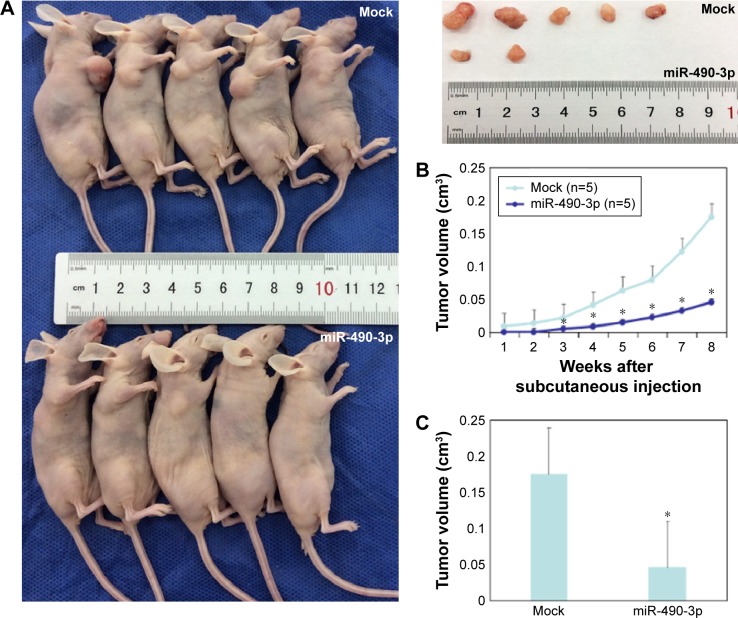
MiR-490-3p inhibits breast cancer tumorigenesis and progression in vivo
To investigate the effects of miR-490-3p on breast cancer tumorigenesis and progression in vivo, we transfected nude mice with miR-490-3p-stable expressing MCF-7 cells. The tumor volumes in the miR-490-3p tumor xenograft mice were smaller than that in the mock tumor xenograft mice (Figure 5A and C, P<0.05). After 2 weeks, the tumor growth in the miR-490-3p tumor xenograft mice was obviously slower than that in the mock tumor xenograft mice (Figure 5B, PWEEK2 <0.05, Pdeviation of the tumor xenografts <0.01).
Figure 5.
MiR-490-3p inhibits breast cancer tumorigenesis and progression in vivo.
Notes: (A) The miR-490-3p tumor xenografts were smaller than the mock tumor xenografts. (B) MiR-490-3p tumor growth was obviously slower than that in the mock tumor xenografts after 2 weeks and (C) the deviation of the tumor xenografts was significantly increased in the following period. *P<0.05, compared with Mock group.
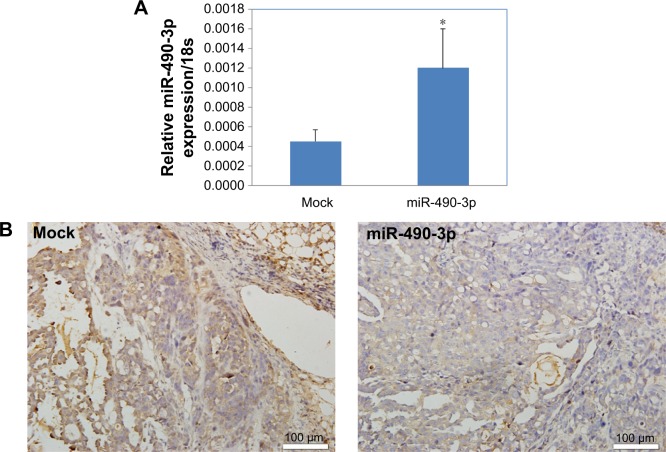
MiR-490-3p downregulates RhoA expression in tumor xenografts
Immunohistochemistry revealed that the RhoA expression levels in the miR-490-3p overexpression tumor xenografts were lower than those in the mock xenografts (Figure 6A and B).
Figure 6.
MiR-490-3p downregulates RhoA expression in tumor xenografts.
Notes: RhoA expression levels in the miR-490-3p overexpression (A) tumor xenografts were lower (B) as compared with that in the mock tumor xenografts. Magnification: (B) ×20. *P<0.05, compared with Mock group.
Abbreviation: RhoA, Ras homolog gene family member A.
Discussion
An increasing amount of evidence suggests that microRNAs can affect multiple cancers by altering base pairing to the 3′ UTR of the target mRNAs leading to mRNA degeneration or inhibition of translation.10 It has been predicted that mRNAs will be diagnostic and prognostic molecular bio-markers and therapeutic targets in cancer in the future.
It has been reported that miR-490 acts as an inhibitor in certain cancers. In osteosarcoma, miR-490-3p overexpression decreased cell proliferation and induced G1 arrest and apoptosis by targeting HMGA2 (high mobility group AT-hook 2).17 MiR-490 is also a novel tumor suppressor that targets c-FOS (cellular FBJ murine osteosarcoma viral oncogene homolog) in human bladder cancer.18 MiR-490-3p targets cyclin-dependent kinase 1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression.19 Epigenetic silencing of miR-490-3p reactivates the chromatin remodeler SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 1 to promote H. pylori-induced gastric carcinogenesis.13 We found that miR-490-3p was markedly downregulated in breast cancer tissues in comparison with paracarcinoma tissues. The cells transfected with miR-490-3p grew markedly slower and PI staining, flow cytometry, wound healing, and cell invasion assays revealed that miR-490-3p transfection induced G1 arrest, early stage apoptosis, and inhibited cell migration and invasion. Tumor xenografting revealed that miR-490-3p transfection reduced tumor volume and speed of growth. Our results indicate that miR-490-3p acts as a tumor suppressor in breast cancer, which is consistent with the other researchers who also reported that miR-490-3p acts as a tumor suppressor.12–14,17–19
The bioinformatics database predicted that miR-490-3p would bind with the 3′ UTR of RHOA mRNA, which revealed that RhoA is a direct target of miR-490-3p. Furthermore, miR-490 overexpression downregulated RhoA expression. The dual-luciferase reporter assay proved that miR-490-3p affects RhoA directly. In vivo, miR-490-3p inhibited tumor growth and reduced RhoA expression in the miR-490-3p tumor xenografts in comparison with the mock tumor xenografts. These results support the premise that miR-490-3p suppresses breast cancer proliferation and progression by targeting RhoA.
Current studies tend to suggest that RhoA affects tumor metastasis, regulating the action of downstream effectors. Silencing RhoA in OVCAR3 cells inhibited cell proliferation, metastasis, invasion, and apoptosis and reduced AKT, P70S6K, Bcl-xL, survivin, and vascular endothelial growth factor mRNA and protein expression.20 Silencing RhoA in tongue cancer cells inhibited cell migration and invasion and proliferation and reduced MMP-9, β-catenin, and cyclin D1/2 expression while increasing p27Kip1 and p21CIP1/WAF1 expression.21
Furthermore, our results also show that miR-490-3p over-expression reduced Bcl-xL, MMP-9, and P70S6K protein expression. Bcl-xL, MMP-9, and P70S6K are downstream activating sites of RhoA.22 The MMP family proteins play roles in the breakdown of extracellular matrix by lysing the basal membrane collagen and inducing MMP synthesis during tissue remodeling in the normal physiological state or in disease development.23,24 These articles reported that MMP-9 protein content in spent medium and mRNA expression activated RhoA and that colonic epithelial cell migration varied inversely in relation to ASB (5′-aminolevulinate synthase 2) expression and thereby directly with C4S.24 RhoA may affect the relevant genes and phosphorylation of the downstream P70S6K. P70S6K is a downstream effector of the phosphati-dylinositol 3-kinase/AKT signal transduction pathway; it affects protein synthesis and cell proliferation. The P70S6K/P85S6K pathway affects the G1/S checkpoint.22 This implies that miR-490-3p may decrease cell proliferation, inducing G1 arrest by regulating P70S6K while inhibiting apoptosis by downregulating the anti-apoptotic protein Bcl-xL.
In conclusion, we demonstrate for the first time that miR-490-3p inhibits breast cancer tumorigenesis and progression by targeting RhoA and regulating Bcl-xL, MMP-9, and P70S6K expression. This is a different and significant understanding of the molecular mechanism of breast cancer. MiR-490-3p is a promising new strategy for breast cancer diagnosis and treatment.
Supplementary material
Table S1.
miR-490 expression in breast cancer and paracarcinoma tissues
| Groups | N | miR-490 expression/18s | P-value |
|---|---|---|---|
| Breast cancer | 137 | 0.000368±0.001433 | 0.05 |
| Paracarcinoma | 137 | 0.000817±0.002862 |
Notes: The fold-difference of para samples to breast cancer was 2.22. Statistically significant values are shown in bold (P<0.05).
Acknowledgments
This work was supported by grants from the Hi-Tech Research Development Program of China (863 Program) (grant number 2006AA02Z493) and the National Natural Science Foundation of China (grant numbers 81071900, 81172199).
Footnotes
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med. 2013;274(2):113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61:409–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 4.Hong W, Dong E. The past, present and future of breast cancer research in China. Cancer Lett. 2014;351:1–5. doi: 10.1016/j.canlet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Yu LL, Gu JY. Advances in the role of Rho sub-family in tumor invasion. Fudan Univ J Med Sci. 2010;37:617–619. [Google Scholar]
- 6.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 7.Der Mardirossian C, Bokoch GM. GDIs: Central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Pasquinelli AE. MicroRNAs and their target: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 9.Bouzahzah B, Albanese C, Ahmed F, et al. Rho family GTPases regulate mammary epithelium cell growth and metastasis through distinguishable pathways. Mol Med. 2001;7(12):816–830. [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang LY, Liu M, Li X, Tang H. miR-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3) J Biol Chem. 2013;288(6):4035–4047. doi: 10.1074/jbc.M112.410506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen J, Xiao Z, Wu WK, et al. Epigenetic silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1 to promote Helico-bacter pylori-induced gastric carcinogenesis. Cancer Res. 2015;75(4):754–765. doi: 10.1158/0008-5472.CAN-14-1301. [DOI] [PubMed] [Google Scholar]
- 14.Gu H, Yang T, Fu S, Chen X, Guo L, Ni Y. MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochem Biophys Res Commun. 2014;444(1):104–108. doi: 10.1016/j.bbrc.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Kuzu OF, Nguyen FD, Noory MA, Sharma A. Current state of animal (Mouse) modeling in melanoma research. Cancer Growth Metastasis. 2015;8(Suppl 1):81–94. doi: 10.4137/CGM.S21214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Xie XB, Chen Q, et al. MiRNA-15a mediates cell cycle arrest and potentiates apoptosis in breast cancer cells by targeting synuclein-γ. Asian Pac J Cancer Prev. 2014;15(16):6949–6954. doi: 10.7314/apjcp.2014.15.16.6949. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Liu H, Li T, Xu G. miR-490-3p decreased cell proliferation, induced G1 arrest and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589(20):3148–3153. doi: 10.1016/j.febslet.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Lan G, Yang L, Xie X, Peng L, Wang Y. MicroRNA-490-5p is a novel tumor suppressor by targeting c-FOS in human bladder cancer. Arch Med Sci. 2015;11(3):561–569. doi: 10.5114/aoms.2015.52359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Chen X, Xiu YL, Sun KX, Zhao Y. MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362(1):122–130. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Wang J, Gou WF, et al. The involvement of RhoA and Wnt-5a in the tumorigenesis and progression of ovarian epithelial carcinoma. Int J Mol Sci. 2013;14(12):24187–24199. doi: 10.3390/ijms141224187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan G, Zou R, Chen Z, et al. Silencing RhoA inhibits migration and invasion through Wnt/β-catenin pathway and growth through cell cycle regulation in human tongue cancer. Acta Biochim Biophys Sin (Shanghai) 2014;46(8):682–690. doi: 10.1093/abbs/gmu051. [DOI] [PubMed] [Google Scholar]
- 22.Brinckerhoff CE, Rutter JL, Benbow U. Interstitial collagenases as markers of tumor progression. Clin Cancer Res. 2000;6:4823–4830. [PubMed] [Google Scholar]
- 23.Van Themsche C, Potworowski EF, St-Pierre Y. Stromelysin-1 (MMP-3) is inducible in T lymphoma cells and accelerates the growth of lymphoid tumors in vivo. Biochem Biophys Res Commun. 2004;315:884–891. doi: 10.1016/j.bbrc.2004.01.144. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharyya S, Tobacman JK. Arylsulfatase B regulates colonic epithelial cell migration by effects on MMP9 expression and RhoA activation. Clin Exp Metastasis. 2009;26(6):535–545. doi: 10.1007/s10585-009-9253-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
miR-490 expression in breast cancer and paracarcinoma tissues
| Groups | N | miR-490 expression/18s | P-value |
|---|---|---|---|
| Breast cancer | 137 | 0.000368±0.001433 | 0.05 |
| Paracarcinoma | 137 | 0.000817±0.002862 |
Notes: The fold-difference of para samples to breast cancer was 2.22. Statistically significant values are shown in bold (P<0.05).