Abstract
The widespread distribution and relapsing nature of Plasmodium vivax infection present major challenges for malaria elimination. To characterise the genetic diversity of this parasite within individual infections and across the population, we performed deep genome sequencing of >200 clinical samples collected across the Asia-Pacific region, and analysed data on >300,000 SNPs and 9 regions of the genome with large copy number variations. Individual infections showed complex patterns of genetic structure, with variation not only in the number of dominant clones but also in their level of relatedness and inbreeding. At the population level, we observed strong signals of recent evolutionary selection both in known drug resistance genes and at novel loci, and these varied markedly between geographical locations. These findings reveal a dynamic landscape of local evolutionary adaptation in P. vivax populations, and provide a foundation for genomic surveillance to guide effective strategies for control and elimination.
P. vivax is the main cause of malarial illness in many parts of the world and it is estimated that over 2.5 billion people are at risk of infection.1–3 It is absent from most of sub-Saharan Africa, where the species appears to have originated, because most of the human population is protected from infection by the Duffy negative blood group, suggesting that P. vivax has been a strong force for human evolutionary selection.4,5 P. vivax is a particularly challenging problem for malaria elimination because of its broad geographical range and its ability to produce hypnozoites, dormant forms of the liver-stage parasite that cause relapsing infection and that are refractory to most classes of antimalarial drugs.6 P. vivax is becoming increasingly resistant to chloroquine, the first-line treatment, and the molecular mechanisms of resistance remain unknown.7
In this study we analysed P. vivax genome variation to investigate how the parasite population varies between locations and how it is evolving. Microsatellite approaches have yielded useful insights into the its epidemiology, population structure and transmission dynamics8–10 but analysis of genome variation has previously been restricted to relatively small numbers of samples.11–18 Practical obstacles to genome sequencing of P. vivax from clinical samples are low levels of parasitaemia and the difficulty of culturing this species of parasite for more than a few days. Our approach was to collect blood samples from patients with P. vivax malaria and perform leucocyte depletion prior to parasite genome sequencing using the Illumina platform.19 Our sampling frame focused on Southeast Asia (Thailand, Cambodia, Vietnam, Laos, Myanmar and Malaysia) and Oceania (Papua Indonesia and Papua New Guinea), with smaller numbers of samples from China, India, Sri Lanka, Brazil and Madagascar (Supplementary Table 1).
In the first stage of analysis, we aligned sequence reads against the Salvador 1 (Sal 1) reference genome11 and used the GATK UnifiedGenotyper to discover 726,077 putative single nucleotide polymorphisms (SNPs). We then applied a series of quality control filters to exclude genomic regions with poor mapping quality, samples with low coverage and SNPs with a high risk of genotypic errors (see Methods). The final dataset contained 303,616 high-quality SNPs called in 228 samples across a core ‘accessible’ genome of 21.4 Mb, comprising 11.1 Mb coding and 10.3 Mb non-coding sequence (Figure 1, Supplementary Figure 1, Supplementary Table 2). For detailed population genetic analyses we used 148 samples from western Thailand (WTH), western Cambodia (WKH) and Papua Indonesia (PID) that had genotype calls for >80% of the high-quality SNPs (Supplementary Table 1). The high-quality SNPs were divided approximately equally between coding and non-coding regions (150,739 vs 152,877) and 58% of the coding SNPs were non-synonymous.
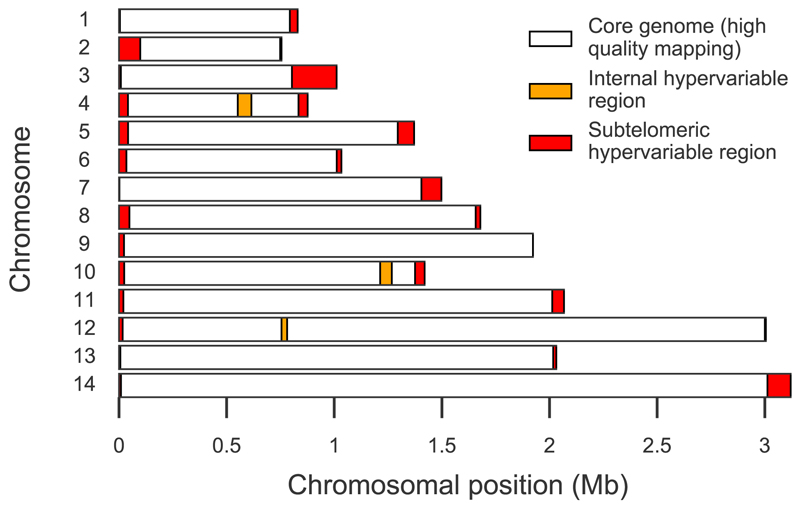
Figure 1. Defining the accessible genome.
When short read sequencing data from clinical samples of Plasmodium vivax are aligned to the 14 chromosomes comprising the Sal1 reference genome, there is low coverage and mapping quality in subtelomeric hypervariable regions (red) and three internal hypervariable regions (orange). Excluding these regions, we defined a core genome (white) which comprises 94.4% of the chromosomal sequence; coordinates are given in Supplementary Table 2. Aggregated across all samples, 99% of nucleotide positions in the core genome are alignable (≤10% reads of mapping quality 0) compared to 86% in subtelomeric and 85% in internal hypervariable regions; and 94% of positions in the core genome have ≥5x read depth compared to 37% in subtelomeric and 54% in internal hypervariable regions. When genome assemblies for other P. vivax strains15 were aligned to the Sal1 reference genome, the genome-wide coverage was 88.5% for India VII, 89.1% for Mauritania I and 89.6% for North Korea strains, whereas the coverage across the core genome was 98.5%, 98.7% and 99.0% respectively.
The allele frequency spectrum was dominated by low frequency variants, with over 50% of high-quality SNPs being at ≤1% minor allele frequency (Supplementary Figures 2 and 3). Nucleotide diversity (π) was estimated to be 1.5×10−3 when all unfiltered SNPs were included, and 5.6×10−4 when restricted to high quality SNPs (Supplementary Table 3). Levels of linkage disequilibrium were extremely low, e.g. r2 decayed to <0.1 within <200 bp in WTH and WKH samples, and within <500 bp in PID samples, after correcting for population structure and other confounders (Supplementary Figure 4). Rates of nucleotide diversity (π), Tajima’s D and the ratio of non-synonymous to synonymous variants (N/S ratio) were estimated for individual genes (Table 1, Supplementary Dataset 1). A striking finding was that π, D and N/S ratio are highly significantly elevated among the >200 genes that lack a known ortholog in P. falciparum, P. yoelii or both. High levels of diversity were also observed in genes expressed in late schizonts, those containing signal peptides or transmembrane domains, vir genes (i.e. those in the accessible genome) and genes encoding reticulocyte binding proteins.
Table 1. Gene categories enriched for high N/S ratio, nucleotide diversity, and Tajima’s D.
Each metric is represented by its median and P value by Mann-Whitney test, comparing genes in a given category versus all others, with bold font indicating significant values (P<0.05 after Bonferroni correction). Rows are ordered by π. N/S=non-synonymous/synonymous ratio. π=nucleotide diversity per base. D=Tajima’s D. No Pf/Py ortholog=genes that lack a known ortholog in P. falciparum/P. yoelii. TM domain=genes containing a transmembrane domain. Max schizont=maximum expression during the intraerythrocytic cycle was in late schizont stage42. Max sporozoite/zygote/ookinete=maximum expression in the sporozoite/zygote/ookinete43. These estimates are based on high-quality SNPs in genes with ≥10 SNPs in the subset of 148 samples used for detailed population comparisons as described in Methods. Estimates for individual genes, including all SNPs or restricted to high-quality SNPs, are given in Supplementary Dataset 1.
| Comparison | Genes | N/S | P(N/S) | π | P(π) | D | P(D) |
|---|---|---|---|---|---|---|---|
| No Pf ortholog | 97 | 2.23 | 6.9×10-18 | 7.3×10-4 | 7.5×10-9 | -1.86 | 2.6×10-4 |
| No Py ortholog | 251 | 1.86 | 1.1×10-20 | 6.7×10-4 | 7.1×10-11 | -1.92 | 5.3×10-8 |
| Max schizont | 844 | 1.60 | 2.0×10-13 | 6.1×10-4 | 5.1×10-7 | -2.04 | 1.7×10-4 |
| Max sporozoite | 422 | 1.43 | 3.6×10-1 | 6.0×10-4 | 3.2×10-2 | -2.03 | 6.9×10-2 |
| Signal peptide | 569 | 1.46 | 6.5×10-2 | 6.0×10-4 | 6.1×10-6 | -1.95 | 1.3×10-12 |
| TM domain | 646 | 1.50 | 1.9×10-2 | 5.9×10-4 | 1.2×10-4 | -1.98 | 1.7×10-13 |
| Max ookinete | 230 | 1.40 | 6.1×10-1 | 5.8×10-4 | 2.0×10-1 | -2.08 | 8.6×10-1 |
| Has paralog | 206 | 1.38 | 3.4×10-1 | 5.7×10-4 | 6.4×10-2 | -2.01 | 5.8×10-3 |
| Max zygote | 339 | 1.35 | 2.6×10-2 | 5.4×10-4 | 7.4×10-1 | -2.10 | 8.9×10-1 |
| All genes | 3062 | 1.43 | 5.5×10-4 | -2.07 | |||
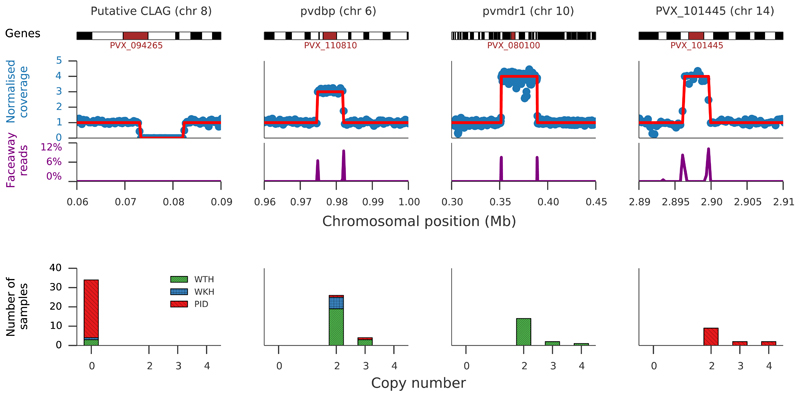
Large copy number variations (CNVs) were identified in nine regions of the core genome (Figure 2, Supplementary Table 4 and Supplementary Dataset 2) and the four most common showed marked geographic variation in frequency. The first was a 9 kb deletion on chromosome 8 (present in 73% PID, 6% WKH, and 3% WTH samples) that includes the first three exons of a gene encoding a cytoadherence-linked asexual protein. The second was a 7 kb duplication on chromosome 6 (5% PID, 35% WKH, 25% WTH) encompassing pvdbp, the gene that encodes the Duffy binding protein which mediates P. vivax invasion of erythrocytes.2 Pvdbp duplications have been shown to be common in Malagasy strains of P. vivax infecting Duffy-negative individuals21, and these findings show they can also reach relatively high frequency in places where nearly all individuals are Duffy-positive22. The third common CNV was a 37 kb duplication on chromosome 10 that includes pvmdr1. Duplication of pvmdr1 duplication has previously been associated with resistance to mefloquine23 and is homologous to the pfmdr1 amplification responsible for mefloquine resistance in P. falciparum. Mefloquine has never been a recommended treatment for P. vivax; it is therefore of considerable interest that pvmdr1 duplication is present in 19% of WTH samples, but not in WKH or PID samples. In Western Thailand, mefloquine has been used extensively as the first-line treatment for P. falciparum, either as a monotherapy or in combination with artesunate, and likely induces high selective pressure on relapsing P. vivax infections, which occur frequently following P. falciparum infection24. The fourth common CNV was a 3kb duplication on chromosome 14 that includes the gene PVX_101445 and was seen only in Papua Indonesia. Notably, this locus also shows signals of recent selection and is discussed further below.
Figure 2. Copy number variation.
Common forms of copy number variation in a region of chromosome 8 with a deletion of the first three exons of PVX_094265; in regions of chromosome 6 and 14 with copy number variations of pvdbp and PVX_101445 respectively; and in a region of chromosome 10 region where multiple genes including pvmdr1 are duplicated. Top panel shows an illustrative sample for each genomic region: upper trace shows GC-normalised coverage with inferred copy number marked by red line; lower trace shows the proportion of read pairs mapping in opposing directions, indicating the presumptive breakpoints of a duplication (note that not all samples have identical breakpoints, Supplementary Dataset 2). Lower panel shows number of samples in each population having a copy number other than one: western Thailand (WTH, n=88), western Cambodia (WKH, n=19) and Papua Indonesia (PID, n=41).
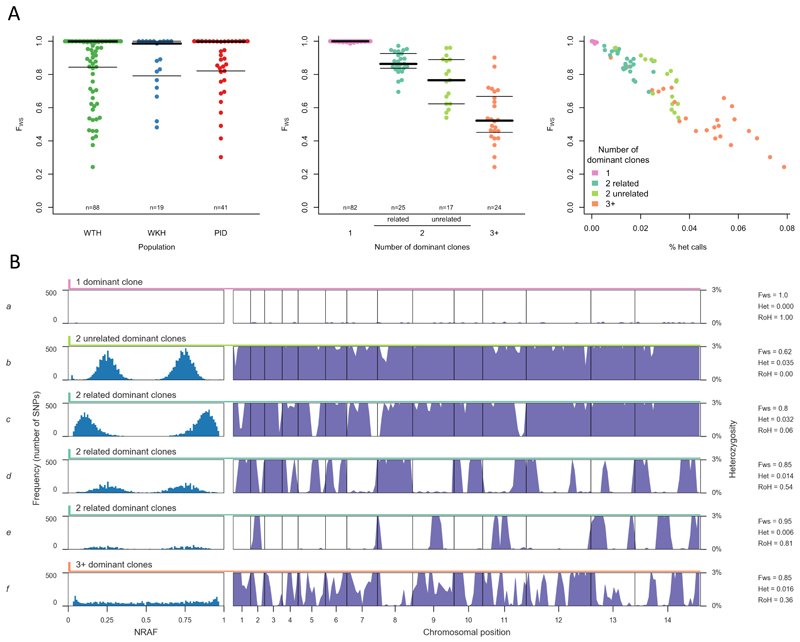
The genetic complexity of P. vivax infection is of particular interest since hypnozoite-induced relapses cause longstanding infections6 which can include sibling parasites inoculated by the same mosquito, or unrelated parasites from separate mosquito bites.16,25,26 Approximately 45% of the samples in this study had genetically mixed infections as determined by the FWS metric27 and within-sample heterozygosity (Figure 3, Supplementary Figure 5). Analysis of heterozygous SNPs revealed that 28% of samples had a strikingly bimodal and symmetrical allele frequency distribution, the signature of two dominant clones, while 16% of samples had a more complex allele frequency distribution indicating the presence of 3 or more dominant clones. These estimates are averaged across WTH, WKH and PID, but broadly similar patterns were observed in each population (Supplementary Table 5).
Figure 3. Genetic structure of mixed infections.
A shows distribution of FWS across all samples. FWS is analogous to an inbreeding coefficient27 and a value of 1 indicates a perfect clone. Left: Distribution of FWS in western Thailand (WTH), western Cambodia (WKH) and Papua Indonesia (PID), showing median (thick line) and inter-quartile range (thin line). Middle: Distribution of FWS stratified by the number of dominant clones in a sample and by whether they are related to each other, showing median (thick line) and inter-quartile range (thin line). Right: Distribution of FWS (vertical axis) and the proportion of heterozygous genotype calls (horizontal axis) in samples with different numbers of dominant clones.
Each row of B shows an illustrative sample. Left: non-reference allele frequency (NRAF) distribution across all heterozygous SNPs. Right: vertical axis is heterozygosity calculated in 20kb bins with the scale truncated (0–0.03) to highlight runs of homozygosity (RoH). Sample a is near-clonal as evidenced by FWS = 1 and lack of heterozygous SNPs. Samples b-e each contain two dominant clones as evidenced by the bimodal NRAF distribution. Sample b contains two unrelated clones (no RoH). Sample c contains two partially related clones (RoH across minority of the genome). Sample d contains two meiotic siblings (RoH extending over ~50% of the genome). Sample e contains two clones that are the product of inbreeding over multiple generations (RoH extending over ~80% of the genome). Sample f appears to contain a complex mixture of related parasites (relatively flat NRAF distribution indicates multiple dominant clones but there is substantial RoH).
To get a more detailed picture of the genetic structure of mixed infections, we analysed long runs of homozygosity (RoH) within heterozygous samples (Figure 3B and Supplementary Figure 5). These RoH are analogous to the long blocks of haplotype-sharing that have been observed by single cell genome sequencing of meiotic sibling parasites isolated from the same infected individual.28 RoH extending across ~50% of the genome indicates that the two clones are meiotic siblings, while less extensive RoH indicates more a distant relationship, and more extensive RoH is indicative of inbreeding over multiple generations. We observed significant RoH in 25 of 43 samples with two dominant clones, covering <40% of the genome in 9 samples, 40-60% in 11 samples, and >60% in 5 samples. A few samples with >2 dominant clones also displayed RoH, suggesting that these infections were dominated by a group of closely related parasites. These data demonstrate the potential utility of deep sequencing data as an epidemiological tool to differentiate mixed infections that are due to separate mosquito bites from those that are due to sibling parasites inoculated by the same mosquito.6,16,25,26,28
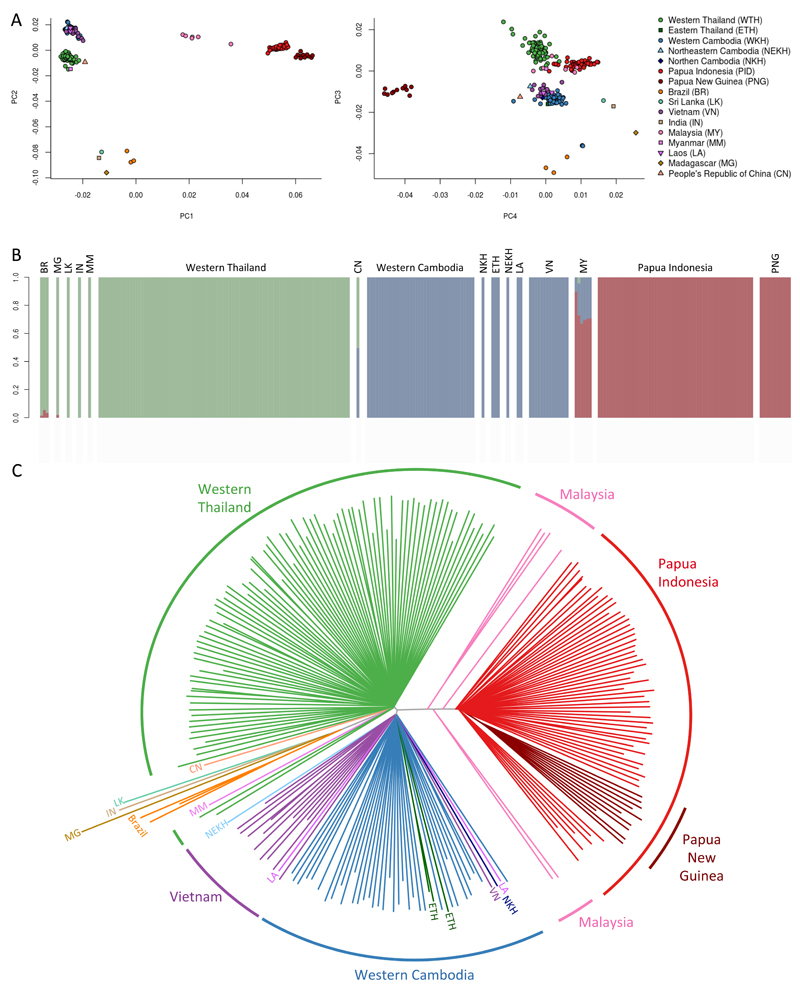
Major geographic divisions of parasite population structure were identified both by principal components analysis and using a model-based approach (ADMIXTURE) which clearly distinguished the three main groups of samples from western Thailand, western Cambodia and Papua Indonesia (Figure 4, Supplementary Figure 6).30 These differences can also be visualised on a neighbour-joining tree (Figure 4) which has three distinct branches separating Western Southeast Asia (Western Thailand, Myanmar and China), Eastern Southeast Asia (Cambodia, Vietnam, Eastern Thailand and Laos) and Southeast Asian and Pacific Islands (Malaysia, Papua Indonesia and Papua New Guinea). The separation of the P. vivax population of Southeast Asia into distinct Western and Eastern groups is consistent with observations in P. falciparum31 and reflects the malaria-free corridor that has been established through central Thailand. Samples from outside Southeast Asia were too disparate and small in numbers to be reliably assigned to specific groups of population structure by this analysis.
Figure 4. Parasite population structure.
Population structure is evident by principal components analysis (panel A), ADMIXTURE (panel B) and on a neighbour joining tree (panel C). ADMIXTURE analysis identifies three major components of population structure which correspond to the three largest groups of samples, i.e. western Thailand (n=88), western Cambodia (n=37) and Papua Indonesia (n=55). The neighbour-joining tree shows how these three major components encompass the Southeast Asian and Pacific Islands (Malaysia, Papua Indonesia, Papua New Guinea), the western part of mainland Southeast Asia (Western Thailand, Myanmar, and China) and the eastern part of the mainland (Cambodia, Vietnam, Eastern Thailand, and Laos). Samples from other parts of the world (India, Sri Lanka, Madagascar, and Brazil) are separated from Southeast Asian samples by long branches.
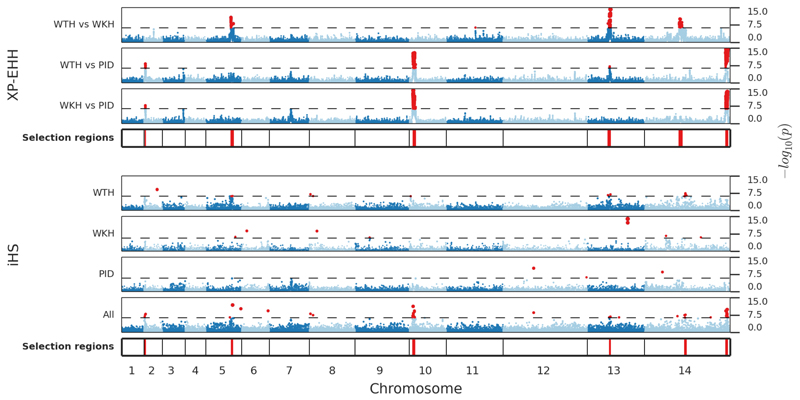
Strong evidence of recent selection was observed in six genomic regions on chromosomes 2, 5, 10, 13 and 14. In all cases there was evidence of geographically localised selection based on the XP-EHH test, with P values of 10−8 to 10−18, supported by other evidence such as the iHS test and highly differentiated SNPs (Figure 5, Supplementary Table 6 and Supplementary Figure 7). Each of these signals of selection encompasses multiple genes, such that we cannot be certain of the specific gene under selection, but several noteworthy candidates are summarised below.
Figure 5. Population-specific signatures of recent positive selection.
Metrics of extended haplotype homozygosity were estimated in 88 samples from western Thailand (WTH), 19 from western Cambodia (WKH) and 41 from Papua Indonesia (PID). The strongest evidence for recent selection was identified by XP-EHH (i.e. by comparing populations) and in most cases this was supported by iHS tests within individual populations. Horizontal axis represents genome position with chromosomes 1-14 shown in alternating colours. Vertical axis shows the results of XP-EHH and iHS tests represented by –log10 P values on a scale of 0 to 15. Dashed line shows the Bonferroni-corrected threshold for genome-wide significance, red points mark significant P values. Loci with ≥2 SNPs with significant P values within 80 kb of each other are marked by red lines in the tracks labelled ‘Selected regions’. The iHS signal on chromosome 13 in WKH was confined to two adjacent SNPs and is therefore not marked as significant. These signatures are described in more detail in Supplementary Table 6 and Supplementary Figure 7.
The signals of selection on chromosome 5 and 14 are strongest in western Thailand, and contain known resistance genes for pyrimethamine (pvdhfr) and sulfadoxine (pvdhps)32,33. Although chloroquine has been the main treatment for P. vivax malaria, sulfadoxine-pyrimethamine was introduced to Thailand in 1973 as first-line treatment for P. falciparum34, and selective pressure on P. vivax may have been considerable because of its widespread use in the private sector and the high frequency of P. vivax relapses following treatment of P. falciparum infection24. Selective sweeps at pvdhfr and pvdhps have also been observed in South America.17,18
The two strongest signals of selection of selection were observed in Papua Indonesia, where high-grade chloroquine resistance of unknown cause is now firmly established.7 Interestingly they did not include pvcrt-o, the P. vivax orthologue of the main chloroquine resistance gene in P. falciparum.36 One of these signals encompassed 22 genes on chromosome 14, of which the strongest candidate appears to be PVX_101445, a hypothetical membrane protein which has a striking pattern of copy number variations seen in PID but not elsewhere (Figure 1, Supplementary Table 4, Supplementary Dataset 2). The other signal encompassed 29 genes on chromosome 10: the peak of the signal was at PVX_079910, a conserved protein of unknown function, and this signal lies close to (but does not include) pvmdr1, which has been implicated in chloroquine resistance in ex-vivo studies in PID37.
Two other notable signals of selection were observed in WTH and WKH on chromosome 2, and in WTH on chromosome 13. The chromosome 2 signal contains four genes including pvmrp1 (PVX_097025) which encodes an ABC transporter that has been implicated as a drug resistance candidate12,18 and whose P. falciparum homologues are associated with resistance to multiple anti-malarial drugs38,39. The chromosome 13 signal includes PVX_084940, which encodes a putative voltage-dependent anion-selective channel containing a porin domain proposed to be implicated in antibiotic resistance35. Further details of the above signals of selection can be found in the Supplementary Note.
SNPs that are highly differentiated between populations can provide additional evidence of evolutionary selection. Pairwise comparisons between WTH, WKH and PID identified 40 SNPs with FST >0.9 (Supplementary Table 7). Half of these were associated with the signals of selection discussed above and the remainder had a significantly higher proportion of non-synonymous changes than the genome-wide average (12/20 vs 87,877/303,616; P=3.3×10−4 by Fisher’s exact test), identifying additional new candidate genes for investigations of drug resistance (Supplementary Note). More generally, this study provides a rich resource of data on the population diversity of P. vivax, which can be explored through a web application (www.malariagen.net/apps/pvgv) which provides summary data on SNP allele frequencies in different populations.
This study demonstrates the feasibility of population-level genome sequencing of P. vivax, despite the low levels of parasitaemia in clinical samples and the lack of an effective culture method. As well as characterising common patterns of genome variation that are the result of ancient events, the present findings reveal a dynamic evolutionary landscape, in which the parasite population is adapting to local selective pressures that reflect ongoing epidemiological processes. The difficulty of investigating P. vivax in the laboratory provides a strong incentive to exploit genomics to address gaps in knowledge of parasite phenotype. Genomic signals of recent selection could help identify local emergences of resistance, both to the drugs used specifically to treat P. vivax and to those that are targeted at P. falciparum. Knowledge of the genetic structure of individual infections is an important step towards understanding local patterns of malaria transmission, the epidemiology of relapsing infection, and the dynamics of genetic recombination in natural populations of P. vivax. Taken together, these findings point to various ways in which genomic analyses might be integrated into future clinical and epidemiological studies of P. vivax, and highlight the importance of translating this information into more effective strategies for malaria control and elimination.
Methods
Ethics statement
All samples used in this study were derived from patient blood samples obtained with informed consent from the patient or a parent or guardian. At each location, sample collection was approved by the appropriate local ethics committee: Eijkman Institute Research Ethics Committee, Jakarta, Indonesia; Human Research Ethics Committee of NT Department of Health and Families and Menzies School of Health Research, Darwin, Australia; Oxford Tropical Research Ethics Committee, Oxford, UK; Ethics Committee, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand; Research Review Committee of the Institute for Medical Research and the Medical Research Ethics Committee (MREC), Ministry of Health Malaysia; Review Board of Jiangsu Institute of Parasitic Diseases, Wuxi, China; National Ethics Committee for Health Research, Phnom Penh, Cambodia; Institutional Review Board, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA; National Ethics Committee for Health Research, Lao Peoples’ Democratic Republic; The Government of the Republic of the Union of Myanmar, Ministry of Health, Department of Medical Research (Lower Myanmar); Institutional Review Board of the Institute of Biomedical Sciences, University of São Paulo, Brazil; Scientific and Ethical Committee of the Hospital for Tropical Diseases in Ho Chi Minh City, Vietnam; Ethics Review Committee, Faculty of Medicine, University of Colombo, Sri Lanka; Papua New Guinea Institute of Medical Research Institutional Review Board, the Medical Research Advisory Committee of Papua New Guinea and the Walter and Eliza Hall Institute Human Research Ethics Committee; National Ethics Committee of Madagascar.
Sample preparation
Samples were collected from patients presenting at hospitals or health centres with symptomatic, uncomplicated P. vivax malaria as determined by microscopy. Venous blood was drawn into tubes coated with ethylenediaminetetraacetic acid (EDTA) or lithium heparin, and leukocyte depletion was carried out to minimise the amount of human DNA in the sample to be sequenced. Methods for leukodepletion included magnetic cell separation technology and filtration using non-woven fabric filters or cellulose-based constructs44,45. Some samples were also cultured ex vivo for up to 48 h to enrich for schizonts45. DNA extraction was typically performed using the QIAamp Blood Midi or Maxi kits (Qiagen) according to the manufacturer’s instructions. Total DNA concentration was measured using the Quant-iT™ dsDNA HS assay (Invitrogen) as per the manufacturer’s protocol, and the proportion of human DNA in each sample was determined by RT-qPCR.45
DNA sequencing
Sequencing was performed on the Illumina GA II or HiSeq 2000 platform at the Wellcome Trust Sanger Institute. Paired-end multiplex or non-multiplex libraries were prepared using the manufacturer’s protocol, with the exception that genomic DNA was fragmented using Covaris Adaptive Focused Acoustics rather than nebulisation. Multiplexes comprised 12 tagged samples. Cluster generation and sequencing were undertaken according to the manufacturer’s protocol for paired-end 75 bp, 76 bp or 100 bp sequence reads. We initially used 272 samples from confirmed cases of P. vivax malaria that had at least 50 ng total gDNA with ≤80% human DNA. At the analysis stage, we included 20 additional samples from presumed cases of P. falciparum malaria, which were found by sequencing to have substantial proportions of reads mapping to the P. vivax reference genome (Supplementary Table 1). Illumina sequence reads have been submitted to the European Nucleotide Archive.
Overview of sequence analysis
We initially aligned sequence reads from 292 samples against the Salvador 1 (Sal 1) reference genome11 using bwa, and then successively applied the Picard tools CleanSam, FixMateInformation and MarkDuplicates, followed by GATK indel realignment. We used GATK CallableLoci to determine a subset of 247 samples for which at least 50% of the 14 chromosomal sequences of the Sal1 reference could be reliably called. After trimming the dataset to remove instances of multiple samples from the same individual, we were left with 228 samples for further analysis.
We used GATK UnifiedGenotyper to discover 726,077 putative single nucleotide polymorphisms (SNPs) amongst the 247 samples. SNPs were annotated using standard GATK annotations as well as a previously described Uniqueness score and a novel HyperHeterozygosity score. We genotyped SNPs using previously define rules and created a Missingness score for each SNP based on the number of missing genotypes across the 247 samples.
We determined SNPs with evidence of genotyping errors by analysis of genotype discordance between technical replicate samples. We masked out subtelomeric regions and three internal chromosome regions (13 SERA family genes on chromosome 4, 11 msp3 family genes on chromosome 10 and 11 msp7 family genes on chromosome 12) which had lower mapping quality, higher levels of missingness, greater SNP density and greater levels of genotype discordance between technical replicates. We also filtered out SNPs in the unmasked regions that had extreme values of the annotation metrics described in the previous paragraph. Thresholds for extreme levels of these metrics were determined using rates of technical replicate discordance.
Read mapping and coverage
Reads mapping to the human reference genome were removed before all analyses, and the remaining reads were mapped to the P. vivax Sal1 reference genome11 using bwa46 version 0.5.9-r16 with default parameters. Standard alignment metrics were generated for each sample using the bamcheck utility from samtools47.
The Picard version 1.110 tools CleanSam, FixMateInformation and MarkDuplicates were successively applied to the bam files of each sample. GATK version 3.1-1 indel realignment48 was applied using default parameters and no list of known indels. The output of this stage was a set of 292 “improved” bam files, one for each sample.
We ran GATK’s CallableLoci49 on each improved sample bam file to determine the proportion of genomic positions callable in each sample using parameters --minDepth 5 --minBaseQuality 27 --minMappingQuality 27. This identifies a site as callable if there are ≥5 reads with base and mapping quality of ≥27 and if ≤10% of reads have mapping quality 0.
The P. vivax Sal1 reference genome11 consists of 14 large chromosomal sequences ranging in size from 0.76-3.12 Mbp, and 2,733 shorter contigs ranging in size from 200-101,928 bases. It is assumed that these shorter contigs are sequences from the subtelomeric ends of the autosomal chromosomes. In all subsequent analyses, we have analysed only those reads that mapped to the 14 large chromosomal sequences, which are named Pv_Sal1_chr01 - Pv_Sal1_chr14.
A total of 247 samples were identified as having at least 50% of Pv_Sal1_chr01 - Pv_Sal1_chr14 positions whose genotypes could be reliably called. After trimming the dataset to remove instances of multiple samples from the same individual, we were left with 228 samples for further analysis (Supplementary Table 1).
SNP discovery and annotation
We discovered potential SNPs by running GATK’s UnifiedGenotyper49 across all 247 sample-level bam files. SNPs were annotated using a number of different methods. Functional annotations were applied using snpEff version 2.0.550, with gene annotations downloaded from GeneDB51. GATK VariantAnnotator was used to create the following standard annotation metrics: BaseQRankSum, DP, Dels, FS, HaplotypeScore, HRun, MQ, MQRankSum, MQ0, QD and ReadPosRankSum.
Because GATK’s UnifiedGenotyper outputs unfiltered allele depths at each SNP for each sample, we created custom Python scripts based on the pyvcf and pysam modules to calculate filtered allele depths (mapping and base quality ≥27). We created a “NonUniqueness” score (UQ)27 for each position in the reference genome and annotated each SNP with this score. Under Hardy–Weinberg equilibrium, it is expected that heterozygosity at a given SNP (the probability of observing multiple alleles in the same sample) is related to its allele frequency in the population and to the inbreeding coefficient of that population by the relationship h = 2(1 − f)p(1 − p), where p is the frequency of the SNP in the population, h its expected heterozygosity, and f the inbreeding coefficient of the population. A substantial divergence from this relationship is likely to arise from alignment artefacts, such as systematic incorrect mappings of reads from paralogous regions. Given that f is unknown and can be influenced by various epidemiological factors, we estimated a surrogate from the data as follows. We used the set of all discovered SNPs with MAF >0.05 to fit a quadratic model of the form y = mx(1 − x), where x represents the allele frequency and y the observed heterozygosity. We obtained a robust estimate of m by using the rq implementation in the R quantreg package and using a median regression (which is more robust to outliers then standard mean regression). The residuals were used as a HyperHeterozgosity score, which was subsequently used in variant filtering.
We imputed the ancestral allele at SNPs by comparison with the closely related species P. cynomolgi. Illumina reads from this species generated in a recent study52 were mapped against the P. vivax reference using bwa46 version 0.6.2-r126. We then selected the SNPs discovered in our P. vivax samples and genotyped (with respect to the P. vivax reference) these positions in the P. cynomolgi data using GATK’s UnifiedGenotyper (version 3.1-1). Where the genotype in the P. cynomolgi was the same as one of the alleles seen in our P. vivax data, the allele was defined as ancestral. In this way we were able to impute ancestral alleles for 30% of the P. vivax SNPs.
Determining SNP genotype
Because many of our samples exhibit evidence of mixed infection, we did not use the GATK genotype calls, as these are made under an assumption of clonality. Instead, genotypes were defined based on filtered allele depths using previously defined rules27. For each SNP, we created a Missingness score, which was the number of samples from all 247 samples that had a missing genotype based on these rules.
Variant filtering
We determined SNPs with evidence of genotyping errors by analysis of genotype discordance between technical replicate samples. We masked out subtelomeric regions and three internal chromosome regions (13 SERA family genes on chromosome 4, 11 msp3 family genes on chromosome 10 and 11 msp7 family genes on chromosome 12) which had lower mapping quality, higher levels of missingness, greater SNP density and greater levels of genotype discordance between technical replicates. We also filtered out SNPs in the unmasked regions that had extreme values of the annotation metrics described in the previous paragraph. Thresholds for extreme levels of these metrics were determined using rates of technical replicate discordance. Further details of the variant filtering process can be found in Supplementary Note 3.
Sequenom analysis of genotyping concordance
The Sequenom® primer-extension mass spectrometry genotyping platform was used to validate SNP genotype calls made by Illumina sequencing. Two separate validation experiments were performed using laboratory procedures described previously27. In the first experiment we assayed 164 SNPs on 142 samples, and in the second experiment we assayed 107 SNPs in 220 samples. After applying quality control filters to the Sequenom data, removing samples with ≥50% missing SNP genotypes, and removing SNPs with artefactual genotype calls in blank control samples, we were left with 111 SNPs that could be reliably compared between Sequenom and the high quality SNPs typed by Illumina sequencing. This gave a concordance rate of 99.98% for homozygous calls and 93.6% when heterozygous calls were included (Supplementary Table 8). Previous work on P. falciparum has shown Illumina sequencing to be generally more reliable than Sequenom for heterozygous calls (see supplementary material to ref 27).
Large copy number variations
Large copy number variations were identified by analysis of read depth after normalisation by GC-content. Coverage in non-overlapping 300bp bins was calculated using pysamstats. Normalisation was undertaken within each sample by dividing the coverage by the median coverage across all bins with the same integer percentage GC content. Copy number variants (CNVs) were called using a hidden Markov model with the Python package sklearn.hmm.GaussianHMM using a similar procedure to that used previously for P. falciparum genetic crosses20. Two samples were removed from this analysis as they had excessive variation in read coverage. Our analysis focused on CNVs >3kbp and those detected by read-depth analysis were further validated by assessment of read pair orientation in the breakpoint regions.
Samples used for population genetic analyses
For population genetic analyses we selected samples that were typable at >80% of the 303,616 high-quality SNPs. They included 88 samples from Western Thailand, 19 from Western Cambodia and 41 from Indonesia. All other locations had <10 eligible samples which was considered too few for detailed population genetic comparisons. This sample size was not pre-determined, but was the largest that we were able to achieve in the timeframe of this study. Supplementary Table 1 identifies the origin of the 148 samples that were used for all population genetic analyses (excepting the PCA and neighbour-joining tree for which we used all 228 samples).
Diversity,Tajima’s D and N/S ratio amongst gene classes
We classified genes using annotations from PlasmoDB. Nucleotide diversity, Tajima’s D and N/S ratio were calculated using custom Python scripts. Statistical analyses were performed using the SciPy stats package.
Population structure
We investigated global population structure and FST using previously applied methods31. To explore the effects on population structure of using a different reference genome, we aligned the same samples to the Papua Indonesia P01 genome assembly (www.genedb.org/Homepage/PvivaxP01) using GATK Best Practices. As shown in Supplementary Figure 9, the neighbour-joining tree was very similar to that obtained with the Sal1 reference genome.
We performed admixture analysis using ADMIXTURE.53 As the ADMIXTURE model assumes perfect linkage equilibrium between markers (i.e. they are independent of each other), we excluded SNP pairs that appeared to be linked. We discarded SNPs according to the observed correlation coefficients by using the PLINK tool set.54 We scanned the genome with a sliding window of 60 SNPs in size, advanced in steps of 10 SNPs, and removed any SNP with a correlation coefficient ≥ 0.1 with any other SNP within the window. Additionally, we removed all SNPs with extremely low minor allele frequency (MAF ≤ 0.005), as these SNPs are less informative for the inference process. We then ran ADMIXTURE 1.3, in haploid mode, using the 76,544 remaining SNPs with 5-fold cross-validation and several K values (i.e. the number of putative populations) ranging from 1 to 12. In order to avoid fluctuations in the likelihood due to the stochasticity of the optimization process we repeated the process 5 times with different random seeds. We assessed the plausible choice for the number of populations by using the delta ΔK metric developed by Evanno and colleagues (Supplementary Figure 6).30
Within host diversity
FWS metrics were calculated as previously described for P. falciparum27. Analysis of heterozygosity within mixed samples was performed using custom Python scripts.
Recombination
We analyzed the decay of LD with genomic distance for each population separately. Complete details are given in Manske et al27.
Signatures of selection
XP-EHH and iHS scores were calculated using previously described methods as per Sabeti et al.55 and Voight et al.56. As described in these studies, the distributions of scores follow an approximately normal distribution and, hence, P values were based on this distribution. Where genotypes exhibited heterozygous calls, the calls were converted to a homozygous call for the allele with the largest number of reads at that position. As a consequence, in mixed samples, haplotype-based analysis was essentially conducted on the majority strain present within each infection.
Supplementary Material
Acknowledgements
We wish to thank the patients and communities that provided samples for this study, and our many colleagues who supported this work in the field. Sequencing, data analysis and project coordination were funded by the Wellcome Trust (098051, 090770/Z/09/Z), the Medical Research Council (G0600718) and the UK Department for International Development (M006212). AEB and IM acknowledge the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. SA and RNP are funded by the Wellcome Trust (Senior Fellowship in Clinical Science awarded to RNP, 091625). This study was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
URLs
MalariaGEN Plasmodium vivax Genome Variation Project: sample information and genotype calls, https://www.malariagen.net/resource/17; SNP information and allele frequencies, https://www.malariagen.net/apps/pvgv/. European Nucleotide Archive (ENA), http://www.ebi.ac.uk/ena; P. vivax Sal1 reference sequence, http://plasmodb.org/common/downloads/release-10.0/PvivaxSal1/fasta/data/PlasmoDB-10.0_PvivaxSal1_Genome.fasta; P. vivax Sal1 gene annotations, ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/P_vivax/2014/May_2014 and http://www.plasmodb.org/common/downloads/release-13.0/PvivaxSal1/txt/PlasmoDB-13.0_PvivaxSal1Gene.txt; P. cynomolgi Illumina reads, ftp://ftp.ddbj.nig.ac.jp/ddbj_database/dra/fastq/DRA000/DRA000196/DRX000265; Picard software, http://picard.sourceforge.net; pyvcf software, https://github.com/jamescasbon/PyVCF; pysam software, https://code.google.com/p/pysam; pysamstats software, https://github.com/alimanfoo/pysamstats; SciPy stats package, http://docs.scipy.org/doc/scipy/reference/stats.html.
Accession codes
A document containing lists of ENA accession codes for all samples used in the present study is available from https://www.malariagen.net/resource/17.
Author Contributions
C.A., S.S., S.M., R.N., H.T., J.M., N.M.A., T.W., M.F.B., C.D., H.T.T., N.J.W., P.M., P.S., L.T., G.H., A.B., I.M., M.U.F., N.K., M.R. and Q.G. carried out field and laboratory work to obtain P. vivax samples for sequencing. C.H., E.D., D.M., M.K., S.C., B.M. and K.A.R. developed and implemented methods for sample processing and sequencing library preparation. R.D.P., L.H., B.J. and M.M. managed data production pipelines. S.A., O.M., V.J.C., B.M., K.A.R., A.M., J.C.R., R.M.F., F.N., R.N.P. and D.P.K. contributed to study design and management. R.D.P., R.A., S.A., O.M., J.A.-G. and D.P.K. performed data analyses. R.D.P., R.A., S.A. and D.P.K. drafted the manuscript, which was reviewed by all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Gething PW, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis. 2012;6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 3.Ménard D, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A. 2010;107:5967–71. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price RN, et al. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 5.Battle KE, et al. The global public health significance of Plasmodium vivax. Adv Parasitol. 2012;80:1. doi: 10.1016/B978-0-12-397900-1.00001-3. [DOI] [PubMed] [Google Scholar]
- 6.White NJ. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 2011;10:297. doi: 10.1186/1475-2875-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price RN, et al. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:982–91. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karunaweera ND, et al. Extensive microsatellite diversity in the human malaria parasite Plasmodium vivax. Gene. 2008;410:105–112. doi: 10.1016/j.gene.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 9.Barry AE, Waltmann A, Koepfli C, Barnadas C, Mueller I. Uncovering the transmission dynamics of Plasmodium vivax using population genetics. Pathog Glob Health. 2015;109:142–152. doi: 10.1179/2047773215Y.0000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koepfli C, et al. Plasmodium vivax diversity and population structure across four continents. PLoS Negl Trop Dis. 2015;9:e0003872. doi: 10.1371/journal.pntd.0003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlton JM, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dharia NV, et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci U S A. 2010;107:20045–50. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hester J, et al. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl Trop Dis. 2013;7:e2569. doi: 10.1371/journal.pntd.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan ER, et al. Whole genome sequencing of field isolates provides robust characterization of genetic diversity in Plasmodium vivax. PLoS Negl Trop Dis. 2012;6:e1811. doi: 10.1371/journal.pntd.0001811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neafsey DE, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 2012;44:1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bright AT, et al. A high resolution case study of a patient with recurrent Plasmodium vivax infections shows that relapses were caused by meiotic siblings. PLoS Negl Trop Dis. 2014;8:e2882. doi: 10.1371/journal.pntd.0002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter DJ, et al. Whole genome sequencing of field isolates reveals extensive genetic diversity in Plasmodium vivax from Colombia. PLoS Negl Trop Dis. 2015;9:e0004252. doi: 10.1371/journal.pntd.0004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flannery EL, et al. Next-generation sequencing of Plasmodium vivax patient samples shows evidence of direct evolution in drug-resistance genes. ACS Infect Dis. 2015;1:367–379. doi: 10.1021/acsinfecdis.5b00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auburn S, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One. 2012;7:e32891. doi: 10.1371/journal.pone.0032891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles A, et al. Genome variation and meiotic recombination in Plasmodium falciparum: insights from deep sequencing of genetic crosses. bioRxiv. 2015 doi: 10.1101/024182. 024182. [DOI] [Google Scholar]
- 21.Menard D, et al. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis. 2013;7:e2489. doi: 10.1371/journal.pntd.0002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howes RE, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suwanarusk R, et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis. 2008;198:1558–1564. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas NM, et al. Plasmodium vivax recurrence following falciparum and mixed species malaria: risk factors and effect of antimalarial kinetics. Clin Infect Dis. 2011;52:612–20. doi: 10.1093/cid/ciq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imwong M, et al. The first Plasmodium vivax relapses of life are usually genetically homologous. J Infect Dis. 2012;205:680–3. doi: 10.1093/infdis/jir806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JT, et al. Using amplicon deep sequencing to detect genetic signatures of Plasmodium vivax relapse. J Infect Dis. 2015;212:999–1008. doi: 10.1093/infdis/jiv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair S, et al. Single-cell genomics for dissection of complex malaria infections. Genome Res. 2014;24:1028–38. doi: 10.1101/gr.168286.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baniecki ML, et al. Development of a single nucleotide polymorphism barcode to genotype Plasmodium vivax infections. PLoS Negl Trop Dis. 2015;9:e0003539. doi: 10.1371/journal.pntd.0003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 31.Miotto O, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–34. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korsinczky M, et al. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob Agents Chemother. 2004;48:2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imwong M, et al. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 2003;47:1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam MT, et al. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agents Chemother. 2011;55:155–164. doi: 10.1128/AAC.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagès J-M, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 36.Pava Z, et al. Expression of Plasmodium vivax crt-o is related to parasite stage but not ex vivo chloroquine susceptibility. Antimicrob Agents Chemother. 2015 doi: 10.1128/AAC.02207-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwanarusk R, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 39.Raj DK, et al. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem. 2009;284:7687–7696. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WWARN. History Of Resistance. 2015 at http://www.wwarn.org/resistance/malaria/history.
- 41.Maguire JD, Marwoto H. Mefloquine is highly efficacious against chloroquine-resistant Plasmodium vivax malaria and Plasmodium falciparum malaria in Papua, Indonesia. Clin Infect {…} 2006;2197:1067–1072. doi: 10.1086/501357. [DOI] [PubMed] [Google Scholar]
- 42.Bozdech Z, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westenberger SJ, et al. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis. 2010;4:e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao Z-Y, Xia H, Cao J, Gao Q. Development and evaluation of a prototype non-woven fabric filter for purification of malaria-infected blood. Malar J. 2011;10:251. doi: 10.1186/1475-2875-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auburn S, et al. Effective preparation of Plasmodium vivax field isolates for high-throughput whole genome sequencing. PLoS One. 2013;8:e53160. doi: 10.1371/journal.pone.0053160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePristo Ma, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Logan-Klumpler FJ, et al. GeneDB--an annotation database for pathogens. Nucleic Acids Res. 2012;40:D98–108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tachibana S-I, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 2012;44:1051–1055. doi: 10.1038/ng.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.