Abstract
Systemic lupus erythematosus (SLE; OMIM 1 152700) is a genetically complex autoimmune disease. Genome-wide association studies (GWASs) have identified more than 50 loci as robustly associated with the disease in single ancestries, but genome-wide transancestral studies have not been conducted. We combined three GWAS data sets from Chinese (1,659 cases and 3,398 controls) and European (4,036 cases and 6,959 controls) populations. A meta-analysis of these studies showed that over half of the published SLE genetic associations are present in both populations. A replication study in Chinese (3,043 cases and 5,074 controls) and European (2,643 cases and 9,032 controls) subjects found ten previously unreported SLE loci. Our study provides further evidence that the majority of genetic risk polymorphisms for SLE are contained within the same regions across both populations. Furthermore, a comparison of risk allele frequencies and genetic risk scores suggested that the increased prevalence of SLE in non-Europeans (including Asians) has a genetic basis.
SLE is a highly complex disease, with occurrence heavily influenced by genetics (heritability of 66% (ref. 1)). SLE incidence varies markedly across populations, with Europeans showing three- to fourfold lower prevalence compared with individuals of African or Asian ancestry2. In recent years, understanding of SLE’s genetic etiology has been transformed by GWASs, with the largest study in Europeans3 (4,036 cases and 6,959 controls) finding evidence of association at 41 autosomal loci. Meanwhile, two published GWASs4,5 in Chinese populations and follow-up studies in Asians6–10 found association at 31 loci, 11 of which are not published for Europeans. Thus 52 SLE disease-susceptibility autosomal loci have been mapped by GWASs in these two populations.
Although fine-mapping of a selected number of known SLE-associated loci11–13 has been successfully undertaken through the combination of genetic results obtained from association mapping in different populations, to date transancestral approaches have not been used at the genome-wide level for SLE. Studies of other diseases14 have also shown the benefit of comparing data from differing ancestries to exploit differences in linkage disequilibrium (LD).
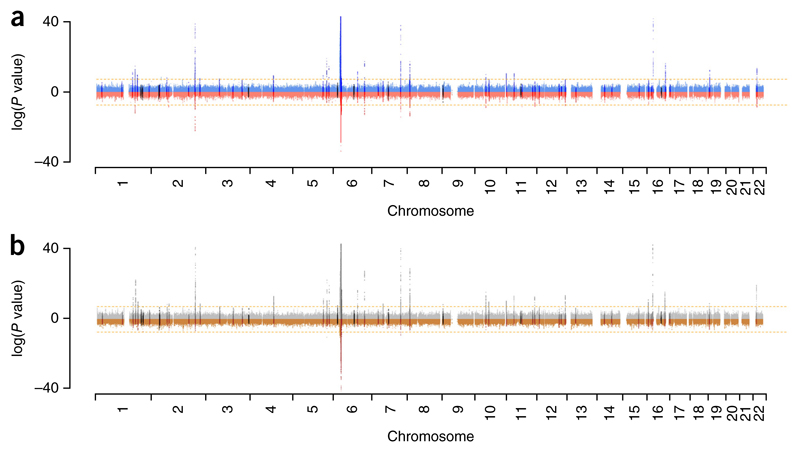
Our initial objective was to compare observed genetic association signals across the genome in Chinese and European subjects. To provide additional power to identify potentially novel SLE-associated loci, we imputed each GWAS (a European study comprising 4,036 cases and 6,959 controls3 (λGC = 1.16 with λ1,000 = 1.02, where λ is a measure of association and “GC” stands for “genomic control”), a study from Anhui Province in mainland China including 1,047 cases and 1,205 controls4 (λGC = 1.05), and a study from Hong Kong including 612 cases and 2,193 controls5,7 (λGC = 1.04)) to the density of the 1000 Genomes Project (1KG) data (Online Methods). Analyses of association results in each population suggested that SLE susceptibility loci were shared extensively. We found that the association signals were mostly mirrored between populations (Fig. 1). Details of the association data for individual SNPs are presented in Supplementary Table 1. When we compared the published genome-wide significant allelic associations for SLE, we saw that many of the alleles previously thought to be associated with SLE in only one population had evidence for association in both European and Chinese SLE cases. By ranking genomic regions on the basis of the strength of association, we also found a significant correlation (P = 2.7 × 10−9, Kendall’s τ = 0.08; Online Methods) between the two populations’ GWASs. These observations suggested that combining GWAS data in a meta-analysis could yield novel association signals. The GWAS meta-analysis results included three associations in novel loci (rs17603856 (6p23), rs1887428 (9p24) and rs669763 (16q13)) with genome-wide levels of significance (P < 5 × 10−8; Fig. 1b). In addition, the major histocompatibility complex (MHC) and, to a lesser extent, the IRF5 locus on chromosome 7 showed significant transancestral heterogeneity (Fig. 1b).
Figure 1.
Comparison of Manhattan plots for the European and Chinese SLE GWASs. (a) Manhattan plot of results from the European (4,036 cases and 6,959 controls) and Chinese (meta-analysis of two Chinese GWASs comprising 1,659 cases and 3,398 controls) association studies. −log10 P values for European subjects are shown in blue, and log10 P values for Chinese subjects are shown in red. The ten novel loci identified as SLE associated by this study are shown in black. (b) −log10 P values for a meta-analysis (using inverse-variance weighting) of European and Chinese GWASs (gray) compared with log10 P values for a test of heterogeneity (using Cochran’s Q statistic) between the European and Chinese GWASs (brown). The 52 loci with published evidence of SLE association are highlighted in dark gray (meta-analysis P values) and dark brown (heterogeneity test); the 10 novel loci identified as SLE associated by this study (after replication) are highlighted in black. The orange dashed lines in both panels indicate the accepted threshold for genome-wide statistical significance, P = 5 × 10−8.
We then carried out a two-stage replication study incorporating rs17603856, rs1887428 and rs669763. We scanned the 1KG imputed data for association at loci independent of those previously published and excluding the MHC. We successfully genotyped a total of 66 SNPs at 56 loci (SNP selection is described in the Online Methods) in an additional 3,043 cases and 5,074 controls of Chinese ancestry recruited from Anhui Province. Eighteen of these SNPs (at 17 independent loci) showed association in this replication study, passing a false discovery rate (FDR) of 0.01. These included rs17603856 and rs1887428 but not rs669763, which failed quality control. We then genotyped these 18 SNPs in a European replication cohort comprising 1,478 cases and 6,925 controls3. Data from an additional European-American GWAS (1,165 independent cases and 2,107 controls) were also included in this final analysis15 (Supplementary Table 2a). Of the 18 candidate SNPs, 11 showed a standard genome-wide level of significance (P < 5 × 10−8) in the combined meta-analysis (11,381 cases and 24,463 controls) of all three main GWASs and the three replication studies (Table 1, Supplementary Fig. 1). The strongest association signal after this meta-analysis was that for rs1887428 (9p24; P = 2.19 × 10−17). Other statistically significant associations were found at rs34889541 (1q31.3; P = 2.44 × 10−12), rs2297550 (1q32.1; P = 1.31 × 10−11), rs6762714 (3q28; P = 4.00 × 10−15), rs17603856 (6p23; P = 3.27 × 10−12), rs597325 (6q15; P = 4.03 × 10−12), rs73135369 (7q11.23; P = 8.77 × 10−14), rs494003 (11q13.1; P = 5.81 × 10−9) and rs1170426 (16q22.1; P = 2.24 × 10−8), and two SNPs at 2p23.1 (rs1732199; P = 2.22 × 10−16 and rs7579944; P = 1.41 × 10−9) were replicated as being independently associated (Online Methods and Table 1). The full set of results for the 18 candidate markers can be found in Supplementary Table 2.
Table 1. Summary of statistical associations for new loci.
| SNP | Chr | Position | Risk allelea | Chinese MAFb |
European MAFb |
Chinese 4,702 cases, 8,472 controls |
European 6,679 cases, 15,991
controls |
Meta-analysis, all 11,381 cases, 24,463
controls |
Gened | Association with other autoimmune diseasesf | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | OR (95% CI)c | P | OR (95% CI) | P | OR (95% CI) | P | ||||||
| rs34889541 | 1q31.3 | 198,594,769 | G | 0.126 0.14 | 0.058 | 0.07 | 0.78 | (0.72–0.84) | 2.96 × 10−10 | 0.86 (0.79–0.94) | 5.34 × 10−4 | 0.81 (0.76–0.86) | 2.44 × 10−12 | PTPRC (CD45) | MS, RA, T1D |
| rs2297550 | 1q32.1 | 206,643,772 | G | 0.577 | 0.546 | 0.14 | 0.12 | 1.14 (1.08–1.20) | 1.73 × 10−7 | 1.18 (1.09–1.27) | 1.43 × 10−5 | 1.16 (1.11–1.21) | 1.31 × 10−11 | IKBKE | |
| rs7579944 | 2p23.1 | 30,445,026 | C | 0.59 | 0.641 | 0.338 | 0.366 | 0.87 (0.82–0.92) | 5.52 × 10−6 | 0.92 (0.88–0.96) | 3.96 × 10−5 | 0.90 (0.87–0.93) | 1.41 × 10−9 | LBH e | RA, AA, IBD, NAR, PSC, SJO, SSC, VIT |
| rs17321999 | 2p23.1 | 30,479,857 | C | 0.16 | 0.164 | 0.161 | 0.191 | 0.82 (0.77–0.88) | 9.55 × 10−9 | 0.84 (0.79–0.89) | 2.26 × 10−9 | 0.83 (0.79–0.87) | 2.22 × 10−16 | LBH e | RA, AA, IBD, NAR, PSC, SJO, SSC, VIT |
| rs6762714 | 3q28 | 188,470,238 | T | 0.848 | 0.825 | 0.421 | 0.392 | 1.20 (1.12–1.29) | 5.56 × 10−7 | 1.14 (1.09–1.19) | 7.97 × 10−10 | 1.16 (1.12–1.20) | 4.00 × 10−15 | LPP, TPRG1-AS1 | ATD, CEL, VIT |
| rs17603856 | 6p23 | 16,630,898 | T | 0.221 | 0.222 | 0.325 | 0.355 | 0.86 (0.80–0.92) | 1.61 × 10−5 | 0.89 (0.85–0.93) | 3.34 × 10−8 | 0.88 (0.85–0.91) | 3.27 × 10−12 | ATXN1 | |
| rs597325 | 6q15 | 91,002,494 | G | 0.485 | 0.52 | 0.357 | 0.385 | 0.84 (0.80–0.89) | 1.05 × 10−10 | 0.92 (0.88–0.96) | 2.65 × 10−4 | 0.89 (0.86–0.92) | 4.03 × 10−12 | BACH2 | AS, ATD, CEL, CRO, MS, T1D, IBD, PSC, VIT |
| rs73135369 | 7q11.23 | 73,940,978 | C | 0.107 | 0.076 | 0.028 | 0.022 | 1.38 (1.26–1.51) | 7.33 × 10−13 | 1.20 (1.05–1.38) | 9.00 × 10−3 | 1.32 (1.23–1.42) | 8.77 × 10−14 | GTF2IRD1-GTF2I | |
| rs1887428 | 9p24 | 4,984,530 | G | 0.372 | 0.346 | 0.398 | 0.373 | 1.24 (1.17–1.31) | 4.49 × 10−14 | 1.11 (1.06–1.16) | 1.25 × 10−6 | 1.16 (1.12–1.20) | 2.19 × 10−17 | JAK2 | CRO, UC IBD, PSC, VIT |
| rs494003 | 11q13.1 | 65,542,298 | A | 0.116 | 0.117 | 0.213 | 0.19 | 1.16 (1.06–1.27) | 8.38 × 10−4 | 1.13 (1.07–1.19) | 1.68 × 10−6 | 1.14 (1.09–1.19) | 5.81 × 10−9 | RNASEH2C | CRO, IBD |
| rs1170426 | 16q22.1 | 68,603,798 | C | 0.198 | 0.176 | 0.252 | 0.235 | 1.20 (1.12–1.28) | 4.36 × 10−8 | 1.06 (1.02–1.10) | 3.59 × 10−3 | 1.12 (1.08–1.17) | 2.24 × 10−8 | ZFP90 | MS, UC, IBD, PSC, VIT |
“Chinese” comprises the two Chinese GWASs (1,659 cases and 3,398 controls) and the Chinese replication study (3,043 cases and 5,074 controls). “European” comprises the European GWAS (4,036 cases and 6,959 controls), the additional European GWAS (1,165 cases and 2,107 controls) and the European replication study (1,478 cases and 6,925 controls).
The risk allele refers to the effect in the overall meta-analysis.
The minor allele frequency (MAF) refers to the allele that is minor in Europeans.
The odds ratio (OR) is with respect to the minor allele. CI, confidence interval.
For the rationale for candidate gene selection at the associated loci, see Table 2.
C6orf1 is also known as LBH, but we chose LBH for our work because it possesses two separate signals. rs7579944 and rs17321999 were found to be independently associated with SLE (Online Methods): rs17321999 was significant (Chinese P = 2.62 × 10−11; European P = 6.14 × 10−6; meta-analysis P = 3.33 × 10−15) when rs7579944 was used as a covariate in logistic regression, and rs7579944 was significant (Chinese P = 1.38 × 10−8; European P = 4.49 × 10−6; meta-analysis P = 4.16 × 10−13) in the meta-analysis when rs17321999 was used as a covariate in logistic regression. The LD between these two SNPs was very weak in all studies (The r2 was as follows in each data set: Anhui GWAS, 0.039; Hong Kong GWAS, 0.024; Anhui replication study, 0.030; European GWAS, 0.005; Hom et al.15 GWAS, 0.007; European replication study, 0.005).
Association for the gene(s) implicated by each SNP in other autoimmune diseases (excluding SLE) in Immunobase (http://www.immunobase.org): type 1 diabetes (T1D), celiac disease (CEL), multiple sclerosis (MS), Crohn’s disease (CRO), primary biliary cirrhosis (PBC), psoriasis (PSO), rheumatoid arthritis (RA), ulcerative colitis (UC), ankylosing spondylitis (AS), autoimmune thyroid disease (ATD), juvenile idiopathic arthritis (JIA), alopecia areata (AA), inflammatory bowel disease (IBD), narcolepsy (NAR), primary sclerosing cholangitis (PSC), Sjögren’s syndrome (SJO), systemic scleroderma (SSC) and vitiligo (VIT).
To highlight potential causal genes at the ten newly described susceptibility loci, we tested the associated SNPs at each locus for correlation with cis-acting gene expression in ex vivo naive CD4+ T cells and CD14+ monocytes in both Asian and European population data16, and in B cells, T cells and monocytes (stimulated and naive) in Europeans only17. We calculated regulatory trait concordance (RTC) scores18 (Online Methods) to test the relationship between expression quantitative trait loci (eQTLs) driven by disease-associated alleles and other, potentially stronger eQTLs, which we identified at each locus. Supplementary Table 3 and Supplementary Figure 2 present results for this analysis in all cell types in circumstances where eQTLs were found in at least one cell type or population. The eQTLs were consistent across cell type and population for LBH (rs19991732), CTSW (rs494003), RNASEH2C (rs494003) and ZFP90 (rs1170426), with carriage of the SLE risk allele correlating with reduced expression (except in lipopolysaccharide-stimulated monocytes for RNASEH2C, for which the eQTL results were not significant and the RTC scores were very low). The SNP rs2297550 was found to be a putative eQTL for IKBKE. The SLE risk allele for this SNP correlated with reduced expression in T cells, interferon-stimulated monocytes, B cells and NK cells, but increased expression in monocytes.
We integrated the results of the eQTL analyses with an in silico survey of murine phenotype data resulting from gene knockouts within the associated SLE loci19–28 (Table 2). These lines of evidence pointed to a single likely causal gene at some loci—IKBKE and JAK2, for example. In other instances, we found evidence supporting the role of multiple genes as candidates at a given locus—for example, CTSW/RNASEH2C and CDH1/ZFP90. Locus Zoom29 plots, created using the European and meta-analyzed Chinese data, for all ten loci can be seen in Supplementary Figure 3. These plots facilitate a comparison of the alignment of the association signals in the two populations. Potential roles of the putative causal genes at the loci mapped in this study are described in Supplementary Table 4.
Table 2. Candidate genes at SLE-associated loci in meta-analysis.
| Associated SNP | Chr | Genes within ±200 kb of SNP | Genes within the same LD block as SNPa | Immune phenotype in murine modelb | Cis-eQTLs with SNP | Likely causal gene at locus |
|---|---|---|---|---|---|---|
| rs34889541 | 1 | ATP6V1G3, PTPRC (CD45), MIR181A1HG | PTPRC | PTPRC | PTPRC (ref. 19) | |
| rs2297550 | 1 | SRGAP2, SRGAP2D, IKBKE, RASSF5, EIF2D, DYRK3 | IKBKE | IKBKE, RASSF5 | IKBKE | IKBKE 20 |
| rs17321999 | 2 | YPEL5, LBH, LOC285043, LCLAT1 | LBH | LBH | LBH 21 | |
| rs6762714 | 3 | LPP, TPRG1-AS1 | LPP | |||
| rs17603856 | 6 | ATXN1 | ATXN1 | |||
| rs597325 | 6 | BACH2 | BACH2 | BACH2 | BACH2 (refs. 22,23) | |
| rs73135369 | 7 | CLIP2, GTF2IRD1, GTF2I, LOC101926943 | GTF2IRD1 | GTF2IRD1/GTF2I 24 | ||
| rs1887428 | 9 | RCL1, JAK2, INSL6 | JAK2 | JAK2 | JAK2 (ref. 25) | |
| rs494003 | 11 | EHBP1L1, KCNK7, MAP3K11, PCNX3, SIPA1, RELA, KAT5, RNASEH2C, AP5B1, OVOL1, OVOL1-AS1, SNX32, CFL1, MUS81, EFEMP2, CTSW, FIBP, CCDC85B, FOSL1, C11orf68, DRAP1, TSGA10IP, SART1 | AP5B1, OVOL1, OVOL1-AS1 | CTSW, MUS81, RELA, SIPA1 | CTSW, FIBP, MUS81, RNASEH2C | RNASEH2C26,27 |
| rs1170426 | 16 | SMPD3, ZFP90, CDH3, CDH1 | ZFP90, CDH3 | CDH1 | ZFP90 | ZFP90 (FIK)28 |
The LD block is defined as SNPs showing a correlation (r2) of 0.75 with the associated SNP.
The immune phenotype designation is taken from http://www.informatics.jax.org/phenotypes.shtml of genes within ±200 kb of the associated SNP.
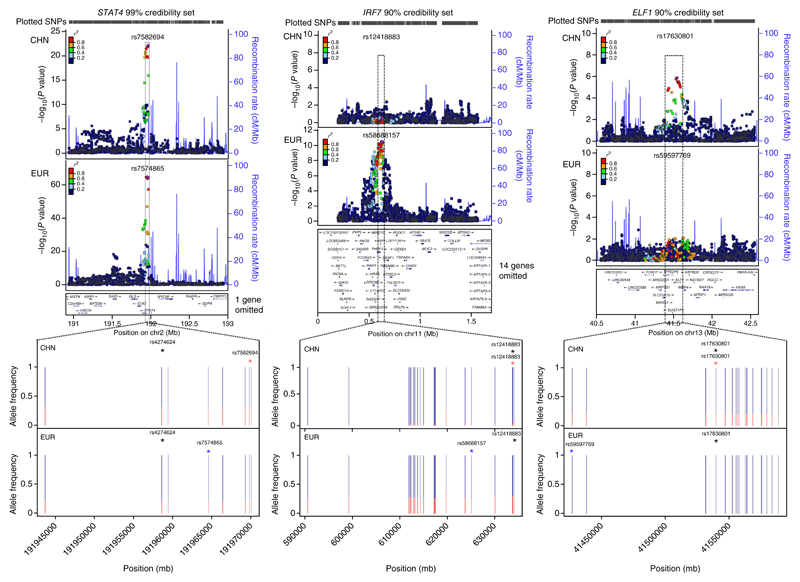
We further exploited the level of shared association we noted in our initial combination of the GWASs for the two populations studied using fine-mapping analyses of all published associated loci (Supplementary Table 1) and the new loci reported here. We derived Bayesian credibility sets in each population for the most likely causal variants using a previously published approach30–32; here we report the intersection of these sets (Online Methods). Supplementary Figure 4 shows the observed cumulative distribution for the number of SNPs in the intersection over a range of levels. When we used the least stringent criterion (75% credibility set), 80% of the mapped loci had sets identifying ten or fewer likely causal SNPs. When we used a very rigorous criterion (99% credibility set), seven of the loci comprised fewer than ten SNPs (Supplementary Table 5). STAT4 is an example of the colocalization of signals from each ancestry; in contrast, in two examples the association arose in one population only: IRF7 (European) and ELF1 (Chinese) (Fig. 2). In each case it is evident that the likely explanation for the discrepant association signal is population-specific differences in allele frequency within the credible SNP set. Supplementary Figure 5 shows fine-mapping data for the novel loci.
Figure 2.
Fine-mapping examples for STAT4, IRF7 and ELF1. The upper plots are LocusZoom plots showing association significance (−log10(P value)) and local LD (r2; color-coded). Circular points represent SNPs contained within the credibility sets, and square points represent SNPs not contained in the sets. The lower plots display the minor allele frequencies for all the SNPs in the intersection of the European (EUR) and Chinese (CHN) credibility sets. The minor allele frequency is plotted in red. The SNPs with the highest posterior probability within the intersection of the confidence intervals are highlighted by blue (highest posterior probability in the EUR data), red (highest posterior probability in the CHN data) and black (highest posterior probability in the CHN–EUR meta-data) asterisks. The credibility set coverage (99% for STAT4, 90% for IRF7 and ELF1) was chosen as the maximum coverage that included a maximum of 30 SNPs.
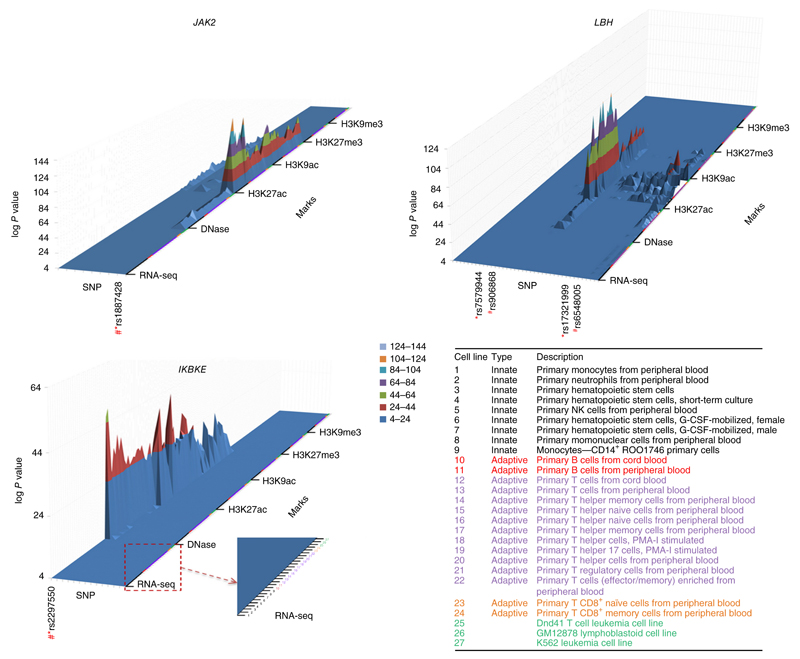
We downloaded epigenetic data covering each of the ten newly associated loci identified by our meta-analysis (Table 1) from the RoadMap Consortium for all blood cell types33. This was done for all SNPs within the credibility set at each locus. Figure 3 shows the results for SNPs at three loci, including the level of RNA expression (RNA-seq), accessibility to DNase, histone modification by acetylation (H3K27ac, H3K9ac) and histone modification by methylation (H3K27me3, H3K9me3). Supplementary Figure 6 shows results for the other seven SNPs (identities of all SNPs are presented in Supplementary Table 6). The histone marks were selected to indicate the activation status of promoter and enhancer regions and regions of repression. This epigenetic annotation provides an interesting point of comparison with the eQTL results. Two intense histone acetylation peaks were observed around the associated SNPs rs2297550 (IKBKE) and rs1887428 (JAK2), yet only the variant in IKBKE showed a significant eQTL in the cells examined (for example, P = 1.5 × 10−8 in naive monocytes in Europeans). Although we did find a significant eQTL for rs1887428 with JAK2 in monocytes, the RTC scores were low (<0.4). At SNPs rs34889541 (CD45) and rs597325 (BACH2), there was local evidence of histone acetylation in lymphocytes, but the two SNPs were not significant eQTLs. In contrast, rs1170426 (ZFP90) was a very significant eQTL (for example, in Europeans, P = 7.2 × 10−22 in CD4+ T cells and P = 4.6 × 10−55 in B cells), but the region around the associated SNP showed little evidence of regulatory function. However, there was strong evidence of epigenetic effects at other SNPs contained in the ZFP90 credibility set. Some of the discrepancies between eQTL and epigenetic annotation probably represent the limited set of activation states (and perhaps samples sizes) of primary immune cells that have been subject to eQTL investigation.
Figure 3.
3D enrichment plots depicting epigenetic modifications of ±50 bp overlapping all SNPs in the credibility sets for the 11 newly identified associated SNPs. The SNPs are shown as individual tracks on the x-axis with the SNP used in the replication study (*) and the SNP that showed the best evidence for colocalization with the most prominent epigenetic mark (#). Other SNP identities are listed in supplementary Table 6. The z-axis represents the log10 P value against the null hypothesis that peak intensity arises from the control distribution. The z-axis is truncated at a lower level (P < 10−4). For each novel associated locus, results are shown for RNA expression (RNA-seq), accessibility to DNase, histone modification by acetylation (H3K27ac, H3K9ac) and histone modification by methylation (H3K27me3, H3K9me3) over 27 immune cells. The data from the blood cell types are consistently ordered on the y-axis according to the annotation in the lower right of the figure: categories 1–9, innate-response immune cells; categories 10–24, adaptive-response immune cells (categories 10 and 11, B cells; categories 12–24, T cells); categories 25–27, cell lines.
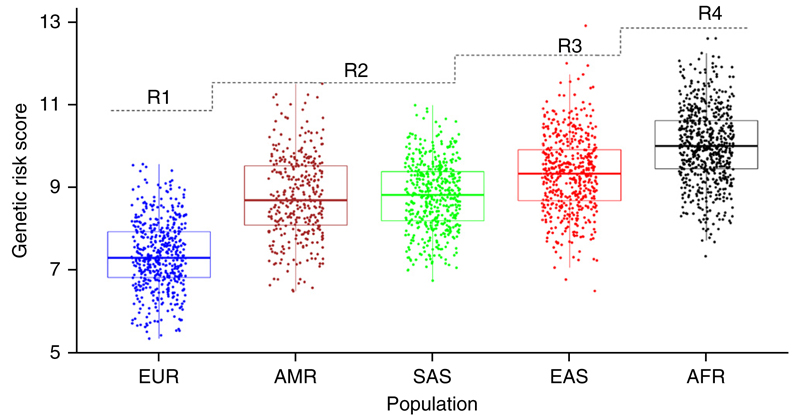
We investigated the amount of shared risk effects between the Chinese and European populations further with a coheritability analysis using LD score regression34 (Online Methods), which showed a significant (P = 4.0 × 10−3, rg = 0.51) correlation between the two populations. This correlation was stronger (P = 4.88 × 10−5, rg = 0.62) after removal of the MHC, which emphasizes its heterogeneity (Fig. 1b). We observed that on average the risk allele frequencies in Chinese control subjects were significantly higher than those in European controls in the respective GWASs (paired t-test, P = 0.02, Supplementary Fig. 7a), whereas the effect sizes (odds ratios) were not statistically different (P = 0.47, Supplementary Fig. 7b), suggesting that the higher prevalence of SLE in Asians (as compared with Europeans) may have a genetic basis. We also compared the genetic risk scores (GRSs)—the joint effect of odds ratios and risk allele frequencies—between the two populations in data from 1KG (phase 3) (Fig. 4) and between the Chinese and European GWAS controls (Supplementary Fig. 8). The GRS for SLE in East Asians (EAS) was significantly higher than that in Europeans (EUR) in the 1KG data (fold (EAS/EUR) = 1.27, P = 4.99 × 10−179; EUR = 7.38, 95% CI 7.31–7.45; EAS = 9.35, 95% CI 9.27–9.43). There was a similar difference in score between the GWAS controls (fold (Chinese/EUR) = 1.28, P = 1.00 × 10−797; EUR = 7.42, 95% CI 7.40–7.44; Chinese = 9.51, 95% CI 9.46–9.55). If more associations are identified in future studies, especially with increased power in non-European populations, including East Asians, the difference in genetic predisposition between populations identified by GWASs might increase further. We note that an analysis of chip heritability (using all genotyped SNPs to calculate heritability explained; Online Methods) in both the Chinese and the European data resulted in 28% (s.e. = 2.6%) explained in Chinese subjects and 27% (s.e. = 1.0%) explained in Europeans.
Figure 4.
Box plots of GRS across the five major population groups. These are standard box plots showing medians, interquartile ranges and whiskers indicating 1.5 times the interquartile range (Tukey box plots). EUR, European, N = 498; AMR, Amerindian, N = 347; SAS, South Asian, N = 487; EAS, East Asian, N = 503; AFR, African, N = 657; from the 1KG phase 3 release. The dashed line represents the increase in prevalence with the rank order (R1 represents the lowest prevalence, and R4 the highest).
Furthermore, we noted correlation among the GRSs across all five major 1KG super-populations and rank of the prevalence2 (Online Methods) of SLE (Fig. 4). A t-test on mean GRS between each pair of population data showed high significance (P < 10−16) for all pairs except Amerindian versus South Asian (P = 0.67), and a linear model with rank of prevalence predicting the GRS was significant (P < 10−16, r2 = 0.39). We excluded the MHC from this analysis because of the difficulty of defining the best model of association in this region, owing to the extensive LD and limited genotyping of SNPs and classical HLA in both populations.
The increased genetic risk load in Chinese individuals would help explain the continued increased prevalence of SLE in Asians after their migration to Western locations2. We acknowledge that the trends we have observed are a snapshot, as all available genotyped SNPs explained <30% of disease heritability, and the comparison of GRSs might not be a full reflection of genetic risk among these populations. A more detailed study of the increased prevalence of SLE in Asians, and in Africans, will require extensive comparisons of genetic and environmental data, including generation of DNA sequence data to exclude European bias in genotyping arrays.
URLs. Department of Twin Research, King’s College London, Twins–UK samples, http://www.twinsuk.ac.uk; Ingenuity Pathway Analysis, http://www.ingenuity.com/; Immunobase, http://www.immunobase.org; Systems Biology and Complex Disease Genetics, http://insidegen.com; RoadMap data, http://egg2.wustl.edu/roadmap/data/byFileType/signal/consolidatedImputed/; 1KG imputed summary statistics, http://insidegen.com/insidegen-LUPUS-data.html.
Online Methods
Study design in brief
We combined summary genome-wide association data from two Chinese GWASs4,5 (Anhui Province, mainland China, 1,047 cases (63 males) and 1,205 controls (673 males), λGC = 1.05; Hong Kong, 612 cases (50 males) and 2,193 controls (919 males), λGC = 1.04) and a European GWAS (4,036 cases (365 males) and 6,959 controls (2,785 males), λGC = 1.16 with λ1,000 = 1.02), after imputing all three studies to the 1KG data density, and conducted a meta-analysis. As the European data comprised 70% of both total cases and total controls, and were therefore the driving force in this meta-analysis, we selected SNPs for replication in an additional set of Chinese samples first. We identified a subset of SNPs in the Chinese replication that passed an FDR of 1% to take forward for replication in European samples. We then carried out replication using a second European GWAS15 independent of our main European GWAS and de novo genotyping in a new data cohort of European ancestry.
Imputation
We pre-phased each of the three studies separately using SHAPEIT35. We then separately imputed the studies (using IMPUTE36) with 1KG reference data (phase 1 integrated set, March 2012, build 37). The three data sets were aligned and meta-analyzed using R37 by the King’s College London group and independently by the groups at Anhui and Hong Kong using METAL38. SNPs with imputation INFO scores of <0.7 in any of the three studies were removed from further analysis. The numbers of SNPs available before and after quality control (QC), per chromosome and per associated locus, are shown in Supplementary Table 7a,f. A summary of INFO scores and imputation cross-validation are presented in Supplementary Table 7b-e for each chromosome and Supplementary Table 7g-j for each associated locus. Supplementary Note 3 presents a discussion of the limitations of using imputed data.
Statistical analysis
Association testing
After imputation, we analyzed each GWAS data set for association (SNPTEST36), fitting an additive model. We used the inverse variance method for meta-analysis, combining data from the three studies for SNPs with an imputation INFO score of >0.7 in all three studies.
Testing for heterogeneity. We tested for heterogeneity between the association signals in the Chinese and European data using Cochran’s Q statistic (1 degree of freedom in this case). The P values on the −log10 scale are plotted in Figure 1b. Q-Q plots (one per chromosome) for the heterogeneity P values can be seen in Supplementary Figure 9a, and Bland–Altman plots for differences in genetic effect (log odds ratio) estimates are in Supplementary Figure 9b.
Assessment of shared association between ancestries
To assess the extent to which genetic association with SLE was shared between the Chinese and European populations, we compared association results in the European GWAS3 with a meta-analysis of both Chinese GWASs, for SNPs published as associated in European3 and/or Chinese studies4,6–9. Association signals were declared as ‘shared’ between the Chinese and European populations if the SNP met any one of the following four criteria:
The locus had a published association in both Chinese and European studies at a genome-wide level of significance (P < 5 × 10−8).
The SNP was published only for Europeans but the association P value in the Chinese meta-analysis was significant (FDR < 0.01 across all SNPs in this group) and the direction of effect in all three GWASs was the same.
The SNP was published only in a Chinese study but the association P value in the European GWAS was significant (FDR < 0.01) and the direction of effect in all three GWASs was the same.
If the SNP failed to meet the requirements for either (2) or (3), we performed a gene-based test (applying the software KGG39–41) on genes within ±1 Mb of the published SNP. The locus was deemed shared if the gene-based P value was significant at the 0.01 FDR level after adjustment for multiple testing across all genes tested.
We also performed a meta-analysis (European GWAS + both Chinese GWASs) of all loci published in either Chinese or European studies (each published SNP ± 1 Mb) and recorded the most associated SNP. For loci published in Europeans, we declared the loci shared if the P value (adjusted for multiple testing over all SNPs tested within the 2-Mb region) in the Chinese data passed an FDR of 0.01 across all the loci published only in Europeans. We performed the reverse test for all loci published only in Chinese. Although this did not identify any additional shared loci (Supplementary Table 1b), there was suggestive evidence for two loci (P < 0.05 after multiple testing adjustment within loci but not after adjusting across loci).
Consistency of association between ancestries
We tested the hypothesis that the genome-wide association signals were consistent between the two populations. Post-1KG imputed association data were used for SNPs with INFO > 0.7. These genome-wide association signals were separated into 1-Mb regions (moving 1-Mb windows across the genome, 2,698 in total). We removed the extended MHC with a conservative buffer zone (chr. 6, from 20 Mb to 40 Mb), leaving 2,678 regions. We also removed regions that had an excessively (more than 2 s.d. from the average) low (N < 1,000) or high (N > 3,000) density of SNPs. This removed only 10% of the regions, leaving 2,338 regions. The lowest P value within each window was taken as the strength of association for that particular window. Each P value within each region was adjusted for multiple testing using a Bonferroni adjustment, to avoid bias in ranking agreement owing to the lowest P value being correlated with the number of statistical tests. The 1-Mb regions within each population’s data were then ranked according to the P value (lowest P value having rank 1). We tested agreement in ranking using Kendall’s τ statistic. Supplementary Figure 7c shows heat maps of the ranks for all 2,338 regions, the top 250 regions and the top 50 regions. The order in the heat maps was determined by the sum of the ranks. For comparison, we also included a simulated ranked data set; we permuted the numbers 1–2,338 in two separate data sets and produced a heat map ordered by the sum of the ranks.
Testing for independent effects within loci
We tested for independent effects of the two SNPs (rs17321999 and rs7579944) within the 2p23.1 locus by fitting a multiple regression model with both SNPs as explanatory variables (results for each SNP in this analysis are conditional on the other SNP as a covariate). We checked LD between the two SNPs in all data sets. We combined the conditional results in meta-analysis in the same way as in the single-marker analysis.
Selection of SNPs for replication study
We used a number of criteria to select SNPs for replication in the Chinese samples. We chose only SNPs that were not within a 1-Mb window of loci that had previously been published as associated with SLE. We selected SNPs that had P value significance levels at meta-analysis of <10−4. Three SNPs in loci not previously reported as associated with SLE had a genome-wide level of significance (P < 5 × 10−8) after meta-analysis. SNPs spanning a 1-Mb window were considered as one region, and we selected only independent SNPs within this region, using LD as a measure of independence. We carried out a gene-based test on the meta-analyzed data, using only SNPs with INFO scores > 0.9, with the software KGG39–41. One SNP from each of the loci that passed a gene-based test at the level of P < 10−5 was chosen; some of these had already been selected as having P < 10−4 in the meta-analysis as single markers. In total, 105 SNPs were selected for replication in the Chinese replication cohort. Of these, 66 passed QC, and 18 SNPs with FDR < 1% were taken forward to the European replication.
Genotyping of replication data
Genotyping of 130 SNPs was carried out for the 3,614 cases and 5,924 controls forming the Chinese replication set, using the Sequenom platform. This set of 130 SNPs included 105 SNPs in loci not previously reported as associated with SLE and 25 SNPs in loci that had previously been published as associated with SLE. The 105 potential new SLE SNPs included, in some cases, multiple SNPs in the same loci where we had some evidence of independence. We carried out several QC steps: we removed SNPs with >10% missing data (25 SNPs), and then subjects with >5% missing data. Two SNPs were monomorphic. Of the remaining 103 SNPs, 77 were in regions of the genome with potential new SLE associations. We removed 13 SNPs after we checked the genotyping allele intensity plots closely for clustering quality and tested for Hardy–Weinberg equilibrium (HWE). SNPs were removed if HWE P < 1.00 × 10−4. After QC, the Chinese replication consisted of 3,043 cases and 5,074 controls with genotyping on 64 SNPs. The European replication data comprised 1,478 cases and 6,925 controls genotyped for 18 SNPs with an FDR of 1% in the Chinese replication study. The cases were of European ancestry and were a subset of those used in the replication study in the European GWAS3; in the current study we carried out new genotyping on these 18 SNPs, and the controls were the same as used in that study (these samples were checked for European ancestry using a principal component analysis spiked with HapMap samples; see the original paper3). One of the 18 SNPs typed in the European replication cohort for this study (rs2297550) failed genotyping, and the remaining 17 SNPs passed QC (<3% missing data, HWE P > 1.00 × 10−4). An additional European GWAS was also used for replication, comprising 1,165 cases and 2,107 controls15.
Gene expression data
Gene expression data came from two sources. We obtained data from Fairfax et al.17 and unpublished data from B. Fairfax and J. Knight for NK cells, naive monocytes, monocytes stimulated by lipopolysaccharide (harvested after 2 h and 24 h), monocytes stimulated by interferon, and B cells. We obtained CD4 (CD4+ T cells) and CD14 (CD14/16+ monocytes) data from a previous study of gene expression in immune-related cells16. We made an adjustment for multiple testing using FDR = 0.01. To test whether observed associations between SNPs and expression levels of cis-acting genes were due to chance, we calculated the RTC score18.
Fine-mapping Bayesian credibility sets
For each of the associated loci in Supplementary Table 1 and Table 1, we calculated a Bayes factor for each SNP within the 2-Mb window. We used the approximate Bayes factor of Wakefield32. We then calculated the posterior probability that each SNP was driving the association, using the Bayes factors, and created credibility sets as recently described32. We created credibility sets using the European data and the Chinese data separately and overlaid the sets (Supplementary Fig. 5). We focused on the intersection of these two sets and determined the SNPs with highest posterior probability within this intersection, along with allele frequencies. We focused on the intersection of the two populations’ sets, as credibility sets calculated from the overall meta-analysis were driven by the European data. This would also be true if we were to use Bayesian updating (where the posterior probabilities from one population are used as priors in the other population). The intersection of the sets gave a subset of each population’s credibility set that was more likely to contain the true casual SNP.
RoadMap data
We downloaded the epigenetic data for SNPs within the credibility intervals (as defined in Supplementary Fig. 5) around each meta-analysis SNP (Table 1) from the RoadMap Consortium for all blood cell types. We chose DNase, RNA-seq, H3K27ac (distinguishing active enhancers/promoters), H3K27me3 (repressive domains), H3K9ac (promoters) and H3K9me3 (constitutive heterochromatin). The files downloaded contained the consolidated imputed epigenetic data based on the P value signals from each of the individual epigenetic marks in each of the cell types within whole blood. We used the UCSC genome browser (hg19) to subset each epigenetic track for regions containing each credibility SNP and then exported the signal data via Galaxy42. In selecting chromatin enrichments at each mark for each SNP within the credibility set, we ensured that no SNP was less than 10 bp away from the edge of the 25-bp epigenetic interval containing it. For SNPs closer to the edge of the chromatin interval, we averaged the enrichment from two adjacent intervals. We plotted 3D enrichment diagrams for each chromatin mark in each cell type for each SNP within the credibility set (Fig. 3 and Supplementary Fig. 6). Figure 3 and Supplementary Figure 6 highlight SNPs contained within peaks of enrichment (log10 P < 1 × 10−4) with tick marks; these SNPs are listed in Supplementary Table 6.
Genetic structure of SLE in European and Asian populations
We calculated the genetic risk score according to the method described by Hughes et al.43, taking the number of risk alleles (i.e., 0, 1 or 2) for a given SNP and multiplying it by the natural log of its odds ratio (OR). We calculated the cumulative risk score in each subject by summing the risk scores from the loci in Supplementary Table 1, excluding the MHC, plus the 11 SNPs newly reported in this paper, which robustly associated with SLE and passed QC in each population:
where m represents the number of SLE risk loci, Oi indicates the OR of risk SNPi, and G is the number of risk alleles at a given SNP. Cumulative risk scores were calculated for 498 founders in Europeans (EUR), 503 in East Asians (EAS), 487 in South Asians (SAS), 347 in the Amerindian group (AMR) and 657 in Africans (AFR) from 1KG phase 3. We tested for differences in GRS using a t-test. A Q-Q plot for each data set satisfied assumptions of normality, and given the large sample sizes, the central limit theorem would satisfy normality for the distribution of sample means. As there was evidence of differences in variances of the GRSs between some pairs of populations (EUR versus AMR, P = 9.97 × 10−5; AMR versus SAS, P = 5.37 × 10−5; SAS versus EAS, P = 4.50 × 10−3), we used a Welch two-sample t-test that does not assume equal variances. The variances in each group were as follows: Chinese controls, 0.75; European controls, 0.69; 1KG EAS, 0.86; 1KG EUR, 0.67; 1KG SAS, 0.66; 1KG AMR, 0.99; 1KG AFR, 0.77. We used the SNPs in Supplementary Table 1a to calculate the GRS for each population. We used the estimated OR from the EUR GWAS for the calculation of the GRS in Europeans (EUR and GWAS controls) and the OR from the Chinese GWAS for the calculation of the GRS in the EAS and Chinese GWAS controls. The OR from the EUR–Chinese meta-analysis was used in calculating the GRS in the AMR, SAS and AFR populations. Supplementary Note 1 presents an assessment of the robustness of our approach. Supplementary Note 2 provides details on SLE prevalence.
Heritability explained
We calculated the heritability explained by all genotyped SNPs in the Chinese and European populations using GCTA44. We assumed that the Chinese have an approximately threefold increase in prevalence compared with the Europeans, so we set the prevalence at 0.0003 in Europeans and 0.001 in Chinese. We used a cutoff for relatedness at 0.05, and we used sex as a covariate. The results were h2 = 28.4% (s.e. = 2.6%) in Chinese and h2 = 27.0% (s.e. = 1.0%) in Europeans for autosomal SNPs. We found that the results were robust to choice of relatedness for the autosomal SNPs (a cutoff of 0.125 resulted in h2 = 28.4% (s.e. = 2.6%) in Chinese and h2 = 27% (s.e. = 1.0%) in Europeans), whereas this was not so for the X chromosome (a cutoff of 0.125 resulted in h2 = 1.2% (s.e. = 0.5%) in Chinese and h2 = 1.1% (s.e. = 0.2%) in Europeans); a cutoff for relatedness at 0.05 resulted in h < 0.015 in both populations.
To compare both populations using the same SNP density, we re-ran the analysis on the overlap of genotyped SNPs (267,005 SNPs with minor allele frequency > 1% in Chinese and 264,833 with minor allele frequency > 1% in Europeans) and found that the heritability explained was higher in the data for the Chinese population: h2 = 30.2% (s.e. = 2.6%) in Chinese versus h2 = 22.7% (s.e. = 0.9%) in Europeans.
Genetic correlation between European and Chinese SLE GWASs
To estimate genetic correlation (rg), we applied LD score regression34 to the summary association data in the European GWAS and the meta-analysis of the Chinese data (the input data were all GWAS summary statistics, not just the SLE risk loci discussed in this paper). Although this methodology is designed to compare the similarity of genetic risk across diseases in the same population, here it served only to illustrate similarity across populations for one disease and to highlight the heterogeneity at the MHC. We used both Asian (rg = 0.49, P = 3.00 × 10−3) and European (rg = 0.51, P = 4.00 × 10−3) reference LD information. This analysis was carried out using summary data on all the SLE risk loci presented in this paper, and a further analysis was conducted after removal of the MHC (Asian (rg = 0.63, P = 6.92 × 10−7) and European (rg = 0.62, P = 4.88 × 10−5)). The increase in rg after removal of the MHC illustrates the major heterogeneity at this locus.
Supplementary Material
Note: Any Supplementary Information and Source Data files are available in the online version of the paper..
Acknowledgments
P.T. is employed by the Biomedical Research Centre. L.C. was funded by the China Scholarship Council (201406380127). The research was funded/supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. T.J.V. was awarded funding to carry out genotyping and analysis from G. Koukis, an Arthritis Research UK Special Strategic Award, and the Wellcome Trust (grant 085492). T.J.V. was awarded funding by the MRC (L002604/1, “Functional genomics of SLE: a transancestral approach”). Y.C., X. Zhang, S.Y. and Y.S. acknowledge support from the Key Basic Research Program of China (2014CB541901, 2012CB722404 and 2011CB512103), the National Natural Science Foundation of China (81402590, 81371722, 81320108016 and 81171505), the Research Project of the Chinese Ministry of Education (213018A), the Program for New Century Excellent Talents in University (NCET-12-0600) and the Natural Science Fund of Anhui Province (1408085MKL27). W.Y. and Y.L.L. acknowledge support from the Research Grant Council of the Hong Kong Government (GRF HKU783813M, HKU 784611M, 17125114 and HKU 770411M). Y.Z. thanks the Health and Medical Research Fund (12133701) from the Food and Health Bureau, Hong Kong. We thank T. Raj and P. De Jager for contributing gene expression data (CD4+ T cells and CD14/16+ monocytes in Asian and European populations; available in the NCBI Gene Expression Omnibus under accession number GSE56035). We thank B. Fairfax and J. Knight for contributing gene expression data on NK cells, naive monocytes, LPS-stimulated monocytes (harvested after 2 h and 24 h), interferon and B cells. We thank S. Daffern for downloading the ChIP-seq data in contribution to the epigenetic analysis.
For the replication study in Europeans, samples were provided by the Swedish SLE Network (led by L.R.). Replication genotyping was performed by the SNP&SEQ Technology Platform in Uppsala, which is part of the Swedish National Genomics Infrastructure (NGI) hosted by Science for Life Laboratory. The controls for the European GWASs and replication were obtained from dbGaP accession phs000428.v1.p1 (a study sponsored by the National Institute on Aging (grants U01AG009740, RC2AG036495 and RC4AG039029) and conducted by the University of Michigan); melanoma study data under dbGaP accession phs000187.v1.p1; a blood clotting study (dbGaP accession phs000304.v1.p1); and prostate cancer study data under dbGaP accession phs000207.v1.
The French cases for the European replication study were provided by the PLUS study, funded by a grant from the French PHRC 2005 Ministère de la Santé (ClinicalTrials.gov: NCT00413361 to N.C-C.). Participants were F. Ackermann, Z. Amoura, B. Asli, L. Astudillo, O. Aumaître, C. Belizna, N. Belmatoug, O. Benveniste, A. Benyamine, H. Bezanahary, P. Blanco, O. Bletry, P. Bourgeois, B. Brihaye, P. Cacoub, E. Chatelus, J. Cohen-Bittan, R. Damade, E. Daugas, C. De-Gennes, J.-F. Delfraissy, A. Delluc, H. Desmurs-Clavel, P. Duhaut, A. Dupuy, I. Durieu, H.-K. Ea, O. Fain, D. Farge, C. Funck-Brentano, C. Frances, L. Galicier, F. Gandjbakhch, J. Gellen-Dautremer, B. Godeau, C. Goujard, C. Grandpeix, C. Grange, G. Guettrot, L. Guillevin, E. Hachulla, J.-R. Harle, J. Haroche, P. Hausfater, J.-S. Hulot, M. Jallouli, J. Jouquan, G. Kaplanski, H. Keshtmand, M. Khellaf, O. Lambotte, D. Launay, P. Lechat, D.L.T. Huong, V. Le-Guern, J.-E. Kahn, G. Leroux, H. Levesque, O. Lidove, N. Limal, F. Lioté, E. Liozon, L.Y. Kim, M. Mahevas, K. Mariampillai, X. Mariette, A. Mathian, K. Mazodier, M. Michel, N. Morel, L. Mouthon, J. Ninet, E. Oksenhendler, T. Papo, J.-L. Pellegrin, L. Perard, O. Peyr, A.-M. Piette, J.-C. Piette, V. Poindron, J. Pourrat, F. Roux, D. Saadoun, K. Sacre, S. Sahali, L. Sailler, B. Saint-Marcoux, F. Sarrot-Reynauld, Y. Schoindre, J. Sellam, D. Sène, J. Serratrice, P. Seve, J. Sibilia, C. Simon, A. Smail, C. Sordet, J. Stirnemann, S. Trad, J.-F. Viallard, E. Vidal, B. Wechsler, P.-J. Weiller, and N. Zahr.
Y.L.L. is thankful for generous donations from Shun Tak District Min Yuen Tong of Hong Kong that partially supported the SLE GWAS in Hong Kong. Y.L.L. and W.Y. thank the doctors who contributed SLE cases and colleagues in the LKS Faculty of Medicine, University of Hong Kong, who provided controls used in the GWASs.
Footnotes
Author Contributions
Y.-F.W., Z.Z. and P.T. contributed equally to this work. T.J.V., X. Zhang, Y.C., Y.L.L. and W.Y. supervised the study. Z.Z., L.W., C.Y., L.L., L.Y., F.L., Y.H., X.Y. and S.Y. performed sample selection and data management, undertook recruitment and collected phenotype data for the Anhui Chinese data. L.R., B.G.F., R.E.V., G.S., N.C.-C. and P.M.G. performed sample selection and data management, undertook recruitment and collected phenotype data for the European data. A.L.R. and Y.S. worked on genotyping of both Chinese and European replication studies. D.L.M., Y.S., Y.Z. and Y.-F.W. carried out statistical analysis of the GWAS data. D.L.M. and P.T. carried out the 1000 Genomes Project imputation in the European GWAS. Y.S., X. Zuo, R.C. and T.W. carried out the 1000 Genomes Project imputation in the Anhui and Hong Kong Chinese GWASs. D.L.M., P.T., Y.S., X. Zuo, Y.-F.W. and Y.Z. carried out statistical analysis for the meta-analysis of the 1000 Genomes Project imputed data. D.L.M., Y.S. and Y.Z. designed the replication studies’ chips. B.G.F. and R.E.V. contributed data to the European replication cohort. D.L.M. and J.B. performed quality control on the European data for the replication study. D.L.M. analyzed European replication data. D.L.M., Y.S. and Y.Z. analyzed Anhui replication data. Y.-F.W. and D.L.M. designed and performed genetic risk score comparison between the populations. Y.-F.W. performed the LD score regression analysis. D.L.M. and L.C. carried out the eQTL analysis. D.L.M. and D.S.C.G. carried out the epigenetic analysis. D.L.M., T.J.V., D.S.C.G., X. Zhang, Y.C., Y.S. and W.Y. wrote the manuscript. All authors read and contributed to the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Lawrence JS, Martins CL, Drake GL. A family survey of lupus-erythematosus. 1. Heritability. J Rheumatol. 1987;14:913–921. [PubMed] [Google Scholar]
- 2.Danchenko N, Satia J, Anthony M. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus. 2006;15:308–318. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 3.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng YJ, et al. Follow-up study identifies two novel susceptibility loci PRKCB and 8p11.21 for systemic lupus erythematosus. Rheumatology. 2011;50:682–688. doi: 10.1093/rheumatology/keq313. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet. 2013;92:41–51. doi: 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. Association analyses identifying two common susceptibility loci shared by psoriasis and systemic lupus erythematosus in the Chinese Han population. J Med Genet. 2013;50:812–818. doi: 10.1136/jmedgenet-2013-101787. [DOI] [PubMed] [Google Scholar]
- 9.Sheng YJ, et al. Association analyses confirm five susceptibility loci for systemic lupus erythematosus in the Han Chinese population. Arthritis Res Ther. 2015;17:85. doi: 10.1186/s13075-015-0602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, et al. ELF1 is associated with systemic lupus erythematosus in Asian populations. Hum Mol Genet. 2011;20:601–607. doi: 10.1093/hmg/ddq474. [DOI] [PubMed] [Google Scholar]
- 11.Fernando MMA, et al. Transancestral mapping of the MHC region in systemic lupus erythematosus identifies new independent and interacting loci at MSH5, HLA-DPB1 and HLA-G. Ann Rheum Dis. 2012;71:777–784. doi: 10.1136/annrheumdis-2011-200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manku H, et al. Trans-ancestral studies fine map the SLE-susceptibility locus TNFSF4. PLoS Genet. 2013;9:e1003554. doi: 10.1371/journal.pgen.1003554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 16.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jury EC, Kabouridis PS, Flores-Borja F, Mageed RA, Isenberg DA. Altered lipid raft-associated signaling and ganglioside expression in T lymphocytes from patients with systemic lupus erythematosus. J Clin Invest. 2004;113:1176–1187. doi: 10.1172/JCI20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, et al. Contribution of IKBKE and IFIH1 gene variants to SLE susceptibility. Genes Immun. 2013;14:217–222. doi: 10.1038/gene.2013.9. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann Rheum Dis. 2015;74:e13. doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plagnol V, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medici M, et al. Identification of novel genetic loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS Genet. 2014;10:e1004123. doi: 10.1371/journal.pgen.1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tantin D, Tussie-Luna MI, Roy AL, Sharp PA. Regulation of immunoglobulin promoter activity by TFII-I class transcription factors. J Biol Chem. 2004;279:5460–5469. doi: 10.1074/jbc.M311177200. [DOI] [PubMed] [Google Scholar]
- 25.Lu LD, et al. Depletion of autoreactive plasma cells and treatment of lupus nephritis in mice using CEP-33779, a novel, orally active, selective inhibitor of JAK2. J Immunol. 2011;187:3840–3853. doi: 10.4049/jimmunol.1101228. [DOI] [PubMed] [Google Scholar]
- 26.Crow YJ, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Günther C, et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J Clin Invest. 2015;125:413–424. doi: 10.1172/JCI78001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C, et al. Cutting Edge: a novel, human-specific interacting protein couples FOXP3 to a chromatin-remodeling complex that contains KAP1/TRIM28. J Immunol. 2013;190:4470–4473. doi: 10.4049/jimmunol.1203561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beecham AH, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell J, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 37.R-Core-Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li MX, Kwan JSH, Sham PC. HYST: A hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91:478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li MX, Gui HS, Kwan JSH, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li MX, Sham PC, Cherny SS, Song YQA. Knowledge-based weighting framework to boost the power of genome-wide association studies. PLoS One. 2010;5:e14480. doi: 10.1371/journal.pone.0014480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes T, et al. Analysis of autosomal genes reveals gene-sex interactions and higher total genetic risk in men with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:694–699. doi: 10.1136/annrheumdis-2011-200385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.