Abstract
Endocrine disruptions induced by environmental toxicants have placed an immense burden on society to properly diagnose, treat and attempt to alleviate symptoms and disease. Environmental exposures during critical periods of development can permanently reprogram normal physiological responses, thereby increasing susceptibility to disease later in life - a process known as developmental reprogramming. During development, organogenesis and tissue differentiation occur through a continuous series of tightly-regulated and precisely-timed molecular, biochemical and cellular events. Humans may encounter endocrine disrupting chemicals (EDCs) daily and during all stages of life, from conception and fetal development through adulthood and senescence. Though puberty and perimenopausal periods may be affected by endocrine disruption due to hormonal effects, prenatal and early postnatal windows are most critical for proper development due to rapid changes in system growth. Developmental reprogramming is shown to be caused by alterations in the epigenome. Development is the time when epigenetic programs are ‘installed’ on the genome by ‘writers’, such as histone methyltransferases (HMTs) and DNA methyltransferases (DNMTs), which add methyl groups to lysine and arginine residues on histone tails and to CpG sites in DNA, respectively. A number of environmental compounds, referred to as estrogenic endocrine disruptors (EEDs), are able to bind to estrogen receptors (ERs) and interfere with the normal cellular development in target tissues including the prostate and uterus. These EEDs, including diethylstilbestrol (DES), bisphenol A (BPA), and genistein (a phytoestrogen derived from soybeans), have been implicated in the malformation of reproductive organs and later development of disease. Due to the lack of fully understanding the underlying mechanisms of how environmental toxicants and their level of exposure affect the human genome, it can be challenging to create clear clinical guidance to address the potential health effects of lower-level exposures commonly experienced within the general population. In addition, human studies concerning environmental exposures are limited in feasibility by ethical concerns for human safety. Therefore, studies in animal models provide great opportunities to reveal links between early-life exposure to EDCs and related diseases. It has been shown that developmental exposure to EDCs, such as diethylstilbestrol (DES) and genistein, during reproductive tract development increases the incidence, multiplicity and overall size of uterine fibroids in the Eker rat model, concomitantly reprogramming estrogen-responsive gene expression. Importantly, EDC exposure represses enhancer of zeste 2 (EZH2) and reduces levels of the histone 3 lysine 27 trimethylation (H3K27me3) repressive mark through Estrogen receptor / Phosphatidylinositide 3-kinases / Protein kinase B non-genomic signaling in the developing uterus. More recent research identified a developmental reprogramming target, Scbg2a1 gene, whose epigenetic status can be altered by early exposure to BPA in the rat prostate. Molecular analyses revealed markedly increased expression (greater than 100 fold) of Scgb2a1, a secretaglobin gene in response to developmental exposure to BPA. This increase in Scgb2a1 expression is concomitantly associated with increased enrichment of acetylated H3K9 (H3K9Ac representing active chromatin status) and hypomethylation of DNA for a CpG island upstream of the transcription start site of Scgb2a1. These data suggest that expression of Scgb2a1 in the adult prostate could be epigenetically reprogrammed by BPA exposure during prostate development. Further studies are needed to create more targeted preventative interventions as well as specific, effective therapeutics to decrease the incidence of diseases.
Keywords: developmental environmental exposure, endocrine disrupting chemicals, epigenetics, prostate, uterine fibroids
Endocrine disruption and diseases
Endocrine disruptions induced by environmental toxicants have placed an immense burden on society to properly diagnose, treat, and attempt to alleviate symptoms and disease (1,2). Though not yet evaluated in the United States, a recent European study reports that the provision of long-term care and treatment for those affected by health conditions in which endocrine disrupting chemicals (EDCs) are suspected exceeds more than (the equivalent of) two billion dollars per year (3). Environmental exposures during critical periods of development can permanently reprogram normal physiological responses, thereby increasing susceptibility to disease later in life—a process known as developmental reprogramming. During development, organogenesis and tissue differentiation occur through a continuous series of tightly-regulated and precisely-timed molecular, biochemical and cellular events. Humans may encounter EDCs daily and during all stages of life, from conception and fetal development through adulthood and senescence. Though puberty and perimenopausal periods may be affected by endocrine disruption due to hormonal effects, prenatal and early postnatal windows are most critical for proper development due to rapid changes in system growth (4). Additionally, EDCs differ from other environmental toxicants and chemicals in that the effects of EDCs are often induced at small doses and vary based on the window of time of exposure. Thus, these seemingly minor levels of exposure exert subtle changes at the molecular and cellular levels that ultimately induce more severe pathophysiologic effects. Exposure to these and other environmental chemicals has been linked with infertility, delayed puberty, and premature birth (5), as well as with later development of several diseases such as diabetes mellitus (6), cardiovascular disease (7), and particularly neoplasia (8,9). Notably, early developmental exposures contribute to transgenerational inheritance of phenotype (10).
Developmental reprogramming and epigenetic regulation
It has been well-established that the genetic makeup of a human being, i.e. the human genome, plays a major role in determining predisposition to developing certain diseases. Family medical histories, for example, may demonstrate trends of increased risk of breast cancer due to family members carrying the BRCA1/2 gene mutation (11). Yet another example includes parent carriers of a mutation in the CFTR gene—offspring from these carriers are more likely to be diagnosed with cystic fibrosis as compared to parents who do not carry a CFTR mutation (12). Classical twin designs can decompose genetic and environmental sources of variance. More difficult to elucidate, however, is how a person’s physical environment alters the expression of his or her genome. The molecular mechanisms as to how exposure to external variables, e.g. diet, exposure to chemicals or radiation, climate, or medications, ultimately affect the regulation of the human epigenome leading to increased risk of developing disease are not well understood.
In recent years, more research has focused on the effects these environmental exposures have on a developing fetus. During fetal development, the human genome’s expression can be adapted to suit proper development of tissues in response to the fluctuating needs of the growing fetus’ body—genetic expression is altered to maintain physiologic conditions that optimize the fetus’ chance for survival and continued growth. Unfortunately, the plasticity of genetic expression during this critical time in fetal development can also negatively impact the developing fetus—as the fetus responds to adverse stimuli. A historical example of this was evidenced in infants born to mothers prenatally administered thalidomide: infants exhibited limb malformation; or diethylstilbestrol (DES): girls developed clear cell carcinoma of the vagina later in life (3,14).
Developmental reprogramming is shown to be caused by alterations in the epigenome (2,13,14). Epigenetic modifications play an important role in ‘programming’ lineage determination and cellular identity during development (15). Several different types of epigenetic modifications are thought to contribute to the alteration of gene expression during development (16–19). Among many type of epigenetic proteins which play a role in epigenetic modification, histone methyltransferases (HMTs) and DNA methyltransferases (DNMTs) function as epigenetic “writers”, which add methyl groups to lysine and arginine residues on histone tails and to CpG sites in DNA, respectively (20,21). For histone modifications, the epigenetic programs that are installed by these writers form a ‘histone code’ that is interpreted by ‘readers’ (effector molecules that recognize histone modifications) and modified by ‘erasers’ such as histone demethylases (20,22,23). Methylated CpG sites are also remodeled during this time via Ten-eleven translocation (Tet) enzymes that function as erasers for DNA methylation, converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and other oxidation products (24,25). The activity of epigenetic enzymes can be altered through specific posttranslational modifications (PTMs) leading to activating or inactivating enzymes, or regulating modifier binding of chromatin indirectly. For example, phosphorylation, one of the common PTMs, occurs via kinase mediators, e.g. cyclin-dependent kinases, (CDKs), protein kinase A (PKA), and protein kinase B (PKB/Akt) on these “readers, writers, and erasers” of histone methyl marks. The specific histone marks can be correlated with the activity of specific effector proteins, methyltransferases, and demethylases which ultimately play a role in epigenome reprogramming (2,26,27). The epigenetic modifiers whose activities are altered via phosphorylation include enhancer of Zeste homolog 2 (EZH2), mixed-lineage leukemia protein 1 (MLL1), and lysine-specific histone demethylase 1A (LSD1) (13). The activity of epigenetic enzymes is particularly important during development, because they play an important role in remodeling the epigenome after fertilization and during gametogenesis (24), as well as in several types of cancer (28).
Environmental exposure and estrogen signaling
A number of environmental compounds, referred to as estrogenic endocrine disruptors (EEDs), are able to bind to estrogen receptors (ERs) and interfere with the normal cellular development in target tissues including the prostate and uterus. These EEDs, including diethylstilbestrol (DES), bisphenol A (BPA), and genistein (a phytoestrogen derived from soybeans), have been implicated in the malformation of reproductive organs and later development of disease (2).
These past mass-exposures to endocrine disrupting chemicals, like DES, during reproductive tract development have been linked with reprogramming of estrogen-responsive gene expression in the uterine myometrium. This leads to tissue hyper-responsiveness to ovarian sex hormones, specifically estrogen and progesterone, later in adult life, and predisposes women to the development of uterine leiomyoma (27). Animal studies have shown that early-life exposure to DES during uterine development (in rats, uterine development occurs post-natally) permanently alters the morphology of the reproductive tract via an “estrogen imprint” despite the readily-metabolized nature of DES and its efficient clearing from the body (3). These experiments have provided evidence of the long-term, permanent pathophysiological effects of even brief exposure to environmental endocrine disruptors, such as DES. The challenge remains, however, to connect exposure to other environmental toxicants which result in permanent epigenetic changes and disease-related outcomes.
Developmental exposures to EDCs increase uterine fibroid development
Due to the lack of fully understanding the underlying mechanisms of how environmental toxicants and their level of exposure affect the human genome, it can be challenging to create clear clinical guidance to address the potential health effects of lower-level exposures commonly experienced within the general population. In addition, human studies concerning environmental exposures are limited in feasibility by ethical concerns for human safety. Therefore, studies in animal models provide great opportunities to reveal links between early-life exposure to EDCs and related human diseases including uterine fibroids. For example, Eker (TscEk/+) rats, heterozygous for the tuberous sclerosis 2 (Tsc2) gene, i.e. carrying one Tsc2 mutant allele, have a 65% incidence of spontaneously developing uterine fibroids, generally around 12 months of age (8). Dr. Walker’s group described the long-term effects of postnatal exposure to EDCs (DES, and genistein) on the uteri of Eker rats after allowing them to develop to 16 months of age (14). Early-life exposure to EDCs including DES or genistein increased tumor penetrance (from 65% to >90%), tumor multiplicity, and overall size. While the molecular mechanisms are still being revealed, these experiments with genistein show that in the developing uterus, genistein induces epigenetic changes of non-genomic estrogen receptor (ER) signaling by way of activation of the PI3K/AKT pathway. This in turn phosphorylates histone methyltransferase Enhancer of Zeste Homolog 2 (EZH2), a potent epigenetic regulator of gene expression and inactivates EZH2. This ultimately reduces levels of H3K27me3 found in chromatin, which are normally a mark of repressed gene expression (14). Thus, the overall expression of estrogen-responsive genes is, in turn, increased (8). Even 16 months following brief exposure to genistein during uterine development, their studies showed increased activity of EZH2 suggesting interference with epigenetic programming in the development of the uterus, leading to permanent alterations that persist into adult life. Further investigation is needed to identify the direct epigenetic link between developmental programming targets and EDC exposures during uteri development.
Developmental exposure to BPA increases the risk of carcinogenesis in the prostate
Besides the uterus, the prostate is another altered organ targeted by adverse developmental exposure. Induction of carcinogenesis in the prostate in response to developmental exposure to BPA has been reported (29). In the animal model, brief developmental exposure to BPA ultimately induces later estrogen-mediated carcinogenesis of the prostate in rats. Prins et al conducted experiments using progenitor cells expressing estrogen receptors (ERs)-α and β, derived from prostate glands of young, disease-free men. These cells, when grafted into a kidney-capsule mouse model for tissue formation, formed normal human prostate epithelium that produced prostate-specific antigen (PSA). When these mice were treated with testosterone and estradiol (T+E), the prostate tissue began to show pathologic progression from normal tissue growth to hyperplasia and finally prostatic intraepithelial neoplasia over a 4-month period (15). These findings suggest that the estrogen-responsiveness of prostate stem and progenitor cells may provide a link to the epigenetic disruption caused by early-life BPA exposure in the human prostate, potentially leading to carcinogenesis of the prostate (29).
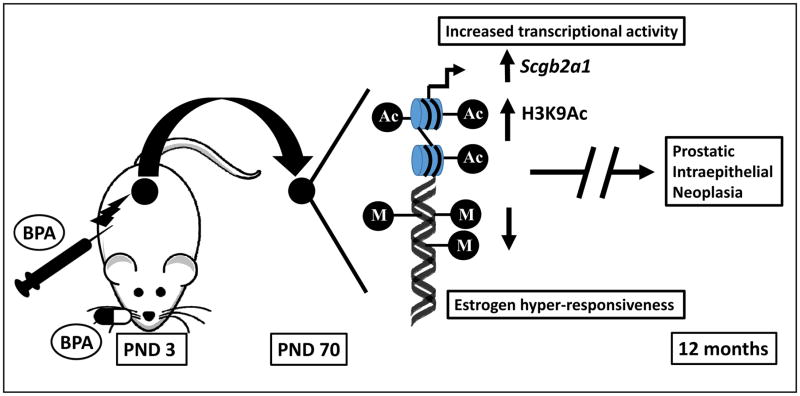
A recent study by Wong et al further supports the link between exposure to environmental toxicants during plastic developmental periods and epigenetic alteration of gene expression (30). This study identified a developmental reprogramming target, Scbg2a1 gene, whose epigenetic status can be altered by early exposure to BPA in the rat prostate. As depicted in the figure, using an animal model of early developmental exposure, rat pups were exposed (either subcutaneously or orally, to mimic the route of exposure likely to occur in humans) to BPA at postnatal days 1, 3, and 5 in three separate populations. The following were analyzed: 1) serum BPA levels; 2) molecular changes in the prostate at 70 days post-BPA-treatment; 3) histopathologic changes in the prostate at 12 months after the rats were implanted with T+E-containing capsules at Day 70 to drive prostate carcinogenesis. The histopathologic results in the population maintained for the 12-month longitudinal demonstrated dysplasia (the rat equivalent of human prostatic intraepithelial neoplasia), with the incidence of dysplasia increasing with increasing oral dose of BPA. Additionally, both BPA-exposed and vehicle groups treated with T+E on and after 70 days demonstrated adenocarcinomas and carcinomas in situ, indicative of these rats’ susceptibility to T+E-promoted carcinogenesis in the prostate (30).
Figure.
Early-life exposure to BPA results in increased enrichment of acetylated H3K9 (H3K9Ac representing active chromatin status) and hypomethylation of DNA for a CpG island upstream of the transcription start site of Scgb2a1. M: Methyl; Ac: Acetyl; PND: postnatal days; BPA: Bisphenol A
Molecular analyses revealed markedly increased expression (greater than 100 fold) of Scgb2a1, a secretaglobin gene in response to developmental exposure to BPA. This increase in Scgb2a1 expression is concomitantly associated with increased enrichment of acetylated H3K9 (H3K9Ac representing active chromatin status) and hypomethylation of DNA for a CpG island upstream of the transcription start site of Scgb2a1. These data suggest that expression of Scgb2a1 in the adult prostate could be epigenetically reprogrammed by BPA exposure during prostate development. Further potential implications include increased risk for cancer in response to chemotherapeutics associated with prostatein binding (30). Though the functional significance of reprogrammed Scgb2a1 has yet to be fully elucidated, there is evidence of its being a marker of carcinogenesis and disease recurrence in ovarian cancer. Its overexpression has also been found in endometrial and lung cancers (16). SCGB2A1 has been proposed to be involved in micrometastasis via the lymph node in abdominal cancers, biliary tract carcinoma, and breast cancer indicating its potential role as a gene that can be reprogrammed. It remains unclear, however, as to whether it serves as a marker of reprogramming rather than a driver of carcinogenesis (16). Discovering genes like Scgb2a1 and understanding the mechanisms connecting them to developmental reprogramming could manifest into libraries of potential biomarkers of epigenetic alterations. These biomarkers could help predict one’s predisposition to developing disease and one’s response to therapeutics as a result of early-life exposure to environmental toxicants, events that would otherwise seem disconnected by the span of time between them. It remains to be determined whether these effects of reprogramming are similarly present in the human prostate gland and whether similar early-life exposure to BPA can be determined to mediate later prostate carcinogenesis in humans.
Concluding remarks
With increasing emphasis on the susceptibility and mechanisms of developmental environmental exposures, greater advances will be made toward identifying the biomarkers useful in predicting human predisposition to disease. Moreover, researchers can then focus on creating more targeted preventative interventions as well as specific, effective therapeutics to decrease the incidence of disease. Though the complexity of signaling pathways, gene expression patterns, and the vast array of environmental toxicants may blur the lines connecting early cause and late effect, each step towards increased knowledge will bring humanity closer to decreasing the burden on society that poor health sets upon it.
Acknowledgments
This work was supported in part by an Augusta University Startup package, the National Institutes of Health grant HD04622811 (to AA), and the Augusta University Intramural Grants Program (QY). We would like to thank Walidah Walker, MPH for editing this manuscript.
Footnotes
Conflict of interest: None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Heindel JJ. Role of exposure to environmental chemicals in the developmental basis of reproductive disease and dysfunction. Seminars in reproductive medicine. 2006;24:168–177. doi: 10.1055/s-2006-944423. [DOI] [PubMed] [Google Scholar]
- 2.Walker CL, Ho SM. Developmental reprogramming of cancer susceptibility. Nature reviews Cancer. 2012;12:479–486. doi: 10.1038/nrc3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trasande L, Zoeller RT, Hass U, Kortenkamp A, Grandjean P, Myers JP, DiGangi J, Bellanger M, Hauser R, Legler J, Skakkebaek NE, Heindel JJ. Estimating burden and disease costs of exposure to endocrine-disrupting chemicals in the European union. The Journal of clinical endocrinology and metabolism. 2015;100:1245–1255. doi: 10.1210/jc.2014-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gore AC, Heindel JJ, Zoeller RT. Endocrine disruption for endocrinologists (and others) Endocrinology. 2006;147:S1–3. doi: 10.1210/en.2005-1367. [DOI] [PubMed] [Google Scholar]
- 5.Crews D, McLachlan JA. Epigenetics, evolution, endocrine disruption, health, and disease. Endocrinology. 2006;147:S4–10. doi: 10.1210/en.2005-1122. [DOI] [PubMed] [Google Scholar]
- 6.Chevalier N, Fenichel P. Endocrine disruptors: new players in the pathophysiology of type 2 diabetes? Diabetes & metabolism. 2015;41:107–115. doi: 10.1016/j.diabet.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Roseboom TJ. Undernutrition during fetal life and the risk of cardiovascular disease in adulthood. Future cardiology. 2012;8:5–7. doi: 10.2217/fca.11.86. [DOI] [PubMed] [Google Scholar]
- 8.Cook JD, Davis BJ, Cai SL, Barrett JC, Conti CJ, Walker CL. Interaction between genetic susceptibility and early-life environmental exposure determines tumor-suppressor-gene penetrance. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8644–8649. doi: 10.1073/pnas.0503218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook JD, Davis BJ, Goewey JA, Berry TD, Walker CL. Identification of a sensitive period for developmental programming that increases risk for uterine leiomyoma in Eker rats. Reprod Sci. 2007;14:121–136. doi: 10.1177/1933719106298401. [DOI] [PubMed] [Google Scholar]
- 10.Bernal AJ, Jirtle RL. Epigenomic disruption: the effects of early developmental exposures. Birth defects research Part A, Clinical and molecular teratology. 2010;88:938–944. doi: 10.1002/bdra.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. American journal of human genetics. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller K. Genetic counseling: DNA testing for the patient. Proceedings. 2005;18:134–137. doi: 10.1080/08998280.2005.11928052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ly L, Chan D, Trasler JM. Developmental windows of susceptibility for epigenetic inheritance through the male germline. Seminars in cell & developmental biology. 2015;43:96–105. doi: 10.1016/j.semcdb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Sandovici I, Hammerle CM, Ozanne SE, Constancia M. Developmental and environmental epigenetic programming of the endocrine pancreas: consequences for type 2 diabetes. Cellular and molecular life sciences : CMLS. 2013;70:1575–1595. doi: 10.1007/s00018-013-1297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nature structural & molecular biology. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Richardson WD. Genetics meets epigenetics: HDACs and Wnt signaling in myelin development and regeneration. Nature neuroscience. 2009;12:815–817. doi: 10.1038/nn0709-815. [DOI] [PubMed] [Google Scholar]
- 17.Stuppia L, Franzago M, Ballerini P, Gatta V, Antonucci I. Epigenetics and male reproduction: the consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clinical epigenetics. 2015;7:120. doi: 10.1186/s13148-015-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen TP. The epigenetics of early development: inferences from stem cells. Molecular reproduction and development. 2014;81:194–201. doi: 10.1002/mrd.22269. [DOI] [PubMed] [Google Scholar]
- 19.Van de Vijver G, Van Speybroeck L, De Waele D. Epigenetics: a challenge for genetics, evolution, and development? Annals of the New York Academy of Sciences. 2002;981:1–6. doi: 10.1111/j.1749-6632.2002.tb04909.x. [DOI] [PubMed] [Google Scholar]
- 20.Trevino LS, Wang Q, Walker CL. Phosphorylation of epigenetic “readers, writers and erasers”: Implications for developmental reprogramming and the epigenetic basis for health and disease. Progress in biophysics and molecular biology. 2015;118:8–13. doi: 10.1016/j.pbiomolbio.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. The New England journal of medicine. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 22.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Developmental cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews Molecular cell biology. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Mas A, Diamond MP, Al-Hendy A. The Mechanism and Function of Epigenetics in Uterine Leiomyoma Development. Reprod Sci. 2015 doi: 10.1177/1933719115584449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bredfeldt TG, Greathouse KL, Safe SH, Hung MC, Bedford MT, Walker CL. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol Endocrinol. 2010;24:993–1006. doi: 10.1210/me.2009-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greathouse KL, Bredfeldt T, Everitt JI, Lin K, Berry T, Kannan K, Mittelstadt ML, Ho SM, Walker CL. Environmental estrogens differentially engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Molecular cancer research : MCR. 2012;10:546–557. doi: 10.1158/1541-7786.MCR-11-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends in genetics: TIG. 2014;30:464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prins GS, Hu WY, Shi GB, Hu DP, Majumdar S, Li G, Huang K, Nelles JL, Ho SM, Walker CL, Kajdacsy-Balla A, van Breemen RB. Bisphenol A promotes human prostate stem-progenitor cell self-renewal and increases in vivo carcinogenesis in human prostate epithelium. Endocrinology. 2014;155:805–817. doi: 10.1210/en.2013-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong RL, Wang Q, Trevino LS, Bosland MC, Chen J, Medvedovic M, Prins GS, Kannan K, Ho SM, Walker CL. Identification of secretaglobin Scgb2a1 as a target for developmental reprogramming by BPA in the rat prostate. Epigenetics : official journal of the DNA Methylation Society. 2015;10:127–134. doi: 10.1080/15592294.2015.1009768. [DOI] [PMC free article] [PubMed] [Google Scholar]