Abstract
The deubiquitinase-encoding gene Cyld displays a dominant genetic linkage to a wide spectrum of skin-appendage tumors, which could be collectively designated as CYLD mutant–syndrome (CYLDm-syndrome). Despite recent advances, little is understood about the molecular mechanisms responsible for this painful and difficult-to-treat skin disease. Here, we generated a conditional mouse model with epidermis-targeted expression of a catalytically deficient CYLDm through K14-Cre–mediated deletion of exon 9 (hereafter refer to CyldEΔ9/Δ9). CyldEΔ9/Δ9 mice were born alive but developed hair and sebaceous gland abnormalities and dental defects at 100% and 60% penetrance, respectively. Upon topical challenge with DMBA/TPA, these animals primarily developed sebaceous and basaloid tumors resembling human CYLDm-syndrome as opposed to papilloma, which is most commonly induced in WT mice by this treatment. Molecular analysis revealed that TRAF6-K63-Ubiquitination (K63-Ub), c-Myc-K63-Ub, and phospho-c–Myc (S62) were markedly elevated in CyldEΔ9/Δ9 skin. Topical treatment with a pharmacological c-Myc inhibitor induced sebaceous and basal cell apoptosis in CyldEΔ9/Δ9 skin. Consistently, c-Myc activation was readily detected in human cylindroma and sebaceous adenoma. Taken together, our findings demonstrate that CyldEΔ9/Δ9 mice represent a disease-relevant animal model and identify TRAF6 and c-Myc as potential therapeutic targets for CYLDm-syndrome.
Cyld tumor suppressor plays a key role in ectodermal morphogenesis and suppression of epidermal neoplasia.
Introduction
CYLD is a deubiquitinase that can remove the K63-linked (K63-Ub) and M1-linked (M1-Ub) polyubiquitin polymers from an array of target proteins involved in signal transduction and gene regulation (1–9). Most notably, CYLD controls NF-κB signaling by hydrolyzing K63-Ub and/or M1-Ub chains from various substrates. Dysregulation of CYLD, as a result of transcriptional and posttranslational downregulation or genetic mutations, is linked to a number of human diseases, including inflammation and cancer. Somatic mutations of CYLD have been detected in spiradenocylindroma of kidney, gastric, and colon cancers (10, 11), while germline Cyld mutations predispose patients to multiple types of adnexal skin tumors, including cylindroma (OMIM 132700), Brooke-Spiegler syndrome (OMIM 605041), and triochoepithelioma (OMIM 601606), as well as sebaceous adenoma and eccrine spiradenoma (hereafter collectively referred to as CLYD mutant–syndrome [CYLDm-syndrome]) (12–20). Over 50 missense and truncation mutations have been characterized in CYLDm-syndrome, and all of them result in expression of a catalytically deficient CYLDm. Tumors of CYLDm-syndrome generally develop after puberty and constitute the primary morbidity in these patients. Approximately 70% of these tumors exhibit loss-of-heterozygosity (LOH) of the WT allele (13, 14, 16, 18). Although mostly benign, CYLDm-syndrome is painful, disfiguring, and difficult to treat due to the diffuse and recurrent nature of the lesions. Additionally, they carry the risk of malignant transformation and metastasis over time (21–24).
Despite the increasing knowledge about the mutation status and disease linkage, little is understood about the molecular mechanisms mediating the multitude of CYLDm-driven human diseases. To date, several animal models have been created to examine the role of CYLD in the immune system and cancer, but none of them mirrors the genetic alterations and the clinical phenotypes observed in patients with CYLDm-syndrome. Cyld-/- mice displayed developmental defects of T-lymphocytes (7, 25), and in response to topical challenges with 7,12-Dimethylbenz(a)anthracene/12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA), they showed increased growth of papilloma without an increase of progression to squamous cell carcinoma (SCC) (25). In contrast, transgenic mice with K14-driven expression of catalytically deficient CYLDm (K14-CYLDm) were prone to the development of highly malignant and metastatic skin tumors following DMBA/TPA induction (26). The results of these studies underscore the importance of CYLD as an immune regulator and skin tumor suppressor but fail to recapitulate the fundamental features of human CYLDm-syndrome.
Since human CYLDm-syndrome is characterized by tumors originating from the hair follicles and is virtually always linked to expression of a catalytically deficient CYLDm, we predicted that CYLDm expressed in its native genetic setting selectively sensitizes mice to hair follicle defects and tumorigenesis. To test this idea, we resorted to the conditional Cyldfl9/fl9 mice, which — upon Cre-mediated deletion of exon 9 — expressed a catalytically deficient mutant (CYLDm) replacing the WT protein. Interestingly, mice with homozygous germline deletion of Cyld exon 9 displayed postnatal lethality due to lung defects (27), prohibiting further skin phenotypic analyses. In this study, we therefore generated K14Cre-Cyldfl9/fl9 mice (hereafter referred to as CyldEΔ9/Δ9) to introduce Cyld mutation exclusively in K14-positive hair follicle and basal epidermal cells. CyldEΔ9/Δ9 mice were born alive but developed skin, hair, and dental defects and were prone to the development of sebaceous adenoma or basaloid tumors that histologically resembled human adnexal skin tumors of CYLDm-syndrome following DMBA/TPA treatment. These results indicate that CyldEΔ9/Δ9 mice represent a human disease-relevant animal model and identify c-Myc as a mediator for CYLDm-syndrome.
Results
CyldEΔ9/Δ9 mice develop hair defects.
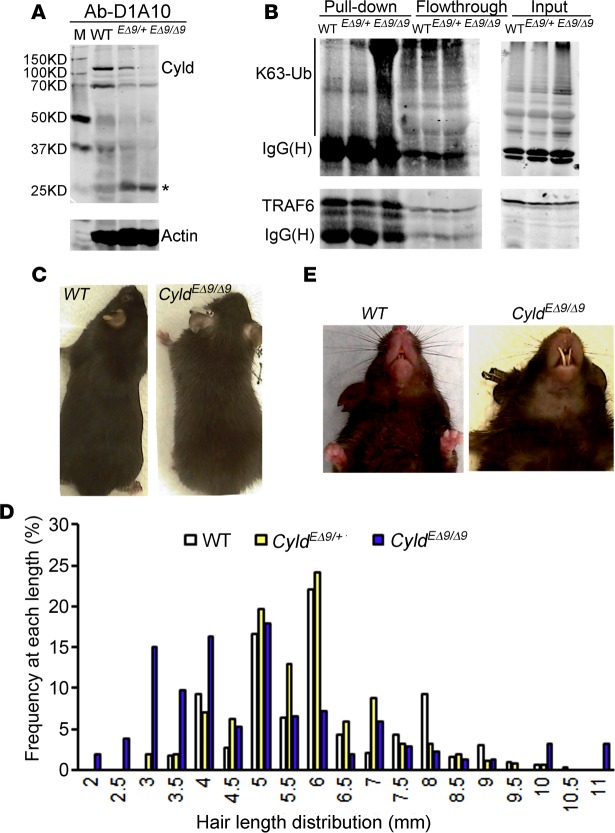
Mice with Cre-recombinase mediated deletion of Cyld exon 9 in germ cells carry a patient-relevant carboxyl-terminal-truncating Cyld mutation (Cyldm), but these animals die postnatally due to lung defects (27), thus prohibiting phenotypic analysis of skin and its appendages where Cyld is ubiquitously expressed (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.86548DS1). To circumvent the lethality issue, we generated a conditional knock-in mouse model (CyldEΔ9/Δ9) that introduced the Cyldm in the epidermal cells. This was achieved by crossbreeding the Cyldfl9 mice with transgenic mice expressing Cre-recombinase under the control of K14, a promoter active specifically in the hair follicle, sebaceous gland, and basal epidermal progenitor cells (28, 29). Epidermis-targeted deletion of exon 9 in the resulting CyldEΔ9/Δ9 mice was confirmed by PCR and reverse transcription PCR (RT-PCR) with genomic DNA and total RNA isolated from 1-month-old mouse epidermal cells, respectively (Supplemental Figure 2, A–C). Immunoblotting of mouse skin extracts with an antibody (Ab-D1A10) that preferentially recognizes the full-length CYLD indicated a marked decrease of the full-length protein in CyldEΔ9/Δ9 skin samples and an increase of a shorter fragment previously characterized as a proteolytic product of Cyld (27) (Figure 1A). On the other hand, immunoblotting of the same skin extracts with an antibody (Ab-H419) that apparently recognizes the carboxyl-terminally truncated Cyldm mutant detected a band at the predicted molecular weight of approximately 60KD in CyldEΔ9/+ and CyldEΔ9/Δ9 mouse skin extracts (Supplemental Figure 2D). To further verify that the expressed CYLDm is catalytically inactive, we analyzed the K63-Ub status of TRAF6, a previously well-characterized CYLD substrate (30). For this, protein extracts isolated from 1-month-old mice were subject to immunoprecipitation with an antibody against TRAF6, followed by immunoblotting for K63-Ub. As shown in Figure 1B, TRAF6-K63-Ub was significantly higher in CyldEΔ9/Δ9 mouse skin than the heterozygous CyldEΔ9/+ and WT control mice, consistent with the catalytic inactivation of CYLDm. As expected, the downstream NF-κB pathway exhibited increased activation in CyldEΔ9/Δ9 mouse skin, as indicated by the elevated levels of pIκBα (Supplemental Figure 2E). In contrast, the basal level of phospho-c–Jun was not significantly changed in these tissues.
Figure 1. Conditional expression of a truncated Cyld mutant (Cyldm) in epidermis leads to hair and dental defects.
(A) Immunoblotting of mouse skin extracts for CYLD (Ab-D1A10) and Actin. (B) IP of mouse epidermal protein extracts with an antibody against TRAF6 followed by immunoblotting with antibodies specific for K63-ubiquitin (K63-Ub) or TRAF6. The precipitated proteins, flow-through supernatants, and inputs were used for immunoblotting. (C) Clinical presentation of 1-month-old WT and CyldEΔ9/Δ9 mice. (D) Hair-length distribution. A total of 300 hairs of 2- to 3-month-old mice of each genotype were measured. (E) Clinical presentation of incisor teeth.
The CyldEΔ9/Δ9 mice were born at the Mendelian ratio and had no apparent developmental problems during the first week after birth. By 2 weeks of age, when hair shafts emerge (31), CyldEΔ9/Δ9 mice developed fuzzy hair coats as compared with those of the WT, CyldEΔ9/+, and Cyldfl/fl mice (Figure 1C, data not shown). Quantitative microscopic analysis of the hair shafts showed that CyldEΔ9/Δ9 mice hair had a normal ratio of guard, zig-zag, and owl/auchene hairs but were generally shorter and thinner than those of control siblings (Figure 1D and Supplemental Figure 3, A and B).
In addition to the hair defects, 60% of the CyldEΔ9/Δ9 adult animals exhibited an overgrowth of the lower incisor teeth that appeared longer but thinner than those of the control mice (Figure 1E), which is consistent with the expression of K14 in dental epithelial cells and a role of TRAF6 in dental development (32, 33). Examination of the upper jaw revealed an absence of the upper incisor teeth, suggesting that the uncontrolled growth of the lower incisor teeth is due to the lack of normal wear caused by grinding actions between the lower and upper incisor teeth. These findings indicate that epidermal cell–targeted Cyld loss-of-function mutation induces hair and dental growth defects.
CyldEΔ9/Δ9 mice develop sebaceous gland hyperplasia.
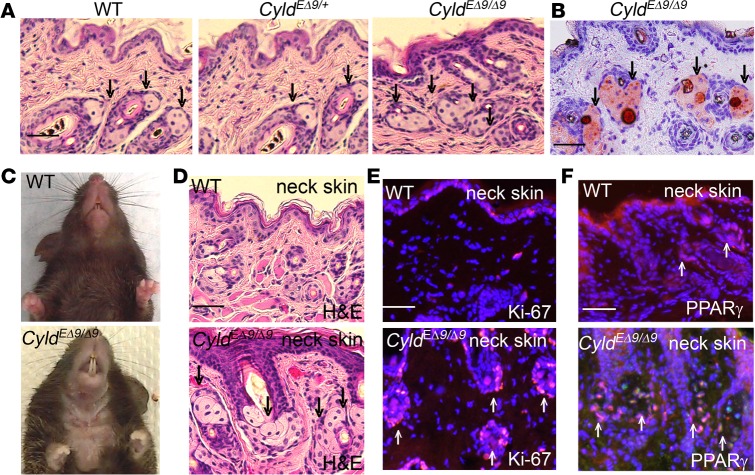
In order to examine the histological features of the epidermal structures, skin biopsies were collected from newborn and adult animals of various ages. At birth, CyldEΔ9/Δ9 mouse skin looked indistinguishable from that of the WT siblings as shown by H&E staining (Supplemental Figure 4A). Within 1 month after birth, the back skins of CyldEΔ9/Δ9 mice showed an increased abundance of negatively stained cell islands typical of K14-positive stem cell–derived and lipid-producing sebaceous glands (Figure 2A and Supplemental Figure 4B) (29, 34). Oil-red staining verified lipid-production in these cells (Figure 2B). The increase of sebaceous gland was also observed in the tail skin and in the Meibomian glands of the lower eyelid as revealed by Oil Red O and H&E staining (Supplemental Figure 4, C–E). In addition, 50% of CyldEΔ9/Δ9 mice suffered complete hair loss in the ventral neck area starting around 5 months of age (Figure 2C). Similar to the back skin, the neck skin of CyldEΔ9/Δ9 mice also contained an increased number of shiny cell islands that were highly proliferative, as indicated by the increased numbers of Ki-67–positive cells (Figure 2, D and E). As expected, those cell clusters expressed PPARγ, a peroxisome proliferator–activated receptor known to be expressed in the nuclei of sebaceous gland cells (35) (Figure 2F). Also in line with sebaceous glands being the sebum producer, the skin of CyldEΔ9/Δ9 mice appeared oily compared with the control mice, perhaps in part contributing to the fuzzy hair appearances. Of note, the skin, hair, and dental phenotypes described above were observed in every generation of offspring examined to date, suggesting that endogenous Cyldm-driven skin defects are penetrable. These results demonstrate that CYLD inactivation induces a hair growth defect and a sebaceous gland overgrowth.
Figure 2. CyldEΔ9/Δ9 mice develop sebaceous gland hyperplasia.
(A) H&E staining of the back skin sections of 1-month-old WT, CyldEΔ9/+, and CyldEΔ9/Δ9 mice. (B) Oil Red O staining of the back skin sections of 1-month-old CyldEΔ9/Δ9 mice. (C) Clinical presentation of 5-month-old mice. (D) H&E staining of the neck skin of 5-month-old mice. (E–F) Immunostaining for Ki-67 and PPARγ (orange) of the neck skin of 5-month-old mice. Nuclei (blue, Hoechst 32558). Scale bars: 50 μm. Arrowheads mark sebaceous glands with negative staining by H&E and positive nuclei staining for Ki-67 and PPARγ.
CyldEΔ9/Δ9 mice develop sebaceous adenoma or basaloid tumors in response to chemical treatment.
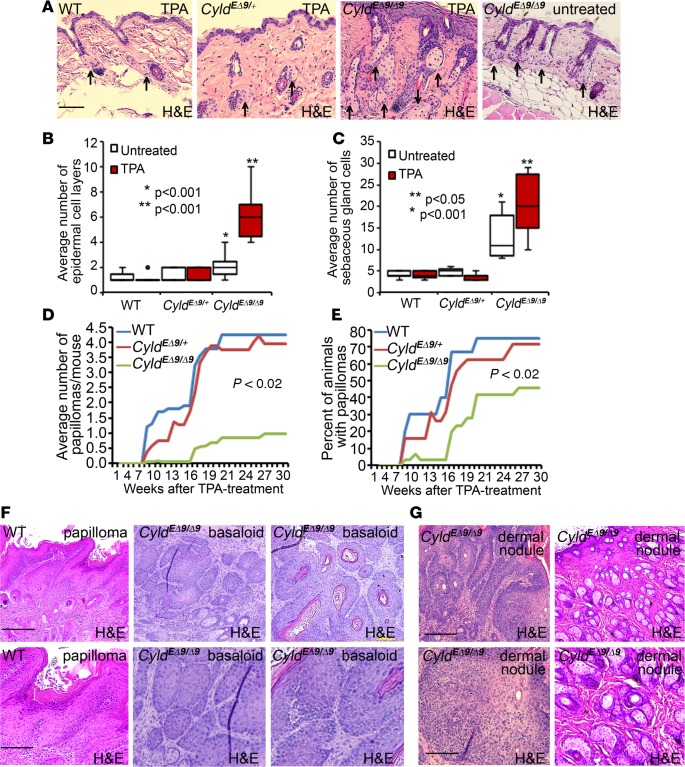
To determine whether CyldEΔ9/Δ9 mice were sensitive to environmental challenges, we subjected animals to biweekly topical treatments with TPA, a chemical promoter of murine keratinocyte cell proliferation. As expected, at 2 weeks after TPA treatment, all animals displayed interfollicular epidermal hyperplasia, and CyldEΔ9/Δ9 mice also showed sebaceous gland hyperplasia (Supplemental Figure 5). Interestingly, after 20 weeks of TPA treatment, WT and CyldEΔ9/+ mice resumed normal epidermal thickness, while CyldEΔ9/Δ9 mice maintained the interfollicular epidermal and sebaceous gland hyperplasia (Figure 3A). Quantitative analysis revealed that epidermal cellularity was increased by 2-fold and 5-fold in untreated and TPA-treated CyldEΔ9/Δ9 mice skin compared with WT counterparts, respectively (Figure 3B). Likewise, sebaceous gland cellularity was increased by about 2.5-fold and 4-fold in untreated and TPA-treated CyldEΔ9/Δ9 mice skin, respectively (Figure 3C). These results indicate that CyldEΔ9/Δ9 mice epidermis and sebaceous glands are sensitive to TPA challenge.
Figure 3. CyldEΔ9/Δ9 mice are prone to the development of basaloid tumor and sebaceous adenoma.
(A) H&E staining. Animals (n = 5/group) were treated with topical applications of TPA (10 ng/100 μl acetone) biweekly for 20 weeks. Histology of an untreated skin section of the same-age animal is included for comparison. Arrows mark sebaceous glands. Scale bar: 100 μm. (B–C) Epidermal and sebaceous gland cellularity. Data represent 25th–75th percentiles (box), median (line), and 5th and 95th percentiles (whiskers). Ten images of each group were counted. P <0.05 were obtained via 2-tiered student t test by (*) comparing mutant to WT skins and (**) TPA-treated to untreated skins. (D–E) Tumor multiplicity and percent of animals with clinically visible tumor following DMBA/TPA challenge. Animals (n = 10–29) were treated with 1 dose of DMBA followed by biweekly applications of TPA. P < 0.02 were obtained between CyldEΔ9/Δ9 and CyldEΔ9/+ mice via Student t test. (F–G) Representative images of H&E staining of (F) a WT tumor and 2 mutant tumors and (G) 2 dermal nodules from CyldEΔ9/Δ9 mice showing papilloma, basaloid, and sebaceous adenoma morphologies, respectively. Scale bar of the top panel: 200 μm; bottom panel: 100 μm.
Next, we challenged animals with DMBA/TPA as described in previous studies (26). We observed that CyldEΔ9/Δ9 animals displayed distinct features of tumor growth. Unlike WT and heterozygous control siblings, which developed typical surface skin tumors (namely papilloma-benign skin tumors arising from keratinocytes), CyldEΔ9/Δ9 mice showed a reduced rate and multiplicity of papilloma development (Figure 3, D and E). Instead, these animals developed numerous nodules that were visible on the dermal side during skin biopsy at the 20th week after DMBA/TPA challenge. In addition, the surface skin tumors developed in CyldEΔ9/Δ9 mice were histologically distinct from papilloma of control siblings. They had basaloid appearance that resembled human cylindroma and trichoepithelioma (Figure 3F). The dermal nodules showed features of (50%) basaloid tumor and (50%) sebaceous adenoma (Figure 3G). Further immunostaining showed that tumors developed on CyldEΔ9/Δ9 mice and control siblings were highly proliferative with minimal apoptosis, as indicated by the abundance of Ki-67–positive cells and the scarcity of cleaved caspase 3–positive cells, respectively (Supplemental Figure 6). Interestingly, proliferative cells were located in the basal and superbasal layers in the WT tumors; such a polarized distribution was not apparent in CyldEΔ9/Δ9 tumors. In addition, all tumors also expressed molecules commonly detected in benign skin tumors, including K5, K17, and E-cadherin, as well as β-catenin and CDK4 (Supplemental Figure 7 and Supplemental Figure 8, A and B). Gli1 was detected in all tumors but appeared a little more intense in some sebaceous adenoma cells of CyldEΔ9/Δ9 mice (Supplemental Figure 8C). Taken together, these results indicate that endogenous expression of Cyldm selectively sensitizes mice to hair follicle–derived epidermal tumorigenesis.
c-Myc displays increased activation and K63-Ub in CyldEΔ9/Δ9 mouse skin.
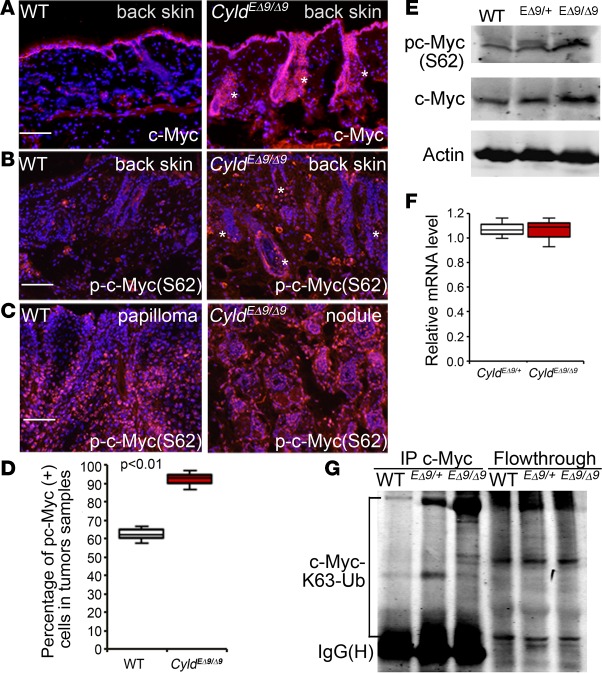
To investigate the molecular mechanisms responsible for the Cyldm-driven skin abnormalities, we examined the status of c-Myc and β-catenin, both of which have been previously characterized as important regulators of hair follicle cell lineage specification (34, 36, 37). By immunostaining, we found that c-Myc but not β-catenin was markedly increased in the epidermal and hair follicle cells of CyldEΔ9/Δ9 mice (Figure 4A and Supplemental Figure 9A). In addition, phospho-c–Myc (S62), which was previously characterized as an essential signal for c-Myc–mediated growth promotion (38), was highly expressed in the nuclei of sebaceous glands of CyldEΔ9/Δ9 mice compared with its basal cell–restricted expression in the WT skin (Figure 4B). As expected, phospho-c–Myc (S62) exhibited significantly increased expression in the basal and suprabasal cells but not in the outer layers of the WT papillomas; such a tissue polarity was absent in CyldEΔ9/Δ9 mouse sebaceous adenoma where phospho-c–Myc was significantly increased (Figure 4, C and D). Consistent with the increased c-Myc activation, CDK4, a known c-Myc target gene (39), was expressed at a high level in CyldEΔ9/Δ9 sebaceous gland cells, as demonstrated by immunostaining (Supplemental Figure 9B).
Figure 4. c-Myc displays increased activation and K63-Ub in CyldEΔ9/Δ9 mouse skin.
(A–C) Immunostaining of mouse back skin or chemically induced skin tumors. c-Myc or phospho-c–Myc(S62) (orange); Nuclei (blue). Asterisks mark c-Myc–positive hair follicles and sebaceous glands. (D) Quantification of phospho-c–Myc–positive cells. Graph shows average percentages of phospho-c–Myc (S62)–positive cells with data representing 25th–75th percentiles (box), median (line), and 5th and 95th percentiles (whiskers). Three images of each group were counted. P value was obtained via student t test. (E) Immunoblotting with mouse epidermal extracts for c-Myc, phospho-c–Myc (S62), and Actin. (F) RT-PCR for c-Myc with total RNA isolated from 3-month-old CyldEΔ9/+ and CyldEΔ9/Δ9 mice skin (n = 3 per group) and GAPDH used as an internal control. Data represent 25th–75th percentiles (box), median (line), and 5th and 95th percentiles (whiskers). (G) IP of mouse epidermal extracts with an antibody against c-Myc and then immunoblotting for K63-Ub or c-Myc. Scale bars: 100 μm.
Further immunoblotting with epidermal protein extracts confirmed the increase of phospho-c–Myc and c-Myc in CyldEΔ9/Δ9 mouse skin (Figure 4E). Since c-Myc expression was shown to be suppressed at a transcriptional level by CYLD through a JNK-dependent manner in liver cells (40), we asked whether c-Myc mRNA was increased in CyldEΔ9/Δ9 skin. By RT-PCR, we detected similar levels of c-Myc mRNA in the skin of CyldEΔ9/+ and CyldEΔ9/Δ9 mice (Figure 4F), which suggests that c-Myc is regulated at a posttranscriptional level in skin. To determine whether c-Myc undergoes K63-Ub, we performed immunoprecipitation for c-Myc followed by immunoblotting for K63-Ub and found that Myc-K63-Ub was markedly elevated in CyldEΔ9/Δ9 epidermis as compared with WT and CyldEΔ9/+ (Figure 4G). These data indicate that c-Myc polyubiquitination by K63-Ub chains is increased in CyldEΔ9/Δ9 mouse epidermal cells and may mediate its activation.
c-Myc is essential for hair follicle and sebaceous gland cell survival, and it is activated in human cylindroma and sebaceous adenoma.
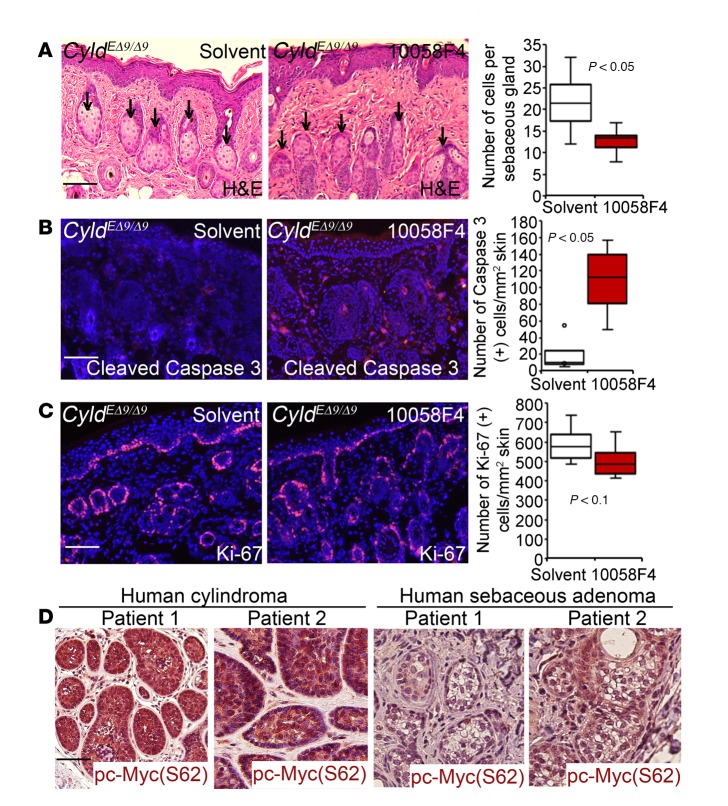
To determine whether c-Myc is important for the observed skin defects, we treated CyldEΔ9/Δ9 mice for 10 days with daily topical applications of 10058-F4, a pharmacological inhibitor for c-Myc/Max dimerization and transcriptional activity (41). By H&E staining, we found that up to 80% of sebaceous glands in tissues treated with 10058-F4 appeared smaller and darker than those of the solvent control group, with an average of 45% reduction of sebaceous gland cellularity (Figure 5A). To determine whether these phenotypic changes were caused by increased cell death or decreased cell proliferation, we performed immunostaining for the apoptotic cell marker cleaved caspase 3 and the cell proliferation marker Ki-67. We found that the number of cells immunostained for the cleaved caspase 3, an apoptotic cell marker, was increased in the treated tissues by about 4-fold (Figure 5B). In contrast, cell proliferation was not significantly affected by 10058-F4, as indicated by quantitative analysis of Ki-67–positive cells (Figure 5C). In agreement with these animal data, phospho-c–Myc (S62) was readily detectable in 90% of human cylindroma and sebaceous adenoma tissue samples examined (n = 10 for each group) (Figure 5D). These results indicate that c-Myc is essential for hair follicle and sebaceous gland cell survival and tumorigenesis.
Figure 5. c-Myc is important for hair follicle–derived cell survival of CyldEΔ9/Δ9 mouse skin and is activated in human cylindroma and sebaceous adenoma.
(A) H&E staining of CyldEΔ9/Δ9 mice skin tissues treated with DMSO solvent or 10058-F4. Arrows mark sebaceous glands. Graph shows averages of sebaceous gland cells counted from 10 images of each condition. Data represent 25th–75th percentiles (box), median (line), and 5th and 95th percentiles (whiskers). (B–C) Immunostaining for cleaved caspase 3 and Ki-67 (orange); nuclei (blue, Hoechst 32558). Graphs show the average number of cells positively stained for cleaved caspase 3 and Ki-67 counted from 6 images of 2 different mice treated with the same condition. Data represent 25th–75th percentiles (box), median (line), and 5th and 95th percentiles (whiskers). P values were obtained via 2-tiered Student t test. (D) Immunoperoxidase staining of human cylindroma and sebaceous adenoma for phospho-c–Myc (S62) (brown) counterstained with hematoxylin. Two representative patient samples were shown for each group (n = 10). Scale bar: 100 μm.
Discussion
We have demonstrated that epidermal cell–targeted homozygous Cyld-mutation induces hair, sebaceous gland, and dental growth defects and sensitizes animals to the development of trichoepithelioma, sebaceous adenoma, and basaloid tumors. These tumor growth phenotypes are characteristic of human CYLDm-syndrome. Additionally, although rare, Brooke-Spiegler syndrome, which overlaps with CYLDm-syndrome both phenotypically and genotypically (42, 43), has been associated with peg-shaped teeth (44). Thus, CyldEΔ9/Δ9 mice represent a multidisease-relevant animal model. Current studies with this model have led to the identification of c-Myc, a new effector of CYLDm-syndrome.
Previous studies have characterized the NF-κB signaling pathway as a predominant CYLD downstream effector (2–4), which has led to the clinical trial of treating cylindroma patients with aspirin (45). The clinical outcome is encouraging but not satisfactory, suggesting that there are other molecular targets involved in the etiology. c-Myc, an oncogene that we found to be activated in human cylindroma and sebaceous adenoma, represents another effector molecule in the pathogenesis. In agreement with this notion, c-Myc has been previously characterized as a dominant regulator that promotes sebaceous gland growth and commitment of epidermal stem cells into hair follicle appendage cells and tumors (34, 36, 46, 47).
c-Myc activation via K63-Ub has been previously characterized in breast cancer and colorectal cancer cells and has been shown to play an essential role in regulation of cancer cell proliferation (48, 49). In breast cancer cells, c-Myc-K63-Ub is catalyzed by the E3 ubiquitin ligase HectH9 and enhances interaction with the coactivator p300 and consequently increased c-Myc transcriptional activity (48). It will be interesting to determine whether HectH9 or another E3 ubiquitin ligase is required for c-Myc activation in keratinocytes.
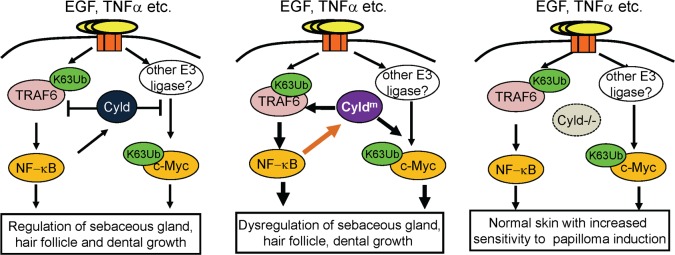
It is intriguing to note that, unlike CyldEΔ9/Δ9 animals, both K14-driven Cyldm transgenic and Cyld–/– animals are sensitive to the development of papilloma but have no apparent abnormalities in the hair follicle (25, 26). Thus, the expression of Cyldm by the endogenous Cyld promoter, as opposed to the K14 promoter, appears to be essential for the hair follicle cell–specific defects. Another important difference between our Cyldm knockin model and the K14-driven Cyldm transgenic mouse model is the fact that, in the latter model, the WT Cyld gene is coexpressed with the Cyldm transgene, raising the possibility of a functional interference between the 2 gene products. Current findings pinpoint a working model in which NF-κB and c-Myc are positively regulated by growth factors such as EGF, cytokines such as TNFα via TRAF6, and likely other E3 ubiquitin ligases in epidermal cells (Figure 6). In the presence of WT Cyld, NF-κB transcriptionally induces CYLD, which in turn inhibits NF-κB activation (3, 4, 50), thereby forming a self-regulated signaling loop. In homozygous Cyld mutant cells, CYLDm exacerbates signal transduction to gene regulators such as NF-κB, c-Myc, and AP-1 through binding to various substrates and forms a self-augmenting feedback loop. Consequently, the elevated actions of c-Myc, NF-κB, AP-1, and possibly other gene regulators collectively promote hair follicle–derived cell growth and survival. Such dominant positive effects on signal transduction and transcriptional feedback loops are absent in Cyld–/– cells. Further, in agreement with a dominant role of CYLDm versus complete CYLD loss in pathogenesis, mice with homozygous germline deletion of Cyld exon 9 (CyldEΔ9/Δ9) display postnatal lethality due to developmental lung defects (27), while CyldEΔ9/+ and Cyld–/– mice exhibit near normal development (7, 25). Similarly, mice with homozygous deletion in hepatocytes (CyldLΔ9/Δ9), but not the heterozygous CyldLΔ9/+ mice, develop liver fibrosis, inflammation, and hepatocarcinoma (51). In agreement with the animal data, heterozygous Cyld mutant carriers have no overt developmental abnormalities, whereas homozygous human carriers have not been reported thus far, presumably due to lethality or low frequency of marriage between carriers (52). Nevertheless, the majority of the CYLDm-syndrome–related tumors show LOH of the WT allele (15, 16, 18, 20), further supporting the importance of establishing a CYLDm-dominated signaling loop in order to observe the relevant skin phenotypic features. An additional possibility for the differences between CyldEΔ9/Δ9 and Cyld–/– animals is the fact that, in the former, the inactivation of CYLD is limited in epidermal cells, whereas in Cyld-/- animals, all cell types are CYLD deficient. Therefore, the phenotype of Cyld–/– animals may be affected by interactions between epidermal and nonepidermal CYLD-deficient cells such as immune cells. Taken together, we demonstrate that epidermal cell–targeted Cyld mutation disrupts a complex genetic and signaling network involving TRAF6, NF-κB, and c-Myc, which together confer cell lineage–specific growth and survival abnormalities.
Figure 6. Working model for Cyld regulation of ectodermal appendages.
Growth factors such as EGF and cytokines such as TNFα increase K63-Ub of TRAF6 and c-Myc, as well as TRAF6 downstream NF-κB signaling. NF-κB transcriptionally induces Cyld, which in turn inhibits TRAF6/NF-κB and c-Myc. In cells with homozygous Cyld mutation, the catalytically deficient CYLDm protein exerts dominant negative effects on K63-Ub deubiquitination of its substrates and consequently uncontrolled activation of NF-κB, c-Myc, and other gene regulators. NF-κB in turn increases Cyldm gene transcription, thereby forming a positive feedback loop to promote dysregulation of sebaceous gland, hair follicle, and dental growth. In Cyld–/– cells, such dominant negative effects of Cyldm on substrates and the transcriptional feedback from NF-κB to Cyldm gene expression are absent.
Our studies also demonstrate that homozygous CYLDm expression in epidermal cells disrupts the morphogenesis of upper incisor teeth, establishing CyldEΔ9/Δ9 mice as an unexpected working model for dental research. Future studies may be directed to understanding the mechanisms mediating CYLDm effects on dental growth and development.
Methods
Animal studies.
Generation of Cyldfl9/+ was reported previously (27). These mice were crossbred with K14-Cre mice (28) (provided by Terry Lechler, Duke University) to generate K14-Cre-Cyldfl9/fl9 mice, namely CyldEΔ9/Δ9 mice. For the treatment of TPA (Sigma-Aldrich), the back skin of 4-week-old animals were shaved and treated daily with 100 μl of 100 ng TPA/ml in acetone for 2 or 20 weeks. For 2-stage carcinogenesis, the back skins of newborn animals were treated with 1 dose of 2.5 μg DMBA (Sigma-Aldrich) in 50 μl acetone followed by twice-weekly applications of 100 ng TPA in 200 μl acetone for 20 weeks. Tumor growth and tissue analyses were performed as previously described (26). For topical treatments, 2 sets (n = 4) of 3-month-old animals were shaved on both left and right sides of the back skin and then treated daily for 10 days with 200 μl solvent (20% dimethyl sulfoxide in acetone) or 10 mM 10058-F4 (Sigma-Aldrich). Animals were then euthanized for histological analyses. No clinical distress was observed throughout the course of treatment.
Quantitative PCR.
Total RNA was isolated from 3-month-old CyldEΔ9/+ and 3 CyldEΔ9/Δ9 mouse skins (n = 3/genotype) using RNAeasy column purification (Qiagen), and 5μg of each sample was used for cDNA synthesis via oligo-dT–directed reverse transcription (Ambion). SYBR green–based quantitative PCR (qPCR) was performed in Bio-Rad iCycler with the following primers for mouse c-Myc (forward 5′-TAACTCGAGGAGGAGCTGGA-3′ and reverse 5′-GCCAAG GTTGTGAGGTTAGG-3′) and GAPDH (forward 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse 5′-TGTAGACCATGTAGTTGAGGTCA-3′).
Protein analysis.
For IP, protein lysates (250 μg/sample) of adult epidermal scrapes were incubated with polyclonal antibodies against c-Myc or TRAF6 for 2 hours at 4°C followed by 2 hours of incubation with protein A-agarose beads (ThermoFisher Scientific). The precipitated proteins were immunoblotted with an antibody against K63-Ubiquitin (Au3, clone 05-1308, EMD Millipore). Immunoblots were detected with an IRDye-688–conjugated secondary antibody (Li-Cor Biotechnology) or a HRP-conjugated anti-rabbit IgG conformation specific secondary antibody (L27A9, Cell Signaling Technology). IHC and immunofluorescent staining were performed with paraffin and frozen tissue sections, respectively, as described (53). The primary antibodies for PPARγ (H-100), TRAF6 (H-274), CYLD (H-419), and Actin (I-19) were obtained from Santa Cruz Biotechnology Inc. Antibodies against Ki-67 (SP6) and the Alexa 555 dye–conjugated secondary antibody were from ThermoFisher Scientific. Antibodies against phospho-c–Myc(62) (E1J4K), CYLD (D1A10), and cleaved caspase 3 were from Cell Signaling Technology. The antibody against c-Myc (Ab32072) was from Abcam. Oil Red O staining was performed with 0.18% Oil Red O and then counterstained with hematoxylin as described (34).
Statistics.
P values were calculated using 2-tailed Student’s t test. A P value of less than 0.05 was considered significant.
Study approval.
Animal studies were conducted in accordance with protocols approved by Duke Animal Care and Use Committee. Paraffin sections of human tissue specimens were obtained from Duke Dermatopathology lab through a protocol approved by Duke IRB.
Author contributions
YJJ performed the animal work, SW performed topical treatments and hair analyses, JC performed plasmid Cyld-gene expression reporter analysis, MAS prepared human tissue section analysis, TW performed dental analysis, GM provided Cyldfl/fl mice and was involved in data interpretation and manuscript preparation, and JYZ designed the experiments and prepared the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by the R03 (AR054556) and R01 (AR057746) grants from NIH/NIAMS and the research grant from American Skin Association to JZ, as well as the P30 grant (AR066527) from NIH/NIAMS to Duke Skin Disease Research Center. We thank Terry Lechler (Duke University) for providing the K14-Cre mice and insightful suggestions. We also thank Marja Mikkola (University of Helsinki, Helsinki, Finland), Russell Hall and Amanda Macleod (Duke University), and Kali Xu (MIT, Cambridge, Massachusetts, USA) for the critical comments of the manuscript, as well as René Bernard (Netherland Cancer Institute, Amsterdam, The Netherlands) for providing CYLD-expression constructs.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(11):e86548. doi:10.1172/jci.insight.86548.
Contributor Information
Sally Wang, Email: sallywang2018@yahoo.com.
Joshua Cho, Email: joshxcho@gmail.com.
M. Angelica Selim, Email: selim001@mc.duke.edu.
Tim Wright, Email: Tim_wright@unc.edu.
George Mosialos, Email: gmosialo@bio.auth.gr.
References
- 1.Regamey A, et al. The tumor suppressor CYLD interacts with TRIP and regulates negatively nuclear factor kappaB activation by tumor necrosis factor. J Exp Med. 2003;198(12):1959–1964. doi: 10.1084/jem.20031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 3.Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424(6950):801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 4.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 5.Reiley W, Zhang M, Sun SC. Negative regulation of JNK signaling by the tumor suppressor CYLD. J Biol Chem. 2004;279(53):55161–55167. doi: 10.1074/jbc.M411049200. [DOI] [PubMed] [Google Scholar]
- 6.Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci USA. 2007;104(21):8869–8874. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reiley WW, et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7(4):411–417. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- 8.Reiley WW, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204(6):1475–1485. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tauriello DV, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37(5):607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 10.Ströbel P, et al. Spiradenocylindroma of the kidney: clinical and genetic findings suggesting a role of somatic mutation of the CYLD1 gene in the oncogenesis of an unusual renal neoplasm. Am J Surg Pathol. 2002;26(1):119–124. doi: 10.1097/00000478-200201000-00016. [DOI] [PubMed] [Google Scholar]
- 11.An CH, et al. Frameshift mutations of ATBF1, WNT9A, CYLD and PARK2 in gastric and colorectal carcinomas with high microsatellite instability. Pathology. 2010;42(6):583–585. doi: 10.3109/00313025.2010.508735. [DOI] [PubMed] [Google Scholar]
- 12.Bignell GR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25(2):160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 13.Poblete Gutiérrez P, et al. Phenotype diversity in familial cylindromatosis: a frameshift mutation in the tumor suppressor gene CYLD underlies different tumors of skin appendages. J Invest Dermatol. 2002;119(2):527–531. doi: 10.1046/j.1523-1747.2002.01839.x. [DOI] [PubMed] [Google Scholar]
- 14.Bowen S, et al. Mutations in the CYLD gene in Brooke-Spiegler syndrome, familial cylindromatosis, and multiple familial trichoepithelioma: lack of genotype-phenotype correlation. J Invest Dermatol. 2005;124(5):919–920. doi: 10.1111/j.0022-202X.2005.23688.x. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi M, et al. Linkage and LOH studies in 19 cylindromatosis families show no evidence of genetic heterogeneity and refine the CYLD locus on chromosome 16q12-q13. Hum Genet. 2000;106(1):58–65. doi: 10.1007/s004399900227. [DOI] [PubMed] [Google Scholar]
- 16.Biggs PJ, Chapman P, Lakhani SR, Burn J, Stratton MR. The cylindromatosis gene (cyld1) on chromosome 16q may be the only tumour suppressor gene involved in the development of cylindromas. Oncogene. 1996;12(6):1375–1377. [PubMed] [Google Scholar]
- 17.Biggs PJ, et al. Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12-q13: evidence for its role as a tumour suppressor gene. Nat Genet. 1995;11(4):441–443. doi: 10.1038/ng1295-441. [DOI] [PubMed] [Google Scholar]
- 18.Thomson SA, Rasmussen SA, Zhang J, Wallace MR. A new hereditary cylindromatosis family associated with CYLD1 on chromosome 16. Hum Genet. 1999;105(1-2):171–173. doi: 10.1007/s004399900077. [DOI] [PubMed] [Google Scholar]
- 19.Saggar S, et al. CYLD mutations in familial skin appendage tumours. J Med Genet. 2008;45(5):298–302. doi: 10.1136/jmg.2007.056127. [DOI] [PubMed] [Google Scholar]
- 20.Leonard N, Chaggar R, Jones C, Takahashi M, Nikitopoulou A, Lakhani SR. Loss of heterozygosity at cylindromatosis gene locus, CYLD, in sporadic skin adnexal tumours. J Clin Pathol. 2001;54(9):689–692. doi: 10.1136/jcp.54.9.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Francesco V, et al. Carcinosarcoma arising in a patient with multiple cylindromas. Am J Dermatopathol. 2005;27(1):21–26. doi: 10.1097/01.dad.0000141548.69423.c7. [DOI] [PubMed] [Google Scholar]
- 22.Wannenmacher M, Schutz J. [Therapeutic consequences from the slow growth of cylindroma and their metastasis] Stoma (Heidelb) 1971;24(4):197–204. [PubMed] [Google Scholar]
- 23.Fukai K, Aida K, Aoki K. [2 Autopsy Cases of So-Called Cylindroma with Widespread Metastasis.] Kita Kanto Igaku. 1963;13:93–106. [PubMed] [Google Scholar]
- 24.Kazakov DV, Benkova K, Michal M, Vanecek T, Kacerovska D, Skalova A. Skin type spiradenoma of the parotid gland with malignant transformation: report of a case with analysis of the CYLD gene. Hum Pathol. 2009;40(10):1499–1503. doi: 10.1016/j.humpath.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fässler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125(4):665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 26.Miliani de Marval P, Lutfeali S, Jin JY, Leshin B, Selim MA, Zhang JY. CYLD inhibits tumorigenesis and metastasis by blocking JNK/AP1 signaling at multiple levels. Cancer Prev Res (Phila) 2011;4(6):851–859. doi: 10.1158/1940-6207.CAPR-10-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trompouki E, et al. Truncation of the catalytic domain of the cylindromatosis tumor suppressor impairs lung maturation. Neoplasia. 2009;11(5):469–476. doi: 10.1593/neo.81424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96(15):8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3(1):33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin W, et al. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118(5):1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller-Röver S, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117(1):3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 32.Duverger O, et al. Hair keratin mutations in tooth enamel increase dental decay risk. J Clin Invest. 2014;124(12):5219–5224. doi: 10.1172/JCI78272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naito A, et al. TRAF6-deficient mice display hypohidrotic ectodermal dysplasia. Proc Natl Acad Sci USA. 2002;99(13):8766–8771. doi: 10.1073/pnas.132636999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horsley V, et al. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126(3):597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trivedi NR, et al. Peroxisome proliferator-activated receptors increase human sebum production. J Invest Dermatol. 2006;126(9):2002–2009. doi: 10.1038/sj.jid.5700336. [DOI] [PubMed] [Google Scholar]
- 36.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11(8):558–568. doi: 10.1016/S0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 37.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105(4):533–545. doi: 10.1016/S0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71(3):925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermeking H, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97(5):2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pannem RR, Dorn C, Ahlqvist K, Bosserhoff AK, Hellerbrand C, Massoumi R. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis. 2014;35(2):461–468. doi: 10.1093/carcin/bgt335. [DOI] [PubMed] [Google Scholar]
- 41.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22(40):6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 42.Blake PW, Toro JR. Update of cylindromatosis gene (CYLD) mutations in Brooke-Spiegler syndrome: novel insights into the role of deubiquitination in cell signaling. Hum Mutat. 2009;30(7):1025–1036. doi: 10.1002/humu.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, et al. Diverse phenotype of Brooke-Spiegler syndrome associated with a nonsense mutation in the CYLD tumor suppressor gene. Exp Dermatol. 2006;15(12):966–970. doi: 10.1111/j.1600-0625.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 44.Carlson RM, Haddad L, Pui JC. Brooke-Spiegler syndrome with associated pegged teeth. Cutis. 2008;82(5):345–349. [PubMed] [Google Scholar]
- 45.Lakhani SR. Putting the brakes on cylindromatosis? N Engl J Med. 2004;350(2):187–188. doi: 10.1056/NEJMcibr032650. [DOI] [PubMed] [Google Scholar]
- 46.Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28(2):165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- 47.Honeycutt KA, Waikel RL, Koster MI, Wang XJ, Roop DR. The effect of c-myc on stem cell fate influences skin tumor phenotype. Mol Carcinog. 2010;49(4):315–319. doi: 10.1002/mc.20617. [DOI] [PubMed] [Google Scholar]
- 48.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123(3):409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 49.Menssen A, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109(4):E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jono H, et al. NF-kappaB is essential for induction of CYLD, the negative regulator of NF-kappaB: evidence for a novel inducible autoregulatory feedback pathway. J Biol Chem. 2004;279(35):36171–36174. doi: 10.1074/jbc.M406638200. [DOI] [PubMed] [Google Scholar]
- 51.Nikolaou K, Tsagaratou A, Eftychi C, Kollias G, Mosialos G, Talianidis I. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell. 2012;21(6):738–750. doi: 10.1016/j.ccr.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 52.Rajan N, et al. Tumor mapping in 2 large multigenerational families with CYLD mutations: implications for disease management and tumor induction. Arch Dermatol. 2009;145(11):1277–1284. doi: 10.1001/archdermatol.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang JY, Adams AE, Ridky TW, Tao S, Khavari PA. Tumor necrosis factor receptor 1/c-Jun-NH2-kinase signaling promotes human neoplasia. Cancer Res. 2007;67(8):3827–3834. doi: 10.1158/0008-5472.CAN-06-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.