Abstract
We present a practical approach for coregistration of bioluminescence tomography (BLT), computed tomography (CT), and magnetic resonance (MR) images. For this, we developed a customized animal shuttle composed of nonfluorescent, MR-compatible Delrin plastic that fits a commercially available MR surface coil. Mouse embryonic stem cells were transfected with the luciferase gene and labeled with superparamagnetic iron oxide nanoparticles. Cells were stereotaxically implanted in the mouse brain and imaged weekly for 4 weeks with bioluminescent imaging (IVIS Spectrum CT scanner) and magnetic resonance imaging (MRI; 11.7 T horizontal bore scanner). Without the use of software coregistration, in vitro phantom studies yielded root-mean-square errors of 7.6 × 10−3, 0.93 mm, and 0.78 mm along the medial–lateral (ML), dorsal–ventral (DV), and anterior–posterior (AP) axes, respectively. Rotation errors were negligible. Software coregistration by translation along the DV and AP axes resulted in consistent agreement between the CT and MR images, without the need for rotation or warping. In vivo coregistered BLT/MRI mouse brain data sets showed a single diffuse region of bioluminescent imaging photon signal and MRI hypointensity. Over time, the transplanted cells formed tumors as histopathologically validated. Disagreement between BLT and MRI tumor location was greatest along the DV axis (1.4 ± 0.2 mm) than along the ML (0.5 ± 0.3 mm) and the AP axes (0.6 mm) because of the uncertainty of the depth of origin of the BLT signal. Combining the high spatial anatomical information of MRI with the cell viability/proliferation data from BLT should facilitate preclinical evaluation of novel therapeutic candidate stem cells.
Keywords: multimodal imaging, stem cells, cell tracking, computed tomography, bioluminescence imaging, magnetic resonance imaging, coregistration
Introduction
Stem cell therapy is a burgeoning area of research for the treatment of a diverse range of diseases. A persistent challenge has been the need to monitor the accuracy of cell injection, survival and migration, and the potential tumorgenicity of undifferentiated stem cells (1). In vivo molecular and cellular imaging modalities that are currently used for tracking cells include bioluminescent imaging (BLI) (2–5), magnetic resonance imaging (MRI) (6–8), and magnetic particle imaging (MPI) (9–11), and nuclear imaging modalities include single photon emission computed tomography (12–14) and positron emission tomography (PET) (15, 16). Each of these techniques has its own advantage and limitation with respect to temporal resolution, anatomical detail, and functional information.
BLI is a widely used preclinical imaging technique that captures the propagation of light produced by luciferase (Luc)-transduced cells following the administration of the substrate luciferin. Because the depth of the light source and hence its tissue attenuation may vary, BLI provides a semi-quantitative planar image, with the signal intensity being proportional to the number of viable or actively expressing cells, but without background anatomical information. In contrast, MRI provides excellent soft tissue anatomical detail while simultaneously allowing tracking of cells that are labeled ex vivo with magnetic resonance (MR) contrast agents (17, 18) or MR reporter genes (19–22). MR-based cell tracking using superparamagnetic iron oxide (SPIO) as the MR contrast agent can localize single cells with high anatomical detail (23, 24). While there have been efforts to develop methods to quantify cell viability or cell number using MRI reporter genes (25), these techniques are not robust and are limited to a detection threshold number of ∼104 cells (18). In comparison, under optimal conditions BLI has been reported to be able to visualize lower numbers of cells in vivo (26, 27), but with a limited spatial resolution in the order of millimeters.
A recent development has been the introduction of bioluminescence tomography (BLT), where the spatial cell distribution in three dimensions can be visualized. A fusion of both BLT and MRI has the potential to compensate for the shortcomings of each method. One approach to fuse BLI/BLT images with other imaging modalities has been to use the coregistered information in an attempt to improve BLT reconstruction accuracy (28–31) or validate BLT results (32). While a growing body of work has examined the coregistration of BLI and MRI in these feasibility studies, an underdeveloped area is the application of coregistered BLT in preclinical or discovery research (33, 34). Among the few examples in the literature, Virostko et al. coregistered BLT and PET images to evaluate three new PET radiotracers for imaging human pancreatic beta cells (35). Deroose et al. reported on using a multimodal BLI–fluorescence–PET reporter gene to provide BLI and coregistered PET–computed tomography (CT) images of tumors (36). As an alternative multimodal approach for imaging brain tumors, fluorescence molecular tomography has been combined with MRI (37) or micro-CT and acoustic tomography (38) to exploit the sensitivity of fluorescence imaging with anatomical imaging at 0.1 mm of spatial resolution.
A more common approach is multimodal imaging without using image fusion. In one example, Zhang et al. used independently acquired MRI and planar BLI to assess stem cell survival in a rat model of myocardial infarction (39). Others have examined the fate of cell transplants in the mouse brain using MRI and BLI (40–43), without using BLT or coregistration. From these studies, it has become evident that a more accurate coregistration of BLT with other anatomical imaging modalities (MRI or CT) for in vivo applications is highly desirable. In this study, we present a protocol for coregistration of reconstructed BLT volumes with MRI anatomical data as exemplified by tracking SPIO-labeled embryonic stem cells in mouse brain.
Materials and Methods
Design of Customized Animal Holder for Multimodal BLI/CT/MR Imaging
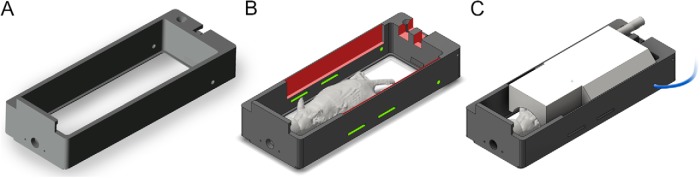
In a preclinical setting, coregistration between MRI and BLI requires transporting the subject between different imaging scanners. Maintaining the subject in a fixed posture between image acquisitions and determining an a priori transformation between the scanner coordinate systems can simplify the registration procedure. We adapted a commercially available animal holder (PerkinElmer Inc., MA, USA) (Figure 1A) into a custom-built shuttle, which was used for animal immobilization and transportation between an IVIS Spectrum CT scanner (PerkinElmer Inc., Massachusetts, USA) and a Bruker Biospec 117/16 (Bruker Corporation, MA, USA) 11.7 T MRI scanner. Two recesses (1-mm depth, 100-mm length, and 10.5-mm height) were milled into the inside surface of the shuttle (Figure 1B) to fit a radiofrequency MR surface coil, suitable for either brain or cervical spinal cord imaging (Figure 1C). The coil rests a few millimeters dorsal to the surface of the animal to avoid disturbing its position between the scanners. The removable MR coil allows the shuttle to maintain an open top during BLI to avoid optical distortion and/or attenuation of photon signal. The original holder is composed of a nonfluorescent, MR-compatible, Delrin plastic and is colored black to minimize light scatter during BLI. Additional slots were cut into the rear of the holder (Figure 1B) to accommodate leads from the MRI coil and a pressure respiration pad, as well as slots to pass restraint straps. We used a set of clear, elastic polyurethane straps to gently restrain the mouse during imaging without detectable autofluorescence or autoluminescence within the wavelengths of interest (580–680 nm). The entire shuttle locks into an imaging platform in the IVIS Spectrum CT and a dedicated support arm in the 11.7 T MRI scanner that provides anesthesia and heating.
Figure 1.
Unmodified commercial mouse imaging shuttle (A). Custom modification (indicated in red) to accommodate the radiofrequency (RF) magnetic resonance imaging (MRI) surface coil with cutouts to hold clear animal restraint straps and a respiration sensor lead (green) (B). Drawing showing the animal holder assembled with the RF MRI coil (white), placed directly above the mouse brain during magnetic resonance (MR) imaging (C).
Registration Procedure and Repeatability Tests
Repeatable positioning of the shuttle in the IVIS Spectrum CT scanner was accomplished by a snap-fit mechanism that locks the shuttle into the stage. For the Bruker 11.7 T scanner, a motorized positioning stage with precision of 0.1 mm was used to position the shuttle along the axis of the bore. To determine the accuracy of the transformation between the coordinate systems of the BLI/CT and MRI scanners, an air–water phantom visible on both CT and MRI was made out of a 15-mL polypropylene tube filled with deionized water. Smaller 0.5-mm tubes filled with either air or water were then placed inside this larger tube. The phantom was imaged using the MRI and CT protocols described below, and repeated three times with the removal of the shuttle, reinsertion, and readjustment of the stage and ion knobs in the MR scanner.
MR and CT data sets were imported into Amira 5.3 (FEI Visualization Sciences Group, Bordeaux, France) and coregistered for each trial by manual positioning followed by automatic registration using a rigid transformation and normalized mutual information metric. The accuracy of the registration was verified by visual inspection. The transformation between the CT and MR coordinate systems was considered as the mean of the transformations obtained from the three trials. Repeatability was computed as the root-mean-square error between the individual transformation components along each axis against the averaged transformation above.
Cell Transfection and Labeling
Mouse embryonic stem cells (mESCs) were purchased from ATCC and cultured on neomycin-resistant primary mouse embryonic fibroblast (PMEF-N; Millipore-Chemicon, MA, USA). mESCs were cultured in embryonic stem (ES) culture medium (ES cell-qualified Dulbecco's Modified Eagle Medium containing ES cell-qualified 15% fetal bovine serum, 0.1mM nonessential amino acid, 2mM L-glutamine, 0.1mM 2-mercaptomethanol, 1% nucleosides, 1% penicillin-streptomycin, and 1000 IU/mL leukemia inhibitory factor). The ES culture medium was changed once daily, and the ES cells were passaged every 4 days.
mESCs were transduced with lentivirus carrying the firefly Luc reporter gene and a neomycin resistance selection gene under control of the ubiquitin promoter (Lenti-Luc) at a multiplicity of infection of 10–30 in 2 mL of medium. After 24 hours, cells were washed with 10mM phosphate-buffered saline (PBS), pH = 7.4, and fresh medium was added. After lentivirus transduction, 100% of mESCs were expressing firefly Luc, achieved by adding 400 μg/mL of G418 antibiotic to the medium, followed for 5 days of culture for G418 selection. There was no adverse effect of lentivirus transduction on the growth of mESCs.
For validation of the Luc gene expression by BLI, the ES medium was removed and 30 μg/mL D-luciferin in PBS was added. The luminescence signal was observed using an IVIS Spectrum CT scanner (Perkin Elmer Inc.).
For MRI labeling, Luc-mESCs were incubated overnight with 25 μg/mL Molday ION-Rhodamine SPIO nanoparticles (BioPal, Inc., MA, USA) before transplantation. Red-channel fluorescence microscopy was used to verify SPIO labeling of the cells.
Cell Transplantation
All animal procedures were performed under an approved protocol from our Institutional Animal Care and Use Committee (IACUC). Three 3-week-old male BALB/c mice (Harlan Laboratories, IN, USA) were anesthetized using 2% isoflurane, were shaved, and then immobilized in a stereotactic frame (Harvard Apparatus, MA, USA). SPIO-labeled Luc-mESCs (5 × 104 cells in 2 μL of volume) were injected into the brain using a 31-gauge needle and a motorized injector (Stoelting Co., IL, USA) at a rate of 0.5 μL/min and the following coordinates: anterior–posterior (AP) = 0 mm, medial–lateral (ML) = 2.0 mm, and dorsal–ventral (DV) = 1.5 mm. The needle was carefully withdrawn 2 minutes after the end of the injection to minimize backflow. Animals were immunosuppressed by intraperitoneal daily administration of a cocktail of (FK-506) + (rapamycin) (1 mg/kg each; LC Laboratories, Woburn, Massachusetts), beginning 3 days before cell transplantation, and then daily until sacrifice.
Imaging
Mice were imaged the next day after cell transplantation, and then weekly for a total of 4 weeks. For each imaging session, anesthesia was induced using 3% isoflurane in oxygen and maintained using 0.5 L/min of 1%–2% isoflurane throughout the imaging session. In the shuttle, the mice, which were in the prone position, were gently restrained using the elastic straps, with the nose fully inserted into the anesthetic nose cone. Anesthetic delivery was briefly interrupted at the end of BLI/CT imaging while the shuttle was transported to the MRI scanner, and resumed within 1 minute before the animals could recover.
MR Imaging
For phantom imaging, a 72-mm-diameter volume coil (Bruker Corporation) and a T2-weighted rapid acquisition with relaxation enhancement sequence were used with the following parameters: repetition time (TR) = 3400 milliseconds, echo time (TE) = 30 milliseconds, field of view (FOV) = 6 × 2 × 1.6 cm, number of slices = 32 with 0.5-mm spacing, matrix = 360 × 128, and number of averages (NA) = 1. For in vivo animal imaging, a 2 × 2 cm of phased array surface coil (Bruker Corporation) was placed into the open top of the shuttle. Mice were imaged using 2 sequences:(1) A T1-weighted fast low-angle shot sequence with the following parameters: TR = 480 milliseconds, TE = 6.3 milliseconds, FOV = 1.6 × 1.6 cm, number of slices = 40 with 0.35-mm spacing, matrix = 196 × 196, NA = 1. (2) A T2-weighted rapid acquisition with relaxation enhancement sequence with TR = 4000 milliseconds, TE = 31.9 milliseconds, flip angle = 180°, FOV = 1.6 × 1.6 cm, matrix = 256 × 256, and NA = 3. Respiration gating was used to suppress motion artifacts.
CT Imaging
CT images for both phantom and mice were acquired using an IVIS Spectrum CT scanner (PerkinElmer Inc.) with the following parameters: 50 kVp at 1 mA current, 50 milliseconds of exposure time, and using an aluminum filter. A total of 720 projections spaced 0.5° apart were acquired, and the CT volume was reconstructed using Living Image software (PerkinElmer Inc.), using an FOV = 12.0 × 12.0 × 3.0 cm with 0.15-mm isotropic resolution.
BLI
BLI was acquired with a cooled charge-coupled device (CCD) camera using the same IVIS Spectrum CT scanner. Each animal was intraperitoneally injected with 150 mg/kg of D-luciferin 10 minutes before imaging. Using emission filters, 4 spectrally resolved images were acquired at 600, 620, 640, and 660 nm with a bandwidth of 20 nm each. The imaging parameters were as follows: exposure time = 180 seconds, aperture = f/1, FOV = 13 × 13 cm, and pixel resolution = 2048 × 2048. Binning was set to 8 × 8 for an effective image resolution of 256 × 256.
BLT Reconstruction
Reconstruction of the bioluminescent source and superposition over the CT volume was performed using the Diffuse Light Imaging Tomography (DLIT) algorithm available in Living Image software 4.3 (44). In brief, the algorithm uses single-view, multispectral bioluminescent images to perform the reconstruction with segmentation of the CT images to provide the mouse body boundary. Bioluminescent source and tissue absorption spectra for the Luc reporter and mouse tissue were predefined in the software. The source distribution was visualized using a voxel size of 0.31 mm and no smoothing, and then exported to Amira for coregistration with MRI.
Histopathology
All animals were euthanized following the last imaging time point at week 4 post-transplantation. Mice were transcardially perfused with 10mM PBS, followed by 4% paraformaldehyde in PBS. The brains were removed, fixated in paraformaldehyde overnight at 4°C, cryopreserved in 30% sucrose, and then snap frozen on dry ice. Serial 30-μm-thick coronal sections were cut using a Thermo Scientific HM 550 cryostat and transferred to glass slides. Sections were stained with hematoxylin and eosin for tissue morphology and Prussian Blue (Perls reagent)/neutral red counter stain for visualizing SPIO labeling (45).
For immunohistochemistry, nonspecific binding was blocked by incubating with a solution of 10% donkey serum and 0.1% Triton X-100-PBS for 2 hours at room temperature. Sections were then incubated overnight at 4°C with rabbit polyclonal antifirefly Luc antibody (1:3000, GeneTex, Inc., CA, USA) in a blocking solution. Goat antirabbit secondary antibodies (1:200, Alexa Fluor-488, Invitrogen-Thermo Fisher Scientific, MA, USA) were then added in the blocking solution for 2 hours at room temperature. Sections were rinsed with 10mM PBS, counterstained with DAPI (4′,6-diamidino-2-phenylindole), and mounted on coverslips with aqueous nonfluorescent medium (Fluoro-gel with Tris Buffer, Electron Microscopy Sciences, PA, USA). Microscopic images were acquired with a Zeiss AX10 (Carl Zeiss Microscopy GmbH, Jena, Germany) fluorescence microscope.
Results
Accuracy of Shuttle Repositioning
Repeatability tests with the phantom showed that errors were greatest in the DV and AP axes without the use of software coregistration, with root-mean-square errors of 7.6 × 10−3 mm, 0.93 mm, and 0.78 mm along the ML, DV, and AP, axes respectively. Rotation errors were negligible. Software coregistration by translation along the DV and AP axes resulted in good agreement between the CT and T1-weighted MR images (Figure 2), with no need for either rotation or warping. Subsequent tests on live mice using the registration procedure showed excellent agreement between the 2 imaging modalities (Figure 3). The soft brain tissue as depicted using the MRI could be successfully overlaid with the radiopaque CT contrast of the bone with an error of <0.1 mm.
Figure 2.
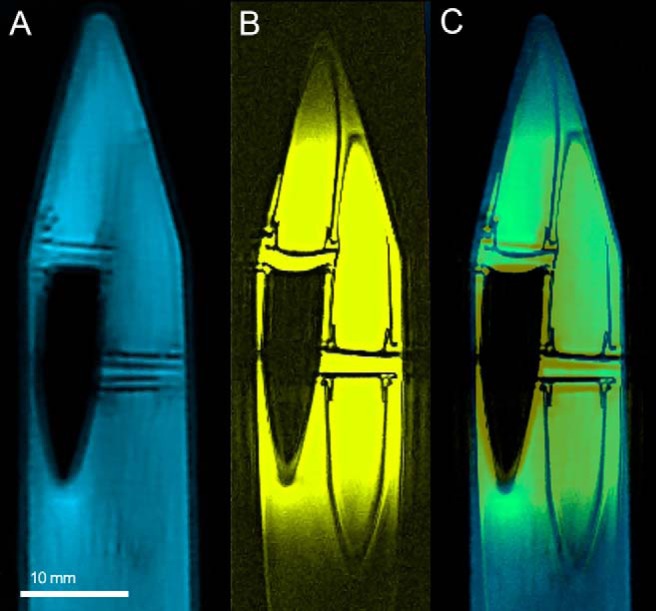
Transaxial images of coregistered air–water phantom images from computed tomography (CT) (A) and MRI (B), and showing excellent agreement between the sample overlay of the 2 modalities (C).
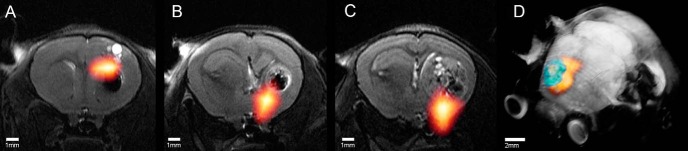
Figure 3.
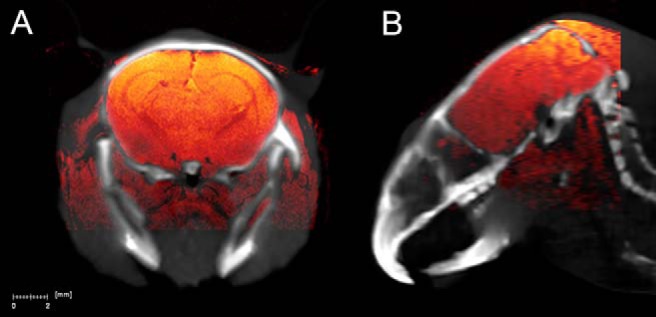
Coronal (A) and sagittal (B) in vivo mouse brain images of coregistered CT (gray scale) and T1-weighted fast low-angle shot (FLASH) MR images (hot color scale).
Comparison of Transplanted Cell Location by Imaging Modality
MR coronal T2-weighted images (Figure 4) showed a spherical hypointense implantation site representing SPIO-labeled cells. Over time, the mESCs formed tumors, as they were transplanted in an immunosuppressed host. The hypointense contrast remained predominantly located at the center of cell implantation, as the rapidly dividing tumorigenic mESCs diluted the SPIO label to undetectable levels. Coregistered BLT data sets, superimposed over the MR images, showed a single diffuse region of viable Luc cells. Histopathological analysis (Figure 5) was used to validate the imaging results from the different modalities. Hematoxylin and eosin staining confirmed the presence of a tumor mass both in the hypointense region and in the area of increased signal intensity seen on MRI. Prussian blue staining confirmed the presence of iron deposits within the hypointense region seen on MRI. Anti-Luc staining indicated the presence of Luc-expressing cells at both the original transplantation site and the distant sites containing migrated cells.
Figure 4.
In vivo coronal images 4 weeks post cell transplantation (A–C). The bioluminescence tomography (BLT) (hot color scale)-reconstructed luciferase (Luc)-mouse embryonic stem cells (mESC) location is superimposed on the T2-weighted rapid acquisition with relaxation enhancement (RARE) MR images for all 3 mice. T2-weighted MR volume-rendered mouse brain from panel (A) at 4 weeks post cell transplantation, showing an overlay of the BLT-reconstructed Luc-mESC location (orange) and the segmented MRI tumor volume (green) (D).
Figure 5.
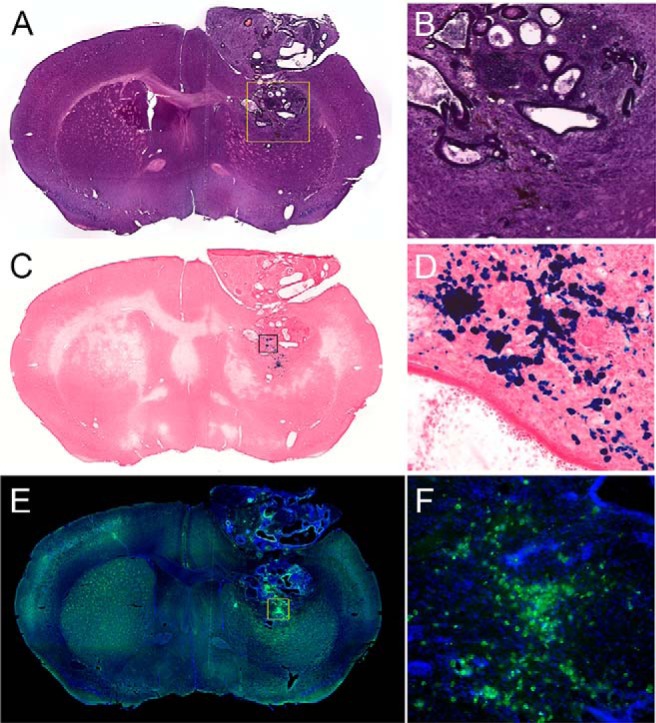
Hematoxylin and eosin (H&E)-stained coronal section showing tumor near implantation site (A, B). Prussian blue-stained section with nuclear fast red counterstain (C, D). Superparamagnetic iron oxide (SPIO) appears as blue deposits in the stain. Immunohistological stain for Luc (green) against DAPI (4′,6-diamidino-2-phenylindole) nuclear counterstain, showing Luc-expressing cells at both the original transplantation site and superficial lesion (E, F).
Comparison of MR- and BLI-Reported Cell Proliferation
The cellular transplant volume as is apparent from the MR data sets showed minimal changes over the first few weeks, and it was not until week 4 post transplantation that a rapid tumor growth could be observed (Figure 6). The hypointense areas containing the original SPIO-labeled cells masked the tumor mass initially, but these became fragmented and dispersed within the growing tumor at a later time point. Focal hyperintensities were also seen, which matched the edematous area of fluid-filled pockets on histology (compare Figure 5A-C). The BLI signal, corresponding to the number of viable cells, increased significantly within 1 week post transplantation (P < .05) (Figure 7A). Quantification of the BLT-reconstructed light source in absolute units (photons/s) showed a similar trend as the BLI (Figure 7B).
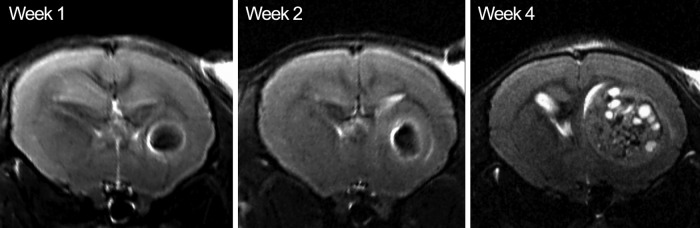
Figure 6.
Coronal MR images at 1, 2, and 4 weeks (left to right) after transplantation. The SPIO-labeled cell hypointensity induces a blooming effect, masking an initial visualization of tumor growth at the 1- and 2-week time points. By week 4, the tumor has considerably expanded, with fragmented pockets of the originally hypointense cells located within the center.
Figure 7.
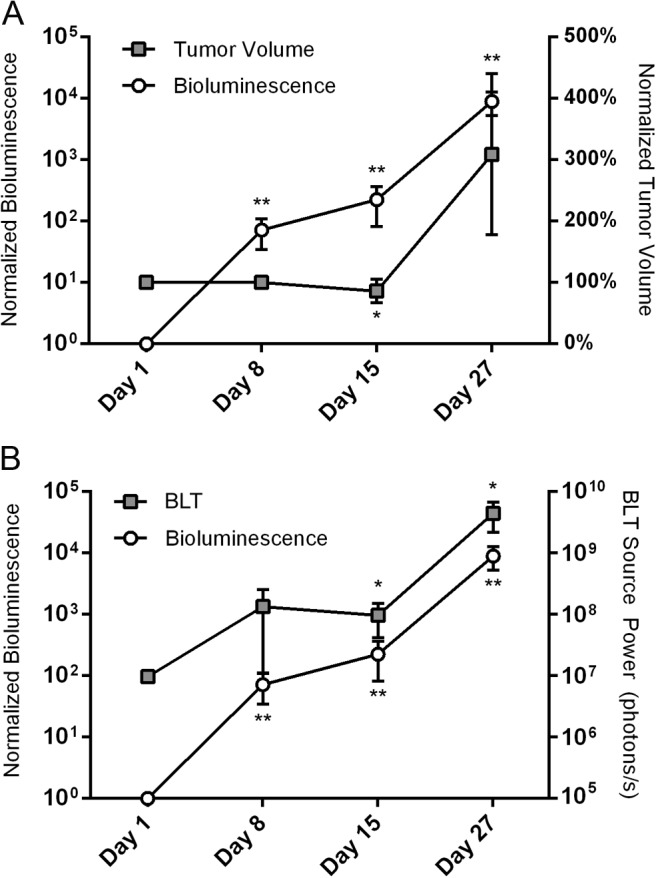
Bioluminescent imaging (BLI) signal and segmented tumor volume values at different time points following transplantation, normalized to the initial values at day 1 (n = 3) (A). Comparison of the total BLI signal, normalized to the initial values at day 1, against the BLT-reconstructed source power (n = 3) (B). Results are presented as mean values ± SD; asterisks denote significance level versus day 1 (*P < .10, **P < .05).
Discussion
We have developed a protocol for noninvasive BLT/MRI tracking of stem cells transplanted in mouse brain. We combined BLI and MRI for longitudinal assessment of both the location and proliferation of engrafted cells. In an ideal case, the use of a prior-determined transformation between the BLI/CT and MR scanner coordinates should eliminate the need for subsequent software registration. However, in the current study, the shuttle repeatability test indicated subvoxel (<0.15 mm) accuracy along the ML axis with the need for software coregistration along the other 2 axes to correct for <1 mm of deviations observed. For comparison, Beattie et al.'s previous work measured coregistered BLI to CT using an animal holder with a mean repeatability error of 0.36 mm (30). Other studies on coregistration of PET and MRI data have cited repeatability in the range of 0.2–0.3 mm (46, 47). We attribute the positioning errors along the AP and DV axes using a prior-determined transformation to the design of the MR scanner stage, which includes manual fine-positioning knobs that translate the stage along the sagittal plane and are necessary to allow the scanner to accommodate stages and inserts of different geometries. In contrast, the MR scanner stage is fixed from lateral motion, which is consistent with the higher repeatability seen in the ML axis. Future improvements may be made using a fixed-position MR scanner stage, trading versatility for increased registration accuracy. In addition, fiducial markers visible on both CT and MRI may be embedded into the animal shuttle, allowing the user to verify registration accuracy without running a separate phantom imaging validation. Nonetheless, we found the use of our current animal holder design in our study to be valuable, in that it maintains the animal in a fixed position, eliminating the need for nonrigid deformation-based registration, greatly simplifying the registration procedure from a 6-degrees-of-freedom problem to a simple translation along 2 axes.
While the use of BLI in the brain is well established, there are fewer studies on the use of BLT; to the best of our knowledge, only two other reports exist. Chaudhari et al. used multiview, multispectral BLT to reconstruct the location of a xenografted U87MG tumor in a nude mouse brain to within ∼0.7 mm of the actual location (n = 1), citing a BLT resolution of 1.5–2.2 mm for depths of up to 6 mm (48). Abdelwahab et al. describe a protocol for BLT imaging of GL261 gliomas in mice, but they do not cite the accuracy of tumor localization (49). For our current results, in vivo imaging of transplanted SPIO-labeled and Luc-transfected mESCs showed an overall good agreement between BLT and MRI on cell location along the AP and ML axes. However, there was a notable discrepancy between the BLT- and MRI-reported cell locations along the DV axis (Figure 4) after software coregistration, with a mean difference of 1.4 ± 0.2 mm, than between the BLT- and MRI-reported cell locations along the ML axis after software coregistration, with a mean difference of 0.5 ± 0.3 mm. Subsequent histological sections confirmed the presence of tumor masses and viable Luc-mESCs at locations corresponding to the MR images. A possible explanation is that a single-view, multispectral BLT reconstruction such as that used in this study depends on accurate knowledge of the optical properties for the tissue transmitting light to localize the luminescent source depth. Depth errors in the reconstruction may have been introduced through SPIO altering the optical properties at the tumor site, which was visible as a darker region in the tissue sections. BLT depth localization accuracy may also be improved in future work by segmentation of the coregistered anatomical images to define regions of different tissue optical properties. Using such an approach, Chaudhari et al. reported accurate localization in the horizontal plane with a residual error of 0.7 mm along the DV axis relative to the true tumor location (48).
In addition to monitoring the precise location of stem cell engraftment and dispersion, the potential tumorigenicity of stem cells is a critical safety factor that must be evaluated before translation to clinical trials (50–52). In our study, BLI showed a significant increase in luminescence by week 2 post transplantation by nearly two orders of magnitude (Figure 7), indicating a rapid proliferation of mESCs and the formation of tumors. The apparent tumor volume did not, however, change appreciably on MRI during the first 2 weeks; an increase in tumor volume in all three animals was only visible at the 4-week time point, although not statistically significant at this sample size. A limitation when measuring the apparent tumor volume from the MR volumes was the blooming artifact of the hypointense SPIO signal, masking the growing tumor, and causing an apparent decrease in tumor volume by 2 weeks post-transplantation as some of the SPIO cleared. Nevertheless, MRI aided the visualization of tumor morphology that could not be inferred by using BLT alone. These results suggest that care must be taken when interpreting MRI- or BLI-derived measures of tumorigenicity independently; a multimodal approach can help compensate for the limitations of each modality. Our findings agree with the work of Jost et al., who noted an increasing discrepancy between BLI- and MRI-derived measurements of glioblastoma tumor growth in mice as the tumor size increased (40).
The calculated BLT-reported source power (Figure 7B), which is a quantitative measure of light output by the Luc-mESCs, followed a similar trend as that seen for BLI but with a higher variability. This may be attributed to errors in the reconstructed source depth (Figure 4), which is a major determinant of source quantification accuracy in BLT (18). Because BLT accounts for the attenuation of light as it travels through the tissue, an incorrectly estimated depth will result in errors in source quantification.
In summary, we have developed a strategy and have shown a preclinical application in which transplanted stem cells can be tracked using coregistered MR and BLT data. This dual-modality imaging approach may aid in obtaining a better understanding of stem cell and stem cell-derived tumor dynamics by providing a more comprehensive analysis of cell fate in vivo.
Acknowledgments
The authors thank Jiadi Xu and Irina Shats for technical support. This work was supported in part by NIH R01 NS045062, NIH S10 OD010744, NMSS RG 4994-A-3, and ALSA 16-IIP-252.
Conflict of Interest: The authors have no disclosures to report.
Footnotes
- BLT
- Bioluminescence tomography
- MR
- magnetic resonance
- MRI
- magnetic resonance imaging
- DV
- dorsal–ventral
- AP
- anterior–posterior
- ML
- medial–lateral
- BLI
- bioluminescent imaging
- PET
- positron emission tomography
- Luc
- luciferase
- SPIO
- superparamagnetic iron oxide
- CT
- computed tomography
- mESCs
- mouse embryonic stem cells
- PBS
- phosphate-buffered saline
- TR
- repetition time
- TE
- echo time
- FOV
- field of view
- NA
- number of averages
References
- 1. Srivastava AK, Bulte JW. Seeing stem cells at work in vivo. Stem Cell Rev. 2014;10(1):127–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Contag CH, Ross BD. It's not just about anatomy: in vivo bioluminescence imaging as an eyepiece into biology. J Magn Reson Imaging. 2002;16(4):378–387. [DOI] [PubMed] [Google Scholar]
- 3. Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt. 2012;17(1):016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang Y, Agren L, Lyczek A, Walczak P, Bulte JW. Neural progenitor cell survival in mouse brain can be improved by co-transplantation of helper cells expressing bFGF under doxycycline control. Exp Neurol. 2013;247:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Srivastava AK, Bulte CA, Shats I, Walczak P, Bulte JW. Co-transplantation of syngeneic mesenchymal stem cells improves survival of allogeneic glial-restricted precursors in mouse brain. Exp Neurol. 2016;275 Pt 1:154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cromer Berman SM, Walczak P, Bulte JW. Tracking stem cells using magnetic nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2011;3(4):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bible E, Dell'Acqua F, Solanky B, Balducci A, Crapo PM, Badylak SF, Ahrens ET, Modo M. Non-invasive imaging of transplanted human neural stem cells and ECM scaffold remodeling in the stroke-damaged rat brain by (19)F- and diffusion-MRI. Biomaterials. 2012;33(10):2858–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rose LC, Kadayakkara DK, Wang G, Bar-Shir A, Helfer BM, O'Hanlon CF, Kraitchman DL, Rodriguez RL, Bulte JW. Fluorine-19 labeling of stromal vascular fraction cells for clinical imaging applications. Stem Cells Transl Med. 2015;4(12):1472–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bulte JW, Walczak P, Gleich B, Weizenecker J, Markov DE, Aerts HC, Boeve H, Borgert J, Kuhn M. MPI cell tracking: What can we learn from MRI? Proc SPIE Int Soc Opt Eng. 2011;7965:79650z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bulte JW, Walczak P, Janowski M, Krishnan KM, Arami H, Halkola A, Gleich B, Rahmer J. Quantitative “hot spot” imaging of transplanted stem cells using superparamagnetic tracers and magnetic particle imaging (MPI). Tomography. 2015;1(2):91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng B, von See MP, Yu E, Gunel B, Lu K, Vazin T, Schaffer DV, Goodwill PW, Conolly SM. Quantitative magnetic particle imaging monitors the transplantation, biodistribution, and clearance of stem cells in vivo. Theranostics. 2016;6(3):291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kraitchman DL, Tatsumi M, Gilson WD, Ishimori T, Kedziorek D, Walczak P, Segars WP, Chen HH, Fritzges D, Izbudak I, Young RG, Marcelino M, Pittenger MF, Solaiyappan M, Boston RC, Tsui BM, Wahl RL, Bulte JW. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation. 2005;112(10):1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang Y, Zhang C, Wang J, Lin X, Zhang L, Yang Y, Wanh Y, Zhang Z, Bulte JW, Yang YG. MRI/SPECT/Fluorescent tri-modal probe for evaluating the homing and therapeutic efficacy of transplanted mesenchymal stem cells in a rat ischemic stroke model. Adv Funct Mater. 2015;25(7):1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng SH, Yu D, Tsai HM, Morshed RA, Kanojia D, Lo LW, Leoni L, Govind Y, Zhang L, Aboody KS, Lesniak MS, Chen CT, Balyasnikova IV. Dynamic in vivo SPECT imaging of neural stem cells functionalized with radiolabeled nanoparticles for tracking of glioblastoma. J Nucl Med. 2016;57(2):279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yaghoubi SS, Campbell DO, Radu CG, Czernin J. Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics. 2012;2(4):374–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daadi MM, Hu S, Klausner J, Li Z, Sofilos M, Sun G, Wu JC, Steinberg GK. Imaging neural stem cell graft-induced structural repair in stroke. Cell Transplant. 2013;22(5):881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bulte JW, Kraitchman DL. Monitoring cell therapy using iron oxide MR contrast agents. Curr Pharm Biotechnol. 2004;5(6):567–584. [DOI] [PubMed] [Google Scholar]
- 18. Ahrens ET, Bulte JW. Tracking immune cells in vivo using magnetic resonance imaging. Nat Rev Immunol. 2013;13(10):755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilad AA, McMahon MT, Walczak P, Winnard PT Jr., Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. [DOI] [PubMed] [Google Scholar]
- 20. Bar-Shir A, Liu G, Liang Y, Yadav NN, McMahon MT, Walczak P, Nimmagadda S, Pomper MG, Tallman KA, Greenberg MM, van Zijl PC, Bulte JW, Gilad AA. Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. J Am Chem Soc. 2013;135(4):1617–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartelle BB, Mana MD, Suero-Abreu GA, Rodriguez JJ, Turnbull DH. Engineering an effective Mn-binding MRI reporter protein by subcellular targeting. Magn Reson Med. 2015;74(6):1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patrick PS, Rodrigues TB, Kettunen MI, Lyons SK, Neves AA, Brindle KM. Development of Timd2 as a reporter gene for MRI. Magn Reson Med. 2016;75(4):1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shapiro EM, Sharer K, Skrtic S, Koretsky AP. In vivo detection of single cells by MRI. Magn Reson Med. 2006;55(2):242–249. [DOI] [PubMed] [Google Scholar]
- 24. Heyn C, Ronald JA, Mackenzie LT, MacDonald IC, Chambers AF, Rutt BK, Foster PJ. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn Reson Med. 2006;55(1):23–29. [DOI] [PubMed] [Google Scholar]
- 25. Gilad AA, Ziv K, McMahon MT, van Zijl PCM, Neeman M, Bulte JW. MRI reporter genes. J Nucl Med. 2008;49:1905–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, Cooper LJ, Gelovani J, Hwu P. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc Natl Acad Sci U S A. 2008;105(38):14342–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aswendt M, Adamczak J, Couillard-Despres S, Hoehn M. Boosting bioluminescence neuroimaging: an optimized protocol for brain studies. PLoS One. 2013;8(2):e55662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allard M, Côté D, Davidson L, Dazai J, Henkelman RM. Combined magnetic resonance and bioluminescence imaging of live mice. J Biomed Opt. 2007;12:034018. [DOI] [PubMed] [Google Scholar]
- 29. Klose AD, Beattie BJ. Bioluminescence tomography with CT/MRI co-registration. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:6327–6330. [DOI] [PubMed] [Google Scholar]
- 30. Beattie BJ, Klose AD, Le CH, Longo VA, Dobrenkov K, Vider J, Koutcher JA, Blasberg RG. Registration of planar bioluminescence to magnetic resonance and x-ray computed tomography images as a platform for the development of bioluminescence tomography reconstruction algorithms. J Biomed Opt. 2009;14(2):024045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J, Chen D, Liang J, Xue H, Lei J, Wang Q, Chen D, Meng M, Jin Z, Tian J. Incorporating MRI structural information into bioluminescence tomography: system, heterogeneous reconstruction and in vivo quantification. Biomed Opt Express. 2014;5(6):1861–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu Y, Machado HB, Bao Q, Stout D, Herschman H, Chatziioannou AF. In vivo mouse bioluminescence tomography with radionuclide-based imaging validation. Mol Imaging Biol. 2011;13(1):53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Darne C, Lu Y, Sevick-Muraca EM. Small animal fluorescence and bioluminescence tomography: a review of approaches, algorithms and technology update. Phys Med Biol. 2014;59:R1–64. [DOI] [PubMed] [Google Scholar]
- 34. Zhang B, Wang KK, Yu J, Eslami S, Iordachita I, Reyes J, Malek R, Tran PT, Patterson MS, Wong JW. Bioluminescence tomography-guided radiation therapy for preclinical research. Int J Radiat Oncol Biol Phys. 2016;94(5):1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Virostko J, Henske J, Vinet L, Lamprianou S, Dai C, Radhika A, Baldwin RM, Ansari MS, Hefti F, Skovronsky D, Kung HF, Herrera PL, Peterson TE, Meda P, Powers AC. Multimodal image coregistration and inducible selective cell ablation to evaluate imaging ligands. Proc Natl Acad Sci U S A. 2011;108(51):20719–20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deroose CM, De A, Loening AM, Chow PL, Ray P, Chatziioannou AF, Gambhir SS. Multimodality imaging of tumor xenografts and metastases in mice with combined small-animal PET, small-animal CT, and bioluminescence imaging. J Nucl Med. 2007;48(2):295–303. [PMC free article] [PubMed] [Google Scholar]
- 37. McCann CM, Waterman P, Figueiredo JL, Aikawa E, Weissleder R, Chen JW. Combined magnetic resonance and fluorescence imaging of the living mouse brain reveals glioma response to chemotherapy. Neuroimage. 2009;45(2):360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deliolanis NC, Ale A, Morscher S, Burton NC, Schaefer K, Radrich K, Razansky D, Ntziachristos V. Deep-tissue reporter-gene imaging with fluorescence and optoacoustic tomography: a performance overview. Mol Imaging Biol. 2014;16(5):652–660. [DOI] [PubMed] [Google Scholar]
- 39. Zhang H, Qiao H, Bakken A, Gao F, Huang B, Liu YY, El-Deiry W, Ferrari VA, Zhou R. Utility of dual-modality bioluminescence and MRI in monitoring stem cell survival and impact on post myocardial infarct remodeling. Acad Radiol. 2011;18:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jost SC, Collins L, Travers S, Piwnica-Worms D, Garbow JR. Measuring brain tumor growth: A combined BLI/MRI strategy. Mol Imaging. 2009;8(5):245–253. [PMC free article] [PubMed] [Google Scholar]
- 41. Tennstaedt A, Aswendt M, Adamczak J, Hoehn M. Noninvasive multimodal imaging of stem cell transplants in the brain using bioluminescence imaging and magnetic resonance imaging. In: Turksen K, ed. Imaging and Tracking Stem Cells. Methods in Molecular Biology. New York, NY: Humana Press; 2013:153–166. [DOI] [PubMed] [Google Scholar]
- 42. Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P. Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med. 2011;65(2):564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rehemtulla A, Stegman LD, Cardozo SJ, Gupta S, Hall DE, Contag CH, Ross BD. Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging. Neoplasia. 2000;2(6):491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuo C, Coquoz O, Troy TL, Xu H, Rice BW. Three-dimensional reconstruction of in vivo bioluminescent sources based on multispectral imaging. J Biomed Opt. 2007;12(2):024007. [DOI] [PubMed] [Google Scholar]
- 45. Bulte JW, Arbab AS, Douglas T, Frank JA. Preparation of magnetically labeled cells for cell tracking by magnetic resonance imaging. Methods Enzymol. 2004;386:275–299. [DOI] [PubMed] [Google Scholar]
- 46. Ng TSC, Procissi D, Wu Y, Jacobs RE. A robust coregistration method for in vivo studies using a first generation simultaneous PET/MR scanner. Med Phys. 2010;37:1995–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bartoli A, Esposito G, D'Angeli L, Chaabane L, Terreno E. MRI and PET compatible bed for direct co-registration in small animals. IEEE Trans Nucl Sci. 2013;60(3):1596–1602. [Google Scholar]
- 48. Chaudhari AJ, Darvas F, Bading JR, Moats RA, Conti PS, Smith DJ, Cherry SR, Leahy RM. Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging. Phys Med Biol. 2005;50(23):5421–5441. [DOI] [PubMed] [Google Scholar]
- 49. Abdelwahab MG, Sankar T, Preul MC, Scheck AC. Intracranial implantation with subsequent 3D in vivo bioluminescent imaging of murine gliomas. J Vis Exp. 2011;(57):e3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19(8):998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blum B, Benvenisty N. The tumorigenicity of human embryonic stem cells. Adv Cancer Res. 2008;100:133–158. [DOI] [PubMed] [Google Scholar]
- 52. Kuroda T, Yasuda S, Sato Y. Tumorigenicity studies for human pluripotent stem cell-derived products. Biol Pharm Bull. 2013;36(2):189–192. [DOI] [PubMed] [Google Scholar]