Abstract
Background
In this study, we investigated the contribution of a gene expression–based signature (composed of BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, SH3BGR) to survival prediction for early-stage lung adenocarcinoma categorized by the new International Association for the Study of Lung Cancer (IASLC)/the American Thoracic Society (ATS)/the European Respiratory Society (ERS) classification. We also aimed to verify whether gene signature improves the risk discrimination of IASLC/ATS/ERS classification in early-stage lung adenocarcinoma.
Patients and methods
Total RNA was extracted from 93 patients with pathologically confirmed TNM stage Ia and Ib lung adenocarcinoma. The mRNA expression levels of ten genes in the signature (BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, and SH3BGR) were detected using real-time polymerase chain reaction. Each patient was categorized according to the new IASLC/ATS/ERS classification by accessing hematoxylin–eosin-stained slides. The corresponding Kaplan–Meier survival analysis by the log-rank statistic, multivariate Cox proportional hazards modeling, and c-index calculation were conducted using the programming language R (Version 2.15.1) with the “risksetROC” package.
Results
The multivariate analysis demonstrated that the risk factor of the ten-gene expression signature can significantly improve the discriminatory value of TNM staging in survival prediction, but not the value of the IASLC/ATS/ERS classification. Further analysis suggested that only BRCA1 and ERBB3 in the signature were independent risk factors after adjusting for the IASLC/ATS/ERS classification by Cox regression. A new algorithm of the two-gene expression signature containing BRCA1 and ERBB3 was generated. Adding the two-gene signature into the IASLC/ATS/ERS classification model further improved the discriminatory c-statistic from 0.728 to 0.756.
Conclusion
The two-gene signature composed of BRCA1 and ERBB3 was an independent risk factor of the IASLC/ATS/ERS classification, which can be used to improve the discriminatory power of prognosis prediction of the IASLC/ATS/ERS classification in early-stage lung adenocarcinoma. The two-gene signature combination with the IASLC/ATS/ERS classification might contribute to better patient stratification for adjuvant chemoradiotherapy or targeted therapy after the surgery.
Keywords: molecular biomarker, prognosis, lung, adenocarcinoma
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related deaths worldwide, accounting for more than 1 million deaths per year.1 Adenocarcinoma is the most common histological subtype of NSCLC, comprising almost half of all lung cancers.2 During the last few years, with the improvement in the knowledge of lung cancer biology, molecular and clinical characteristics, the International Association for the Study of Lung Cancer (IASLC), the American Thoracic Society (ATS), and the European Respiratory Society (ERS) integrated the clinical, radiological, molecular, and pathologic approaches to classify various types of lung adenocarcinoma. The new IASLC/ATS/ERS classification established histological categories with prognostic differences to identify the candidates for adjunctive therapy after/before surgery.2
The goal of the new classification of lung adenocarcinoma is to provide accurate diagnosis and obtain predictive and prognostic data for the patients’ outcomes. The stronger association with long-term survival for the IASLC/ATS/ERS classification has affect on TNM staging of early-stage lung adenocarcinomas. In addition, several gene expression-based signatures predicting survival with the use of microarrays or polymerase chain reaction (PCR) have also been demonstrated to predict the prognosis of patients with lung cancer accurately after resection of early-stage NSCLC.3–5 Kratz et al3 developed a practical gene signature for early-stage lung nonsquamous carcinoma that improved risk stratification compared with conventional staging; our research team participated in the multicenter validation in this study. In this gene set, BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, and SH3BGR, which are intricately related to lung cancer proliferation, are known elements of classical oncogenic molecular pathways (Table 1). This gene expression-based signature can better predict survival and disease recurrence compared to clinical stage among early-stage adenocarcinoma patients, but the predictive power of this prognostic gene signature has never been evaluated in the circumstances of the IASLC/ATS/ERS classification.
Table 1.
Algorithm genes
| Official symbol | Name | Biological functions | Aliases | Reference sequence |
|---|---|---|---|---|
| BAG1 | BCL2-associated athanogene | Blocks a step in a pathway leading to apoptosis or programmed cell death | HAP, BAG-1, RAP46 | NM_004323 |
| BRCA1 | Breast cancer 1, early onset | Induction of apoptosis, protein ubiquitination, regulation of G2/M transition DNA damage checkpoints, regulation of DNA repair | IRIS, PSCP, BRCA1, BRCC1, PNCA4, RNF53, BROVCA1, PPP1R53 | NM_007294 (672) |
| CDC6 | Cell division cycle 6 homologue | Interferes with the expression of INK4/ARF tumor suppressor genes through a mechanism involving the epigenetic modification of chromatin at the INK4/ARF locus | CDC18L, HsCDC18, HsCDC6 | NM_001254 |
| CDK2AP1 | Cyclin-dependent kinase 2 associated protein 1 | Regulation of S phase of mitotic cell cycle, epigenetic regulation | DOC1, DORC1, ST19, doc-1, p12DOC-1 | NM_004642 |
| ERBB3 | V-erb-b2 erythroblastic leukemia viral oncogene homologue 3 | Encodes a member of the EGFR family of receptor tyrosine kinases, transmembrane receptor protein tyrosine kinase signaling, regulation of phosphoinositide 3-kinase cascade, regulation of cell proliferation | ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | NM_001982 |
| FUT3 | Fucosyltransferase 3 | Carbohydrate metabolism, protein amino acid glycosylation | CD174, FT3B, FucT-III, LE, Les | NM_000149 |
| IL-11 | Interleukin-11 | Stimulates T-cell-dependent development of immunoglobulin-producing B cells | AGIF, IL-11 | NM_000641 |
| LCK | Lymphocyte-specific protein tyrosine kinase | Regulation of T-cell receptor signaling, leukocyte migration | LSK, YT16, IMD22, p56lck, pp58lck | NM_001042771 |
| RND3 | Rho family GTPase 3 | A negative regulator of cytoskeletal organization | ARHE, Rho8, RhoE, memB | NM_005168 |
| SH3BGR | SH3 domain binding glutamic acid-rich protein | Protein coding | 21-GARP | NM_007341 |
Abbreviations: BCL2, B-cell lymphoma 2; EGFR, epidermal growth factor receptor.
In this study, we investigated the relationship between this gene set and the histological categories according to the IASLC/ATS/ERS classification, and we tried to clarify whether the gene signature is a suitable molecular biomarker to provide improved risk stratification by the IASLC/ATS/ERS classification criteria in patients with early-stage lung adenocarcinoma.
Patients and methods
Patients
The human sample collection procedure was approved by the ethical committee of the Shanghai Chest Hospital, Shanghai Jiaotong University, Shanghai Pulmonary Hospital, Shanghai Tongji University, Shanghai Huadong Hospital, and Shanghai Fudan University. Between April 2002 and April 2007, 2,696 patients underwent surgical lobectomy of NSCLC at the Shanghai Pulmonary Hospital, including 469 cases with pathologically confirmed post-operative-TNM stage Ia (T1a, b N0M0) and Ib (T2a N0M0) lung adenocarcinoma. We reviewed the clinical data and formalin-fixed, paraffin-embedded samples of all 469 patients. The exclusion criteria were: 1) patients undergoing preoperative chemotherapy or radiation treatment; 2) patients undergoing lobectomy, but without systematic lymph node dissection; 3) patients with tumors >5 cm in diameter; and 4) badly preserved paraffin-embedded specimens. Ultimately, 105 patients with adequate clinical and histological characteristics and TNM staging were included in the study. All patients or significant others signed informed consent statements for scientific research using the resection specimen, data collection, and follow-up telephone contact or other interviews. Twelve of the 105 patients were excluded due to inadequate RNA quality, and 93 patients were included in the final statistics. Information on clinical variables and patient follow-ups was obtained from Shanghai Pulmonary Hospital’s patient database, with the end point of follow-up being April 2011 or date of death. The overall survival (OS) was calculated from the day of operation to the day of death or the final follow-up.
TNM staging was unified according to the seventh edition of the IASLC’s system for NSCLC before being subjected to subsequent statistical analysis.6
IASLC/ATS/ERS classification of lung adenocarcinoma and grouping methods
The new classification strategy of lung adenocarcinoma is based on the criteria proposed by the IASLC, ATS, and ERS in 2011.2 Four subtypes were included: adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), invasive adenocarcinoma (IA, including the subtypes of lepidic predominant, acinar predominant, papillary predominant, micropapillary predominant, and solid predominant), and variants of IA (including the subtypes of invasive mucinous adenocarcinoma, colloid, fetal, and enteric). Two pathologists (L Hou and H Xie) assessed all hematoxylin and eosin-stained slides independently.
According to the prognosis of survival of the IASLC/ATS/ERS classification subtypes reported by Yoshizawa et al7 and Van Schil et al,8 the selected 93 patients were assigned to three groups: low, intermediate, and high grade (Table 2).
Table 2.
Groups by prognostic value of different subtypes of the IASLC/ATS/ERS classification
| Groups | Relevant histological type | Prognostic implication |
|---|---|---|
| Low grade | AIS | Excellent |
| MIA | Excellent | |
| Intermediate grade | Lepidic predominant | Intermediate |
| Papillary predominant | Intermediate | |
| Acinar predominant | Intermediate | |
| High grade | Solid predominant | Poor |
| Micropapillary predominant | Poor | |
| Colloid predominant | Poor | |
| Invasive mucinous adenocarcinoma, mixed mucinous/nonmucinous | Poor |
Abbreviations: AIS, adenocarcinoma in situ; ATS, American Thoracic Society; ERS, European Respiratory Society; IASLC, International Association for the Study of Lung Cancer; MIA, minimally invasive adenocarcinoma.
Total RNA extraction and real-time PCR quantification of the selected gene
To identify the correlation between gene expressions and the clinical outcome, we analyzed the expression of eleven genes, BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, SH3BGR, and YAP1, in 105 surgical specimens using real-time PCR.
For RNA extraction from the formalin-fixed, paraffin-embedded samples, we used kits (MasterPure RNA Purification Kit; Epicentre Biotechnologies, Madison, WI, USA) and followed the manual’s instruction. Reverse transcription was done for the extracted RNA with gene-specific priming (iScript Select cDNA Synthesis Kit; Bio-Rad Laboratories Inc., Hercules, CA, USA) followed by cDNA preamplification (TaqMan PreAmp Master Mix; Thermo Fisher Scientific, Waltham, MA, USA). Taqman real-time PCR was performed for cDNA in 384-well plates using Prism the 7900HT machine (Thermo Fisher Scientific). Quantitative PCR was performed for each sample in triplicate. The −ΔΔCt method was used to analyze the relative quantitative expression levels of mRNAs with YAP1 as the reference gene. The expression of ten genes (BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, and SH3BGR) was analyzed.
Statistical analysis
The data were expressed as median and interquartile range. A nonparametric Wilcoxon Mann–Whitney U-test was used to calculate the difference between the gene expression of two groups, and Kruskal–Wallis H test was used for several independent samples. A chi-square test was used to compare proportional data.
The risk score of the ten-gene signatures (RS10) for each patient was calculated as a linear combination of the −ΔΔCt of BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, and SH3BGR with the same weights, as described by Kratz et al3 (RS10 =39.39747*(0.00237*BAG1 +0.14607* BRCA1 +0.08335*CDC6 +0.18653*CDK2AP1 −0.06638* ERBB +0.03458*FUT3 +0.01383*IL11 -0.20988*LCK +0.08849*RND3 -0.19821*SH3BGR) −16.94965+1). The resulting predicted risk scores were dichotomized (at the median), and the corresponding Kaplan–Meier survival analysis was conducted by log-rank statistic and Cox proportional hazards modeling using the programming language R (Version 2.15.1) with the “risksetROC” package (Patrick J Heagerty, Washington, USA).
Results
Patients’ survival analysis grouped by TNM stage, IASLC/ATS/ERS classification, or ten-gene signatures
The demographic and clinical characteristics of 93 patients with early-stage lung adenocarcinoma are listed in Table 3. Follow-up information was obtained from 91 cases, with 97.8% follow-up rate. Survival analysis was conducted in 91 cases with complete follow-up information. To evaluate the discriminatory value of the TNM stage, IASLC/ATS/ERS classification, or ten-gene signatures in the prediction of survival, the patients were grouped into low, intermediate, or high grade according to the IASLC/ATS/ERS classification, into Ia and Ib according to the World Health Organization TNM staging, and into lower risk score (<61.62) and higher risk score (≥61.62) according to the ten-gene signature.
Table 3.
Demographic and clinical characteristics of 93 NSCLC patients
| Characteristics | n (%) |
|---|---|
| Age, years | |
| Median (IQR) | 60 (49.5–68) |
| >60 | 43 (46.3) |
| ≤60 | 50 (53.7) |
| Sex | |
| Male | 45 (48.4) |
| Female | 48 (51.6) |
| Smoking history | |
| Nonsmoking | 68 (73.1) |
| Smoking (current/ever) | 25 (26.9) |
| Tumor location | |
| Right upper lobe | 33 (35.5) |
| Right middle lobe | 15 (16.1) |
| Right lower lobe | 9 (9.7) |
| Left upper lobe | 20 (21.5) |
| Left lower lobe | 16 (17.2) |
| Tumor size | |
| >2.5 cm | 40 (43.0) |
| ≤2.5 cm | 53 (57.0) |
| TNM stage | |
| Ia | 55 (59.1) |
| Ib (with visceral pleural involvement) | 38 (40.9) |
| IASLC/ATS/ERS classification | |
| AIS | 12 (12.9) |
| MIA | 12 (12.9) |
| IA | 59 (63.4) |
| Lepidic predominant | 6 (6.5) |
| Acinar predominant | 26 (28.0) |
| Papillary predominant | 10 (10.8) |
| Micropapillary predominant | 6 (6.5) |
| Solid predominant with mucin | 11 (11.8) |
| VIA | 10 (10.8) |
| Invasive mucinous and mixed mucinous/nonmucinous | 8 (8.6) |
| Colloid predominant | 2 (2.1) |
| Risk score of ten-gene signature | |
| Low | 76 (81.7) |
| High | 17 (18.3) |
| Risk score of two-gene signature | |
| Low | 47 (50.5) |
| High | 46 (49.5) |
| Follow-up (months) | |
| Median (IQR) | 66 (56–79) |
Abbreviations: AIS, adenocarcinoma in situ; ATS, American Thoracic Society; ERS, European Respiratory Society; IA, invasive adenocarcinoma; IASLC, International Association for the Study of Lung Cancer; IQR, interquartile range; MIA, minimally invasive adenocarcinoma; NSCLC, non-small-cell lung cancer; VIA, variants of invasive adenocarcinoma.
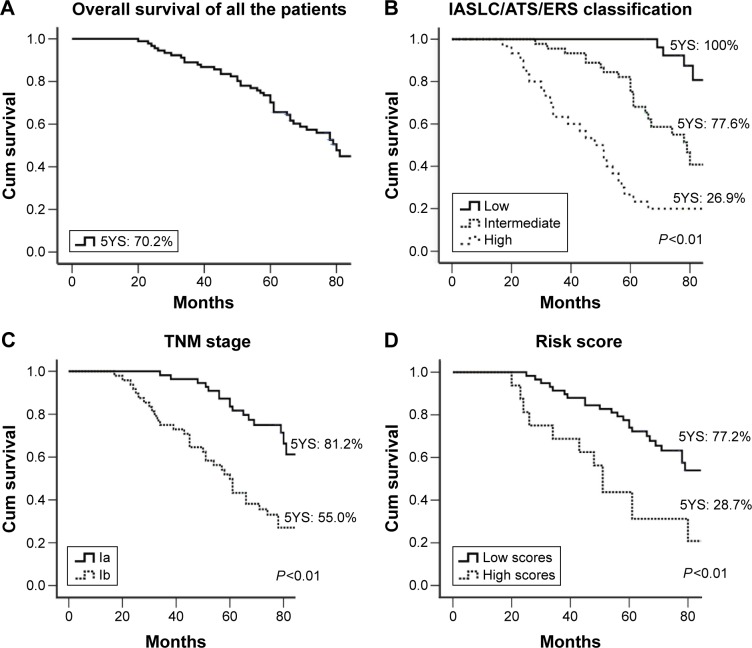
In the univariate Cox regression (Table 4), tumor size, TNM stage, IASLC/ATS/ERS classification, and the risk score predicted survival, whereas sex, age, and smoking status did not. The median survival for the 91 patients with early-stage lung adenocarcinoma was 80 months (95% confidence interval [CI]: 72.4–87.6) (Table 4; Figure 1A). The IASLC/ATS/ERS classification with a c-index of 0.704 was better in discriminating patients’ prognosis, compared to TNM staging with a c-index of 0.640 (Table 4; Figure 1B and C). The ten-gene risk score was also related to prognosis with a c-index of 0.587. The median survival time was 81.0 months for the lower risk score (95% CI: 67.2–94.8), while it was 51.0 months (95% CI: 38.2–63.8) for the higher risk score, which is significantly longer compared to the group with the lower risk score (P<0.001) (Table 4; Figure 1D).
Table 4.
Univariate Cox analysis of the risk factors of overall survival in the 91 patients with pulmonary adenocarcinoma
| n (%) | mOS, months (95% CI) | HR (95% CI) | P-value | c-Statistics | |
|---|---|---|---|---|---|
| Overall | 91 | 80 (72.4–87.6) | NA | ||
| Sex | 0.61 (0.34–1.11) | 0.099 | 0.559 | ||
| Male | 43 (47.2) | 74 (61.0–97.0) | |||
| Female | 48 (52.8) | 88 (75.4–100.6) | |||
| Age, years | 1.47 (0.81–2.66) | 0.207 | 0.547 | ||
| ≤60 | 50 (54.9) | 88 (74.7–101.3) | |||
| >60 | 41 (45.1) | 74 (56.7–91.3) | |||
| Smoking status | 0.82 (0.41–1.67) | 0.585 | 0.516 | ||
| Nonsmoking | 66 (72.5) | 78 (68.2–87.8) | |||
| Smoking (current/ever) | 25 (27.5) | 81 (60.2–101.8) | |||
| Tumor size | 3.21 (1.75–5.87) | 1.65*10−4 | 0.643 | ||
| ≤2.5 cm | 53 (58.2) | 101+ | |||
| >2.5 cm | 38 (41.8) | 61 (51.5–65.5) | |||
| TNM stage | 3.14 (1.72–5.74) | 1.89*10−4 | 0.640 | ||
| Ia | 53 (58.2) | 101+ | |||
| Ib | 38 (41.8) | 61 (51.5–70.5) | |||
| IASLC/ATS/ERS classification | 3.06 (1.98–4.73) | 5.33*10−7 | 0.704 | ||
| Low | 24 (26.4) | 101+ | |||
| Intermediate | 41 (45.0) | 79 (71.9–86.1) | |||
| High | 26 (28.6) | 48 (34.3–61.7) | |||
| RS10 | 3.42 (1.74–6.72) | 3.81*10−4 | 0.587 | ||
| Low | 75 (82.4) | 81 (67.2–94.8) | |||
| High | 16 (17.6) | 51 (38.2–63.8) | |||
| RS2 | 3.2 (1.74–6.07) | 2.11*10−4 | 0.644 | ||
| Low | 47 (51.6) | 101+ | |||
| High | 44 (48.4) | 61 (56.8–65.2) |
Note:
P<0.05.
Abbreviations: ATS, American Thoracic Society; CI, confidence interval; ERS, European Respiratory Society; HR, hazard ratio; IASLC, International Association for the Study of Lung Cancer; mOS, median overall survival; NA, not available; RS2, risk score of two-gene signature; RS10, risk score of ten-gene signature.
Figure 1.
The discriminatory value of the IASLC/ATS/ERS classification, TNM-stage, or ten-gene signatures predicting the survival of the 91 patients with early-stage lung adenocarcinoma.
Notes: (A) Overall survival curve for the 91 cases of early-stage lung adenocarcinoma; (B) overall survival curves for patients grouped by the IASLC/ATS/ERS classification; (C) overall survival curves for patients grouped by the TNM stage; (D) overall survival curves for patients grouped by the risk score of ten-gene signatures.
Abbreviations: 5YS, 5-years survival; ATS, American Thoracic Society; ERS, European Respiratory Society; IASLC, International Association for the Study of Lung Cancer.
The ten-gene signature does not predict survival of early-stage pulmonary adenocarcinoma independently of the IASLC/ATS/ERS classification.
To clarify the interaction between the ten-gene risk score and TNM staging or IASLC/ATS/ERS classification in survival prediction, the hazard ratio of the ten-gene risk score was adjusted in the TNM-based or IASLC/ATS/ERS classification-based models using multivariable Cox regression (Table 5). The ten-gene risk score is still an independent prognostic factor after adjusting for TNM staging, suggesting that the ten-gene risk score can improve the discriminatory value of TNM staging in survival prediction. However, the hazard ratio of the ten-gene risk score was not statistically significant after adjusting for the IASLC/ATS/ERS classification, which implies the collinearity of predictive variables between the ten-gene risk score and the IASLC/ATS/ERS classification.
Table 5.
TNM-based or IASLC/ATS/ERS classification-based models adjusted for risk score and other clinical parameters among the 91 patients with early-stage pulmonary adenocarcinoma
| TNM-based model
|
IASLC/ATS/ERS-based model
|
||||
|---|---|---|---|---|---|
| Variables | Adjusted HR (95% CI) | P-value | Variables | Adjusted HR (95% CI) | P-value |
| RS10 | 3.41 (1.61–7.26) | 0.001* | RS10 | 2.01 (0.93–4.34) | 0.070 |
| TNM stage | 2.72 (1.47–5.05) | 0.001* | IASLC/ATS/ERS | 2.57 (1.60–4.12) | 8.8E-5* |
| Age | 1.41 (0.77–2.57) | 0.270 | Age | 1.14 (0.62–2.10) | 0.690 |
| Sex | 0.54 (0.29–1.02) | 0.053 | Sex | 0.58 (0.31–1.10) | 0.094 |
| Smoking c-index =0.715 | 0.52 (0.24–1.12) | 0.087 | Smoking c-index =0.735 | 0.54 (0.25–1.16) | 0.113 |
Note:
P<0.01.
Abbreviations: ATS, American Thoracic Society; CI, confidence interval; ERS, European Respiratory Society; HR, hazard ratio; IASLC, International Association for the Study of Lung Cancer; RS10, risk score of ten-gene signature.
Two-gene signature containing BRCA1 and ERBB3 can improve the discriminatory value of prognostic prediction of IASLC/ATS/ERS classification in early-stage lung adenocarcinoma.
Because the algorithm of the ten-gene signature may not be optimal for the IASLC/ATS/ERS classification, we reanalyzed the association of survival with the expression levels of the ten genes separately, adjusting for the IASLC/ATS/ERS classification using the Cox regression. The expression levels of BRCA1 and ERBB3 emerged as independent predictive factors of survival after adjusting for the IASLC/ATS/ERS classification (Table 6). Therefore, we developed a new algorithm of two-gene signature containing BRCA1 and ERBB3, namely, the risk score of the two-gene signature (RS2) =0.475*BRCA1 +0.048*ERBB3, to improve the discriminatory value of prognosis prediction of the IASLC/ATS/ERS classification. RS2 was calculated for each patient, and it was dichotomized at the median of 4.98.
Table 6.
Multivariate COX regression analysis of the ten-gene expression levels to predict survival adjusted with the IASLC/ATS/ERS classification in the 91 patients
| Variables | HR (95% CI) | P-value |
|---|---|---|
| Age (≧60 vs <60) | 1.95 (0.95–4.00) | 0.071 |
| Sex (F vs M) | 0.77 (0.36–1.62) | 0.483 |
| Smoking (ever vs never) | 0.39 (0.16–0.92) | 0.032* |
| IASLC/ATS/ERS classification (H vs I vs L) | 3.64 (1.99–6.68) | 2.90E-5* |
| BAG1 (H vs L) | 1.16 (0.50–2.71) | 0.732 |
| BRCA1 (H vs L) | 4.50 (1.64–12.32) | 0.003* |
| CDC6 (H vs L) | 2.55 (0.26–25.45) | 0.426 |
| CDK2AP1 (H vs L) | 0.66 (0.08–5.35) | 0.698 |
| ERBB3 (H vs L) | 3.52 (1.40–8.80) | 0.007* |
| FUT3 (H vs L) | 0.84 (0.42–1.72) | 0.639 |
| IL11 (H vs L) | 2.19 (0.96–5.00) | 0.062 |
| LCK (H vs L) | 0.84 (0.39–1.79) | 0.647 |
| RND3 (H vs L) | 1.68 (0.42–6.76) | 0.465 |
| SH3BGR (H vs L) | 0.81 (0.32–2.10) | 0.668 |
Note: P<0.01.
Abbreviations: ATS, American Thoracic Society; CI, confidence interval; ERS, European Respiratory Society; F, female; H, high; I, intermediate; HR, hazard ratio; IASLC, International Association for the Study of Lung Cancer; L, low; M, male; RS10, risk score of ten-gene signature.
To evaluate the improvement of the prognostic value by the risk score of the two-gene signature consisting of BRCA1 and ERBB3, the discriminatory power of the predictive models of the TNM stage or IASLC/ATS/ERS classification with risk score was evaluated using c-statistics or areas under the receiver operating characteristic curve. The TNM stage alone achieved a discriminatory c-statistic of 0.640 (Table 4). Adding the clinical parameters only slightly improved the discriminatory value to 0.685 (Table 7). The IASLC/ATS/ERS classification alone achieved a c-statistic of 0.704 (Table 4), and adding the clinical parameters improved the c-statistic to 0.728 (Table 7). Adding the risk score of the two-gene signature to the IASLC/ATS/ERS classification model further improved the discriminatory c-statistic to 0.756, which gave the best performance of the predictive model (Table 7).
Table 7.
Two-gene signatures can improve the risk discrimination of the IASLC/ATS/ERS classification
| Models | Model based on TNM | Model based on IASLC/ATS/ERS classification | Improved model based on IASLC/ATS/ERS classification and RS2 |
|---|---|---|---|
| Predictors | Age | Age | Age |
| Sex | Sex | Sex | |
| Smoking | Smoking | Smoking | |
| TNM stage | IASLC/ATS/ERS classification | IASLC/ATS/ERS classification RS2 | |
| c-Statistics | 0.685 | 0.728 | 0.756 |
Abbreviations: ATS, American Thoracic Society; ERS, European Respiratory Society; IASLC, International Association for the Study of Lung Cancer; RS2, risk score of two-gene signature.
Overall, the prognostic value of the risk score of the two-gene signature improved the IASLC/ATS/ERS classification for assessing the prognosis of early-stage lung adenocarcinoma. The time-dependent area under the curve plot of the model based on the IASLC/ATS/ERS classification and improved model based on risk score confirmed that the risk score of the two-gene signature together with IASLC/ATS/ERS classification showed a better overall discriminatory ability over time compared to the IASLC/ATS/ERS classification alone (Figure 2).
Figure 2.
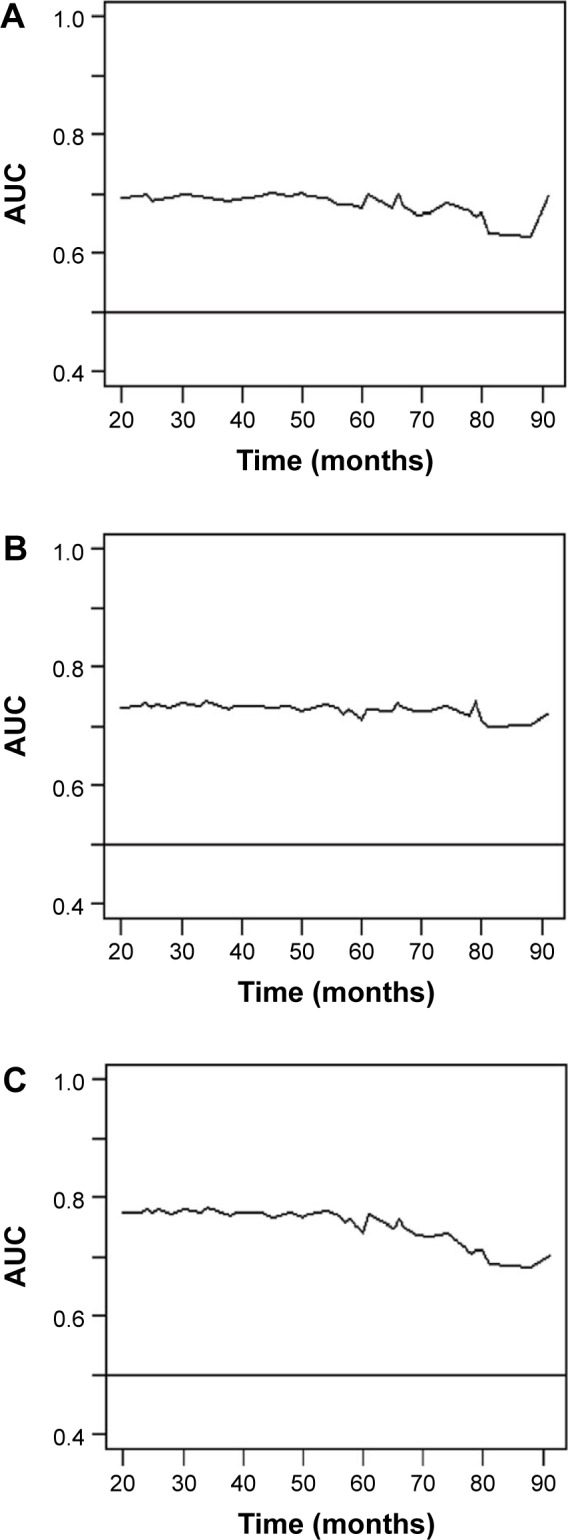
The time-dependent AUC plot of the three models.
Notes: (A) Model based on the TNM stage; (B) model based on the IASLC/ATS/ERS classification; (C) improved model based on the IASLC/ATS/ERS classification and RS2.
Abbreviations: ATS, American Thoracic Society; AUC, area under the curve; ERS, European Respiratory Society; IASLC, International Association for the Study of Lung Cancer; RS2, risk score of two-gene signature.
Discussion
The novel IASLC/ATS/ERS classification of lung adenocarcinoma has markedly promoted the pathologic diagnosis of lung cancer. The new criteria introduce a new concept of early-stage lung cancer consisting of AIS and MIA and categorize IAs by the predominant histological pattern. Importantly, the IASLC/ATS/ERS classification serves as a rapid and efficient discriminator of patient prognosis, and it can support patient stratification for adjuvant chemoradiotherapy or targeted therapy after surgery.9 Several gene expression-based signatures were also believed to be useful in identifying the candidates for adjunctive therapy after/before surgery. Although some gene signatures were believed to predict survival better compared to traditional TNM classification, whether these gene signatures are independent of the IASLC/ATS/ERS classification has seldom been evaluated.
In this study, we proposed that a two-gene signature composed of BRCA1 and ERBB3 was a risk factor independent of the IASLC/ATS/ERS classification, which would improve the discriminatory power of prognosis prediction for IASLC/ATS/ERS classification. BRCA1 (breast cancer 1, early onset) is a tumor suppressor gene involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks.10 Germline mutations of BRCA1 have been associated with an increased risk of breast, ovarian, and prostate cancer.11 The overexpression of BRCA1 mRNA was strongly associated with poor survival in NSCLC patients.12,13 The ERBB3 gene encodes a member of the epidermal growth factor receptor family of receptor tyrosine kinases. The overexpression of ERBB3 (HER3) has been reported both in primary cancers and in cultured cells, including the carcinomas of lung, breast, ovarian, prostate, colon, pancreas, stomach, and oral cavity,14,15 and it has been shown that ERBB3 (HER3)’s overexpression was associated with worse OS at both 3- and 5-year period.16 We verified that the ten-gene signature was an independent prognostic factor in the TNM-based system, but not in the IASLC/ATS/ERS classification model. Based on the novel IASLC/ATS/ERS scheme, the adjusted model with two-gene signatures showed improved discriminatory power in patient prognostic models in early-stage lung adenocarcinoma.
As a heterogeneous disease, NSCLC will have different statistical outcomes even for patients with similar clinical and pathological features. The prognosis of surgically treated lung adenocarcinoma differed significantly across different subtypes. The survival rate of surgically treated AIS is 100% and MIA is near 100%. Lepidic predominant adenocarcinomas have a 5-year survival rate of 90%. Acinar and papillary predominant adenocarcinomas have a 5-year survival rate of 85%–85.5%.7,17 In addition, micropapillary predominant adenocarcinomas have a 5-year survival rate of only 67% in a series of stage I adenocarcinomas, and solid adenocarcinoma with mucin components has a 5-year survival rate of only 34.6%.7 Based on the prognostic value of the subtype of this new classification, early-stage lung adenocarcinoma was divided into low-, intermediate-, and high-grade groups.7,8 Our findings revealed similar results to those reported previously, with 5-year survival rates of 100%, 77.6%, and 26.9% for the three groups, respectively. Because AIS and MIA types are curable with a complete resection, they may provide a selection for surgeons considering limited resection over standard lobectomy and mediastinal lymph node dissection as treatment options. Therefore, it is important to recognize different subtypes of the IASLC/ATS/ERS classification in early-stage lung adenocarcinomas.
Several previous clinical trials have documented that stage I NSCLC patients did not gain a statistically clinical benefit from adjuvant chemotherapy.18,19 The current therapy for stage I NSCLC patients is surgical resection without adjuvant chemotherapy. The evidence suggests that the IASLC/ATS/ERS classification is able to identify patients in the high-risk group with very poor long-term survival, such as those subtypes with micropapillary and solid predominant lung adenocarcinoma. Subsets with a poor prognosis could be selected for inclusion in clinical trial testing for additional therapeutic agents. We found that the two-gene signature provided better prognostic stratification compared to the IASLC/ATS/ERS classification alone. Thus, we believe two-gene signatures combined with the new classification system could help identify stage I lung adenocarcinoma patients at a high risk for recurrence who may benefit from adjuvant chemotherapy or novel treatment approaches. This prognostic tool would improve their OS rate.
Acknowledgments
This study was supported by the Science and Technology Commission of Shanghai Municipality (No 14411950800) and Shanghai Municipal Commission of Health and Family Planning (No 2013ZYJB0004, No 201540139).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379(9818):823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356(1):11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 5.Raz DJ, Ray MR, Kim JY, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res. 2008;14(17):5565–5570. doi: 10.1158/1078-0432.CCR-08-0544. [DOI] [PubMed] [Google Scholar]
- 6.Rusch VW, Asamura H, Watanabe H, Giroux DJ, Rami-Porta R, Goldstraw P. Members of IASLC Staging Committee. The IASLC lung cancer staging project: a proposal for a new international lymph node map in the forthcoming seventh edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4(5):568–577. doi: 10.1097/JTO.0b013e3181a0d82e. [DOI] [PubMed] [Google Scholar]
- 7.Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24(5):653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 8.Van Schil PE, Asamura H, Rusch VW, et al. Surgical implications of the new IASLC/ATS/ERS adenocarcinoma classification. Eur Respir J. 2012;39(2):478–486. doi: 10.1183/09031936.00027511. [DOI] [PubMed] [Google Scholar]
- 9.Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol. 2012;30(13):1438–1446. doi: 10.1200/JCO.2011.37.2185. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Canc Sci. 2004;95(11):866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4(9):665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 12.Rosell R, Skrzypski M, Jassem E, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One. 2007;2(11):e1129. doi: 10.1371/journal.pone.0001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiseo M, Bordi P, Bortesi B, et al. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108(8):1695–1703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15(7):413–448. doi: 10.1038/cgt.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mujoo K, Choi BK, Huang Z, Zhang N, An Z. Regulation of ERBB3/HER3 signaling in cancer. Oncotarget. 2014;5(21):10222–10236. doi: 10.18632/oncotarget.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E. HER3 overexpression and survival in solid tumors: a meta-analysis. J Natl Cancer Inst. 2013;105(4):266–273. doi: 10.1093/jnci/djs501. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Wu J, Tan Q, Zhu L, Gao W. Why do pathological stage IA lung adenocarcinomas vary from prognosis?: A clinicopathologic study of 176 patients with pathological stage IA lung adenocarcinoma based on the IASLC/ATS/ERS classification. J Thorac Oncol. 2013;8(9):1196–1202. doi: 10.1097/JTO.0b013e31829f09a7. [DOI] [PubMed] [Google Scholar]
- 18.Felip E, Rosell R, Maestre JA, et al. Preoperative chemotherapy plus surgery versus surgery plus adjuvant chemotherapy versus surgery alone in early-stage non-small-cell lung cancer. J Clin Oncol. 2010;28(19):3138–3145. doi: 10.1200/JCO.2009.27.6204. [DOI] [PubMed] [Google Scholar]
- 19.Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9,633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26(31):5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]