Abstract
Background
The objective of this study was to characterize the mRNA expression profile related to rumen epithelial inflammation through the in vivo and in vitro experiments. In the in vivo experiment, rumen papillae were collected from four dairy cows adapted to either a 40 % (LC) or 70 % (HC) concentrate feeds for microarray analysis.
Results
Results showed that 245 differentially expressed genes (DEGs) were detected in the cows fed the HC relative to the LC diet. The DEGs were first annotated, and results revealed that the expression of inflammation-related genes, including IL-1β, IL-2, IL-22, CCL19, CCL8, CX3CR1, CXCL6, INHBE, LEPR, PRL, and TNFRSF9 found in the cytokine-cytokine receptor pathway were up-regulated in the HC-fed cows, indicating local inflammation in the rumen epithelium was triggered. The expression of IL-1β, IL-2, and IL-6 was further validated by qRT-PCR. To demonstrate whether there were relationships between cytokine mRNA expression and ruminal factors (pH and LPS), the isolated ruminal epithelial cells were cultured in vitro. Results showed that the mRNA expression of IL-1β, IL-2, IL-6, and IL-8 increased after the LPS treatment, while low-pH treatment elevated the mRNA expression of TNF-α, suggesting that low-pH coupled with higher levels of LPS in rumen of cows fed the HC may be mainly responsible for the triggered local ruminal inflammation.
Conclusions
Our results indicate that ruminal local inflammation response might be triggered during HC feeding, and these findings also enhance the knowledge of rumen epithelial adaptation to HC at the molecular level.
Electronic supplementary material
The online version of this article (doi:10.1186/s40104-016-0100-1) contains supplementary material, which is available to authorized users.
Keywords: Dairy cows, Gene expression, Microarray, Subacute ruminal acidosis
Background
Feeding ruminants diets rich in fermentable carbohydrates alters ruminal microbiota composition and metabolism, resulting in the accumulation of volatile fatty acids (VFAs) and leading to a dramatic depression of the rumen pH [1, 2]. Long-term high-concentrate feeding likely causes the rumen metabolic disorder commonly termed subacute ruminal acidosis (SARA), which is defined as the daily duration time of ruminal pH remains below 5.6 or 5.8 [3, 4]. SARA has been shown to depress ruminal diet digestibility, feed intake, milk production, and milk fat and has become a significant economic issue in the dairy industry [3, 5].
Dairy cattle health can be compromised by grain-induced SARA since it has been associated with rumenitis, metabolic acidosis, lameness and hepatic abscesses [3, 5]. Many previous reports revealed that grain-induced SARA increased the levels of acute phase proteins such as haptoglobin (Hp) and serum amyloid A (SAA), which indicate a systemic inflammatory response in peripheral blood [6, 7]. This systemic inflammation is considered to be related with the diet-induced massive disruption of rumen epithelial barrier function [8] and increased permeability of the rumen epithelium [9], which aids microbes and immunogenic compounds to transmigrate into portal circulation [7], and further induces systemic inflammation. The principle mechanisms responsible for disrupting the barrier function of the rumen epithelium during SARA are not clear. A study that used isolated rumen and colon tissue from steers demonstrated that LPS and decreased pH may act synergistically to disrupt the epithelial barrier function and then breach the epithelial barrier [10]. Once the epithelium has been breached, the cells of mucosa-associated lymphoid tissue, which are found throughout the digestive mucosa and consist of clusters of white blood cells including innate lymphoid cells and mast cells [11, 12], respond by triggering local inflammation of rumen epithelium and altering cytokine production [11, 13]. Currently the structural transformation of the rumen epithelium during grain-induced SARA has been characterized [8, 14], but inflammation of rumen and the molecular mechanisms that trigger these events are not very clear.
In this study, we hypothesized that a local inflammatory response in the rumen epithelium occurred when cows were fed a high-concentrate diet, and therefore, mRNA expression of genes involved in inflammation would be up-regulated. The first objective of the this study was to characterize the mRNA expression of genes involved in inflammation in the rumen papillae from dairy cows fed high-concentrate diets using bovine genome microarray. The second objective of this study was to explore the relationship between expression of genes related to cytokines and ruminal factors.
Methods
Animals, experimental design, and diets
The experimental procedures and animal care conditions were approved by the Experimental Animal Welfare Ethics Committee of Nanjing Agricultural University.
This study was part of a larger experiment investigating the impact of subacute ruminal acidosis adaptation on rumen health in dairy cattle. A detailed description of the experimental design has been previously described [15]. Briefly, four ruminally cannulated Holstein cattle (average body weight, 460 ± 16.4 kg; 84 ± 25 d in milk at the beginning of the trial) were used in a 2 × 2 crossover design trial with 2 cows in each square and 2 periods. All animals were fed a low-concentrate diet (30 % concentrate feed, DM basis) for 3 weeks before the start of this experiment. The dietary treatments were a low-concentrate diet (LC; 40 % concentrate feed, DM basis) and a high-concentrate diet (HC; 70 % concentrate feed, DM basis) (Additional file 1: Table S1). Each experimental period was 21 d, with the first 11 d used for diet adaptation and 10 d of measurements. For the HC group, the dietary concentrate level was stepped up (by approximately 15 percentage units/d compared with the LC diet) during the first 2 d. Throughout the experimental period, the cows received a total mixed ration (TMR), ad libitum to about 5 % orts. Cows were housed in tie stalls and had free access to water.
Blood and rumen papillae samples
Blood samples were taken from the coccygeal vein at 0 and 4 h following the morning feeding on d 12, 17, and 21 of each experimental period. Blood hematological analysis was carried out using an automated haematological analyser (Sysmex K-1000D; Sysmex, Tokyo, Japan), and blood chemistry was measured by Vitros 250 Chemistry System (Ortho Clinical Diagnostics, Markham, Canada).
Rumen papillae samples were collected at 4 h after the morning feeding on d 21 of each experimental period. The ventral sac of the rumen was chosen for collected site [16]. The rumen papillae were excised (approximately 150 mg) from the rumen, washed 20 times in ice-cold PBS, and then frozen immediately in liquid nitrogen until the RNA was isolated.
Total RNA preparation
Total RNA was isolated from about 100 mg of frozen rumen tissues using TRIzol (Takara Bio, Otsu, Japan) following the manufacturer’s instructions. Qualified total RNA was further purified with the RNeasy mini kit (Qiagen, Hilden, Germany) and RNase-Free DNase Set (Qiagen, Hilden, Germany). The RNA concentration was determined using a NanoDrop spectrophotometer ND-1000UV-Vis (Thermo Fisher Scientific, Madison, WI). The RNA quality was routinely checked by measuring the optical density (260/280 and 260/230 ration). The total RNA from the rumen papillae was amplified and labelled with the Low Input Quick Amp Labeling Kit, One-Color (Agilent technologies, Santa Clara, CA), and the labelled cRNA was purified with RNeasy mini kit (Qiagen, Hilden, Germany).
Microarray hybridizations and data analysis
The hybridization of the cRNA was performed with Agilent Whole Bovine Genome Oligo (4 × 44 K) Microarrays using the Gene Expression Hybridization Kit (Agilent Technologies) according to the manufacturer’s protocols. After washing in staining dishes, hybridized slides were scanned with the Agilent Microarray Scanner (Agilent Technologies) with the default setting. Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent technologies).
Isolation and culturing of ruminal epithelial cells
Ruminal epithelial cells were isolated using the modified serial tryptic digestion procedure as described previously [17]. Rumen papillae were taken from the ventral blind sac of healthy lactating Holstein cows (n = 3). The cows were fed grass hay. During isolation, each fraction was examined under a phase contrast microscope. The fractions that contained increasing numbers of cells from the stratum basal and the stratum spinosum of the rumen epithelium were pooled. The collected cells were seeded into 25-mL cell culture flasks and cultured with DMEM (Corning Cellgro, Manassas, VA) supplemented with 10 % fetal bovine serum (Corning Cellgro, Manassas, VA) and 1 % antibiotic/antimycotic solution at 38 °C under 5 % CO2 for 24 h. Thereafter, the cells were allocated for the following treatments: (1) pH 7.4; (2) pH 5.5; (3) pH 7.4 + 10 μg/mL LPS; (4) pH 5.5 + 10 μg/mL LPS. After incubation for 24 h, the cells were collected and stored at - 80 °C for further analysis. All reagents used were purchased from Sigma (St. Louis, MO) unless indicated otherwise.
Quantitative RT- PCR and data analysis
Microarray results were validated by quantitative RT-PCR, and the relative expression of cytokines in an in vitro experiment was also conducted by qRT-PCR. Steele et al. [16] reported that GAPDH always displayed a minor variation in rumen epithelium samples of dairy cows. Thus, in the present study, the gene GAPDH was selected and used as a housekeeping gene. The sequences, amplicon sizes, Gene ID and references of primers used in the present study are listed in Table 1. The dissociation curves obtained from PCR amplification of each target genes (including GAPDH) exhibited a unique peak to verify the presence of a single product. Extracted RNA (1 μg) previously used for the microarray analyses was converted into cDNA using a ®PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Otsu, Japan). PCR analyses were performed in triplicate in a final volume of 20 μL reactions containing 10 μL ®SYBR Premix Ex Taq (Takara Bio, Otsu, Japan), 2 μL RT product, 0.4 μmol/L of each forward and reverse primers for the target genes, 0.4 μL ROX Reference Dye (Takara Bio, Otsu, Japan) and 6.8 μL dH2O. Reactions were run in a StepOne Plus Realtime PCR System (Applied Biosystems). The thermal cycling parameters were as follows: 30 s at 95 °C, 40 cycles of denaturation at 95 °C for 5 s followed by 30 s annealing at 60 °C. Gene expression of cytokines was normalized to housekeeping gene GAPDH (ΔCt = Cttarget - CtGAPDH), and the relative expressions, compared with the control group, were calculated using the 2-ΔΔCt method.
Table 1.
Primers for real-time quantitative PCR
| Gene Name | Gene ID | Primer sequence (5′ → 3′) | Amplicon Size,bp | Reference |
|---|---|---|---|---|
| IL-1β | NM_174093.1 | For: AACCGAGAAGTGGTGTTCTGC | 167 | [18] |
| R: TTGGGGTAGACTTTGGGGTCT | ||||
| IL-2 | NM_180997.2 | For: ACATTTGACTTTTACGCGCCCAAG | 307 | [19] |
| R: AATGAGAGGCACTTAGTGATC | ||||
| IL-6 | NM_000600.3 | For: GGAGGAAAAGGACGGATGCT | 227 | [18] |
| R: GGTCAGTGTTTGTGGCTGGA | ||||
| IL-8 | NM_173925 | For: CCTCTTGTTCAATATGACTTCCA | 170 | [18] |
| R: GGCCCACTCTCAATAACTCTC | ||||
| IL-12β | U11815 | For: AGGTCGTGGTAGAAGCTGTG | 276 | [20] |
| R: CCTTGTGGCATGTGACTTTG | ||||
| TNF-α | NM_173966.2 | For: CTTCTGCCTGCTGCACTTCG | 156 | [18] |
| R: GAGTTGATGTCGGCTACAACG | ||||
| GAPDH | NM001034034 | For: GGGTCATCATCTCTGCACCT | 176 | [21] |
| R: GGTCATAAGTCCCTCCACGA |
Statistical analyses
Data for blood chemistry and hematology parameters were analysed by using the general linear model (GLM) procedure in SPSS Version 18 (SPSS, Chicago, IL, USA), according to the model shown below: yijkmn = μ + Si + C(S)ij + Pk + Tm + (ST)im + eijkmn, where μ is the overall mean, Si is the diet treatment (i = 1–2), C(S)ij is the random effect of cow j (j = 1–4) nested within the diet i, Pk is the fixed effect of period k (k = 1–2), Tm is the fixed effect of day m (m = 1–3), (ST)im is the fixed effect of the diet by day interaction, and eijkmn is the random residual error. Significance was declared at P < 0.05.
Statistical calculations for the microarray data were conducted by GLM procedure in SPSS Version 18 (SPSS, Chicago, IL, USA), according to the model shown below: yijk = μ + Di + Pj + (DP)ij + eijk, where μ is the overall mean, Di is the diet treatment (i = 1–2), Pj is the fixed effect of period j (j = 1–2), (DP)ij is the interaction between treatment and experimental period, and eijk is the random residual error. Significance was declared at P < 0.05, and a tendency was considered to exist at 0.05 ≤ P < 0.10. Differentially expressed genes (DEGs) were defined by genes whose fold-change in expression (the LC group vs. the HC group) was equal to or higher than 1.5-fold and P < 0.05. DEGs and heat-map analysis were performed by SAS software from SHANGHAI BIOTECHNOLOGY CORPORATION eBioService (http://www.ebioservice.com). For better understanding of DEGs, overrepresentation analysis of Gene Ontology (GO) terms and pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was performed. Correlation analysis was performed using GraphPad Prism 5 software (Graphpad Software, San Diego, CA).
Data obtained from the in vitro cell culture experiment were analyzed by GLM procedure of SPSS Version 18 (SPSS, Chicago, IL, USA) to determine the effects of LPS and pH on the mRNA expression of cytokines, according to the model shown below: yijk = μ + Li + Pj + (LP)ij + eijk, where μ is the overall mean, Li is the fixed effect of LPS (i = 1–2 for dosage), Pj is the fixed effect of pH (j = 1–2), (LP)ij is the fixed effect of the LPS treatment × pH, and eijk is the random residual error. Differences with P < 0.05 were considered significant.
Results
Ruminal pH, SCFA and LPS
The results presented here must be interpreted in light of the overall effects of HC feeding on rumen fermentation in the experimental cattle reported in Mao’s prior study [15]. Briefly, the ruminal pH of the HC-fed cattle was lower (P < 0.001) than that of the LC-fed cattle, and the duration of time for which the ruminal pH was less than 5.8 was about 5.1 h after the first feeding in the HC group. The HC feeding increased (P < 0.001 to P = 0.018) the concentrations of propionate (24.32 mmol/L vs. 20.39 mmol/L), butyrate (14.39 mmol/L vs. 11.28 mmol/L), valerate (7.27 mmol/L vs. 4.81 mmol/L), isovalerate (2.64 mmol/L vs. 2.08 mmol/L), total volatile fatty acid (124.85 mmol/L vs. 111.23 mmol/L), lactic acid (0.24 mmol/L vs. 0.18 mmol/L) and LPS (26,266.88 EU/mL vs. 14,741.13 EU/mL) compared with the LC group.
Blood chemistry and hematology parameters
Compared with LC group, HC feeding led to an increase (P < 0.05) in white blood cells (WBC) and lymphocytes (Table 2), whereas HC treatments had no significant effects (P > 0.05) on the number of neutrophils and monocytes in blood. Concentrations of serum total protein, globulin, cholesterol and low density lipoprotein were lesser (P < 0.05) and the albumin concentration were greater (P < 0.05) in the HC group compared with the LC group. There were no differences (P > 0.05) in the urea nitrogen, glucose triglyceride, and high density lipoprotein between the two treatments.
Table 2.
Mean values for blood chemistry and hematology for cows fed two different diets
| Item | Diet | P-value | ||||
|---|---|---|---|---|---|---|
| LCa | HCb | SEM | Diet | Day | Diet x Day | |
| White blood cells, 1012/L | 7.30 | 8.82 | 0.326 | 0.004 | 0.001 | 0.091 |
| Neutrophils, 109/L | 2.27 | 2.58 | 0.171 | 0.212 | 0.412 | 0.164 |
| Lymphocytes, 109/L | 3.81 | 4.90 | 0.242 | 0.005 | <0.001 | 0.084 |
| Monocytes, 109/L | 1.22 | 1.08 | 0.075 | 0.185 | 0.029 | 0.295 |
| Total protein, g/L | 85.16 | 78.55 | 1.537 | 0.006 | 0.091 | 0.484 |
| Globulin, g/L | 54.75 | 46.69 | 1.280 | 0.000 | 0.091 | 0.513 |
| Albumin, g/L | 30.40 | 31.86 | 0.350 | 0.008 | 0.295 | 0.306 |
| Urea nitrogen, mmol/L | 8.11 | 7.56 | 0.276 | 0.173 | 0.542 | 0.215 |
| Glucose, mmol/L | 3.48 | 3.45 | 0.074 | 0.804 | 0.474 | 0.234 |
| Triglyceride, mmol/L | 0.10 | 0.09 | 0.003 | 0.091 | 0.124 | 0.110 |
| Cholesterol, mmol/L | 2.40 | 2.04 | 0.106 | 0.024 | 0.594 | 0.857 |
| Low density lipoprotein, mmol/L | 0.39 | 0.32 | 0.024 | 0.045 | 0.431 | 0.958 |
| High density lipoprotein, mmol/L | 1.15 | 1.13 | 0.041 | 0.757 | 0.471 | 0.967 |
aLow-concentrate diet
bHigh-concentrate diet
x means the interaction between two factors
Annotation of DEGs using Gene Ontology
To gain new insights into the underlying biological functions of DEGs, we used GO to analyze the identified DEGs. GO is a well-documented and widely used annotation system that assigns molecular function, biological process, and cellular component information to gene products [22, 23]. A total of 246 DEGs were detected (Additional file 2: Table S2), among them, 141 genes were significantly up-regulated and 105 genes were down-regulated in the HC group compared with the LC group (Fig. 1). Among these DEGs, 87, 77, and 74 DEGs were involved in molecular function, biological process, and cellular component, respectively (Table 3). Within molecular function, DEGs were mainly distributed in categories catalytic activity (31 genes) and binding (67 genes). For further analysis, most proportion of DEGs fell in category protein binding, hydrolase activity, ion binding and nucleic acid binding in GO molecular function. Enrichment analysis indicated category carbohydrate binding, auxiliary transport protein activity, channel regulator activity and cyclase regulator activity within molecular function were affected (P < 0.05) by the different diets. Within the cellular component, DEGs were mainly distributed in the categories cell (61 genes) and organelle (33 genes). Only two enriched categories (P < 0.05) were found in cellular component, namely extracellular region and extracellular region part. DEGs in the category biological process were mainly distributed in the categories cellular process (65), metabolic process (43 genes), biological regulation (41 genes) and regulation of biological process (37 genes). Our analysis indicated that 24 categories in biological process (at levels 2 and 3) were enriched and included genes associated with the following functions: establishment of localization in cell, immune response, cellular localization, cytokine production, immune system process, etc. (Table 3). The number of genes up- or down- regulated in each GO term and included genes within molecular function, biological process and cellular component is presented in Fig. 2 and Additional file 3: Table S3.
Fig. 1.
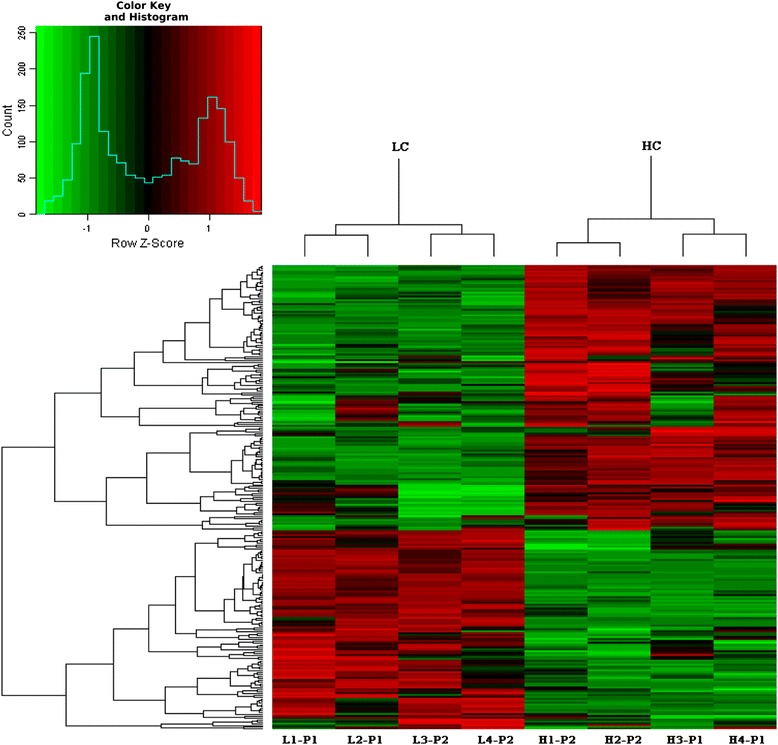
The hierarchical cluster analysis of DEGs. 246 differentially expressed (P < 0.05 and fold-change > 1.5) were included in the analysis. The red color indicated the high expression and the green color indicated low expression
Table 3.
Enriched GO terms within molecular function, cellular component and biological process
| GO ID | Level | Name | Hits | Enrichment test p-value | |
|---|---|---|---|---|---|
| GO:0030246 | 3 | carbohydrate binding | 5 | 0.026 | MF |
| GO:0015457 | 2 | auxiliary transport protein activity | 2 | 0.046 | MF |
| GO:0016247 | 3 | channel regulator activity | 2 | 0.046 | MF |
| GO:0010851 | 3 | cyclase regulator activity | 1 | 0.045 | MF |
| GO:0044421 | 2 | extracellular region part | 13 | 0.014 | CC |
| GO:0005576 | 2 | extracellular region | 18 | 0.015 | CC |
| GO: 0051649 | 3 | establishment of localization in cell | 15 | 0.001 | BP |
| GO: 0006955 | 3 | immune response | 11 | 0.002 | BP |
| GO: 0051641 | 3 | cellular localization | 15 | 0.003 | BP |
| GO: 0001816 | 3 | cytokine production | 6 | 0.005 | BP |
| GO: 0001776 | 3 | leukocyte homeostasis | 3 | 0.007 | BP |
| GO: 0019725 | 3 | cellular homeostasis | 8 | 0.008 | BP |
| GO: 0032879 | 3 | regulation of localization | 10 | 0.010 | BP |
| GO: 0051239 | 3 | regulation of multicellular organismal process | 13 | 0.011 | BP |
| GO: 0042330 | 3 | taxis | 4 | 0.011 | BP |
| GO: 0002376 | 2 | immune system process | 13 | 0.012 | BP |
| GO: 0051283 | 3 | negative regulation of sequestering of calcium ion | 2 | 0.016 | BP |
| GO: 0032844 | 3 | regulation of homeostatic process | 3 | 0.025 | BP |
| GO: 0001659 | 3 | temperature homeostasis | 2 | 0.028 | BP |
| GO: 0051093 | 3 | negative regulation of developmental process | 8 | 0.038 | BP |
| GO: 0006810 | 3 | transport | 25 | 0.040 | BP |
| GO: 0051707 | 3 | response to other organism | 5 | 0.041 | BP |
| GO: 0051235 | 3 | maintenance of location | 3 | 0.041 | BP |
| GO: 0032501 | 2 | multicellular organismal process | 27 | 0.042 | BP |
| GO: 0051234 | 2 | establishment of localization | 25 | 0.043 | BP |
| GO: 0021700 | 3 | developmental maturation | 3 | 0.044 | BP |
| GO: 0065008 | 3 | regulation of biological quality | 15 | 0.047 | BP |
MF Molecular Function, CC Cellular Component, BP Biological process
Fig. 2.
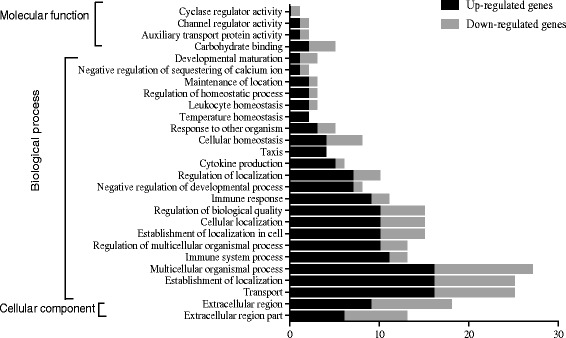
The distribution of DEGs in ontology terms for molecular function, biological process, and cellular component. The black bars indicated the number of up-regulated genes, while the grey bars indicated the number of down-regulated genes
Assignments of DEGs based on KEGG
For increased understanding of the effects of the HC diet on rumen tissue functional changes, the identified DEGs were subjected to pathway analysis (Additional file 4: Table S4). Combined with enrichment analysis, 11 KEGG pathways were enriched (P < 0.05) by the diets (Fig. 3), including cytokine-cytokine receptor interaction (bta04060), graft-versus-host disease (bta05332), Jak-STAT signaling pathway (bta04630), intestinal immune network for IgA production (bta04672), type I diabetes mellitus (bta04940), hematopoietic cell lineage (bta04640), NOD-like receptor signaling pathway (bta04621), calcium signaling pathway (bta04020), chemokine signaling pathway (bta04062), prion diseases (bta05020) and allograft rejection (bta05330). The number of up- or down- regulated DEGs involved in each KEGG pathway are presented in Fig. 3.
Fig. 3.
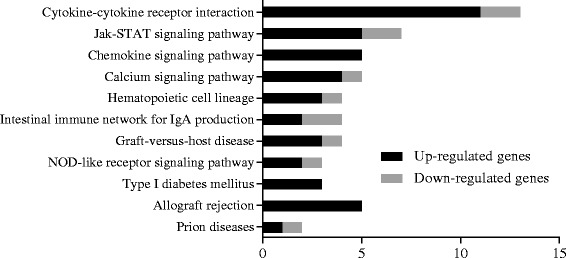
The distribution of DEGs in pathways, only enriched pathways were presented. The black bars indicated the number of up-regulated genes, while the grey bars indicated the number of down-regulated genes
Functions of inflammatory cytokines in response to ruminal environment variations
Since cytokines always mediated inflammatory responses, the cytokine-cytokine receptor interaction pathway attracted our attention. Of the 13 DEGs involved in this pathway, 11 genes (such as IL-1β, IL-2, CCL19, CCL8, CX3CR1, CXCL6, IL-22, INHBE, LEPR, PRL, and TNFRSF9) were significantly up-regulated, and only two genes (IL-6 and IL15RA) were significantly down-regulated in the HC group compared with the LC group. For further analysis, these 13 DEGs were classified as multi-functional genes that involved in more than two enriched KEGG pathways or GO terms (Table 4). It is worth noting that the inflammation-related genes IL-1β, IL-2, and IL-6 were participated five, seven, and eight pathways, respectively. The three genes were simultaneously involved in four pathways or GO terms, namely graft-versus-host disease (bta05332), extracellular region (GO: 0005576), immune system process (GO: 0002376) and multicellular organismal process (GO: 0032501). Due to the importance of multi-functional genes, the expression of IL-1β (up-regulated), IL-2 (up-regulated) and IL-6 (down-regulated) was validated by qRT- PCR (Table 5).
Table 4.
Relationships between DEGs involved in Cytokine-cytokine receptor interaction and other enriched pathways
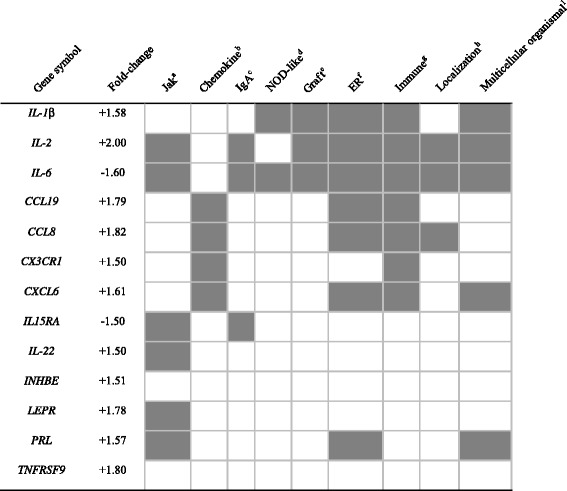
The grey indicated that this gene was also included in this pathway
aJak-STAT signaling pathway (bta04630)
bChemokine signaling pathway (bta04062)
cIntestinal immune network for IgA production (bta04672)
dNOD-like receptor (bta04621)
eGraft-versus-host disease (bta05332)
fExtracellular region part (GO: 0044421)
gImmune system process (GO: 0002376)
hEstablishment of localization (GO: 0051234)
iMulticellular organismal (GO: 0032501)
Table 5.
Validation of microarray gene expressions (IL-1β, IL-2 and IL-6) using Realtime-PCR method
| Gene symbol | qRT-PCR relative expression | Microarray expression | |||
|---|---|---|---|---|---|
| LC | HC | P-value | fold change | P value | |
| IL-1β | 1.15 ± 0.164 | 2.40 ± 0.159 | P < 0.05 | +1.58 | P < 0.05 |
| IL-2 | 0.80 ± 0.142 | 1.58 ± 0.135 | P < 0.05 | +2.20 | P < 0.05 |
| IL-6 | 1.31 ± 0.010 | 0.66 ± 0.068 | P < 0.05 | - 1.76 | P < 0.05 |
The relationship between the ruminal pH, LPS levels, and cytokines mRNA expression level
To explore whether there were possible relationships between the cytokines’ mRNA expression level and ruminal factors (pH and LPS), correlation analysis was conducted (Fig. 4). Results revealed that ruminal pH exhibited a positive correlation (P < 0.05) with IL-1β and IL-2 mRNA expressions but a negative correlation (P < 0.05) with IL-6 mRNA expression. There was a trend toward correlation (P = 0.067) and a negative correlation (P < 0.05) between the concentration of ruminal LPS and IL-1β, IL-6 relative mRNA expression, respectively. However, no significant correlation between ruminal LPS and IL-2 mRNA expression was observed.
Fig. 4.
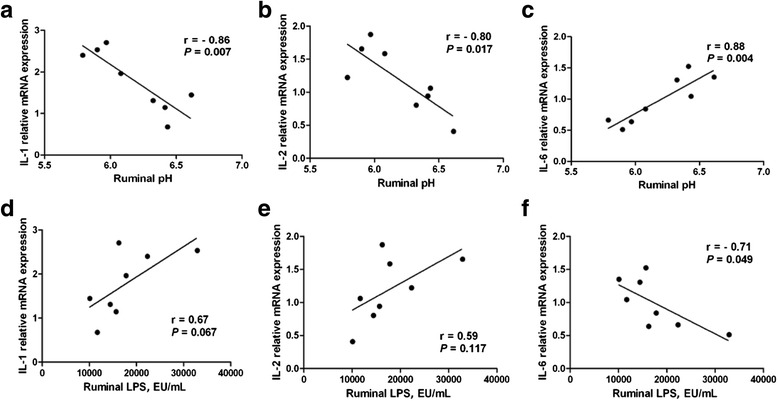
Correlations between cytokine mRNA expression (derived from Realtime-PCR) and ruminal factors. a-c: Correlation analysis between IL-1, IL-2, IL-6 relative mRNA expressions and ruminal pH. d-f: Correlation analysis between IL-1, IL-2, IL-6 relative mRNA expressions and ruminal LPS
Effects of pH and LPS on selected cytokines’ mRNA expression in cultured ruminal epithelial cells
As shown in Table 6, the effects of pH and LPS on these pro-inflammatory mediators in vitro appear to be selective. There was no significant interaction (P < 0.05) in the expression of IL-1β, IL-2, IL-6, IL-8, IL-12 and TNF-α between LPS and pH. Compared with the control group (group pH = 7.4), the mRNA expression of IL-1β, IL-2, IL-6, and IL-8 increased (P < 0.05) after the LPS treatment. However, the mRNA expression of IL-12 and TNF-α in the cultured ruminal epithelial cells remained unchanged (P > 0.05) between the control and LPS treatment groups. Low-pH treatment significantly elevated (P < 0.05) the mRNA expression of TNF-α, and had no significant effects (P > 0.05) on the mRNA expression of IL-1β, IL-2, IL-6 and IL-8 compared with the control group.
Table 6.
The relative expression of cytokines after low pH and lipopolysaccharides treatments in vitro
| Genes | pH 7.4 | pH 5.5 | SEM | P value | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | LPS, 10 μg/mL | 0 | LPS, 10 μg/mL | pH | LPS | pH x LPS | ||
| IL-1β | 1.13 | 2.75 | 0.62 | 2.36 | 0.259 | 0.104 | 0.001 | 0.103 |
| IL-2 | 1.08 | 2.06 | 0.81 | 2.91 | 0.280 | 0.357 | 0.001 | 0.093 |
| IL-6 | 0.91 | 1.66 | 1.43 | 1.62 | 0.106 | 0.107 | 0.007 | 0.068 |
| IL-8 | 0.89 | 4.34 | 1.46 | 3.23 | 0.455 | 0.551 | <0.001 | 0.088 |
| IL-12 | 1.04 | 1.25 | 0.65 | 0.94 | 0.115 | 0.153 | 0.292 | 0.847 |
| TNF | 0.87 | 1.62 | 2.04 | 2.57 | 0.255 | 0.031 | 0.155 | 0.783 |
x which means the interaction between two factors
Discussion
The overall goal of this study was to characterize the profile of gene expression especially target genes associated with inflammation after SARA challenge in vivo, and to ascertain to find out how environmental factors in rumen affect the mRNA expression of these cytokines in vitro. Therefore, defining SARA was imperative. The ruminal pH, SCFA, and LPS data have been reported previously by our team [15], and the duration time for ruminal pH below 5.8 was about 5.1 h in the HC group during the diurnal period, and the a higher LPS level was observed in the HC group, suggesting that SARA was successfully induced in cows fed the HC diet. Many studies have confirmed that the grain based-SARA challenge can cause translocation of lipopolysaccharide and triggers systemic inflammation leading to an increase in acute phase proteins in blood [7, 24, 25]. In the present study, we cannot absolutely certify the occurrence of systemic inflammation in cows fed HC diet in the absence of detecting acute phase proteins or inflammatory cytokines in blood, as our research centred on local inflammation of the rumen epithelium. However, the increase in the serum white blood cells, lymphocytes, and albumin and the decrease in total protein and globulin in HC-fed cows, indicating that the inflammatory responses may be activated and the synthesis ability of globulin and albumin in liver were affected by HC feeding [26, 27].
In the present study, the analyses of GO categories revealed that most changes centralized in biological processes. In this category, our study showed that GO terms transport, establishment of localization, and cellular localization were enriched by the HC feeding, and these terms were defined as the process or result of the movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location. Considerable evidence has accumulated describing the changes in ruminal epithelial nutrient and ion transport (VFA, glucose, Na+), as well as the gene expression of transporters regulating these processes during the adaptation of HC diets [28]. Moreover, HC feeding also induced accelerated differentiation and cellular migration in the stratum basale, spinosum and granulosum layers of the rumen epithelium [14]. It is well established that rumen fermentation profile altered dramatically in response to highly fermentable diets; therefore the transports of these substances and cellular migrations may be driven or stimulated by ruminal fermentation products, and the exact mechanism warrants further exploration. In addition, our study also showed that GO term immune system process and its child terms leukocyte homeostasis and immune response were enriched by the different diet treatments. Indeed, several studies have confirmed that immune system is involved in the adaptation of the rumen epithelium to HC diets, and these immune responses are thought to be initiated from the stimulation of ruminal microbiota or toxin compounds [29, 30]. Correspondingly, the present study demonstrated that the level of some harmful or pro-inflammatory compounds such as LPS, biogenic amines, ethanolamine and glutaric acid in rumen were elevated in the cows fed HC diet (data not published). It is noticeable that a certain degree of immune response may be the self-protective behavior which prevents the further ruminal epithelial injury, and thus, future studies should evaluate the relationship between the degree of immune response and the severity of ruminal acidosis, as well as the process of local inflammation in the rumen epithelium.
To date, relatively little information is available about the global effects of HC feeding on the molecular functions and pathways of the ruminal epithelium. However, a recent study revealed that the alteration of multiple pathways including calcium signaling pathway, gap junction, and Jak-STAT signaling pathway contributes to the changes in ruminal fermentation induced by HC diets [31]. Consistent with the above report, calcium signaling pathway and Jak-STAT signaling pathway were both found enriched in the present study. Enriched calcium pathway in the present study also supported and confirmed the previously observed physiological change that HC feeding clearly altered calcium transport [32], and this alteration in the rumen epithelium may be associated with the stimulation of accumulated VFA in the rumen [33]. Thus, some calcium-dependent events occurred in the rumen epithelium, such as cytokine release and signal transduction process may be affected when cows were fed HC diets. Jak-STAT pathway is one of a handful of pleiotropic cascades used to transduce a multitude of signals for development and homeostasis in mammals [34]. Jak-STAT signaling pathway was reported to be closely related with the activation of inflammation, and the activate JAK and STAT proteins usually played a positive role in inflammation progresses [35]. In the present study, even though the key genes encoded JAK kinases (JAK1, JAK2, JAK3 and Tyk2) and STATs were not changed, enriched Jak-STAT signaling pathway may to some extent reflect that the functional homeostasis of rumen epithelium were impaired after HC feeding.
In the present study, aside from the changes in Calcium signaling pathway and Jak-STAT signaling pathway, inflammation related pathways including Cytokine-cytokine receptor interaction and Chemokine signaling pathway were also enriched by the different diet treatments. Inflammation is a complex set of synergistic actions of inflammatory mediators (such as cytokines, chemokines, and nitric oxide) and immune cells (such as macrophages, lymphocytes, and monocytes), as the first response of the immune system to infection or tissue injury [36, 37]. Although some review articles reported HC feeding may lead to rumenitis [3, 38] local inflammation in the rumen epithelium, there is still a lack of understanding of how inflammatory-related genes respond to HC diet at the molecular level. Our study revealed that the expression of genes found in cytokine-cytokine receptor pathway, including IL-1β, IL-2, IL-22, CCL19, CCL8, CX3CR1, CXCL6, INHBE, LEPR, and PRL, were up-regulated in HC-fed cows; only IL-6 and IL-15RA were down-regulated. It was well known that IL-1β, IL-2 and IL-22 have the potential to stimulate pro-inflammatory responses, and mediated inflammatory diseases [39]. Therefore, the higher expression of IL-1β, IL-2 and IL-22 mRNA in the rumen epithelium indicates that local inflammation might occur in the rumen epithelium of the HC group. Chemokines are chemotactic cytokines that can participate in regulating inflammatory leukocyte recruitment, lymphocyte recruitment and homing [40]. Chemokines were categorized as inflammatory or homeostatic chemokine according to the functional role. Results from previous studies indicated that bovine mammary epithelial cells can express CXCL6 (also called GCP2) and CCL8 (also called MCP2) in response to certain bacterial cell components during the simulation course of mastitis in vitro [41, 42]. In addition, CX3CR1, the receptor of chemokine CX3CL1, was also classified as an inflammatory chemokine [43]. In the present study, up-regulated genes (CCL19, CCL8, and CX3CR1) were observed in HC-fed cows, further indicating that local inflammation in the rumen epithelium might be triggered.
Inflammation may be the interaction result of multiple pathways or biological processes. In this study, the GO term immune system process was enriched in the HC group, and most affected inflammatory cytokines such as IL-1β, IL-2 and IL-6 were also in the category immune system process, indicating that the immune system activation might be involved in the adaptation of the rumen epithelium to the HC diet. Of the affected pro-inflammatory cytokines, there is a paucity of information on the expression of IL-1β in ruminants fed HC diet. However, IL-1β has been shown to increase the permeability of the intestinal tight junctions of Caco-2 cells [44]. If IL-1β acts similarly in the rumen epithelium, the interleukin may play a role in reducing the barrier function of the rumen epithelium in cattle fed the HC diet. Previous studies showed that inducers of IL-1β expression in the gut include hyperosmolarity, T and B cells, LPS, and other interleukin proteins [44, 45] indicating that these factors may have played a role in IL-1β up-regulation in cattle fed high grain diets. This is also consistent with our in vitro results that LPS treatment significantly increased the expression of IL-1β. The role of IL-2 in the inflammatory process is complex and involves pro-inflammatory as well as regulatory aspects, and dysregulation seem to contribute to various immune system-related diseases in humans [46, 47]. A previous study showed that the aberrant production of IL-2 can disrupt primary human T lymphocytes function by leading to the loss of T cell anergy and induction of autoimmunity [48]. Thus, it is reasonable to postulate that an increase in the expression of IL-2 during HC feeding might have a negative impact on the innate immune response in the rumen epithelium. IL-6 is a multifunctional, pleiotropic cytokine involved in regulating immune responses, acute-phase responses and inflammation [49, 50]. Previous studies revealed that IL-6 is involved in tissue repair and cytoprotection in the human gut, and preventing IL-6 production could favor wound healing and remodelling following injury [39, 50, 51]. In this study, the true cause of the observed effects in the down-regulation of IL-6 mRNA in the HC group is unclear. Nevertheless, since the reduction of pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α can reduce the length and severity of the immune response [52], it follows that decreasing the expression of IL-6 during HC feeding can have beneficial effects.
To validate the results of the in vivo experiment and get a better understanding of the inflammation response during HC feeding, the rumen factors (LPS and low pH) which are most associated with SARA were selected for the in vitro experiment. In vitro, the mRNA expression of tested pro-inflammatory mediators IL-1β, IL-2, and IL-8 was up-regulated after LPS was stimulated. These results further strengthened the well-documented theory of the pro-inflammation ability of LPS, and demonstrated that the higher levels of LPS in rumen fluid during HC feeding could contribute to local inflammation in the rumen epithelium. Our in vitro results also showed that the expression of IL-6 mRNA was increased by the LPS challenge, and this is not consistent with the down-regulation results observed in vivo in the present study. These differences indicate that IL-6 mRNA expression might be affected by factors other than LPS. Indeed, recent progress in the understanding of the role butyrate plays in inflammation confirmed that n-butyrate can down-regulate the level of IL-6 during LPS-induced inflammation response but not affect the level of TNF-α [41]. In this study, a higher concentration of butyrate was observed in the HC group compared with the LC group. Therefore, the stimulation effect of LPS on the production of IL-6 might be concealed by the inhibition effect of butyrate, and this might finally result in the lower mRNA expression of IL-6 observed in the in vivo tests.
Although it has been well known that feeding dairy cows diets containing high proportions of grain is associated with a rapid decline in ruminal pH, and low ruminal pH can lead to rumenitis and eventually to ruminal parakeratosis, erosion, and ulceration of the rumen epithelium [30], there is little information on the effects of extracellular pH on rumen immune function. Clinical studies of organic acidosis in humans revealed clinical acidaemias are usually accompanied by immune deficiency, including a decrease in white cell numbers, globulins, and mitogenic responses, a diminution of the inflammatory response and delayed phagocytosis [53]. In this study, our findings revealed that low-pH treatment significantly elevated the mRNA expression of gene TNF-α compared with the high pH group. These results suggest that lower pH may contribute to local inflammation of the rumen epithelium by up-regulating certain genes. However, the mechanism underlying the up-regulation of TNF-α expression induced by lower pH during HC feeding is unclear, and more research is needed to evaluate the effects of extracellular pH on the immunity function of the rumen epithelium.
Conclusions
Our findings revealed that the biological functional changes in the rumen epithelium were mainly focused on biological processes in dairy cattle fed a high-concentrate diet, and the local inflammation response was triggered during HC feeding in the present study. The lower pH and high level of LPS in rumen during HC feeding may play an important role in the process of local inflammation in the rumen epithelium, and their pro-inflammatory effects may weaken the permeability barrier of the rumen epithelium. Future research is required to explore the mechanism of rumen epithelium inflammation as well as the role of lower ruminal pH in the damage to the rumen tissue of dairy cattle during SARA.
Abbreviations
DEGs, differentially expressed genes; HC, high-concentrate; Hp, haptoglobin; LC, low-concentrate; SAA, serum amyloid A; SARA, subacute ruminal acidosis; TMR, a total mixed ration; VFA, volatile fatty acids
Acknowledgments
The authors thank Dongsheng Wang for the collection of biopsy samples.
Funding
This study was carried out with the support of the National Basic Research Program of China (2011CB100801). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets generated during the current study are available in the Gene Expression Omnibus (GEO) repository. The accession number is GSE82272 (http://www.ncbi.nlm.nih.gov/geo/query/ acc.cgi?acc = GSE82272).
Authors’ contributions
The authors’ contributions are as follows: RYZ carried out the majority of the animal studies including animal care, RNA isolation and real-time PCR; RYZ and SYM carried out data interpretation and manuscript preparation. SYM and WYZ were responsible for the conception of the project and the oversight of the experiment. All authors read and approved this manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The experimental procedures and animal care conditions were approved by the Experimental Animal Welfare Ethics Committee of Nanjing Agricultural University.
Additional files
Ingredients and chemical composition of diets. (DOCX 17 kb)
List of differentially expressed genes. (PDF 254 kb)
Enriched GO terms (only significantly enriched GO terms were listed). (PDF 20 kb)
DEGs were assigned to KEGG pathway. (PDF 29 kb)
Contributor Information
Ruiyang Zhang, Email: zhangruiyangnjau@163.com.
Weiyun Zhu, Email: zhuweiyun@njau.edu.cn.
Shengyong Mao, Email: maoshengyong@163.com.
References
- 1.Ametaj BN, Zebeli Q, Saleem F, Psychogios N, Lewis MJ, Dunn SM, et al. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics. 2010;6:583–94. doi: 10.1007/s11306-010-0227-6. [DOI] [Google Scholar]
- 2.Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, Wright AG, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. 2011;78:275–84. doi: 10.1111/j.1574-6941.2011.01154.x. [DOI] [PubMed] [Google Scholar]
- 3.Plaizier JC, Krause DO, Gozho GN, McBride BW. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J. 2008;176:21–31. doi: 10.1016/j.tvjl.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Zebeli Q, Dijkstra J, Tafaj M, Steingass H, Ametaj BN, Drochner W. Modeling the adequacy of dietary fiber in dairy cows based on the responses of ruminal pH and milk fat production to composition of the diet. J Dairy Sci. 2008;91:2046–66. doi: 10.3168/jds.2007-0572. [DOI] [PubMed] [Google Scholar]
- 5.Dionissopoulos L, Laarman AH, AlZahal O, Greenwood SL, Steele MA, Plaizier JC, et al. Butyrate-mediated genomic changes involved in non-specific host defenses, matrix remodeling and the immune response in the rumen epithelium of cows afflicted with subacute ruminal acidosis. Am J Anim Vet Sci. 2013;8:8–27. doi: 10.3844/ajavsp.2013.8.27. [DOI] [Google Scholar]
- 6.Gozho GN, Krause DO, Plaizier JC. Ruminal lipopolysaccharide concentration and inflammatory response during grain-induced subacute ruminal acidosis in dairy cows. J Dairy Sci. 2007;90:856–66. doi: 10.3168/jds.S0022-0302(07)71569-2. [DOI] [PubMed] [Google Scholar]
- 7.Khafipour E, Krause DO, Plaizier JC. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. J Dairy Sci. 2009;92:1060–70. doi: 10.3168/jds.2008-1389. [DOI] [PubMed] [Google Scholar]
- 8.Liu JH, Xu TT, Liu YJ, Zhu WY, Mao SY. A high-grain diet causes massive disruption of ruminal epithelial tight junctions in goats. Am J Physiol Regul Integr Comp Physiol. 2013;305:R232–41. doi: 10.1152/ajpregu.00068.2013. [DOI] [PubMed] [Google Scholar]
- 9.Klevenhusen F, Hollmann M, Podstatzky-Lichtenstein L, Krametter-Frötscher R, Aschenbach JR, Zebeli Q. Feeding barley grain-rich diets altered electrophysiological properties and permeability of the ruminal wall in a goat model. J Dairy Sci. 2013;96:2293–302. doi: 10.3168/jds.2012-6187. [DOI] [PubMed] [Google Scholar]
- 10.Emmanuel DG, Madsen KL, Churchill TA, Dunn SM, Ametaj BN. Acidosis and Lipopolysaccharide from Escherichia coli B:055 Cause Hyperpermeability of Rumen and Colon Tissues. J Dairy Sci. 2007;90:5552–7. doi: 10.3168/jds.2007-0257. [DOI] [PubMed] [Google Scholar]
- 11.Kurashima Y, Goto Y, Kiyono H. Mucosal innate immune cells regulate both gut homeostasis and intestinal inflammation. Eur J Immunol. 2013;43:3108–15. doi: 10.1002/eji.201343782. [DOI] [PubMed] [Google Scholar]
- 12.Pearson C, Uhlig HH, Powrie F. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 2012;33:289–96. doi: 10.1016/j.it.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Mani V, Weber TE, Baumgard LH, Gabler NK. Growth and Development Symposium: Endotoxin, inflammation, and intestinal function in livestock. J Anim Sci. 2012;90:1452–65. doi: 10.2527/jas.2011-4627. [DOI] [PubMed] [Google Scholar]
- 14.Steele MA, Croom J, Kahler M, AlZahal O, Hook SE, Plaizier K, et al. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am J Physiol Regul Integr Comp Physiol. 2011;300:R1515–23. doi: 10.1152/ajpregu.00120.2010. [DOI] [PubMed] [Google Scholar]
- 15.Mao SY, Zhang RY, Wang DS. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. 2013;24:12–9. doi: 10.1016/j.anaerobe.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Steele MA, Vandervoort G, AlZahal O, Hook SE, Matthews JC, McBride BW. Rumen epithelial adaptation to high-grain diets involves the coordinated regulation of genes involved in cholesterol homeostasis. Physiol Genomics. 2011;43:308–16. doi: 10.1152/physiolgenomics.00117.2010. [DOI] [PubMed] [Google Scholar]
- 17.Galfi P, Neogrády S, Kutas F. Culture of epithelial cells from bovine ruminal mucosa. Vet Res Commun. 1981;4:295–300. doi: 10.1007/BF02278507. [DOI] [PubMed] [Google Scholar]
- 18.Günther J, Petzl W, Zerbe H, Schuberth H, Koczan D, Goetze L, et al. Lipopolysaccharide priming enhances expression of effectors of immune defence while decreasing expression of pro-inflammatory cytokines in mammary epithelia cells from cows. BMC Genomics. 2012;13:17. doi: 10.1186/1471-2164-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alluwaimi AM. Detection of IL-2 and IFN-γ m RNA expression in bovine milk cells at the late stage of the lactation period with RT-PCR. Res Vet Sci. 2000;69:185–7. doi: 10.1053/rvsc.2000.0411. [DOI] [PubMed] [Google Scholar]
- 20.Riollet C, Rainard P, Poutrel B. Cell subpopulations and cytokine expression in cow milk in response to chronic Staphylococcus aureus infection. J Dairy Sci. 2001;84:1077–84. doi: 10.3168/jds.S0022-0302(01)74568-7. [DOI] [PubMed] [Google Scholar]
- 21.Wang A, Gu Z, Heid B, Akers RM, Jiang H. Identification and characterization of the bovine G protein-coupled receptor GPR41 and GPR43 genes. J Dairy Sci. 2009;92:2696–705. doi: 10.3168/jds.2009-2037. [DOI] [PubMed] [Google Scholar]
- 22.Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, et al. The comparative toxicogenomics database: update 2011. Nucleic Acids Res. 2011;39:D1067–72. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Butler H, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo WJ, Zhu WY, Mao SY. Effects of feeding increasing proportions of corn grain on concentration of lipopolysaccharide in the rumen fluid and the subsequent alterations in immune responses in goats. Asian-Australas J Anim Sci. 2013;26:1437–45. doi: 10.5713/ajas.2013.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zebeli Q, Ametaj BN. Relationships between rumen lipopolysaccharide and mediators of inflammatory response with milk fat production and efficiency in dairy cows. J Dairy Sci. 2009;92:3800–9. doi: 10.3168/jds.2009-2178. [DOI] [PubMed] [Google Scholar]
- 26.Clark VL, Kruse JA. Clinical Methods: The history, physical, and laboratory examinations. J Am Med Assoc. 1990;264:2808–9. doi: 10.1001/jama.1990.03450210033015. [DOI] [Google Scholar]
- 27.Geffken DF, Cushman M, Burke GL, Polak JF, Sakkinen PA, Tracy RP. Association between physical activity and markers of inflammation in a healthy elderly population. Am J Epidemiol. 2001;153:242–50. doi: 10.1093/aje/153.3.242. [DOI] [PubMed] [Google Scholar]
- 28.Penner GB, Steele MA, Aschenbach JR, McBride BW. Ruminant Nutrition Symposium: Molecular adaptation of ruminal epithelia to highly fermentable diets. J Anim Sci. 2011;89:1108–19. doi: 10.2527/jas.2010-3378. [DOI] [PubMed] [Google Scholar]
- 29.Dionissopoulos L, Steele MA, AlZahal O, McBride BW. Adaptation to high grain diets proceeds through minimal immune system stimulation and differences in extracellular matrix protein expression in a model of subacute ruminal acidosis in non-lactating dairy cows. Am J Anim Vet Sci. 2012;7:84–91. doi: 10.3844/ajavsp.2012.84.91. [DOI] [Google Scholar]
- 30.Chen Y, Oba M. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet Microbiol. 2012;159:451–9. doi: 10.1016/j.vetmic.2012.04.032. [DOI] [PubMed] [Google Scholar]
- 31.Taniguchi M, Penner GB, Beauchemin KA, Oba M, Guan LL. Comparative analysis of gene expression profiles in ruminal tissue from Holstein dairy cows fed high or low concentrate diets. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:274–9. doi: 10.1016/j.cbd.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Uppal SK, Wolf K, Martens H. The effect of short chain fatty acids on calcium flux rates across isolated rumen epithelium of hay-fed and concentrate-fed sheep. J Anim Physiol Anim Nutr. 2003;87:12–20. doi: 10.1046/j.1439-0396.2003.00401.x. [DOI] [PubMed] [Google Scholar]
- 33.Leonhard-Marek S, Becker G, Breves G, Schröder B. Chloride, gluconate, sulfate, and short-chain fatty acids affect calcium flux rates across the sheep forestomach epithelium. J Dairy Sci. 2007;90:1516–26. doi: 10.3168/jds.S0022-0302(07)71637-5. [DOI] [PubMed] [Google Scholar]
- 34.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–3. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 35.Hanada T, Yoshimura A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 2002;13:413–21. doi: 10.1016/S1359-6101(02)00026-6. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Liu Y. Regulation of innate immune response by MAP kinase phosphatase-1. Cell Signal. 2007;19:1372–82. doi: 10.1016/j.cellsig.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–62. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause KM, Oetzel GR. Understanding and preventing subacute ruminal acidosis in dairy herds: A review. Anim Feed Sci Tech. 2006;126:215–36. doi: 10.1016/j.anifeedsci.2005.08.004. [DOI] [Google Scholar]
- 39.Akdis M, Burgler S, Crameri R, Eiwegger T, Fujita H, Gomez E, et al. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2011;127:701–21. doi: 10.1016/j.jaci.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 40.Nomiyama H, Osada N, Yoshie O. The evolution of mammalian chemokine genes. Cytokine Growth F R. 2010;21:253–62. doi: 10.1016/j.cytogfr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Mount JA, Karrow NA, Caswell JL, Boermans HJ, Leslie KE. Assessment of bovine mammary chemokine gene expression in response to lipopolysaccharide, lipotechoic acid + peptidoglycan, and CpG oligodeoxynucleotide 2135. Can J Vet Res. 2009;73:49–57. [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Shi ZR, Chu CY, Lee KH, Zhao X, Lee JW. Expression of bovine granulocyte chemotactic protein-2 (GCP-2) in neutrophils and a mammary epithelial cell line (MAC-T) in response to various bacterial cell wall components. Vet J. 2010;186:89–95. doi: 10.1016/j.tvjl.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Widdison S, Coffey TJ. Cattle and chemokines: evidence for species-specific evolution of the bovine chemokine system. Anim Genet. 2011;42:341–53. doi: 10.1111/j.1365-2052.2011.02200.x. [DOI] [PubMed] [Google Scholar]
- 44.Al-Sadi RM, Ma TY. IL-1β causes an increase in intestinal epithelial tight junction permeability. J Immunol. 2007;178:4641–9. doi: 10.4049/jimmunol.178.7.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–147. [PubMed] [Google Scholar]
- 46.Hanisch UK, Quirion R. Interleukin-2 as a neuroregulatory cytokine. Brain Res Rev. 1996;21:246–84. doi: 10.1016/0165-0173(95)00015-1. [DOI] [PubMed] [Google Scholar]
- 47.Lan RY, Selmi C, Gershwin ME. The regulatory, inflammatory, and T cell programming roles of interleukin-2 (IL-2) J Autoimmun. 2008;31:7–12. doi: 10.1016/j.jaut.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953–61. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 49.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–52. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tebbutt NC, Giraud AS, Inqlese M, Jenkins B, Waring P, Clay FJ, et al. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat Med. 2002;8:1089–97. doi: 10.1038/nm763. [DOI] [PubMed] [Google Scholar]
- 51.Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127:514–25. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 52.Troutman TD, Hu W, Fulenchek S, Yamazaki T, Kurosaki T, Bazan JF, et al. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc Natl Acad Sci U S A. 2012;109:273–8. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lardner A. The effects of extracellular pH on immune function. J Leukoc Biol. 2001;69:522–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available in the Gene Expression Omnibus (GEO) repository. The accession number is GSE82272 (http://www.ncbi.nlm.nih.gov/geo/query/ acc.cgi?acc = GSE82272).


