ABSTRACT
The spindle assembly checkpoint (SAC) ensures accurate chromosome segregation during mitosis by delaying the activation of the anaphase-promoting complex/cyclosome (APC/C) in response to unattached kinetochores. The Mad2 protein is essential for a functional checkpoint because it binds directly to Cdc20, the mitotic co-activator of the APC/C, thereby inhibiting progression into anaphase. Mad2 exists in at least 2 different conformations, open-Mad2 (O-Mad2) and closed-Mad2 (C-Mad2), with the latter representing the active form that is able to bind Cdc20. Our ability to dissect Mad2 biology in vivo is limited by the absence of monoclonal antibodies (mAbs) useful for recognizing the different conformations of Mad2. Here, we describe and extensively characterize mAbs specific for either O-Mad2 or C-Mad2, as well as a pan-Mad2 antibody, and use these to investigate the different Mad2 complexes present in mitotic cells. Our antibodies validate current Mad2 models but also suggest that O-Mad2 can associate with checkpoint complexes, most likely through dimerization with C-Mad2. Furthermore, we investigate the makeup of checkpoint complexes bound to the APC/C, which indicate the presence of both Cdc20-BubR1-Bub3 and Mad2-Cdc20-BubR1-Bub3 complexes, with Cdc20 being ubiquitinated in both. Thus, our defined mAbs provide insight into checkpoint signaling and provide useful tools for future research on Mad2 function and regulation.
KEYWORDS: APC/C, Cdc20, conformational specific, kinetochore, Mad2, SAC
Introduction
Proper partitioning of sister chromatids to the 2 new daughter cells is one of the most important aspects of mitosis. This outcome is ensured by the correct attachment of kinetochores to microtubules of the mitotic spindle resulting in biorientation of the sister chromatids and their subsequent partitioning at anaphase.1,2 Unattached kinetochores will result in the activation of the spindle assembly checkpoint (SAC) that inhibits anaphase entry until proper biorientation of all sisters have been established.3-5 SAC activation results in the generation of the mitotic checkpoint complex (MCC) that inhibits Cdc20, the mitotic co-activator of the anaphase-promoting complex/cyclosome (APC/C). The MCC is composed of the checkpoint proteins Mad2 and BubR1-Bub3 that bind directly to Cdc20. 6-11 The MCC binds stably to the APC/C, and recent work shows that the MCC can bind and inhibit a second molecule of Cdc20 already bound to the APC/C, which is required for checkpoint signaling.12 The rate-limiting step in the formation of the MCC is the kinetochore-catalyzed binding of Mad2 to Cdc20 because Mad2 has to undergo a large structural change for this to occur.13 Once the Mad2-Cdc20 complex has formed, BubR1-Bub3 can bind to Cdc20, and subsequently Mad2 might dissociate to form a Cdc20-BubR1-Bub3 complex that has been referred to as the BBC complex.10,14-16 The removal of Mad2 from Cdc20 is stimulated by p31comet and the AAA-ATPase TRIP13, but the exact details of this are still to be determined.14,17-22
The Mad2 protein almost exclusively consists of a HORMA domain that can adopt 2 different conformations referred to as open-Mad2 (O-Mad2) and closed-Mad2 (C-Mad2).23-27 The conversion from O-Mad2 to C-Mad2 requires a large movement of both the N terminus and the C terminus of Mad2 (Fig. 1A). C-Mad2 is the active form of Mad2 that binds to a short sequence in the N terminus of Cdc20, and, in the Mad2-Cdc20 complex, the C terminus of Mad2 crosses Cdc20 like a seatbelt.25 Interestingly, a similar sequence is present in the Mad1 checkpoint protein that also binds C-Mad2 in a structurally similar way.26 The Mad1-Mad2 complex is a very stable complex and localizes to unattached kinetochores through Mad1 interactions with outer kinetochore proteins.28-33 Given the fact that C-Mad2 can dimerize with O-Mad2, the template model proposes that kinetochore-localized Mad1-Mad2 recruits soluble O-Mad2 to kinetochores and this converts O-Mad2 into C-Mad2.34 This model is supported by many elegant biochemical experiments but data also suggest that unknown aspects of the kinetochore environment could contribute to Mad2-Cdc20 complex formation.13,16,35-39 To further understand Mad2 function in vivo, it is critical to have well-defined tools to probe the different Mad2-containing complexes that are formed during an active checkpoint. A mouse monoclonal C-Mad2-specific antibody has been generated by the Nigg lab, but mouse monoclonal O-Mad2 and pan-Mad2 antibodies have not been reported.40 Here, we describe 3 novel Mad2 mouse monoclonal antibodies (mAbs) that recognize C-Mad2, O-Mad2 and pan-Mad2, and use these to probe Mad2 complexes during mitosis.
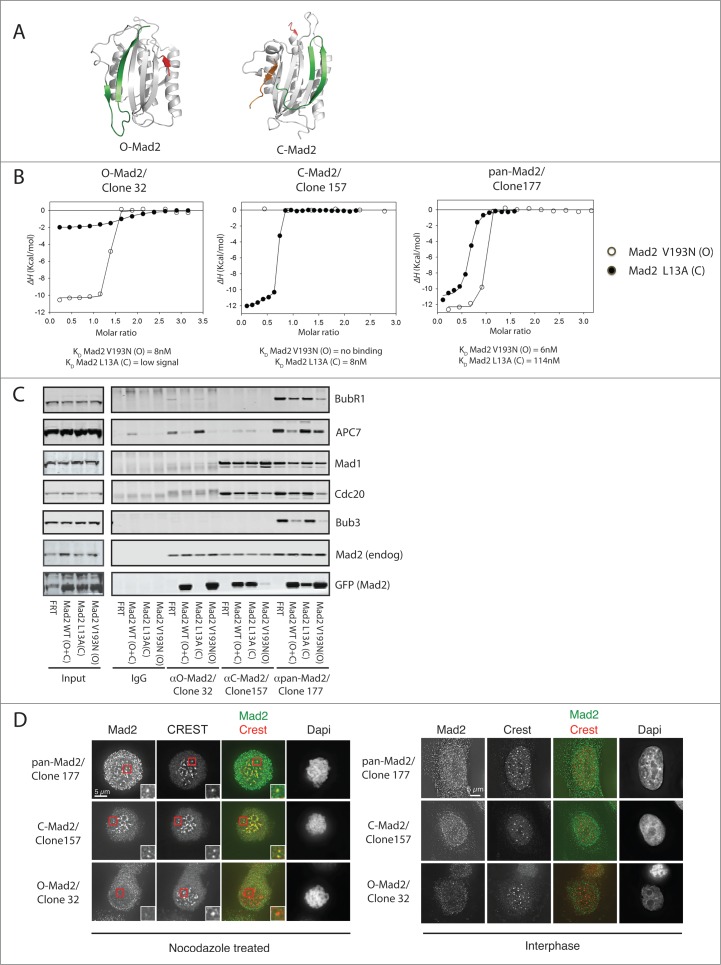
Figure 1.
Characterization of Mad2 monoclonal antibodies. (A) Structure of O-Mad2 and C-Mad2 with the C- and N-terminus colored in green and red, respectively. In the C-Mad2 structure, a ligand (orange) is bound mimicking the binding of Cdc20 and Mad1. O-Mad2 modified from PDB 1DUJ and C-Mad2 modified from PDB 1KLQ. (B) ITC measurements using purified Mad2 monoclonal antibodies and recombinant Mad2 L13A/R133A (C-Mad2) and Mad2 V193N/R133A (O-Mad2) to determine binding specificity and affinity of the antibodies. (C) Stable HeLa cell lines expressing Mad2-Venus, Mad2 L13A-Venus or Mad2 V193N-Venus or a control cell line (FRT) were treated with nocodazole and cells were harvested by mitotic shake-off. A mitotic extract was prepared from each cell line and Mad2 complexes were immunoprecipitated with the different Mad2 monoclonal antibodies. Samples were subsequently analyzed by SDS-PAGE and western blot and probed for the indicated proteins. (D) Immunofluorescence images of cells stained with the indicated Mad2 conformational specific antibodies. Cells were arrested in prometaphase using nocodazole or left untreated to obtain interphase cells.
Results
Characterization of conformation-specific anti-Mad2 mAbs
To investigate Mad2 biology in vivo, we aimed at generating Mad2-specific mAbs to address more precisely the composition of checkpoint complexes. Mice were immunized with bacterially produced His-tagged full-length Mad2, which contains a mixture of different Mad2 conformations. Following fusion with hybridoma cells, single clones were isolated and screened by ELISA for reactivity toward recombinant Mad2. Supernatants from positive clones were subsequently screened for their ability to immunopurify Mad2 from mitotic cells, which resulted in 3 positive clones (clone numbers 32,157,177) that were characterized further.
First, we used isothermal titration calorimetry (ITC) to measure the affinity of the antibodies for a mutant of Mad2 locked in its closed conformation (Mad2 L13A) or a mutant locked in its open conformation (Mad2 V193N)27,41 (see ref 39 for characterization of these recombinant Mad2 mutants). In addition, both Mad2 mutants harbored the R133A mutation to avoid any chance of Mad2 dimerization. All three antibodies bound with nanomolar (nM) affinity to Mad2 with a molar ratio of 2, but there was a clear difference in their recognition of the different Mad2 conformations (Fig. 1B and Fig. S1A). Clone 32 bound very specifically to O-Mad2 while clone 157 bound very specifically to C-Mad2. In contrast, clone 177 bound tightly to both O-Mad2 and C-Mad2. None of the antibodies recognized Mad2 by western blot (data not shown), clearly suggesting that they were recognizing structural rather than linear epitopes.
We further analyzed the specificity of the 3 antibodies in immunopurifications from mitotic HeLa cells stably expressing Mad2 WT-Venus, Mad2 L13A-Venus and Mad2 V193N-Venus (Fig. 1C). Cells were arrested with nocodazole, a microtubule-depolymerizing agent, to activate the checkpoint and then harvested by mitotic shake-off. From the mitotic cell extracts, we immunopurified Mad2 with the different antibodies and analyzed samples by protein gel blot. Clone 177 bound all 3 forms of exogenous Mad2 and co-purified known Mad2 interactors, which is in agreement with the ITC measurements. Clone 32 only precipitated Mad2-Venus and Mad2 V193N-Venus, while clone 157 precipitated Mad2-Venus and Mad2 L13A-Venus but not Mad2 V193N-Venus. Thus, the specificity of the antibodies determined by ITC was confirmed by these immunopurification experiments.
The Mad1-Mad2 complex localizes to the nuclear pores in interphase and to kinetochores during mitosis; we thus analyzed the ability of the different antibodies to detect Mad2 localization. Cells treated with nocodazole were stained with a centromere marker (CREST) and the different Mad2 antibodies and analyzed by immunofluorescence. Clone 157 specifically stained the kinetochore of mitotic cells and stained the nuclear envelope of interphase cells, while clone 32 did not stain any cellular structures (Fig. 1D). Clone 177 stained the kinetochores, but the signal was not as strong as for clone 157.
Based on these tests, we conclude that clone 177 is a pan-Mad2-specific antibody, clone 157 is a C-Mad2-specific antibody and clone 32 is an O-Mad2-specific antibody. We use these names rather than clone numbers in the sections below.
Analysis of Mad2 complexes in mitotic cells
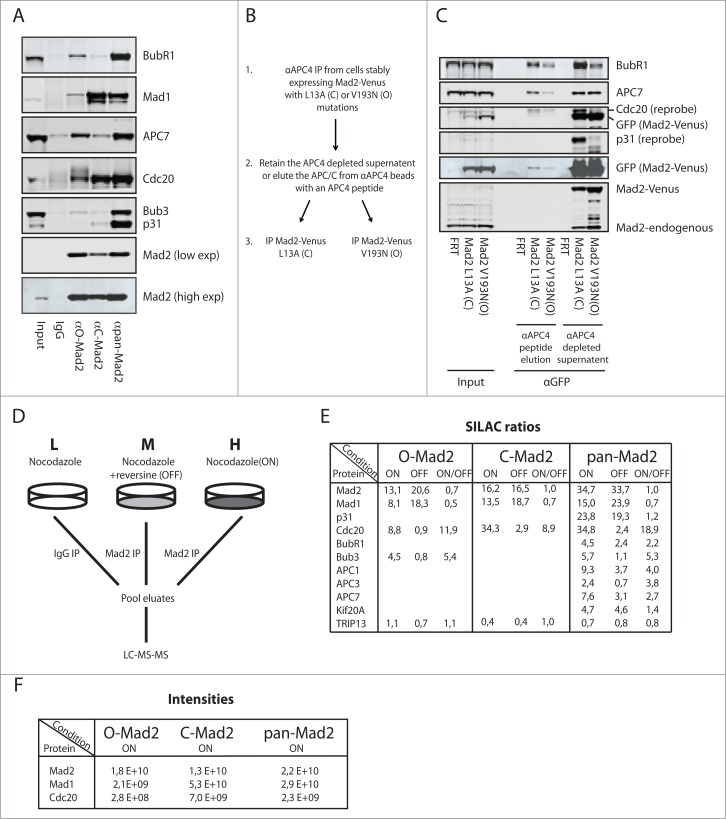
To explore the different complexes formed by Mad2 during mitosis, we precipitated endogenous Mad2 from nocodazole-arrested cells with the 3 different antibodies and analyzed their ability to co-purify known interactors (Fig. 2A). The pan-Mad2 antibody co-purified all known Mad2 interactors efficiently suggesting that the epitope recognized by this antibody does not interfere with Mad2 interactions. In contrast, the C-Mad2 antibody strongly co-purified Cdc20 and Mad1, but very little BubR1, APC/C and p31comet. As p31comet and BubR1 binds the dimerization surface of C-Mad2,7,42 we suspect that the C-Mad2 antibody is recognizing a similar region on Mad2. This was also reported for the C-Mad2-specific antibody generated by the Nigg lab and a different C-Mad2 clone we have described, potentially reflecting the dimerization surface as a strong antigen in C-Mad2.39,40 Although the O-Mad2 antibody was specific for O-Mad2 in our previous tests, we did detect some binding to known C-Mad2 interactors, such as Cdc20 and APC/C components. This could either reflect O-Mad2 interaction with C-Mad2 in these complexes or the weak affinity of the antibody for C-Mad2 that we can detect by ITC (Fig. 1B). We note that a sheep polyclonal antibody produced by the Taylor lab might also be a O-Mad2-specific antibody, and this antibody also co-purifies small amounts of Cdc20 and Mad1.14 Since the O-Mad2 antibody did not purify any Mad2 L13A-Venus from cells, we believe that this antibody is specific for O-Mad2 under our standard immunoprecipitation (IP) conditions. To further analyze if O-Mad2 can associate with checkpoint complexes, most likely through dimerization with C-Mad2, we purified APC/C complexes from nocodazole-arrested cells stably expressing Mad2 L13A-Venus and Mad2 V193N-Venus well below the endogenous Mad2 levels (Fig. 2B-C). Following peptide release of APC/C complexes from an APC4 affinity column, the samples were subjected to a second purification using a Venus affinity resin. In these experiments, APC/C and MCC components were co-purified by both Mad2 L13A-Venus and Mad2 V193N-Venus. The supernatant remaining after the APC4 depletion was also subjected to purification with a Venus affinity resin and again we could detect some APC/C and MCC components in both Mad2 purifications (Figs. 2B–C). Importantly, only Mad2 L13A-Venus co-purified p31comet as we expected. Based on this, we favor that the reason why our O-Mad2 antibody can co-purify small amounts of APC/C and checkpoint proteins is due to dimerization of O-Mad2 with C-Mad2 in these complexes.
Figure 2.
Analysis of Mad2 complexes using conformation specific antibodies. (A) HeLa cells arrested in mitosis with nocodazole were harvested by mitotic shake-off and Mad2 complexes purified with the different Mad2 monoclonal antibodies. Representative of 3 independent experiments. (B) Schematic of the sequential purification of Mad2-Venus complexes. (C) HeLa FRT/Trex cells stably expressing Mad2 L13A-Venus or Mad2 V193N-Venus were collected by mitotic shake-off after nocodazole treatment. APC/C complexes were purified with a monoclonal APC4 antibody and then eluted by the antigenic peptide. The eluate containing APC/C and the supernatant after APC/C depletion was then used for purification of Mad2-Venus complexes using GFP-Trap beads (chromotek) and analyzed by protein gel blot. Input is total cell extract prior to APC4 purification. Representative of 2 independent experiments. (D) Schematic of the triple SILAC experimental set-up. L: light condition, M: medium condition, H: heavy condition. (E) SILAC ratios determined for known Mad2 interactors in the SAC ON and SAC OFF condition. Only known interactors having a SILAC ratio higher than 2 are included except for TRIP13, which is included despite not being enriched above IgG control levels. (F) Peptide intensities for Mad2, Mad1 and Cdc20 in the indicated conditions to illustrate that the O-Mad2 antibody purifies much lower levels of Mad1 and Cdc20.
Mass spectrometry analysis of Mad2 complexes in checkpoint-arrested cells
To investigate, in an unbiased manner, the proteins interacting with the different conformations of endogenous Mad2 during an active checkpoint, we used SILAC quantitative mass spectrometry. HeLa cells were grown in the presence of different isotopes of Arg and Lys to obtain 3 different populations (light (L), medium (M), heavy (H)), the peptides of which can be distinguished in the mass spectrometer (Fig. 2D).43 The heavy population was arrested in mitosis with nocodazole and the medium population was in addition incubated with the Mps1 inhibitor reversine for one hour to silence the SAC.44 Cells treated with reversine were also treated with the proteasome inhibitor MG132 to avoid mitotic exit. The light population was arrested with nocodazole and a mouse IgG was used for control purification. We used the 3 Mad2-specific mAbs to purify Mad2 from the heavy and medium conditions to get a quantitative comparison of “SAC ON” and “SAC OFF” complexes and referenced this to the IgG control (Fig. 2D). In agreement with the above analysis, the pan-Mad2 antibody enriched all known Mad2 interactors, and there was a clear reduction in Cdc20, BubR1, Bub3 and APC/C components when the checkpoint was turned off (Fig. 2E). The association with Mad1 did not change much as expected and similarly p31comet association with Mad2 was not responsive to SAC status in agreement with p31comet constantly antagonizing Mad2 function.14,20 In contrast, the C-Mad2 and O-Mad2 antibodies mainly precipitated Cdc20 and Mad1, and again Cdc20 showed the expected reduction in the SAC OFF condition. It should be noted that the intensities for Cdc20 and Mad1 was highest in pan-Mad2 and C-Mad2 purifications and much lower in O-Mad2 purifications, as expected (Fig. 2F). Our pan-Mad2 purification also validated Kif20A as an endogenous Mad2 interactor45 but TRIP13 was not enriched above background in any of these purifications, likely reflecting a transient interaction of this protein with checkpoint complexes.
When combined, the mass spectrometry analysis supports current models of Mad2 checkpoint complexes and their dynamics.
Distribution of Mad2 in different complexes
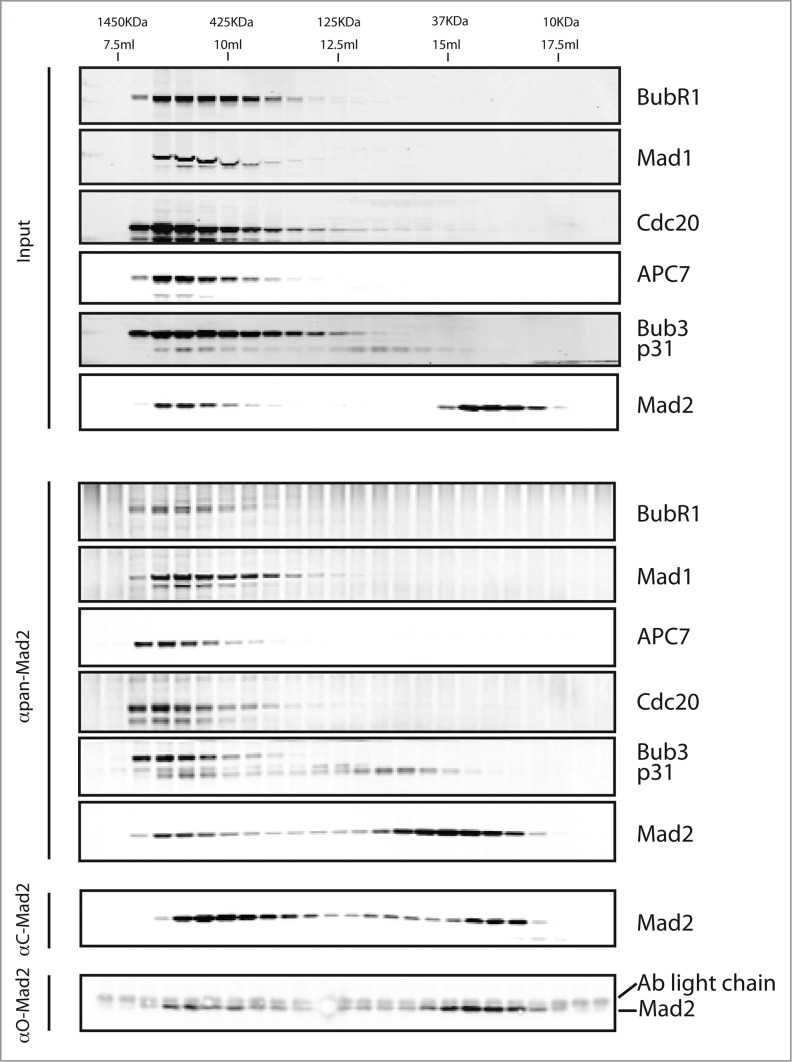
The above analysis provided a detailed analysis of Mad2 interactors but it did not enable the separation of different checkpoint complexes containing Mad2. To do this, we combined size-exclusion chromatography and the pan-Mad2 antibody. A nocodazole extract was prepared and separated on a Superdex 200 column, and from each fraction we immunoprecipitated Mad2 using the pan-Mad2 antibody and analyzed these by western blot (Fig. 3). As expected for a pan-Mad2 antibody, the migration profile of immunoprecipitated Mad2 resembled the total Mad2 profile in the extract. Two distinct populations of Mad2 were evident, one peaking around 1000 kDa and one around 25 kDa, likely corresponding to free Mad2. The Mad2 population at 1000 kDa represented approximately 20% of the Mad2 and precipitated MCC components, Mad1 and APC/C explaining their large Stokes radius. The Mad2 migrating at 25 kDa corresponds in size to monomeric Mad2, and we also detected some p31comet-Mad2 complex migrating around 50 kDa. The presence of a p31comet-Mad2 complex suggests that C-Mad2 can exist unbound to its known ligands Mad1 and Cdc20.
Figure 3.
Analysis of Mad2 complexes during an active checkpoint. HeLa cells arrested with nocodazole were harvested by mitotic shake-off and a cell extract was prepared. The total extract was loaded onto a Superdex 200 column following clarification by centrifugation and 500 μl fractions collected and analyzed by SDS-PAGE and western blot (Input). Mad2 complexes were isolated from each fraction by immunopurification with the pan-Mad2 antibody and analyzed by protein gel blot (αpan-Mad2). A similar purification was done with the C-Mad2 specific antibody (αC-Mad2) or O-Mad2 specific antibody (αO-Mad2).
When we purified O-Mad2 and C-Mad2 from the fractions, there was a clear difference between Mad2 migration in the input and Mad2 enrichment in the immunopurifications. The O-Mad2 antibody primarily precipitated Mad2 in low molecular weight complexes and a small fraction in high molecular weight complexes. In contrast, the C-Mad2 antibody largely precipitated Mad2 in the high molecular weight complexes and some Mad2 migrating at 25 kDa on the column. This is in agreement with C-Mad2 being the active form binding to MCC and Mad1 but it indicated that unliganded C-Mad2 might be present during an active checkpoint. We cannot, however, exclude antibody induction of O-Mad2 to C-Mad2 conversion in these experiments.
Analysis of checkpoint complexes associated with the APC/C
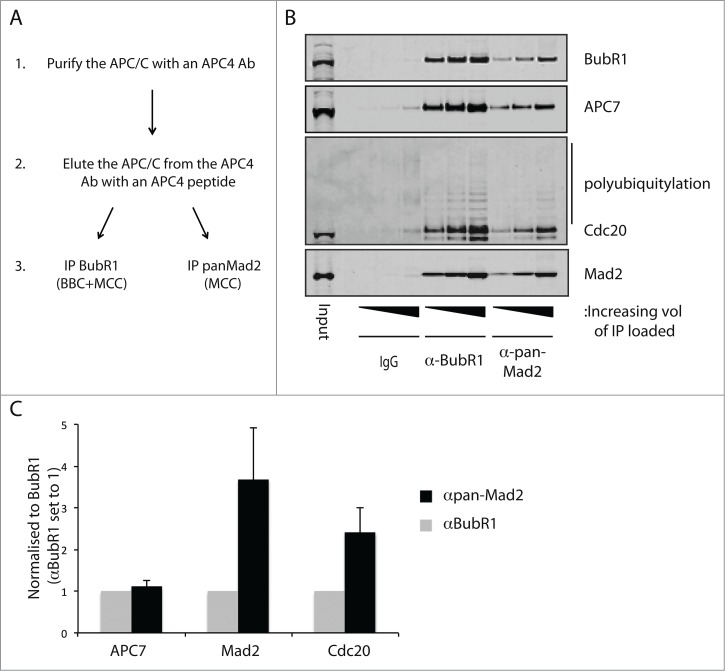
To analyze the composition of checkpoint complexes associated with the APC/C, we used a sequential purification approach (Fig. 4A). From a nocodazole extract, we first immunopurified the APC/C using a monoclonal APC4 antibody and eluted the complexes by antigen peptide competition. The purified APC/C was then split in 3 and one-third was then incubated with a BubR1 mAb recognizing residues 116–127 in the TPR domain (Fig. S1B), another one-third was incubated with the pan-Mad2 antibody and the last one-third was incubated with a mouse IgG. The complexes were then analyzed by quantitative protein gel blot and signals normalized to the BubR1 level (Fig. 4). As the BubR1 and Mad2 antibodies precipitated equal levels of APC7, it clearly indicates that very little Mad2-Cdc20 is present on the APC/C in agreement with Mad2 inhibiting and BubR1 stimulating the interaction with the APC/C.46-48 It was clear that the ratio of BubR1 to Mad2 was lower in the BubR1 purification, suggesting that indeed BBC complexes are present on the APC/C in addition to the MCC. In both the BubR1 and Mad2 purifications, Cdc20 was clearly ubiquitinated, as revealed by the ubiquitination pattern recognized by the Cdc20 antibody. The level of Cdc20 ubiquitination appeared similar in the 2 purifications, and thus ubiquitination of Cdc20 does not correlate with Mad2 dissociation in endogenous complexes.
Figure 4.
Analysis of checkpoint complexes associated with the APC/C. (A) Schematic of the sequential purification of the APC/C complexes. HeLa cells arrested with nocodazole were harvested by mitotic shake-off and APC/C complexes purified with a monoclonal APC4 antibody and subsequently eluted by the antigenic peptide. This purified APC/C was subsequently used in BubR1 and pan-Mad2 purifications and analyzed by western blot. (B) Analysis of the APC/C complexes purified with the BubR1 antibody and the pan-Mad2 antibody. Representative of 3 independent experiments. (C) Quantification using Licor technology of the level of APC7, Mad2 and Cdc20 in the 2 purifications. Values were normalized to the level of BubR1 and the αBubR1 samples set to one. Average and standard deviation of 3 independent experiments indicated.
Discussion
The ability of Mad2 to convert between O-Mad2 and C-Mad2 is critical for the checkpoint and understanding how this is controlled is fundamental for understanding checkpoint signaling. Beautiful biochemical work have provided important models for how this is regulated but our ability to study Mad2 biology in vivo is still lagging behind. Here, we provide important tools in the form of defined Mad2-specific mAbs that are useful for dissecting endogenous Mad2 complexes and the conformation of Mad2 in these.
Our C-Mad2 specific antibody data is in agreement with the data from the Nigg lab showing that C-Mad2 mainly exists in complex with its known ligands Mad1 and Cdc20.40 However, in our size-exclusion chromatography experiments, we do detect a small amount of unliganded C-Mad2 and p31comet-Mad2 complexes, which could be C-Mad2 that has been removed from Cdc20. Recent work from the Cleveland lab proposes that C-Mad2 removed from the MCC could facilitate cytoplasmic amplification of the checkpoint signal by binding a new Cdc20 molecule, and potentially this is the C-Mad2 pool that we observe here.15
A puzzling observation is that while the Mad2-Cdc20 interaction is very dynamic and responds to SAC status, 2 other interactors that use a similar motif for binding Mad2, Mad1 and Kif20A, are stable (Fig. 2). Currently, we do not know the details of how C-Mad2 is removed from Cdc20 but it does not appear to depend on BubR1 because we recently showed that Mad2 is more efficiently removed from a Cdc20 mutant that does not bind BubR1.48 p31comet binding is also not specific for the Mad2-Cdc20 complex because it also associates with Mad1-Mad2.40,49 Mad2 removal might require that it is bound to an unstructured region as in the case for Cdc20, and could explain why C-Mad2 is not removed from Mad1. However, the Mad2 binding site of Kif20A is at the extreme C terminus of Kif20A and one would assume that it should be very easy to remove C-Mad2.45 Understanding the details of specific Mad2 removal from Cdc20 is an important goal.
We find that Cdc20 is ubiquitinated in both BBC and MCC complexes and our data do not support that Cdc20 ubiquitination in itself dissociates the Mad2-Cdc20 complex,50 which is in agreement with observations from the Morgan lab.46 This does not rule out a subsequent role of ubiquitination on Cdc20 in SAC silencing. Another possibility is that Cdc20 ubiquitination is a bystander effect from the stable association of the MCC with the APC/C, and that p31comet dissociation of the MCC is required for subsequent Cdc20 degradation.20 The pan-Mad2 antibody described here will be useful in further investigations of Mad2 interactions since it efficiently purifies all Mad2 interactors and ubiquitinated Cdc20.
Materials and methods
Cell culture
HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS, HyClone) and 1% penicillin-streptomycin (Life Technologies). Protein expression was induced in stable cell lines by treatment with doxycycline (5 ng/ml).
Cloning and stable cell lines
Mad2 was cloned into pcDNA5/FRT/TO Venus (C-terminal tag). Stable cell lines were generated using the Flp-In system (Invitrogen) and clones were kept under selection by supplementing the growth media with 200 mg/ml hygromycin B and 5 mg/ml blasticidin S.
Antibodies
The following antibodies were used for Western blotting as indicated: APC7 1:1000 WB (rabbit, Bethyl A302-551A-1), Bub3 1:1000 WB (mouse, BD Bioscience 611731), BubR1 1:1000 WB (rabbit, raised against the TPR domain), Cdc20 1:1000 WB (mouse, Santa Cruz sc-13162), GFP 1:500 WB (mouse, Roche 11814460001), Mad1 1:1000 WB (mouse, Sigma M8069), Mad2 1:1000 WB (rabbit, Bethyl A300-301A), and p31 1:1000 WB (mouse, raised against full length protein).
Size-exclusion chromatography
Cells were treated with nocodazole (200ng/ml) for 18 hrs and mitotic cells were collected by shake-off. Cells were lysed for 30 mins in 20 mM Tris-HCl pH 7.8, 100 mM NaCl, 0.1% IGEPAL, 1 mM dithiothreitol (DTT), 0.5 mM EDTA, 1x protease inhibitor cocktail (Roche) and 1x phosphatase inhibitor cocktail (Roche). Lysates were clarified at 13000 rpm for 15 mins and then for 20 mins at 55000 rpm. 500 μl of lysate (8 mg/ml) was loaded onto a Superdex 200 10/300 GL column. The column was equilibrated using 1.5x column volumes of 20 mM Tris-HCl pH 7.6, 100 mM NaCl, 0.1% IGEPAL, 1 mM DTT, 0.5 mM EDTA. Fractions were collected in 500 μl volumes and Mad2 purifications were performed using 400 ul of each fraction.
Immunofluorescence
HeLa cells were seeded on coverslips and treated with 200 ng/ml nocodazole overnight. The cells were permeablized in 0.5% Triton-X100 for 5 minutes and then fixed in 4% formaldehyde for 20 minutes. Upon staining, the cells were first quenched with 25 mM glycine for a minimum of 20 minutes and then blocked with 3% bovine serum albumin in phosphate-buffered saline (PBS) + 0.1% Tween for 30 minutes. The cells were incubated with primary antibodies for 2 hours at room temperature (primary antibodies were used at the following concentrations: pan-Mad2 (16 μg/ml), C-Mad2 (4 μg/ml), O-Mad2 (4 μg/ml) and CREST (Antibodies Inc., 1:400)), followed by 45 minutes of incubation with appropriate secondary antibodies and DAPI (1:1000). Images were acquired taking z stacks of 200 nm using a 100X/1.4NA objective on a DeltaVision Elite Microscope (GE Healthcare). Figures were generated using ImageJ.
Immunopurification experiments
Cells were treated with nocodazole (200 ng/ml) for 18 hours, harvested by shake-off and lysed in 50 mM Tris-HCl pH 7.8, 100 mM NaCl, 0.1% NP40, 1 mM DTT, 0.5 mM EDTA, 1x protease inhibitor cocktail (Roche) and 1x phosphatase inhibitor cocktail (Roche). Lysates were clarified at 13000 rpm for 15 mins and then for 20 mins at 55000rpm. 800 μg of protein was used for each IP in Fig 1C and 2 mgs in Fig 2A. Immunoprecipitations were performed with 10 μl of Protein-G sepharose 4B beads crosslinked to antibodies (4 μg antibody/10 μl beads) for 2 hours and then washed in lysis buffer by inversion 3 times.
For the reciprocal IPs in Figs 2C and 4B, the cell lysates were obtained as described in the above paragraph. In Fig. 2C, the APC/C was then immunoprecipitated from 6 mgs of protein, whereas in Fig. 4B the IP was preformed from 30 mg. IPs were performed for 2 hours using an APC4 Ab. IPs were then washed 3 times by inversion and the APC/C was eluted form the beads for 2 hours using 1 ml of APC4 peptide dissolved in lysis buffer (1mg/ml).51 After pelleting of the beads, the supernatant was divided into 300 μl aliquots and used to perform IPs with GFP-Trap beads (Chromotek) (Fig. 2C), or panMad2 or BubR1 antibodies (Fig. 4B). IPs were washed in lysis buffer by inversion 3 times.
Isothermal titration calorimetry measurements
All proteins and antibodies were dialyzed extensively in PBS prior to the ITC experiments. Protein concentrations were determined by UV spectroscopy and a molar extinction coefficient at 280 nm of 28545 M−1 cm−1 for both MD2L1-L13A-R133A and MD2L1-V193N-R133A. ITC experiments were performed at 25°C using an ITC200 instrument (Microcal). Three µL volumes of either MD2L1-L13A-R133A or MD2L1-V193N-R133A, at approximately 150 µM, were titrated into the ITC sample cell containing approximately 15 µM of antibody, until saturation was achieved. The heat of the reaction was obtained by integrating each peak after the injection of protein and fit to a model describing a single binding site using software provided by the ITC200 manufacturer.
Silac labeling
For quantitative proteomic experiments, cells were grown in SILAC DMEM (PAA) supplemented with 10% dialyzed FBS (PAA), penicillin-streptomycin and glutamax (Life Technologies) and isotope labeled L-arginine and L-lysine. HeLa cells were grown in light “L” SILAC medium supplemented with normal arginine (R0) and lysine (K0) (Sigma). For the medium “M” condition cells were grown in SILAC DMEM supplemented with U-13C6 arginine (K6) and 4, 4, 5, 5-D4 lysine (K4) and for the heavy “H” labeling cells were grown with U-13C6, U-15N4 arginine (R10) and U-13C6, U-15N2 lysine (K8) (Cambridge Isotope Laboratories). Cells were grown in SILAC medium for at least 5 divisions before harvest.
Mass spectrometry analysis
The eluate from the beads was reduced, alkylated and loaded onto an SDS–PAGE gel. The lane was cut into 7 pieces and subjected to in-gel tryptic digestion and subsequent sample desalting and concentration. The resulting peptide mixture was analyzed by nano-HPLC–MS/MS using an easy-nLC nanoflow system (Thermo Scientific, Odense, Denmark) connected to a Q Exactive mass spectrometer (Thermo Scientific, Bremen, Germany) through a nano-electrospray ion source. The column length was 150 mm with an inner diameter of 75 µm, and packed in-house with 1.9 µm C18 beads (Reprosil-AQ Pur, Dr. Maisch). Each sample was analyzed using a 140 min gradient from 4 to 36% acetonitrile in 0.5% acetic acid. The mass spectrometer was run as described for sensitive acquisition. Data analysis was performed in the MaxQuant environment configured for triple SILAC and the Andromeda search engine. The database searched was the UniProt complete human proteome concatenated with a list of commonly observed background contaminants.
Protein production and purification
Mad2 DNA and mutants thereof were cloned into pET30 and expressed in BL21(DE3) cells at 37 degrees. Cells were harvested and Mad2 affinity purified using Ni-NTA affinity resin (Qiagen) and subsequently purified on a Superdex 75 column.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Nina Sejthen with help in generating the monoclonal antibodies and the protein production facility at NNF CPR for producing recombinant Mad2. We thank Stephen Taylor for the HeLa FRT cell line.
Funding
The Novo Nordisk Foundation Center for Protein Research, University of Copenhagen is supported financially by the Novo Nordisk Foundation (Grant agreement NNF14CC0001). GS was supported by a grant from the Danish Council for Independent research and the work was supported by grants to JN from the Danish Cancer Society and the Lundbeck Foundation.
References
- 1.Nilsson J. Looping in on Ndc80 - how does a protein loop at the kinetochore control chromosome segregation? Bioessays 2012; 34:1070-7; PMID:23154893; http://dx.doi.org/ 10.1002/bies.201200096 [DOI] [PubMed] [Google Scholar]
- 2.Cheeseman IM. The Kinetochore. Cold Spring Harb Perspect Biol 2014; 6(7):a015826; PMID:24984773; http://dx.doi.org/ 10.1101/cshperspect.a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond, B, Biol Sci 2011; 366:3595-604; PMID:22084386; http://dx.doi.org/ 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sacristan C, Kops GJPL. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol 2015; 25:21-8; PMID:25220181; http://dx.doi.org/ 10.1016/j.tcb.2014.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol 2012; 22:R966-80; PMID:23174302; http://dx.doi.org/ 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 6.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 2001; 154:925-36; PMID:11535616; http://dx.doi.org/ 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao WCH, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature 2012; 484:208-13; PMID:22437499; http://dx.doi.org/ 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- 8.Tipton AR, Wang K, Link L, Bellizzi JJ, Huang H, Yen T, Liu S-T. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J Biol Chem 2011; 286:21173-9; PMID:21525009; http://dx.doi.org/ 10.1074/jbc.M111.238543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrow CJ, Tighe A, Johnson VL, Scott MIF, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci 2005; 118:3639-52; PMID:16046481; http://dx.doi.org/ 10.1242/jcs.02487 [DOI] [PubMed] [Google Scholar]
- 10.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol 2008; 10:1411-20; PMID:18997788; http://dx.doi.org/ 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol 2000; 148:871-82; PMID:10704439; http://dx.doi.org/ 10.1083/jcb.148.5.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 2014; 517:631-34; PMID:25383541; http://dx.doi.org/ 10.1038/nature13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonetta M, Manzoni R, Mosca R, Mapelli M, Massimiliano L, Vink M, Novak B, Musacchio A, Ciliberto A. The influence of catalysis on mad2 activation dynamics. PLoS Biol 2009; 7:e10; PMID:19143472; http://dx.doi.org/ 10.1371/journal.pbio.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci 2011; 124:3905-16; PMID:22100920; http://dx.doi.org/ 10.1242/jcs.093286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han JS, Holland AJ, Fachinetti D, Kulukian A, Cetin B, Cleveland DW. Catalytic Assembly of the Mitotic Checkpoint Inhibitor BubR1-Cdc20 by a Mad2-Induced Functional Switch in Cdc20. Mol Cell 2013; 51:92-104; PMID:23791783; http://dx.doi.org/ 10.1016/j.molcel.2013.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell 2009; 16:105-17; PMID:19154722; http://dx.doi.org/ 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teichner A, Eytan E, Sitry-Shevah D, Miniowitz-Shemtov S, Dumin E, Gromis J, Hershko A. p31comet Promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci USA 2011; 108:3187-92; PMID:21300909; http://dx.doi.org/ 10.1073/pnas.1100023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagan RS, Manak MS, Buch HK, Meier MG, Meraldi P, Shah JV, Sorger PK, Doxsey SJ. p31comet acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol Biol Cell 2011; 22:4236-46; PMID:21965286; http://dx.doi.org/ 10.1091/mbc.E11-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J 2004; 23:3133-43; PMID:15257285; http://dx.doi.org/ 10.1038/sj.emboj.7600322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell 2011; 44:710-20; PMID:22152475; http://dx.doi.org/ 10.1016/j.molcel.2011.11.014 [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Sturt-Gillespie B, Hittle JC, Macdonald D, Chan GK, Yen TJ, Liu S-T. Thyroid Hormone Receptor Interacting Protein 13 (TRIP13) AAA-ATPase Is a Novel Mitotic Checkpoint-silencing Protein. Journal of Biological Chemistry 2014; 289:23928-37; PMID:25012665; http://dx.doi.org/ 10.1074/jbc.M114.585315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eytan E, Wang K, Miniowitz-Shemtov S, Sitry-Shevah D, Kaisari S, Yen TJ, Liu S-T, Hershko A. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31comet. Proc Natl Acad Sci USA 2014; 111:12019-24; PMID:25092294; http://dx.doi.org/ 10.1073/pnas.1412901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol 2000; 7:224-9; PMID:10700282; http://dx.doi.org/ 10.1038/73338 [DOI] [PubMed] [Google Scholar]
- 24.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol 2004; 11:338-45; PMID:15024386; http://dx.doi.org/ 10.1038/nsmb748 [DOI] [PubMed] [Google Scholar]
- 25.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell 2002; 9:59-71; PMID:11804586; http://dx.doi.org/ 10.1016/S1097-2765(01)00435-X [DOI] [PubMed] [Google Scholar]
- 26.Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang K-T, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a “safety belt” binding mechanism for the spindle checkpoint. EMBO J 2002; 21:2496-506; PMID:12006501; http://dx.doi.org/ 10.1093/emboj/21.10.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell 2007; 131:730-43; PMID:18022367; http://dx.doi.org/ 10.1016/j.cell.2007.08.049 [DOI] [PubMed] [Google Scholar]
- 28.Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J 2001; 20:6371-82; PMID:11707408; http://dx.doi.org/ 10.1093/emboj/20.22.6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev 2014; 28:140-52; PMID:24402315; http://dx.doi.org/ 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol 2000; 10:675-8; PMID:10837255; http://dx.doi.org/ 10.1016/S0960-9822(00)00515-7 [DOI] [PubMed] [Google Scholar]
- 31.Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol 2014; 204:647-57; PMID:24567362; http://dx.doi.org/ 10.1083/jcb.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 2005; 15:856-61; PMID:15886105; http://dx.doi.org/ 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- 33.Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol 1998; 143:283-95; PMID:9786942; http://dx.doi.org/ 10.1083/jcb.143.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al.. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 2005; 15:214-25; PMID:15694304; http://dx.doi.org/ 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- 35.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 2004; 14:942-52; PMID:15182667; http://dx.doi.org/ 10.1016/j.cub.2004.05.046 [DOI] [PubMed] [Google Scholar]
- 36.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol 2000; 150:1233-50; PMID:10995431; http://dx.doi.org/ 10.1083/jcb.150.6.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung E, Chen R-H. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell 2002; 13:1501-11; PMID:12006648; http://dx.doi.org/ 10.1091/mbc.02-01-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol 2011; 13:475-82; PMID:21394085; http://dx.doi.org/ 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kruse T, Larsen MSY, Sedgwick GG, Sigurdsson JO, Streicher W, Olsen JV, Nilsson J. A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO Rep 2014; 15:282-90; PMID:24477933; http://dx.doi.org/ 10.1002/embr.201338101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J 2011; 30:3322-36; PMID:21772247; http://dx.doi.org/ 10.1038/emboj.2011.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, et al.. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J 2006; 25:1273-84; PMID:16525508; http://dx.doi.org/ 10.1038/sj.emboj.7601033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell 2007; 131:744-55; PMID:18022368; http://dx.doi.org/ 10.1016/j.cell.2007.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ong S-E, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods 2003; 29:124-30; PMID:12606218; http://dx.doi.org/ 10.1016/S1046-2023(02)00303-1 [DOI] [PubMed] [Google Scholar]
- 44.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol 2010; 190:73-87; PMID:20624901; http://dx.doi.org/ 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SH, McCormick F, Saya H. Mad2 inhibits the mitotic kinesin MKlp2. J Cell Biol 2010; 191:1069-77; PMID:21149564; http://dx.doi.org/ 10.1083/jcb.201003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster SA, Morgan DO. The APC/C Subunit Mnd2/Apc15 Promotes Cdc20 Autoubiquitination and Spindle Assembly Checkpoint Inactivation. Mol Cell 2012; 47:921-32; PMID:22940250; http://dx.doi.org/ 10.1016/j.molcel.2012.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol 2012; 199:27-37; PMID:23007648; http://dx.doi.org/ 10.1083/jcb.201205170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lischetti T, Zhang G, Sedgwick GG, Bolanos-Garcia VM, Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat Commun 2014; 5:5563; PMID:25482201; http://dx.doi.org/ 10.1038/ncomms6563 [DOI] [PubMed] [Google Scholar]
- 49.Hutchins JRA, Toyoda Y, Hegemann B, Poser I, Hériché J-K, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al.. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010; 328:593-9; PMID:20360068; http://dx.doi.org/ 10.1126/science.1181348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 2007; 446:921-5; PMID:17443186; http://dx.doi.org/ 10.1038/nature05734 [DOI] [PubMed] [Google Scholar]
- 51.Sedgwick GG, Hayward DG, Di Fiore B, Pardo M, Yu L, Pines J, Nilsson J. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J 2013; 32:303-14; PMID:23288039; http://dx.doi.org/ 10.1038/emboj.2012.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.