ABSTRACT
The β common-signaling cytokines interleukin (IL)-3, granulocyte-macrophage colony stimulating factor (GM-CSF) and IL-5 stimulate pro-inflammatory activities of haematopoietic cells via a receptor complex incorporating cytokine-specific α and shared β common (βc, CD131) receptor. Evidence from animal models and recent clinical trials demonstrate that these cytokines are critical mediators of the pathogenesis of inflammatory airway disease such as asthma. However, no therapeutic agents, other than steroids, that specifically and effectively target inflammation mediated by all 3 of these cytokines exist. We employed phage display technology to identify and optimize a novel, human monoclonal antibody (CSL311) that binds to a unique epitope that is specific to the cytokine-binding site of the human βc receptor. The binding epitope of CSL311 on the βc receptor was defined by X-ray crystallography and site-directed mutagenesis. CSL311 has picomolar binding affinity for the human βc receptor, and at therapeutic concentrations is a highly potent antagonist of the combined activities of IL-3, GM-CSF and IL-5 on primary eosinophil survival in vitro. Importantly, CSL311 inhibited the survival of inflammatory cells present in induced sputum from human allergic asthmatic subjects undergoing allergen bronchoprovocation. Due to its high potency and ability to simultaneously suppress the activity of all 3 β common cytokines, CSL311 may provide a new strategy for the treatment of chronic inflammatory diseases where the human βc receptor is central to pathogenesis. The coordinates for the βc/CSL311 Fab complex structure have been deposited with the RCSB Protein Data Bank (PDB 5DWU).
KEYWORDS: Affinity maturation, asthma, β common receptor, cytokine, eosinophil, GM-CSF, IL-3, IL-5, myeloid, phage display, therapeutic antibody
Abbreviations
- mAb
monoclonal antibody
- IL-3
interleukin-3
- IL-5
interleukin-5
- GM-CSF
granulocyte-macrophage colony stimulating factor
- STAT-5
Signal Transducer and Activator of Transcription-5
- SPR
Surface Plasmon Resonance
- D
domain
- CDR
Complementarity-Determining Region; CFU, Colony Forming Unit
- WT
wild type
- Eo/B
Eosinophil/Basophil
- IC50
half maximal inhibitory concentration
- KD
dissociation constant
- ECmax
maximal effective concentration
- EC80
80% maximal effective concentration
- NAMC
Non-adherent mononuclear cell
Introduction
The pleiotropic cytokines interleukin (IL)-3, IL-5 and granulocyte-macrophage colony stimulating factor (GM-CSF) play critical and overlapping roles in the differentiation and function of certain myeloid cells. While IL-3 and IL-5 production is largely T-cell restricted, GM-CSF is produced by many cells, including T and B cells, endothelial cells, fibroblasts, mast cells, macrophages, and eosinophils.1 They are important mediators of host defense and innate immunity, but can also contribute significantly to the development and progression of inflammatory pathologies, including chronic inflammatory airway diseases such as asthma. Asthma is a common but complex and heterogeneous chronic disorder in humans that is characterized by airway inflammation, reversible airway obstruction, mucus hypersecretion, airway wall remodeling and airway hyperresponsiveness (AHR).2 There is considerable evidence that multiple haematopoietic cell types, including eosinophils, neutrophils and basophils, contribute to the chronic inflammation seen in severe asthma phenotypes. The airway inflammation in different asthma phenotypes can be characterized, based on induced sputum analysis, as being either eosinophilic, i.e., sputum eosinophils greater than 3.0%, neutrophilic i.e., sputum neutrophils greater than 76%, mixed granulocytic (increased sputum eosinophils and neutrophils) or paucigranulocytic (normal levels of eosinophils and neutrophils).3 However, individual patients with severe asthma can have overlapping and heterogeneous inflammatory profiles with diverse symptoms and variable responses to therapy.4 The exact mechanisms of pathogenesis are incompletely understood, and no cure currently exists. Current management relies on the use of steroids and long-acting β adrenergic receptor agonists; however, many patients exhibit steroid-resistant responses, and in others steroid effectiveness needs to be carefully balanced with their significant side-effects. Elevated levels of cytokines, particularly the β common (βc, βcR, CD131, CSF2RB), receptor family of cytokines, potently stimulate haematopoietic cell function, and, together with thymic stromal lymphopoietin (TSLP), interleukins-4, -13, -17 -25 and -33, these molecules co-ordinate an inflammatory infiltration of lymphoid and myeloid cells, extensive mucosal remodeling and mucus hypersecretion in the affected tissue.5
GM-CSF, which is abundantly produced by lung epithelial cells, promotes the differentiation and proliferation of granulocyte/macrophage progenitor haematopoietic cells and regulates the survival and function of neutrophils, eosinophils, macrophages and dendritic cells.5 In asthma and chronic obstructive pulmonary disease (COPD), GM-CSF expression is elevated in sputum and bronchoalveolar lavage fluid (BALF).6 IL-3 acts at the early stages of hematopoiesis and synergizes with other growth factors for haematopoietic development.1 It also modulates the activity of mature cell types such as monocytes, dendritic cells, megakaryocytes, mast cells and can activate eosinophils and prime basophils to release histamine. Increased levels of IL-3 in BALF are typically present after allergen challenge.7 IL-5 is more cell type-specific, regulating the production and release of mature eosinophils from the bone marrow into the circulation. Elevated levels of IL-5 have been found in the serum and airway fluid of patients with asthma.8 In asthmatic subjects, IL-5 inhalation increased AHR as well as the recruitment of activated eosinophils to the airways.9 An early stage in IL-5-driven development of eosinophils is upregulation of the IL-5 receptor α chain (IL-5Rα) expression on the surface of CD34+ progenitor cells.10 An increase in the percentage of bone marrow CD34+/IL-5Rα+ cells 24 hours after inhaled allergen challenge has been noted in dual responder allergic subjects.11
IL-3, IL-5, and GM-CSF bind with low affinity to their specific α chain receptor and the responsiveness of myeloid cells to a particular cytokine depends on which α chain is expressed. Upon cytokine binding to an α chain receptor, a shared common β receptor, which on its own is unable to bind ligand or initiate signaling, is recruited to the low-affinity complex and converts cytokine binding to a high-affinity interaction.12 Studies of mouse models of allergic inflammation, using mice deficient in βc and an IL-3-specific β homodimer (βIL-3) unique to mice (βc−/−and βIL-3−/− designated βc−/−mice), demonstrated that signaling through βc by IL-3, IL-5 and GM-CSF was critical for the development and function of immune cells associated with the inflammatory response and detrimental pathology.13 In βc−/− mice, allergen-induced expansion and accumulation of eosinophils in the peripheral blood, peribronchial tissue and airway lumen is abolished.13 In addition, βc deficiency resulted in a significant reduction in airway hypersensitivity, mucus hypersecretion and production of antigen-specific IgE. In contrast, studies targeting individual ligands such as IL-5 and GM-CSF resulted in only partial ablation of myeloid cell function and diminution of the hallmark features of asthma.14
As a consequence of the evidence supporting a key role for haematopoietic cells, particularly those of the myeloid lineage, and the βc-signaling cytokines in the development and progression of inflammatory airways disease, a number of therapeutic monoclonal antibodies (mAbs) targeting individual βc-signaling cytokines or their receptor α chains are or have been in clinical development for asthma.15-17 While these agents, including those targeting human GM-CSF,18 human IL-5 (mepolizumab, reslizumab)19 and human IL-5Rα (benralizumab),20 are useful in selected subsets of asthma patients, it is likely that their broader application will be limited by the cellular redundancy of the βc cytokines and the variable nature of the inflammatory cell infiltrate that can underpin the asthma phenotype. These major limitations have been recently highlighted by the failure of KB003, an anti-GM-CSF mAb to meet its primary clinical endpoints in a Phase 2 asthma study21 (ClinicalTrials.gov Identifier: NCT01603277). Recent studies have confirmed that treatment with mepolizumab, reduced reliance on glucocorticoids, reduced exacerbations and improved several measures of asthma control in a subpopulation of patients stratified for severe eosinophilic asthma.15,22,23 However, since eosinophils only partially contribute to asthma, modest improvements in AHR have been observed.24 Ideally, a therapeutic agent able to effectively target the common cytokine binding site in βc could inhibit the activity of all 3 βc-signaling cytokines and affect the functional activity of the haematopoietic cells that they regulate. A βc-specific mAb, BION-1, that simultaneously inhibits IL-3, GM-CSF and IL-5 activities has been described previously, but its low potency would limit any therapeutic applications.25
In this study, we used phage display to identify a rare positive mAb, which after multiple rounds of combinatorial affinity maturation, resulted in a βc-specific, fully human mAb designated CSL311. Through directly blocking the ability of each cytokine to interact with βc, CSL311 is a potent inhibitor of IL-3-, GM-CSF- and IL-5-mediated functions at therapeutic doses. We further showed that CSL311 is a potent inhibitor of the combined effects of IL-3, GM-CSF and IL-5 on the survival of eosinophils, key effector cells involved in the pathogenesis of asthma. Notably, we demonstrated, using cells cultured ex vivo from mild atopic asthmatic subjects undergoing inhaled allergen challenge, that CSL311 can block eosinophil, basophil and neutrophil survival and significantly reduce the numbers of eosinophil (Eo)/basophil (B)-colony forming units (CFU) and granulocyte macrophage (GM)-CFU derived from peripheral blood and bone marrow CD34+ progenitors. The unique mode of action of CSL311 and ability to target 3 cytokines simultaneously offers distinct advantages for the treatment of inflammatory disorders that involve the βc-signaling cytokines, including severe asthma, nasal polyposis, and hyper-eosinophilic syndromes.26
Results
Generation of a neutralizing, fully human mAb with specificity for the human βc homodimer
The human βc receptor exists as a preformed intertwined homodimer with a unique arrangement for the cytokine binding site (Site 2) that comprises residues from domain 1 (D1) of one βc monomer and domain 4 (D4) of the second βc monomer.27 Previous studies of BION-1, a hybridoma-derived anti-βc mAb isolated from mice immunized with the extracellular D4 of βc, and thus only part of the Site 2 cytokine binding region, is of relatively high affinity (KD = 5 nM) but low potency.25 Its weak antagonist activity was confirmed in IL-3-, GM-CSF- and IL-5-induced TF-1 erythroleukemic cell28 proliferation assays (Fig. 1A). We therefore screened a semi-synthetic human Fab phage library with a diversity of 1010, using the entire recombinant extracellular region of the βc homodimer, to facilitate the identification of Fabs targeting an epitope spanning regions of both D1 and D4, thus enabling the simultaneous antagonism of IL-3, GM-CSF and IL-5 with high potency. Of the 3548 individual phage clones selected, 209 unique clones were specific to βc. Competitive phage ELISA was used to screen for the highest affinity clones.
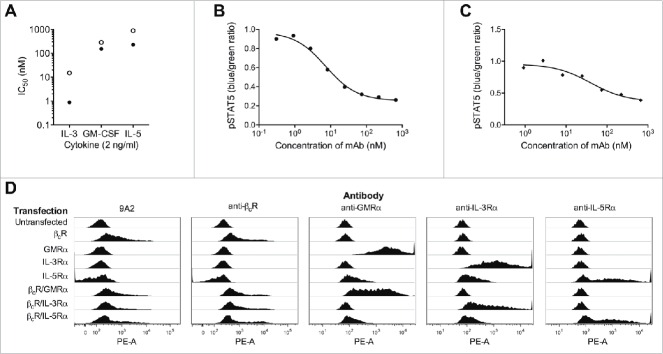
Figure 1.
9A2 inhibits IL-3/GM-CSF/IL-5 induced TF-1 proliferation and STAT5 phosphorylation, and binds specifically to the human βc receptor. (A) TF-1 cells were starved of growth factor for 18 h and then treated with test antibodies BION-1 (○) and 9A2 (•) for 30 min prior to the addition of cytokines (IL-3, GM-CSF, IL-5) for 72 hr. Proliferation was assayed by [H3]-thymidine incorporation and the IC50 values plotted. TF-1-bla cells were starved of growth factor for 18 hours and then treated with 9A2 for 30 minutes prior to the addition of cytokines. For pStat5 assays TF-1-bla cells were incubated with (B) IL-3 (10 ng/ml) or (C) GM-CSF 3 ng/ml) at 37°C for 5 h and FRET B/G substrate was added for 2.5 hours prior to reading the output on an Envision plate reader. Histograms show mean and standard error of technical replicates. Representative experiments are shown. (D) FreeStyle™ 293-F cells were transiently transfected with cDNAs encoding the βcR, the GMRα, the IL-3Rα and the IL5Rα and analyzed by flow cytometry. Control antibodies to the human βcR, GMRα, the IL-3Rα or the IL-5Rα confirmed expression of these proteins in the transfected cells.
From these studies, 29 unique clones were chosen and reformatted as whole human IgG4-kappa antibodies (IgG4pK) containing a hinge mutation to increase stability,29 and screened for their ability to inhibit GM-CSF-induced TF-1 cell proliferation. Remarkably, only a single antibody, 9A2, was identified that could dose-dependently inhibit the downstream proliferative response of TF-1 cells to GM-CSF. 9A2 was also able inhibit TF-1 proliferative responses to IL-3, and IL-5 (Fig. 1A), and IL-3- and GM-CSF-induced STAT-5 phosphorylation (Fig. 1B and Fig. 1C, Table S1). As expected, 9A2 bound specifically to cells expressing βc, but not to cells expressing either human IL-3Rα, human GM-CSFRα (GMRα ) or human IL-5Rα alone (Fig. 1D). TF-1 cells also proliferate in response to IL-6, IL-4, Epo and SCF.30 9A2 did not block TF-1 proliferation in response to any of these factors (data not shown), confirming that 9A2 inhibited the biological activity of only the βc-signaling cytokines.
Although 9A2 completely inhibited IL-3 activity, it was a very weak antagonist of GM-CSF and IL-5 activity (Fig. 1A) with an IC50 of ˜456 nM in the GM-CSF-mediated TF-1 cell proliferation assay (Table S1). Surface plasmon resonance (SPR) analysis of a 9A2 Fab fragment binding to immobilized extracellular domain of the βc homodimer determined an equilibrium dissociation constant (KD) of 49 nM (Table S1). We hypothesized that the low potency of 9A2 was a consequence of its low affinity due to a relatively fast off-rate (Table S1 and Fig. 2A). Affinity maturation of this unique antibody was therefore undertaken in order to identify variants with a higher affinity for βc and the capacity to completely inhibit IL-3-, GM-CSF- and IL-5-mediated receptor activation at clinically relevant concentrations.
Figure 2.
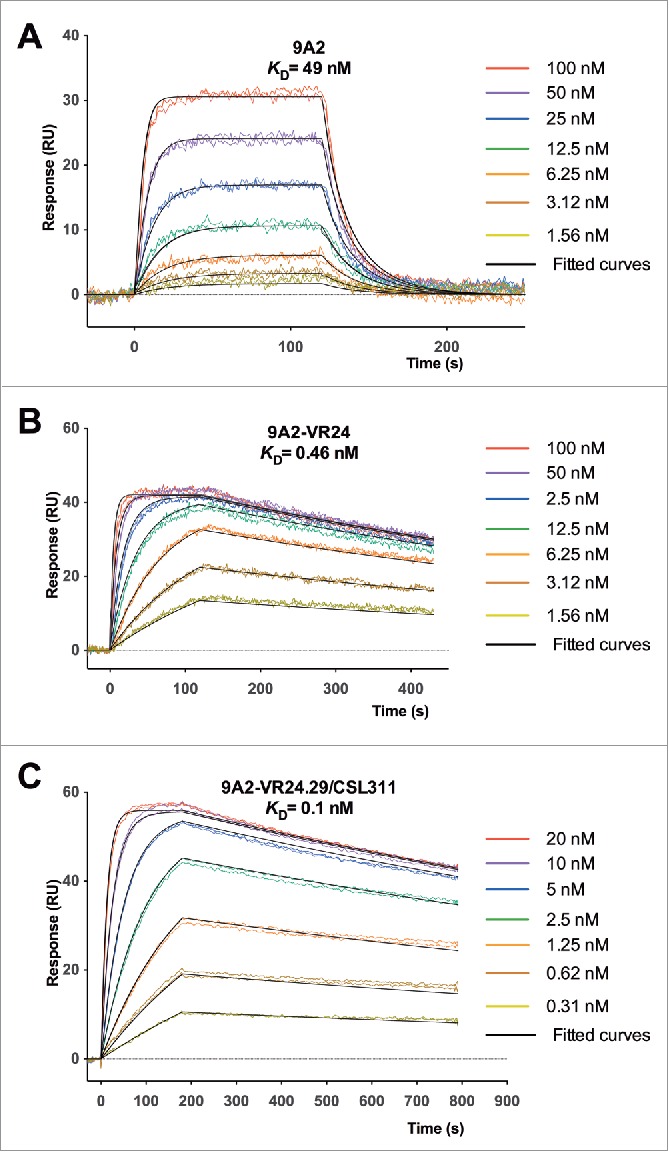
Affinity maturation of 9A2 results in Fabs with improvements in dissociation rate (kd) and affinity (KD). Binding of (A) 9A2 Fab, (B) 9A2-VR24 Fab and (C) 9A2-V24.29/CSL311 Fab to immobilized βc was determined by surface plasmon resonance. Sensorgram (colored) and fitted curves (black) for each concentration are shown. The kinetic constants can be found in Tables S1 and S2.
Seven phage libraries each covering 6 amino acid residues and randomized for all 19 possible amino acids (excluding cysteine) were generated to systematically analyze the contribution of each 9A2 complementarity-determining region (CDR) to the binding of the antibody to βc. CDR2 of the light chain was omitted from the analysis as its interaction with antigens is relatively infrequent.31 Following 5 rounds of selection, 24 randomly selected clones from each library were sequenced. Clones from the L3.2 library failed to form a consensus sequence and were excluded from further study. Unique variants from all other libraries were converted to human IgG4pK molecules, expressed, purified and screened in TF-1 proliferation assays. Variants from all libraries were either similar to or showed an improvement in potency against GM-CSF compared to parental 9A2 (Table S1 and Fig. 3A), with the exception of those targeting CDR3 from the light chain of parental 9A2. Improved dissociation rates were observed for all variants that showed increased affinities (Table S1 and Fig. 2B). Potency improvements against GM-CSF were greatest for clones derived from heavy chain CDR1 (H1.1) and CDR2 (H2.1) libraries (Fig. 3A). Variant 9A2-VR24 originated from the heavy chain CDR1 library (H1.1) and compared with parental 9A2 had affinity (KD = 0.46 nM, Fig. 2B) and GM-CSF potency improvements of 106-fold and 182-fold, respectively (Table S1, and Fig. 3A). The most potent CDR2 (H2.1) variant (9A2-VR39) showed ˜180-fold improvement in binding affinity (KD = 0.27 nM) and 518-fold improvement in GM-CSF potency over parental 9A2 (Table S1, Fig. 3A).
Figure 3.
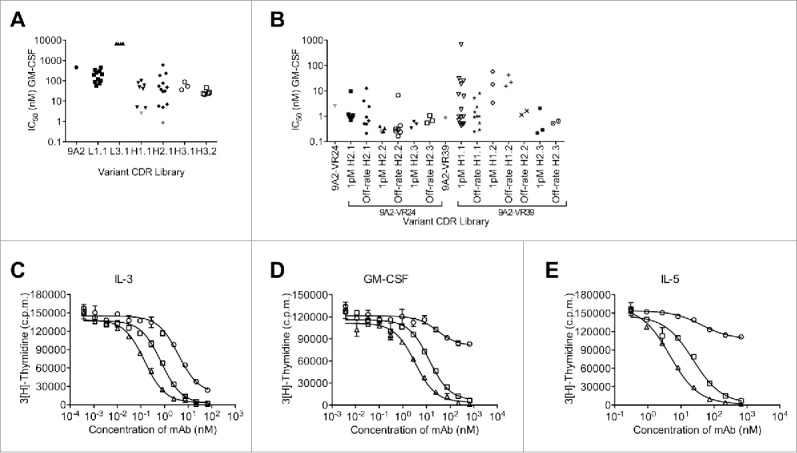
Affinity maturation of 9A2 results in mAbs with improved potency against IL-3, GM-CSF and IL-5. (A) For the first round of affinity maturation 6 different CDR libraries were subjected to 5 rounds of selection in solution with immobilized βc-Fc protein, where the concentration was sequentially reduced with each round. Affinity-matured phage were isolated and unique variants were re-formatted into full length IgG4 antibodies. Variants 9A2-VR24 (▾) and 9A2-VR39 (♦) were selected for the second round of affinity maturation. (B) Affinity-matured phage were isolated and unique variants re-formatted into full length IgG4 antibodies. All affinity-matured variants from each library were tested for potency against GM-CSF in TF-1 proliferation assays and individual IC50 values plotted. 9A2-VR-24.29/CSL311 (▴). TF-1 cells were starved of growth factor for 18 h and then treated with test antibodies 9A2 (○), 9A2-VR24 (□) and 9A2-VR24.29/CSL311 (Δ)) for 30 min prior to the addition of cytokines. (C) IL-3. (D) GM-CSF. (E) IL-5. Cells were incubated at 37°C for 72 h and pulsed with 3[H]-thymidine for the final 6 h then harvested onto glass filters and read on a β-counter. Histograms show mean and standard error of technical replicates. Experiments were repeated at least 3 times. Representative experiments are shown.
With the aim of achieving further potency improvements, variants 9A2-VR24 and 9A2-VR39 (Fig. 3A) were chosen for additional affinity optimization. We undertook a combinatorial approach where the sequence for either 9A2-VR24 or 9A2-VR39 was fixed and libraries were generated targeting all residues for CDR H2 with the 9A2-VR24 amino acid sequence (VR24-H2.1–2.3), and all residues in CDR H1 and residues 57-65 in CDR H2 with the 9A2-VR39 amino acid sequence (VR39-H1.1, H1.2, H2.1, H2.3). An additional “off-rate” selection step was included as all previous potency improvements correlated with improvements in antibody off-rates. Unique variants from all selection strategies were converted to IgG4pK molecules for further analysis in TF-1 proliferation assays. Fig. 3B shows that variants with improved potency against GM-CSF were generated from each of the libraries. Improvements in dissociation rate were observed in those antibodies showing increased affinities (Table S2 and Fig. 2C). Interestingly, an antibody variant comprising the combination of the VR24 CDR1 sequence and the VR39 CDR2 sequence was not identified as a high potency inhibitor from this second round of affinity maturation. We generated an antibody containing these sequences, but there was no potency improvement compared to parental 9A2 (data not shown), supporting the combinatorial approach that was undertaken.
Variant 9A2-VR24.29 (Fig. 3B) was selected for detailed study and is hereafter referred to as CSL311. We compared the ability of CSL311 to inhibit the proliferation of TF-1 cells in response to IL-3, GM-CSF and IL-5 with parental antibodies 9A2 and 9A2-VR24. CSL311 was significantly more potent than both parental antibodies at inhibiting the proliferation of TF-1 cells in response to all 3 βc-signaling cytokines (Fig. 3C-E) with an IC50 of 0.29 nM against GM-CSF (Table S2). This equates to a 8.6-fold improvement compared to 9A2-VR24 (2.5 nM) and a 1572-fold improvement over the parental antibody, 9A2. Improvements in potency of CSL311 compared to parental 9A2 were also observed for IL-3 (41-fold, IC50 = 0.144 nM) and IL-5 (310-fold, IC50 = 4.67 nM) (Table S1, Table S2). CSL311 bound to soluble βc (KD = 100 pM) with a 4.6-fold greater affinity than 9A2-VR24 due to a further 2.6-fold improvement in dissociation rate (kd), (Table S2 and Fig. 2C). Human neutrophils and eosinophils are potently activated by the βc family of cytokines with neutrophils expressing only GM-CSF receptors and eosinophils expressing receptors for GM-CSF, IL-3 and IL-5.32,33 The human cell line TF-1 expresses receptors for GM-CSF, IL-3 and IL-5. In cell surface receptor binding assays we found that 125I-labeled CSL311 bound βc-expressing cells with comparable binding affinity, in the range of KD = 250–730 pM with an average KD= 426 ±79 pM (Table S3). A recombinant Fab fragment of CSL311 bound with similar or slightly higher affinity than the intact CSL311, KD = 380–670 pM, suggesting that the IgG form of CSL311 binds monovalently to cells expressing βc (Table S3).
CSL311 inhibits IL-3, GM-CSF and IL-5 function by binding to residues in site 2 of βc
We assessed the ability of CSL311 to compete for the binding of IL-3, GM-CSF, and IL-5 to cells expressing receptors for these cytokines. Pre-incubation of human eosinophils with CSL311 but not an irrelevant isotype control mAb abolished the binding of 125I-labeled IL-3 and reduced 125I-labeled IL-5 binding by 85% (Fig. 4A and Fig. 4C). Pre-incubation of human neutrophils with CSL311, but not an irrelevant isotype control mAb, completely abolished 125I-labeled GM-CSF binding (Fig. 4B).
Figure 4.
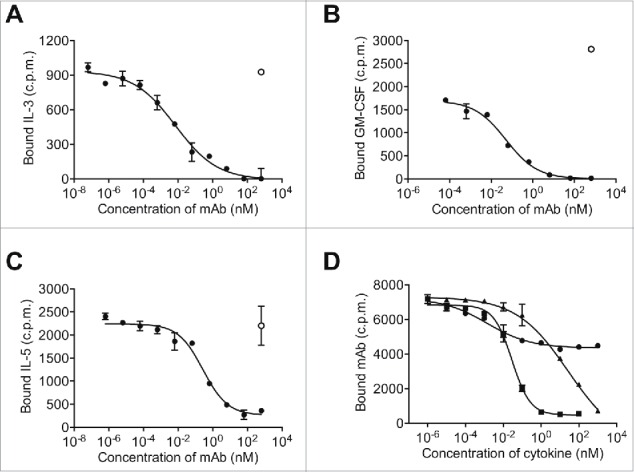
CSL311 competes with IL-3, GM-CSF, and IL-5 for binding to primary human myeloid cells. Purified human eosinophils (A,C) or neutrophils (B) were pre-incubated with CSL311 (•) or a human IgG4 control antibody (○). Cells were then equilibrated with (A) 340 pM radio-iodinated IL-3, (B) 40 pM radio-iodinated GM-CSF (C) 200 pM radio-iodinated IL-5. Each point is the mean of duplicate determinations of cell-bound radioiodinated cytokine and error bars represent the standard deviation. Data from representative experiments is shown, n=2. (D) TF-1 cells were pre-incubated with IL-3 (▴), GM-CSF (▪) or IL-5 (•) then cells were equilibrated with 85 pM radio-iodinated CSL311. Each point is the mean of duplicate determinations of cell-bound radio-iodinated CSL311 and error bars represent the standard deviation. Data from a representative experiment is shown, n = 3.
We next investigated the ability of IL-3, GM-CSF and IL-5 to compete with radio-labeled CSL311 for binding to TF1 cells. We found that pre-incubation of TF1 cells with IL-3 and GM-CSF reduced the binding of 125I-labeled CSL311 by 90% at the highest concentration tested (Fig. 4D). IL-5 pre-incubation also reduced CSL311 binding but only by 40% (Fig. 4D). The reason for this is most likely a consequence of the relatively high affinity of the IL-5/Il-5Rα complex, which allows significant IL-5 binding even when the contribution of βc is excluded.34,35
Structural analysis of the CSL311 binding epitope on the βc receptor
We determined the crystal structure of the Fab fragment of CSL311 bound to the extracellular domain of βc27 to gain further insights into the mechanism of action of CSL311. As expected, CSL311 binds to Site 2 on the βc receptor, in particular to a non-contiguous interface composed of the A-B and E-F loops of D1 from one βc monomer and the B-C and F-G loops of D4 from the other βc monomer.12,27,34,36-39 Superimposition of the GM-CSF receptor ternary structure (PDB 4NKQ)27 on the βc/CSL311 complex reveals that CSL311 exerts its inhibitory function on the βc receptor by direct blockade of the cytokine binding site (Fig. 5A). A surface area of 933 Å2 is buried in the βc/CSL311 complex and the surface complementarity of the βc/CSL311 interface (Sc=0.59)40 is consistent with the tight binding affinity of the complex with a KD of 100 pM as measured by SPR (Table S2). The structure reveals that majority of the CSL311 contacts with βc are mediated through heavy chain CDR loops (CDR H1 to H3) (Fig. 5B), consistent with changes in these CDRs providing the greatest improvements in affinity.
Figure 5.
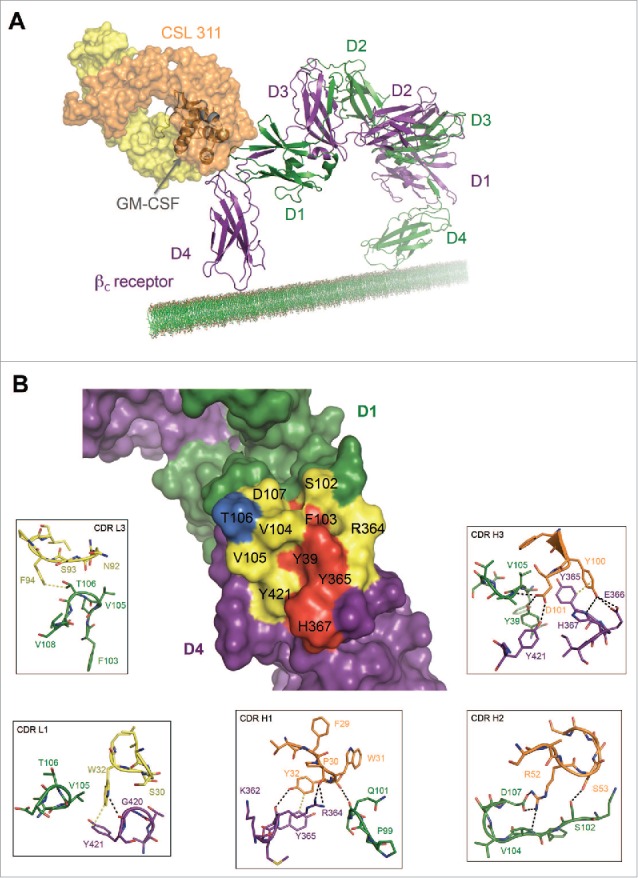
The Site 2 Interaction interface between CSL311 and the βc receptor. (A) Superimposition of the βc/CSL311 Fab complex on the GM-CSF receptor ternary structure, showing the overlap of the interaction interface between CSL311 Fab (heavy chain shown in orange and light chain in yellow) and GM-CSF (gray). The βc dimer from the GM-CSF ternary structure is colored in green and purple. (B) A surface representation of the key residues on the βc dimer that interact with CSL311 is shown. Individual residues are colored to indicate the effect of specific alanine substitution mutations on CSL311 binding affinity: mutations that lead to no binding or negligible binding are colored in red, mutations that reduce binding to the 10−5 to 10−7 M range are shown in yellow and mutations that improve binding are shown in blue. The detailed interactions involving CDRs H1-H3 and CDR L1 and L3 are shown in the adjoining zoom-in panels. Polar interactions are shown as black broken lines and key van der Waals interactions are shown in yellow. All figures were made using PyMOL.
Both hydrophobic and polar contacts contribute to the binding of CSL311 to βc. CDR H1 main-chain carbonyls of P30 and W31 make polar contacts with the guanidine side chain of R364 and main-chain carbonyl of Q101 on the βc receptor respectively. The side-chain of Y32 from CDR H1 can hydrogen bond with the main-chain carbonyl of R364 and can potentially form π-π interactions with Y365 from βc. S53 from CDR H2 makes polar contacts with S102 from D1 of the βc receptor. The side-chain of R52 forms a salt bridge with D107 and it is also within hydrogen bond distance from the main-chain amine of V104 on the βc receptor. The side-chain of Y100 from the CDR H3 of CSL311 hydrogen bonds with βc residues E366 and H367 and forms π-π interactions with Y365 on the βc receptor. The side-chain of D101 from the CDR H3 makes polar contacts with the side-chains of Y39, Y421 and the main-chain amine of V105 on the βc receptor. By comparison the CSL311 light chain appears to play a more limited role in the interaction with βc. The side-chain of W32 on CDR L1 makes a polar contact with the main-chain carbonyl of G420 and van der Waals contact with the side-chain of Y421. No residues from CDR L2 make contact with βc. F94 on CDR L3 is within van der Waals contact of the side-chain T106 from D1 of the βc receptor. In addition to these contacts, the aromatic side-chains of Y32 from CDR H1 and Y100 from CDR H3, are buried in a cleft in the Site 2 interface of βc and form π-π interactions with the side-chains of Y365 and H367 from the βc receptor, further stabilizing the βc/CSL311 complex (Fig. S1).
Site-directed mutagenesis studies were conducted to determine the importance of the various interactions between CSL311 and βc observed in the crystal structure. A total of 32 alanine point mutations of residues in and proximal to the cytokine-binding site on βc were generated. Using SPR, we measured the affinities of a purified Fab fragment of 9A2 to these mutants relative to βc (Table S4). The binding of a purified recombinant Fab fragment of CSL311 to key mutants was evaluated and shown to be similar (Table S4). The most prominent effects were noted for F103A, Q339A and I424A mutants, which resulted in negligible binding and the I338A mutation, which resulted in no binding of 9A2/CSL311. Structurally, F103 and I338 are involved in stabilizing the side chain of Y39, which makes an important hydrogen bond with the side-chain of D101 on CDR H3 (Fig. S2). Therefore mutating these residues may have an effect on the orientation of Y39, which in turn affects binding of CSL311. Q339 and I424 do not make any interactions with CSL311, in contrast they are buried deep in the D1-D4 interface of the βc dimer, therefore the loss of binding of CSL311 observed for these mutants may be due to a structural effect on the βc dimer.
The Y39A mutation from D1 of βc resulted in weak binding of 9A2/CSL311 further emphasizing the importance of Y39 for high affinity binding to 9A2/CSL311. Mutation of residues Y365, H367 and I368 from D4 of βc also resulted in weak binding of 9A2/CSL311. Of these, Y365 and H367 are involved in making important hydrogen bonds with D101 and Y100 on CDR H3 respectively. These residues also form a part of the π-π interaction network comprising Y32 and Y100 from CDRs H1 and H3, which further emphasizes their role in CSL311 engagement (Fig. S1). Although I368 does not interact with CSL311 directly, it is part of the hydrophobic groove at the Site 2 interface and may be structurally important for maintaining proper orientation of the surrounding aromatic side chains of Y39, Y421, Y365 and H367.
CSL311 inhibits survival of primary human eosinophils stimulated with IL-3, GM-CSF and IL-5
Eosinophils are the predominant immune effector cells in atopic asthma, and, through their activation and degranulation, they are proposed to contribute substantially to airway inflammation. The importance of eosinophils in the pathogenesis of asthma has been confirmed with the reduction of symptoms seen in patients treated with antibodies to IL-5 or the IL-5 receptor.15,16,41,42 Human eosinophils also respond to GM-CSF and IL-3 in addition to IL-5.1 In particular, administration of recombinant IL-3 or GM-CSF in humans causes a rise in levels of circulating eosinophils.43 Therefore, in diseases where eosinophilia is a pathogenic feature, all 3 cytokines in combination may contribute to enhanced eosinophil numbers by mediating their recruitment from the bone marrow and their increased survival at the site of pathology. The response of eosinophils to stimulation with IL-3, GM-CSF and IL-5 is shown in Fig. S3 A-C. We assessed the ability of CSL311 to block eosinophil survival in response to a cocktail of all 3 cytokines (at EC80 for each cytokine). We also assessed individual antagonists directed against the IL-3Rα,44 GMRα,45 or IL-5Rα,46 alone or in combination in the same assay. The potency of each individual receptor antagonist in response to stimulation with their corresponding cytokine was determined and complete inhibition was observed at ˜10 nM for each antagonist (Fig. S3 D-E). However, in response to a cocktail of all 3 cytokines (at EC80 for each cytokine) inhibition by individual α chain-specific antagonists at concentrations above the ICmax in the same assay was partial. Although the GMRα antagonist was the most potent inhibitor in this assay the combination of all 3 anti-α chain mAbs provided even greater inhibition of the pro-survival activity of the cytokine cocktail. In contrast, treatment of eosinophils with CSL311 at concentrations as low as 200 nM completely blocked their survival in the presence of the cocktail of IL-5, GM-CSF and IL-3 (Fig. 6).
Figure 6.
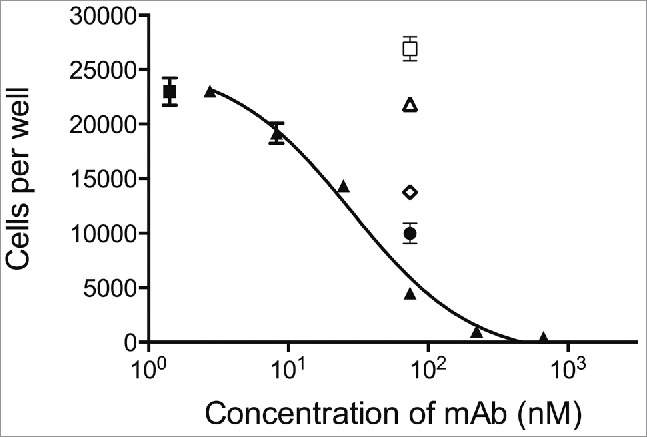
CSL311 inhibits the survival of primary human eosinophils cells following stimulation with βc-signaling cytokines. Eosinophils were pre-treated with CSL311 (▴), anti-IL-5Rα (□, 66.7 nM), anti-IL-3Rα (▵, 66.7 nM), anti-GMRα (◊, 66.7 nM), or anti-IL-5Rα/anti-IL-3Rα/anti-GMRα, (•, 66.7 nM each mAb) before treatment with IL-3/GM-CSF/IL-5, for 72h and cell survival determined. No antibody added (▪). Data from representative experiments are shown, n = 5. All values expressed as mean +SEM. Experiments were repeated at least 4 times with representatives shown.
CSL311 reduces survival of primary cells isolated and cultured ex vivo from human airway disease tissue
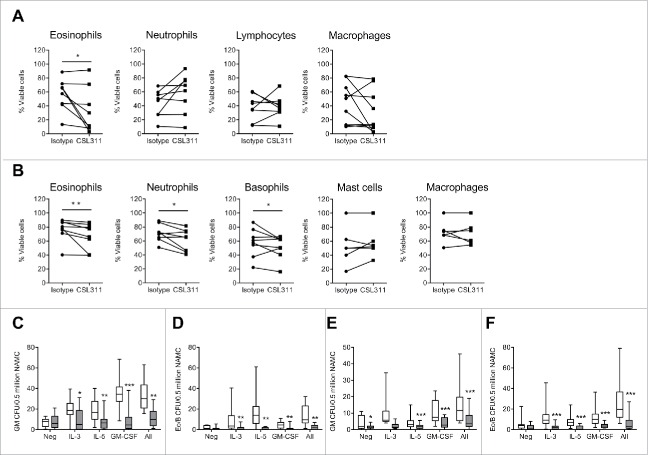
We next sought to determine whether CSL311 had the same efficacy on immune cells from asthmatics as it did on cells derived from healthy donors. We first employed the inhaled allergen bronchoprovocation/challenge model47 to study the effect of CSL311 on human allergic asthma sputum cells cultured ex vivo. The subject characteristics on screening are shown in Table S5. In agreement with previous studies, differential cell counting of the mixed cell population indicated that the predominant cell types were neutrophils and macrophages, with a smaller proportion of eosinophils, lymphocytes, and bronchial epithelial cells (data not shown) and an increase in sputum eosinophil count is observed 24 h post-allergen challenge.47 The sputum cells (combined from baseline and 24 h post-allergen challenge) were cultured ex vivo without exogenous IL-3, IL-5 and GM-CSF, in the presence of CSL311 or isotype control mAb, and the effect on their survival investigated by flow cytometry. CSL311 caused a significant decrease in survival of sputum eosinophils in 5 of 8 subjects (P value = 0.0391) as detected by annexin V staining (Fig. 7A). By contrast, survival of neutrophil, lymphocyte or macrophage populations was unaffected by CSL311. Interestingly, when isolated sputum cells from the same donors were also incubated with recombinant human IL-3, IL-5 and GM-CSF (1 ng/ml of each), CSL311 significantly decreased the survival of sputum neutrophils (6 out of 8 subjects, P value = 0.0391) and basophils (5 out of 8 subjects, P value = 0.3828) as well as eosinophils (6 of 8 subjects, P value = 0.0078) (Fig. 7B).
Figure 7.
CSL311 inhibits survival of cells isolated from human inflammatory airway disease tissue. Sputum cells from samples induced at baseline and 24 h after inhaled allergen challenge from allergic asthmatic subjects were incubated with (A) CSL311 (100 µg/ml) or an isotype control mAb (100 µg/ml) at 37°C for 24 h or (B) incubated with CSL311 (100 µg/ml) or an isotype control mAb (100 µg/ml) at 37°C for 24 h in the presence of 1 ng/ml each IL-3, GM-CSF and IL-5. Cells were analyzed for viability by flow cytometry. Data are expressed as percent cell viability and compared to the isotype control for each donor. Samples where the pre-incubation viability was<10% were excluded from the analysis. Non-adherent mononuclear cells (NAMC) from (C-D) bone marrow and (E-F) blood samples collected at baseline and 24 h after inhaled allergen challenge from allergic asthmatic subjects were incubated with CSL311 (100 µg/ml, filled bars) or an isotype control mAb (100 µg/ml, open bars) at 37°C for 24 h in the presence of diluent (neg) or 1 ng/ml each IL-3, GM-CSF and IL-5, or cytokines combined (All) and (C,E) GM CFU and (D,F) Eo/Baso CFU were enumerated. Data expressed as median ± range and 95% confidence intervals. A Wilcoxon matched pairs signed rank 2 tailed t-test was used to determine statistical significance: ns, not significant p > 0.05; * p < 0.05; ** p < 0.01; *** p < 0.005.
Eosinophil and basophil progenitors are found in increased numbers in peripheral blood and bone marrow in response to allergen challenge in atopic subjects, a process that can be regulated by IL-3, GM-CSF and IL-5.48 We examined if CSL311 affected the number of CFUs arising from CD34+ progenitors for these populations in bone marrow and blood samples obtained from asymptomatic allergic asthmatics pre- and 24 h post- allergen challenge. In the presence of either IL-3, GM-CSF, IL-5 individually or a combination of all 3 cytokines, reduced numbers of GM-CFU and Eo/B CFU were counted when cultured in the presence of CSL311 compared to isotype control mAb (Fig. 7C-F).
Discussion
IL-3, GM-CSF and IL-5 are a group of pro-inflammatory cytokines that signal via a receptor complex composed of a cytokine-specific α chain and the βc homodimer. They are implicated in the pathogenesis of chronic inflammatory diseases of the airways such as asthma through their ability to recruit and activate myeloid cells. Here, we describe the generation and characterization of an antibody that is capable of binding the βc homodimer and potently inhibiting the functions of IL-3, GM-CSF and IL-5. Through screening a human Fab phage library against the entire recombinant extracellular domain of the βc homodimer combined with a strategy of 2 rounds of affinity maturation targeting all CDRs, apart from the light chain CDR2, we identified a number of high affinity, biologically potent mAbs. CSL311 was selected as a lead candidate and demonstrated to effectively neutralize the biological activities of IL-3, GM-CSF and IL-5 on primary human eosinophils at clinically appropriate concentrations.
Evidence from the crystal structure of the βc/CSL311 complex, competitive binding experiments and mutagenesis studies of key ligand binding residues in the βc receptor confirmed that CSL311 interfered with the assembly of an active signaling complex by directly competing with cytokine for binding to βc. The binding affinity of each cytokine to its cognate receptor α chain is different for each cytokine, with IL-3 binding with the lowest affinity (KD = 20-100 nM), GM-CSF with intermediate affinity (KD = 2–12 nM) and IL-5 with the highest affinity (KD = 0.8-1 nM).39 Co-expression of βc increases the binding affinity of each cytokine/α chain complex to different extents. Thus, IL-5 binding affinity is increased 2-5 fold, GM-CSF is increased 20–100 fold and IL-3 is increased 500–1000 fold,39,49-53 resulting in a high affinity complexes of KD = 110 pM, 130 pM and 290 pM, respectively.39 These differences are reflected in the ability of CSL311 to compete with cytokine, where increasing concentrations of CSL311 were required to compete IL-5 compared to IL-3 and GM-CSF. The differences in the ability of CSL311 to compete with each of the cytokines translated to biological assays where, although CSL311 demonstrated complete inhibition of all 3 cytokines, it did so with different potencies.
The lack of any appreciable neutralizing activity of CSL311 against the βc receptor from other species (unpublished observations) led us to undertake a novel translational medicine approach utilizing relevant human inflammatory airway disease tissue to assess the potential of this antibody to be effective in disease situations such as allergic asthma. The widely used inhaled allergen bronchoprovocation/challenge exacerbation model of allergic asthma induces many clinical and pathophysiological features of asthma observed in allergic subjects, including bronchoconstriction, increased numbers of eosinophils in the sputum and peripheral blood, and increased numbers of CD34+ progenitors in bone marrow and peripheral blood. Although relatively few studies have employed sputum cell culture, it is likely that cells from the airways will provide highly relevant data due to their proximity to the site of asthmatic inflammation. We used the techniques of allergen challenge and induced sputum to isolate and culture ex vivo induced sputum cells from asymptomatic mild allergic asthmatic subjects before and after allergen challenge. CSL311 was able to significantly decrease the survival of unstimulated eosinophils cultured ex vivo from mild atopic asthmatic sputum before and after inhaled allergen challenge, suggesting that the cells were producing factors in vitro that enhanced their survival that could be blocked by CSL311. This is consistent with previous observations of delayed eosinophil apoptosis in asthma attributed to the presence of GM-CSF.54 Although eosinophils are key contributors to the pathogenesis of asthma, other effector cells, including those from the myeloid lineage contribute to the inflammation and the dysregulated environmental milieu associated with asthma and similar chronic inflammatory conditions. Furthermore, considerable heterogeneity in inflammatory cell measures has been reported within severe asthma phenotype clusters.55 Interestingly, when a combination of IL-3, GM-CSF and IL-5 was added to the culture medium together with CSL311, decreased neutrophil and basophil survival was also observed. This observation shows that one or more of these cytokines are able to enhance neutrophil and basophil survival above that provided by endogenous factors present in the sputum. These data also point to the specificity of action of CSL311 as other survival factors in the unstimulated sputum cell culture, most likely Granulocyte-Colony Stimulating Factor (G-CSF) in the case of neutrophil survival, are not affected by CSL311.
In addition to mature eosinophils, circulating common progenitors of eosinophils and basophils are detected in greater numbers in the peripheral blood of asymptomatic atopic patients in response to allergen challenge. This involves production of Eo/B progenitors in the bone marrow, the release of both progenitors and progeny from the bone marrow compartment and then recruitment to the respiratory mucosa and other tissues after which locally produced haematopoietic growth factors provide further hematopoiesis.56 We examined the ex vivo effect of CSL311 on early GM and Eo/B progenitors by measuring the number of peripheral blood or bone-marrow-derived GM- or Eo/B CFUs grown in methylcellulose culture. CSL311 reduced the numbers of GM- and Eo/B-CFU, as defined by the number of CFU arising from CD34+ progenitors from blood and bone marrow pre- and post- allergen challenge. By comparison, clinical studies indicated that while mepolizumab caused partial maturational arrest of the eosinophil lineage it did not fully deplete bone marrow or bronchial eosinophils. There was no change in the total number of blood or bone marrow CD34+ cells after mepolizumab therapy nor were there effects on CD34+/IL-5Rα+ cells.57 A plausible explanation for persistence of progenitors and their progeny is the actions of IL-3 and GM-CSF, which are unaffected by mepolizumab. Some reports have suggested that sputum eosinophils may differ from their circulating counterparts due to a decrease in IL-5Rα expression accompanied by a lack of responsiveness to IL-5 following allergen challenge and a concomitant increase in GMRα expression with retained responsiveness to GM-CSF.58 Other studies have also suggested that airway eosinophils are unresponsive to IL-5 due to down-regulation of IL-5Rα by all 3 βc cytokines whereas they retain their responsiveness to IL-3.59 In human clinical studies mepolizumab reduced blood and BALF eosinophils by 100% and 80% respectively but there was only a 55% reduction in airway mucosal eosinophils.60 In our in vitro studies, CSL311 blocked eosinophil survival stimulated with a cocktail of IL-3, GM-CSF and IL-5 to a greater extent than any of the individual α-chain antagonists alone, and to a similar extent as the combination of neutralizing mAbs to the individual α-chains of the IL-3, GM-CSF and IL-5 receptors. Although CSL311 has a lower IC50 against IL-5 in the TF-1 assay (4.67 nM) compared to the reported IC50 of mepolizumab (IC50 = 75 pM),61 it is still a complete antagonist of IL-5. CSL311 therefore may provide a more complete reduction of sputum eosinophilia through its broader spectrum of activity compared to anti-IL-5 antagonists through its ability to also potently inhibit IL-3 and GM-CSF.
The success of corticosteroids as anti-inflammatory agents for a number of diseases including asthma is due to their significant pleiotropic effects on a range of key inflammatory pathways and cell types. One known mechanism of action of corticosteroids is the inhibition of pro-survival signals from cytokines such as IL-5, GM-CSF and IL-3 in eosinophils.62 Although single agents targeting each of these cytokines or their receptor α chains are currently in clinical development44,45,63,64 and could conceivably be combined to treat patients with severe asthma, such an approach would be expected to encounter substantial practical, regulatory and economic impediments. Our data provides exciting evidence that a single antibody targeting the βc homodimer may prove to be a potent and more directed anti-inflammatory therapy in diseases such as asthma when compared to current treatments including corticosteroids. In support of this view, recent clinical studies have shown that preventing the expression of human βc using antisense oligonucleotides (TPI ASM8) inhibits the function of eosinophils as well as basophils and mast cells and is of benefit in attenuating allergen-induced responses in subjects with mild allergic asthma.65 An antibody-based approach directed at βc would be expected to offer a number of potential advantages over an oligonucleotide-based therapy, including route of delivery, improved pharmacokinetics and the ability to target βc cytokine responsive cells within a broader range of tissue types.
Mice lacking the GM-CSF gene or βc/βIL-3 homodimers develop a pulmonary syndrome of abnormal surfactant accumulation resembling human Pulmonary Alveolar Proteinosis (PAP).66 However, the absence of significant adverse events in trials using TPI ASM865 and also with antagonists targeting human GM-CSF (KB003, ClinicalTrials.gov Identifier: NCT01603277),18 human GMRα (mavrilimumab),18 human IL-5Rα (benralizumab) and human IL-5 (mepolizumab, reslizumab)19 is encouraging and suggests that targeting βc is unlikely to have significant side-effects.
Our study provides the first demonstration that targeting βc with a potent therapeutic mAb (CSL311) can directly inhibit the survival of multiple primary human cells, including eosinophils, neutrophils and basophils in the presence of more than one cytokine as occurs in an in vivo inflammatory setting such as asthma. Its range of action may also provide advantages compared with other specific inhibitors of the IL-3/GM-CSF/IL-5 pathway. We anticipate that CSL311 will have broad therapeutic utility in the treatment of heterogeneous and relatively poorly understood diseases including, but not limited to, severe asthma, severe nasal polyposis, COPD, eosinophilic disorders such as Churg-Strauss syndrome, Hyper-eosinophilic syndrome and Eosinophilic Esophagitis and other inflammatory disorders where the βc-signaling cytokines are implicated in their pathogenesis.
Methods
Cell culture
FreeStyle™ 293-F (FS293F) cells and the mammalian expression vector pcDNA3.1 were obtained from Invitrogen™, Thermo Fisher Scientific (R790-07, V790-20). Cells were cultured in GIBCO®FreeStyle™ 293 Expression Medium (Invitrogen™, Thermo Fisher Scientific.12338-018). All tissue culture media were supplemented with Antibiotic-Antimycotic (GIBCO®, Thermo Fisher Scientific 15240-096) and cells were maintained at 37°C in incubators with an atmosphere of 8% CO2.
Cytokines and antibodies
Recombinant human GM-CSF encoding the P6YY substitution to facilitate radio-iodination was prepared from E.coli as previously described.67 Recombinant human IL-3 comprising residues 13–121 and the W13Y substitution to facilitate radio-iodination was also expressed and purified from E.coli.68 Carrier-free recombinant human IL-5 was purchased from R&D Systems (205-IL-005/CF). Cytokines (IL-3, GM-CSF) for cellular assays were purchased from R&D Systems (203-IL-010, 215-GM-010). Anti-IL3Rα antibody44 and anti-GMRα antibody69 were prepared as previously described.
Generation of hybridomas
Fifty µg of Hexa-His tagged βc extracellular domain protein was injected intraperitoneally into BALB/c mice deficient in both βc and βIL-3.13 The mice received a further 2 × 4 weekly injections of the same dose at the same site. Spleen cells were fused with Sp2/0 myeloma cells and hybridomas generated according to standard methods. Individual hybridomas (3H3), which was directed against a C-terminal Hexa-His tag, and anti-human βc mAbs 7H12 and 3F1) were re-cloned by limiting-dilution. Hybridomas were cultured in Hybridoma SFM (GIBCO®, Thermo Fisher Scientific.12045–076) supplemented with 0.5–1.0 % low IgG FBS (GIBCO®, Thermo Fisher Scientific16250–078) in roller bottles at 37°C.
Generation of cDNA expression plasmids
Human βc cDNA (GenBank Accession no. P32927) and amino acid mutants of the βc receptor were codon-optimized for human expression and synthesized by Geneart® (Invitrogen™, Thermo Fisher Scientific) each with a Kozak consensus sequence70 (GCCACC) immediately upstream of the initiating methionine (+1). Full-length transmembrane βc mutants and soluble βc variants (truncated after Ser438 with C-terminal 6x Histidine-tags fused in-frame) were generated using standard PCR-based mutagenesis techniques. Once each cDNA was completed, it was digested with NheI and XhoI and ligated into pcDNA3.1 (Invitrogen™, Thermo Fisher Scientific R790-07, V790-20).
Human IL-3R cDNA (GenBank Accession no. NP_002174), Human GMRα cDNA (GenBank Accession no. NP_006131) and Human IL-5Rα isoform 1 cDNA (GenBank Accession no NP_000555) were obtained either from Dr H. Ramshaw (Center for Cancer Biology, Adelaide, Australia) or Geneart® (Invitrogen™, Thermo Fisher Scientific) and cloned as described above. Anti-IL-5Rα chain antibody cDNA46,71 was synthesized by (Geneart®, Invitrogen™, Thermo Fisher Scientific).
Recombinant Fab fragments of 9A2 and affinity matured variants were generated by cloning the entire light chain and a truncated heavy chain, where a stop codon was introduced after amino acid 241, separately into pcDNA3.1 as described above.
Large-scale preparations of plasmid DNA were carried out using QIAGEN Plasmid Giga Kits (12191) according to the manufacturer's instructions. The nt sequences of all plasmid constructs were verified by sequencing both strands using BigDye™ Terminator Version 3.1 Ready Reaction Cycle Sequencing (Invitrogen™, Thermo Fisher Scientific.4337455) and an Applied Biosystems 3130xl Genetic Analyzer.
Transient transfections for generation of recombinant proteins
Transient transfections of expression plasmids using FS293F cells were performed using 293fectin™ transfection reagent (Invitrogen™, Thermo Fisher Scientific. 12347–019) as previously described.72
Antibody purification
All mAbs and recombinant Fab fragments were affinity-purified using HiTrap® MabSelect™ SuRe™ or KappaSelect™ (1 ml, GE Healthcare Life Sciences, 11-0034-93, 17-5458-03) chromatography resins, respectively, and then desalted with a HiPrep™ 26/10 Desalting column (GE Healthcare Life Sciences 17–5087–01) on an ÄKTAxpress high throughput chromatography system (GE Healthcare Life Sciences). Fab fragments were generated by digestion of the purified antibodies using immobilized Papain Agarose (Sigma P4406) and purified using Protein A and size exclusion chromatography. The filtered cell culture media (500 ml) was applied to the column that had been equilibrated 1 X MTPBS buffer, at a rate of 1 ml/min and washed sequentially with 1 X MTPBS pH 7.3 (10 ml) and 10 mM Tris, 150 mM NaCl pH 7.2 (80 ml) in the presence of 0.5 M arginine to facilitate endotoxin removal.73 The bound antibody was then eluted with 8 ml 0.1 M sodium acetate pH 3.0 (or 0.1 M glycine pH 2.5) and immediately applied to the desalting column. Protein fractions were pooled and concentrated using an Amicon® UltraCel 50K centrifugal device (Merck-Millipore) prior to sterile filtration using 0.22 µm filters. Antibody purity was assessed by SDS-PAGE and protein visualized using PlusOne™ Coomassie™ Blue PhastGel™ R-350 Stain (GE Healthcare Life Sciences 17-0518-01), as per the manufacturer's instructions, and antibody concentration was determined chromatographically by comparison to control antibody standards.
His-tagged protein purification
Soluble βc and βc mutants were purified by tandem Nickel and size exclusion chromatography on an AKTA™ express (GE Healthcare Life Sciences) purification system. Column chromatography was generated as per manufacturer's instructions. Post-elution samples were applied directly to a Superdex 200 pg 26/60 column (GE Healthcare Life Sciences 28-9893-36) at 4 ml/min in PBS and fractions collected. Peak fractions containing βc fractions were pooled after additional size exclusion analysis and sterile-filtered for subsequent testing.
Antibody generation
A human semi-synthetic Fab phage display library was screened for phagemids that bound the recombinant extracellular domain of hβc fused to the Fc region of human IgG1 (hβc-Fc, Human CRC-β-Fc Chimera, Apollo Cytokine Research 1103H) immobilized on Dynabeads® M-280 Streptavidin (Invitrogen™, Thermo Fisher Scientific 11205D) by biotin-anti-human Fc antibody capture (Jackson ImmunoResearch Laboratories 109–065–098). The selection was done following methods described previously.74 Three rounds of selection were performed by incubating the streptavidin bead-depleted phage input with decreasing concentrations of immobilized hβc-Fc (15 µg, 10 µg and 5 µg) in 2% milk/PBST (MTPBS, 0.1% Tween-20) for 20 min at room temperature and then washed 12 times. Selected phage clones were amplified in log phase E. coli TG1 cells and the Fab-phagemid rescued by super-infection with M13K07 helper phage prior to purification using standard protocols.75 Individual clones were picked after the second and third round of selection and the Fab cassettes and light chains were PCR-amplified and sequenced essentially as described.74 Competitive phage ELISA was used to screen for high affinity clones. Twenty nine unique antibody clones were reformatted to express full-length IgG4 antibodies with the serine 241 to proline hinge region mutation29 and a kappa light chain by cloning the entire light chain (variable and constant domains) and the variable domain of the heavy chain from the selected phage-derived Fab constructs into the pRhG4 vector.76
Affinity maturation of 9A2
Clone 9A2 was affinity-matured by randomization of CDRs (excluding CDR2 of the light chain) with primers that included a 19 amino acid combination (without cysteine). Seven different libraries were constructed using methods previously described using “stop template” versions of pTac-geneIII-9A2 Fab where for each phagemid, a germline stop template was created by replacing 18 codons (6 amino acid residues) in all CDRs with TAA stop codons.77 Each stop template was used as a template for the Kunkel mutagenesis method.78 The mutagenesis reactions were electroporated into E. coli SS320 then phage production initiated with addition of M13-KO7 helper phage prior to incubation at 30°C for 18 h. Phage were purified using standard protocols. Mutagenesis efficiencies ranged from 27% to 100% as assessed by sequencing of 12 clones picked randomly from each library. Each library contained 4 × 109 – 1.05 × 1010 individual clones.
Libraries were subjected to 5 rounds of selection in solution with decreasing concentration of immobilized biotinylated hβc-Fc. The target concentration was reduced 10-fold with each round, from 100 nM in Round 1 to 10 pM in Round 5 for all libraries, except H3.1. In the case of library H3.1, the target concentration was reduced 10-fold from 100 nM in Round 1 to 10 nM in Round 2, and kept constant at 10 nM in all subsequent panning rounds due to a dramatic reduction in phage output titres at lower target concentrations. Affinity matured phage were isolated from the libraries essentially as described above for antibody generation. Unique phage clones were identified by sequencing 20 randomly selected clones from each library. Unique variants were reformatted into full-length human IgG4/Kappa antibodies as described above.
Variants 9A2-VR24 and 9A2-VR39 were selected for further affinity maturation and libraries constructed using the methods described above with mutagenic oligonucleotides. The first set of 3 libraries for the second round of affinity maturation of 9A2 was based on clone VR24 sequence, by generating stop templates targeting 6 residues at a time in CDR H2. Similarly, the second set of 4 libraries was based in the VR39 clone sequence, with 2 libraries in CDR H1 and further 2 in CDR H2. The mutagenesis efficiencies ranged from 50% to 90% as assessed by sequencing 12 clones selected randomly from each library. Each library contained 0.25 × 109 − 2.5 × 109 individual clones. Libraries were subjected to 4 rounds of selection in solution with decreasing concentration of immobilized hβc-Fc, using methods essentially as described above. The target concentration was reduced 10-fold with each round, from 100 pM down to 1 pM in Round 4.
Following Round 4 of selection, beads with 1 pM output phage and the corresponding blank sample were washed as described above and resuspended in PBS. Half was subsequently eluted with 50 mM dithiothreitol (DTT) and the eluted and neutralized phage were designated as 1 pM output. The second half was used to select for variants with improved off-rates, designated Off-rate output, by incubation at room temperature for 1 h in the presence of excess immobilized hβc-Fc (1 nM) followed by washing, elution and neutralization as described above. Unique variants from each library were identified by sequencing and reformatted into fully human IgG4 /kappa antibodies as described above.
ELISA
Phagemid-Fab clones were tested for target binding by ELISA. Purified hβc-Fc protein or irrelevant human IgG antibody were coated at 2 µg/ml in MTPBS; pH 7.3 onto 96-well MaxiSorp™ ELISA (Sigma M9140) plates overnight at 4°C. Plates were blocked for 2 h at 37°C with 200 µl/well of 5% skim milk/PBST, washed twice with PBST before incubation with phage supernatant (100 µl/well) for 90 min at room temperature. Plates were washed x 5 with PBST prior to incubation with anti-M13-HRP antibody (GE Healthcare Life Sciences 27942101) diluted 1:10,000 in PBST. Plates were washed x6 with PBST and signal developed with 100 µl TMB/E substrate (Merck Millipore ES001-500ML). The reaction was stopped with 2 M phosphoric acid (50 µl/well) and measured at 450 nm. To determine approximate binding affinities of phagemid clones to hβc-Fc protein, competition ELISAs were performed. The phage supernatant was diluted with 2% skim milk/PBST to give an absorbance 450 nm value of 1.5 from extrapolating phage titration ELISA results. Prior to addition of 50 µl/well of appropriately diluted phage supernatant, an equal volume of competitor βc-Fc protein was added per well at a starting concentration of 2.5 µM and subsequently diluted 4-fold in 2% skim milk/PBST.
Binding affinity determination
Binding kinetics were measured using SPR with a BIAcore® A-100 instrument (GE Healthcare Life Sciences). An anti-C-terminal Hexa-His antibody (manufactured in-house) was immobilized on spots 1, 2, 4 and 5 of each flow cell of a CM-5 sensor chip (GE Healthcare Life Sciences) using amine-coupling chemistry. The typical immobilization level was between 13000 and 15000 response units (RU).
As βc is a dimer, avidity effects were avoided by performing kinetics assays with βc extracellular domain captured on the sensor surface, and purified Fab injected as the analyte. βc was captured on spots 1 and 5 of each flow cell for 2 min at a concentration of 0.4 µg/ml. For 9A2 analysis, purified Fab was injected over each flow cell for 2 min and dissociation was monitored for a further 5 min. For CSL311 analysis, purified Fab was injected over each flow cell for 3 min and dissociation was monitored for a further 10 min. Regeneration of the surface was performed after each cycle with a 40 second injection of 25 mM glycine, pH 2.0. The analysis was performed with Fabs at several concentrations between 100 and 0.31 nM, with each concentration analyzed twice and in random order. The analysis was performed at a flow rate of 30 µl/min in HBS-EP buffer (10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20, pH 7.4) at 37°C. Responses from spots 2 and 4 of each flow cell, (in which βc was not captured, but otherwise treated identically), were subtracted from those of spots 1 and 5, respectively, to produce reference-subtracted data. Reference subtracted responses from a blank injection comprising buffer alone were subtracted from the resultant sensorgrams to produce double referenced data suitable for kinetic analysis. Double-referenced sensorgrams were fitted using non-linear regression to a model describing 1:1 kinetics, including a term for mass transport limitation. The Rmax value was fitted locally to account for slight deviations in the level of βc captured, with association rate (ka), dissociation rate (kd) and equilibrium dissociation constant (KD) fitted globally.
Epitope mapping
Mapping of the CSL311 epitope on βc was performed by measurement of the affinity of 9A2 and CSL311 Fabs for various alanine point mutants of βc essentially as described above, with the following exceptions. Each βc mutant was captured for 120 seconds at concentrations between 1 and 5 µg/ml. For kinetic analysis, purified recombinant Fabs of 9A2/ CSL311 were injected for 2 min, and dissociation was monitored for a further 2 min. For steady-state affinity analysis, Fab was injected for 2 or 3 min and dissociation monitored for 60 seconds. Fab concentrations ranged from 16 µM to 3.9 nM in two-fold dilutions. The assays were conducted at 25°C. All sensorgrams were reference subtracted as described above. For determination of steady-state affinity (KD only), the response at the end of the binding phase was used to fit the data to a single-site binding model. Kinetic parameters were determined as described above. The kinetics of 2 control anti-human βc Fabs (7H12 and 3F1) with epitopes distinct from 9A2 were also determined for each point mutant to establish whether the mutation had caused structural perturbations to βc. 7H12 and 3F1 bind epitopes within domain 4 and 3 of βc, respectively (data not shown).
Crystallization of the βc/CSL311 Fab complex
The βc/CSL311 Fab complex was purified from a mixture of monomeric components27 by size exclusion chromatography. All crystallization trials of the complex were carried out at 18°C using commercially available protein crystallization screens with the protein at a concentration of 6 mg/ml. Initial crystallization trials were set up using a Art Robbins liquid handler (Gryphon) in 96-well sitting drop format. Rectangular plate shaped crystals appeared between 2-3 d in 40% PEG 200 and 100 mM Tris pH 8.5 from the PEGs Suite (Qiagen 130904). Crystals were optimized by the hanging-drop vapor-diffusion method. Although several crystal forms of the βc/CSL311 Fab complex were obtained, these crystals typically diffracted to 8Å resolution. Several pre-crystallization and post crystallization methods were used to optimize crystal diffraction. Finally treatment of crystals with 0.3% to 1 % of gluteraldehyde (Sigma-Aldrich 111-30-8) for 15 minutes to 1 hr drastically improved diffraction from 8 Å to 4 Å. After screening several crystals, 3 partial datasets were combined to obtain complete data set at 3.9 Å resolution (Table S6).
Structure determination of βc/CSL311 Fab complex
Data collection was carried out with a 20% attenuated beam at the MX2 beam line at the Australian Synchrotron.79 The crystals belonged to the C2 space group with unit cell dimensions of a = 99.95, b = 71.28, c = 221.23. Data were scaled and processed using XDS80 and Aimless81 and molecular replacement was carried out using Phaser (CCP4 suite).82 The βc ectodomain structure (PDB 2GYS)83 was modified to a partial βc dimer consisting of domains D4 and D1 only and was used as a search model to locate the βc molecule. The CSL311 Fab molecule was located using PDB 3HI5 as the search model (sequence similarity of ˜84% for heavy chain of CSL311)84 after deleting the CDR loops. A single solution comprising a partial βc dimer (comprising domains D1 and D2 from monomer A and domains D3 and D4 from monomer B) and one molecule of the CSL311 Fab comprising heavy (H) and light (L) chains was obtained with an Rwork/Rfree of 0.38/0.45 after first round of rigid body refinement. Initial electron density was clearest for the Fab molecule and domains D4 and D1 of βc, while density for βc domains D2 and D3 was the weakest. The initial βc/CSL311 model was further improved by iterative cycles of model building in Coot and model refinement using Phenix.85-87
Although the electron density maps for domains D4 and D1 of βc and the variable domains of Fab H and L chains was continuous, no electron density was observed for parts of D2 and D3 of βc and parts of the constant domains of the Fab H and L chains even at the end of refinement, suggesting that they might be either highly flexible or adopt multiple conformations. The structure was refined to a final Rwork/Rfree of 0.30/0.34. The structure was validated using Molprobity.88 The coordinates for the structure have been deposited with the RCSB Protein Data Bank (PDB 5DWU).
Cell surface receptor binding assays
Recombinant cytokines and antibodies were radio-iodinated using Pierce Pre-Coated Iodination tubes (Thermo Scientific™, Thermo Fisher Scientific 28601) according to the manufacturer's instructions. MAb binding to cells expressing βc was determined by incubating 1-2 ×106 cells with radio-iodinated mAb at a range of concentrations at 23°C for 1-2 h with gentle mixing. Cell suspensions were then centrifuged through fetal calf serum (FCS) and radioactivity associated with the cell pellets was assessed by counting in a Wizard2 2470 Automatic Gamma Counter. Non-specific binding was assessed for each radio-iodinated mAb in the presence of at least a 500-fold excess of unlabeled mAb. Dissociation constants and receptor numbers were calculated using the EBDA and LIGAND programs89 (KELL Radlig, Biosoft). Competition binding assays were performed essentially as previously described.25 Briefly 1-2 ×106 cells were incubated with mAb at a range of concentrations at 4°C for 45 min with gentle mixing. Radio-iodinated cytokine was then added and the mixture incubated at 23°C for a further 1-2 h with gentle mixing. Cell suspensions were then centrifuged through FCS and radioactivity associated with the cells pellets assessed by counting.
TF-1 proliferation assays
TF-1 cells were maintained in RPMI media with 10% FCS, 1 x glutamine, 1 x Antibiotic Antimycotic and 2 ng/ml hGM-CSF at 37°C and 5% CO2. Cells were starved of growth factor for 18 h, plated in 96 well flat bottom plates at 1 × 104 cells/well then treated with test antibodies for 30 min prior to the addition of IL-3, IL-5 or GM-CSF. Cells were incubated at 37°C and 5% CO2 for 72 h and pulsed with 3[H]-thymidine for the final 6 h before harvesting to glass filters. 3[H]-thymidine incorporation was determined by liquid-scintillation counting with a Beckman β-counter.
Cell signaling assays
GeneBLAzer® TF-1 bla pStat5 assay
TF-1 bla cells (Invitrogen™, Thermo Fisher Scientific K1637) were cultured in RPMI media 1640 (Gibco® Thermo Fisher Scientific 72400–047) with 10% FCS, 0.1 mM Non-Essential Amino Acids (NEAA, Gibco® Thermo Fisher Scientific 11140–050) 1 mM sodium pyruvate, Blasticidin (Invitrogen™, Thermo Fisher Scientific.A11139–02) (5 µg/ml) and hGM-CSF (2 ng/ml). Prior to assay, cells were washed 3 x with PBS with 0.1% FCS to remove growth factor then resuspended in assay media (Opti-MEM®, Gibco® Thermo Fisher Scientific 11058–021) with 0.5% FCS, 0.1 mM NEAA, 1 mM sodium pyruvate, Antimycotic-Antibiotic, and incubated at 37°C and 5% CO2 for 18 h. Cells were plated in assay media at 1.2 × 105 cells per well in 96-well flat, clear bottom, black-walled plates then treated with test antibodies for 30 min prior to the addition of 1 ng/ml IL-3 or GM-CSF Cells were incubated at 37°C and 5% CO2 for 5 h then FRET B/G substrate (Invitrogen™, Thermo Fisher Scientific, K1095, K1096, K1030) added for 2.5 h before reading (EnVision, Perkin Elmer).
Isolation of neutrophils and eosinophils
Eosinophils were isolated from the buffy coats from healthy donors (Australian Red Cross Blood Service (ARCBS), Melbourne, Victoria and Adelaide, South Australia). Peripheral blood mononuclear cells were separated from granulocytes and red blood cells by centrifugation over Ficoll-Paque™ PLUS (GE Healthcare Life Sciences 71-7167-00-AG) density gradients. Neutrophils were separated from the red blood cell pellet by dextran sedimentation. Neutrophils were washed with cold PBS and red-blood cells were lysed by hypotonic shock. The red blood cell pellet containing granulocytes was lysed with Ammonium Chloride Solution (Stem Cell Technologies 07850) and eosinophils were isolated using a MACS® Eosinophil Isolation Kit (Miltenyi Biotec 130–092–010).
Eosinophil survival assays
Purified eosinophils were plated at 1 × 104/well in flat bottom 96-well plates then test antibodies were added for 30 minutes prior to the addition of IL-5, IL-3 and GM-CSF. Cells were incubated at 37°C and 5% CO2 for 5 d and cell number determined with the ViaLight®Plus Cell Proliferation and Cytotoxicity BioAssay Kit (Lonza LT17–221).
Inhaled allergen challenge assays
Subjects with stable, mild atopic asthma who were also non-smokers, free of other lung diseases and not pregnant with baseline FEV1 ≥70 % of predicted were chosen for allergen challenge. The allergen challenge study was approved by the McMaster Faculty of Health Sciences/Hamilton Health Sciences Research Ethics Board (project 12-583) and signed informed consent was obtained from subjects. Allergen challenge was performed as previously described.90 Sputum samples were mixed in PBS (without DTT) to disperse the cells, and the mixture filtered to remove mucus before centrifugation. The cell pellet was resuspended at a concentration of 1 × 106 cells/ml in RPMI with100 U/ml pen/strep (Gibco, 15140-122) and 10% FCS (Sigma, C8056). A cytospin was made for differential cell counting, including percentage eosinophils, neutrophils, macrophages, lymphocytes, and bronchial epithelial cells. The mixed cell population was cultured either with no growth factors or in the presence of 1 ng/ml each recombinant human IL-3 (R&D Systems, 203-IL-010), GM-CSF (R&D Systems, 215-GM-010) and IL-5 (R&D Systems, 205-IL-005) and incubated at 37°C in a humidified incubator with 5% CO2 with CSL311 at a final concentration of 100 µg/ml. Outcomes were compared to incubation with an irrelevant isotype control mAb at a final concentration of 100 µg/ml. After 24 h, the cells were removed from the wells, washed, and re-suspended in Binding Buffer (BD PharMingen, 556454). The cells were co-stained with specific cell lineage markers using the following antibodies and isotype controls: anti-CD16 FITC (BD PharMingen, 555406), anti-IgG1 FITC (BD PharMingen, 555786), anti-CD3 PeCy7 (BD PharMingen, 563423), anti-IgG1 PeCy7 (BD PharMingen, 561298), anti-CD68 APC (Biolegend, 333809), anti-IgG2b APC (Biolegend, 401209), Siglec 8-PE (Biolegend, 347103) and anti-IgG1 PE (Biolegend, 406607), and the viability of specific populations evaluated at baseline and 24 h post allergen-challenge. Cultures where the baseline viability was less than 10% were excluded from the analysis.
Preparation of bone marrow and peripheral blood cells for quantification of Eo/Baso-CFU and GM-CFU from CD34+ cells populations
Blood samples (80 ml) and bone marrow aspirates (5 ml) were collected as previously described from mild atopic asthmatic subjects pre- and 24 h post-allergen challenge.91 Low-density mononuclear cells (MNCs) were isolated by sedimentation on Accuprep™ density gradients (Cedarlane, AN551).92 Non-adherent mononuclear cells (NAMNCs) were resuspended in Iscove's 2+ (Iscove's modified Dulbecco's medium with 1% pen/strep (Gibco, 15140-122) and 1% 2-mercaptoethanol (Sigma, M3148) and placed in Methocult® cultures (StemCell Technologies, Inc., 04236) in the presence of 16% fetal bovine serum (Sigma, 13G210) and IL-5 (10 ng/ml) (R&D Systems, 205-IL-005), IL-3 (25 ng/mL) (R&D Systems, 203-IL-010), GM-CSF (10 ng/ml) (R&D Systems, 215-GM-010), or a combination of all 3 growth factors. The NAMNC cells were cultured at a concentration of 0.5 × 106 cells/ml for 2 weeks at 5% CO2, with high humidity at 37°C. The number of Eo/B CFU was quantified in duplicate plates using an inverted light microscope at 40x magnification and the average number of colony-forming units per plate was calculated. A colony was defined as a cluster of eosinophils/basophils with a minimum density of 40 cells.
Statistics
For the comparison of matched non-parametric samples we used the Wilcoxon rank-sum t test. P values of less than .05 were considered statistically significant. Unless otherwise specified all data are presented as means ±SEM, and data are from at least 3 independent experiments (all measurements for each in vitro experiment performed in at least duplicate). Statistical analyses were calculated using GraphPad Prism version 6.0.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank CSL Limited Research staff (Recombinant Proteins Group, Antibody Technologies Group, Protein Technologies Group, Analytical Biochemistry Group), Karen Howie (McMaster University) and Kay Wycherley (WEHI Antibody Facility). This research was partly undertaken on the MX2 beamline at the Australian Synchrotron, Victoria, Australia and we thank the beamline staff for their assistance. This crystallography work was supported by a grant from the National Health and Medical Research Council of Australia (NHMRC) to M.W.P and A.F.L. and from the Australian Cancer Research Foundation to M.W.P. Funding from the Victorian Government Operational Infrastructure Support Scheme to St Vincent's Institute is acknowledged. U.D. and M.W.P. are NHMRC Postdoctoral and Research Fellows, respectively.
References
- 1.Martinez-Moczygemba M, Huston DP. Biology of common β receptor-signaling cytokines: IL-3, IL-5, and GM-CSF. J Allergy Clin Immunol 2003; 112:653-65; PMID:14564341; http://dx.doi.org/ 10.1016/j.jaci.2003.08.015 [DOI] [PubMed] [Google Scholar]
- 2.Hamelmann E, Gelfand EW. IL-5-induced airway eosinophilia–the key to asthma? Immunol Rev 2001; 179:182-91; PMID:11292022; http://dx.doi.org/ 10.1034/j.1600-065X.2001.790118.x [DOI] [PubMed] [Google Scholar]
- 3.Dente FL, Carnevali S, Bartoli ML, Cianchetti S, Bacci E, Di Franco A, Vagaggini B, Paggiaro P. Profiles of proinflammatory cytokines in sputum from different groups of severe asthmatic patients. Ann Allergy Asthma Immunol 2006; 97:312-20; PMID:17042136; http://dx.doi.org/ 10.1016/S1081-1206(10)60795-8 [DOI] [PubMed] [Google Scholar]
- 4.Jarjour NN, Erzurum SC, Bleecker ER, Calhoun WJ, Castro M, Comhair SA, Chung KF, Curran-Everett D, Dweik RA, Fain SB, et al.. Severe asthma: lessons learned from the National Heart, Lung, and Blood Institute Severe Asthma Research Program. Am J Respir Crit Care Med 2012; 185:356-62; PMID:22095547; http://dx.doi.org/ 10.1164/rccm.201107-1317PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuijs MJ, Willart MA, Hammad H, Lambrecht BN. Cytokine targets in airway inflammation. Curr Opin Pharmacol 2013; 13:351-61; PMID:23643194; http://dx.doi.org/ 10.1016/j.coph.2013.03.013 [DOI] [PubMed] [Google Scholar]
- 6.Saha S, Doe C, Mistry V, Siddiqui S, Parker D, Sleeman M, Cohen ES, Brightling CE. Granulocyte-macrophage colony-stimulating factor expression in induced sputum and bronchial mucosa in asthma and COPD. Thorax 2009; 64:671-6; PMID:19213775; http://dx.doi.org/ 10.1136/thx.2008.108290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoskins A, Roberts JL 2nd, Milne G, Choi L, Dworski R. Natural-source d-α-tocopheryl acetate inhibits oxidant stress and modulates atopic asthma in humans in vivo. Allergy 2012; 67:676-82; PMID:22435735; http://dx.doi.org/ 10.1111/j.1398-9995.2012.02810.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker C, Bauer W, Braun RK, Menz G, Braun P, Schwarz F, Hansel TT, Villiger B. Activated T cells and cytokines in bronchoalveolar lavages from patients with various lung diseases associated with eosinophilia. Am J Respir Crit Care Med 1994; 150:1038-48; PMID:7921434; http://dx.doi.org/ 10.1164/ajrccm.150.4.7921434 [DOI] [PubMed] [Google Scholar]
- 9.Shi HZ, Xiao CQ, Zhong D, Qin SM, Liu Y, Liang GR, Xu H, Chen YQ, Long XM, Xie ZF. Effect of inhaled interleukin-5 on airway hyperreactivity and eosinophilia in asthmatics. Am J Respir Crit Care Med 1998; 157:204-9; PMID:9445301; http://dx.doi.org/ 10.1164/ajrccm.157.1.9703027 [DOI] [PubMed] [Google Scholar]
- 10.Tavernier J, Van der Heyden J, Verhee A, Brusselle G, Van Ostade X, Vandekerckhove J, North J, Rankin SM, Kay AB, Robinson DS. Interleukin 5 regulates the isoform expression of its own receptor α-subunit. Blood 2000; 95:1600-7; PMID:10688814 [PubMed] [Google Scholar]
- 11.Sehmi R, Wood LJ, Watson R, Foley R, Hamid Q, O'Byrne PM, Denburg JA. Allergen-induced increases in IL-5 receptor α-subunit expression on bone marrow-derived CD34+ cells from asthmatic subjects. A novel marker of progenitor cell commitment towards eosinophilic differentiation. J Clin Invest 1997; 100:2466-75; PMID:9366561; http://dx.doi.org/ 10.1172/JCI119789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hercus TR, Broughton SE, Ekert PG, Ramshaw HS, Perugini M, Grimbaldeston M, Woodcock JM, Thomas D, Pitson S, Hughes T, et al.. The GM-CSF receptor family: Mechanism of activation and implications for disease. Growth Factors 2012; 30:63-75; PMID:22257375; http://dx.doi.org/ 10.3109/08977194.2011.649919 [DOI] [PubMed] [Google Scholar]
- 13.Asquith KL, Ramshaw HS, Hansbro PM, Beagley KW, Lopez AF, Foster PS. The IL-3/IL-5/GM-CSF common receptor plays a pivotal role in the regulation of Th2 immunity and allergic airway inflammation. J Immunol 2008; 180:1199-206; PMID:18178860; http://dx.doi.org/ 10.4049/jimmunol.180.2.1199 [DOI] [PubMed] [Google Scholar]
- 14.Trifilieff A, Fujitani Y, Coyle AJ, Kopf M, Bertrand C. IL-5 deficiency abolishes aspects of airway remodelling in a murine model of lung inflammation. Clin Exp Allergy 2001; 31:934-42; PMID:11422160; http://dx.doi.org/ 10.1046/j.1365-2222.2001.01084.x [DOI] [PubMed] [Google Scholar]
- 15.Pavord ID, Korn S, Howarth P, Bleecker ER, Buhl R, Keene ON, Ortega H, Chanez P. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet 2012; 380:651-9; PMID:22901886; http://dx.doi.org/ 10.1016/S0140-6736(12)60988-X [DOI] [PubMed] [Google Scholar]
- 16.Ghazi A, Trikha A, Calhoun WJ. Benralizumab-a humanized mAb to IL-5Ralpha with enhanced antibody-dependent cell-mediated cytotoxicity-a novel approach for the treatment of asthma. Expert Opin Biol Ther 2012; 12:113-8; PMID:22136436; http://dx.doi.org/ 10.1517/14712598.2012.642359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh GM. Profile of reslizumab in eosinophilic disease and its potential in the treatment of poorly controlled eosinophilic asthma. Biologics 2013; 7:7-11; PMID:23326187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton JA. GM-CSF as a target in inflammatory/autoimmune disease: current evidence and future therapeutic potential. Expert Rev Clin Immunol 2015; 11:457-65; PMID:25748625; http://dx.doi.org/ 10.1586/1744666X.2015.1024110 [DOI] [PubMed] [Google Scholar]
- 19.Hilvering B, Xue L, Pavord ID. Evidence for the efficacy and safety of anti-interleukin-5 treatment in the management of refractory eosinophilic asthma. Ther Adv Respir Dis 2015; 9:135-45; PMID:25900924; http://dx.doi.org/ 10.1177/1753465815581279 [DOI] [PubMed] [Google Scholar]
- 20.Molfino NA. Targeting of eosinophils in asthma. Expert Opin Biol Ther 2012; 12:807-9; PMID:22500988; http://dx.doi.org/ 10.1517/14712598.2012.674938 [DOI] [PubMed] [Google Scholar]
- 21.Kalobios Pharmaceuticals Inc. KaloBios Reports Top Line Data for Phase 2 Study of KB003 in Severe Asthma. Study Does Not Meet Primary Endpoint. Media release, January 29, 2014. 2014:http://www.kalobios.com/. [Google Scholar]
- 22.Ortega HG, Liu MC, Pavord ID, Brusselle GG, FitzGerald JM, Chetta A, Humbert M, Katz LE, Keene ON, Yancey SW, et al.. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med 2014; 371:1198-207; PMID:25199059; http://dx.doi.org/ 10.1056/NEJMoa1403290 [DOI] [PubMed] [Google Scholar]
- 23.Bel EH, Wenzel SE, Thompson PJ, Prazma CM, Keene ON, Yancey SW, Ortega HG, Pavord ID; SIRIUS Investigators . Oral glucocorticoid-sparing effect of mepolizumab in eosinophilic asthma. N Engl J Med 2014; 371:1189-97; PMID:25199060; http://dx.doi.org/ 10.1056/NEJMoa1403291 [DOI] [PubMed] [Google Scholar]
- 24.Nair P. Anti-interleukin-5 monoclonal antibody to treat severe eosinophilic asthma. N Engl J Med 2014; 371:1249-51; PMID:25197762; http://dx.doi.org/ 10.1056/NEJMe1408614 [DOI] [PubMed] [Google Scholar]
- 25.Sun Q, Jones K, McClure B, Cambareri B, Zacharakis B, Iversen PO, Stomski F, Woodcock JM, Bagley CJ, D'Andrea R, et al.. Simultaneous antagonism of interleukin-5, granulocyte-macrophage colony-stimulating factor, and interleukin-3 stimulation of human eosinophils by targetting the common cytokine binding site of their receptors. Blood 1999; 94:1943-51; PMID:10477723 [PubMed] [Google Scholar]
- 26.Wechsler ME, Fulkerson PC, Bochner BS, Gauvreau GM, Gleich GJ, Henkel T, Kolbeck R, Mathur SK, Ortega H, Patel J, et al.. Novel targeted therapies for eosinophilic disorders. J Allergy Clin Immunol 2012; 130:563-71; PMID:22935585; http://dx.doi.org/ 10.1016/j.jaci.2012.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, et al.. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell 2008; 134:496-507; PMID:18692472; http://dx.doi.org/ 10.1016/j.cell.2008.05.053 [DOI] [PubMed] [Google Scholar]
- 28.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao YF, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol 1989; 140:323-34; PMID:2663885; http://dx.doi.org/ 10.1002/jcp.1041400219 [DOI] [PubMed] [Google Scholar]
- 29.Angal S, King DJ, Bodmer MW, Turner A, Lawson AD, Roberts G, Pedley B, Adair JR. A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol Immunol 1993; 30:105-8; PMID:8417368; http://dx.doi.org/ 10.1016/0161-5890(93)90432-B [DOI] [PubMed] [Google Scholar]
- 30.Sautina L, Sautin Y, Beem E, Zhou Z, Schuler A, Brennan J, Zharikov SI, Diao Y, Bungert J, Segal MS. Induction of nitric oxide by erythropoietin is mediated by the β common receptor and requires interaction with VEGF receptor 2. Blood 2010; 115:896-905; PMID:19965681; http://dx.doi.org/ 10.1182/blood-2009-04-216432 [DOI] [PubMed] [Google Scholar]
- 31.Wilson IA, Stanfield RL. Antibody-antigen interactions. Curr Op Struct Biol 1993; 3:113-8; http://dx.doi.org/ 10.1016/0959-440X(93)90210-C [DOI] [PubMed] [Google Scholar]
- 32.Lopez AF, Vadas MA, Woodcock JM, Milton SE, Lewis A, Elliott MJ, Gillis D, Ireland R, Olwell E, Park LS. Interleukin-5, interleukin-3, and granulocyte-macrophage colony-stimulating factor cross-compete for binding to cell surface receptors on human eosinophils. J Biol Chem 1991; 266:24741-7; PMID:1761568 [PubMed] [Google Scholar]
- 33.Hercus TR, Thomas D, Guthridge MA, Ekert PG, King-Scott J, Parker MW, Lopez AF. The granulocyte-macrophage colony-stimulating factor receptor: linking its structure to cell signaling and its role in disease. Blood 2009; 114:1289-98; PMID:19436055; http://dx.doi.org/ 10.1182/blood-2008-12-164004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy JM, Ford SC, Wiedemann UM, Carr PD, Ollis DL, Young IG. A novel functional epitope formed by domains 1 and 4 of the human common β-subunit is involved in receptor activation by granulocyte macrophage colony-stimulating factor and interleukin 5. J Biol Chem 2003; 278:10572-7; PMID:12525483; http://dx.doi.org/ 10.1074/jbc.M211664200 [DOI] [PubMed] [Google Scholar]
- 35.Scibek JJ, Evergren E, Zahn S, Canziani GA, Van Ryk D, Chaiken IM. Biosensor analysis of dynamics of interleukin 5 receptor subunit β(c) interaction with IL5:IL5R(α) complexes. Anal Biochem 2002; 307:258-65; PMID:12202242; http://dx.doi.org/ 10.1016/S0003-2697(02)00043-X [DOI] [PubMed] [Google Scholar]
- 36.Murphy JM, Ford SC, Olsen JE, Gustin SE, Jeffrey PD, Ollis DL, Young IG. Interleukin-3 binding to the murine betaIL-3 and human betac receptors involves functional epitopes formed by domains 1 and 4 of different protein chains. J Biol Chem 2004; 279:26500-8; PMID:15060062; http://dx.doi.org/ 10.1074/jbc.M402705200 [DOI] [PubMed] [Google Scholar]
- 37.Woodcock JM, Bagley CJ, Zacharakis B, Lopez AF. A single tyrosine residue in the membrane-proximal domain of the granulocyte-macrophage colony-stimulating factor, interleukin (IL)-3, and IL-5 receptor common β-chain is necessary and sufficient for high affinity binding and signaling by all three ligands. J Biol Chem 1996; 271:25999-6006; PMID:8824238; http://dx.doi.org/ 10.1074/jbc.271.42.25999 [DOI] [PubMed] [Google Scholar]
- 38.Lock P, Metcalf D, Nicola NA. Histidine-367 of the human common β chain of the receptor is critical for high-affinity binding of human granulocyte-macrophage colony-stimulating factor. Proc Natl Acad Sci USA 1994; 91:252-6; PMID:8278375; http://dx.doi.org/ 10.1073/pnas.91.1.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodcock JM, Zacharakis B, Plaetinck G, Bagley CJ, Qiyu S, Hercus TR, Tavernier J, Lopez AF. Three residues in the common β chain of the human GM-CSF, IL-3 and IL-5 receptors are essential for GM-CSF and IL-5 but not IL-3 high affinity binding and interact with Glu21 of GM-CSF. EMBOJ 1994; 13:5176-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol 1993; 234:946-50; PMID:8263940; http://dx.doi.org/ 10.1006/jmbi.1993.1648 [DOI] [PubMed] [Google Scholar]
- 41.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, et al.. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med 2009; 360:973-84; PMID:19264686; http://dx.doi.org/ 10.1056/NEJMoa0808991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O'Byrne PM. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med 2009; 360:985-93; PMID:19264687; http://dx.doi.org/ 10.1056/NEJMoa0805435 [DOI] [PubMed] [Google Scholar]
- 43.Lindemann A, Ganser A, Herrmann F, Frisch J, Seipelt G, Schulz G, Hoelzer D, Mertelsmann R. Biologic effects of recombinant human interleukin-3 in vivo. J Clin Oncol 1991; 9:2120-7; PMID:1960553 [DOI] [PubMed] [Google Scholar]
- 44.Nievergall E, Ramshaw HS, Yong AS, Biondo M, Busfield SJ, Vairo G, Lopez AF, Hughes TP, White DL, Hiwase DK. Monoclonal antibody targeting of IL-3 receptor α with CSL362 effectively depletes CML progenitor and stem cells. Blood 2014; 123:1218-28; PMID:24363400; http://dx.doi.org/ 10.1182/blood-2012-12-475194 [DOI] [PubMed] [Google Scholar]
- 45.Burmester GR, Feist E, Sleeman MA, Wang B, White B, Magrini F. Mavrilimumab, a human monoclonal antibody targeting GM-CSF receptor-α, in subjects with rheumatoid arthritis: a randomised, double-blind, placebo-controlled, phase I, first-in-human study. Ann Rheum Dis 2011; 70:1542-9; PMID:21613310; http://dx.doi.org/ 10.1136/ard.2010.146225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koike M, Furuya A, Nakamura K, Iida A, Anazawa H, Hanai N, et al.. Antibody against human interleukin-5-receptor α chain. United States: US patent 6,018,032, 1997.
- 47.Diamant Z, Gauvreau GM, Cockcroft DW, Boulet LP, Sterk PJ, de Jongh FH, Dahlén B, O'Byrne PM. Inhaled allergen bronchoprovocation tests. J Allergy Clin Immunol 2013; 132:1045-55 e6; PMID:24119772; http://dx.doi.org/ 10.1016/j.jaci.2013.08.023 [DOI] [PubMed] [Google Scholar]
- 48.Gauvreau GM, Ellis AK, Denburg JA. Haemopoietic processes in allergic disease: eosinophil/basophil development. Clin Exp Allergy 2009; 39:1297-306; PMID:19622087; http://dx.doi.org/ 10.1111/j.1365-2222.2009.03325.x [DOI] [PubMed] [Google Scholar]
- 49.Tavernier J, Devos R, Cornelis S, Tuypens T, Van der Heyden J, Fiers W, Plaetinck G. A human high affinity interleukin-5 receptor (IL5R) is composed of an IL5-specific α chain and a β chain shared with the receptor for GM-CSF. Cell 1991; 66:1175-84; PMID:1833065; http://dx.doi.org/ 10.1016/0092-8674(91)90040-6 [DOI] [PubMed] [Google Scholar]
- 50.Hayashida K, Kitamura T, Gorman DM, Arai K, Yokota T, Miyajima A. Molecular cloning of a second subunit of the receptor for human granulocyte-macrophage colony-stimulating factor (GM-CSF): reconstitution of a high-affinity GM-CSF receptor. Proc Natl Acad Sci USA 1990; 87:9655-9; PMID:1702217; http://dx.doi.org/ 10.1073/pnas.87.24.9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitamura T, Hayashida K, Sakamaki K, Yokota T, Arai K, Miyajima A. Reconstitution of functional receptors for human granulocyte/macrophage colony-stimulating factor (GM-CSF): evidence that the protein encoded by the AIC2B cDNA is a subunit of the murine GM-CSF receptor. Proc Natl Acad Sci USA 1991; 88:5082-6; PMID:1828890; http://dx.doi.org/ 10.1073/pnas.88.12.5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitamura T, Sato N, Arai K, Miyajima A. Expression cloning of the human IL-3 receptor cDNA reveals a shared β subunit for the human IL-3 and GM-CSF receptors. Cell 1991; 66:1165-74; PMID:1833064; http://dx.doi.org/ 10.1016/0092-8674(91)90039-2 [DOI] [PubMed] [Google Scholar]
- 53.Kitamura T, Miyajima A. Functional reconstitution of the human interleukin-3 receptor. Blood 1992; 80:84-90; PMID:1377056 [PubMed] [Google Scholar]
- 54.Kankaanranta H, Lindsay MA, Giembycz MA, Zhang X, Moilanen E, Barnes PJ. Delayed eosinophil apoptosis in asthma. J Allergy Clin Immunol 2000; 106:77-83; PMID:10887309; http://dx.doi.org/ 10.1067/mai.2000.107038 [DOI] [PubMed] [Google Scholar]
- 55.Moore WC, Fitzpatrick AM, Li X, Hastie AT, Li H, Meyers DA, Bleecker ER. Clinical heterogeneity in the severe asthma research program. Ann Am Thorac Soc 2013; 10 Suppl:S118-24; PMID:24313761; http://dx.doi.org/ 10.1513/AnnalsATS.201309-307AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sehmi R, Baatjes AJ, Denburg JA. Hemopoietic progenitor cells and hemopoietic factors: potential targets for treatment of allergic inflammatory diseases. Curr Drug Targets Inflamm Allergy 2003; 2:271-8; PMID:14561146; http://dx.doi.org/ 10.2174/1568010033484007 [DOI] [PubMed] [Google Scholar]
- 57.Menzies-Gow A, Flood-Page P, Sehmi R, Burman J, Hamid Q, Robinson DS, Kay AB, Denburg J. Anti-IL-5 (mepolizumab) therapy induces bone marrow eosinophil maturational arrest and decreases eosinophil progenitors in the bronchial mucosa of atopic asthmatics. J Allergy Clin Immunol 2003; 111:714-9; PMID:12704348; http://dx.doi.org/ 10.1067/mai.2003.1382 [DOI] [PubMed] [Google Scholar]
- 58.Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, Kelly EA. Decreased expression of membrane IL-5 receptor α on human eosinophils: II. IL-5 down-modulates its receptor via a proteinase-mediated process. J Immunol 2002; 169:6459-66; PMID:12444155; http://dx.doi.org/ 10.4049/jimmunol.169.11.6459 [DOI] [PubMed] [Google Scholar]
- 59.Gregory B, Kirchem A, Phipps S, Gevaert P, Pridgeon C, Rankin SM, Robinson DS. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor α-chain expression by cytokines: IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor α expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor α expression. J Immunol 2003; 170:5359-66; PMID:12759409; http://dx.doi.org/ 10.4049/jimmunol.170.11.5359 [DOI] [PubMed] [Google Scholar]
- 60.Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am J Respir Crit Care Med 2003; 167:199-204; PMID:12406833; http://dx.doi.org/ 10.1164/rccm.200208-789OC [DOI] [PubMed] [Google Scholar]
- 61.Hart TK, Cook RM, Zia-Amirhosseini P, Minthorn E, Sellers TS, Maleeff BE, Eustis S, Schwartz LW, Tsui P, Appelbaum ER, et al.. Preclinical efficacy and safety of mepolizumab (SB-240563), a humanized monoclonal antibody to IL-5, in cynomolgus monkeys. J Allergy Clin Immunol 2001; 108:250-7; PMID:11496242; http://dx.doi.org/ 10.1067/mai.2001.116576 [DOI] [PubMed] [Google Scholar]
- 62.Fulkerson PC, Rothenberg ME. Targeting eosinophils in allergy, inflammation and beyond. Nat Rev Drug Discov 2013; 12:117-29; PMID:23334207; http://dx.doi.org/ 10.1038/nrd3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolbeck R, Kozhich A, Koike M, Peng L, Andersson CK, Damschroder MM, Reed JL, Woods R, Dall'acqua WW, Stephens GL, et al.. MEDI-563, a humanized anti-IL-5 receptor α mAb with enhanced antibody-dependent cell-mediated cytotoxicity function. J Allergy Clin Immunol 2010; 125:1344-53 e2; PMID:20513525; http://dx.doi.org/ 10.1016/j.jaci.2010.04.004 [DOI] [PubMed] [Google Scholar]
- 64.Smith DA, Minthorn EA, Beerahee M. Pharmacokinetics and pharmacodynamics of mepolizumab, an anti-interleukin-5 monoclonal antibody. Clin Pharmacokinet 2011; 50:215-27; PMID:21348536; http://dx.doi.org/ 10.2165/11584340-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 65.Gauvreau GM, Pageau R, Seguin R, Carballo D, Gauthier J, D'Anjou H, Campbell H, Watson R, Mistry M, Parry-Billings M, et al.. Dose-response effects of TPI ASM8 in asthmatics after allergen. Allergy 2011; 66:1242-8; PMID:21605124; http://dx.doi.org/ 10.1111/j.1398-9995.2011.02638.x [DOI] [PubMed] [Google Scholar]
- 66.Robb L, Drinkwater CC, Metcalf D, Li R, Kontgen F, Nicola NA, Begley CG. Hematopoietic and lung abnormalities in mice with a null mutation of the common β subunit of the receptors for granulocyte-macrophage colony-stimulating factor and interleukins 3 and 5. Proc Natl Acad Sci USA 1995; 92:9565-9; PMID:7568173; http://dx.doi.org/ 10.1073/pnas.92.21.9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hercus TR, Bagley CJ, Cambareri B, Dottore M, Woodcock JM, Vadas MA, Shannon MF, Lopez AF. Specific human granulocyte-macrophage colony-stimulating factor antagonists. Proc Natl Acad Sci USA 1994; 91:5838-42; PMID:8016076; http://dx.doi.org/ 10.1073/pnas.91.13.5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hercus TR, Barry EF, Dottore M, McClure BJ, Webb AI, Lopez AF, Young IG, Murphy JM. High yield production of a soluble human interleukin-3 variant from E. coli with wild-type bioactivity and improved radiolabeling properties. PLoS One 2013; 8:e74376; PMID:23991218; http://dx.doi.org/ 10.1371/journal.pone.0074376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minter RR, Cohen ES, Wang B, Liang M, Vainshtein I, Rees G, Eghobamien L, Harrison P, Sims DA, Matthews C, et al.. Protein engineering and preclinical development of a GM-CSF receptor antibody for the treatment of rheumatoid arthritis. Br J Pharmacol 2013; 168:200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucl Acids Res 1987; 15:8125-48; PMID:3313277; http://dx.doi.org/ 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koike M, Nakamura K, Furuya A, Iida A, Anazawa H, Takatsu K, Hanai N. Establishment of humanized anti-interleukin-5 receptor α chain monoclonal antibodies having a potent neutralizing activity. Hum Antibodies 2009; 18:17-27; PMID:19478395 [DOI] [PubMed] [Google Scholar]
- 72.Spanevello MD, Tajouri SI, Mirciov C, Kurniawan N, Pearse MJ, Fabri LJ, Owczarek CM, Hardy MP, Bradford RA, Ramunno ML, et al.. Acute delivery of EphA4-Fc improves functional recovery after contusive spinal cord injury in rats. J Neurotrauma 2013; 30:1023-34; PMID:23557244; http://dx.doi.org/ 10.1089/neu.2012.2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ritzen U, Rotticci-Mulder J, Stromberg P, Schmidt SR. Endotoxin reduction in monoclonal antibody preparations using arginine. J Chromatogr B Analyt Technol Biomed Life Sci 2007; 856:343-7; PMID:17644450; http://dx.doi.org/ 10.1016/j.jchromb.2007.06.020 [DOI] [PubMed] [Google Scholar]
- 74.Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H, et al.. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nature Biotechnol 2005; 23:344-8; http://dx.doi.org/ 10.1038/nbt1067 [DOI] [PubMed] [Google Scholar]
- 75.Barbas CF 3rd, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA 1991; 88:7978-82; PMID:1896445; http://dx.doi.org/ 10.1073/pnas.88.18.7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jostock T, Vanhove M, Brepoels E, Van Gool R, Daukandt M, Wehnert A, Van Hegelsom R, Dransfield D, Sexton D, Devlin M, et al.. Rapid generation of functional human IgG antibodies derived from Fab-on-phage display libraries. J Immunol Methods 2004; 289:65-80; PMID:15251413; http://dx.doi.org/ 10.1016/j.jim.2004.03.014 [DOI] [PubMed] [Google Scholar]
- 77.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol 2000; 328:333-63; PMID:11075354; http://dx.doi.org/ 10.1016/S0076-6879(00)28406-1 [DOI] [PubMed] [Google Scholar]
- 78.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 1987; 154:367-82; PMID:3323813; http://dx.doi.org/ 10.1016/0076-6879(87)54085-X [DOI] [PubMed] [Google Scholar]
- 79.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, et al.. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat 2002; 9:401-6; PMID:12409628; http://dx.doi.org/ 10.1107/S0909049502015170 [DOI] [PubMed] [Google Scholar]
- 80.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr 2010; 66:125-32; PMID:20124692; http://dx.doi.org/ 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 2013; 69:1204-14; PMID:23793146; http://dx.doi.org/ 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658-74; PMID:19461840; http://dx.doi.org/ 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carr PD, Conlan F, Ford S, Ollis DL, Young IG. An improved resolution structure of the human β common receptor involved in IL-3, IL-5 and GM-CSF signalling which gives better definition of the high-affinity binding epitope. Acta Crystallogr Sect F Struct Biol Cryst Commun 2006; 62:509-13; PMID:16754968; http://dx.doi.org/ 10.1107/S1744309106016812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang H, Liu JH, Yang W, Springer T, Shimaoka M, Wang JH. Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1. Proc Natl Acad Sci U S A 2009; 106:18345-50; PMID:19805116; http://dx.doi.org/ 10.1073/pnas.0909301106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Collaborative Computational Project N . The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 50:760-3; PMID:15299374; http://dx.doi.org/ 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 86.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213-21; PMID:20124702; http://dx.doi.org/ 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126-32; PMID:15572765; http://dx.doi.org/ 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 88.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66:12-21; PMID:20057044; http://dx.doi.org/ 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Munson PJ, Rodbard D. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal Biochem 1980; 107:220-39; PMID:6254391; http://dx.doi.org/ 10.1016/0003-2697(80)90515-1 [DOI] [PubMed] [Google Scholar]
- 90.O'Byrne PM, Dolovich J, Hargreave FE. Late asthmatic responses. Am Rev Resp Dis 1987; 136:740-51; PMID:3115156; http://dx.doi.org/ 10.1164/ajrccm/136.3.740 [DOI] [PubMed] [Google Scholar]
- 91.Wood LJ, Sehmi R, Gauvreau GM, Watson RM, Foley R, Denburg JA, O'byrne PM. An inhaled corticosteroid, budesonide, reduces baseline but not allergen-induced increases in bone marrow inflammatory cell progenitors in asthmatic subjects. Am J Respir Crit Care Med 1999; 159:1457-63; PMID:10228111; http://dx.doi.org/ 10.1164/ajrccm.159.5.9808123 [DOI] [PubMed] [Google Scholar]
- 92.Cyr MM, Baatjes AJ, Dorman SC, Crawford L, Sehmi R, Foley R, Alam R, Byrne PO, Denburg JA. In vitro effects of budesonide on eosinophil-basophil lineage commitment. Open Respir Med J 2008; 2:60-6; PMID:19343093; http://dx.doi.org/ 10.2174/1874306400802010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeLano, W.L 2002. DeLano Scientific (San Carlos, CA, USA). http://pymol.sourceforge.net/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.