ABSTRACT
In 10–20% of the cases of chronic lymphocytic leukemia of B-cell phenotype (B-CLL), the IGHV1-69 germline is utilized as VH gene of the B cell receptor (BCR). Mouse G6 (MuG6) is an anti-idiotypic monoclonal antibody discovered in a screen against rheumatoid factors (RFs) that binds with high affinity to an idiotope expressed on the 51p1 alleles of IGHV1-69 germline gene encoded antibodies (G6-id+). The finding that unmutated IGHV1-69 encoded BCRs are frequently expressed on B-CLL cells provides an opportunity for anti-idiotype monoclonal antibody immunotherapy. In this study, we first showed that MuG6 can deplete B cells encoding IGHV1-69 BCRs using a novel humanized GTL mouse model. Next, we humanized MuG6 and demonstrated that the humanized antibodies (HuG6s), especially HuG6.3, displayed ∼2-fold higher binding affinity for G6-id+ antibody compared to the parental MuG6. Additional studies showed that HuG6.3 was able to kill G6-id+ BCR expressing cells and patient B-CLL cells through antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Finally, both MuG6 and HuG6.3 mediate in vivo depletion of B-CLL cells in NSG mice. These data suggest that HuG6.3 may provide a new precision medicine to selectively kill IGHV1-69-encoding G6-id+ B-CLL cells.
KEYWORDS: B-cell chronic lymphocytic leukemia, cytotoxicity, GTL mice, humanization, immunotherapy, monoclonal antibody, precision medicine
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is the most common leukemia in the United States, accounting for approximately 30% of all adult leukemia cases.1,2 Over 14,620 individuals develop B-CLL annually, there are circa 4,650 deaths 3 and no curative therapies.4 The small molecule inhibitors, such as BTK and PI3-kinase inhibitors, and Bcl-2 family inhibitors, as well as purine nucleoside analogs (PNAs), that are used as the standard treatment for B-CLL patients have shown response rates ranging between 19 to 70%.5-9 However, treatment with such agents is invariably associated with side effects that range from mild to severe, and eventual emergence of drug resistance. In addition, immunotherapies against B-CLL, including ofatumumab (Arzerra®, anti-CD20), rituximab (Rituxan®, anti-CD20), and alemtuzumab (Campath®, anti-CD52), show a common caveat that the targeted cell surface markers are indistinctively expressed on both normal and malignant B cells or very poorly expressed on B-CLL cells.10-13 Therefore, there is an immediate need for an alternative therapy that is specific toward malignant cells in B-CLL patients.
One characteristic of B-CLL is that their B cell receptors (BCR) have shown biased immunoglobulin (Ig) heavy chain (IGHV) usage.14 In addition, current understanding of pathological mechanisms involved in B-CLL suggest that there are two distinct subgroups of patients based on the absence or presence of somatic mutations in the variable regions of the IGHV genes.14-16 It is estimated that half of B-CLL cases express variable heavy chain (VH) gene mutations, compared to the patient's VH germline gene sequence, whereas the other half express essentially unmutated VH genes (<2 % deviation from germline sequence).17 Patients with unmutated VH genes have a distinctly more aggressive and malignant disease with much shorter survival rates.15,16 In addition, a high proportion of IGHV-unmutated B-CLL cases often carry complementarity-determining region (CDR) 3 stereotyped rearrangement of the V, D, and J segments. Both the biased germline gene usage and the stereotyped VH CDR3 (CDR-H3) rearrangements present a highly restricted set of BCRs that may serve as distinctive immunological markers or tumor-associated antigens for targeted immunotherapy.18-22
The IGHV1-69 germline gene was among the first of the genes shown to be overrepresented in B-CLL patients with unmutated IGHV genes, and has the highest frequencies (>40 %) of stereotyped VHDHJH rearrangements.19,21 Several studies have reported that 10 to 20% of B-CLL patients overexpress IGHV1-69 encoded BCRs by their leukemic cells.12,14,23-25 The IGHV1-69 locus shows extensive allelic polymorphism, with evidence of gene duplication and deletions and 14 alleles that are broadly characterized as belonging to either the 51p1 and hv1263 allelic groups. An individual can utilize one or more copies of either or both allelic groups for BCR expression. The 51p1 alleles of the IGHV1-69 germline gene are distinctly expressed by the BCRs of B-CLL cells.23,24,26,27
Murine G6 (MuG6) is a heavy chain directed anti-IGHV1-69 idiotypic antibody that is generally not affected by the structure of the CDR-H3.28 The IGHV1-69 idiotype (G6-id+) is expressed by the eight 51p1 alleles, but not the six hv1263 alleles of the IGHV1-69 germline gene that differ by 3 residues in CDR-H2 and one residue in framework (FRW)-H3.29 MuG6 preferentially recognizes antibodies encoded by IGHV1-69 51p1 alleles that are expressed in their germline configuration, and is particularly sensitive to amino acid substitutions in its distinctly hydrophobic CDR-H2 loop.30
Since circa 6.16±0.55 of the circulating IGH repertoire from healthy individuals express IGHV1-69 BCRs, an anti-cancer reagent that targets this B cell subset will not lead to global B cell depletion, and therefore offers the potential for a new precision medicine that a more selective therapeutic agent for this aggressive subset of B-CLL.30,31 In this study, we report the humanization of MuG6 and experiments performed to investigate its therapeutic potential to immunodeplete malignant B cells from B-CLL patients that utilize IGHV1-69-expressing BCRs. The most potent humanized antibody HuG6.3 showed higher binding affinity than the parental MuG6 and exhibited potent killing activity on multiple G6-id+ IGHV1-69 expressing cells and human B-CLL tumor cells in vitro and in a humanized mouse model in vivo.
Results
MuG6 antibody mediates systemic depletion of IGHV1-69 encoding B cells in GTL mice
To determine if MuG6 could mediate in vivo depletion of IGHV1-69 G6-id+ expressing lymphocytes, we utilized a GTL mouse model (NOD.Cg-PrkdcscidIl2rgtm1wjl/SzJ (NSG) mice engrafted with human fetal bone marrow, liver, and thymus tissue for generation of human immunity) to investigate the in vivo immunodepletion activity of MuG6 on the reconstituted B cell population. First, the peripheral blood from GTL mice was stained for human CD45+ mononuclear cells at 16 weeks post immune reconstruction to verify levels of engraftment. These GTL mice were then randomly assigned into different groups and treated with MuG6 or control antibody. Seven days after treatment, mouse blood was harvested to analyze serum MuG6 levels, as well as the total CD20+ B cell population and the G6-id+ B cells by fluorescence-activated cell sorting (FACS). Serum MuG6 levels at days 7, 9 and 10 were 12.4, 7.2 and 5.4 ng/ml, respectively, after correcting for background binding by normal mouse serum (Fig. S1). As shown in Fig. 1A, MuG6 treatment did not result in a change in the total B-cell population; however, the G6-id+ B cell subpopulation dramatically decreased in MuG6-treated mice compared to the other groups (Fig. 1B). While it is possible that this result could be due to saturation of 51p1 allele encoded BCRs by serum MuG6, this is unlikely because the day 7 serum MuG6 level (0.08 nM) was significantly below the equilibrium dissociation constant (KD) of MuG6 for the IGHV1-69 idiotype (discussed below). Moreover, the expression of the cognate IgM and IgG G6-id+ antibodies in the plasma of the MuG6-treated but not control IgG-treated mice was markedly decreased at day 7 (Fig. 1C and 1D) and day 21 (Fig. S2). The dramatic loss of G6-id+ B cells and loss of 51p1 allele encoding IgM and IgG in MuG6-treated mouse plasma demonstrate that MuG6 has the capacity to deplete IGHV1-69 G6-id+ B cells in vivo.
Figure 1.
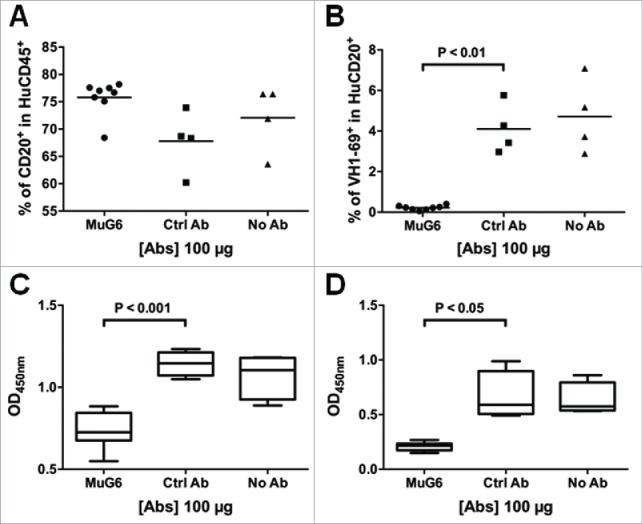
The in vivo function of MuG6 in the humanized GTL mice model. (A) GTL mice were injected with MuG6, control mouse IgG, or the equal amount of PBS. After 7 days, mouse blood were harvested and the percentage of human B cells in the total human lymphocytes was analyzed via flow cytometry. (B) The percentage of IGHV1-69 expressing cells was measured in the total human B cells from GTL mice blood. (C) IGHV1-69 encoded human IgM and (D) IgG responses as detected by ELISA in GTL plasma samples obtained on day 7 after antibody injection. Each symbol is representative of a single GTL mouse. P value is determined by two-tailed Mann–Whitney U-test to analyze significant differences between median values of the datasets. Individual plasma samples were tested at a 1:100 dilution.
Humanization of MuG6
The MuG6 heavy and light chain variable regions (VH and VL) genes from the hybridoma cell line were individually recovered by RT-PCR using specific primers for mouse antibody variable genes. The MuG6 VH and VL belong to mouse VH1 (IGHV1-5*01), JH4 (IGHJ4*01) and DH2 (IGHD2-12*01) and VK14 (IGKV14-100*01) and JK5 (IGKJ5*01) segments, respectively (Fig. 2). Next, the structure-guided CDR grafting approach was employed to humanize MuG6. For selection of the human acceptor FRW template for CDR-grafting, the VH and VL amino acid sequences of MuG6 were separately compared to human antibody sequences in the IMGT database to identify the most similar human antibody and Ig germline VH and VL sequences. The best-matched human Ig germline V sequences were IGHV1-46*13 (68.4% homology to MuG6-VH) and IGKV1-16*01 (67.4% homology to MuG6-VL). Using IgG protein sequence blast, MuG6 was compared with other humanized or human monoclonal antibodies (mAbs), including 5c8 (anti-CD40L), Fab 7G10 (anti-IL23), hATR-5 (anti-Tissue factor), 1C12 (anti-musk odorant traseolide), CAMPATH-1H (anti-CD52), and a mouse Fab 64M-2 (anti-DNA t(6-4) T photoproduct), and revealed variable degree of sequence homology. Subsequently, Fab 64M-2 structure (PDB-ID: 1EHL) was chosen as a template for VH chain, with 87.5% sequence identity and 92% similarity to MuG6. As such, the FRW residues of 1EHL VH chain were unchanged, but the CDRs were mutated to reflect MuG6 CDRs. Closer examination of this modified structure did not reveal any major steric clashes. In addition, 1C12 structure was chosen as a template for VL chain, with 94% sequence identity and 96% similarity to MuG6 sequence. Further analysis in the structure revealed one steric clash with Val89 (CDR-L3) when Leu4 (FRW-L1) was replaced with Met4 (humanized). The final humanized MuG6 was designed through multiple sequence alignment (CLUSTALX) and the most conserved FWR residues. In addition, we manually exchanged the residues in both VH and VL FWRs to the consensus human germline sequence and then generated HuG6 version 1 (HuG6.1). Sequence alignment of amino acids between the MuG6 and HuG6.1 is shown in Fig. 2, with 18 and 18 amino acids mutations in the VH and VL, respectively.
Figure 2.
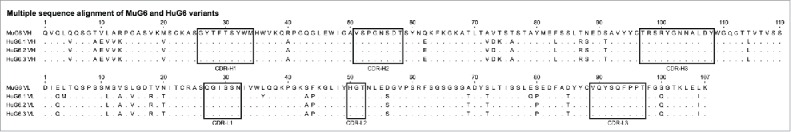
Amino acid sequence alignment of the rearranged mouse and humanized variable heavy (VH) and variable light kappa (VK) domains. The complementarity determining regions (CDRs) of heavy and light chains are marked in black rectangles. There are 18 mutations in the heavy chain and 18 mutations in light chain between HuG6.1 and MuG6. One residue is changed (VH-Threonine73 to Lysine73) between HuG6.2 and HuG6.3.
Structure-based ab-initio generation of antibody homology model for humanization
The ab-initio generated homology model of MuG6 was used to identify surface accessible (solvent exposed) residues via WAM 32 server and visualized through DeepView program (Swiss-PdbViewer, http://www.expasy.org/spdbv/) 33 (Fig. 3A). These residues were identical to the human germline sequence in the FWR regions. In-silico mutations were performed via PyMOL mutagenesis tool to replace framework residues of mouse to human HuG6.1 sequence (Delano scientific-www.pymol.org). GROMOS force field energy minimization parameter was then applied to the homology model HuG6.1 using DeepView program with default settings, and displayed certain residues with high entropy in their side chain rotamers (Fig. 3B).34,35 Examination of this energy minimized homology model of HuG6.1 revealed residues that had distorted geometry or steric clashes with other residues. These anomalies upon closer examination with either distorted geometry or steric clashes were further visualized in PyMOL (Fig. 3C-F). Residues with steric clashes included (Fig. 3C, left) Lys73 (FRW-H3) with Gly54 (CDR-H2), (Fig. 3D, left) Met4 (FRW-L1) with Cys88 (FRW-L3), (Fig. 3E, left) Tyr36 (FRW-L2) with Leu100b (CDR-H3), and (Fig. 3F, left) Gln79 (FRW-L3) with Arg61 (FRW-L3). Based on the conserved homologous sequence alignment and structural analysis, the residues that caused steric clashes were back mutated to the mouse counterpart, including (1) Lys73 to Thr73 (Fig. 3C, right), (2) Met4 to Leu4 (Fig. 3D, right), (3) Tyr36 to Leu36 (Fig. 3E, right), and (4) Gln79 to Glu79 (Fig. 3F, right). In summary, four identified residues were mutated back to the original mouse residues, including one residue in VH (Thr73) and three residues in VL (Leu4, Leu36, Glu79), creating HuG6 version 2 (HuG6.2). Furthermore, to test the contribution of threonine (mouse germline VH chain) as opposed to lysine (human germline) in binding to the target, this T73K mutation (Fig. 3C) was made as HuG6 version 3 (HuG6.3). The sequence alignment highlighted the differences in MuG6 with different versions of HuG6 (HuG6.1, HuG6.2, and HuG6.3) as shown in Fig. 2.
Figure 3.
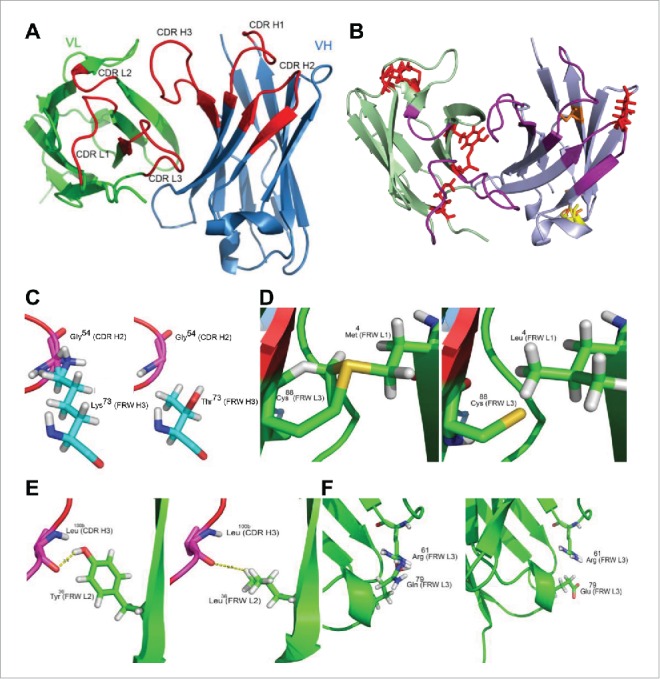
Antibody structural homology model. (A) The 3D structure of MuG6 is generated from web antibody modeling program (WAM). Light chain, heavy chain, and CDRs are colored in green, blue, and red, respectively. (B) The energy homology model of HuG6.1 is minimized with GROMOS force field energetics (identical orientation as the MuG6 homology model). Residues with steric clashes, bad field problems, and buried side chain with no hydrogen bonds are colored in red, yellow, and orange, respectively. (C-F) The residues with steric clashes are identified in the humanized G6.1 structural homology model. (C) Lys73 (FRW-H3) steric clashed with Gly54 (CDR-H2) (left) is mutated back to Thr73 (right). (D) Met4 (FRW-L1) steric clashed with Cys88 (FRW-L3) (left) is mutated back to Leu3 (right). (E) Tyr36 (FRW-L2) has a potential non-relevant hydrogen bond with Leu100B (CDR-H3) (left) and is mutated back to Leu36 (right). (F) Gln79 (FRW-L3) is steric clashed with Arg61 (FRW-L3) (left). Arg residue is conserved across different homologous antibodies but Gln79 is not. Thus, Gln79 was back mutated to Glu79 (right).
Binding affinities of MuG6 and HuG6 variants antibodies for G6-id+ D80 mAb
The humanized VH and VL genes of HuG6 variants (HuG6s) were de novo synthesized and codon-optimized for mammalian cell expression. The binding affinities of MuG6 and HuG6s scFv-Fcs to G6-id+ D80 IgG were further analyzed by ELISA. The results in Fig. 4A showed that HuG6.1 lost its binding ability; however, HuG6.2 and HuG6.3 exhibited even better binding affinity than the parental MuG6. Next, we used BIAcore to interrogate the binding kinetics of MuG6 and HuG6s scFv-Fcs against D80 scFv. As shown in Fig. 4B and Fig. S3 the KD of MuG6, HuG6.2, and HuG6.3 against D80 scFv were 0.35, 0.23, and 0.16 nM, respectively. These results were consistent with the apparent higher affinity of HuG6.2 and HuG6.3 over MuG6 by ELISA.
Figure 4.
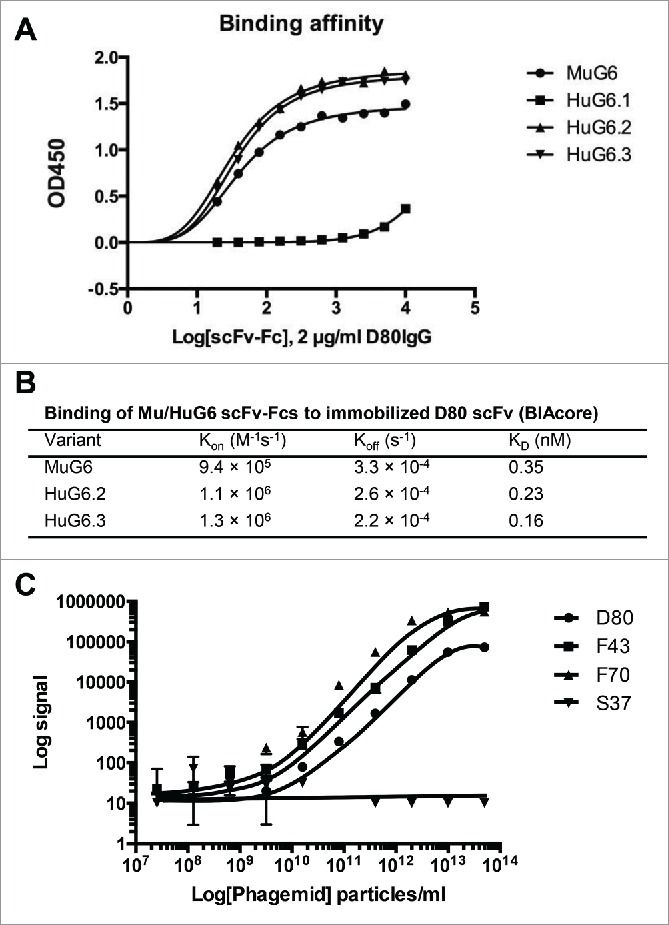
Binding affinity and kinetics of MuG6 and HuG6s. (A) Qualitative binding analysis of MuG6 and HuG6s scFv-Fc antibodies (0–10 µg/ml) to D80 IgG (2 µg/ml) through ELISA. (B) BIAcore surface plasmon resonance kinetic data for MuG6, HuG6.2, and HuG6.3 scFv-Fcs binding to immobilized D80 scFv. (C) Relative binding relationships between HuG6.3 and IGHV1-69 encoding scFvs, D80, F43 and F70, were measured by MSD.
Interestingly, only one residue difference between HuG6.2 (Thr73) and HuG6.3 (Lys73) influenced the binding affinity, suggesting a definitive role of lysine in modulating the binding pattern. The in-silico modeling suggested that residue Lys73 (FRW-H2) has a steric clash with Gly54 (CDR-H2), and thus the Lys73 was back mutated to mouse residue Thr73 (Fig. 3C). However, this resulted in loss of affinity, albeit small, indicating that Lys73 may cause subtle changes in the binding site to position CDR-H2 in a conformation that enables HuG6.3 to increase its binding affinity to D80. These results showed that humanization of MuG6 was successful and HuG6.2 and HuG6.3 have better binding affinity than MuG6.
HuG6.3 binds with higher affinity to two IGHV1-69 antibodies with unmutated VH segments
Three IGHV1-69 encoding scFvs (D80, F43, and F70) and one control IGHV1-18 encoding scFv (S37) were investigated the binding affinity to HuG6.3 using Meso Scale Discovery (MSD) immunoassay. Interestingly, HuG6.3 bound more strongly to F43 and F70, which are scFvs with 100% identity to IGHV1-69*01 germline gene, compared to D80 (89% identity). There was no reactivity of HuG6.3 to S37 scFv (Fig. 4C). To further test the binding affinity, a HuG6.3-labeled sensor system was used to measure the association and dissociation kinetics against IGHV1-69 encoding D80 and F43 scFv-Fcs coated on the SA sensor tip surfaces (Fig. S4). The Octet-Red assay results showed specific binding between the HuG6.3 and IGHV1-69 encoding scFv-Fcs, whereas it did not bind the control scFv-Fc. Moreover, there was a 3.5-fold slower Kon for HuG6.3 binding to germline F43 compared to D80 scFv-Fc and a circa 600-fold slower Koff with a resulting 50-fold increase in binding affinity. These results provide additional support that HuG6.3 can specifically bind to IGHV1-69 encoding immunoglobulins with higher affinity binding for unmutated IGHV1-69 51p1 alleles.
MuG6 and HuG6s mediates killing of IGHV1-69 encoded G6-id+ cells through CDC and ADCC
To delineate the biological mechanism(s) by which MuG6 treatment caused depletion of G6-id+ B cells, CDC and ADCC assays were performed. To mimic G6-id+ B-CLL cells, we used IGHV1-69 encoding D80, F70, and F43 scFvs (Fig. S5A) that showed 89, 100, and 100% identity to IGHV1-69*01 germline gene, respectively, to construct G6-id+ 293T cells by transfection (Fig. S5A). To anchor the surface-expressed G6-id+ scFv to the cell membrane, scFv-Fc proteins were fused, in frame, to a transmembrane moiety.36 To follow transfection efficiency, ZsGreen was co-expressed in the bicistronic message and was visualized by fluorescence microscope (Fig. S5B). Surface expression of the G6-id+ and G6-id− (11A-scFv) antibodies was analyzed by FACS stained with allophycocyanin (APC)-conjugated MuG6 and ZsGreen (Fig. S5C).
IGHV1-69 and non-IGHV1-69 transduced 293T cells were used as the target cells for CDC and ADCC assays. The HuG6.2 and HuG6.3 scFv-Fcs were further evaluated for their capacity to mediate CDC activity on the G6-id+ 293T cells using rabbit serum. As shown in Fig. 5A-C, circa 20% of target cells were killed when treated with Mu/HuG6s. HuG6.3 exhibited a slightly better potent CDC activity than HuG6.2 against the three G6-id+ 293T cells, and had comparable CDC activity as MuG6. The specificity of the CDC-mediated killing was shown by the lack of cytotoxicity on negative control G6-id− 11A-293T cells (Fig. 5D). We further tested the activity of mouse anti-human Fc IgG2a and showed 25% killing activity among all transduced cells (Fig. S6A). We subsequently focused on HuG6.3 IgG1 alone in the evaluation of ADCC activity. As shown in Fig. 6A-C, HuG6.3 maintained potent ADCC activity against the three G6-id+ cell lines at 20 µg/ml, and more variable killing activity compared to MuG6 at lower antibody concentrations. Again, neither MuG6 nor HuG6.3 killed negative control G6-id− 11A-293T cells by ADCC (Fig. 6D). A mouse anti-human Fc IgG2a was used in the ADCC assay and showed 40% killing activity on all transduced 293T cells (Fig. S6B).
Figure 5.
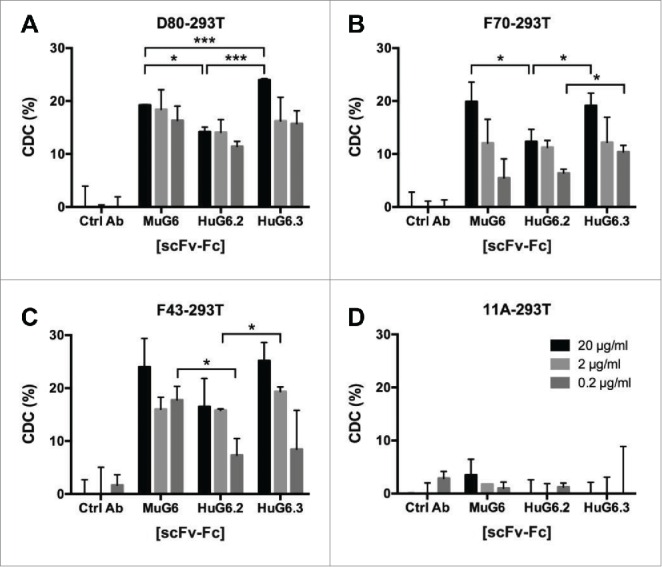
Mu/HuG6s mediate killing of IGHV1-69-expressing cells via CDC. 293T cells were transfected to express the IGHV1-69 G6-id+ idiotype, including (A) D80-293T, (B) F70-293T, and (C) F43-293T, and (D) non-IGHV1-69 encoded 11A-293T. IGHV1-69-G6-id+ expressing cells were incubated with rabbit serum and either Mu/HuG6s or a control human IgG. Percent of cell death was measured using a LDH kit. Bars represent ± standard deviation. “*,” p < 0.05; “**,” p < 0.01.
Figure 6.
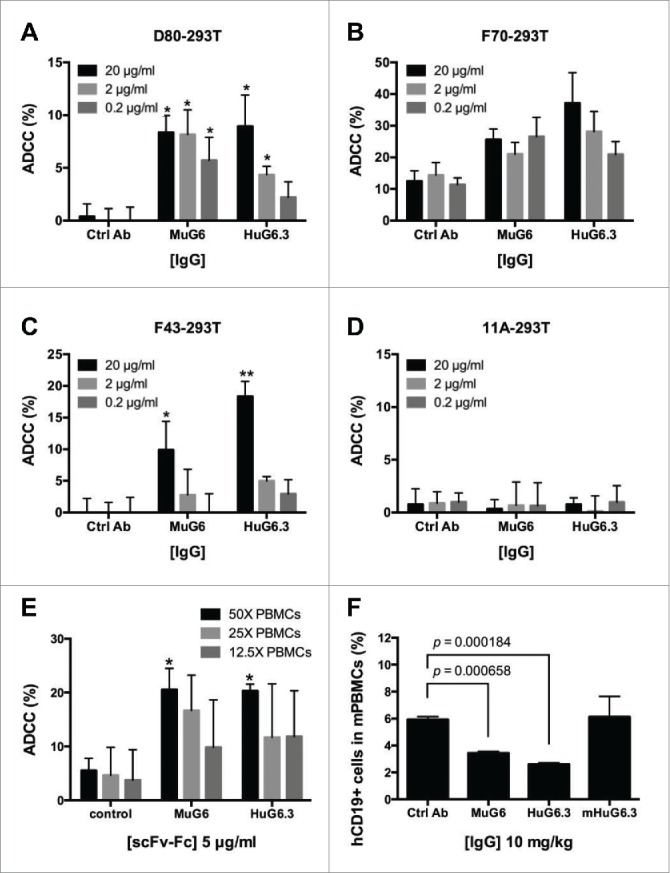
MuG6 and HuG6s mediate killing of IGHV1-69-expressing cells and B-CLL cells via ADCC. MuG6 and HuG6.3 induce specific ADCC activities against IGHV1-69 G6-id+ cells when compared with control antibody. Freshly isolated human PBMC cells were used as the effector cells in the ADCC assay. (A) D80-293T, (B) F70-293T, (C) F43-293T, and (D) 11A-293T (non-IGHV1-69 encoding 293T cells as negative target cells) were incubated with PBMCs at the ratio of 25 (effector (E) cells) to 1 (target (T) cells). In panel (E) IGHV1-69 G6-id+ B cell from CLL patients were used as target (T) cells and mixed with the PBMCs at an E/T ratio of 25 to 1. Antibodies over a concentration range of 0.2 to 20 µg/mL were tested. (F) Patient B-CLL cells were injected into mice treated with or without G6 antibodies for 16 hours. Mouse blood were harvested and stained with human CD19. The percentage of human CD19 as well as B-CLL cells was quantified. Graph represents data were performed in three individual experiments. mHuG6.3, a L234A and L235A mutation version of HuG6.3. Values are mean (± SEM) of triplicate measurements. “*,” p < 0.05; “**,” p < 0.01.
MuG6 and HuG6s mediate killing of IGHV1-69 G6-id+ B-CLL cells
We investigated whether HuG6.3 could induce ADCC of IGHV1-69 G6-id+ B-CLL cells obtained from peripheral blood samples of B-CLL patients. The in vitro lactate dehydrogenase (LDH) release assay was performed by co-incubation of B-CLL target cells with peripheral blood mononuclear cells (PBMCs) from healthy donors in the presence of different antibodies. The results showed that both MuG6 and HuG6.3, but not control antibody, could mediate ADCC in a dose-dependent manner (Fig. 6E). Next, an in vivo examination of HuG6.3-mediated B-CLL cell killing was performed in which G6-id+ and G6-id− B-CLL cells (Fig. S7A) were injected with human natural killer (NK) cells and MuG6/HuG6s into NSG mice intravenously. To further confirm whether HuG6.3 could functionally deplete G6-id+ B-CLL cells, we generated a double mutant (L234A, L235A) HuG6.3 (mHuG6.3, that did not bind either FcγR or C1q) for in vivo examination. After 16-hour circulation, mouse blood was harvested and circulating cells were detected by FACS. The results in Fig. 6F show that G6-id+ B-CLL cells were depleted in vivo in both MuG6-treated and HuG6.3-treated mice, but not in mice treated with control and Fc-mutated HuG6.3 (mHuG6.3). In contrast to G6-id+ patients 1–3, G6-id− patient 4 did not show depletion of CD19 B-CLL cells (Fig. S7B). Taken together, HuG6.3 demonstrates ADCC- and CDC-mediated killing of G6-id+ 293T cells and IGHV1-69-encoding G6-id+ B-CLL cells in vitro and in vivo.
Discussion
In this study, we sought to humanize murine anti-IGHV1-69 idiotope antibody MuG6 because of its unique binding properties and potential broad clinical application. Studies in humanized mice demonstrated that MuG6 IgG1 was able to selectively deplete G6-id+ B cells, which lead to prolonged loss of serum IGHV1-69-encoded IgM and IgG over the 21-day study. Successful humanization of MuG6 was achieved using an in silico modeling approach combined with structure aided antibody design. Two versions of humanized G6 resulted in improved binding affinities compared to parental MuG6. In comparative in vitro biological studies of ADCC and CDC activities, HuG6.3 proved its ability to kill G6-id+ cells and G6-id+ B-CLL cells irrespective of CDR-H3 diversity and light chain pairing (Figs. 2 and 5).
To develop a de novo antibody homology-based structure, the ab-initio generation of antibody homology model in WAM program provided us with the canonical structure of MuG6.32 Our approach using this uniform conformational sampling combined with iterative CONGEN algorithm and visualized by DEEPVIEW further predicted the conformation of side chain in the loops and yielded the most energetically minimized conformation and modeling of non-canonical loop regions from the structures in the protein data bank.37 In addition, this model is energy minimized and sorted in a root mean square deviation screen, which has the advantage over the sequence-based design to reveal any potential steric clashes or distorted geometry in the “humanized” homology model.38-41 This ideally led to a humanized homology model with an appropriate scaffold, which is most conformationally compatible with the engrafted CDRs, and retains the affinity similar to the equivalent mouse antibody. In this study, the sequence-based homology approach was not sufficient to generate an ideal humanized antibody, which resulted in the completely lost binding affinity of HuG6.1. To overcome this problem, we further used CDR grafting combined with certain conserved key residues to further create two humanized versions via selection of suitable scaffold surrounding the CDRs.42-44 Consequently, the humanized antibodies HuG6.2 and HuG6.3 revealed increased binding affinity (∼1.5–2 folds) and slower off rates over the parental molecule. In addition, the 100% IGHV1-69-encoding F43 scFv-Fc has exhibited more than 10-fold higher affinity to HuG6.3 compared to D80 scFv-Fc (89% IGHV-encoding immunoglobulin) (Fig. S8). It is well known that affinity loss is a frequent side effect of humanization; 45 however, our data demonstrate that this multi-parameter modeling strategy resulted in successful humanization of MuG6 with higher affinity.
Current small-animal models have limited usefulness in analyzing the therapeutic effects of antibody treatment to tumor-associated antigens expressed on tumor cells and in developing antibody treatments for B-CLL.46 The experimental evaluation of specificity of anti-idiotypic therapies in vivo is even more challenging due to the low percentage of id+ human B cells. In this study, we utilized two different humanized mouse models in our evaluation of humanized G6 mAb. To reconstruct a natural human B cell repertoire in mice, we employed the GTL mouse model that utilizes human fetal liver and thymus tissues on the NSG background strain in addition to CD34+ haematopoietic stem cells to support the development of an immune system that is more capable of developing a robust adaptive B cell response. This model allowed us to use mAb G6 to monitor the reconstruction of the G6-id+ population of BCR-bearing B cells that utilize the 51p1 alleles of IGHV1-69 germline gene. Donor CD34+ stem cells can encode 2–4 copies of IGHV1-69 germline gene through chromosomal gene duplication and deletion, which leads to a proportional variation in the percentage of B cells in the circulating repertoire, respectively.47 We demonstrated rapid and efficient depletion of G6-id+ IGHV1-69 BCR-bearing B cells that resulted in markedly reduced human IGHV1-69-encoded IgG and IgM for an extended period of time. The mechanism(s) of anti-idiotype depletion in this mouse model is presumably mediated through ADCC by mouse myeloid cells, as we have demonstrated with other human mAbs.48,49 In addition, while complement component C5a is also defective,50 some level of CDC may still occur that could lead to depletion of G6-id+ B cells.
In a second mouse model of B-CLL, we directly co-injected NSG mice with B-CLL patient PBMCs that were almost entirely composed of the malignant clones (data not shown) due to the very little G6-id− B-CLL cells (Fig. S7) and CD56+ NK cells from a healthy donor, and depletion of the G6-id+ malignant B cells was evaluated 16 hours later. This early time point was chosen for several reasons, including the limited number of B-CLL cells that were injected into the mice (10-fold lower than in other mouse models 51), their expected rapid killing by NK cells and the well-known limited long-term viability of the malignant cells in these mice. This NSG model may be further improved upon as reported by Bagnara 52 by use of patient PBMCs instead of purified B cells together with allogeneic antigen-presenting cells. These authors were able to demonstrate B-CLL proliferation in vivo over several doublings that was mediated by proliferating autologous CD4+ T cells that were activated in response to alloantigens. While the B-CLL cells ultimately died in these mice over several weeks, the authors noted more robust B-CLL proliferation and survival in the mouse spleen compared to peripheral blood, a finding that has been seen by other investigators.53,54 There was also evidence of proliferation center formation and T cells in and around follicular structures. The addition of allogeneic human NK cells or CD14+ antigen-presenting cells to this model, as we and others 46,55 have used, may provide the opportunity to further improve the evaluation of mAb HuG6.3 with additional patient samples.
Most patients with B-CLL and other B cell malignancies remain incurable with currently available therapies, and therefore the discovery of novel agents to treat these disorders is urgently needed. The use of anti-idiotypic mAbs as anti-cancer agents is not a new concept, nor has it gained wide acceptance, in part due to the success of other pan-immunodepleting mAbs such as anti-CD20 (rituximab) and anti-CD52 (alemtuzumab), and to the perception that cross-reactive idiotypes are not conserved on malignant B cells from different patients. B cell tumors can be targeted more specifically with antibodies against idiotopes encoded by their unique surface immunoglobulins.56,57 Active vaccination of follicular lymphoma patients with the id protein from their tumors has shown some promising effects, particularly in patients that developed anti-id antibody responses.58 In this new era of precision cancer medicine, HuG6.3 immunotherapy may be able to fill a void for the 10–20% of B-CLL patients in whom the selective depletion of the IGHV1-69-encoding G6-id+ BCR-expressing leukemic cells is the treatment goal. This may also serve as an alternative targeted immunotherapy for G6-id+ patients who may have suboptimal clinical responses to rituximab due to down-regulation of CD20 expression 59 or due to heterogeneity of CD20 expression.60 In addition, given the involvement of IGHV1-69 germline gene in several disorders involving pathophysiologic B-cell clonal expansion, including idiopathic thrombocytopenic purpura, mixed cryoglobulinemia, and hepatitis C virus-induced B cell clonal disorder,61-64 a broader range of potential therapeutic applications for this humanized anti-idiotypic mAb should be investigated.
Material and methods
Cells
293T (CRL-11268) cell line was purchased from American Type Culture Collection and incubated in 10% FBS Dulbecco's Modified Eagle's Medium. 293F cell line was purchased from Invitrogen™ and incubated in 293 FreeStyle serum-free medium (Life Technologies, Carlsbad, CA). IGHV1-69 positive B-CLL cells were isolated from B-CLL patients, obtained from Dr. Jennifer Brown (Dana-Farber Cancer Institute, Department of Medical Oncology) and cultured in IMDM, GlutaMAX medium (Life Technologies) supplemented with 10% Human AB Serum (Gemini Bio-Products, West Sacramento, CA), penicillin and streptomycin at concentrations of 100 U/ml and 100 mg/ml (Sigma-Aldrich, St. Louis, MO), 50 µg/ml Transferrin (Roche, Mannheim, Germany) and 5 µg/ml Human Insulin (Roche). All patients had signed written informed consent to an institutional review board-approved tissue acquisition protocol.
Expression and purification of antibodies
MuG6, HuG6 variants, IGHV1-69 idiotypic antibodies (D80, F70, and F43) and control antibody (11A) were produced as described previously.48 Briefly, scFv-Fcs were constructed by cloning the scFv into pcDNA3.1-Hinge vector in frame with human IgG1 Fc region without CH1 domain. IgG1s were generated by cloning heavy chain variable region (VH) and light chain variable region (VL) into TCAE5.3 vector.65 Fc-mutated HuG6.3 was constructed in TCAE5.3 with Leu234Ala and Leu235Ala mutations on CH2 domain. Antibodies (scFv-Fc and IgG1) were produced in 293F cells and MuG6 was harvested from the supernatant of MuG6 hybridoma. All the antibodies were further purified by protein A sepharose affinity chromatography (GE Healthcare, Newark, NJ). In addition, D80 scFv was cloned into C-terminal histidine tagged pET22b(+) bacterial expression vector (Novagen, Madison, WI), expressed in E. coli BL21(DE3) (Novagen), and purified from supernatant of lysed cells using Nickel affinity chromatography via ÄKTAPurifier FPLC (GE Healthcare).66
Construction of the humanized GTL mouse model and the verification of G6 in vivo activity using flow cytometry
The humanized mice were constructed in 5- to 7-week-old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Jackson Laboratories, Bar Harbor, ME, as described previously,67-69 after sublethal whole body irradiation (325 rads) with a Gammacell 40 Exactor (Best Theratronics, Ottawa, ON, Canada) and by injecting CD34+ haematopoietic stem cells intravenously and implanting 1 mm3 pieces of human fetal thymus and liver tissues under the kidney capsule. Each cohort was produced with tissues from a single donor. CD34+ HSC were isolated from the remaining portion of the same fetal liver using anti-human CD34 microbeads (Miltenyi, Auburn, CA) with > 98% purity determined by flow cytometry after staining with anti-CD34-PE (Miltenyi). All engrafted mice were housed under Biosafety Level-2 conditions and provided with autoclaved food and water supplemented with Baytril (Bayer, Shawnee Mission, KS). All animal experiments were approved by the Institutional Animal Care and Research Committee and the Office of Human Subjects Research at the Dana-Farber Cancer Institute, Boston, MA.
After 16 weeks post-engraftment, the levels of human immune reconstitution were measured by flow cytometry. The GTL mice were randomly assigned into different groups and treated with recombinant human CD40 ligand (rCD40L for maintaining B cell population).70,71 The mice were additionally treated in the presence or absence of 10 µg MuG6 or control antibody by intravenously injection. All the mice were bled out 7 d post-injection; blood samples were centrifuged to separate the plasma fraction, and then the cell pellets were treated with ammonium chloride (ACK) lysing solution (Life Technologies) to enrich for PBMCs. PBMCs and mouse plasma were further stained with fluorochrome-conjugated antibodies to different cell surface markers, followed by multi-color flow cytometry using a LSRII (BD Biosciences, San Jose, CA). The following fluorochrome-conjugated antibodies were used: anti-human CD45-APC (clone H130), CD20-phycoerythrin (PE) (MB19-1) (both from eBioscience, San Diego, CA), and MuG6-FITC (conjugated using Pierce FITC antibody labeling kit, Thermo Scientific, Hudson, NH). Gating was performed on viable lymphoid cells based on the forward and side scatter profiles of the total cells, and stained cells were analyzed within the lymphoid gate. A comparison between the percentages of human CD45+ and endogenous mouse CD45+ was performed to measure the level of immune reconstitution in GTL mice, and other markers were used to analyze the different human B-lymphocyte subsets. Background staining was determined using the corresponding isotype controls or staining cells isolated from non-engrafted animals. Data were analyzed using flowjo version 8.6.3 (Tree Star, Ashland, OR).
ELISA binding assay
The binding activities of MuG6 and HuG6 variants (HuG6s, including HuG6.1, HuG6.2, and HuG6.3) were tested and compared using an IGHV1-69 cognate antibody. D80 IgG1 was chosen as an antigen (cross-reactive idiotope) that uses the 51p1 sequence with IGHV1-69 germline configuration. Biotinylation of MuG6 and HuG6s were done with a commercial biotinylation kit (Pierce) and ELISA analysis was performed. Briefly, D80 IgG1 (2 µg/ml) were coated on to a 96-well Maxisorp plate and incubated overnight at 4°C. Unbound protein was washed away with PBST (0.05% Tween-20 in PBS) and blocked with 2% milk for 1 hour at 25°C. Diluted biotinylated MuG6 and HuG6s as scFv-Fc format were added to the wells and incubated at 25°C for 1 hour. Plates were washed with PBST, added streptavidin-HRP (1:1000), and then incubated at 25°C for 30 minutes. The results were measured at OD450 using an ELISA reader by developing with tetramethylbenzidine solution. For human IgG and IgM ELISA, plates were coated overnight at 4°C with capture antibody MuG6 (100 µg/ml) and developed with detection antibodies, anti-human IgM and IgG (Bethyl Labs), conjugated with HRP.
BIAcore biosensor assay
The binding of MuG6, HuG6.2, or HuG6.3 scFv-Fcs to D80 scFv was compared using surface plasmon resonance instrument BIAcore T100 optical biosensor (BIAcore AB, Uppsala, Sweden). The experiments were performed at 4°C in HBS-P buffer (150 mM NaCl, 10 mM HEPES, 0.05% surfactant P20, and 50 µM EDTA). Briefly, the C-terminal histidine-tagged D80 scFv was first immobilized on a NTA sensor chip with around 100 RU captured level. MuG6 and HuG6s were then injected at various concentrations (0.1, 0.5, 1.5, 4.5, and 14.5 nM) via single cycle kinetics wizard program. The capture surface was regenerated using 0.35 M EDTA, followed by the injection of the running buffer. Double reference subtraction of the data was performed to correct for the buffer contribution to the instrument signal to noise ratio. After this initial subtraction, kinetics analysis of the data was performed using the BIAevaluation software version 2.0.3 (Biacore AB) assuming a simple 1:1 analyte binding model.
Meso scale discovery immunoassay
MSD plates were coated overnight at 4°C with 6 μl of 1.042 μg/ml HuG6.3 IgG1 in PBS. Coated plates were washed and blocked with 75 μl 2% BSA in PBS at 37°C for 1 h. D80, F43, F70, and S37 (IGHV1-18-encoding scFv) phagemids were diluted in 2% milk PBST and added to the wells. The plates were incubated at 37°C for 1 h. Plates were washed three times with PBST and then incubated with 10 µl of diluted sulfo-tagged-anti-M13 mAb (6 µg/ml) at 37°C for 1 h. After another wash step, 1X Read Buffer was added and the electrochemiluminescence was measured with a MSD Sector Imager 2400 or 6000.
CDC and ADCC assays
The IGHV1-69 cognate antibodies (D80, F70, and F43) and an irrelevant control antibody (11A) were constructed into an expression vector, pHAGE.36 The scFv of antibodies were inserted between the leader peptide (LP) and the Fc region of a human IgG1 molecule. The Fc domain was linked in-frame to a short segment of extracellular domain of CD28, followed by the respective transmembrane domain and cytoplamic domain of CD28 and the incorporation motif of HIV-1 gp41, forming a sequence as LP-scFvs-CD28-gp41. A reporter gene, IRES-ZsGreen, was further constructed into the vector after the gp41. 293T cells were transfected with DNA encoding for scFvs-Fc-CD28-gp41-IRES-ZsGreen plasmids. At 48 hours post transfection, the expression of D80, F70, F43, and 11A on 293T cells were analyzed by a fluorescence microscope and flow cytometry using APC-conjugated G6 or anti-human IgG (Biolegend, San Diego, CA) antibodies. The ZsGreen positive cells were further sorted (via FACS) as target cells.
The LDH release assay was described previously.48 Briefly, D80-, F70-, F43-, and 11A-expressed 293T cells were used as target cells (4×104 cells/well) and incubated with medium containing rabbit serum complement (Cedarlane Laboratories, Hornby, Ontario, Canada) in the absence and presence of MuG6 and HuG6s. After 6 hours incubation, the supernatants were harvested and measured using non-radioactive cytotoxicity assay kits (Promega, Madison, WI) at 490 nm.
For ADCC, human PBMCs were used as effector cells and incubated with target cells (2×104 cells/well). Cells were plated into 96-well plates, incubated with antibodies at different concentrations, and then effector cells were added at an effector/target (E/T) ratio of 25:1 for 4 hours incubation at 37°C. The supernatants were harvested and detected by LDH release assay.
In vivo activity of HuG6.3 on xenogeneic B-CLL from patients
Eight-week-old NSG mice received 1×106 PBMCs from B-CLL patients and 5×106 human nature killer (NK) cells through intravenous injection. PBMCs from B-CLL patients consented as described above were isolated by Ficoll-Paque PLUS (GE Healthcare Life Sciences, Pittsburgh, PA). NK cells were isolated using EasySep™ Human NK Cell Enrichment Kit (StemCell Technologies, Vancouver, British Columbia, Canada). Mice were further treated with 10 mg/kg control IgG1, MuG6, HuG6.3, and mHuG6.3 (a mutation version of HuG6.3 with L234A and L235A mutation in Fc domain 72) intravenously. After 16 hours, mouse blood samples were harvested for further staining, including human CD45, CD19, and CD56, and then analyzed by flow cytometry.
Statistics
Data were analyzed using One-Way ANOVA or two-sided unpaired Student's t-test. The difference was considered statistically significant if P value < 0.05. “*," “**," and “***” indicate p < 0.05, 0.01 and 0.001, respectively. All values and bars are represented as mean ± standard deviation (SD).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank patients that contributed to this study for their generosity and support. JRB would like to acknowledge funding from the American Cancer Society (RSG-13-002-01-CCE) and the Leukemia Lymphoma Society (TRP#6289-13).
Funding
This work was also funded by the Defense Advanced Research Projects Agency's “7-Day Biodefense” program under contract # W911NF-10-1-0266 to WAM.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med 2005; 352:804-15; PMID:15728813; http://dx.doi.org/ 10.1056/NEJMra041720 [DOI] [PubMed] [Google Scholar]
- 2.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, et al.. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111:5446-56; PMID:18216293; http://dx.doi.org/ 10.1182/blood-2007-06-093906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Cancer Society : Cancer Facts and Figures 2015. Atlanta, Ga: American Cancer Society, 2015. [Google Scholar]
- 4.Byrd JC, Marcucci G, Parthun MR, Xiao JJ, Klisovic RB, Moran M, Lin TS, Liu S, Sklenar AR, Davis ME, et al.. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 2005; 105:959-67; PMID:15466934; http://dx.doi.org/ 10.1182/blood-2004-05-1693 [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Escobar I, Sepulveda J, Castellano D, Cortes-Funes H. Therapeutic management of chronic lymphocytic leukaemia: state of the art and future perspectives. Crit Rev Oncol Hematol 2011; 80:100-13; PMID:21146422; http://dx.doi.org/ 10.1016/j.critrevonc.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 6.Robak T. The role of nucleoside analogues in the treatment of chronic lymphocytic leukemia-lessons learned from prospective randomized trials. Leuk Lymphoma 2002; 43:537-48; PMID:12002757; http://dx.doi.org/ 10.1080/10428190290012029 [DOI] [PubMed] [Google Scholar]
- 7.Byrd JC, Lin TS, Dalton JT, Wu D, Phelps MA, Fischer B, Moran M, Blum KA, Rovin B, Brooker-McEldowney M, et al.. Flavopiridol administered using a pharmacologically derived schedule is associated with marked clinical efficacy in refractory, genetically high-risk chronic lymphocytic leukemia. Blood 2007; 109:399-404; PMID:17003373; http://dx.doi.org/ 10.1182/blood-2006-05-020735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman SE, Gordon AL, Wagner AJ, Heerema NA, Zhao W, Flynn JM, Jones J, Andritsos L, Puri KD, Lannutti BJ, et al.. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood 2010; 116:2078-88; PMID:20522708; http://dx.doi.org/ 10.1182/blood-2010-02-271171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin TS. New agents in chronic lymphocytic leukemia. Curr Hematol Malig Rep 2010; 5:29-34; PMID:20425394; http://dx.doi.org/ 10.1007/s11899-009-0039-9 [DOI] [PubMed] [Google Scholar]
- 10.Byrd JC, Murphy T, Howard RS, Lucas MS, Goodrich A, Park K, Pearson M, Waselenko JK, Ling G, Grever MR, et al.. Rituximab using a thrice weekly dosing schedule in B-cell chronic lymphocytic leukemia and small lymphocytic lymphoma demonstrates clinical activity and acceptable toxicity. J Clin Oncol 2001; 19:2153-64; PMID:11304767 [DOI] [PubMed] [Google Scholar]
- 11.Hale G, Bright S, Chumbley G, Hoang T, Metcalf D, Munro AJ, Waldmann H. Removal of T cells from bone marrow for transplantation: a monoclonal antilymphocyte antibody that fixes human complement. Blood 1983; 62:873-82; PMID:6349718 [PubMed] [Google Scholar]
- 12.Lanasa MC, Allgood SD, Slager SL, Dave SS, Love C, Marti GE, Kay NE, Hanson CA, Rabe KG, Achenbach SJ, et al.. Immunophenotypic and gene expression analysis of monoclonal B-cell lymphocytosis shows biologic characteristics associated with good prognosis CLL. Leukemia 2011; 25:1459-66; PMID:21617698; http://dx.doi.org/ 10.1038/leu.2011.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause G, Patz M, Isaeva P, Wigger M, Baki I, Vondey V, Kerwien S, Kuckertz M, Brinker R, Claasen J, et al.. Action of novel CD37 antibodies on chronic lymphocytic leukemia cells. Leukemia 2012 Mar; 26(3):546-549 [DOI] [PubMed] [Google Scholar]
- 14.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, et al.. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998; 102:1515-25; PMID:9788964; http://dx.doi.org/ 10.1172/JCI3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, et al.. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94:1840-7; PMID:10477712 [PubMed] [Google Scholar]
- 16.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94:1848-54; PMID:10477713 [PubMed] [Google Scholar]
- 17.Lanasa MC. Novel insights into the biology of CLL. Hematology Am Soc Hematol Educ Program 2010; 2010:70-6; PMID:21239773; http://dx.doi.org/9788964 10.1182/asheducation-2010.1.70 [DOI] [PubMed] [Google Scholar]
- 18.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, et al.. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998; 102:1515-25; PMID:9788964; http://dx.doi.org/ 10.1172/JCI3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messmer BT, Albesiano E, Messmer D, Chiorazzi N. The pattern and distribution of immunoglobulin VH gene mutations in chronic lymphocytic leukemia B cells are consistent with the canonical somatic hypermutation process. Blood 2004; 103:3490-5; PMID:14695232; http://dx.doi.org/ 10.1182/blood-2003-10-3407 [DOI] [PubMed] [Google Scholar]
- 20.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stella S, Guida G, Michel A, Crespo M, Laoutaris N, Montserrat E, et al.. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3-21 gene. Blood 2005; 105:1678-85; PMID:15466924; http://dx.doi.org/ 10.1182/blood-2004-07-2606 [DOI] [PubMed] [Google Scholar]
- 21.Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T, Belhoul L, Stella S, Stavroyianni N, Crespo M, et al.. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 2007; 109:259-70; PMID:16985177; http://dx.doi.org/ 10.1182/blood-2006-03-012948 [DOI] [PubMed] [Google Scholar]
- 22.Messmer BT, Albesiano E, Efremov DG, Ghiotto F, Allen SL, Kolitz J, Foa R, Damle RN, Fais F, Messmer D, et al.. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med 2004; 200:519-25; PMID:15314077; http://dx.doi.org/ 10.1084/jem.20040544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potter KN, Orchard J, Critchley E, Mockridge CI, Jose A, Stevenson FK. Features of the overexpressed V1-69 genes in the unmutated subset of chronic lymphocytic leukemia are distinct from those in the healthy elderly repertoire. Blood 2003; 101:3082-4; PMID:12480699; http://dx.doi.org/ 10.1182/blood-2002-08-2432 [DOI] [PubMed] [Google Scholar]
- 24.Widhopf GF 2nd, Kipps TJ. Normal B cells express 51p1-encoded Ig heavy chains that are distinct from those expressed by chronic lymphocytic leukemia B cells. J Immunol 2001; 166:95-102; PMID:11123281; http://dx.doi.org/ 10.4049/jimmunol.166.1.95 [DOI] [PubMed] [Google Scholar]
- 25.Darzentas N, Hadzidimitriou A, Murray F, Hatzi K, Josefsson P, Laoutaris N, Moreno C, Anagnostopoulos A, Jurlander J, Tsaftaris A, et al.. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia 2010; 24:125-32; PMID:19759557; http://dx.doi.org/ 10.1038/leu.2009.186 [DOI] [PubMed] [Google Scholar]
- 26.Widhopf GF 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood 2004; 104:2499-504; PMID:15217828; http://dx.doi.org/ 10.1182/blood-2004-03-0818 [DOI] [PubMed] [Google Scholar]
- 27.Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C, Laoutaris N, Karlsson K, Baran-Marzsak F, Tsaftaris A, et al.. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 2008; 111:1524-33; PMID:17959859; http://dx.doi.org/ 10.1182/blood-2007-07-099564 [DOI] [PubMed] [Google Scholar]
- 28.Mageed RA, Dearlove M, Goodall DM, Jefferis R. Immunogenic and antigenic epitopes of immunoglobulins. XVII–Monoclonal antibodies reactive with common and restricted idiotopes to the heavy chain of human rheumatoid factors. Rheumatol Int 1986; 6:179-83; PMID:2431452; http://dx.doi.org/ 10.1007/BF00541285 [DOI] [PubMed] [Google Scholar]
- 29.Potter KN, Li Y, Mageed RA, Jefferis R, Capra JD. Molecular characterization of the VH1-specific variable region determinants recognized by anti-idiotypic monoclonal antibodies G6 and G8. Scand J Immunol 1999; 50:14-20; PMID:10404046; http://dx.doi.org/ 10.1046/j.1365-3083.1999.00524.x [DOI] [PubMed] [Google Scholar]
- 30.Forconi F, Potter KN, Wheatley I, Darzentas N, Sozzi E, Stamatopoulos K, Mockridge CI, Packham G, Stevenson FK. The normal IGHV1-69-derived B-cell repertoire contains stereotypic patterns characteristic of unmutated CLL. Blood 2010; 115:71-7; PMID:19887677; http://dx.doi.org/ 10.1182/blood-2009-06-225813 [DOI] [PubMed] [Google Scholar]
- 31.Brezinschek HP, Brezinschek RI, Dorner T, Lipsky PE. Similar characteristics of the CDR3 of V(H)1-69/DP-10 rearrangements in normal human peripheral blood and chronic lymphocytic leukaemia B cells. Br J Haematol 1998; 102:516-21; PMID:9695967; http://dx.doi.org/ 10.1046/j.1365-2141.1998.00787.x [DOI] [PubMed] [Google Scholar]
- 32.Whitelegg NR, Rees AR. WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng 2000; 13:819-24; PMID:11239080; http://dx.doi.org/ 10.1093/protein/13.12.819 [DOI] [PubMed] [Google Scholar]
- 33.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18:2714-23; PMID:9504803; http://dx.doi.org/ 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- 34.Stocker U, van Gunsteren WF. Molecular dynamics simulation of hen egg white lysozyme: a test of the GROMOS96 force field against nuclear magnetic resonance data. Proteins 2000; 40:145-53; PMID:10813839; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 35.Daura X, Oliva B, Querol E, Aviles FX, Tapia O. On the sensitivity of MD trajectories to changes in water-protein interaction parameters: the potato carboxypeptidase inhibitor in water as a test case for the GROMOS force field. Proteins 1996; 25:89-103; PMID:8727321; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 36.Taube R, Zhu Q, Xu C, Diaz-Griffero F, Sui J, Kamau E, Dwyer M, Aird D, Marasco WA. Lentivirus display: stable expression of human antibodies on the surface of human cells and virus particles. PLoS One 2008; 3:e3181; PMID:18784843; http://dx.doi.org/ 10.1371/journal.pone.0003181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruccoleri RE, Karplus M. Prediction of the folding of short polypeptide segments by uniform conformational sampling. Biopolymers 1987; 26:137-68; PMID:3801593; http://dx.doi.org/ 10.1002/bip.360260114 [DOI] [PubMed] [Google Scholar]
- 38.Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins 1988; 4:31-47; PMID:3054871; http://dx.doi.org/ 10.1002/prot.340040106 [DOI] [PubMed] [Google Scholar]
- 39.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285-9; PMID:1350088; http://dx.doi.org/ 10.1073/pnas.89.10.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mader A, Kunert R. Humanization strategies for an anti-idiotypic antibody mimicking HIV-1 gp41. Protein Eng Des Sel 2010; 23:947-54; PMID:21037278; http://dx.doi.org/ 10.1093/protein/gzq092 [DOI] [PubMed] [Google Scholar]
- 41.Presta LG, Chen H, O'Connor SJ, Chisholm V, Meng YG, Krummen L, Winkler M, Ferrara N. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res 1997; 57:4593-9; PMID:9377574 [PubMed] [Google Scholar]
- 42.Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986; 321:522-5; PMID:3713831; http://dx.doi.org/ 10.1038/321522a0 [DOI] [PubMed] [Google Scholar]
- 43.Villani ME, Morea V, Consalvi V, Chiaraluce R, Desiderio A, Benvenuto E, Donini M. Humanization of a highly stable single-chain antibody by structure-based antigen-binding site grafting. Mol Immunol 2008; 45:2474-85; PMID:18313757; http://dx.doi.org/ 10.1016/j.molimm.2008.01.016 [DOI] [PubMed] [Google Scholar]
- 44.Haidar JN, Yuan QA, Zeng L, Snavely M, Luna X, Zhang H, Zhu W, Ludwig DL, Zhu Z. A universal combinatorial design of antibody framework to graft distinct CDR sequences: A bioinformatics approach. Proteins 2012 Mar; 80(3):896-912 http://dx.doi.org/ 10.1002/prot.23246 [DOI] [PubMed] [Google Scholar]
- 45.Hwang WY, Almagro JC, Buss TN, Tan P, Foote J. Use of human germline genes in a CDR homology-based approach to antibody humanization. Methods 2005; 36:35-42; PMID:15848073; http://dx.doi.org/ 10.1016/j.ymeth.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 46.Bertilaccio MT, Scielzo C, Simonetti G, Ten Hacken E, Apollonio B, Ghia P, Caligaris-Cappio F. Xenograft models of chronic lymphocytic leukemia: problems, pitfalls and future directions. Leukemia 2013; 27:534-40; PMID:23041721; http://dx.doi.org/ 10.1038/leu.2012.268 [DOI] [PubMed] [Google Scholar]
- 47.Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, et al.. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol 2010; 184:6986-92; PMID:20495067; http://dx.doi.org/ 10.4049/jimmunol.1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang DK, Sui J, Geng S, Muvaffak A, Bai M, Fuhlbrigge RC, Lo A, Yammanuru A, Hubbard L, Sheehan J, et al.. Humanization of an anti-CCR4 antibody that kills cutaneous T-cell lymphoma cells and abrogates suppression by T-regulatory cells. Mol Cancer Ther 2012; 11:2451-61; PMID:22869555; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han T, Abdel-Motal UM, Chang DK, Sui J, Muvaffak A, Campbell J, Zhu Q, Kupper TS, Marasco WA. Human anti-CCR4 minibody gene transfer for the treatment of cutaneous T-cell lymphoma. PLoS One 2012; 7:e44455; PMID:22973452; http://dx.doi.org/ 10.1371/journal.pone.0044455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shultz LD, Schweitzer PA, Christianson SW, Gott B, Schweitzer IB, Tennent B, McKenna S, Mobraaten L, Rajan TV, Greiner DL, et al.. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol 1995; 154:180-91; PMID:7995938 [PubMed] [Google Scholar]
- 51.Chen SS, Chiorazzi N. Murine genetically engineered and human xenograft models of chronic lymphocytic leukemia. Seminars Hematol 2014; 51:188-205; PMID:25048783; http://dx.doi.org/ 10.1053/j.seminhematol.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 52.Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R, Chum P, Yan XJ, Allen SL, Kolitz JE, et al.. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood 2011; 117:5463-72; PMID:21385850; http://dx.doi.org/ 10.1182/blood-2010-12-324210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durig J, Ebeling P, Grabellus F, Sorg UR, Mollmann M, Schutt P, Göthert J, Sellmann L, Seeber S, Flasshove M, et al.. A novel nonobese diabetic/severe combined immunodeficient xenograft model for chronic lymphocytic leukemia reflects important clinical characteristics of the disease. Cancer Res 2007; 67:8653-61; PMID:17875705; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1198 [DOI] [PubMed] [Google Scholar]
- 54.Aydin S, Grabellus F, Eisele L, Mollmann M, Hanoun M, Ebeling P, Moritz T, Carpinteiro A, Nückel H, Sak A, et al.. Investigating the role of CD38 and functionally related molecular risk factors in the CLL NOD/SCID xenograft model. Eur J Haematol 2011; 87:10-9; PMID:21692849; http://dx.doi.org/ 10.1111/j.1600-0609.2011.01626.x [DOI] [PubMed] [Google Scholar]
- 55.Xing D, Ramsay A, Decker W. Dramatic reduction of chronic lymphocytic leukemia cells following adoptive transfer of cord blood natural killer cells using cord blood-engrafted NOD/SCID IL2Rgnull mice as a model. Blood 2009; 114:936 [Google Scholar]
- 56.George AJ, Tutt AL, Stevenson FK. Anti-idiotypic mechanisms involved in suppression of a mouse B cell lymphoma, BCL1. J Immunol 1987; 138:628-34; PMID:3491853 [PubMed] [Google Scholar]
- 57.Timmerman JM, Czerwinski DK, Davis TA, Hsu FJ, Benike C, Hao ZM, Taidi B, Rajapaksa R, Caspar CB, Okada CY, et al.. Idiotype-pulsed dendritic cell vaccination for B-cell lymphoma: clinical and immune responses in 35 patients. Blood 2002; 99:1517-26; PMID:11861263; http://dx.doi.org/ 10.1182/blood.V99.5.1517 [DOI] [PubMed] [Google Scholar]
- 58.Levy R, Ganjoo KN, Leonard JP, Vose JM, Flinn IW, Ambinder RF, Connors JM, Berinstein NL, Belch AR, Bartlett NLet al. Active idiotypic vaccination versus control immunotherapy for follicular lymphoma. J Clin Oncol 2014; 32:1797-803; PMID:24799467; http://dx.doi.org/ 10.1200/JCO.2012.43.9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jilani I, O'Brien S, Manshuri T, Thomas DA, Thomazy VA, Imam M, Naeem S, Verstovsek S, Kantarjian H, Giles F, et al.. Transient down-modulation of CD20 by rituximab in patients with chronic lymphocytic leukemia. Blood 2003; 102:3514-20; PMID:12893761; http://dx.doi.org/ 10.1182/blood-2003-01-0055 [DOI] [PubMed] [Google Scholar]
- 60.Golay J, Lazzari M, Facchinetti V, Bernasconi S, Borleri G, Barbui T, Rambaldi A, Introna M. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood 2001; 98:3383-9; PMID:11719378; http://dx.doi.org/ 10.1182/blood.V98.12.3383 [DOI] [PubMed] [Google Scholar]
- 61.Pos W, Luken BM, Kremer Hovinga JA, Turenhout EA, Scheiflinger F, Dong JF, Fijnheer R, Voorberg J. VH1-69 germline encoded antibodies directed towards ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. J Thrombosis Haemostasis 2009; 7:421-8; PMID:19054323; http://dx.doi.org/ 10.1111/j.1538-7836.2008.03250.x [DOI] [PubMed] [Google Scholar]
- 62.Landau DA, Saadoun D, Calabrese LH, Cacoub P. The pathophysiology of HCV induced B-cell clonal disorders. Auto Immunity Rev 2007; 6:581-7; PMID:17854753; http://dx.doi.org/ 10.1016/j.autrev.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 63.Marasca R, Vaccari P, Luppi M, Zucchini P, Castelli I, Barozzi P, Cuoghi A, Torelli G. Immunoglobulin gene mutations and frequent use of VH1-69 and VH4-34 segments in hepatitis C virus-positive and hepatitis C virus-negative nodal marginal zone B-cell lymphoma. Am J Pathol 2001; 159:253-61; PMID:11438472; http://dx.doi.org/ 10.1016/S0002-9440(10)61691-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanovski M, Silvestri F, Pozzato G, Anand S, Mazzaro C, Burrone OR, Efremov DG. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood 1998; 91:2433-42; PMID:9516143 [PubMed] [Google Scholar]
- 65.Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, Newman RA, Hanna N, Anderson DR. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood 1994; 83:435-45; PMID:7506951 [PubMed] [Google Scholar]
- 66.Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, et al.. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A 2004; 101:2536-41; PMID:14983044; http://dx.doi.org/ 10.1073/pnas.0307140101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Long BR, Stoddart CA. Alpha interferon and HIV infection cause activation of human T cells in NSG-BLT mice. J Virol 2012; 86:3327-36; PMID:22238321; http://dx.doi.org/ 10.1128/JVI.06676-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang H, Biswas S, Tallarico AS, Sarkis PT, Geng S, Panditrao MM, Zhu Q, Marasco WA. Human B-cell ontogeny in humanized NOD/SCID gammac(null) mice generates a diverse yet auto/poly- and HIV-1-reactive antibody repertoire. Genes Immun 2012; 13:399-410; PMID:22592523; http://dx.doi.org/22044090 10.1038/gene.2012.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biswas S, Chang H, Sarkis PT, Fikrig E, Zhu Q, Marasco WA. Humoral immune responses in humanized BLT mice immunized with West Nile virus and HIV-1 envelope proteins are largely mediated via human CD5+ B cells. Immunology 2011; 134:419-33; PMID:22044090; http://dx.doi.org/ 10.1111/j.1365-2567.2011.03501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becker PD, Legrand N, van Geelen CM, Noerder M, Huntington ND, Lim A, Yasuda E, Diehl SA, Scheeren FA, Ott M, et al.. Generation of human antigen-specific monoclonal IgM antibodies using vaccinated “human immune system” mice. PLos One. 2010 Oct 4;5(10). pii. e13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayashi T, Treon SP, Hideshima T, Tai YT, Akiyama M, Richardson P, Chauhan D, Grewal IS, Anderson KC. Recombinant humanized anti-CD40 monoclonal antibody triggers autologous antibody-dependent cell-mediated cytotoxicity against multiple myeloma cells. Br J Haematol 2003; 121:592-6; PMID:12752100; http://dx.doi.org/ 10.1046/j.1365-2141.2003.04322.x [DOI] [PubMed] [Google Scholar]
- 72.Hezareh M, Hessell AJ, Jensen RC, van de Winkel JG, Parren PW. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J Virol 2001; 75:12161-8; PMID:11711607; http://dx.doi.org/ 10.1128/JVI.75.24.12161-12168.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


