Abstract
Numerous cellular functions depend on actin filament (F-actin) disassembly. The best-characterized disassembly proteins, the ADF/cofilins/twinstar, sever filaments and recycle monomers to promote actin assembly. Cofilin is also a relatively weak actin disassembler, posing questions about mechanisms of cellular F-actin destabilization. Here we uncover a key link to targeted F-actin disassembly by finding that F-actin is efficiently dismantled through a post-translational-mediated synergism between cofilin and the actin-oxidizing enzyme Mical. We find that Mical-mediated oxidation of actin improves cofilin binding to filaments, where their combined effect dramatically accelerates F-actin disassembly compared to either effector alone. This synergism is also necessary and sufficient for F-actin disassembly in vivo, magnifying the effects of both Mical and cofilin on cellular remodeling, axon guidance, and Semaphorin/Plexin repulsion. Mical and cofilin, therefore, form a Redox-dependent synergistic pair that promotes F-actin instability by rapidly dismantling F-actin and generating post-translationally modified actin that has altered assembly properties.
Multiple cellular behaviors depend on the rapid assembly and disassembly of the actin filament (F-actin) cytoskeleton1. Under cellular conditions, F-actin assembly is favored2,3, making it critical to clarify how targeted and rapid F-actin disassembly occurs. In addition, specific extracellular cues including repellents such as ephrins, slits, semaphorins, myelin-associated inhibitors, and Wnts selectively collapse F-actin networks4–6, but their direct effectors are still enigmatic. The best-known F-actin disassembly proteins, the ubiquitous ADF/cofilins, sever actin filaments and recycle monomers with a net effect of promoting new actin assembly2,3,7,8. Moreover, cofilin’s relatively weak disassembly of actin9–11 further complicates the current understanding of cellular F-actin destabilization.
Recently, we identified an unusual class of F-actin regulatory proteins, the MICALs, which are multidomain Redox enzymes that induce F-actin disassembly via the direct post-translational oxidation of actin12,13. Notably, this Mical-modified actin no longer assembles normally13,14, differentiating Mical’s effects from that of other F-actin disassembly proteins2,3. Cellular and in vivo work has also revealed that MICALs are widely-expressed in different tissues5,15–18 and control multiple cellular behaviors including motility, axon guidance, synaptogenesis, immune responses, cardiovascular integrity, muscle function, and tumorigenesis 12–14,18–25. The MICALs have also been identified as working with different growth factors, adhesion molecules, and repulsive guidance cues to exert their effects12,19,21,22,24,26,27. Yet, nothing is known of how MICALs integrate with other better-known actin regulatory proteins to direct actin cytoskeletal reorganization and cellular functions.
We now find that Mical synergizes with the ubiquitous actin regulatory protein cofilin to dramatically enhance the dismantling of actin filaments. This coupling between Mical and cofilin depends on the Redox-mediated post-translational alteration of actin. Mical oxidation of actin improves cofilin binding to filaments accelerating F-actin severing and disassembly by over an order of magnitude compared to either effector alone. This synergism also regulates F-actin disassembly in vivo and serves to remodel cells, wire the nervous system, and orchestrate Semaphorin/Plexin repulsive signaling. The Redox-dependent synergy between Mical and cofilin, therefore, rapidly disassembles F-actin and also generates oxidized actin that reassembles abnormally. This collective action has a net effect of promoting F-actin instability, revealing a previously unknown pathway of cellular F-actin disassembly.
RESULTS
Cofilin modulates Mical Redox-mediated F-actin disassembly
Mical Redox enzymes are a new type of actin regulator – one that controls filament dynamics via the direct post-translational oxidation of actin12,13. Specifically, the enzyme activity of MICALs is activated in the presence of their substrate F-actin, which triggers consumption of Mical’s coenzyme NADPH and stereospecific oxidation of actin’s methionine (M) 44 and M47 residues to induce F-actin disassembly (Fig. 1a;12–14,20,21,28–31). Mical’s characteristic consumption of NADPH in an F-actin dependent manner has thus provided a simple biochemical test for proteins that may affect Mical’s activity. We found that the well-known actin regulatory protein – cofilin7,8 – strongly suppressed the ability of F-actin to trigger Mical-mediated NADPH consumption (Figs. 1b–c; Supplementary Fig. 1a).
Figure 1. Mical/F-actin dynamics are modulated by cofilin.
(a) Mical (1) physically associates with its substrate F-actin (2), which triggers Mical’s conversion/consumption of its co-enzyme NADPH to NADP+ (3). Mical then oxidizes F-actin subunits on their M44 and M47 residues (4) triggering F-actin disassembly. For simplicity, the presence of molecular oxygen (O2) and flavin adenine dinucleotide (FAD) have been excluded from this diagram. (b–c) Mical’s enzymatic activity (as determined by conversion of NADPH to NADP+, which is measured by a change in absorbance at 340 nm [NADPH Consumption]) is markedly accelerated by F-actin, but not when cofilin is present. [Mical]=600nM, [NADPH]=200μM, actin and cofilin were used at equal molar concentrations. n=3 independent experiments per condition. Mean +/− standard error of the mean (SEM). (d) Sedimentation/Association of Mical with F-actin is not altered by the addition of cofilin. S, soluble (G-actin); P, pellet (F-actin). [Actin]=4.6 μM; [Cofilin]=4.6 μM; [Mical]=2.4 μM. No NADPH present; n=3 independent experiments per condition. Mean +/− SEM. (e–h) Pyrene–actin assays, where the fluorescence (407 nm) is higher in the polymerized state. (e) Cofilin alone (at 1:10 mole ratio to actin) has minimal effects on F-actin disassembly (pH 6.8). (f–h) Mical/NADPH-mediated F-actin disassembly (f) is rapidly accelerated by cofilin (at 1:10 molar ratio to actin) (g), resulting in a substantial increase in the change in pyrene-actin fluorescence/min (h). (i) Sedimentation of F-actin following short incubation times (3 minutes) with Mical/NADPH and/or cofilin. Sedimentation of actin shows an increase in the soluble (disassembled) actin amount following Mical/NADPH+cofilin treatment in comparison to Mical/NADPH treatment alone. For (e–i), [Actin]=2.5 μM; [Cofilin]=0.25 μM; [Mical]=10 nM; [NADPH]=100 μM. n=3 independent experiments per condition. Mean +/− SEM. See also Supplementary Fig. 7 for uncropped gels of d and i.
The ubiquitous actin depolymerizing/severing factor cofilin is known to change the conformation of the D-loop of actin32, which harbors Mical’s substrate residues M44 and M4713. These results, coupled with the observation that non-muscle human cofilin-1 is a relatively weak severer of F-actin9–11, prompted our investigation of a possible interrelation between Mical and cofilin effects on actin. In light of our NADPH consumption results (Fig. 1b–c), we first wondered if cofilin affected Mical’s ability to bind to its substrate F-actin. However, using co-sedimentation assays we did not observe any difference in the ability of Mical to associate with F-actin in the presence or absence of cofilin (Fig. 1d). Therefore, we tested if cofilin affected Mical’s ability to disassemble F-actin. Strikingly, we found that preincubation of F-actin with cofilin, which alone only minimally affects F-actin disassembly under these conditions (Fig. 1e), dramatically enhanced Mical-mediated F-actin disassembly (Fig. 1f–g). The rate of disassembly was greater than the combined rates with cofilin and Mical added individually (Fig. 1h), which was also confirmed by co-sedimentation (Fig. 1i). This cooperation was not observed in the absence of NADPH (see Supplementary Fig. 1b–e), which rules out the possibility that cofilin and Mical without its NADPH coenzyme form a complex that is more efficient in F-actin dismantling than its individual components. Thus, cofilin enhances Mical-mediated actin filament disassembly and their synergistic effect requires the NADPH-dependent Redox activity of Mical.
Cofilin synergizes with Mical to accelerate F-actin disassembly
We therefore reasoned that cofilin might enhance Mical-mediated F-actin disassembly by allowing Mical to more efficiently oxidize its M44 and M47 substrate residues on actin (and thereby consume less NADPH in the process). To test for this possibility it was important to develop an independent assay for M44/M47 actin oxidation, since NADPH consumption is not an accurate measure of Mical-mediated F-actin oxidation and occurs to some extent even in the absence of F-actin (Fig. 1c and13). We found that the enzyme subtilisin, which under limited proteolysis conditions cleaves unoxidized actin between M47 and G48 (33), does not cleave Mical-oxidized actin under such conditions (Fig. 2a, Supplementary Figs. 1f, 2a–c). Using this observation as an assay, we found that cofilin strongly decreased Mical’s rate of F-actin oxidation (Fig. 2b). Furthermore, generating antibodies that specifically recognized the wild-type (unoxidized) M44 residue of actin (Supplementary Fig. 2d) and the Mical stereospecifically oxidized M44 residue of actin (MetO-44) (Fig. 2c), allowed us to confirm that cofilin does not increase the efficiency of Mical-mediated F-actin oxidation, but actually suppresses it (Figs. 2d, Supplementary Fig. 2e). Comparison of the time courses of Mical-mediated F-actin oxidation (Fig. 2b and d) and F-actin disassembly (Fig 2e, left; and13), indicated that Mical rapidly (~ 1 min) oxidizes F-actin but it takes hundreds of seconds for Mical-oxidized actin to disassemble. Strikingly, the addition of cofilin dramatically accelerated the disassembly of Mical-oxidized actin filaments (Fig. 2e, right). Thus, Mical rapidly oxidizes but only relatively slowly disassembles filaments, and cofilin markedly accelerates this disassembly. These results are also consistent with cofilin’s suppressive effects on Mical-mediated NADPH consumption and actin oxidation (Figs. 1c, 2b and 2d), because they reveal that Mical and cofilin combine to rapidly disassemble (i.e., deplete) F-actin – which is Mical’s substrate and triggers Mical’s NADPH consumption and actin oxidation activities (Fig. 1a).
Figure 2. Cofilin slows F-actin oxidation by Mical but accelerates filament disassembly.
(a) Subtilisin digestion of actin to assess its oxidation by Mical. (Top): Schematic representation of limited proteolysis of unmodified and Mical-oxidized actin with subtilisin. (Bottom): Subtilisin cleavage occurs between residues 47 and 48 in the D-loop of actin in unmodified actin monomers (red arrowhead), but not in Mical-oxidized actin (ox-G-actin). Cleavage time (0–15 min) is indicated; n=8 preps of Mical-oxidized actin. (b) Cofilin decreases Mical-mediated oxidation of F-actin, as assayed by limited proteolysis with subtilisin. Top panel (Mical/NADPH): Mical oxidation of bare F-actin. Subtilisin cleavage of bare actin (actincleaved), which is diagnostic for unoxidized actin, was abolished within 1 min of the addition of Mical/NADPH (oxidation time) due to the accumulation of oxidized actin. Bottom panel (+Cofilin): Mical oxidation of F-actin-cofilin complex (1:1 molar ratio). Significant amounts of subtilisin-cleaved actin (unoxidized actin) were detected even 30 min after the addition of Mical/NADPH indicating that cofilin strongly suppresses Mical-mediated actin oxidation. Conditions: [Actin]=3.5μM; [Mical]=25nM; [NADPH]=100μM; [Cofilin]=3.5μM; zero time points correspond to the limited proteolysis of unoxidized (non Mical/NADPH-treated) actin using this approach. (c) Characterization of an antibody that specifically recognizes Mical-oxidized actin (actinMetO-44). This antibody recognizes Mical-treated actin but not untreated actin or Mical-treated actin following incubation with SelR, a reductase enzyme that reverses Mical-mediated actin oxidation14. SelRC124S is an enzymatically-dead version of SelR that does not reduce Mical-oxidized actin14. Specifically, 2.3μM of actin (Drosophila actin 5C) was polymerized with either 600nM Mical alone (untreated actin) or 600nM Mical/100μM NADPH (Mical-treated actin) for 1 hour at room temperature. Mical-treated actin was then incubated with 2.4μM of SelR or 2.4μM of SelRC124S and samples were subjected to SDS-PAGE and Western blotting with the actinMetO-44 antibody (see also Supplementary Fig. 2d). Similar amounts of actin (lower panel) are present in all experiments. (d) Cofilin suppresses Mical-mediated oxidation of actin, as observed using the actinMetO-44 antibody. [Actin]=1.15μM; [Cofilin]=1.15μM; [Mical]=50nM; [NADPH]=100μM. (e) Mical induces F-actin disassembly (left), while the addition of cofilin (right) rapidly accelerates Mical/NADPH-mediated F-actin disassembly. [Actin]=2.5μM; [Cofilin]=0.25μM; [Mical]=10nM; [NADPH]=100μM. See also Supplementary Fig. 7 for uncropped gels/blots of a–d.
Mical-mediated oxidation of actin weakens the mechanical properties of filaments
To more directly monitor and quantify the effect of Mical oxidation of actin and its disassembly by cofilin, we purified Mical-oxidized actin (Methods and13,14). We found that Mical-oxidized actin forms filaments, but such filaments have altered polymerization kinetics and a critical concentration of at least an order of magnitude higher than that of unmodified actin (≥1μM) (Fig. 3a–e, Supplementary Fig. 2f; see also 13,14). Specifically, purified Mical-oxidized actin did not exhibit noticeable polymerization at 1.1 μM (Fig. 3a; 13,14), but did polymerize to increasing levels when incubated at 2.2 μM, 3.3 μM and 4.4 μM (Fig. 3b–d). However, we found that polymerization of Mical-oxidized actin proceeded after a longer nucleation phase than normal and (consistent with the higher critical concentration) reached lower plateau levels than observed for unmodified actin (Fig. 3b–d). Notably, re-treating the purified Mical-oxidized actin with Mical/NADPH did not alter its polymerization properties (Fig. 3d), indicating that Mical-oxidized actin is not significantly reduced during purification and storage. Thus, above its critical concentration values, Mical-oxidized actin polymerizes but with abnormal kinetics indicative of the inhibited nucleation phase.
Figure 3. Mical-mediated oxidation of actin alters polymerization and weakens the mechanical properties of filaments.
(a–d) Purified Mical-oxidized actin can be induced to polymerize when incubated at high-enough concentrations, although with altered kinetics and extent. Pyrene-actin and cosedimentation (insets, a–b) assays show that purified Mical-oxidized actin (ox-actin) does not polymerize at 1.1μM (a; see also 13,14), but does polymerize to increasing levels when at concentrations of 2.2μM and 4.4μM (b–c). (d) Re-treating purified Mical-oxidized actin with Mical/NADPH (lower arrowhead) does not alter its polymerization state (compare with untreated actin [green curve], upper arrowhead). [Mical]=600nM; [NADPH]=100μM. Representative SDS-PAGE gels: S, soluble (G-actin); P, pellet (F-actin). (e) Critical concentration (Cc) of Mical-oxidized actin (ox-actin) is at least one order of magnitude higher than that of unoxidized actin. For ox-actin, intersects of linear plots of concentrations of pelleted F-actin versus total actin with the abscissa yielded a Cc value at pH 7 of 1.1μM±0.25 standard deviation (SD) (n=3 independent ox-actin preps) (red circles) and had similar values at pH 6.8–8. Unoxidized actin Cc was close to 0.1μM. Linear fits are shown for Mical-oxidized and unoxidized actin in zoomed inset. (f) Quantification of copolymers content using the subtilisin limited proteolysis assay reveals that polymerization of actin mixtures containing unoxidized actins and 25% and 50% of Mical-oxidized actin (ox-actin) yielded copolymers with 10.8±3.2% and 27.7±1.6% ox-actin, respectively (mean+/−SD). (g–h) Copolymers of Mical-oxidized (11%) and unoxidized actin show decreased mechanical stability compared to unmodified actin. (g) No statistically significant differences in average length of non-oxidized versus 11% Mical-oxidized F-actin were observed when filaments were assembled in flow chambers (no mixing). n=3 independent measurements of 26–129 filaments per condition per repeat; Mean+/−SEM; NS (not significant) using Student’s t-test (two-tailed). (h) In contrast, even minimal handling (gentle pipetting and mixing) decreases the average length of Mical-oxidized actin (11%) copolymers much more than unoxidized F-actin. Filament length distributions. Student’s t-test (two-tailed). P=0.0059. Mean+/−SD. n=3 independent measurements of 69–188 unoxidized actin filaments (top panel) and 107–182 11% Mical-oxidized copolymer (bottom). See Supplementary Fig. 7 for uncropped gels of a–b, f and Supplementary Table 1 for source data for e, g–h.
Further analysis of purified Mical-oxidized actin revealed that it also copolymerized with unoxidized actin monomers (Fig. 3f; see also Fig. S11C of 13). We employed subtilisin digestion to quantify the extent of Mical-oxidized actin incorporation into such copolymers (Fig. 3f). This allowed us to form and examine copolymers containing different and well-determined fractions of Mical-oxidized actin. Our results revealed that unlike unoxidized filaments, Mical-oxidized actin filaments easily fragment upon minimal handling (gentle pipetting and mixing). Even copolymers composed of low amounts of Mical-oxidized actin (11%) had a significantly lower mechanical stability than non-oxidized actin filaments (Fig. 3g–h). Therefore, Mical-oxidized actin copolymers have different mechanical properties than non-oxidized actin.
Cofilin accelerates the dismantling of Mical-oxidized actin filaments
To directly assess the effect of cofilin on the disassembly dynamics of filaments composed of Mical-oxidized actin, we polymerized purified Mical-oxidized actin and employed time-lapse TIRF microscopy. We first grew filaments composed of 100% Mical-oxidized actin from unoxidized F-actin seeds. Dramatically, such Mical-oxidized actin filaments were rapidly dismantled by the addition of cofilin within the solution exchange time (~30 s) (Fig. 4a, lower right) but not upon addition of buffer (Fig. 4a, lower middle panel). Under the same conditions, F-actin severing in the presence of Mical/NADPH or cofilin only was much weaker (see Supplementary Fig. 3a, Fig. 4a [compare upper right to lower right]). Thus, these results confirmed our observations using both pyrene-actin and actin sedimentation assays (Figs. 1e-i, 2e) and demonstrated that cofilin markedly accelerates Mical-mediated F-actin disassembly.
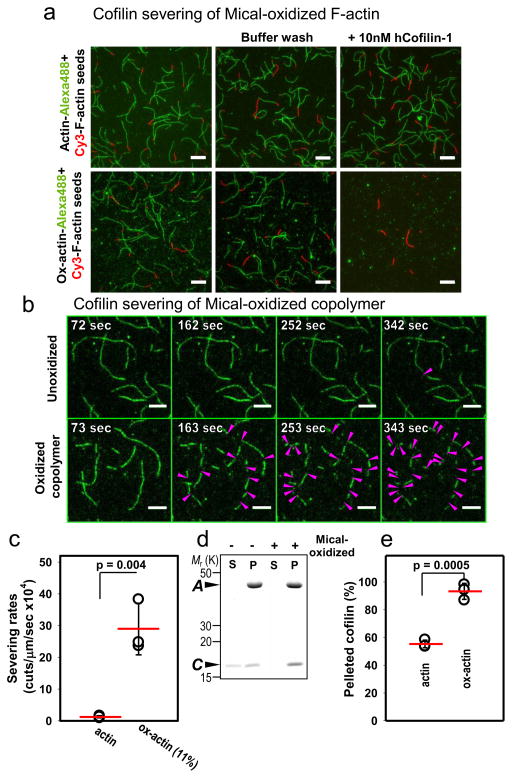
Figure 4. Mical oxidation of F-actin improves cofilin binding and results in accelerated filament severing.
(a) Rapid disassembly of fully oxidized actin by human cofilin. Unoxidized Cy3-F-actin seeds were introduced on the slide surface (red filaments, unoxidized) and extended with 100% Mical oxidized actin (ox-actin) labeled with Alexa488 (green stretches, oxidized). Addition of 10 nM human cofilin-1 (but not buffer) to such filaments resulted in full dismantling of Mical ox-actin stretches (green) within the mixing time (~30 sec), but unoxidized actin (red stretches) was not disassembled and stayed on the surface (bottom panel). No cofilin severing/fragmentation of control (unoxidized 2-colored filaments) was observed under identical conditions (top panel). Scale bar=10 μm. (b–c) Enhanced cofilin severing of Mical-oxidized actin containing filaments. (b) Severing events are indicated with magenta arrowheads. Top panel: Severing of F-actin with human cofilin-1 (100 nM) over time. Bottom panel: Severing of F-actin copolymers containing Mical-oxidized actin (11%) by human cofilin-1 (100 nM) over time. Scale bar=5 μm. (c) Quantification of cofilin severing of unoxidized F-actin and 11% Mical-oxidized (ox-actin) copolymers. Mean +/− standard deviation (SD). Number of filaments analyzed is 43–45 copolymers and 31–42 unoxidized polymers from each of 3 independent experiments (n=132 copolymers analyzed and 112 unoxidized polymers analyzed). The result of Student’s t-test (two-tailed) is shown (p=0.004). (d–e) Improved binding of cofilin to Mical-oxidized Cu2+-cross-linked Q41C F-actin, which is disassembly-resistant. (d) Representative SDS-PAGE gel of Mical-oxidized actin pelleted with cofilin. Before gel analysis disulfide cross-linking in Q41C actin was reversed with beta-mercaptoethanol. Actin (A). The bottom bands in the gel are cofilin (C). S, soluble (G-actin); P, pellet (F-actin). (e) Quantification of cofilin that co-sedimented with unoxidized and Mical-oxidized (ox-actin) Q41C cross-linked F-actin. Mean +/− SD. n=3 independent experiments per condition. The result of Student’s t-test (two-tailed) is shown (p=0.0005). Also see Supplementary Fig. 5. See also Supplementary Fig. 7 for uncropped gel of d.
Mical-mediated oxidation of actin increases cofilin’s binding and severing of filaments
We also examined the effects of partial Mical-oxidation on cofilin-mediated F-actin disassembly by employing copolymers with known amounts of Mical-oxidized actin incorporated. We found that even “lightly oxidized” F-actin copolymers (11% Mical-oxidized actin) accelerated cofilin severing by more than an order of magnitude (22-fold) compared to that of unmodified control F-actin (Fig. 4b–c, Supplementary Fig. 3e, Supplementary Movies 1–2). Increasing the content of the Mical-oxidized actin in the copolymers further accelerated cofilin severing and disassembly (Supplementary Movies S3–S4, Fig. 4a, compare upper right to lower right), and this effect was not cofilin isoform specific since we also observed it with yeast cofilin (Supplementary Fig. 3b–d). Thus, the presence of Mical-oxidized actin makes cofilin much more efficient at F-actin disassembly. Furthermore, when assisted by cofilin, partial oxidation of actin filaments by Mical is sufficient for their fast disassembly.
Additional analysis using two-color TIRF microscopy and co-sedimentation also indicated improved cofilin binding to filaments containing Mical-oxidized actin when compared to unoxidized control filaments (Fig. 4d–e, Supplementary Movies 5–6, Supplementary Figs. 4–5). In light of the extremely rapid nature of cofilin severing of Mical-oxidized F-actin, we quantified this improved cofilin binding by employing F-actin composed of either Q41C actin (yeast) or ANP-modified (skeletal) F-actin, since they both become disassembly-resistant when cross-linked between residues 41 and 374 (Supplementary Fig. 5). Using this disassembly-resistant F-actin, we found that more cofilin co-sediments with filaments containing Mical-oxidized actin in comparison to unoxidized cross-linked control filaments (Figs. 3d–e, Supplementary Fig. 5) Therefore, Mical-oxidized actin increases both cofilin binding to filaments and the rate and extent of cofilin-mediated F-actin disassembly.
Cofilin modulates Mical-mediated Redox-dependent F-actin disassembly and cellular remodeling in vivo
In view of these results, we wondered if Mical and cofilin might also work together in vivo. Both cofilin and Mical have widespread effects on the organization of actin in vivo (reviewed in 5,7,8,15–18). For instance, Mical is required to shape Drosophila bristles, which are well-characterized cells (Fig. 5a) that provide a high-resolution model to study actin organization and dynamics in vivo5,34,35. Cofilin (which is encoded by the twinstar gene in Drosophila) is also required for shaping Drosophila bristles36. Thus we employed the bristle model to assay the interaction between Mical and cofilin in vivo.
Figure 5. Cofilin enhances Mical-mediated F-actin alterations in vivo.
(a–b) Drosophila bristles are unbranched (a) but become branched as the result of F-actin disassembly and remodeling (b; arrowheads and drawings) when Mical is overexpressed specifically within them12–14. (c) Mical (red; see also12) and cofilin/twinstar (green) are both expressed in bristle processes in overlapping patterns. Note also that cofilin is more widely distributed than Mical, which shows its highest distribution at the tip of the process. Scale bars=10 μm (d) Decreasing the levels of cofilin (cofilin/twinstar heterozygote genetic background [cofilin+/−]) suppresses Mical-induced F-actin reorganization/bristle branching (arrowheads and drawings). (e) Increasing the levels of cofilin/twinstar (bristle specific expression of a hyperactive cofilinS3A transgene [cofilin+++], which has no bristle effects on its own) enhances Mical-induced F-actin reorganization/bristle branching (arrowheads). (f–g) Quantification of the data from b, d and e. n=20 bristle cells accessed across 20 animals per genotype, Mean +/− SEM, Student’s t-test (two-tailed); ***p=0.0008, ****p<0.0001. (h) Employing the genetic background described in (e), we find that mutating Mical’s substrate residue on actin, the Met-44 residue, and expressing this mutant actin in bristles (actinM44L) suppresses cofilin’s effects on Mical. Likewise, expressing SelR (SelR+++), but not an enzyme dead version of SelR (SelRC124S), suppresses cofilin’s effects on Mical. n=40 bristle cells accessed across 10 animals per genotype. Mean +/− SEM, Student’s t-test (two-tailed); ****p<0.0001. All genotypes are heterozygous (B11-GAL4/+, UAS:Mical/+, mutations/+, and/or transgenes/+). One copy of UAS:GFPactin was used when visualizing F-actin. B11-GAL4, UAS:Mical, UAS:ActinM44L, UAS:SelR, UAS:GFPactin, and UAS:SelRC124S lines were as previously described12–14. For Mical/cofilin expression analysis, we crossed UAS:mCherryMical, B11-GAL4 flies to cofilin/twinstar (tsr) GFP-trap lines and Mical/cofilin expression and localization was imaged in pupal progeny. We used the following cofilin (twinstar [tsr]) publicly available fly lines: tsrN121 (a loss-of-function/“knockout” adult lethal mutant due to P-element mediated imprecise excision in the tsr gene;48–51) and UAS:tsrS3A (a constitutively active tsr transgene;51,52). We also used the following GFP-trap tsr lines, all of which showed similar expression patterns: ZCL2393, tsrCPTI002237, and CC01393.
Elevating the levels of Mical specifically in bristle cells results in F-actin disassembly and cellular remodeling (Fig. 5b) that is dependent on Mical’s Redox activity and its M44 substrate residue within actin12–14. Notably, cofilin and Mical exhibited overlapping localization patterns within developing bristles (Fig. 5c) and removing even a single copy of cofilin (cofilin heterozygous mutants) significantly suppressed the F-actin reorganization and bristle remodeling effects that are dependent on Mical (compare Fig. 5d with Fig. 5b; Figs. 5f–g, Supplementary Fig. 6a). Moreover, raising the levels of cofilin significantly enhanced Mical-mediated effects on F-actin and cellular morphology (compare Fig. 5e with Fig. 5b; Fig. 5f–g). Further analysis revealed that cofilin’s effects on Mical-mediated F-actin reorganization in vivo were dependent on Mical’s M44 substrate residue within actin (Fig. 5h). Similarly, SelR (MsrB), which is an enzyme that reverses Mical-mediated oxidation of actin14,20, reversed cofilin’s ability to enhance Mical’s effects on F-actin reorganization (Fig. 5h). Thus, Mical-mediated F-actin alterations in vivo, as in vitro, are modulated by cofilin.
Mical and cofilin synergize to drive Semaphorin-Plexin repulsive signaling and axon guidance
In light of our in vitro and in vivo results demonstrating a synergistic action between Mical and cofilin, it is notable that Mical and cofilin exhibit widespread overlapping expression patterns18,37 and both mediate the effects of growth factors, adhesion molecules, and guidance cues on diverse cellular behaviors (reviewed in 5,7,8,15–18). For instance, Mical associates with Plexins, which are receptors for one of the largest families of guidance cues – the Semaphorins (Semas), and plays critical roles in Semaphorin/Plexin repulsive signaling (reviewed in 5). Cofilin has also been linked to Semaphorin repulsion5,38–42, but its role and mechanisms of action in this regard have remained poorly understood. Since Mical-mediated bristle actin remodeling occurs in response to Semaphorin/Plexin repulsive guidance signaling12,14, we wondered if cofilin could also be linked with Mical in mediating Semaphorin/Plexin repulsion.
To test this hypothesis, we first employed the bristle system and our genetic experiments demonstrated that cofilin was necessary for Semaphorin/Plexin/Mical-mediated effects on cellular remodeling (Supplementary Fig. 6b–c). Next, we turned to in vivo axon guidance assays using the Drosophila model nervous system, where Semaphorins-Plexins (Sema-1a and Plexin A) serve as repulsive axon guidance cues-receptors and were first functionally linked to Mical19. Notably, we found that cofilin (tsr) mutants exhibit axon guidance defects that are similar to loss of Sema-1a, Plexin A, and Mical (Fig. 6a–c, Supplementary Fig. 6d; 19,43,44). Furthermore, we observed transheterozygous genetic interactions between cofilin and Mical mutants (Fig. 6c), indicating they function in the same signaling pathway to mediate axon guidance. Moreover, we found that increasing the levels of cofilin enhanced Sema-Plexin-Mical repulsive axon guidance, while decreasing the levels of cofilin suppressed these guidance effects (Fig. 6d–f). These results further support that Mical and cofilin work together in vivo, as in vitro, and indicate that their synergistic effects are also instrumental for Semaphorin-Plexin repulsive signaling and axon guidance.
Figure 6. Cofilin enhances Sema-Plexin-Mical repulsive axon guidance.
(a) In wild-type embryos, Drosophila intersegmental nerve b (ISNb) motor axons innervate muscles 6 and 7 (filled arrowhead) and muscles 12 and 13 (open arrowhead). This normal pattern of innervation is also depicted in the drawing. (b) In a cofilin (twinstar) homozygous mutant embryo (cofilin−/−, adult-lethal genotype), note the absence (filled arrowheads) or abnormal (open arrowhead) innervation of these muscles. (c) Quantification of the data from a–b, reveals that cofilin−/− mutant embryos exhibit significant ISNb axon guidance defects (left graph). Embryos with heterozygous mutations for both cofilin and Mical (cofilin+/− and Mical+/−) also exhibit significant ISNb guidance defects in comparison to either heterozygote alone (right graph). Examination of another motor axon pathway (segmental nerve a [SNa]) revealed similar significant differences. n=100 hemisegments assessed across 10 animals per genotype, Mean +/− SEM, Chi-Square Test; ****p<0.0001. (d) Wild-type Drosophila central nervous system (CNS) axons exhibit a characteristic organizational pattern including three longitudinal connectives (1, 2, 3) composed of bundled Fasciclin II (1D4)-positive axons. Increasing the levels of PlexA in combination with Mical in neurons (Neuronal PlexA [PlexA+++] and Neuronal Mical [Mical+++]) alters the pathfinding of these longitudinal axons (e.g., arrow, arrowhead) and creates a sensitive genetic background to quantify CNS axonal pathfinding defects including discontinuous or missing 1st, 2nd, or 3rd CNS longitudinals and/or axons crossing the midline (see also 14,53–55). Increasing the levels of cofilin (+Neuronal cofilin [cofilin+++]) enhances these PlexA-Mical dependent effects, while decreasing the levels of cofilin (+cofilin+/−) suppresses these PlexA-Mical dependent effects. Scale bar applies to all images. (e–f) Quantification of the data from d. n = 480 longitudinals accessed in 160 hemisegments within 10 animals per genotype, Mean +/− SEM, Student’s t-test (two-tailed); ****p<0.0001. All genotypes are heterozygous (ELAV-GAL4/+, UAS:HAPlexA/+, UAS:Mical/+, mutations/+, and/or transgenes/+). (g) A model based on our in vitro and in vivo results that Mical and cofilin form a Redox-driven synergistic pair to negatively affect the stability of the actin cytoskeleton and direct F-actin dismantling, cellular remodeling, axon guidance, and Semaphorin-Plexin repulsion. See also main text.
DISCUSSION
Here we have found that Mical and cofilin function as a pair – synergizing in a Redox-dependent post-translational manner to disassemble F-actin and to control different cellular behaviors. Specifically, cofilin is a well-established actin regulatory protein and a relatively weak severer of F-actin9–11. In contrast, Mical family Redox enzymes have only recently emerged downstream of Semaphorin-Plexin repellents as actin disassembly factors acting via the direct post-translational oxidation of actin12–14,18–21,28–31. Previous work has also revealed that Mical, whose C-terminus associates with the intracellular portion of the Semaphorin transmembrane receptor plexin12,19, binds with its N-terminal NADPH-dependent Redox domain to F-actin and selectively oxidizes actin’s methionine-44 and 47 residues (Fig. 6g, left panel; 12–14). We propose that Mical oxidation-induced changes in filament structure and/or dynamics improve cofilin’s binding to actin filaments (Fig. 6g, middle panel). Herein, we also find that Mical-oxidized actin copolymers have different properties than unoxidized actin filaments. It is also known that the severing of actin filaments by cofilin is related to the mechanical properties of F-actin10,45,46. Our results support the idea that Mical uses oxidation to weaken the inter-actin (inter-protomer) contacts within filaments (13, present study) and these alterations dramatically speed up cofilin’s ability to break/dismantle filaments (Fig. 6g, right panel). These results, therefore, uncover a previously unknown pathway of cellular F-actin disassembly and also present an unusual type of biological synergistic interaction – one involving two different types of proteins (Mical and cofilin) and the Redox-dependent post-translational modification of a third protein (polymerized actin).
Our results also shed new light on the mechanisms of action of both Mical and cofilin. They support a model that Mical and cofilin have been evolutionarily selected to work in tandem to ensure that even a low level of Mical activity in the presence of cofilin would facilitate F-actin disassembly, and vice versa. Moreover, unlike F-actin disassembly by cofilin, which promotes actin turnover by recirculation of monomers for polymerization2,47, Mical post-translationally modifies actin, decreasing its capacity for re-polymerization until the oxidation is reversed (Fig. 6g, right panel). Thus, the Redox-driven synergy between Mical and cofilin not only rapidly disassembles F-actin but also generates post-translationally modified actin that re-assembles abnormally with a net effect of promoting F-actin instability. These results, therefore, provide important insights into how actin-based structures are rapidly and specifically dismantled in cells. Given their widespread overlapping expression patterns (reviewed in 18,37) and diverse effects on cellular behaviors (reviewed in 5,7,8,15–18), this synergistic interaction between Mical and cofilin provides the molecular framework to rapidly dismantle multiple actin-based cellular structures.
Methods
Protein purification
Drosophila MicalredoxCH construct (referred to as Mical in this study)12,56 rabbit skeletal actin57, Drosophila actin mutant M44L/M47L (13), yeast actin58, and human cofilin-1 (59) were expressed and purified as previously described. Yeast cofilin was expressed and purified essentially as described60. In brief, yeast cofilin expression was induced at OD600=0.8 with 1mM IPTG and carried-out for 4 hours at 37°C. Cell lysate was loaded on QAE-52 column equilibrated with 20mM Tris–HCl (pH7.5 at 4°C) containing 1mM DTT, 0.2mM PMSF and cofilin was eluted with linear gradient of NaCl (0–500mM) in 5 column volumes. Cofilin containing fractions were then gel-filtered on HiLoad 16/60 Superdex 75 (Amersham Biosciences) column equilibrated with 20mM Tris–HCl (pH7.5 at 4°C), 200mM NaCl, 1mM DTT, 0.2mM PMSF. Purified cofilin was stored at −80°C.
Mical-oxidized actin purification
Rabbit skeletal G-actin was polymerized at 20μM for 1 hour at room temperature (RT) (buffer composition: 5mM Tris, 0.2mM CaCl2, 0.5mM DTT, 0.2mM ATP, 2mM MgCl2, 50mM KCl, pH8). Polymerized F-actin was then diluted to 2μM and supplemented with NADPH (0.4mM) and Mical 0.2μM (10:1 molar ratio, actin to Mical, unless stated otherwise). Mical-oxidation of actin was carried out for 2 hours at RT. Oxidation efficiency under chosen conditions was confirmed by mass spectrometry. After 2 hours any residual F-actin was pelleted at 100,000g for 20 min at 4°C. Resulting supernatant containing Mical-oxidized actin (ox-actin) was dialyzed overnight into buffer G (GB2): 2mM Tris, 0.2mM CaCl2, 0.5mM DTT, 0.2mM ATP, pH 8. Actin was gel filtered using Superdex S200 16/60 column. Efficiency of oxidation was confirmed in subtilisin digestion assay (Supplementary Fig. 2a).
Mical-treated/oxidized pyrene actin was purified as described13,14. To examine repolymerization of Mical-treated actin, the purified actin was resuspended to 2.3μM in GB5, and polymerization was initiated with 2X polymerization buffer (10mM Tris-HCl pH 7.5, 0.1M KCl, 4mM MgCl2, 2mM EGTA, 0.4mM ATP, 1mM DTT) to get a final concentration of 1.15μM actin. To further test the ability of Mical-treated actin to repolymerize, the purified actin was resuspended to 4.4μM or 8.8μM in GB5, and polymerization was initiated as described above with 2X polymerization buffer (to yield a final concentration of actin at 2.2μM and 4.4μM, respectively). To determine whether Mical-oxidized actin might be reduced during its purification and storage, purified Mical-treated actin was polymerized (as described above), and then re-treated with 600nM of Mical and 200μM NADPH. Polymerization was monitored using either fluorescence or sedimentation assays (described below).
Critical concentration (Cc) determination
To determine the Cc, Mg-ATP-ox-actin was polymerized for 1 hour at RT by adding 10X polymerizing buffer, pH 6.8, 7.0, 7.5 or 8.0. Samples were diluted then into their corresponding 1X polymerizing buffer (pH 6.8 – 8.0), followed by 4°C overnight incubation. Supernatants and pellets were separated by ultracentrifugation (TLA100, 62K, 30 min, 4°C) and analyzed by SDS-PAGE. Gels were stained with Coomassie Blue and densitometry was performed using Scion Image software. The intersects of these linear plots of pelleted actin ([F-actin]) versus total actin ([Actin(total)]) with the abscissa yielded Cc in μM. The following buffers were used for Cc experiments: pH 6.8: 20mM imidazole, pH 6.8, 2mM MgCl2, 0.2mM EGTA, 50mM KCl, 0.2mM ATP, 0.5mM DTT; pH 7.0: 10mM Hepes, pH 7.0, 2mM MgCl2, 0.4mM EGTA, 50mM KCl, 0.2mM ATP, 1mM DTT; pH 7.5: 5mM Tris, pH 7.5, 2mM MgCl2, 0.2mM EGTA, 50mM KCl, 0.2mM ATP, 0.5mM DTT; pH 8.0: 10mM Tris, pH 8.0, 2mM MgCl2, 50mM KCl, 1mM EGTA, 0.2mM ATP, 0.5mM DTT.
Protein labeling
Pyrene-labeled rabbit skeletal actin (RSA) (obtained from Cytoskeleton, Inc). RSA was labeled with Cy3-maleimide in thiol-free GB5 (5mM Tris, pH 8, 0.2mM CaCl2, 0.2mM ATP) using standard approach that included 1) actin polymerization with 2mM MgCl2 and 50mM KCl for 30–60 min at RT; 2) labeling with Cy3 dye (1:1.5 (actin:dye) molar ratio for 2 hours on ice or overnight followed by addition of 1mM DTT; 3) pelleting (TLA110 rotor at 85,000 rpm for 20 min at 4°C); 4) depolymerization on dialysis followed by gel-filtration (Superdex S200 10/300 GL). Extent of labeling was calculated using extinction coefficient ε550=130,000 M−1cm−1. Actin labeling with Alexa488-succinimidyl ester (SE) (Molecular Probes) was done essentially as described61 but Alexa488SE dye was added to F-actin in 3–7 fold excess (overnight, 4°C) and then carried out as described above for Cy3 actin. Actin concentration was measured by Bradford assay or by quantitative gels (Coomassie staining) employing known concentrations of unlabeled RSA as standard. Alexa488SE-actin was used to obtain 100% oxidized labeled ox-actin (GB2, 70:1 (actin:Mical) molar ratio, 100μM NADPH, 1 hour at RT). The resulting actin was dialyzed overnight against GB2 then centrifuged (TLA100 rotor, 90,000 rpm, 30 min, 4°C). Oxidation was confirmed by limited proteolysis with subtilisin.
Yeast cofilin-KCK construct (for C-terminal labeling) was modified with Cy5-maleimide in buffer C: 5mM Tris, pH 7, 0.2mM CaCl2, 50μM TCEP (1:1.5 (cofilin:Cy5-maleimide) molar ratio, 15 min at RT). Excess dye was removed using Zeba Desalt Spin Column (Pierce) equilibrated buffer C supplemented with 1mM DTT. Extinction coefficient of Cy5 was corrected for the solvent conditions (DMF vs [buffer C+1mM DTT]) as described62 and was estimated ε643=121,420 M−1cm−1. Total concentration of labeled cofilin was measured by Bradford assay, using unlabeled WT yeast cofilin as a standard.
NADPH consumption
Different RSA concentrations (unlabeled; Cytoskeleton, Inc.) were polymerized as described13. Each polymer sample (or actin buffer only) was then preincubated with cofilin (human cofilin-1; Cytoskeleton, Inc.) or cofilin buffer (10mM Tris pH 8.0, 10mM NaCl, 5% sucrose, 1% dextran, 1mM DTT) at pH of 6.8. NADPH consumption was measured essentially as described13 with the decrease in the reduced form of NADPH determined from the decreased light absorption at 340 nm or alternatively (Supplementary Fig. 2b), by the decreased fluorescence signal at 460 nm (when excited at 340 nm).
Actin disassembly assays
Standard pyrene-actin and co-sedimentation assays using RSA (pyrene-labeled or unlabeled; Cytoskeleton, Inc.) were performed as described12–14 with minor modifications to adjust sample pH. Actin in GB5 buffer was mixed with 10X polymerization buffer (pH 6.8, 200mM Imidazole, 500mM KCl, 20mM MgCl2, 2mM EGTA) to yield 10μM actin. This mixture was then incubated on ice overnight for actin polymerization and diluted the next day to 2.5μM actin. Then, each polymer sample was incubated with cofilin (or cofilin buffer), Mical (or Mical buffer), and/or NADPH, at pH of 6.8. In some cases, as described in the figures, the polymers were preincubated with cofilin (or cofilin buffer) or Mical (or Mical buffer). For pyrene-labeled actin, the fluorescence intensity was monitored immediately and over time at 407 nm (excitation at 365 nm) by a fluorescence spectrophotometer (Spectra max M2, Molecular Devices) as described13. For co-sedimentation assays, the intensity of each of the stained bands in the pellet and soluble fraction was quantified by densitometry using Image J (NIH) 13 or Scion Image software.
Subtilisin limited proteolysis assay
Actin was polymerized at pH 6.8 for 1 hour at RT. Next, F-actin (3.5μM) was mixed with hCofilin-1 (3.5μM) or buffer (control) to form complexes. Samples were supplemented with NADPH (0.1mM). After removing unoxidized controls, reactions were started by addition of 25–50nM Mical. Aliquots of the samples were removed at selected time points and oxidation was stopped by addition of 1.5mM NADP+ and 3.5μM of Kabiramide C (KabC, marine macrolide toxin, a kind gift from Dr. Gerard Marriott)63. NADPH and KabC were also added to the control unoxidized actin/complexes. Samples were incubated overnight at 4°C for complete actin depolymerization. Then, reaction mixtures were digested with subtilisin (limited proteolysis conditions) at 1:200 subtilisin:actin w/w ratio for 20–30 min at RT. Subtilisin stock was prepared in 2mM Tris, 0.2mM CaCl2 and used within 9 min. Limited digestion was started by adding 1μl of subtilisin solution to the samples (25μl) arranged in random order and stopped with PMSF (1mM). Samples were analyzed by SDS-PAGE (Coomassie stain). Densitometry analysis was performed using Scion Image software. Increased amounts of uncleaved actin reported on the accumulation of Mical-oxidized actin. After making corrections for undercleaved actin in unoxidized controls (~5–14%), the amount of Mical-oxidized actin was plotted vs oxidation time. We elected to use subtilisin:actin ratio that yields slightly undercleaved preparation in order to restrict proteolysis to a single site (47/48) on actin. We have found that a higher ratio of subtilisin to actin is needed for limited digestion of actin samples depolymerized under F-buffer conditions. For limited digestion of G-actin/G-actin-KabC in GB (pH 8) by subtilisin, we routinely used 1:1000 (w/w) ratio of subtilisin:actin.
Using this assay we quantified the amount of Mical-oxidized actin incorporated into copolymers under conditions closely mimicking those of our TIRF experiments (pH 6.8, 1μM of total actin, 30 min polymerization at RT). After polymerization, F-actin was pelleted (TLA110 rotor, 150,000g, 30 min, 4°C). Pellets were resuspended in 100μl of GB2 and depolymerized overnight by dialysis (against GB2), followed by a second high speed spin (TLA100 rotor, 150,000g, 30 min, 4°C). Alternatively, samples were depolymerized by adding KabC (30μM) followed by a 3 hour dialysis against GB2. Limited proteolysis with subtilisin was carried out at 1:1000 subtilisin:actin w/w ratio, for 15–20 min, at RT and stopped with PMSF (1mM). The resulting samples were analyzed by SDS-PAGE (Coomassie staining). The fraction of ox-actin in the sample was determined (Fig. 3f) as described above.
Actin Met-44 and MetO-44 Specific Antibodies
We generated an antibody that preferentially recognized the unoxidized Met-44 residue of actin (Supplementary Fig. 2d1–d3). We also generated an antibody that specifically recognized the oxidized Met-44 residue on actin (MetO-44) (Fig. 2c). These antibodies were used to observe the effect of cofilin on Mical-mediated oxidation of actin, by incubating 1.15μM F-actin for 1 hour at RT with 1.15μM cofilin or cofilin buffer only. Then, 50nM Mical and 100μM NADPH were added and the reaction was stopped at 1, 3, 5, and 10 minutes (or 1 hour) by adding loading buffer containing β-mercaptoethanol and boiling samples for 5 minutes. For Western blotting, all samples were loaded into a 12% SDS-PAGE, transferred to PVDF membrane, blocked with 5% non-fat milk/TBST buffer for 1 hour and then incubated for 1 hour with antiserum (pan actin antibody [C4; Millipore, 1:1000]; Actin Met-44 and Actin MetO-44 antibodies [1:500]).
TIRF microscopy assays
Copolymers of Mical-oxidized and unmodified actin were formed in flow chambers assembled with 25×75×1 glass slides (Fisherfinest, Premium Slides, Superfrost, 12-544-7) and 22×30–1.5 glass coverslips (Fisherbrand, 12-544-A). Coverslips were treated with polylysine PEG solution (1.25mg/ml in H2O) for 4 min, rinsed 3 times with water and air-dried. Single flow chambers (V~30μl) were assembled using two layers of permanent double-sided Scotch tape. Before each experiment the flow chamber was treated with 2 chamber volumes (CV) of 1% Pluronic F127 solution (Sigma, P2443)64 for 3 min then equilibrated with 5 CV of 1xTIRF imaging buffer (20mM imidazole, 2mM MgCl2, 50mM KCl, 0.2mM EGTA (pH 6.8) supplemented with 50mM DTT, 0.2mM ATP, 0.05mg/ml caseine, 20mM glucose, 0.25mg/ml glucose oxidase, 50μM catalase, 0.5% methyl cellulose). G-actin mixtures (10% Cy3b-maleimide labeled) were incubated for 3 min at RT with Mg/EGTA exchange buffer (0.1mM EGTA, 50μM MgCl2) and the resulting mixture (3 CV) was introduced into the flow chamber. After 15 min of on-slide polymerization, the excess of actin monomers was removed with 1 CV of 1xTIRF imaging buffer. Since cofilin binds weaker to ADP-Pi-F-actin (compared to ADP-F-actin)45,65, filaments were aged on the surface to allow for phosphate release. For severing experiments with yeast actin, at least 3 fields were imaged between minutes 29 and 30 to determine the average filaments length before cofilin severing. At 30 min time point from the start of actin polymerization, cofilin/buffer (2 CV) was introduced into the flow chamber and movies of severing were recorded. Copolymers containing 11% ox-actin weren’t severed upon buffer additions within the monitoring time. Images were acquired every 5 sec. Filaments were imaged using DMI6000 TIRF microscope (Leica).
Average filament length of on-slide grown ox-actin copolymers (0 and 11% oxidized) was compared to those pre-polymerized in tubes and applied to the polyK surface (related to Fig. 3h). On-slide polymerization was carried out as described above. Images were collected after 16–17 min from the beginning of polymerization (immediately after the buffer wash, at 15 min of on-slide polymerization) (Fig. 3g). Experiments with pre-polymerized ox-actin copolymers (0% and 11% oxidized) were performed as follows. Coverslips were treated with 1mg/ml polyK for 3 min, rinsed with mQ water and air-dried. Mg-ATP-G-actin (15% Cy3b labeled) was polymerized at 10μM by 1xKMEI6.8 buffer (20mM imidazole, 2mM MgCl2, 50mM KCl, 0.2mM EGTA (pH 6.8)). F-actin samples were diluted to 8μM and mixed by pipetting up and down 2 times. The resulting mixtures were incubated 5 min at RT, followed by one step dilution into 1xKMEI6.8 buffer supplemented with 100mM DTT and 1μM phalloidin and mixing by inversion. Filaments’ length was measured manually using JFilament plugin to Fiji (JFilament 2D).
Severing of fully oxidized actin by human cofilin-1 was examined as follows (related to Fig. 3a). Flow chambers were assembled as described above. Untethered filaments were imaged on Pluronic F127-coated surface64 as described. F-actin-Cy3-maleimide (15% labeled) was polymerized in 1xKMEI6.8 buffer overnight at 4°C and used as F-actin seeds. Mical-oxidized (15% Alexa488SE) or unoxidized actin was mixed with 10nM of Cy3-F-actin seeds in 1xTIRF imaging buffer and polymerized for 20 min on slides. After 20 min, unpolymerized monomers were washed with 1xTIRF buffer. To test for severing, 10nM of human cofilin-1 in 1xTIRF buffer was introduced into the flow chambers. Filaments fragmentation induced by Mical in the presence of NADPH was monitored under the same conditions (Supplementary Fig. 3a). Unoxidized filaments (15% Alexa488SE) were grown in the flow chambers then Mical (55nM)/NADPH (100μM) were introduced into the flow chamber simultaneously washing out the remaining actin monomers.
Cofilin clustering on intact filaments and copolymers with ox-actin (11%) was imaged using two-color TIRF microscopy. Actin (Alexa488-SE, 23% labeled) and yeast cofilin (Cy5-maleimide labeled) were copolymerized in flow chambers prepared as described above and imaged, unattached, on Pluronic F127-coated surface64. Co-polymerization was started by simultaneous addition of cofilin-Cy5 and polymerizing salts to Mg-ATP-G-actin.
Analysis of cofilin severing
Fiji (Image J) software was employed for movie processing. Background subtraction was done using rolling ball radius algorithm (routinely 10 pxls). Total filaments’ length (L0) was estimated using the first frame recorded after cofilin addition (L0 (μm)=sum of the lengths of all filaments in frame #1). Filaments’ length was measured manually using JFilament plugin to Fiji (JFilament 2D). Bundled filaments were excluded from the analysis. To quantify cofilin severing of F-actin, the number of severing events (cuts) was counted manually for each frame, starting from frame #2. Cumulative number of cuts divided by L0 (cuts/μm) was plotted versus time. Linear segments of the obtained dependencies were used to determine the rates of F-actin severing by cofilin (Supplementary Fig. 3e). Between 31 and 45 filaments were analyzed in each movie.
F-actin cross-linking
Disulfide cross-linking of Q41C yeast actin mutant was carried out as follows. DTT-free Ca-ATP-Q41C actin was polymerized by adding 0.1 volume of polymerizing buffer (20mM imidazole, 50mM KCl, 2mM MgCl2) for 1 hour at RT. Disulfide cross-linking in QC-F-actin was triggered by addition of CuSO4 solution (in water) to F-actin in 1:1.5 (actin:Cu) molar ratio and carried out for 1 hour at RT. Cross-linked samples were supplemented with 1mM EGTA and dialyzed against 20mM imidazole, 50mM KCl, 2mM MgCl2, 1mM EGTA, 0.2mM ATP for ≥3 hours. Efficiency of cross-linking was confirmed by SDS-PAGE analysis under non-reducing conditions in the presence of NEM. Cross-linked and uncross-linked QC-F-actin and its cofilin complexes were subjected to Mical-mediated oxidation (140:1, molar ratio Actin:Mical, thiol-free Mical preparation) in the presence of 0.1mM NADPH under non-reducing (DTT-free) conditions for 1 hour at RT. Co-sedimentation with cofilin was performed as described above (TLA100 rotor, 150,000g, 30 min, 4°C).
N-(4-azido-2-nitrophenyl) putrescine (ANP) cross-linking was carried out as described66. In brief, thiol-free skeletal G-actin was incubated with ANP (1:8, actin:ANP molar ratio) and transglutaminase (2 units per 1mg of actin) in DTT-free GB2 (pH 8) for 2 hours at RT. Actin was centrifuged to remove any aggregates (21,000g, 10 min, 4°C) and then polymerized (20mM imidazole, pH 6.8, 50mM KCl, 2mM MgCl2, 0.2mM ATP, 100μM NADPH). Mical-mediated oxidation was started with the addition of thiol-free Mical to ANP-F-actin (1:140, Mical:actin molar ratio) for 1 hour at RT. Prior to photoactivation and cross-linking, the extent of Mical-mediated oxidation was assessed by subtilisin digestion. Cross-linking (between Gln 41-Cys 374) in Mical-oxidized and unoxidized ANP-F-actin was triggered by UV exposure (20 min at RT) and stopped with 1mM DTT. Mical-oxidized and unoxidized ANP-F-actin yielded the same cross-linking patterns. Co-sedimentation with human cofilin-1 and gel analysis was performed as described above.
In vivo data
Expression analysis, F-actin organization, and bristle cell remodeling was examined and quantified as described12,14. Embryos were collected, processed, staged, dissected, and analyzed for axon guidance defects using an antibody to Fasciclin II (1:4, 1D4 supernatant67, Developmental Studies Hybridoma Bank) as described19,44,55,68. Males and females of Drosophila embryos, pupae, and adults were used.
Statistics and reproducibility
For each representative image, gel, immunoblot, graph, movie, or in vivo experiment, the experiments were repeated at least two separate independent times and there were no limitations in repeatability. At least two independent protein purifications and multiple independent actin biochemical experiments were performed with similar results including reproducing the effects in Fig. 1b and Fig. 1g–h independently in both of our labs using non-overlapping/independent sets of reagents. No statistical method was used to predetermine the sample size, which was based on what is published in the field. Differences between experimental and control animal conditions were large, with little variability – and so the sample size was larger than needed to ensure adequate power to detect an effect. Animal studies were based on pre-established criteria to compare against age-matched animals. Animal experiments were not randomized. Animals of the correct genotype were determined and those collected of that genotype were included as data. For genetic experiments, in which the genotype needed to be determined based on different Drosophila genetic/chromosome markers, blinding was not employed. The figure legends list the sample size for each experiment. To the best of our knowledge the statistical tests are justified as appropriate. No cell lines were used in this study.
Data availability
Source data for Fig. 3e and 3g–h have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgments
We thank H. Kramer, M. Quinlan, and M. Rosen for comments, G. Marriott for the kind gift of Kabiramide C, and J. Merriam, the Bloomington Stock Center, the Kyoto Drosophila Genetic Resource Center, the Vienna Drosophila Resource Center, and FlyTrap for flies. This work was supported by US Public Health Service grants to E.R (GM077190) and J.R.T (NS073968) and a Welch Foundation grant (I-1749) to J.R.T.
Footnotes
Author contributions
E.E.G. characterized Mical/cofilin NADPH consumption, collected and analyzed all TIRF microscopy data, developed and carried out subtilisin proteolysis assays, performed and analyzed experiments with Q41C and ANP-modified actin; H.G.Y. performed and analyzed NADPH consumption assays, bulk actin disassembly, co-sedimentation and immunochemistry experiments; H.G.Y., S.K.R. and J.R.T. collected and analyzed the in vivo data; R-J.H. and J.R.T. developed the strategy and characterization of the MetO-44 and wild-type Met-44 antibodies; R-J.H., E.E.G. and H.G.Y. characterized Mical-oxidized actin; all authors designed experiments; E.E.G., H.G.Y., J.R.T., and E.R. wrote the paper; S.K.R. and R-J.H. edited the paper; J.R.T. and E.R. oversaw all aspects of the project in their respective groups.
This manuscript also contains 6 Supplementary Figures, 6 Supplementary Movies, 1 Supplementary Table, and Methods.
Authors declare no conflicts of interest.
References
- 1.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiological Reviews. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 2.Brieher W. Mechanisms of actin disassembly. Molecular Biology of the Cell. 2013;24:2299–2302. doi: 10.1091/mbc.E12-09-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rottner K, Stradal TE. Actin dynamics and turnover in cell motility. Current Opinion in Cell Biology. 2011;23:569–578. doi: 10.1016/j.ceb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Bashaw GJ, Klein R. Signaling from axon guidance receptors. Cold Spring Harbor Perspectives in Biology. 2010;2 doi: 10.1101/cshperspect.a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung RJ, Terman JR. Extracellular inhibitors, repellents, and semaphorin/plexin/MICAL-mediated actin filament disassembly. Cytoskeleton. 2011;68:415–433. doi: 10.1002/cm.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodkin AL, Tessier-Lavigne M. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein BW, Bamburg JR. ADF/Cofilin: a functional node in cell biology. Trends in Cell Biology. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bravo-Cordero JJ, Magalhaes MAO, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/Cofilin. Molecular Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 10.McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophysical Journal. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin SM, Jansen S, Goode BL. TIRF microscopy analysis of human Cof1, Cof2, and ADF effects on actin filament severing and turnover. Journal of Molecular Biology. 2016;428:1604–1616. doi: 10.1016/j.jmb.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol. 2013;15:1445–1454. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giridharan SSP, Caplan S. MICAL-family proteins: complex regulators of the actin cytoskeleton. Antioxidants & Redox Signaling. 2013;20:2059–2073. doi: 10.1089/ars.2013.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanoni MA, Vitali T, Zucchini D. MICAL, the flavoenzyme participating in cytoskeleton dynamics. Int J Mol Sci. 2013;14:6920–6959. doi: 10.3390/ijms14046920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Gunput RA, Adolfs Y, Pasterkamp RJ. MICALs in control of the cytoskeleton, exocytosis, and cell death. Cell Mol Life Sci. 2011;68:4033–4044. doi: 10.1007/s00018-011-0787-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson C, Terman JR, Gonzalez-Billault C, Ahmed G. Actin filaments - a target for redox regulation. Cytoskeleton. 2016 doi: 10.1002/cm.21315. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 20.Lee BC, et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell. 2013;51:397–404. doi: 10.1016/j.molcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundquist MR, et al. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell. 2014;156:563–576. doi: 10.1016/j.cell.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Battum EY, et al. The intracellular redox protein MICAL-1 regulates the development of hippocampal mossy fibre connections. Nat Commun. 2014;5 doi: 10.1038/ncomms5317. [DOI] [PubMed] [Google Scholar]
- 23.Beuchle D, Schwarz H, Langegger M, Koch I, Aberle H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mechanisms of Development. 2007;124:390–406. doi: 10.1016/j.mod.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Hou ST, et al. Semaphorin3A elevates vascular permeability and contributes to cerebral ischemia-induced brain damage. Sci Rep. 2015;5:7890. doi: 10.1038/srep07890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirilly D, et al. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci. 2009;12:1497–1505. doi: 10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal PK, et al. Semaphorin3a promotes advanced diabetic nephropathy. Diabetes. 2015;64:1743–1759. doi: 10.2337/db14-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt EF, Shim SO, Strittmatter SM. Release of MICAL autoinhibition by semaphorin-plexin signaling promotes interaction with collapsin response mediator protein. J Neurosci. 2008;28:2287–2297. doi: 10.1523/JNEUROSCI.5646-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald CA, Liu YY, Palfey BA. Actin stimulates reduction of the MICAL-2 monooxygenase domain. Biochemistry. 2013;52:6076–6084. doi: 10.1021/bi4008462. [DOI] [PubMed] [Google Scholar]
- 29.Zucchini D, Caprini G, Pasterkamp RJ, Tedeschi G, Vanoni MA. Kinetic and spectroscopic characterization of the putative monooxygenase domain of human MICAL-1. Archives of Biochemistry and Biophysics. 2011;515:1–13. doi: 10.1016/j.abb.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Alqassim SS, et al. Modulation of MICAL monooxygenase activity by its calponin homology domain: structural and mechanistic insights. Sci Rep. 2016;6:22176. doi: 10.1038/srep22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitali T, Maffioli E, Tedeschi G, Vanoni MA. Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and modulation by its CH, LIM and C-terminal domains. Archives of Biochemistry and Biophysics. 2016;593:24–37. doi: 10.1016/j.abb.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proceedings of the National Academy of Sciences. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwyter D, Phillips M, Reisler E. Subtilisin-cleaved actin: polymerization and interaction with myosin subfragment 1. Biochemistry. 1989;28:5889–5895. doi: 10.1021/bi00440a027. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland JD, Witke W. Molecular genetic approaches to understanding the actin cytoskeleton. Current Opinion in Cell Biology. 1999;11:142–151. doi: 10.1016/s0955-0674(99)80018-0. [DOI] [PubMed] [Google Scholar]
- 35.Tilney LG, DeRosier DJ. How to make a curved Drosophila bristle using straight actin bundles. Proc Natl Acad Sci. 2005;102:18785–18792. doi: 10.1073/pnas.0509437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, et al. Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat Cell Biol. 2001;3:204–209. doi: 10.1038/35055120. [DOI] [PubMed] [Google Scholar]
- 37.Van Troys M, et al. Ins and outs of ADF/cofilin activity and regulation. European Journal of Cell Biology. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Aizawa H, et al. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat Neurosci. 2001;4:367–373. doi: 10.1038/86011. [DOI] [PubMed] [Google Scholar]
- 39.Bribián A, et al. Sema3E/PlexinD1 regulates the migration of hem-derived Cajal-Retzius cells in developing cerebral cortex. Nat Commun. 2014;5 doi: 10.1038/ncomms5265. [DOI] [PubMed] [Google Scholar]
- 40.Hu H, Marton TF, Goodman CS. Plexin B mediates axon guidance in Drosophila by simultaneously inhibiting active Rac and enhancing RhoA signaling. Neuron. 2001;32:39–51. doi: 10.1016/s0896-6273(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 41.Myster F, et al. Viral semaphorin inhibits dendritic cell phagocytosis and migration but is not essential for gammaherpesvirus-induced lymphoproliferation in malignant catarrhal fever. J Virol. 2015;89:3630–3647. doi: 10.1128/JVI.03634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witherden DA, et al. The CD100 receptor interacts with its plexin B2 ligand to regulate epidermal γδ T cell function. Immunity. 2012;37:314–325. doi: 10.1016/j.immuni.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winberg ML, et al. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 1998;95:903–916. doi: 10.1016/s0092-8674(00)81715-8. [DOI] [PubMed] [Google Scholar]
- 44.Yu HH, Araj HH, Ralls SA, Kolodkin AL. The transmembrane Semaphorin Sema I is required in Drosophila for embryonic motor and CNS axon guidance. Neuron. 1998;20:207–220. doi: 10.1016/s0896-6273(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 45.Suarez C, et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Current Biology. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngo KX, Kodera N, Katayama E, Ando T, Uyeda TQP. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife. 2015;4:e04806. doi: 10.7554/eLife.04806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. The Journal of Cell Biology. 2007;177:465–476. doi: 10.1083/jcb.200610005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren N, Charlton J, Adler PN. The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-Actin disassembly in pupal epidermal cells. Genetics. 2007;176:2223–2234. doi: 10.1534/genetics.107.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson RI, Seppa MJ, Cagan RL. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. The Journal of Cell Biology. 2008;180:1191–1204. doi: 10.1083/jcb.200706108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schottenfeld-Roames J, Rosa J, Ghabrial A. Seamless tube shape is constrained by endocytosis-dependent regulation of active Moesin. Current Biology. 2014;24:1756–1764. doi: 10.1016/j.cub.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng J, Luo L. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 2004;44:779–793. doi: 10.1016/j.neuron.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 52.Stephan D, et al. Drosophila Psidin regulates olfactory neuron number and axon targeting through two distinct molecular mechanisms. The Journal of Neuroscience. 2012;32:16080–16094. doi: 10.1523/JNEUROSCI.3116-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayoob JC, Yu HH, Terman JR, Kolodkin AL. The Drosophila receptor guanylyl cyclase Gyc76C is required for semaphorin-1a-plexin A-mediated axonal repulsion. The Journal of Neuroscience. 2004;24:6639–6649. doi: 10.1523/JNEUROSCI.1104-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He H, Yang T, Terman JR, Zhang X. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc Natl Acad Sci U S A. 2009;106:15610–15615. doi: 10.1073/pnas.0906923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang T, Terman JR. 14-3-3ε couples protein kinase A to semaphorin signaling and silences plexin RasGAP-mediated axonal repulsion. Neuron. 2012;74:108–121. doi: 10.1016/j.neuron.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, Hung RJ, Terman JR. A simple and efficient method for generating high-quality recombinant Mical enzyme for in vitro assays. Protein Expression and Purification. doi: 10.1016/j.pep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spudich JA, Watt S. The Regulation of Rabbit Skeletal Muscle Contraction. I Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 58.Grintsevich EE, et al. Mapping the cofilin binding site on yeast G-actin by chemical cross-linking. Journal of Molecular Biology. 2008;377:395–409. doi: 10.1016/j.jmb.2007.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grintsevich EE, Reisler E. Drebrin inhibits cofilin-induced severing of F-actin. Cytoskeleton. 2014;71:472–483. doi: 10.1002/cm.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bobkov AA, et al. Structural effects of cofilin on longitudinal contacts in F-actin. Journal of Molecular Biology. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 61.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grintsevich EE, et al. Antiparallel dimer and actin assembly. Biochemistry. 2010;49:3919–3927. doi: 10.1021/bi1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klenchin VA, et al. Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin. Nat Struct Mol Biol. 2003;10:1058–1063. doi: 10.1038/nsb1006. [DOI] [PubMed] [Google Scholar]
- 64.Gurel PS, et al. INF2-mediated severing through actin filament encirclement and disruption. Curr Biol. 2014;24:156–164. doi: 10.1016/j.cub.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muhlrad A, Pavlov D, Peyser YM, Reisler E. Inorganic phosphate regulates the binding of cofilin to actin filaments. FEBS Journal. 2006;273:1488–1496. doi: 10.1111/j.1742-4658.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- 66.Kim E, Bobkova E, Hegyi Gr, Muhlrad A, Reisler E. Actin cross-linking and inhibition of the actomyosin motor. Biochemistry. 2002;41:86–93. doi: 10.1021/bi0113824. [DOI] [PubMed] [Google Scholar]
- 67.Van Vactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- 68.Huang Z, Yazdani U, Thompson-Peer KL, Kolodkin AL, Terman JR. Crk-associated substrate (Cas) signaling protein functions with integrins to specify axon guidance during development. Development. 2007;134:2337–2347. doi: 10.1242/dev.004242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Fig. 3e and 3g–h have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.