Abstract
Pollen–stigma interactions are essential for pollen germination. The highly regulated process of pollen germination includes pollen adhesion, hydration, and germination on the stigma. However, the internal signaling of pollen that regulates pollen–stigma interactions is poorly understood. KINβγ is a plant-specific subunit of the SNF1-related protein kinase 1 complex which plays important roles in the regulation of plant development. Here, we showed that KINβγ was a cytoplasm- and nucleus-localized protein in the vegetative cells of pollen grains in Arabidopsis. The pollen of the Arabidopsis kinβγ mutant could not germinate on stigma, although it germinated normally in vitro. Further analysis revealed the hydration of kinβγ mutant pollen on the stigma was compromised. However, adding water to the stigma promoted the germination of the mutant pollen in vivo, suggesting that the compromised hydration of the mutant pollen led to its defective germination. In kinβγ mutant pollen, the structure of the mitochondria and peroxisomes was destroyed, and their numbers were significantly reduced compared with those in the wild type. Furthermore, we found that the kinβγ mutant exhibited reduced levels of reactive oxygen species (ROS) in pollen. The addition of H2O2 in vitro partially compensated for the reduced water absorption of the mutant pollen, and reducing ROS levels in pollen by overexpressing Arabidopsis CATALASE 3 resulted in compromised hydration of pollen on the stigma. These results indicate that Arabidopsis KINβγ is critical for the regulation of ROS levels by mediating the biogenesis of mitochondria and peroxisomes in pollen, which is required for pollen–stigma interactions during pollination.
Author Summary
After landing on the stigma, pollen grains germinate and create pollen tubes following adhesion and hydration processes, during which pollen–stigma interactions determine whether the pollen grains can germinate on the stigma. In recent years, the interaction mechanisms between the pollen and stigma have been studied extensively at the cellular and molecular level in self-incompatibility systems. However, few studies have focused on pollen–stigma interactions during self-compatible pollination. Arabidopsis thaliana provides an excellent system to study the interaction mechanisms between the pollen and stigma during self-compatible pollination. KINβγ is a plant-specific subunit of the SNF1-related protein kinase 1 complex. In this study, we identified an Arabidopsis kinβγ mutant showing defective pollen germination on the surface of the stigma but not on the culture medium, which resulted from the compromised hydration of pollen on the stigma. Further analysis revealed that the biogenesis of mitochondria and peroxisomes was impaired in this mutant, which reduced the levels of reactive oxygen species (ROS) in pollen. Application of H2O2 recovered the capability of pollen to undergo hydration in vitro. These results suggest that ROS signaling is involved in the regulation of pollen–stigma interactions during pollination. This study provides new insights into the mechanism underlying pollen–stigma interactions in self-compatible plant species.
Introduction
In flowering plants, pollen tube growth provides a route for the delivery of nonmotile sperm to the female gamete. The growth of the pollen tube begins during the germination of the pollen grain after it lands on a receptive stigma via pollination. Pollen–stigma interactions are crucial for determining the germination of compatible pollen on the stigma [1, 2]. During the interactions between the pollen and stigma, a series of events occur in succession, including pollen adhesion onto the stigma, pollen hydration and germination, and the polarized growth of pollen tubes across stigmatic tissue. In recent years, the interactions between the pollen and stigma have been studied in a number of plant species [3, 4]. However, determining the mechanisms underlying these interactions remains a major challenge.
The stigmatic exudates at the stigma surface of wet stigma species provide the basis for pollen capture, adhesion, and hydration, which are considered passive, unregulated processes in these plant species. By contrast, there are no stigmatic exudates on the stigma surface in dry stigma species, such as Arabidopsis, maize, and Brassica. The pollen wall, which is thought to be important for pollen adhesion, hydration, and germination at the stigma surface during pollination [2], is involved in the formation of the pollen–stigma interface and promotes pollen adhesion and water flow from the stigma to the pollen grain. For example, the exine and exinic outer layer of pollen function in the initial adhesion of pollen to dry stigmas [5, 6]. Following exine-mediated adhesion, pollen coat (“foot”) formation between the pollen wall and stigma surface might further contribute to adhesion [1]. However, studies of the Arabidopsis eceriferum6 (cer6) mutant revealed that the pollen coat is not essential for initial adhesion, but it may contribute to the magnitude of adhesion after hydration [6, 7]. Some studies suggest that the pollen coat is critical for pollen hydration and germination on the stigma. For example, the pollenkitt of sunflower pollen mediates pollen adhesion on the hydrophilic surface in a humidity-dependent manner [8]. Arabidopsis cer mutants (e.g., cer1, cer3, cer6) have defective pollen coats, which results from the lack of a class of long-chain lipids; these mutants demonstrate defective pollen hydration at wild-type stigma and cannot give rise to pollen tubes, thereby leading to male sterility [9].
Various proteins have been identified in the pollen coat and apoplast of Arabidopsis and maize [10–12], some of which are important for pollen–stigma interactions during pollination. For example, the oleosin-domain protein GRP17 and the lipase EXL4 are Arabidopsis pollen coat proteins that function in pollen hydration on the stigma [13, 14]. In maize, xylanase released from the pollen coat after pollination facilitates pollen tube penetration into the silk via enzymatic xylan hydrolysis [15]. These findings demonstrate that the molecules located at the surface of the pollen grain are critical for the interactions between pollen and the stigma during pollination; by contrast, there is little evidence supporting the role of signaling from the interior of pollen grains in self-compatible pollination. Recently, an Arabidopsis dayu mutant was identified, which has impaired pollen germination in vivo and reduced jasmonic acid levels in pollen [16]. DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a regulator of peroxisome biogenesis in pollen and is expressed in pollen vegetative cells, indicating that jasmonic acid signaling from the interior of pollen grains regulates pollen germination on the stigma in Arabidopsis.
The yeast sucrose nonfermenting 1 (SNF1)/mammal AMP-activated kinase (AMPK)/plant Snf1-related kinase1 (SnRK1)-related protein kinase complex belongs to a family of highly conserved heterotrimeric serine/threonine kinase complexes. This complex can be activated when ATP production decreases relative to increases in AMP or ADP levels in cells that are involved in cellular responses to various nutritional and environmental stresses [17–19]. The SNF1/AMPK/SnRK1 complex is composed of catalytic α subunits and two regulatory noncatalytic subunits: β and γ [20]. The β subunit functions in determining the subcellular localization of the complex and recognition between the kinase and its targets, acting as a scaffold to keep the α and γ subunits together; the γ subunit is involved in the regulation of SNF1/AMPK/SnRK1 kinase activity. The α subunit of Arabidopsis SnRK1 complex, is encoded by KIN10, KIN11, and KIN12, but only the expression of KIN10 and KIN11 has been detected in Arabidopsis [21, 22]. Recently, Arabidopsis SnRK1 was identified as an atypical member of the SNF1/AMPK/SnRK1 family due to its insensitivity to AMP and ADP, resistance to T-loop dephosphorylation, and inability to bind to starch or similar oligo/polysaccharides [23]. Genetic analyses revealed that SnRK1 plays roles in the regulation of seedling, embryo, pollen, and lateral organ development and is also involved in sugar, stress, and hormonal signaling [21, 24–29]. Three classes of putative γ subunits of the SnRK1 complex, i.e., KINγ, KINβγ/HOMOLOG OF YEAST SUCROSE NONFERMENTING 4 (SNF4), and PV42-type proteins, have been identified in plants [30, 31]. Arabidopsis AtPV42a and AtPV42b belong to the PV42 class of γ subunits. Artificial microRNA (amiRNA)-mediated knockdown mutants of both atpv42a and atpv42b exhibit defects in late stamen development and pollen tube growth to the female gametophyte [31]. KINβγ is an atypical γ subunit since it contains a glycogen-binding domain/kinase interaction sequence (GBD/KIS) in its N-terminal region that is normally found in canonical β subunits besides four cystathionine-β-synthase (CBS) domains in its C-terminal region and a pre-CBS domain located between the four CBSs and GBD domains [32]. Arabidopsis KINβγ is a functional homolog of the yeast γ subunit SNF4 and is assembled into a plant-specific SnRK1 complex by interacting with both the α and β subunits [23, 33–36]. As a unique regulatory subunit of the SnRK1 complex, KINβγ might have some plant-specific functions. KINβγ can form an active complex with the α-catalytic KIN10 and KIN11 subunits [37] and may be involved in plant–pathogen interactions by interacting with two leucine-rich repeat proteins [34]. Consistent with the lethal phenotypes of the kin10,11 double mutant in Arabidopsis, no homozygous kinβγ knockout mutant could be isolated [21, 32], thereby indicating that KINβγ is critical to plant development.
In this study, we isolated heterozygous kinβγ knockout mutants in Arabidopsis and found that the pollen of these mutants exhibited abnormal biogenesis of mitochondria and peroxisomes and reduced reactive oxygen species (ROS) levels. Pollen grains of the kinβγ mutants germinated in vitro but not on the surface of the stigma, which resulted from the reduced ROS levels in the mutant pollen. This study provides new insights into the mechanism underlying the interactions between the pollen and the stigma.
Results
Arabidopsis kinβγ mutant exhibits defective pollen
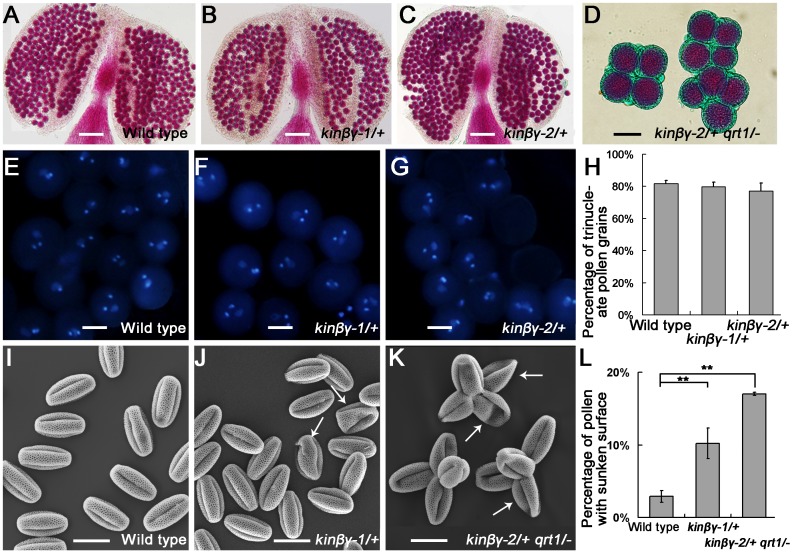
In a previous study, homozygous kinβγ knockout mutant plants could not be identified from the progenies of kinβγ mutants in Arabidopsis [32]. To analyze the reason for the lack of kinβγ homozygous mutants, we obtained two KINβγ T-DNA insertion mutant lines, GK-346E09 and SALK_074210, from the European Arabidopsis Stock Centre and the Arabidopsis Biological Resource Center, respectively. Although no homozygous T-DNA insertion mutant plants were identified from the progenies of these mutant plants according to PCR-based genotype identification, two heterozygous mutant progenies from GK-346E09 and SALK_074210 were identified and designated as kinβγ-1/+ and kinβγ-2/+, respectively (S1 Fig). Segregation analysis of the selfed progenies of these two mutants revealed that the ratios of wild-type/heterozygous plants were 1:1 (S1 Table), indicating that the T-DNA insertions in KINβγ led to gametophytic defects. Analysis of reciprocal crosses between the wild-type plants and the heterozygous mutants showed that there were significant decreases in the transmission of the mutant allele via the male gametophytes (S2 Table). The Arabidopsis quartet1 (qrt1) mutant contains pollen tetrads instead of separate pollen grains, which is convenient for the identification of pollen phenotypes resulted from a gametophytic mutation [38]. To make a comparison of the pollen grains between the wild type and kinβγ mutant in the following analysis, we introduced qrt1 into kinβγ/+ mutant plants. The viability and nuclei of mature pollen were examined in the heterozygous kinβγ-1/+ and kinβγ-2/+ mutants using Alexander staining and 4',6-diamidino-2-phenylindole (DAPI)-labeling procedures, respectively. All of the mature pollen grains appeared red after Alexander staining, and most of them harbored three nuclei (Fig 1A–1H). Next, we observed the morphology of the mature pollen grains by scanning electron microscopy (SEM). There were no detectable differences in pollen size or sculpturing patterns in the exine between the mutants and wild type, but approximately 10.2% and 17.0% of grains in kinβγ-1/+ and kinβγ-2/+ qrt1/- had sunken surfaces, respectively. By contrast, only 2.9% of grains with sunken surfaces were identified in the wild type (Fig 1I–1L).
Fig 1. Viability and morphology of mature kinβγ/+ pollen grains.
(A–D) Alexander staining revealing the viability of mature pollen of the wild type (A), kinβγ-1/+ (B), kinβγ-2/+ (C), and kinβγ-2/+ qrt1/- (D). (E–G) DAPI labeling of the nuclei in mature pollen grains of the wild type (E), kinβγ-1/+ (F), and kinβγ-2/+ (G). (H) Statistical analysis of the trinucleate pollen grains of the wild type (E), kinβγ-1/+ (F), and kinβγ-2/+ (G). (I–K) SEM analysis of the morphology of mature pollen grains of the wild type (I), kinβγ-1/+ (J), and kinβγ-2/+ qrt1/- (K). Arrows indicate the pollen grains with sunken surfaces. (L) Statistical analysis of pollen grains with sunken surfaces. Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). Bars, 100 μm in (A–C) and 20 μm in (D–G) and (I–K).
To confirm that the kinβγ mutation led to pollen defects, we performed a complementary analysis using the SNF4-YFP transgenic line, in which the genomic sequence of KINβγ (including the promoter and coding sequences) was fused with the yellow fluorescent protein (YFP) sequence and transformed into wild-type Arabidopsis [36]. We obtained SNF4-YFP/+ kinβγ-1/+ plants by crossing the kinβγ-1/+ plant with the SNF4-YFP transgenic plant. We determined the ratios of plants with and without T-DNA insertions (T-DNA+ and T-DNA-, respectively) in the offspring plants of both kinβγ-1/+ and SNF4-YFP/+ kinβγ-1/+. The T-DNA+ to T-DNA- ratio was 0.81:1 (93:115) in the offspring of the kinβγ-1/+ plant, whereas it was 2.70:1 (165:61) in the offspring of the SNF4-YFP/+ kinβγ-1/+ plant. Next, we observed the pollen morphology of SNF4-YFP/+ kinβγ-1/- and wild-type plants. The majority of pollen grains exhibited the normal morphology of the wild type (S2 Fig). The percentage of pollen grains with a sunken surface decreased to 5.2% in SNF4-YFP/+ kinβγ-1/-; by contrast, the percentage was approximately 10.2% in kinβγ-1/+ pollen grains. These results confirm that the defective phenotype of kinβγ/+ pollen resulted from mutation of the KINβγ gene.
Localization of KINβγ in pollen grains and pollen tubes
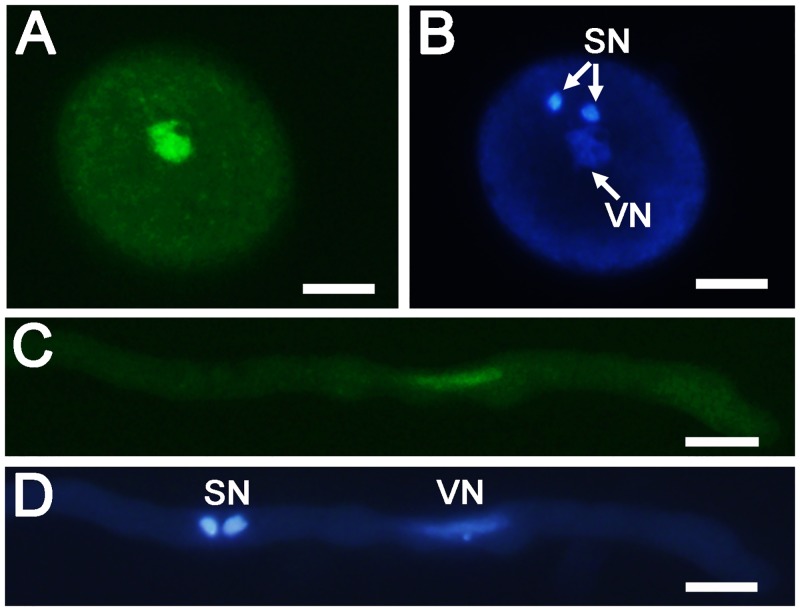
Arabidopsis KINβγ is a pollen-expressed protein [36]. To obtain detailed information about Arabidopsis KINβγ expression and its localization in pollen grains and pollen tubes, we observed YFP signals in the pollen grains and pollen tubes of the SNF4-YFP transgenic line. In mature pollen grains and pollen tubes, diffuse SNF4-YFP signals were distributed in the cytoplasm and concentrated signals were located in a nucleus-like structure (Fig 2A and 2C), which is consistent with the previous observation [36]. To confirm the localization of KINβγ in pollen grains and pollen tubes, we stained these structures with DAPI and found that KINβγ was mainly localized to the vegetative nuclei, in addition to the cytoplasm of vegetative cells, in pollen grain and pollen tube (Fig 2A–2D).
Fig 2. Localization of KINβγ in pollen and pollen tube.
(A) and (B) Localization of KINβγ in the mature pollen grain expressing SNF4-YFP. (B) The pollen grain in (A) labeled with DAPI to visualize the nucleus. (C) and (D) Localization of KINβγ in pollen tube expressing SNF4-YFP. (D) The pollen tube in (C) labeled with DAPI to visualize the nucleus. SN, sperm nuclei; VN, vegetative nucleus. Bars, 10 μm
Biogenesis and ultrastructure of mitochondria and peroxisomes are impaired in kinβγ mutant pollen
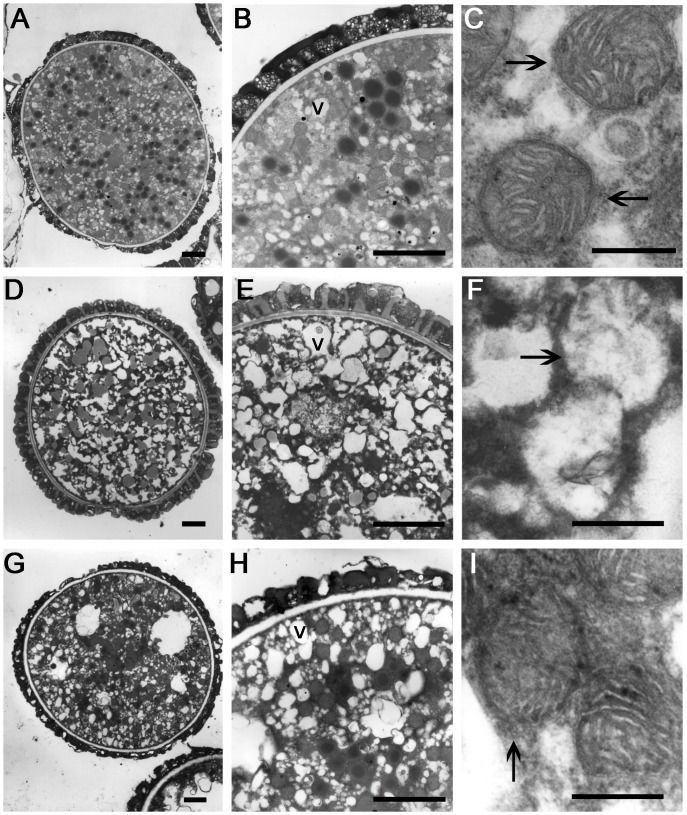
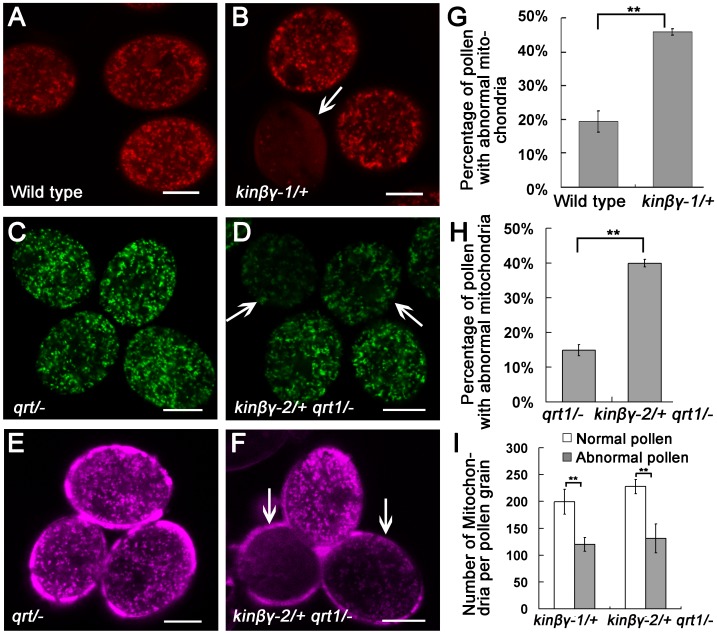
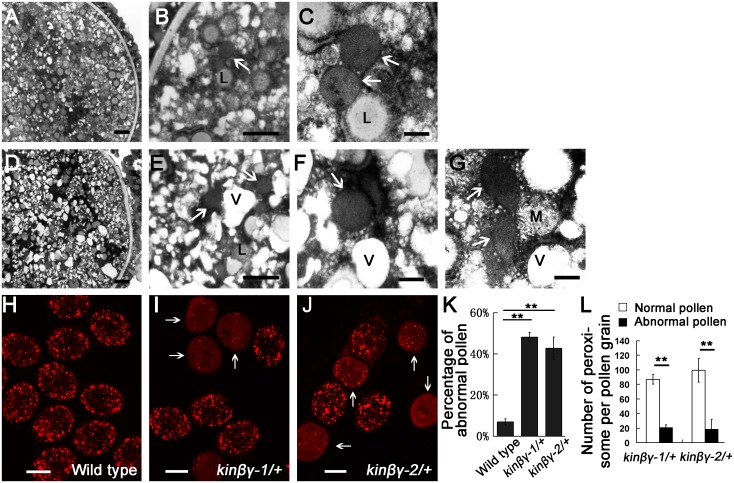
To investigate the reason for the defective transmission of the kinβγ alleles through pollen, we observed the ultrastructure of pollen grains of the kinβγ/+ mutants. Transmission electron microscopy (TEM) did not reveal any differences in pollen coat, exine, or intine morphology between wild-type and kinβγ/+ mutant pollen grains (S3 Fig). However, many enlarged vesicles were found in the cytoplasm of some mutant pollen grains (approximately 47.4% in kinβγ-1/+ and 50.7% in kinβγ-2/+ qrt1/-); by contrast, many small vesicles were detected in the cytoplasm of mature wild-type pollen grains (Fig 3A, 3B, 3D, 3E, 3G and 3H). Interestingly, the mitochondria in wild-type pollen grains contained an electron-dense matrix and well-developed cristae inside the outer membrane (Fig 3C); however, mutant pollen grains with enlarged vesicles harbored mitochondria with incomplete outer membranes and disorganized cristae (Fig 3F and 3I). We then labeled the mitochondria in the pollen grains using two strategies: expressing mitochondria-targeted Vegetative Cell GFP/RFP (VC-mtGFP/RFP) [39] and staining the pollen grains with MitoTracker Deep Red. A number of kinβγ-1/+ and kinβγ-2/+ qrt1/- pollen grains had reduced number of mitochondria (Fig 4A–4F). Approximately 19.4% and 46.0% of pollen grains harbored abnormal mitochondria in wild-type and kinβγ-1/+ plants expressing VC-mtRFP compared with 14.9% and 40.0% in qrt1/- and kinβγ-2/+ qrt1/- plants expressing VC-mtGFP, respectively (Fig 4G and 4H). Moreover, diffuse VC-mtG(R)FP signals were detected in the cytosol of these abnormal pollen grains, or the mitochondria exhibited weaker fluorescence after labeling with MitoTracker Deep Red (Fig 4B, 4D and 4F). Furthermore, we determined the number of mitochondria in mutant pollen grains labeled with VC-mtG(R)FP by counting the mitochondria in a single central optical section via laser scanning confocal microscopy (LSCM). As shown in Fig 4I, there were approximately 120 and 131 mitochondria in the abnormal pollen grains of the kinβγ-1/+ and kinβγ-2/+ qrt1/- mutants, respectively; by contrast, there were approximately 200 and 228 mitochondria in the normal pollen grains of the kinβγ-1/+ and kinβγ-2/+ qrt1/- mutants, respectively.
Fig 3. Vesicles and mitochondria in kinβγ/+ pollen.
(A-C) Wild-type mature pollen grain. (B) and (C) The magnification of the pollen grain in (A) showing the vesicles (B) and mitochondria (C, arrows), respectively. (D–F) Mature pollen grain of kinβγ-1/+ with enlarged vesicles. (E) and (F) The magnification of the pollen grain in (D) showing the enlarged vesicles (E) and abnormal mitochondria (F, arrow). (G–I) Mature pollen grain of kinβγ-2/+ qrt1/- with enlarged vesicles. (H) and (I) The magnification of the pollen grain in (G) showing the enlarged vesicles (H) and abnormal mitochondria (I, arrow), respectively. V, vesicle; Bars, 5 μm in (A, B, D, E, G, H) and 0.2 μm in (C, F, I).
Fig 4. Fluorescence labeling analysis of pollen mitochondria in the kinβγ/+ mutant.
(A) and (B) VC-mtRFP labeling of mitochondria in the pollen of the wild type (A) and kinβγ-1/+ (B), respectively. Arrow in (B) indicates a pollen grain with few mitochondria and the diffuse localization of VC-mtRFP in the cytoplasm. (C) and (D) VC-mtGFP labeling of the pollen mitochondria of qrt1/- (C) and kinβγ-2/+ qrt1/- (D), respectively. Arrows in (D) indicate pollen grains with few mitochondria and diffuse localization of VC-mtGFP in the cytoplasm. (E) and (F) MitoTracker Deep Red labeling of mitochondria in the pollen of qrt1/- (E) and kinβγ-2/+ qrt1/- (F), respectively. Arrows in (F) indicate mutant pollen grains with few mitochondria. (G) and (H) Percentages of pollen grains with abnormal mitochondria in the wild type, kinβγ-1/+ (G), and kinβγ-2/+ (H). Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). (I) Average numbers of mitochondria in kinβγ/+ pollen grains with normal and abnormal mitochondria. Only the mitochondria in the central optical sections of pollen grains observed under LSCM were counted for analysis. Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). Bars, 10 μm.
Since KINβγ and the α-catalytic KIN10 and KIN11 subunits can form an active complex [37], we determined whether the defective mitochondrion phenotype was related to the KINα subunit by employing an amiRNA strategy to reduce KIN10 and KIN11 expression in pollen [40]. We generated a late anther tomato gene52 (Lat52)::amiRNA-KIN10,11 construct and transferred it into Arabidopsis. Twenty-seven transgenic lines were obtained, four of which showed different expression levels of KIN10,11 were chosen for further analysis, including examination of pollen viability, morphology, and mitochondria. Green pollen grains were identified in the transgenic lines after Alexander staining, indicating that the knockdown of KIN10 and KIN11 resulted in some pollen abortion (S4A Fig). Under SEM, some pollen grains of the transgenic lines had sunken surfaces, which were similar to those of the kinβγ mutant (S4B Fig). Furthermore, there were fewer mitochondria in some pollen grains of the transgenic lines, as revealed by MitoTracker Deep Red labeling (S4C Fig), which was also observed in the kinβγ mutant. The similar pollen phenotypes in both these lines suggest that the KINβγ works in coordination with KIN10 and KIN11 in SnRK1 complex for the regulation of Arabidopsis pollen development and mitochondrion biogenesis.
The mitochondria and peroxisomes are metabolically linked and share components of division machinery [41]. Thus, we examined the ultrastructure and the number of peroxisomes in kinβγ/+ pollen grains using 3,3'-diaminobenzidine tetrahydrochloride and fluorescent protein labeling, respectively. In the mutant pollen grains, some peroxisomes lacked an obvious membrane or exhibited irregular morphology compared with the round peroxisomes with single-layered membranes observed in wild-type pollen grains (Fig 5A–5G). We then labeled the peroxisomes in the pollen grains using the peroxisome-targeted marker mCherry-PTS1 [16]. In the kinβγ/+ mutant, many pollen grains harbored few peroxisomes and exhibited diffuse, red fluorescent mCherry-PTS1 signals in the cytoplasm (Fig 5H–5J). Approximately 48.0% of the pollen grains in kinβγ-1/+ and 43.7% in kinβγ-2/+ exhibited abnormal fluorescence patterns in their peroxisomes; by contrast, only 6.8% of wild-type pollen produced abnormal mCherry-PTS1 signals (Fig 5K). In the abnormal pollen grains of the kinβγ-1/+ and kinβγ-2/+ mutants, there were approximately 20 and 18 peroxisomes, respectively, in a single optical section under LSCM, whereas there were approximately 87 and 99 peroxisomes in the normal pollen grains of the kinβγ-1/+ and kinβγ-2/+ mutants, respectively (Fig 5L). These results indicate that the kinβγ mutation led to defects in the structure and biogenesis of mitochondria and peroxisomes in pollen grains.
Fig 5. Peroxisomes in the pollen of kinβγ/+ and the wild type.
(A–C) Wild-type pollen under TEM. (B) and (C) The magnification of the pollen grain in (A), showing normal peroxisomes (arrows). (D–G) The kinβγ-2 pollen under TEM. (E, F and G) The magnification of the pollen grain in (D), showing peroxisomes with difficult-to-detect membranes (arrows, in E and F) or irregular morphology (arrows, in G). (H–J) Pollen labeled with mCherry-PTS1 in the wild type (H), kinβγ-1/+ (I), and kinβγ-2/+ (J), showing the pollen grains with reduced numbers of peroxisomes (arrows) and diffused mCherry-PTS1 signal. (K and L) Statistical analysis of pollen grains with reduced numbers of peroxisomes (K) and the average number of peroxisomes in normal and abnormal pollen grains in kinβγ/+ (L); only peroxisomes in the central optical sections of pollen grains were counted for analysis. Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). L, lipid; M, mitochondrion; V, vesicle. Bars, 1 μm in (A, B, D, E), 200 μm in (C, F, G), and 10 μm in (H, I, J).
Pollen germination on stigma is defective in the kinβγ/+ mutant
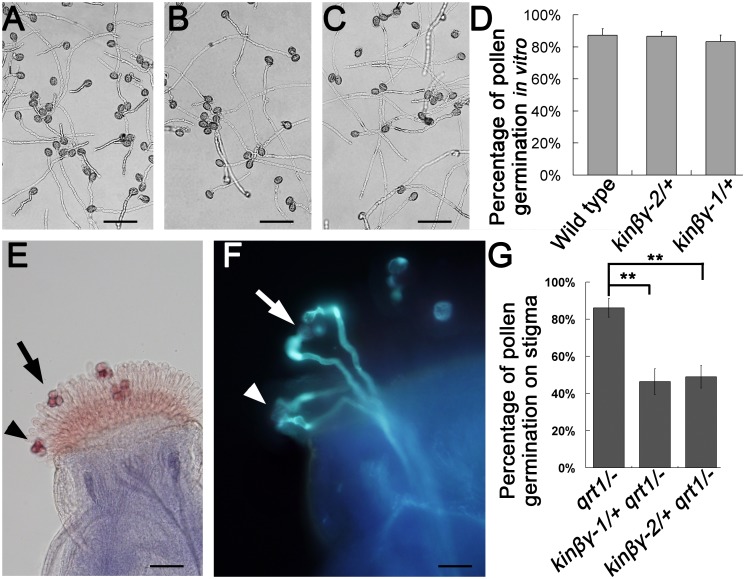
To further examine the pollen defects in the kinβγ/+ mutant, we performed both in vitro and in vivo pollen germination analyses. The pollen germination and pollen tube morphology of the kinβγ/+ mutant were no different from those of the wild type when cultured on germination medium (Fig 6A–6D). We also hand-pollinated the pollen from qrt1/-, kinβγ-1/+ qrt1/-, and kinβγ-2/+ qrt1/- onto emasculated wild-type stigmas, respectively, and analyzed pollen germination after 4 h using aniline blue staining. The pollen germination ratios of the kinβγ-1/+ qrt1/- and kinβγ-2/+ qrt1/- mutants were 46.4% and 49.0%, respectively, but the ratio of qrt1/- pollen was approximately 86.1% (Fig 6E–6G). Therefore, the presence of the kinβγ mutation caused more pollen to exhibit defective germination on the stigma. These results indicate that the in vivo germination of pollen grains with the kinβγ mutation was compromised, although the in vitro germination of mutant pollen did not show significant abnormality compared with that of wild-type pollen.
Fig 6. Pollen germination of the kinβγ mutant in vitro and in vivo.
(A-C) Pollen germination of the wild type (A), kinβγ-1/+ (B), and kinβγ-2/+ (C) on medium. (D) Percentage of pollen germination of the wild type, kinβγ-1/+ and kinβγ-2/+. (E) and (F) Pollen germination of kinβγ-2/+ qrt1/- on the stigma. (E) A bright field photograph. (F) A fluorescence photograph of amplified (E) labeled with aniline blue. Arrows and arrowheads indicate two qrt1/- tetrad pollen grains on the stigma. (G) Statistical analysis of the pollen germination of qrt1/-, kinβγ-1/+ qrt1/-, and kinβγ-2/+ qrt1/- on wild-type stigmas. Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). Bars, 60 μm in (A–C, E); 40 μm in (F).
The adhesion and hydration of kinβγ mutant pollen on the stigma is compromised
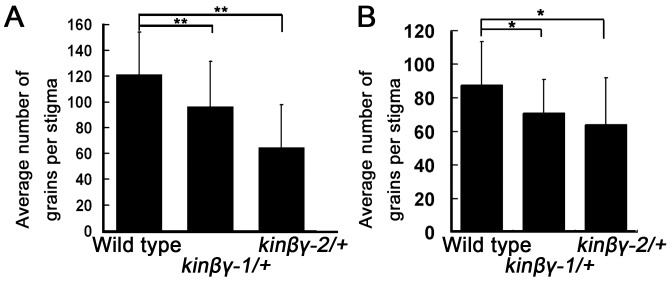
Since pollen adhesion and hydration are required for germination on the stigma, we examined whether adhesion and hydration of pollen on the stigma were affected in the kinβγ mutant. To analyze pollen adhesion, we collected kinβγ-1/+, kinβγ-2/+, and wild-type pistils at 9 h after flowering and counted the number of pollen grains on stigmas that had been washed with 0.01% NP-40 solution, which does not disturb the binding of Arabidopsis pollen on the stigma but removes weakly adhering pollen [7]. The average number of pollen grains on the mutant stigmas (97 in kinβγ-1/+, 65 in kinβγ-2/+) was much less than that on wild-type stigmas (121 in the wild type) after washing (Fig 7A). In addition, we hand-pollinated the pollen grains of the wild-type, kinβγ-1/+, and kinβγ-2/+ onto the wild-type stigmas, and collected the pollinated stigmas after 4 h, respectively. Then, the pollen grains on the stigmas washed with 0.01% NP-40 solution were counted. The average number of the pollen grains of the mutants (71 in kinβγ-1/+, 62 in kinβγ-2/+) was also less than that of the wild type (88 in the wild type) (Fig 7B). The results indicate that the kinβγ mutations result in defective pollen adhesion on the stigmas.
Fig 7. Statistical analysis of kinβγ/+ pollen adhesion on the stigma.
The number of pollen grains of the wild-type, kinβγ-1/+, and kinβγ-2/+ on washed stigmas were counted in the natural pollination (A) and the hand pollination (B). Data were collected from three independent experiments. Nine or ten stigmas were analyzed in each experiment. Double asterisks and asterisk indicate significant difference at P<0.01 and P<0.05 (Student’s t-test), respectively.
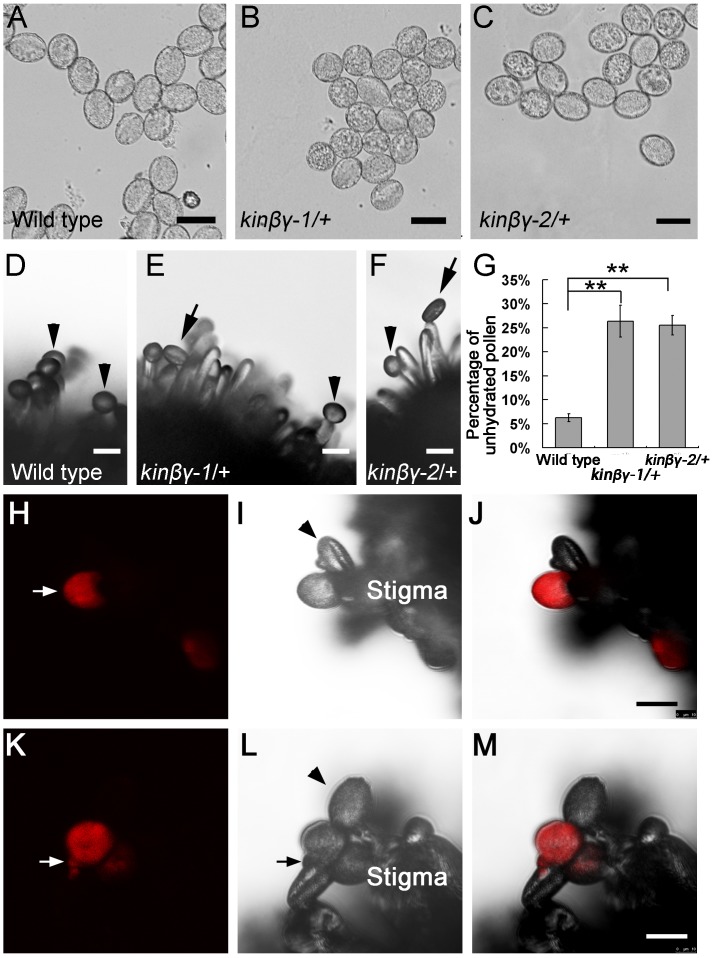
To determine whether pollen hydration was disturbed in the kinβγ mutant, we evaluated the in vitro and in vivo hydration of wild-type and mutant pollen grains. First, we observed pollen hydration on germination medium and found that the mutant and wild-type pollen became rounded at 5 min after hydration. No difference in morphology was identified in both mutant and wild-type pollen grains (Fig 8A–8C). We then investigated pollen hydration on the stigma by observing mutant and wild-type pollen grains on wild-type stigmas. Because the mutant pollen grains could hydrate in the water-contained environment, no water was added in the mounting medium on slide for in vivo hydration and germination analysis. The wild-type pollen completed the hydration process within 5 min on the stigma; however, only some pollen grains of the kinβγ-1/+ and kinβγ-2/+ mutants became hydrated (Fig 8D–8F). Statistical analysis revealed that approximately 25% of the pollen grains of the kinβγ-1/+ and kinβγ-2/+ mutants failed to become hydrated on the stigma; by contrast, <7% of wild-type pollen remained unhydrated (Fig 8G). The fluorescent-tagged line 1262 (FTL1262) is an Arabidopsis transgenic line expressing LAT52::DsRed under qrt1 background in which DsRed is a single-locus insertion in chromosome 1 near the KINβγ locus (At1g09020) [42]. We obtained kinβγ-2/+ dsred/+ qrt1/- plants by crossing kinβγ-2/+ qrt1/- and DsRed/+ qrt1/- (FTL1262) plants, in which the red marker co-segregates with the wild-type allele of KINβγ. We hand-pollinated the kinβγ-2/+ dsred/+ qrt1/- grains on the wild-type stigmas and observed their hydration and germination at 5 min and 25 min after pollination under LSCM, respectively. As shown in Fig 8H–8M, the pollen grains with red fluorescence signal could hydrate and germinate on the wild-type stigmas. Occasionally, the pollen grain without red signal could hydrate on the stigma (Fig 8M).
Fig 8. Pollen hydration in vitro and in vivo.
(A–C) Pollen of the wild type (A), kinβγ-1/+ (B) and kinβγ-2/+ (C) at 5 min after scattering onto the medium. (D-F) Pollen grains of the wild type (D), kinβγ-1/+ (E), and kinβγ-2/+ (F) at 5 min after pollinated on the wild-type stigmas. Arrowheads and arrows indicate hydrated and unhydrated pollen grains, respectively. (G) Statistical analysis of the number of unhydrated pollen grains of the wild type and kinβγ/+ at 5 min after pollinated on the wild-type stigmas. Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). (H–J) Hydration of the kinβγ-2/+ dsred/+ qrt1/- pollen grains on the wild-type stigmas at 5 min after hand-pollination. Arrow in (H) and arrowhead in (I) indicate a hydrated (red fluorescence signal) and an unhydrated (no red fluorescence signal) pollen grains in a tetrad, respectively. (H–J) The photographs of fluorescence channel, bright-field channel, and the merged channel, respectively. (K–M) Germination of the kinβγ-2/+ dsred/+ qrt1/- pollen grains on the wild-type stigmas at 25 min after hand-pollination. Arrows in (K, L) and arrowhead in (L) indicate a pollen tube with red fluorescence signal and a hydrating pollen grain without red signal, respectively. (K), (L), and (M) The photograph of fluorescence channel, bright-field photograph, and the merged photograph, respectively. No mounting medium was used in the preparation of the temporary slides for the observations and photographs of (D–F, H–M). Bars, 20 μm.
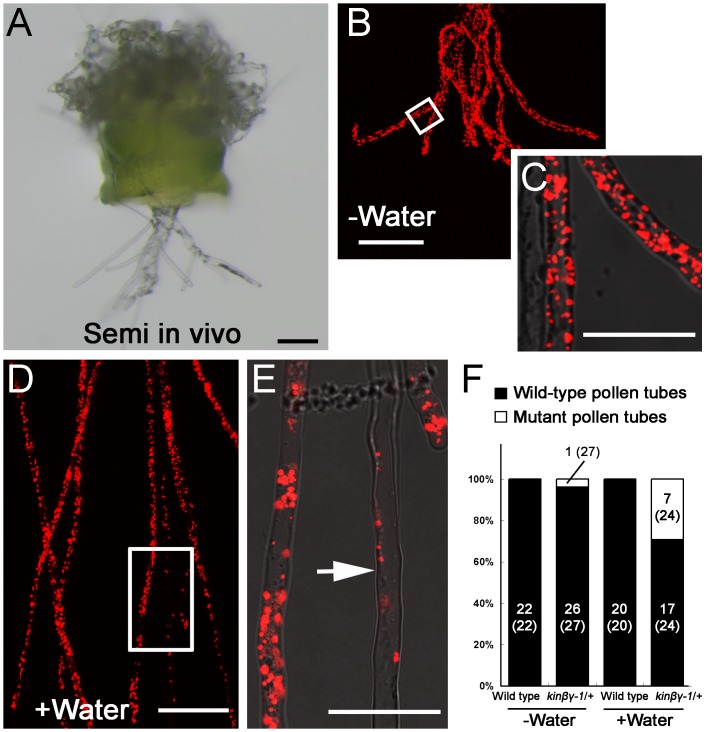
Hydration is a prerequisite for pollen germination on the stigma. To determine whether compromised hydration caused the defective germination of kinβγ pollen on stigma, we analyzed the role of water in the germination of mutant pollen on stigmas. We pollinated kinβγ-1/+ mutant pollen expressing VC-mtRFP onto wild-type stigmas in the presence of water in a semi-in vivo system [43] and identified mutant pollen tubes that had grown through pistil tissues based on their reduced number of mitochondria (Fig 9A–9E). As shown in Fig 9F, 7 of 24 pollen tubes from kinβγ-1/+ VC-mtRFP that penetrated the pistil tissue in the added water condition exhibited the reduced number of mitochondria; by contrast, only 1 of 27 pollen tubes that had penetrated the no water-added stigmas was a mutant pollen tube. No pollen tube showed the reduced number of mitochondria when the wild-type pollen grains were pollinated on the stigma with and without added water (Fig 9F). These results suggest that defective hydration, at least in part, results in the compromised germination of kinβγ pollen on stigmas.
Fig 9. Germination of kinβγ-1 pollen on a wild-type stigma moistened with water in semi-in vivo system.
(A) A semi-in vivo pollen tube growth system. (B) Pollen tubes of kinβγ-1/+ expressing VC-mtRFP penetrated the pistil tissue without added water. (C) Magnification of the box in (B). (D) Pollen tubes of kinβγ-1/+ expressing VC-mtRFP penetrated the pistil tissue after adding water onto the stigma. (E) Magnification of the box in (D), showing a mutant kinβγ-1 pollen tube (arrow). (F) Counting the pollen tubes of the wild type and the kinβγ-1/+ that penetrated pistil tissue with and without added water. The number in the columns represents the number of pollen tubes. The number in the brackets is the total number of counted pollen tubes. Bars, 50 μm in (A), (B) and (D), and 25 μm in (C) and (E).
Reactive oxygen species levels are reduced in kinβγ pollen
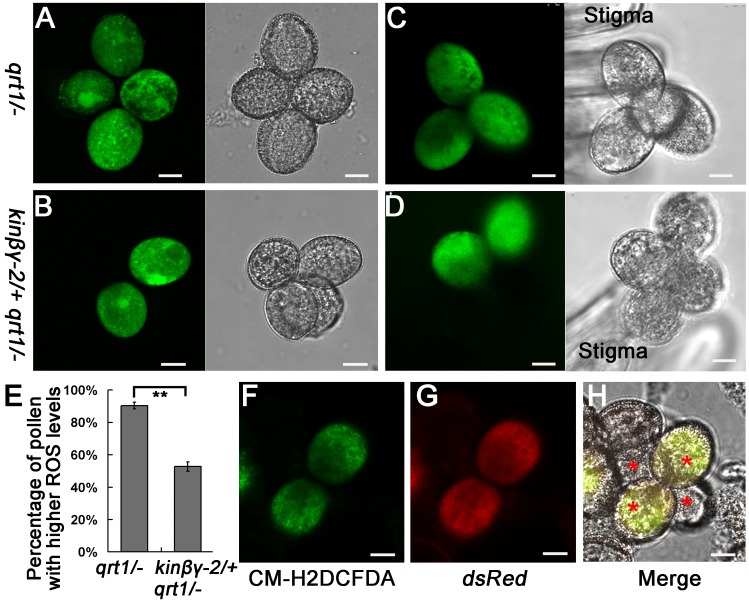
The mitochondria, peroxisomes, and chloroplasts are the major sites of intracellular ROS production [44]. We hypothesized that ROS levels might be reduced in mutant kinβγ pollen because of its defects in mitochondrial and peroxisomal biogenesis. Thus, we investigated the ROS levels in pollen in vitro by labeling it with chloromethyl derivative of 2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA), which revealed high ROS levels in most qrt1/- pollen grains (90.3%); however, only approximately half of the kinβγ-2/+ qrt1/- pollen grains (52.5%) showed high ROS levels (Fig 10A, 10B and 10E). When the pollen grains were pollinated onto the stigma (in vivo), a similar phenotype, i.e., high ROS levels in pollen, was observed (Fig 10C and 10D). We also detected the ROS levels by CM-H2DCFDA-labeling in the pollen from kinβγ-2/+ dsred/+ qrt1/- plants: green fluorescent signals in these plants indicated that the pollen had high ROS levels, and red signals represented the wild-type pollen. Both red and green fluorescent signals were detected together in two pollen grains of a pollen tetrad (Fig 10F–10H), confirming that the ROS levels in pollen grains with the kinβγ mutation were significantly reduced.
Fig 10. ROS analysis in pollen grains of qrt1/- and kinβγ-2/+ qrt1/-.
(A–D) CM-H2DCFDA labeling of ROS in mature pollen grains (A, B) and pollinated grains (C, D) of qrt1/- (A, C) and kinβγ-2/+ qrt1/- (B, D). (E) Statistical analysis of pollen with high ROS levels in qrt1/- and kinβγ-2/+ qrt1/-. ImageJ software was used for the analysis of the images. First, the format of images was changed into 8-bit and inverted into image color. Then, the threshold levels of the images were set between 0.21 and 0.02. The pollen grains were marked by red color with high ROS levels. Data were collected from three independent experiments. Asterisks indicate significant difference (Student’s t-test, P<0.01). (F–H) ROS labeled by CM-H2DCFDA in kinβγ-2/+ dsred/+ qrt1/- pollen grains. Green fluorescence signals (F) and red fluorescence signals (G) originate from the CM-H2DCFDA and LAT52::DsRed, respectively. (H) Merged image of (F) and (G). Asterisks in (H) indicate qrt1/- tetrad pollen. Bars, 10 μm.
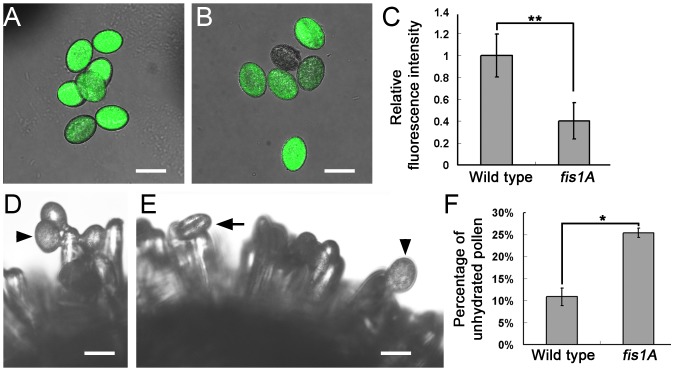
Arabidopsis FISSION1A (FIS1A) is a mitochondrion- and peroxisome-targeted protein required for the division of both organelles. In the knockout mutant of FIS1A (fis1A), the numbers of mitochondria and peroxisomes are less than those of the wild type [45]. To determine whether the defect in mitochondrial and peroxisomal biogenesis is correlated with the reduced ROS level, we detected the ROS levels and hydration of the pollen grains from the fis1A mutant plants. The ROS levels in the pollen grains of fis1A were significantly decreased compared with those of the wild type (Fig 11A–11C). Furthermore, about 25.4% of the fis1A pollen grains on the stigmas could not complete their hydration for 5 min; in contrast, only 10.5% wild-type pollen grains did not hydrate for 5 min (Fig 11D–11F). These results confirmed that the defect in the mitochondrial and peroxisomal biogenesis contributes to the reduced levels of ROS in pollen grains and the compromised hydration of pollen grains.
Fig 11. ROS levels and in vivo hydration of the pollen grains of the fis1A mutant.
(A, B) CM-H2DCFDA labeling to determine the ROS levels of pollen grains in the wild type (A) and fis1A mutant (B). (C) Relative ROS levels in pollen grains of the wild type and fis1A mutant. Data were collected from three independent experiments. Double asterisks indicate significant difference at P<0.01 (Student’s t-test). (D, E) In vivo hydration of the pollen grains of the wild type (D) and fis1A mutant (E). (F) Percentages of the unhydrated pollen grains of the wild type and fis1A mutant. Data were collected from three independent experiments. Asterisk indicates significant difference at P<0.05 (Student’s t-test). Arrowheads and arrow indicate the hydrated and unhydrated pollen grains in (D) and (E). Bars, 20 μm in (A, B, D, and E).
ROS is essential for pollen hydration on stigmas
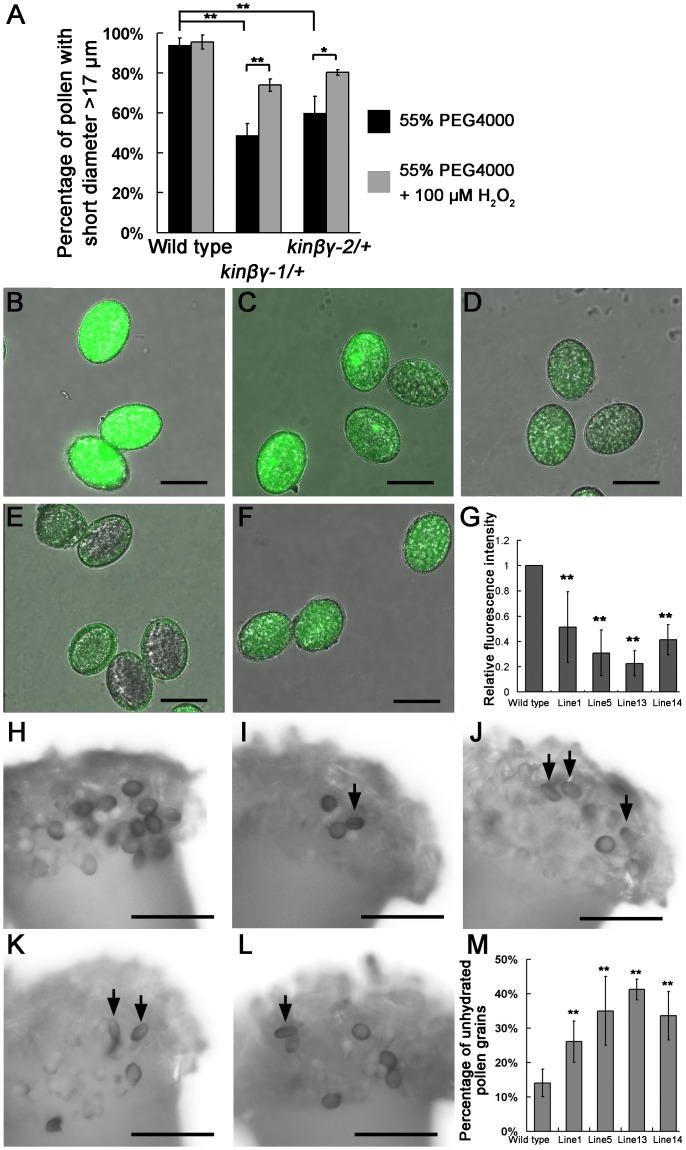
To determine whether the reduced ROS levels resulted in defective pollen hydration, we determined the water absorption capacity of mutant pollen in the modified pollen germination medium containing H2O2 (please see “Methods and Materials” in detail). We added polyethylene glycol 4000 (PEG4000) in medium to mimic a low water potential environment and investigated pollen hydration by measuring the short diameters of pollen grains at 15 min. The difference of pollen percentages with short diameter <17 μm between the wild type and the kinβγ-2/+ mutant in the 55% PEG4000 is more obvious than that in 35% PEG4000 (S6 Fig). Furthermore, we found that 100 μM H2O2 was the most appropriate concentration for the analysis of the pollen hydration of kinβγ-1 mutant in the 55% PEG4000-containing medium (S6 Fig). Thus, we used the medium with 55% PEG4000 and 100 μM H2O2 for further analysis of pollen hydration. The short diameter of approximately 94.0% of the wild-type pollen grains was >17 μm; by contrast, only 48.8% and 59.9% of the pollen grains from kinβγ-1/+ and kinβγ-2/+, respectively, had a short diameter >17 μm. However, when we added 100 μM H2O2 in the medium, the percentage of pollen grains with a short diameter >17 μm was 95.43%, 73.97%, and 80.30% in wild-type, kinβγ-1/+, and kinβγ-2/+ pollen, respectively (Fig 12A), suggesting that ROS promote water absorption in kinβγ mutant pollen.
Fig 12. ROS functions in pollen hydration.
(A) H2O2 promotes water absorption of kinβγ/+ pollen in vitro. 55% PEG4000 was added to the pollen medium (please see “Methods and Materials” in detail) to mimic a low water-potential environment. At 15 min after the pollen grains were scattered in the medium with or without 100 μM H2O2, the short diameters of pollen grains were measured. Data were collected from three independent experiments. Double asterisks and single asterisk indicate significant difference at P<0.01 and <0.05 (Student’s t-test), respectively. (B–F) CM-H2DCFDA labeling to determine the ROS levels of pollen from the wild type (B), and the transgenic lines 1 (C), 5 (D), 13 (E), and 14 (F) overexpressing Arabidopsis CAT3. (G) Relative ROS levels of pollen grains labeled with CM-H2DCFDA in different transgenic lines overexpressing Arabidopsis CAT3. ** indicates significant difference. (H–L) Hydration of pollen grains of the wild type (H), and the transgenic lines 1 (I), 5 (J), 13 (K), and 14 (L) overexpressing Arabidopsis CAT3 on the wild-type stigmas at 10 min after pollination. (M) Percentage of unhydrated pollen grains on the surface of wild-type stigmas at 10 min after pollination. ** indicates the significant difference. Bars, 20 μm in (B–F), and 100 μm in (H–L).
To determine whether ROS are involved in the regulation of pollen hydration on the stigma, we reduced the ROS levels in the pollen grains by overexpressing Arabidopsis CATALASE 3 (CAT3, At1g20620), which catalyzes the decomposition of hydrogen peroxide to water and oxygen [46]. We produced a LAT52::CAT3 construct and obtained 22 Arabidopsis transgenic lines. There were some shrunken and nonviable pollen grains in four lines, suggesting the overexpression of CAT3 in these lines resulted in abnormal pollen development (S5 Fig). Then, four transgenic lines that showed different expression levels of CAT3 and did not exhibit abnormal pollen morphology were chosen for the following analysis (S5 Fig). Alexander staining revealed that the pollen viability of these transgenic lines was similar to that of the wild-type control (S5 Fig). The ROS levels of the pollen grains in these four lines were significantly reduced, as revealed by CM-H2DCFDA labeling and semiquantitative analysis (Fig 12B–12G). As expected, a number of pollen grains in the transgenic lines did not become rounded at 10 min after pollination; by contrast, the majority of wild-type grains (>85%) on the stigmas were round at this time (Fig 12H–12M). Thus, ROS levels in pollen are involved in the hydration of pollen on stigmas.
Discussion
KINβγ regulates the biogenesis of mitochondria and peroxisomes
The KINβγ subunit is a plant-specific component of the SnRK1 heterotrimeric complexes. This subunit contains four CBS domains, as well as a GBD/KIS domain that is normally identified in canonical β subunits [32]. In Arabidopsis, KINβγ can form an active complex with either of the catalytic subunits (KIN10, KIN11) [37]. It was recently demonstrated that the carbohydrate-binding module in the GBD/KIS domain of Arabidopsis KINβγ does not bind to starch or similar oligo/polysaccharides [23], although the GBD/KIS domains in the γ subunits of the SNF1/AMPK complex in yeast and mammals can bind to glycogen to regulate complex processes. Two Arabidopsis leucine-rich proteins that interact with KINβγ are involved in plant–pathogen interactions [34]. Here, we found that the kinβγ mutation led to defective biogenesis of mitochondria and peroxisomes in Arabidopsis pollen, which resulted in compromised pollen germination on the surface of the stigma. Indeed, a previous study showed that the homozygous knockout mutant of KINβγ in Arabidopsis could not survive [32], perhaps due to the abnormal biogenesis of its mitochondria and peroxisomes. In this study, we found the knockdown expression of the Arabidopsis KIN10,11 also led to abnormal pollen morphology and reduced mitochondrial number in pollen, similar to the phenotypes of kinβγ knockout mutant. Additionally, the pollen viability was severely impaired in the knock-down mutant of the KIN10,11, which is consistent with the phenotypes of the small, non-functional pollen of the barley (Hordeum vulgare) transgenic lines expressing an antisense construct of barley SnRK1b (BKIN12), encoding a homolog of SnRK1 kinase subunit [24]; in contrast, kinβγ mutations did not result in obvious loss of pollen vitality in Alexander staining. Thus, it is likely that other γ subunits of SnRK1 complex also play roles for the activity regulation of the kinase subunit in determining pollen viability.
In mammals, AMPK promotes the biogenesis of mitochondria through the action of the transcriptional coactivator PGC-1α (peroxisome proliferator-activated receptor gamma coactivator 1-alpha) [47] and destroys defective mitochondria through unc-51-like autophagy activating kinase 1 (ULK1)-dependent autophagy [48, 49], by which new, functional mitochondria replace the defective mitochondria [19]. However, the number of mitochondria in mouse liver cells significantly increases under AMPK deficit [48], which differs from our observation that defective expression of KIN10/11 and KINβγ reduced the number of mitochondria. These results suggest that the plant SnRK1 complex utilizes different regulatory mechanisms for mitochondrial proliferation from that of its counterparts in mammals, which is consistent with the finding that Arabidopsis SnRK1 is an atypical member of the SNF1/AMPK/SnRK1 family [23]. Identifying SnRK1 complex-interacting molecules in plants might help elucidate the different ways in which the SnRK1 complex regulates the biogenesis of mitochondria and peroxisomes. There is some evidence that KIN10/11 interacts with transcription factors and other proteins in Arabidopsis [50–52]. Recently, the proteins interacting with Arabidopsis KIN10/11 were identified using a yeast two-hybrid system [53], in which several proteins containing a domain of unknown function (DUF) 581, such as DUF581-5, DUF581-13, DUF581-18, and DUF581-19, are mitochondrial-localized proteins based on the analysis at subcellular localization database for Arabidopsis proteins (http://suba.plantenergy.uwa.edu.au/) [54]. Whether these mitochondrial-localized proteins mediate the SnRK1 complex regulation in the biogenesis of mitochondria and peroxisomes in Arabidopsis remains to be investigated.
ROS signaling in pollen mediates pollen–stigma interactions
After pollen grains land on the stigma during pollination, the ability of the pollen grains to germinate in dry stigma species depends on their interactions with the papilla cells, a highly regulated process [2]. The pollen exine and coat are the first components that come in contact with the stigmatic papilla, which is involved in regulating pollen adhesion, hydration, and germination on the surface of the stigma. Many molecules located in the pollen coat function in these interaction processes, such as long-chain lipids [9], GRP17 [13], and EXL4 [14] in Arabidopsis, xylanase in maize [15], and S-locus protein 11 (SP11) in Brassica [55]. In general, the components of the exine and coat originate in the tapetum, the innermost sporophytic cell layer of the anther [56]. In this study, we did not detect any structural changes in the exine or coat of kinβγ mutant pollen grains, although the adhesion and hydration of the mutant pollen on the surface of the stigma were compromised. Nonionic detergent-washing analysis previously demonstrated that the lipophilic molecules in the exine mediate pollen adhesion on the stigma [7]. We found that NP-40 disrupted the adhesion of kinβγ mutant pollen grains to the stigma, suggesting that some lipophilic molecules localized on the surface of kinβγ mutant pollen were altered, or perhaps their functions varied compared with the wild type. However, the wild-type pollen produced by the kinβγ/+ mutant did not exhibit detectable changes in hydration or germination compared with that from wild-type plants, suggesting that some molecules in the exine and coat involved in pollen adhesion and hydration are regulated by factors in the pollen rather than in sporophytic tissue. The internal molecules in the exine and pollen coat and molecules involved in pollen signaling may not readily interact during the interactions between the pollen and stigma because they do not directly come in contact with each other. In the present study, we found that ROS signaling originating from the interior of pollen grains is important for the interactions between the pollen and stigma. A study of the Arabidopsis dayu mutant demonstrated that jasmonic acid signaling in the pollen grain controls pollen germination on the stigma [16]. DAYU and KINβγ are expressed in the vegetative pollen cell. These findings indicate that the internal signaling of pollen grains is involved in pollen–stigma interactions.
ROS, which are ubiquitous regulatory molecules, have been implicated in many pollen-related processes in sexual plant reproduction, such as tapetum and pollen development [57–59], in vitro pollen germination [60], pollen tube tip growth [61–63], pollen tube rupture for sperm release [64], and the self-incompatibility response [65]. Nicotinamide adenine dinucleotide phosphate oxidase, which is localized to the plasma membrane, is involved in ROS production, except in rice tapetum, where the mitochondria are essential for maintaining ROS homeostasis [58]. Our results show that the mitochondria and peroxisomes contribute to ROS production in mature pollen of the kinβγ and fis1A mutants, which regulates the interactions between the pollen and stigma during pollination. However, the regulatory mechanisms remain to be investigated. A possible role for internal signaling in pollen, which functions in pollen–stigma interactions, is the control of the addition of the pollen coat and exine molecules; indeed, some maize pollen coat proteins are thought to be synthesized within the pollen interior [66]. We propose that ROS signaling might control the addition of lipophilic or other molecules in the exine or on the surface of pollen, which is important for pollen–stigma interactions. Alternatively, internal pollen signaling might give rise to the transport of some mobile signaling molecules from the pollen interior to the surface of pollen grains to regulate pollen–stigma interactions following pollination. Therefore, identifying potential ROS targets that regulate the interactions between the pollen and stigma is important for elucidating the role of ROS signaling in pollination interactions. ROS signaling is intimately linked to the regulation of Ca2+ and K+ levels and, consequently, turgor homeostasis in pollen tubes [62–64]. The Ca2+ and K+ levels in pollen might be regulated by internal pollen ROS signaling, which adjusts the water potential and turgor in pollen during its hydration on the stigma. Additionally, we found some pollen grains of the kinβγ mutants showing the shrunken morphology under SEM. However, this abnormal morphology could not be detected after pollen hydration. In deed, various degrees of defective ultrastructures of vesicles and mitochondria in the mutant pollen were observed in our study. The severe defect of the ultrastructures of mutant pollen grains might correlate with their severe defect in osmotic adjustment ability. Thus, an artificial dry environment was imposed during the SEM sample preparation, which resulted in some of the mutant pollen showing the sunken surfaces under this condition.
Another way in which internal signaling in pollen may regulate pollen–stigma interactions is that it may initiate a signaling cascade to activate stigma cells during their interaction. For example, in Brassica, during self-incompatibility, SP11 is secreted from the pollen coat and interacts with its cognate S-receptor kinase in the papilla cell of the stigma to elicit the self-incompatibility response [55]. The pollen coat of Brassicaceae can induce the expression of Ca2+-ATPase 13, a Ca2+ transporter in the papillae cell that functions in the export of Ca2+ to compatible pollen tubes [67]. Similarly, the Arabidopsis pollen coat can induce the expression of many stigma genes, suggesting that the upregulation of these genes is most likely involved in pollen–stigma interactions. The stigma contains water on its surface, providing water for pollen hydration; the water is covered by a cuticle. However, it is currently unclear whether the stigma cuticle is constitutively permeable to water or if its permeability is modulated by pollen signaling [2]. Compatible pollen can induce the hydration and germination of normally incompatible pollen in mixed-pollen populations [9], suggesting that the permeability of the cuticle to water is regulated by pollen signaling. Thus, ROS signaling is essential for pollen hydration on the surface of the stigma, which might modulate the water permeability of the cuticle.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis thaliana T-DNA insertion mutants GABI_346E09, SALK_074210 and fis1A were obtained from European Arabidopsis Stock Centre and Arabidopsis Biological Resource Center, respectively. Wild-type Arabidopsis (Columbia-0) and T-DNA insertion mutants were grown in a 22°C greenhouse under a 10-h light/14-h dark cycle (short-day condition) for 2 weeks and then a 16-h light/8-h dark cycle (long-day condition). The genotypes of the T-DNA insertion line plants were identified using a PCR-based method with the following primers: o8409, kinβγ-1LP, and kinβγ-1RP for GABI_346E09; and LBb1.3, kinβγ-2LP, and kinβγ-2RP for SALK_074210. All of the sequences of the primers are listed in the S3 Table.
Pollen viability and germination analysis
The mature pollen grains or mature anthers were soaked in Alexander stain for 1–2 days to determine pollen viability. DAPI (1 μg/ml; w/v) (Sigma-Aldrich) was used to stain the nuclei of the pollen grains. Alexander stain, DAPI and aniline blue staining of pollen grain/tube were performed as previously described [68]. The semi in vivo and in vitro pollen germination assay was performed as described as Li et al. [16] and Xu et al. [69], respectively. All samples were observed and photographed under a BX51 microscope (Olympus, Tokyo, Japan).
Pollen adhesion assay
The pollen adhesion assay was performed as described by Zinkl et al. [7] with minor modification. The self-pollinated pistils at the flowering day were collected at 7:00 PM. The hand-pollinated wild-type pistils which were emasculated two days before anthesis were collected at 4 hours after pollination. After washing with 0.05% NP-40 in 50 mM phosphate buffer (pH 7.4), the pistils were centrifuged for 30 s at 300 g. The adhering pollen grains on the washed pistils were photographed and counted under a BX51 microscope (Olympus, Tokyo, Japan).
Pollen hydration assay
For the in vivo assay of pollen hydration, the stigmas of the emasculated flowers were hand pollinated with mature pollen grains in a greenhouse and observed after 5–10 min. For the in vitro assay of pollen hydration, the mature pollen grains were spread in the modified pollen germination medium (1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 0.01% (w/v) H3BO3, pH 7.0) by removing the sucrose and agar [69], at 28°C and observed after 15 min. To test the in vitro pollen hydration capacity, we used 35% and 55% (w/v) PEG4000 solution to mimic the low water potential environment. Fifteen min after the pollen grains incubated in the medium, the short-axis diameters of the pollen grains were measured. To analyze the role of ROS in pollen hydration of pollen in vitro, 0, 50, 100, and 150 μM H2O2 were added to the PEG4000-containing medium, and the short-axis diameters of the pollen grains were measured after 15 min. All pollen grains were observed under a BX51 microscope (Olympus, Tokyo, Japan) or LEICA TCS SP5 II laser scanning confocal microscope (Leica, Wetzlar, Germany).
Fluorescent labeling of mitochondria and peroxisomes
Two approaches were used to label mitochondria. One was fluorescence staining using 500 mM MitoTracker Deep Red FM (Cat# M22426; Molecular Probes), which was excited with 633 nm laser and the signal were collected 650–700 nm. Another method for detecting mitochondria was using the Arabidopsis mitochondria marker lines—VC-mtGFP and VC-mtRFP—which were excited at 488 and 561 nm, and emissions were collected at 505–530 and 570–660 nm, respectively. The Lat52::mCherry-PTS1 transgenic line was used to label peroxisome. mCherry-PTS1 was excited by a 561 nm laser, and the emissions were observed at 600–630 nm. All images were captured using LEICA TCS SP5 II LSCM (Leica, Wetzlar, Germany). The numbers of mitochondria and peroxisomes in the central optical sections of the pollen grains were counted for the statistical analysis.
Scanning and transmission electron microscopy (SEM and TEM)
Sample preparation and observation for routine SEM and TEM were performed as previously described [69]. To visualize the peroxisomes of the pollen under TEM, 3,3'-diaminobenzidine tetrahydrochloride (DAB) staining was used to detect the catalase activity, which marks the peroxisome, as described by Li et al. [16]. The anthers were fixed at 4°C for 10 h with 2.5% glutaraldehyde and 1% osmium tetroxide at 4°C for 5 h in 0.1 M cacodylate buffer (pH 7.2). After washing 3 times with cacodylate buffer, the anthers were incubated for 2 h at 28°C in the dark in a solution containing 0.2% DAB (Sigma-Aldrich) and 0.02% H2O2 in 50 mM Tris-HCl (pH 7.6) and washed another 5 times using cacodylate buffer. The samples were then embedded in Epon812 for sectioning.
Plasmid construction and plant transformation
To specifically decrease the ROS level in pollen, Arabidopsis CAT3 was amplified using the CAT3LP and CAT3RP primers and cloned into the pROKII vector to create the Lat52::CAT3 construct for pollen-specific expression. This construct was transformed into wild-type Col-0 Arabidopsis plants by agrobacterium-mediated transformation. The amiRNA construct, which targets KIN10,11, was constructed as previously described [40]. The amiRNA precursor was amplified by overlapping PCR from the pRS300 template using the ImiR-s, IImiR-a, IIImiR*-s, IVmiR*-a primers designed by WMD3 Web microRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) (S3 Table). The sequenced fragment containing the amiRNA foldback in the cloning vector was subsequently subcloned into the pROKII vector under the control of Lat52 promoter to form LAT52::amiRNA-KIN10,11 for pollen-specific expression inhibition. This construct was transformed into the wild-type Col-0 Arabidopsis plants by Agrobacterium-mediated transformation. All of the sequences of primers are listed in S3 Table.
ROS staining for pollen grains
To label ROS, pollen grains were incubated in 10 μM CM-H2DCFDA (Cat# C6827; Molecular Probes) dissolved in 20 mM HEPES buffer (pH 7.2) for 5–10 min at room temperature. After washing with HEPES buffer, the pollen grains were observed under a LEICA TCS SP5 II LSCM (Leica, Wetzlar, Germany) using a 488-nm laser as the excitation light, and the emissions were collected at 505–530 nm. To analyze the relative ROS level in the pollen labeled by CM-H2DCFDA, photos were taken under the same LSCM microscopic settings. The relative fluorescence intensities of the pollen grains indicate the relative ROS levels analyzed by ImageJ software (http://rsb.info.nih.gov).
Quantitative real-time-PCR (qRT-PCR) analysis
The mature pollen grains of wild-type plants, kinβγ-1/+ and kinβγ-2/+ mutant, Lat52::amiRNA-KIN10,11 and Lat52::CAT3 transgenic lines were collected. Total RNAs of pollen grains were extracted according to the manufacture using Ultrapure RNA Kit (TIANGEN, China) and cDNAs were got by reverse transcription using PrimerScript RT reagent Kit with gDNA Eraser-Perfect Real Time (TAKARA, Dalian). qRT-PCR reaction was performed with BIO-RAD CFX96 real-time system using iQ SYBR Green Supermix (BIO-RAD, Singapore) using the corresponding primer pairs (S3 Table). The RNA levels were normalized to that of TUBULIN2 with three biological replicates and three technical replicates.
Supporting Information
(A) Schematic representation of the KINβγ gene and the positions of T-DNA insertion in kinβγ-1/+ and kinβγ-2/+. (B) Identification of kinβγ-1/+ and kinβγ-2/+ mutants using a PCR-based method with gene-specific primers. (C) The relative expression levels of KINβγ in pollen of the wild type, kinβγ-1/+ and kinβγ-2/+ determined by qRT-PCR analysis. The expression level in the wild type was set to 1.0. The error bars represent the SD of three biological replicates.
(DOC)
(A) Pollen grains of the SNF4-YFP/+ observed by SEM. (B) Pollen grains of SNF4-YFP/+ kinβγ-1/- observed by SEM. Arrow indicates the pollen grain with sunken surface. (C) Statistical analysis of abnormal pollen with sunken surfaces in the SNF4-YFP/+ and SNF4-YFP/+ kinβγ-1/-. Data were collected from three independent experiments. No significant difference was detected (Student’s t-test, P = 0.464). Bars, 50 μm.
(DOC)
No significant difference in appearance was detected between the coat and wall of the wild type (A) and the mutants, kinβγ-1/+ (B) and kinβγ-2/+ qrt1/- (C). Ba, bacula; In, intine; Ne, nexine; PC, pollen coat; Tc, tectum. Bars, 5 μm.
(DOC)
(A) Alexander staining to detect the viability of mature pollen. (B) Pollen morphology under SEM. Arrows indicate the pollen grains with abnormal morphology. (C) Mitochondria in pollen stained with MitoTracker Deep Red. (D) The relative expression levels of KIN10 and KIN11 in the wild type and the four transgenic lines determined by qRT-PCR analysis. The expression level in the wild type was set to 1.0. The error bars represent the SD of three biological replicates. Bars, 50 μm in (A), and 20 μm in (B) and (C).
(DOC)
(A–E) Alexander staining of mature pollen of the wild-type (A) and transgenic lines overexpressing Arabidopsis CAT3, including line 1 (B), line 5 (C), line 13 (D), and line 14 (E). (F) qRT-PCR analysis of the relative expression levels of CAT3 in the wild type and the four transgenic lines. The expression level in the wild type was set to 1.0. The error bars represent the SD of three biological replicates. (G, H) Alexander staining of mature pollen of two transgenic lines, line 3 (G), and line 9 (H), showing the shrunken and nonviable pollen grains. Bars, 25 μm in (A–E) and 200 μm in (G, H).
(DOC)
(A) Percentages of the pollen (short diameter < 17 μm) of the wild type and the kinβγ-2/+ mutant in the medium containing 35% and 55% PEG4000, respectively. (B) Percentages of the kinβγ-1/+ pollen (short diameter > 17 μm) in the medium containing 55% PEG4000 and H2O2. The error bars represent the SD of three biological replicates.
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We thank all the people who provided materials for this study. The SNF4-YFP transgenic line was gifted from Dr. Csaba Koncz in Max-Planck Institute for Plant Breeding Research, Germany. The peroxisome marker line (LAT52::mCherry-PTS1) was gifted from Dr. Wei-Cai Yang in Chinese Academy of Sciences, China. The mitochondria marker lines, VC-mtGFP and VC-mtRFP, were gifted from Dr. Sodmergen in Peking University, China. The Arabidopsis fluorescent-tagged line FTL1262 was gifted from Dr. Gregory P. Copenhaver in University of North Carolina, USA. The Arabidopsis qrt1 mutant was gifted from Dr. Yan Zhang in Shandong Agricultural University, China. The Arabidopsis T-DNA insertion mutants GABI_346E09, SALK_074210 and fis1A were obtained from European Arabidopsis Stock Centre and Arabidopsis Biological Resource Center, respectively.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Major Research Plan from the Ministry of Science and Technology of China (No. 2013CB945100) http://www.most.gov.cn/eng/programmes1/200610/t20061009_36223.htm, and by the National Natural Science Foundation of China (No. 31270358 and No. 31300267) http://www.nsfc.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Edlund AF, Swanson R, Preuss D. Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell. 2004; 16: S84–S97. 10.1105/tpc.015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiscock SJ, Allen AM. Diverse cell signalling pathways regulate pollen-stigma interactions: the search for consensus. New Phytol. 2008; 179: 286–317. 10.1111/j.1469-8137.2008.02457.x [DOI] [PubMed] [Google Scholar]
- 3.Dresselhaus T, Franklin-Tong N. Male–female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol Plant. 2013; 6: 1018–1036. 10.1093/mp/sst061 [DOI] [PubMed] [Google Scholar]
- 4.Indriolo E, Safavian D, Goring DR. Signaling events in pollen acceptance or rejection in the Arabidopsis species In Sawada H, Inoue N, Iwano M Editors. Sexual Reproduction in Animals and Plants. Springer; 2014; pp. 255–271. ISBN: 978-4-431-54588-0 [Google Scholar]
- 5.Gaude T, Dumas C. A membrane-like structure on the pollen wall surface in Brassica. Ann Bot. 1984; 54: 821–825. [Google Scholar]
- 6.Zinkl GM, Preuss D. Dissecting Arabidopsis pollen-stigma interactions reveals novel mechanisms that confer mating specificity. Ann Bot. 2000; 85: 15–21. 1999.1066 [Google Scholar]
- 7.Zinkl GM, Zwiebel BI, Grier DG, Preuss D. Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 1999; 126: 5431–5440. [DOI] [PubMed] [Google Scholar]
- 8.Lin H, Lizarraga L, Bottomley LA, Carson MJ. Effect of water absorption on pollen adhesion. J Colloid Interface Sci. 2015; 442: 133–139. 10.1016/j.jcis.2014.11.065 [DOI] [PubMed] [Google Scholar]
- 9.Hülskamp M, Kopczak SD, Horejsi TF, Kihl BK, Pruitt RE. Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 1995; 8: 703–714. 10.1046/j.1365-313X.1995.08050703.x [DOI] [PubMed] [Google Scholar]
- 10.Mayfield JA, Fiebig A, Johnstone SE, Preuss D. Gene families from the Arabidopsis thaliana pollen coat proteome. Science. 2001; 292: 2482–2485. 10.1126/science.1060972 [DOI] [PubMed] [Google Scholar]
- 11.Ge W, Song Y, Zhang C, Zhang Y, Burlingame AL, Guo Y. Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim Biophys Acta. 2011; 1814: 1964–1973. 10.1016/j.bbapap.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, Cai G, Gong F, An S, Cresti M, Wang W. Proteome profiling of maize pollen coats reveals novel protein components. Plant Mol Biol Rep. 2015; 33: 975–986. 10.1007/s11105-014-0812-3 [DOI] [Google Scholar]
- 13.Mayfield JA, Preuss D. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nature Cell Biol. 2000; 2: 128–130. 10.1038/35000084 [DOI] [PubMed] [Google Scholar]
- 14.Updegraff EP, Zhao F, Preuss D. The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex Plant Reprod. 2009; 22: 197–204. 10.1007/s00497-009-0104-5 [DOI] [PubMed] [Google Scholar]
- 15.Suen DF, Huang AH. Maize pollen coat xylanase facilitates pollen tube penetration into silk during sexual reproduction. J Biol Chem. 2007; 282: 625–636. 10.1074/jbc.M608567200 [DOI] [PubMed] [Google Scholar]
- 16.Li XR, Li HJ, Yuan L, Liu M, Shi DQ, Liu J, et al. Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell. 2014; 26: 619–635. 10.1105/tpc.113.121087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carling D, Mayer FV, Sanders MJ, Gamblin SJ. AMP-activated protein kinase: nature's energy sensor. Nature Chem Bio. 2011; 7: 512–518. 10.1038/nchembio.610 [DOI] [PubMed] [Google Scholar]
- 18.Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, et al. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J. 2011; 278: 3978–3990. 10.1111/j.1742-4658.2011.08315.x [DOI] [PubMed] [Google Scholar]
- 19.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature Cell Biol. 2011; 13: 1016–1023. 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emanuelle S, Doblin MS, Stapleton DI, Bacic A, Gooley PR. Molecular insights into the enigmatic metabolic regulator, SnRK1. Trends Plant Sci. 2016; 21: 341–353. 10.1016/j.tplants.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 21.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signaling. Nature. 2007; 448: 938–942. 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- 22.Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE. Regulation of Sucrose non-Fermenting Related Kinase 1 genes in Arabidopsis thaliana. Front Plant Sci. 2014; 5 10.3389/fpls.2014.00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emanuelle S, Hossain MI, Moller IE, Pedersen HL, Meene AM, Doblin MS, et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015; 82:183–192 10.1111/tpj.12813 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG. Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 2001; 28: 431–441. 10.1046/j.1365-313X.2001.01167.x [DOI] [PubMed] [Google Scholar]
- 25.Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2006; 140: 263–278. 10.1104/pp.105.071167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, et al. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 2010; 61: 324–338. 10.1111/j.1365-313X.2009.04057.x [DOI] [PubMed] [Google Scholar]
- 27.Tsai AYL, Gazzarrini S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012; 69: 809–821. 10.1111/j.1365-313X.2011.04832.x [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell. 2013; 25: 3871–3884. 10.1105/tpc.113.114066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, et al. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci. 2014; 5:190 10.3389/fpls.2014.00190 eCollection 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007; 12: 20–28. 10.1016/j.tplants.2006.11.005 [DOI] [PubMed] [Google Scholar]
- 31.Fang L, Hou X, Lee LYC, Liu L, Yan X, Yu H. AtPV42a and AtPV42b redundantly regulate reproductive development in Arabidopsis thaliana. PloS One. 2011; 6: e19033 10.1371/journal.pone.0019033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. The hybrid four-CBS-domain KINβγ subunit functions as the canonical γ subunit of the plant energy sensor SnRK1. Plant J. 2013; 75: 11–25. 10.1111/tpj.12192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C. Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 2000; 23: 115–122. 10.1046/j.1365-313x.2000.00809.x [DOI] [PubMed] [Google Scholar]
- 34.Gissot L, Polge C, Jossier M, Girin T, Bouly J, Kreis M, et al. AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiol. 2006; 142: 931–944. 10.1104/pp.106.087718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Paz C, Vilela B, Riera M, Pagès M, Lumbreras V. Maize AKINβγ dimerizes through the KIS/CBM domain and assembles into SnRK1 complexes. FEBS Lett. 2009; 583: 1887–1894. 10.1016/j.febslet.2009.05.022 [DOI] [PubMed] [Google Scholar]
- 36.Bitrián M, Roodbarkelari F, Horváth M, Koncz C. BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 2011; 65: 829–842. 10.1111/j.1365-313X.2010.04462.x [DOI] [PubMed] [Google Scholar]
- 37.Avila-Castaneda A, Gutierrez-Granados N, Ruiz-Gayosso A, Sosa-Peinado A, Martinez-Barajas E, Coello P. Structural and functional basis for starch binding in the SnRK1 subunits AKINbeta2 and AKINbetagamma. Front Plant Sci. 2014; 5: 199 10.3389/fpls.2014.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson-Brousseau SA, McCormick S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 2004; 39: 761–775. 10.1111/j.1365-313X.2004.02147.x [DOI] [PubMed] [Google Scholar]
- 39.Matsushima R, Hamamura Y, Higashiyama T, Arimura S, Sodmergen, Tsutsumi N, et al. Mitochondrial dynamics in plant male gametophyte visualized by fluorescent live imaging. Plant Cell Physiol. 2008; 49:1074–1083. 10.1093/pcp/pcn084 [DOI] [PubMed] [Google Scholar]
- 40.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006; 18: 1121–1133. 10.1105/tpc.105.039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, et al. Plant peroxisomes: biogenesis and function. Plant Cell. 2012; 24: 2279–2303. 10.1105/tpc.112.096586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Nat Acad Sci USA. 2007; 104: 3913–3918. 10.1073/pnas.0608936104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palanivelu R, Preuss D. Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol. 2006; 6: 7 10.1186/1471-2229-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrzaczek M, Brosché M, Kangasjärvi J. ROS signaling loops—production, perception, regulation. Curr Opin Plant Biol. 2013; 16: 575–582. 10.1016/j.pbi.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 45.Zhang XC, Hu JP. FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant. 2008; 1: 1036–1047. 10.1093/mp/ssn056 [DOI] [PubMed] [Google Scholar]
- 46.Frugoli JA, Zhong HH, Nuccio ML, McCourt P, McPeek MA, Thomas TL, et al. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996; 112: 327–336. 10.1104/pp.112.1.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, et al. In vivo correction of COX deficiency by activation of the AMPK/PGC-1α axis. Cell Metab. 2011; 14: 80–90. 10.1016/j.cmet.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011; 331: 456–461. 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Kundu M, Viollet B, Guan K. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 2011; 13: 132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Brien M, Kaplan-Levy RN, Quon T, Sappl PG, Smyth DR. PETAL LOSS, a trihelix transcription factor that represses growth in Arabidopsis thaliana, binds the energy-sensing SnRK1 kinase AKIN10. J Exp Bot. 2015; 66: 2475–2485. 10.1093/jxb/erv032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai AY, Gazzarrini S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012; 69: 809–821. 10.1111/j.1365-313X.2011.04832.x [DOI] [PubMed] [Google Scholar]
- 52.Mair A, Pedrotti L, Wurzinger B, Anrather D, Simeunovic A, Weiste C, et al. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. Elife. 2015; 4:e05828 10.7554/eLife.05828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nietzschea M, Landgraf R, Tohge T, Börnke F. A protein–protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in Arabidopsis thaliana. Curr Plant Biol. 2016; 5: 36–44. 10.1016/j.cpb.2015.10.004 [DOI] [Google Scholar]
- 54.Hooper CM, Tanz SK, Castleden IR, Vacher MA, Small ID, Millar AH. SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics. 2014; 30: 3356–3364. 10.1093/bioinformatics/btu550 [DOI] [PubMed] [Google Scholar]
- 55.Takayama S, Shimosato H, Shiba H, Funato M, Che FS, Watanabe M, et al. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature. 2001; 413: 534–538. 10.1038/35097104 [DOI] [PubMed] [Google Scholar]
- 56.Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol. 2011; 62: 437–460. 10.1146/annurev-arplant-042809-112312 [DOI] [PubMed] [Google Scholar]
- 57.Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell. 2011; 23: 515–533. 10.1105/tpc.110.074369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo D, Xu H, Liu Z, Guo J, Li H, Chen L,et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013; 45: 573–577. 10.1038/ng.2570 [DOI] [PubMed] [Google Scholar]
- 59.Xie H, Wan Z, Li S, Zhang Y. Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell. 2014; 26: 2007–2023. 10.1105/tpc.114.125427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speranza A, Crinelli R, Scoccianti V, Geitmann A. Reactive oxygen species are involved in pollen tube initiation in kiwifruit. Plant Biol. 2012; 14: 64–76. 10.1111/j.1438-8677.2011.00479.x [DOI] [PubMed] [Google Scholar]
- 61.Potocký M, Jones MA, Bezvoda R, Smirnoff N, Žárský V. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phyto. 2007; 174: 742–751. 10.1111/j.1469-8137.2007.02042.x [DOI] [PubMed] [Google Scholar]
- 62.Kaya H, Nakajima R, Iwano M, Kanaoka MM, Kimura S, Takeda S, et al. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell. 2014; 26: 1069–1080. 10.1105/tpc.113.120642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lassig R, Gutermuth T, Bey TD, Konrad KR, Romeis T. Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J. 2014; 78: 94–106. 10.1111/tpj.12452 [DOI] [PubMed] [Google Scholar]
- 64.Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu H, et al. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nature Commun. 2014; 5: 3129 10.1038/ncomms4129 [DOI] [PubMed] [Google Scholar]
- 65.Wilkins KA, Bancroft J, Bosch M, Ings J, Smirnoff N, Franklin-Tong VE. Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of papaver. Plant Physiol. 2011; 156: 404–416. 10.1104/pp.110.167510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong F, Wu X, Wang W. Diversity and function of maize pollen coat proteins: from biochemistry to proteomics. Front Plant Sci. 2015; 6: 199 10.3389/fpls.2015.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwano M, Igarashi M, Tarutani Y, Kaothien-Nakayama P, Nakayama H, Moriyama H, et al. A pollen coat–inducible autoinhibited Ca2+-ATPase expressed in stigmatic papilla cells is required for compatible pollination in the Brassicaceae. Plant Cell. 2014; 26: 636–649. 10.1105/tpc.113.121350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai XR, Gao XQ, Chen GH, Tang LL, Wang H, Zhang XS. ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyltransferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR BIOSYNTHESIS B in human, is required for Arabidopsis pollen tube micropylar guidance and embryo development. Plant Physiol. 2014; 165: 1544–1556. 10.1104/pp.114.236133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu N, Gao XQ, Zhao XY, Zhu DZ, Zhou LZ, Zhang XS. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol Biol. 2011; 77: 251–260. 10.1007/s11103-011-9806-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of the KINβγ gene and the positions of T-DNA insertion in kinβγ-1/+ and kinβγ-2/+. (B) Identification of kinβγ-1/+ and kinβγ-2/+ mutants using a PCR-based method with gene-specific primers. (C) The relative expression levels of KINβγ in pollen of the wild type, kinβγ-1/+ and kinβγ-2/+ determined by qRT-PCR analysis. The expression level in the wild type was set to 1.0. The error bars represent the SD of three biological replicates.
(DOC)
(A) Pollen grains of the SNF4-YFP/+ observed by SEM. (B) Pollen grains of SNF4-YFP/+ kinβγ-1/- observed by SEM. Arrow indicates the pollen grain with sunken surface. (C) Statistical analysis of abnormal pollen with sunken surfaces in the SNF4-YFP/+ and SNF4-YFP/+ kinβγ-1/-. Data were collected from three independent experiments. No significant difference was detected (Student’s t-test, P = 0.464). Bars, 50 μm.
(DOC)
No significant difference in appearance was detected between the coat and wall of the wild type (A) and the mutants, kinβγ-1/+ (B) and kinβγ-2/+ qrt1/- (C). Ba, bacula; In, intine; Ne, nexine; PC, pollen coat; Tc, tectum. Bars, 5 μm.
(DOC)
(A) Alexander staining to detect the viability of mature pollen. (B) Pollen morphology under SEM. Arrows indicate the pollen grains with abnormal morphology. (C) Mitochondria in pollen stained with MitoTracker Deep Red. (D) The relative expression levels of KIN10 and KIN11 in the wild type and the four transgenic lines determined by qRT-PCR analysis. The expression level in the wild type was set to 1.0. The error bars represent the SD of three biological replicates. Bars, 50 μm in (A), and 20 μm in (B) and (C).
(DOC)
(A–E) Alexander staining of mature pollen of the wild-type (A) and transgenic lines overexpressing Arabidopsis CAT3, including line 1 (B), line 5 (C), line 13 (D), and line 14 (E). (F) qRT-PCR analysis of the relative expression levels of CAT3 in the wild type and the four transgenic lines. The expression level in the wild type was set to 1.0. The error bars represent the SD of three biological replicates. (G, H) Alexander staining of mature pollen of two transgenic lines, line 3 (G), and line 9 (H), showing the shrunken and nonviable pollen grains. Bars, 25 μm in (A–E) and 200 μm in (G, H).
(DOC)
(A) Percentages of the pollen (short diameter < 17 μm) of the wild type and the kinβγ-2/+ mutant in the medium containing 35% and 55% PEG4000, respectively. (B) Percentages of the kinβγ-1/+ pollen (short diameter > 17 μm) in the medium containing 55% PEG4000 and H2O2. The error bars represent the SD of three biological replicates.
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.