Abstract
Fibroblast growth factors (Fgfs) regulate critical biological processes such as embryonic development, tissue homeostais, wound healing, and tissue regeneration. In zebrafish, Fgf signaling plays an important role in the regeneration of the spinal cord, liver, heart, fin, and photoreceptors, although its exact mechanism of action is not fully understood. Utilizing an adult zebrafish extraocular muscle (EOM) regeneration model, we demonstrate that blocking Fgf receptor function using either a chemical inhibitor (SU5402) or a dominant-negative transgenic construct (dnFGFR1a:EGFP) impairs muscle regeneration. Adult zebrafish EOMs regenerate through a myocyte dedifferentiation process, which involves a muscle-to-mesenchyme transition and cell cycle reentry by differentiated myocytes. Blocking Fgf signaling reduced cell proliferation and active caspase 3 levels in the regenerating muscle with no detectable levels of apoptosis, supporting the hypothesis that Fgf signaling is involved in the early steps of dedifferentiation. Fgf signaling in regenerating myocytes involves the Mapk/Erk pathway: inhibition of Mek activity with U0126 mimicked the phenotype of the Fgf receptor inhibition on both muscle regeneration and cell proliferation, and activated ERK (p-ERK) was detected in injured muscles by immunofluorescence and western blot. Interestingly, following injury, ERK2 expression is specifically induced and activated by phosphorylation, suggesting a key role in muscle regeneration. We conclude that the critical early steps of myocyte dedifferentiation in EOM regeneration are dependent on Fgf signaling.
Keywords: strabismus, cell cycle, heat-shock, MMT, cell reprogramming
Graphical Abstract
1. Introduction
De novo regeneration of injured or degenerated tissues and organs carries great promise for curing a multitude of debilitating disorders, potentially replacing the need for organ transplantation and creating new opportunities for treating many chronic disorders. Unfortunately, mammals in general, and humans in particular, have a rather limited capacity to regenerate. The reasons for that are not well understood, since most mammalian tissues contain resident stem cells that at least in vitro can give rise to progenitor cells with regenerative capabilities. Muscle injury and degeneration is a particularly important cause of morbidity and mortality, and the ability to regenerate muscle, whether skeletal or cardiac, would completely alter the therapeutic landscape. Despite the presence of skeletal muscle stem cells (i.e. satellite cells) that can repair and maintain muscles (at least until they exhaust in degenerative diseases, such as Duchenne Muscular Dystrophy [1, 2]), mammalian skeletal muscles cannot regenerate and replace muscle tissue following extirpation.
Fish and amphibians have a much greater capacity to regenerate, which appears to rely less on resident stem cells and much more on cell reprogramming and dedifferentiation [3]. Indeed, adult zebrafish are able to regenerate both skeletal [4] and cardiac muscle [5, 6], as well as retina [7], spinal cord [8], liver [9] and fin [10, 11]. Our lab recently characterized a robust regenerative pathway in adult zebrafish extraocular muscles (EOMs) –a form of skeletal muscle– which is driven by myocyte dedifferentiation with no significant contribution of satellite cell [4]. Thus, we can now utilize a novel model for studying the mechanistic underpinnings of myocyte reprogramming via dedifferentiation.
Fibroblast growth factors (Fgfs) are a family of polypeptide growth factors composed, in vertebrates, by over twenty members [12]. Fgfs exert their biological pleiotropic functions by activating the Fgf receptor (Fgfr) tyrosine kinase, except for Fgf11–Fgf14 which function intracellularly via Fgfr-independent mechanisms [13]. Fgf signaling controls embryonic development and morphogenesis, maintains tissue homeostasis, regulates organ function, and promotes wound healing [14]. The roles of Fgf in skeletal muscle development, repair, and regeneration have been extensively studied. Thus, Fgfs regulate muscle development through autocrine and paracrine mechanisms in vitro [15] and several steps of myogenesis in limb development [16]. Specifically in zebrafish embryos, Fgf signaling plays a role in morphogenesis and cell fate determination of myotomes [17], and is required for fast muscle differentiation [18]. Furthermore, it has been shown that Fgfs promote satellite cell proliferation [19–21] and, therefore, plays important roles in the muscle response to injury [22, 23].
Among their many functions, Fgfs also play a key role in tissue repair and regeneration in zebrafish [9, 10, 24–26], amphibians [27, 28] and even mammals [29, 30]. Since Fgfs play a universal role in regulating tissue regeneration and zebrafish EOMs regenerate by myocyte reprogramming and dedifferentiation, we directly tested whether Fgfs are important in EOM regeneration, and whether their role is primarily early, during cell reprogramming and proliferation, or late during migration and redifferentiation. Here we report that blocking Fgfr resulted in severely delayed regeneration, and that Fgf signaling is most important during the early events of myocyte dedifferentiation. We further demonstrate that Fgf exerts its effects through the Mapk/Erk signaling pathway, and that ERK2 is a candidate for a critical role in the process.
2. Materials and methods
2.1. Zebrafish (Danio rerio) rearing and surgeries
All animal work was performed in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, and approved by the University of Michigan Committee on the Use and Care of Animals, protocol 06034. Sexually mature adult (4–18 month old) zebrafish were spawned in our fish facility and raised per standard protocol at 28°C with a 14-h light/10-h dark alternating cycle.
Adult zebrafish were anesthetized (0.05% Tricaine Methanosulfate) and about 50% of the LR muscle was surgically excised, i.e. myectomy [4]. The remaining muscle following surgery (48.42% ± 4.9%, average ± S.D.) is represented in the figures as a grey area. The length of the regenerating muscle was quantified by craniectomy as described previously [4]. Regeneration is represented as the relative size of the injured LR muscle normalized to the length of the uninjured LR muscle (representing 100%). All experiments were performed using 5 fish per experimental group and/or time point, unless stated otherwise in the text and/or figure legend.
To label muscles with a marker other than EGFP, transgenic α-actin-mcherry and double transgenic α-actin-mcherry/hsp70-dnFGFR1a:EGFP fish [31] were generated by co-injecting 100 pg acta1-mCherry plasmid (kindly provided by Dr. Thomas Hall) with 50 pg transposase mRNA into 1-cell stage wild type or hsp70-dnFGFR1a:EGFP embryos. For each fish line, mCherry positive fish were crossed to one another to generate F1 embryos, which were raised and used for experiments.
2.2. Drug treatments
SU5402 (Sigma-Aldrich, St. Louis, MO) was dissolved in DMSO as a 17 mM stock and added to fish water at a final concentration of 17 μM as described [10], tanks were kept in the dark. U0126 (Fisher Scientific, Pittsburgh, PA) was dissolved in DMSO as a 25 mM stock and added to fish water at a final concentration of 25 μM [32]. Up to 5 fish were treated in 1 liter of water, tanks were maintained at 28.5°C and drug solutions were replaced every 24 h. Drug treatments were performed immediately after surgery and no significant mortality was noted.
2.3. Adult heat induction experiments
Heat-shock treatments were performed on adult zebrafish directly in the housing racks using our customized system, as described [33]. Fish were exposed to warm water (38.0 – 38.3ºC) for 1 hour daily to drive the ectopic expression of dnFGFR1a:EGFP [31]. The transgenic fish line hsp70-GFP fish [34] was used as a technical control. All experiments were performed using 5 fish per experimental group and/or time point, unless stated otherwise in the text and/or figure legend.
2.4. Specimen processing
Zebrafish heads were excised and fixed in 4% paraformaldehyde (PFA) overnight at 4ºC. Decalcification was performed using either Morse’s solution or 10% ethylenediamine-tetraacetic acid (EDTA) in phosphate-buffered saline (PBS, pH 7.2–7.4). Fixed and decalcified tissues were cryopreserved with 20% sucrose in PBS, embedded in OCT (Fisher Scientific), frozen and evaluated microscopically using coronal frozen sections (12 μm) as described previously [4]
2.5. Staining and Biochemical Analysis
Immunostaining was performed as described [4]. Briefly, slides were washed in PBS for 5 minutes and placed in blocking solution (5% goat serum in PBS +0.2% Tween, PBST) for 30 minutes. Slides were incubated in a humid chamber for overnight at 4ºC in primary antibody (rabbit anti-phospho-p44/42 MAPK [Erk 1/2], #4370 from Cell Signaling Technology, Danvers, MA) diluted to 1:200 in PBST + 1% goat serum. Slides were again washed 4 times for 5 minutes in PBST and then incubated in the dark with Alexafluor 647-conjugated goat anti-rabbit secondary antibody (Invitrogen) diluted 1:1000 in PBST + 1% goat serum. Nuclei were stained with DAPI and coverslipped with ProLong Gold Antifade Reagent. Negative control experiments with no primary or secondary antibodies were performed. Sections from 5 different fish per experiment were stained.
Western blots were performed following standard protocols. Injured or uninjured LR muscles from 10 to 15 fish were pooled and homogenized in lysis buffer containing protease (cOmplete, Roche Diagnostics Corporation, Indianapolis, IN) and phosphatase (PhosSTOP, Roche Diagnostics Corporation) inhibitors. The transgenic α actin-EGFP fish were used to visualize the muscles. Samples were sonicated and centrifuged at 10,000 g for 10 min at 4ºC. Supernatant was collected and protein concentration determined using the Pierce™ BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) and bovine serum albumin (BSA) as standard. Equal amounts of protein (20 μg) were loaded on 7.5% (Erk) or 12.5% (Caspase 3) SDS polyacrylamide gels covered with a 3.9% stacking polyacrylamide gel and separated at 130 V for 1 h using Mini-Protean III (Bio-Rad, Hercules, CA). Proteins were then electroblotted into PVDF membranes (Bio-Rad) by wet transfer (Mini Trans-Blot® Cell, Bior-rad) at 100 W for 1 h. Membranes were blocked for 1 h at room temperature with 5% BSA in TBST and incubated overnight at 4ºC with primary antibody diluted in blocking solution. Anti-γ-tubulin antibody (1:10000, T5326) was obtained from Sigma-Aldrich, active caspase 3 antibody (1:500, ab13847) from abcam (Cambridge, MA), anti-p44/42 MAPK (Erk 1/2) (1:1000, #9107) and anti-phospho-p44/42 MAPK (Erk 1/2) (1:2000, #4370) were purchased from Cell Signaling Technology. Membranes were washed in TBST and incubated with IgG-horseradish peroxidase conjugate secondary antibody (1:10000, anti-mouse #7076 and anti-rabbit #7074 from Cell Signaling Technology) at room temperature for 1 h. Detection of signal was done using WesternBright ECL HRP substrate (advansta, Menlo Park, CA) and an Azure c500 Western Blot Imaging System (azure biosystems, Dublin, CA). Densitometric quantification of the bands was done with ImageJ software (http://rsbweb.nih.gov/ij/). The intensity of the protein of interest is normalized to the intensity of the tubulin band and represented in arbitrary units.
2.6. EdU Incorporation Assays
Cellular proliferation was assessed by intra-peritoneal (IP) injections of 5-ethynyl-2′-deoxyuridine (EdU) and standard detection methods [35]. Fish were anesthetized and injected with EdU (20 μl, 10 mM EdU in PBS) at 26 hours post injury (hpi) and sacrificed four hours later (30 hpi). For each experiment, 3 fish per group were analyzed. The injured muscle of each fish was analyzed and EdU+ or total (DAPI) nuclei were counted from 3 nonconsecutive sections per muscle, representing approximately 1800 total nuclei (range 812 – 3016) per muscle. Cell proliferation is represented as the percentage of EdU+ nuclei in the injured muscle.
2.7. Cell death analysis
Terminal Transferase dUTP Nick End Labeling (TUNEL) assay was performed on frozen sections using the TUNEL Apo-Green Detection Kit (Biotool, Houston, TX), with some minor modifications. The tissue sections were permeabilized in 0.5% Triton X-100 PBS for 10 minutes before TdT reaction. TdT reaction was performed at 37 °C for 1 h per manufacturer’s suggestion. Positive controls were generated by treating tissue sections with DNase I (3000 U/ml in 50 mM TRIS, 10 mM MgCl2, 1 mg/ml BSA buffer) for 10 minutes at room temperature before performing the TdT reaction. Negative controls were performed with no enzyme in the TdT reaction mix (Fig. 2D). Five fish per group were analyzed.
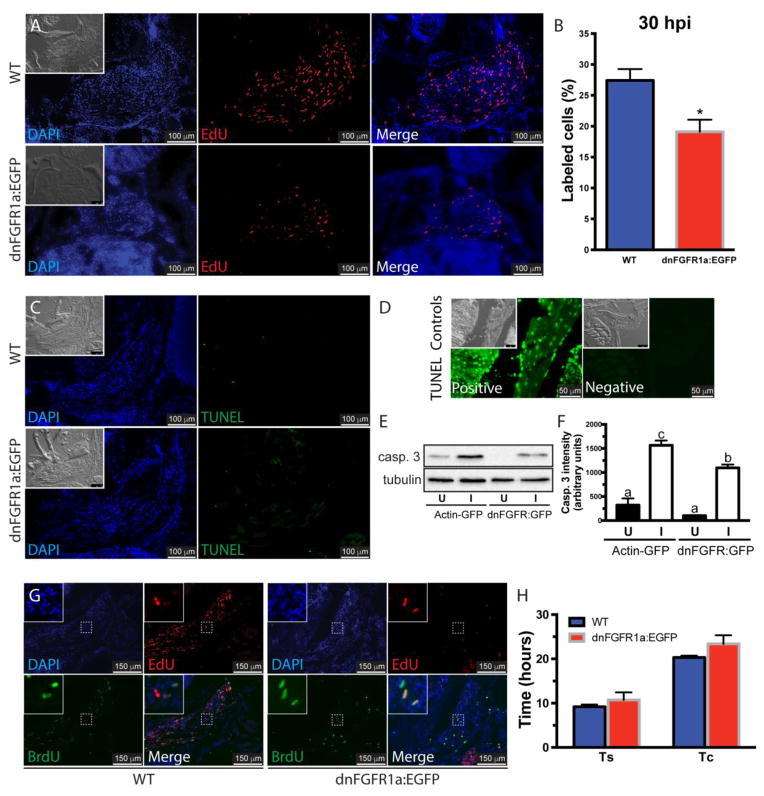
Figure 2. Inhibition of Fgf signaling reduces proliferation in the regenerating muscle.
The role of Fgf in cell proliferation was assessed using WT and hsp70-dnFGFR1a:EGFP fish at 30 hours post-injury (A). DAPI, blue; EdU, red; the insets show the differential interference contrast image. Cell proliferation in the injured muscles from hsp70-dnFGFR1a:EGFP fish was significantly lower than in WT fish (B). Values represent average ± SEM (Student’s t-test, *P < 0.05, n=3). Cell death was assessed by TUNEL assay in WT and hsp70:dnFGFR1a-EGFP fish at 48 hpi showing no apoptosis activation (pictures are representative examples of 5 fish per group), the insets show the differential interference contrast image (C). TUNEL staining DNase treated positive and negative controls, the insets show the differential interference contrast image (D). Ectopic expression of the dominant negative receptor in hsp70:dnFGFR1a-EGFP fish reduced the levels of activated caspase 3 (casp. 3) in the injured muscle compared to α actin-EGFP fish, suggesting a role of caspase 3 in myocyte dedifferentiation during muscle regeneration. Tubulin was used as a loading control (E). Densitometric quantification of the bands in 3 independent experiments (F). Different letters indicate significant differences among groups (P < 0.05, Newman-Keuls multiple comparisons test). The role of Fgf in the length of the S phase (Ts) and the cell cycle (Tc) was measured using the same fish lines (G). DAPI, blue; EdU, red; BrdU, green. In WT and hsp70-dnFGFR1a:EGFP fish, Ts and Tc were not different (H). Values represent average ± SEM (Student’s t-test; Ts, P=0.49; Tc, P=0.24; n=3). U, uninjured muscle; I, injured muscle.
2.8. Cell cycle length quantification
The cell cycle length (Tc) and the length of S-phase (Ts) were estimated following using sequential labeling with EdU and BrdU. A published protocol [36] was followed. EdU and BrdU were injected and detected as described [4]. A 3-hour interval was used for determining Ts (EdU injection at 24 hpi and BrdU at 27 hpi) and an 18-hour interval for determining Tc (EdU at 24 hpi and BrdU injection at 42 hpi). Single- and double-labeled nuclei were counted, and Ts and Tc were calculated as described previously [36]. Calculations were based on 3 fish per group (WT or hsp70-dnFGFR1a:EGFP) and per experiment (Ts or Tc).
2.9. Statistics
Comparisons between 2 groups were analyzed by Student t-test (*P < 0.05; **P < 0.01; ***P< 0.001). When more than 2 groups were compared, one-way analysis of variance (ANOVA, P < 0.05) followed by Newman-Keuls multiple comparisons test (P < 0.05) was performed. Thus, in the time course experiments, differences between fish groups for each time point were analyzed by Student t-test and differences among time points for each fish group were analyzed by ANOVA. All tests were performed using the statistical software Prism 6.03 (GraphPad, LaJolla, CA, USA) for Mac OS X (Apple, Cupertino, CA, USA).
3. Results
3.1. Inhibition of Fgfr impairs adult zebrafish muscle regeneration
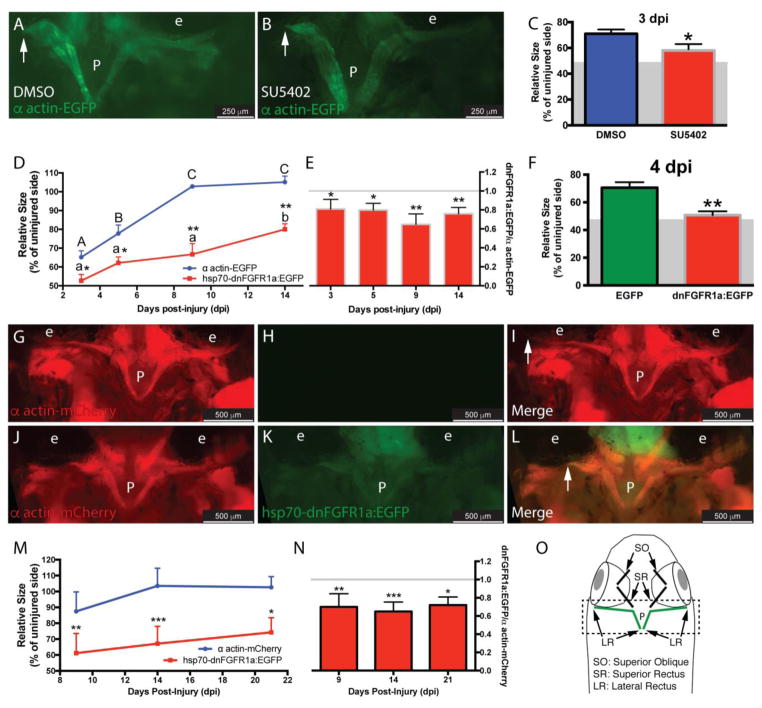
Based on its known roles in tissue regeneration in different species, including zebrafish, we tested whether Fgfs participate in the EOM regeneration of adult zebrafish. As a first step, we used the drug SU5402, an inhibitor that disrupts Fgfr signaling by binding to the tyrosine kinase domain [37] and is effective in adult zebrafish [10]. Transgenic α actin-EGFP fish underwent LR myectomy and treatment with SU5402 for 3 days (Fig. 1A–B). The measured length of the regenerating muscle [4] was significantly lower in SU5402 treated fish compared with control (Fig. 1C - the residual muscle left following myectomy surgery [48.42 ± 4.9%, average ± S.D.] is represented as a grey area in the figure), revealing a role in EOM regeneration.
Figure 1. Inhibition of Fgfr impairs muscle regeneration.
Myectomized α actin-EGFP fish were treated with the Fgfr inhibitor SU5402 (B) or DMSO (A) for 3 days. The length of the regenerating muscle (C) was significantly lower in treated fish; values are averages ± SD (Student’s t-test, *P < 0.05, n=5). To confirm this finding, α actin-EGFP and hsp70-dnFGFR1a:EGFP fish were myectomized and daily heat-shocked, at selected time points (3, 5, 9, and 14 dpi) the regenerating muscle was measured as before (D); values are averages ± SD. For each time point, differences between α actin-EGFP and hsp70-dnFGFR1a:EGFP fish were analyzed by Student’s t-test (*P < 0.05; **P < 0.01, n=5). For each group (α actin-EGFP or hsp70-dnFGFR1a:EGFP), differences among time points were analyzed by ANOVA. Different letters (lowercase for hsp70-dnFGFR1a:EGFP and uppercase for α actin-EGFP) indicate significant differences among positions (P < 0.05, Newman-Keuls multiple comparisons test). Values were normalized to the average of the α actin-EGFP group at each time point revealing that the regeneration of the hsp70-dnFGFR1a:EGFP (no differences among different time points, P= 0.09, ANOVA) was around 75% of the control group (represented with a gray line) (E); values are averages ± SD (Student’s t-test, *P < 0.05, **P < 0.01, n=5). The technical control experiment comparing hsp70-EGFP and hsp70-dnFGFR1a:EGFP fish showed similar results; values are averages ± SD (Student’s t-test, **P < 0.01, n=5) (F). To further confirm our results, a double transgenic α-actin-mCherry/hsp70-dnFGFR1a:EGFP fish line (J-L, 9 dpi) was generated and used in a heat-shock experiment with α-actin-mCherry fish as control group (G-I, 9 dpi). The length of the regenerating muscle was measured at selected time points (9, 14, and 21 dpi) as described (M); values are averages ± SD. For each time point, differences between α actin-mcherry and α actin-mCherry/hsp70-dnFGFR1a:EGFP fish were analyzed by Student’s t-test (*P < 0.05; **P < 0.01, ***P<0.001, n=5–6). There were no differences among the time points for any of the fish groups analyzed (ANOVA, P=0.08 and P=0.31, respectively). Values were normalized to the average of the α actin-mCherry group at each time point revealing that the regeneration of the α actin-mCherry/hsp70-dnFGFR1a:EGFP (no differences among different time points, P= 0.64, ANOVA) was around 70% of the control group (represented with a gray line) (N); values are averages ± SD (Student’s t-test, *P < 0.05, **P < 0.01, n=5). Diagram of a craniectomized zebrafish head (O); muscles visualized by the craniectomy technique are shown, and LR muscles are highlighted in green. The dashed box represents the pictures used to measure the muscle length. The residual muscle left following myectomy surgery (48.42 ± 4.9%, average ± S.D.) is represented as a grey area in C and F. The white arrows in panels A, B, I and L mark the end of the regenerating muscle (fully regenerated at 9 dpi in I). P, pituitary; e, eye.
To further test whether Fgf signaling plays a role in muscle regeneration, we utilized a genetic approach. We took advantage of the hsp70-dnFGFR1a:EGFP fish line [31] which, following a heat-shock, expresses a dominant-negative Fgf receptor that blocks Fgf signaling by competitive inhibition. Thus, α actin-EGFP (control group) or hsp70-dnFGFR1a:EGFP (experimental group) fish underwent myectomy followed by a daily heat-shock over the course of 14 days. At selected time points, regeneration was assessed by measuring the length of the regenerated muscle (Fig. 1D). Expression of dnFGFR1a-EGFP reduced the regeneration of the injured muscle at all the time points, even at 14 days post injury - about twice the time that WT fish usually require [4]. A control experiment using the hsp70-EGFP fish as a control group instead of α actin-EGFP fish validated the results (Fig. 1F - the residual muscle left following myectomy surgery [48.42 ± 4.9%, average ± S.D.] is represented as a grey area in the figure).
To better assess the temporal response, a double transgenic α actin-mCherry/hsp70-dnFGFR1a:EGFP fish line was generated (Fig. 1J–L). Using this double transgenic line in an extended heat-shock time-course experiment revealed that the injured muscle did not fully regenerate even after 21 days (Fig. 1M), a powerful demonstration of the persistent importance of Fgf signaling in the regeneration process.
3.2. Ectopic expression of dnfgfr1a-EGFP reduced cell proliferation during muscle regeneration
The regeneration of adult zebrafish extraocular muscles requires myocyte dedifferentiation to myoblasts, resulting in cell cycle reentry and a proliferative burst at 24–48 hpi. This is followed by a gradual decline in proliferation, migration of cells into the regenerating field, and myocyte redifferentiation [4]. Fgf signaling could be important either in the early dedifferentiation steps leading to proliferation, and/or in the subsequent steps of migration and redifferentiation. In order to assess whether Fgf signaling is important for dedifferentiation, we utilized the endpoint of proliferation. Zebrafish were treated with EdU at 26 hpi to label cells in S-phase, and EdU incorporation into dedifferentiated myocytes was assessed at 30 hpi. The percentage of EdU labeled cells in the regenerating muscle was significantly reduced by the ectopic expression of dnFGFR1a-EGFP (Fig. 2A–B). This loss of proliferating cells was not due to cell death as measured by TUNEL assay (Fig. 2C). Active caspase 3 was detected in intact extraocular muscles suggesting a role in maintaining tissue homeostasis. Interestingly, the regenerating muscle of control fish showed higher levels of active caspase 3 that were reduced by ectopic expression of dnFGFR1a-EGFP (Fig 2. E, F), suggesting a role for caspase 3 in myocyte reprogramming after injury. Overall these experiments reveal that Fgf signaling plays a role in the control of cytoplasmic remodeling and cell proliferation during the early time points following a partial lateral rectus muscle extirpation.
Next, the role of Fgf in regulating cell cycle dynamics was assessed by measuring the length of S-phase (Ts) and the cell cycle (Tc) via sequential injections of EdU and BrdU (Fig. 2G) as described elsewhere [4, 36]. The ectopic expression of dnFGFR1a:EGFP did not alter the length of either both Ts or Tc (Fig. 2H). Therefore, we conclude that Fgf signaling is primarily affecting cell cycle reentry rather than dynamics, supporting again the hypothesis that Fgf signaling functions during myocyte dedifferentiation following partial muscle extirpation.
3.3. Fgf promotes EOM regeneration through the Mapk/Erk signaling pathway
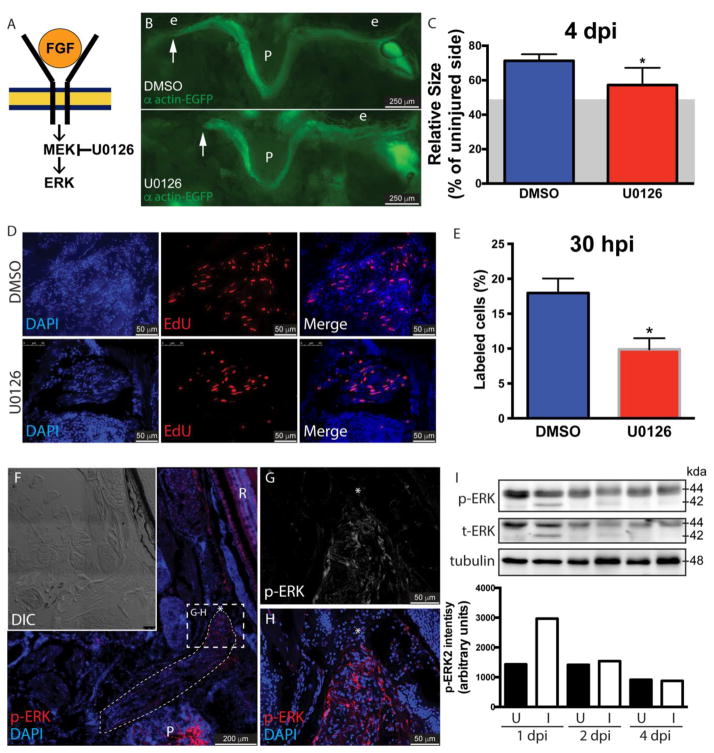
The Mapk/Erk pathway is considered the main signaling pathway downstream of Fgf receptor. Upon ligand binding, Fgfr activates Ras (a G protein) to activate Raf (a MAPKKK). Active Raf phosphorylates and activates Mek (a MAPKK), which phosphorylates Erk (extracellular signal-regulated kinase or MAPK, mitogen-activated protein kinase) [13]. To determine whether this pathway is involved in the regeneration process several approaches were followed. First, α actin-EGFP fish underwent LR myectomy and treatment with the Mek inhibitor U0126 [38] for 4 days (Fig. 3B), as previously described [32]. The length of the regenerating muscle was significantly lower in fish treated with Mek inhibitor (Fig. 3C - the residual muscle left following myectomy surgery [48.42 ± 4.9%, average ± S.D.] is represented as a grey area in the figure). This effect was comparable to treatments with the Fgfr inhibitor SU5402 (Fig. 1C) and the ectopic expression of a dominant-negative Fgf receptor (Fig. 1D, M). Next, wild type fish underwent myectomy and were treated with U0126 to assess cell proliferation at 26 – 30 hpi (Fig. 3D). As with Fgf inhibition (Fig. 2B), Mek inhibition caused a significant reduction in proliferation (Fig. 3E).
Figure 3. Fgf promotes muscle regeneration through its canonical signaling pathway.
Schematic diagram of the Fgf canonical signaling pathway (A). Myectomized α actin-EGFP fish were treated with the MEK inhibitor U0126 or DMSO (B) for 4 days. See Fig. 1O for a diagram of a craniectomized zebrafish head. The length of the regenerating muscle (C) was significantly lower in treated fish; values are averages ± SD (Student’s t-test, *P < 0.05, n=5). The residual muscle following surgery (48.42 ± 4.9%, average ± S.D.) is represented as a grey area. The role of Mek in cell proliferation at 30 hours post-injury was assessed treating WT fish with DMSO or U0126 (D). EdU, red; DAPI, blue. Cell proliferation in the injured muscles from U0126 treated fish was significantly lower than in control fish (E). Values represent average ± SEM (Student’s t-test, *P < 0.05, n=3). The presence of activated Erk (p-Erk) in the injured muscle at 48 hpi was assessed by immunofluorescence (F). The position of the regenerating muscle has been highlighted and the growing end is marked by an asterisk. Higher magnification images showed the presence of p-Erk in the most distal part of the regenerating muscle (G, H), the regenerating end is marked by an asterisk. Images are representative examples from 5 fish. The activation of Erk in the injured muscle was further assessed by western blot in a time course experiment (I). Immunoblotting was performed with anti-Erk1/2 antibody. Total amounts of Erk1/2 were monitored by reprobing membranes with anti-Erk1/2 antibody. Note that Erk2 (p42 MAPK), both phosphorylated and total, was rapidly induced in the injured muscle. Tubulin was used as a loading control. The densitometric quantification of the Erk2 (p42 MAPK) bands is shown. U, uninjured muscle; I, injured muscle; P, pituitary; R, retina; e, eye. The white arrow in panels B and C.
Subsequently, we assayed the regenerated muscle for the presence of activated phospho-ERK1/2 (p-ERK1/2) using immunofluorescence at 48 hpi (Fig. 3F–H). The presence of p-ERK1/2 was detected in all the regenerated muscle but accumulated in the regenerating end of the muscle (Fig. 3G–H). Finally, the time course of ERK1/2 activation in the injured muscle was assessed by western blot (Fig. 3I). Both uninjured and injured EOMs expressed ERK1 (p44 MAPK), and both total and phosphorylated ERK1 could be detected by western blot (Fig. 3I). Following myectomy, phosphorylated and total ERK2 (p42 MAPK) forms were rapidly induced at 1 dpi in the injured muscles, consistent with an early role in the regeneration process.
4. Discussion
Fgf is involved in the regulation of multiple critical biological processes that control cell proliferation and maintenance [39]. The importance of Fgf in tissue regeneration has been demonstrated for species with high regenerative capacity like fish [9, 10, 24–26] and amphibians [27, 28, 40], as well as in the few cases in which mammals also regenerate lost tissues [29, 30]. Furthermore, Fgf also promotes muscle repair in response to injury in mammals [22, 23]. As such, Fgf appears to be a universal regulator of tissue repair and regeneration.
In this report, we took advantage of our in vivo model of myocyte dedifferentiation in adult zebrafish EOMs to test for the first time the role of Fgf in zebrafish skeletal muscle regeneration. Using both pharmacologic and genetic approaches, we demonstrate that Fgfr inhibition greatly impaired regeneration (Fig. 1). Importantly, since the regeneration of adult zebrafish EOMs does not depend on satellite cells but instead requires myocyte dedifferentiation to myoblasts [4], we were able to ascertain whether Fgf signaling plays a role in the dedifferentiation process. Caspases are considered the main mediators apoptosis [41] but also play non-apoptotic roles promoting skeletal muscle differentiation [42, 43] and a development-dependent decrease of skeletal muscle caspase 3 levels has been reported [44]. Caspase 3 also plays a non-apoptotic role in EOM biology [45] supporting our results in uninjured muscles (Fig. 2).
In the context of tissue regeneration, it has been shown that caspase-generated signals from apoptotic cells can stimulate the proliferation of neighbour cells after tissue injury [46, 47]. However, we found that reprogramming myocytes contained higher active caspase 3 levels with no detectable levels of apoptosis (Fig. 2). Our findings suggest that caspase 3 plays a non-apoptotic role in myocyte reprogramming that is regulated, at least in partially, by Fgf. Caspase signal transduction is considered irreversible or long-lasting due to their proteolytic activity [48]. Among their many targets [49], several key cell cycle regulators [50–52] and cytoskeletal components [53] have been identified, and caspases can even induce expression of specific genes [42, 54]. Therefore, caspases are suitable candidates to manipulate the cytoskeletal architecture and rearrange the cytoplasm in complex processes such as cell reprogramming. Our present findings and previous reports showing that autophagy activation is promoted by Fgf in regenerating LR muscles to degrade the sarcomeres [55] indicate that Fgf promotes myocyte reprogramming and dedifferentiation during muscle regeneration in zebrafish (Fig. 4).
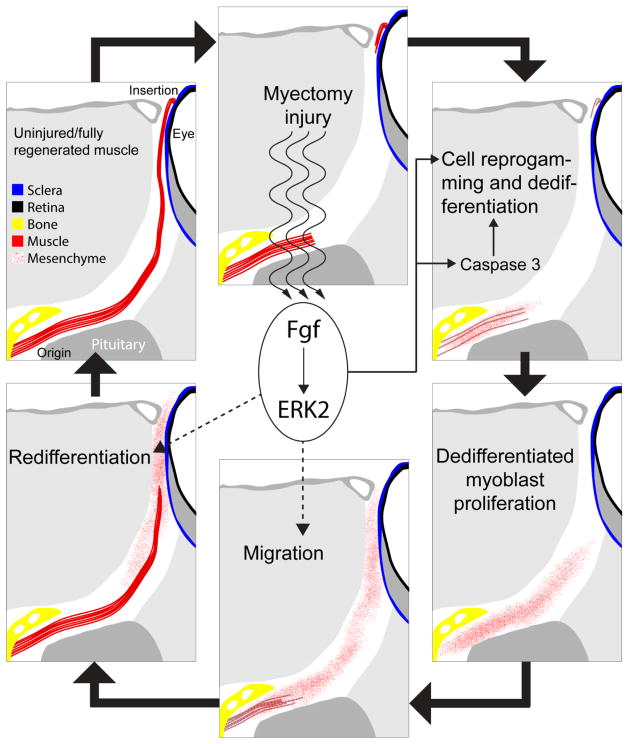
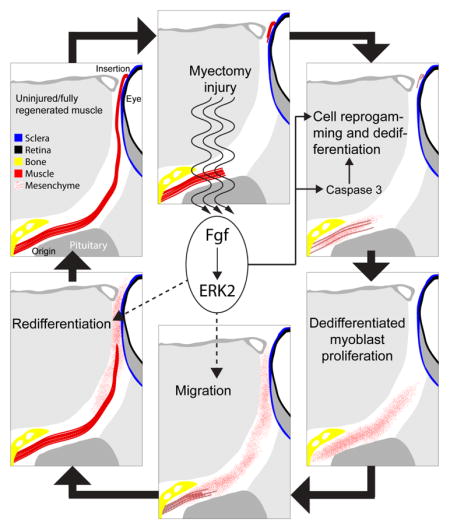
Figure 4. Fgfs roles during EOM regeneration.
Following myectomy injury, Fgf signaling promotes myocyte cell reprogramming and dedifferentiation by activating caspase 3, among others. The known roles of Fgf and our preliminary results (Supplemental Figure 1) suggest that Fgf may also play a role in the regulation of cell migration and redifferentiation. Fgf signal through the Mek/Erk pathway to activate, at early stages of muscle regeneration, ERK2.
Indeed, genetic inhibition of Fgfr resulted in less proliferation (Fig. 2), and treatments with the Mek inhibitor U0126 (Fig. 3) recapitulated the phenotype of Fgfr inhibition (Fig. 1, 2). Inassomuch as proliferation by “post-mitotic” myocytes represents the culmination of the reprogramming and dedifferentiation process we conclude that Fgf signaling through the Mek/Erk pathway is mechanistically important for cellular reprogramming during dedifferentiation. While Fgf signaling has been known to activate satellite cells during skeletal muscle tissue repair in mice [56], its role in cell dedifferentiation has only been clearly demonstrated in human tissue cultures of chondrocytes [57], where it can act as both activator and inhibitor of dedifferentiation, depending on the receptor. Hence, this is the first in vivo demonstration of the role of Fgf signaling in myocyte reprogramming and dedifferentiation to a proliferating myoblast state (Fig. 4).
Fgfr1 and Fgfr4 are known to have important roles in skeletal muscle differentiation and organization during embryogenesis [58, 59]. Indeed, while Fgfr1 is thought to be important for maintaining myoblast cell number and subsequent myotube density (and hence, myoblast proliferation) [58], Fgfr4 is thought to regulate myogenic differentiation [59]. In our model, downregulation of all Fgf receptor functions either pharmacologically or with a dominant negative allele revealed an important role in facilitating myoblast proliferation following dedifferentiation. These findings are consistent with the known role of Fgf receptors during embryogenesis, supporting the hypothesis that embryogenesis and regeneration utilize similar biological pathways even if the exact regulatory programs are somewhat different.
Fgfs were initially discovered for their ability to induce fibroblast proliferation, and the role of Fgf signaling in promoting proliferation in cells and tissues beyond fibroblasts has been bolstered by data from embryogenesis, tissue regeneration and cancer [reviewed in 60, 61]. Fgf signaling has since been shown to act through the MAPK pathway to regulate a variety of cellular processes that also include epithelial to mesenchymal transition and cell migration [13]. In our zebrafish model, myectomy is followed cellular reprogramming that includes a robust muscle-to-mesenchyme transition, culminating in proliferation of dedifferentiated myoblasts [4]. ERK2, a common mediator of Fgf signaling, has been shown to regulate epithelial-mesenchymal transition (EMT) and cell migration in cell culture [62, 63]. Interestingly, we identified ERK2 as the likely mediator of Fgf signaling in EOM regeneration based on its temporal expression and activation by phosphorylation (Fig. 3), linking Fgf signaling with the observed muscle-to-mesenchyme transition. Furthermore, Fgf ligands have shown to be crucial for proper migration of limb muscle precursors [66] as well as in different models from drosophila [64] to mammals [65]. Thus, when assessing muscle regeneration by our whole-mount head craniectomy technique (Fig. 1D), we found an altered morphology of the muscle of dnFGFR1a:EGFP fish, with fibers attached to different parts of the sclera in aberrant orientations (Supplemental Figure 1B, D). In contrast, control fish showed a compact muscle attaching to the sclera in the correct point of the eye (Supplemental Figure 1A, C). This may reflect that Fgf signaling also regulates cell migration during EOM regeneration (Fig. 4), and may be a worthwhile subject for future research.
In conclusion, using an in vivo model of adult myocyte dedifferentiation, we report a new non-apoptotic role for caspase 3 in muscle reprogramming and dedifferentiation during regeneration identifying Fgf signaling as a key regulator of the process. Future efforts will focus on the critical steps of dedifferentiation that require Fgf signaling, the role of ERK2, and on the development of pharmacologic approaches to regulate Fgf signaling and promote tissue regeneration.
Supplementary Material
Pictures of the regenerating muscle of α actin-EGFP fish (A) or hsp70-dnFGFR1a:EGFP fish (B) at 14 dpi (from the experiment shown in Fig. 1D). The arsterisks mark the muscle attachments of the sclera; e, eye. C and D show diagrams of the muscles in A or B, respectively.
Highlights.
Fgf promotes dedifferentiation of regenerating muscles to proliferative myoblasts
Fgf promotes caspase 3 activation with no apoptosis in regenerating muscles
Caspase 3 plays a role in cellular reprogramming of regenerating zebrafish muscles
ERK2 mediates Fgf actions in dedifferentiating zebrafish injured muscles
Acknowledgments
We thank Dr. Thomas Hall for kindly sharing the acta1-mCherry plasmid,
Abbreviations
- EdU
5-ethynyl-2′-deoxyuridine
- EOM
extraocular muscle
- hpi
hours post injury
- dpi
days post injury
- Fgf
Fibroblast growth factor
- Fgfr
Fibroblast growth factor receptor
- LR
lateral rectus
- Tc
cell cycle length
- Ts
length of S-phase
- MAPK
mitogen-activated protein kinase
- MAPKK
MAP kinase kinase
- MAPKKK
MAP kinase kinase kinase kinase
Footnotes
Conflict of interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Webster C, Blau HM. Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somat Cell Mol Genet. 1990;16(6):557–65. doi: 10.1007/BF01233096. [DOI] [PubMed] [Google Scholar]
- 2.Morgan JE, Zammit PS. Direct effects of the pathogenic mutation on satellite cell function in muscular dystrophy. Exp Cell Res. 2010;316(18):3100–8. doi: 10.1016/j.yexcr.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Jopling C, Boue S, Belmonte JCI. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Bio. 2011;12(2):79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 4.Saera-Vila A, Kasprick DS, Junttila TL, Grzegorski SJ, Louie KW, Chiari EF, Kish PE, Kahana A. Myocyte Dedifferentiation Drives Extraocular Muscle Regeneration in Adult Zebrafish. Invest Ophth Vis Sci. 2015;56(8):4977–4993. doi: 10.1167/iovs.14-16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 6.Raya A, Koth CM, Buscher D, Kawakami Y, Itoh T, Raya RM, Sternik G, Tsai HJ, Rodriguez-Esteban C, Izpisua-Belmonte JC. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. P Natl Acad Sci USA. 2003;100:11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitchcock PF, Raymond PA. The Teleost Retina as a Model for Developmental and Regeneration Biology. Zebrafish. 2004;1(3):257–271. doi: 10.1089/zeb.2004.1.257. [DOI] [PubMed] [Google Scholar]
- 8.Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377(4):577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 9.Kan NG, Junghans D, Belmonte JCI. Compensatory growth mechanisms regulated by BMP and FGF signaling mediate liver regeneration in zebrafish after partial hepatectomy. Faseb J. 2009;23(10):3516–3525. doi: 10.1096/fj.09-131730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poss FD, Shen JX, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222(2):347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- 11.Pfefferli C, Jaźwińska A. The art of fin regeneration in zebrafish. Regeneration. 2015;2(2):72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh N. The Fgf families in humans, mice, and zebrafish: Their evolutional processes and roles in development, metabolism, and disease. Biol Pharm Bull. 2007;30(10):1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- 13.Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth F R. 2005;16(2):233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Wang C, Xiao J, McKeehan WL, Wang F. Fibroblast growth factors, old kids on the new block. Semin Cell Dev Biol. 2016 doi: 10.1016/j.semcdb.2015.12.014. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannon K, Kudla AJ, McAvoy MJ, Clase KL, Olwin BB. Differentially expressed fibroblast growth factors regulate skeletal muscle development through autocrine and paracrine mechanisms. J Cell Biol. 1996;132(6):1151–1159. doi: 10.1083/jcb.132.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pownall ME, Isaacs HV. FGF Signalling in Vertebrate Development. San Rafael (CA): 2010. [PubMed] [Google Scholar]
- 17.Nguyen-Chi ME, Bryson-Richardson R, Sonntag C, Hall TE, Gibson A, Sztal T, Chua W, Schilling TF, Currie PD. Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome. Plos Genet. 2012;8(10) doi: 10.1371/journal.pgen.1003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groves JA, Hammond CL, Hughes SM. Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development. 2005;132(19):4211–22. doi: 10.1242/dev.01958. [DOI] [PubMed] [Google Scholar]
- 19.Allen RE, Dodson MV, Luiten LS. Regulation of skeletal muscle satellite cell proliferation by bovine pituitary fibroblast growth factor. Exp Cell Res. 1984;152(1):154–60. doi: 10.1016/0014-4827(84)90239-8. [DOI] [PubMed] [Google Scholar]
- 20.Allen RE, Rankin LL. Regulation of satellite cells during skeletal muscle growth and development. Proc Soc Exp Biol Med. 1990;194(2):81–6. doi: 10.3181/00379727-194-43060. [DOI] [PubMed] [Google Scholar]
- 21.Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Dev Biol. 1986;115(1):129–39. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- 22.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84(1):209–38. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 23.Karalaki M, Fili S, Philippou A, Koutsilieris M. Muscle regeneration: cellular and molecular events. In Vivo. 2009;23(5):779–96. [PubMed] [Google Scholar]
- 24.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 25.Qin Z, Kidd AR, Thomas JL, Poss KD, Hyde DR, Raymond PA, Thummel R. FGF signaling regulates rod photoreceptor cell maintenance and regeneration in zebrafish. Exp Eye Res. 2011;93(5):726–734. doi: 10.1016/j.exer.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldshmit Y, Sztal TE, Jusuf PR, Hall TE, Nguyen-Chi M, Currie PD. Fgf-Dependent Glial Cell Bridges Facilitate Spinal Cord Regeneration in Zebrafish. J Neurosci. 2012;32(22):7477–7492. doi: 10.1523/JNEUROSCI.0758-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen LM, Bryant SV, Torok MA, Blumberg B, Gardiner DM. Nerve dependency of regeneration: The role of Distal-less and FGF signaling in amphibian limb regeneration. Development. 1996;122(11):3487–3497. doi: 10.1242/dev.122.11.3487. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama H, Ide H, Tamura K. FGF-10 stimulates limb regeneration ability in Xenopus laevis. Dev Biol. 2001;233(1):72–79. doi: 10.1006/dbio.2001.0180. [DOI] [PubMed] [Google Scholar]
- 29.Padrissa-Altes S, Bachofner M, Bogorad RL, Pohlmeier L, Rossolini T, Bohm F, Liebisch G, Hellerbrand C, Koteliansky V, Speicher T, Werner S. Control of hepatocyte proliferation and survival by Fgf receptors is essential for liver regeneration in mice. Gut. 2015;64(9):1444–1453. doi: 10.1136/gutjnl-2014-307874. [DOI] [PubMed] [Google Scholar]
- 30.Takeo M, Chou WC, Sun Q, Lee W, Rabbani P, Loomis C, Taketo MM, Ito M. Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature. 2013;499(7457):228–232. doi: 10.1038/nature12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132(23):5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 32.Varga M, Sass M, Papp D, Takacs-Vellai K, Kobolak J, Dinnyes A, Klionsky DJ, Vellai T. Autophagy is required for zebrafish caudal fin regeneration. Cell Death Differ. 2014;21(4):547–556. doi: 10.1038/cdd.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saera-Vila A, Kish PE, Kahana A. Automated Scalable Heat Shock Modification for Standard Aquatic Housing Systems. Zebrafish. 2015;12(4):312–314. doi: 10.1089/zeb.2015.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halloran MC, Sato-Maeda M, Warren JT, Su FY, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127(9):1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 35.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. P Natl Acad Sci USA. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandt MD, Hubner M, Storch A. Brief report: Adult hippocampal precursor cells shorten S-phase and total cell cycle length during neuronal differentiation. Stem Cells. 2012;30(12):2843–7. doi: 10.1002/stem.1244. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276(5314):955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 38.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273(29):18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Wang C, Xiao J, McKeehan WL, Wang F. Fibroblast growth factors, old kids on the new block. Seminars in Cell & Developmental Biology. 2016 doi: 10.1016/j.semcdb.2015.12.014. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama H. Initiation of limb regeneration: The critical steps for regenerative capacity. Dev Growth Differ. 2008;50(1):13–22. doi: 10.1111/j.1440-169X.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 41.Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22(4):526–539. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. P Natl Acad Sci USA. 2002;99(17):11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray TV, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, Zwacka R, Fearnhead HO. A non-apoptotic role for caspase-9 in muscle differentiation. J Cell Sci. 2008;121(Pt 22):3786–93. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- 44.Ruest LB, Khalyfa A, Wang E. Development-dependent disappearance of caspase-3 in skeletal muscle is post-transcriptionally regulated. J Cell Biochem. 2002;86(1):21–28. doi: 10.1002/jcb.10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLoon LK, Rowe J, Wirtschafter J, McCormick KM. Continuous myofiber remodeling in uninjured extraocular myofibers: myonuclear turnover and evidence for apoptosis. Muscle Nerve. 2004;29(5):707–15. doi: 10.1002/mus.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal. 2010;3(145):re8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boland K, Flanagan L, Prehn JH. Paracrine control of tissue regeneration and cell proliferation by Caspase-3. Cell Death Dis. 2013;4:e725. doi: 10.1038/cddis.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuranaga E. Beyond apoptosis: caspase regulatory mechanisms and functions in vivo. Genes Cells. 2012;17(2):83–97. doi: 10.1111/j.1365-2443.2011.01579.x. [DOI] [PubMed] [Google Scholar]
- 49.Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14(4):641–50. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto T, Kikkawa U, Kamada S. Contribution of Caspase(s) to the Cell Cycle Regulation at Mitotic Phase. Plos One. 2011;6(3) doi: 10.1371/journal.pone.0018449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Kabra NH, Cado D, Kang C, Winoto A. FADD-deficient T cells exhibit a disaccord in regulation of the cell cycle machinery. J Biol Chem. 2001;276(32):29815–29818. doi: 10.1074/jbc.M103838200. [DOI] [PubMed] [Google Scholar]
- 52.Frost V, Al-Mehairi S, Sinclair AJ. Exploitation of a non-apoptotic caspase to regulate the abundance of the cdkI p27(KIP1) in transformed lymphoid cells. Oncogene. 2001;20(22):2737–2748. doi: 10.1038/sj.onc.1204367. [DOI] [PubMed] [Google Scholar]
- 53.Helfer B, Boswell BC, Finlay D, Cipres A, Vuori K, Kang TB, Wallach D, Dorfleutner A, Lahti JM, Flynn DC, Frisch SM. Caspase-8 promotes cell motility and calpain activity under nonapoptotic conditions. Cancer Res. 2006;66(8):4273–4278. doi: 10.1158/0008-5472.CAN-05-4183. [DOI] [PubMed] [Google Scholar]
- 54.Larsen BD, Megeney LA. Parole terms for a killer: Directing caspase3/CAD induced DNA strand breaks to coordinate changes in gene expression. Cell Cycle. 2010;9(15):2940–2945. doi: 10.4161/cc.9.15.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saera-Vila A, Kish PE, Louie KW, Grzegorski SJ, Klionsky DJ, Kahana A. Autophagy Regulates Cytoplasmic Remodeling During Cell Reprogramming in a Zebrafish Model of Muscle Regeneration. Autophagy. 2016 doi: 10.1080/15548627.2016.1207015. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Floss T, Arnold HH, Braun T. A role for FGF-6 in skeletal muscle regeneration. Gene Dev. 1997;11(16):2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaoka H, Nishizawa S, Asawa Y, Fujihara Y, Ogasawara T, Yamaoka K, Nagata S, Takato T, Hoshi K. Involvement of fibroblast growth factor 18 in dedifferentiation of cultured human chondrocytes. Cell Prolif. 2010;43(1):67–76. doi: 10.1111/j.1365-2184.2009.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flanagan-Steet H, Hannon K, McAvoy MJ, Hullinger R, Olwin BB. Loss of FGF receptor 1 signaling reduces skeletal muscle mass and disrupts myofiber organization in the developing limb. Dev Biol. 2000;218(1):21–37. doi: 10.1006/dbio.1999.9535. [DOI] [PubMed] [Google Scholar]
- 59.Marics I, Padilla F, Guillemot JF, Scaal M, Marcelle C. FGFR4 signaling is a necessary step in limb muscle differentiation. Development. 2002;129(19):4559–4569. doi: 10.1242/dev.129.19.4559. [DOI] [PubMed] [Google Scholar]
- 60.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 61.Boilly B, Vercoutter-Edouart AS, Hondermarck H, Nurcombe V, Le Bourhis X. FGF signals for cell proliferation and migration through different pathways. Cytokine Growth F R. 2000;11(4):295–302. doi: 10.1016/s1359-6101(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 62.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but Not ERK1 Induces Epithelial-to-Mesenchymal Transformation via DEF Motif-Dependent Signaling Events. Mol Cell. 2010;38(1):114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amatangelo MD, Goodyear S, Varma D, Stearns ME. c-Myc expression and MEK1-induced Erk2 nuclear localization are required for TGF-beta induced epithelial-mesenchymal transition and invasion in prostate cancer. Carcinogenesis. 2012;33(10):1965–75. doi: 10.1093/carcin/bgs227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kadam S, Ghosh S, Stathopoulos A. Synchronous and symmetric migration of Drosophila caudal visceral mesoderm cells requires dual input by two FGF ligands. Development. 2012;139(4):699–708. doi: 10.1242/dev.068791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Gene Dev. 1999;13(14):1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Itoh N, Mima T, Mikawa T. Loss of fibroblast growth factor receptors is necessary for terminal differentiation of embryonic limb muscle. Development. 1996;122(1):291–300. doi: 10.1242/dev.122.1.291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pictures of the regenerating muscle of α actin-EGFP fish (A) or hsp70-dnFGFR1a:EGFP fish (B) at 14 dpi (from the experiment shown in Fig. 1D). The arsterisks mark the muscle attachments of the sclera; e, eye. C and D show diagrams of the muscles in A or B, respectively.