D1bLIC is a subunit of the retrograde IFT motor. Knockdown or knockout of D1bLIC has dose-dependent effects on flagellar assembly, length, motility, and signaling. iTRAQ-based proteomics identifies novel proteins altered in d1blic mutant flagella. TIRF microscopy reveals the kinetics and remodeling of the retrograde motor at the flagellar tip.
Abstract
The assembly of cilia and flagella depends on the activity of two microtubule motor complexes, kinesin-2 and dynein-2/1b, but the specific functions of the different subunits are poorly defined. Here we analyze Chlamydomonas strains expressing different amounts of the dynein 1b light intermediate chain (D1bLIC). Disruption of D1bLIC alters the stability of the dynein 1b complex and reduces both the frequency and velocity of retrograde intraflagellar transport (IFT), but it does not eliminate retrograde IFT. Flagellar assembly, motility, gliding, and mating are altered in a dose-dependent manner. iTRAQ-based proteomics identifies a small subset of proteins that are significantly reduced or elevated in d1blic flagella. Transformation with D1bLIC-GFP rescues the mutant phenotypes, and D1bLIC-GFP assembles into the dynein 1b complex at wild-type levels. D1bLIC-GFP is transported with anterograde IFT particles to the flagellar tip, dissociates into smaller particles, and begins processive retrograde IFT in <2 s. These studies demonstrate the role of D1bLIC in facilitating the recycling of IFT subunits and other proteins, identify new components potentially involved in the regulation of IFT, flagellar assembly, and flagellar signaling, and provide insight into the role of D1bLIC and retrograde IFT in other organisms.
INTRODUCTION
Eukaryotic cilia and flagella are conserved, microtubule-based organelles that play critical roles in cell motility and signaling. Defects in ciliary assembly, motility, or signaling result in a broad spectrum of diseases known as ciliopathies (Yuan and Sun, 2013; Brown and Witman, 2014). Ciliary motility is essential for the determination of the left–right body axis, development of the heart, movement of cerebrospinal fluid, clearance of mucus and particles in the respiratory tract, and sperm motility. Defects in motility can lead to situs inversus or heterotaxy, congenital heart defects, hydrocephalus, respiratory disease, and male infertility, also known as primary ciliary dyskinesia (Li et al., 2015; Praveen et al., 2015; Werner et al., 2015). Defects in ciliary assembly or signaling are linked to other diseases, including include polycystic kidney disease (PKD), Bardet–Biedl syndrome (BBS), and several disorders of the nervous and skeletal systems (Yuan and Sun, 2013; Hou and Witman, 2015).
Ciliary assembly and signaling depend on the intraflagellar transport (IFT) of large particles and associated cargoes driven by kinesin-2 motors to the ciliary tip (anterograde IFT) and dynein 2/1b motors to the cell body (retrograde IFT; Ishikawa and Marshall, 2011; Scholey, 2013; Bhogaraju et al., 2013). Both kinesin-2 and dynein 2/1b are large, multisubunit complexes. In most organisms, kinesin-2 consists of a heterotrimeric complex of two motor subunits and a kinesin-associated protein known as FLA10, FLA8, and FLA3 in Chlamydomonas and KIF3A, KIF3B, and KAP3 in vertebrates. Some organisms contain an additional homodimeric complex, known as OSM3 in Caenorhabditis elegans and KIF17 in vertebrates, that cooperates with heterotrimeric kinesin-2 to build cilia and flagella (reviewed in Scholey, 2013).
The dynein 2/1b motor required for retrograde IFT is considerably larger and more complex than the kinesin-2 motors, and the specific functions of the individual subunits are not well understood (reviewed in Hou and Witman, 2015). Dynein 2/1b is usually a homodimer of two dynein heavy chains (DHCs; known as DYNC2H1 in mammals and dynein 1b heavy chain [DHC1b] in Chlamydomonas) associated with a series of intermediate, light intermediate, and light chain subunits (Supplemental Table S1; Hao et al., 2011; Asante et al., 2014; Schmidts et al., 2015). In humans, mutations in dynein 2 subunits have been linked to a subset of skeletal ciliopathies known as Jeune asphyxiating thoracic dystrophy and short rib polydactyly syndrome (SRPS; Dagoneau et al., 2009; Merrill et al., 2009; Huber et al., 2013; McInerney-Leo et al., 2013; Schmidts et al., 2013a, b, 2015; Okamoto et al., 2014; Taylor et al., 2015; Kessler et al., 2015). Symptoms include shortened limbs, thoracic constriction, and occasionally cystic kidneys and retinal defects. Some of these symptoms are the result of defects in Hedgehog signaling (Huangfu and Anderson, 2005; May et al., 2005; Ocbina and Anderson, 2008; Taylor et al., 2015), but the effect of mutations in different subunits on ciliary assembly can be highly variable.
Null mutations in the DHC in Chlamydomonas, C. elegans, and mice result in short, stumpy (<1 μm) cilia filled with IFT particles, consistent with a defect in recycling of the IFT machinery (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999; Wicks et al., 2000; Huangfu and Anderson, 2005). Temperature-sensitive (ts) mutations in Chlamydomonas DHC1b are more subtle; retrograde IFT is significantly reduced, but flagellar length is maintained at 21°C (Iomini et al., 2001; Engel et al., 2012; Lin et al., 2013; Lechtreck et al., 2013a). However, defects in flagellar gliding can be observed (Shih et al., 2013). Shifting dhc1b ts mutant strains to 32°C reduces the stability of the DHC, decreases both the frequency and velocity of retrograde IFT, and results in shorter flagella, but the kinetics and extent of flagellar loss vary with each allele (Lechtreck et al., 2013a; Engel et al., 2012; Lin et al., 2013).
Several dynein 2/1b light chains (LCs) are subunits of other complexes in the axoneme, making their mutant phenotypes more challenging to interpret. LC8 is a dimerization factor that functions to stabilize the outer dynein arm, I1 inner arm dynein, and radial spokes (Yang et al., 2009). Null mutations in LC8 in Chlamydomonas result in short flagella with reduced retrograde IFT and defects in the assembly of multiple structures within the axoneme (Pazour et al., 1998). However, other LC8 mutations have more limited effect (Yang et al., 2009). Tctex2b is also a subunit of both dynein 2/1b and the I1 dynein, and null mutations in Tctex2b/TCTEX1D2 reduce the stability of both dynein complexes (Dibella et al., 2004; Schmidts et al., 2015). Retrograde IFT and flagellar motility are decreased, but steady-state flagellar length is essentially wild type (WT; Dibella et al., 2004; Schmidts et al., 2015).
Considerable interest has been focused on the roles of the intermediate chain (IC) and light intermediate chain (LIC) subunits in the regulation of cargo binding and ciliary length. Null mutations in the LIC have been identified in C. elegans (xbx-1), Chlamydomonas (d1blic), mice, and Tetrahymena (KO-D2LIC), and most result in shorter cilia that accumulate IFT particles (Schafer et al., 2003; Hou et al., 2004; Rana et al., 2004; Rajagopalan et al., 2009). Similar results have been reported using RNA interference (RNAi) silencing of DLI1 in trypanosomes (Blisnick et al., 2014). However, a recent report of DYNC2LI1 mutations in humans indicated that the length of primary cilia in patient fibroblasts is highly variable, with a substantial increase in the number of hyperelongated cilia (Taylor et al., 2015). Another study using small interfering RNA (siRNA) to knock down DYNC2LI1 reported shorter cilia (Kessler et al., 2015). Mutations in IC subunits (WDR34 and WDR60) have also been associated with variable effects on ciliary assembly and length (Huber et al., 2013; McInerney-Leo et al., 2013; Patel-King et al., 2013; Schmidts et al., 2013b; Asante et al., 2014). One model is that partial loss of IC or LIC function leads to longer cilia, whereas greater loss of function leads to shorter cilia (Asante et al., 2014).
To better understand the function of the LIC subunit in flagellar assembly, motility, and signaling, we reduced expression of dynein 1b–associated LIC (D1bLIC) in Chlamydomonas and compared the resulting phenotypes to those described for mutations in other subunits of the dynein 1b complex. Our results demonstrate that D1bLIC plays a critical role in the stability of DHC1b and that the flagellar phenotypes are extremely sensitive to the amount of active dynein 1b motor in the cell. We also analyzed the composition of the flagellar proteome in a d1blic-knockdown strain to identify the subset of proteins affected by a reduction in D1bLIC. Finally, we generated a D1bLIC–green fluorescent protein (GFP) construct that rescues the d1blic-null and permits detailed examination of the behavior of the retrograde motor during IFT and turnaround at the flagellar tip. These studies provide new insights into the role of D1bLIC in ciliary and flagellar function.
RESULTS
Isolation of strains expressing reduced amounts of D1bLIC
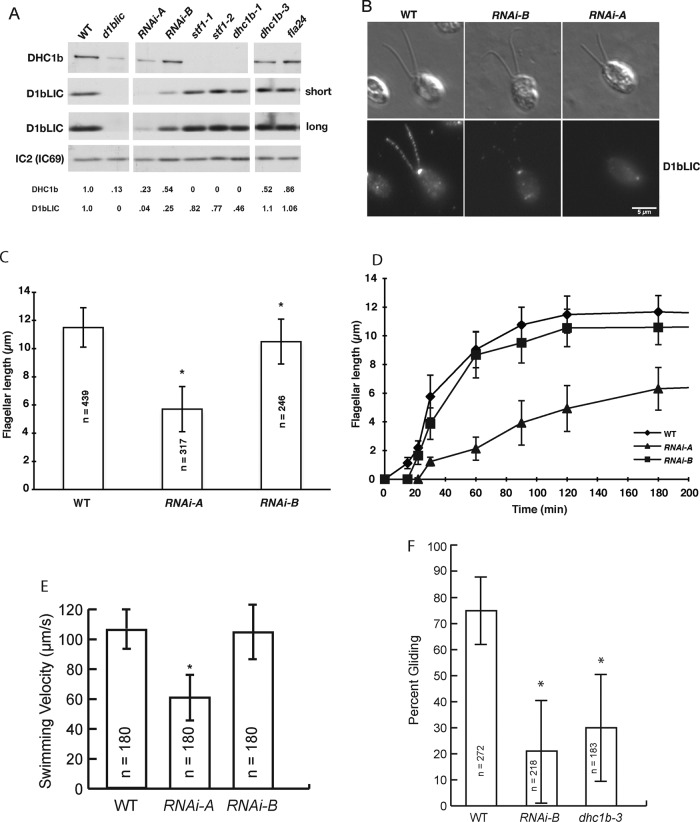
The retrograde IFT motor in Chlamydomonas is composed of multiple subunits (Supplemental Table S1), but the specific contributions of the IC and LIC subunits to IFT, flagellar assembly, and flagellar signaling are poorly understood, in part because very few mutant alleles are available. To isolate strains expressing different amounts of the LIC subunit, we transformed two WT strains with constructs designed to knock down expression of D1bLIC (Supplemental Table S2 and Supplemental Figure S1, A and B). Transformants were screened on Western blots of cell extracts to identify strains with reduced amounts of D1bLIC. Potential knockdowns were rescreened at least twice, and the extent of knockdown was estimated by densitometry. Ten strains with reduced amounts of D1bLIC were identified in 747 transformants, and two strains, 4a2 and 4e11, were retained for further study (Supplemental Figure S1C). The two strains were compared with a d1blic-null strain (Hou et al., 2004) and several dhc1b mutants (Pazour et al., 1999; Porter et al., 1999; Engel et al., 2012; Lin et al., 2013) on Western blots (Figure 1A). The 4a2 strain, now known as RNAi-B, expressed <25% of the WT amount of D1bLIC, and the 4e11 strain, now known as RNAi-A, expressed <5% of the WT amount of D1bLIC. Staining of fixed cells showed that the D1bLIC normally concentrated in the basal body region and flagella of WT cells was greatly reduced in the RNAi strains (Figure 1B). Western blots further demonstrated that reductions in D1bLIC were associated with significant decreases in the amount of DHC1b (Figure 1A), although the decreases were not as severe as that observed in the d1bli-null (Figure 1A; Hou et al., 2004). D1bLIC was also reduced (18–54%) in three dhc1b-null strains (Figure 1A; Perrone et al., 2003; Hou et al., 2004) but not significantly reduced in two dhc1b (ts) mutants, dhc1b-3 and fla24, at the permissive temperature (Figure 1A). Knockdown or knockout of D1bLIC or DHC1b had only minor effects on the stability of dynein 1b–associated IC2 (D1bIC2)/flagellar-associated polypeptide 133 (FAP133) (Supplemental Figure S1D).
FIGURE 1:
Knockdown of D1bLIC alters DHC1b stability, flagellar length, assembly, motility, and gliding. (A) A Western blot of whole-cell extracts was probed with antibodies to DHC1b and D1bLIC to compare the relative amounts expressed in each strain. Two different exposures of the D1bLIC blot are shown here. The outer arm dynein IC2(IC69) served as a loading control. The numbers on the bottom indicated the amount of DHC1b or D1bLIC present in each lane relative to the WT control as determined by densitometry. Knockout (d1blic) or knockdown (RNAi-A and RNAi-B) of D1bLIC had a significant effect on the stability of DHC1b. Knockout of DHC1b (stf1-1, stf1-2, dhc1b-1) was associated with reduced levels of D1bLIC, but the ts dhc1b mutations (dhc1-3, fla24) did not significantly alter D1bLIC at permissive temperatures. (B) WT and the two knockdown strains (RNAi-B, RNAi-A) were fixed and stained with the D1bLIC antibody. Top, DIC images; bottom, fluorescence images. The D1bLIC signal was reduced in the basal body region and flagella of both RNAi strains. (C) The average length of flagella from WT cells compared with that of two d1blic knockdown strains. Only cells with measurable flagella are shown (n = number of flagella, *p < 0.005). (D) Time course of flagellar regeneration after pH shock. Flagellar assembly occurred more slowly in RNAi-A than in WT or RNA-B cells (n = 15–50 cells/time point; only cells with measurable flagella are shown). (E) The average forward swimming velocity of RNAi-A cells was reduced relative to WT cells (n = cells, *p < 0.005). (F) Knockdown of D1bLIC in RNAi-B cells reduced the number of cells gliding on a glass surface. The proportion of RNAi-B and dhc1b-3 ts cells that underwent flagellar gliding was significantly reduced relative to WT cells (p < 0.05) but not significantly different from one another.
Effect of reduced D1bLIC expression on flagellar assembly
Given the extent of D1bLIC knockdown, we anticipated that flagellar assembly and maintenance might be significantly compromised in both strains. However, both the average flagellar length and rate of flagellar regeneration after pH shock were only slightly reduced in the RNAi-B strain (Figure 1, C and D). The phenotype was sufficiently subtle that this strain would not be recovered in most screens for flagellar assembly defects. In contrast, the RNAi-A strain was significantly compromised with respect to both flagellar length and flagellar regeneration. The average flagellar length was less than half the WT length (Figure 1C), and the rate of flagellar regeneration after pH shock was very slow (Figure 1D). WT cells regenerated half-length flagella within 30 min and full-length flagella within 90 min, whereas the RNAi-A strain took >180 min to regenerate half-length flagella (Figure 1D). However, both the average flagellar length and number of flagellated cells (82–95%) of the RNAi-A strain were significantly higher than observed with the d1blic-null strain (<70%; Supplemental Figure S2). Chlamydomonas cells can therefore tolerate a significant reduction (>75%) in D1bLIC levels with only minor effects on flagellar length or assembly, but once D1bLIC levels are reduced below ∼5%, both are severely compromised.
Effect of reduced D1bLIC expression on flagellar motility and signaling
To assess how reductions in D1bLIC might affect other aspects of flagellar behavior, we analyzed the phenotypes of the RNAi strains with respect to forward swimming velocity, photoaccumulation, mating, and gliding. With the RNAi-B strain, forward swimming velocities and photoaccumulation were not significantly different from WT values (Figure 1E and Table 1). In contrast, the average swimming velocity of the RNAi-A strain was significantly reduced, and photoaccumulation in dish assays was also compromised (Figure 1E and Table 1). We then tested the ability of both strains to mate to WT cells of the opposite mating type and form quadriflagellate dikaryons and mature zygotes. Both dikaryons and mature zygotes were readily detected with the RNAi-B strain but infrequently observed with the RNAi-A strain (Table 1). These results might suggest that flagellar signaling was reduced in RNAi-A but not in RNAi-B. However, decreases in swimming velocity, photoaccumulation, and mating could all be indirect consequences of the reduced flagellar length in the RNAi-A strain (Figure 1C).
TABLE 1:
Comparison of flagellar phenotypes in Chlamydomonas retrograde motor mutant strains.
| Strain | DHC1b (%) | Percentage aflagellate | Flagellar length (% WT) | Swimming velocity (% WT) | Phototaxis | Gliding | Mating | IFT velocity (%) (A, R) | IFT frequency (%) (A, R) |
|---|---|---|---|---|---|---|---|---|---|
| WT | 100 | ∼5 | 100 | 100 | Normal | Normal | WT | 100, 100 | 100, 100 |
| DHC1b related | |||||||||
| dhc1b-2 (ts) | ND | 35 | 86 | ND | ND | ND | ND | 150, 68 | 49, 12 |
| dhc1b-3 (ts) | |||||||||
| 21°C | 52 | ∼12 | 100 | 100 | Normal | ND | WT | 100, 75 | 94, 75 |
| 34°C | ND | ∼15 (12–30 h) | 100 (12–30 h) | 50 (12 h) | Change in sign | Reduced | ND | 90, 48 (6 h) | 63, 35 (6 h) |
| ∼55 (36 h) | 95 (36 h) | 89, 40 (12 h) | 58, 20 (12 h) | ||||||
| fla24 (ts) | |||||||||
| 21°C | 86 | ∼35 | 100 | 100 | Normal | ND | WT | 99, 40 | 83, 46 |
| 32°C | ND | ∼95 (5 h) | ∼22 (3.5 h) | ND | Normal (6 h) | ND | ND | ND | ND |
| D1bLIC related | |||||||||
| d1blic mt+ | 13 | ∼30 | ∼14 | Immotile | NA | ND | ND | 105, 47 | 65, 24 |
| d1blic mt− | 13 | ∼30 | ∼22 | 50 | ND | ND | ND | ND | ND |
| RNAi-B | 54 | ∼5 | ∼91 | 100 | Normal | Reduced | WT | 100, 72 | 80, 72 |
| RNAi-A | 23 | ∼11 | ∼49 | 50 | Reduced | NA | Reduced | 85, 44 | 50, 20 |
| LC related | |||||||||
| fla14-1 | ND | ND | 36 | Immotile | NA | ND | ND | 99, 57 | 74, 17 |
| tctex2b | 40–60 | ND | 100 | 70 | ND | ND | ND | 95, ∼50 | 84, <25 |
A, anterograde; R, retrograde. NA, not applicable; ND, not described.
The data used to calculate the values in this table were taken from the following references: dhc1b-2 (Lechtreck et al., 2013a); dhc1b-3 (Engel et al., 2012; Shih et al., 2013); fla24 (Iomini et al., 2001; Lin et al., 2013; this study); d1blic mt+ (T8D9 or TBD9) and d1blic mt− (YH43) (Hou et al., 2004; this study); RNAi-A and RNAi-B (this study); fla14-1 (Pazour et al., 1998; Yang et al., 2009; this study); and tctex2b (Dibella et al., 2004; Schmidts et al., 2015). Flagella of dhc1b-null strains (stf1-1, stf1-2, dhc1b-1) were too short (<1–2 mm) for most assays (Porter et al., 1999; Pazour et al., 1999). The relative concentration of DHC1b present in cell extracts was determined by Western blots as shown in Figure 1A. Samples with ≥30% aflagellate cells are highlighted in bold, as are samples whose flagellar lengths are <50% of WT length. Phototaxis was measured by monitoring photoaccumulation in dishes. Mating tests were performed the absence of added 3-isobutyl-1-methylxanthine (IBMX) or papaverine and dibutyryl-cAMP. Changes in anterograde and retrograde IFT >50% are highlighted in bold. Most mutations had a greater effect on retrograde IFT frequency than retrograde IFT velocity, but reducing retrograde frequency by 50% had only minor effects on flagellar length or assembly. However, larger reductions in retrograde IFT typically resulted in shorter flagella and/or more aflagellate cells.
We therefore tested the ability of both RNAi strains to adhere to a glass surface and undergo flagellar gliding. Previous study showed that flagellar gliding was reduced in the dhc1b-3 (ts) mutant, even though the cells assembled full-length flagella (Engel et al., 2012; Shih et al., 2013). We found that the majority of RNAi-A cells did not adhere tightly to the glass surface, even in the presence of motility inhibitors, and we detected very few gliding cells. In contrast, WT, RNAi-B, and dhc1b-3 cells adhered to the glass in the presence of motility inhibitors. However, the percentage of RNAi-B cells that exhibited robust gliding was reduced relative to WT cells and similar to that observed with dhc1b-3 cells (Figure 1F; Shih et al., 2013). Thus adhesion-induced flagellar signaling (as assayed by gliding) is more sensitive to decreased retrograde motor activity than flagellar assembly or motility.
Effect of reduced D1bLIC expression on intraflagellar transport
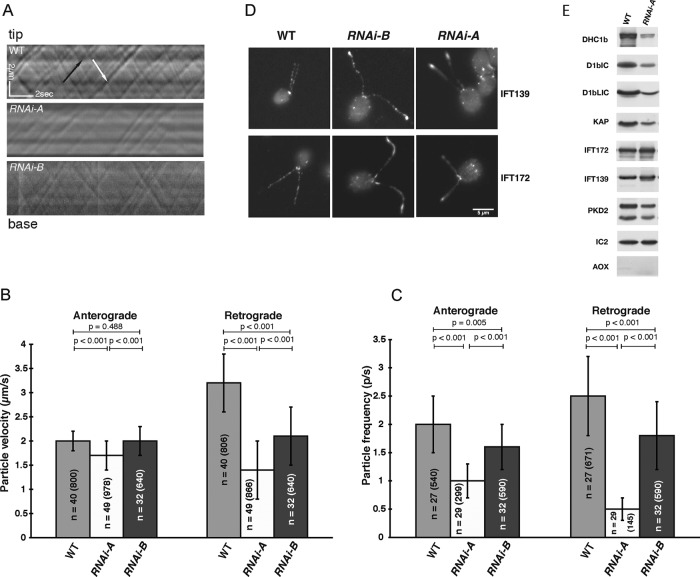
To determine the effect of D1bLIC knockdown on IFT, we analyzed the frequency and velocity of particle movement by differential interference contrast (DIC) microscopy (Figure 2A and Supplemental Videos S1 and S2). Knockdown of D1bLIC below 25% reduced the velocity of retrograde IFT by 28%, with no significant effect on the velocity of anterograde IFT (Figure 2B, RNAi-B). The frequency of retrograde and anterograde IFT was reduced by 28 and 20%, respectively (Figure 2C). Knockdown below 5% reduced the velocity of retrograde IFT by 56%, with only minor effects on the velocity of anterograde IFT (Figure 2B, RNAi-A). However, the frequency of retrograde and anterograde IFT was reduced by 80 and 50% respectively (Figure 2C, RNAi-A). Apparently the failure to efficiently recycle IFT particles or some other factor back to the cell body reduces the pool of particles actively engaged in anterograde IFT (see Discussion). If the defect is severe enough, as in the case of the RNAi-A strain, steady-state flagellar length is reduced (Figure 1C).
FIGURE 2:
Knockdown of D1bLIC alters intraflagellar transport in a dose-dependent manner. (A) Kymographs of IFT obtained by DIC microscopy from WT, RNAi-A, and RNAi-B flagella. Anterograde particles (black arrow) move from base to tip of the flagellum, and retrograde particles (white arrow) move from tip to base. See Supplemental Videos S1 and S2. (B) Average velocities of IFT particles moving in the anterograde and retrograde directions (n = number of flagella and particles measured). Knockdown of D1bLIC primarily affected the velocity of retrograde particles. (C) Average frequencies of IFT particles experiencing anterograde or retrograde transport. Knockdown of D1bLIC had a greater effect on retrograde frequency than anterograde frequency. (D) Fluorescence images of fixed cells stained with antibodies against IFT subunits revealed that both complex A (IFT139) and complex B (IFT172) proteins accumulated in the flagella of the RNAi strains, especially near the flagellar tips. Scale bar, 5 μm. (E) A Western blot of flagella samples used for iTRAQ labeling and mass spectrometry was probed with antibodies to different flagellar proteins. The outer arm dynein IC2(IC69) served as a loading control, and alternative oxidase (AOX) served as a control for potential cell body contamination.
To test this hypothesis more directly, we stained fixed cells with antibodies against IFT particle subunits and analyzed the polypeptide content of isolated flagella by Western blotting (Figure 2, D and E). Staining with antibodies against the IFT139 subunit of complex A and the IFT172 subunit of complex B demonstrated that both subunits were localized in the basal body region and the two flagella of the WT strain (Figure 2D), as previously described (Cole et al., 1998). However, in the RNAi strains, both IFT139 and IFT172 became more concentrated in the two flagella, especially at the flagellar tips (Figure 2D). Analysis of isolated flagella by Western blotting confirmed that IFT139 and IFT172 were increased in RNAi-A flagella (Figure 2E). However, subunits of the retrograde motor (DHC1b, D1bLIC, D1bIC2/FAP133) and the KAP subunit of the anterograde motor were decreased (Figure 2E).
Identification of proteins altered in d1blic flagella by isobaric tag for relative and absolute quantitation labeling and mass spectrometry
To obtain a more quantitative understanding of the changes in total flagellar polypeptide composition, we isolated flagella from WT and RNAi-A cells and subjected them to isobaric tag for relative and absolute quantitation (iTRAQ) labeling and mass spectrometry. Samples of WT and RNAi-A flagella were digested with trypsin, labeled in duplicate with four different iTRAQ tags, fractionated by ion-exchange and capillary liquid chromatography, and analyzed by mass spectrometry. Searching with latest version of the Chlamydomonas database (V5.5) identified 1010 and 1131 proteins with high confidence in two different experiments at a false discovery rate of 5%, and for most proteins, no significant differences in protein levels were observed between the WT and RNAi-A samples. However, a small number of proteins significantly (p < 0.05) increased or decreased in both experiments (Table 2). As expected, nearly all of the IFT and BBSome subunits increased >50% in both experiments. The flagellar TPR protein FAP256 also increased, consistent with its enrichment at the flagellar tip in short flagella (Satish Tammana et al., 2013). Other proteins increased in RNAi-A flagella include six small proteins (22–46 kDa) potentially involved in stress responses (IPY3, glutathione S-transferase [GST], PRX2, FAP179, ICL1, GDP-d-mannose phosphorylase), three proteins involved in RNA binding and translation (RB47, alanine tRNA synthetase, ABC4/EF-3), one uncharacterized protein (Cre05.g239500), the chaperone HSP90α2, and a predicted cNTP-regulated protein kinase known as FAP295 (Tables 2 and 3).
TABLE 2:
Polypeptide ratios in WT and d1blic (RNAi-A) flagella determined by iTRAQ analysis.
| Experiment 1 | Experiment 2 | ||||||
|---|---|---|---|---|---|---|---|
| Polypeptide | Chlamydomonas genome ID (V5.5) | Peptides | WT/WT | d1blic/WT | Peptides | WT/WT | d1blic/WT |
| Decreased | |||||||
| Retrograde motor | |||||||
| DHC1b | Cre06.g250300 | 89 (241) | 1.01 | 0.39, 0.34 | 191 (248) | 1.04 | 0.27, 0.29 |
| FAP133/D1bIC2 | Cre02.g110950 | 4 (7) | 1.00 | 0.61, 0.51 | 11 (13) | 1.05 | 0.35, 0.36 |
| FAP163/D1bIC1 | Cre10.g428664 | 7 (10) | 0.97 | 0.44, 0.51 | 17 (18) | 1.02 | 0.37, 0.36 |
| D1bLIC | Cre09.g398882 | 11 (32) | 1.00 | 0.31, 0.29 | 18 (25) | 1.07 | 0.32, 0.29 |
| Anterograde motor | |||||||
| FLA10 | Cre17.g730950 | 14 (37) | 1.02 | 0.81, 0.70 | 56 (55) | 1.07 | 0.48, 0.48 |
| FLA8 | Cre12.g522550 | 17 (36) | 1.03 | 0.82, 0.88 | 46 (57) | 1.07 | 0.53, 0.52 |
| KAP | Cre10.g449250 | 10 (26) | 1.00 | 0.84, 0.66 | 31 (35) | 1.01 | 0.47, 0.48 |
| Other | |||||||
| RSP17 | Cre07.g340400 | 20 (52) | 1.02 | 0.35, 0.35 | 36 (48) | 1.04 | 0.42, 0.34 |
| WNK1 | Cre17.g700133 | 6 (14) | 1.00 | 0.61, 0.65 | 6 (7) | 1.07 | 0.31, 0.33 |
| FAP24 | Cre02.g081050 | 12 (34) | 1.02 | 0.70, 0.46 | 35 (35) | 1.01 | 0.45, 0.52 |
| PKD2 | Cre17.g715300 | 13 (39) | 1.03 | 0.71, 0.48 | 32 (34) | 1.05 | 0.75, 0.73 |
| FAP5 | Cre16.g687966 | 34 (126) | 1.00 | 0.74, 0.61 | 57 (67) | 1.06 | 0.34, 0.36 |
| FAP208 | Cre11.g482001 | 43 (116) | 1.04 | 0.77, 0.71 | 82 (108) | 1.05 | 0.37, 0.36 |
| TPR protein | Cre17.g747247 | 58 (121) | 1.02 | 0.79, 0.64 | 109 (123) | 1.05 | 0.41, 0.41 |
| FAP26 | Cre04.g215250 | 21 (42) | 1.02 | 0.87, 0.58 | 45 (48) | 1.04 | 0.77, 0.72 |
| FAP48/IP3R | Cre16.g665450 | 61 (199) | 1.00 | 0.92, 0.78 | 122 (139) | 1.03 | 0.79, 0.79 |
| Increased | |||||||
| IFT complex A | |||||||
| IFT144 (FAP66) | Cre13.g572700 | 56 (176) | 1.02 | 1.56, 1.28 | 92 (101) | 1.00 | 1.93, 1.82 |
| IFT140 | Cre08.g362650 | 50 (159) | 1.04 | 1.62, 1.36 | 99 (99) | 0.99 | 1.90, 1.83 |
| IFT139 (FAP60) | Cre06.g268800 | 68 (214) | 1.02 | 1.56, 1.45 | 99 (98) | 0.99 | 1.89, 1.77 |
| IFT121 (FAP118) | Cre11.g475000 | 42 (118) | 1.04 | 1.55, 1.40 | 62 (61) | 0.99 | 2.09, 1.99 |
| IFT43 | Cre06.g251200 | 4 (9) | 0.96 | 1.43, 1.00 | 17 (22) | 1.01 | 1.78, 1.66 |
| IFT complex B | |||||||
| IFT172 | Cre17.g703900 | 86 (244) | 1.04 | 1.59, 1.44 | 171 (158) | 0.98 | 1.76, 1.67 |
| IFT88 | Cre07.g335750 | 36 (115) | 1.05 | 1.59, 1.76 | 71 (86) | 0.99 | 1.80, 1.70 |
| IFT81 | Cre17.g723600 | 38 (123) | 1.02 | 1.60, 1.66 | 68 (88) | 1.03 | 1.71, 1.58 |
| IFT80 | Cre03.g204150 | 29 (80) | 1.01 | 1.58, 1.21 | 41 (42) | 0.99 | 1.83, 1.85 |
| IFT74/72 | Cre01.g027950 | 41 (149) | 1.02 | 1.61, 1.56 | 88 (101) | 1.00 | 1.82, 1.70 |
| IFT70/TPR5 | Cre07.g342200 | 26 (77) | 1.03 | 1.57, 1.78 | 45 (59) | 1.00 | 1.84, 1.85 |
| IFT57 | Cre10.g467000 | 18 (60) | 1.00 | 1.61, 1.82 | 36 (38) | 0.98 | 1.74, 1.55 |
| IFT56 (DYF13) | Cre11.g467616 | 25 (50) | 1.00 | 1.59, 1.33 | 27 (48) | 0.95 | 1.94, 2.04 |
| IFT54 (FAP116) | Cre11.g467739 | 14 (57) | 1.00 | 1.62, 1.76 | 59 (68) | 1.03 | 1.78, 1.68 |
| IFT52 (BLD1) | Cre04.g219250 | 22 (60) | 0.98 | 1.66, 1.45 | 58 (66) | 1.00 | 1.80, 1.80 |
| IFT46 (FAP32) | Cre05.g241637 | 20 (51) | 0.99 | 1.63, 1.30 | 34 (41) | 1.01 | 1.77, 1.66 |
| IFT27 (FAP156) | Cre01.g047950 | 8 (24) | 0.99 | 1.53, 1.67 | 8 (16) | 0.93 | 1.56, 1.44 |
| IFT25 (FAP232) | Cre10.g450350 | 7 (18) | 0.97 | 1.73, 0.88 | 6 (7) | 0.98 | 1.92, 1.95 |
| IFT22/RabL5 | Cre01.g039200 | 9 (16) | 1.03 | 1.49, 0.91 | 22 (21) | 0.97 | 1.60, 1.43 |
| IFT20 | Cre02.g089950 | 9 (27) | 0.99 | 1.60, 1.33 | 19 (21) | 1.05 | 1.75, 1.64 |
| FAP22 (qilin) | Cre17.g721250 | 14 (70) | 0.98 | 1.78, 1.74 | 35 (41) | 1.00 | 1.96, 1.78 |
| BBSome | |||||||
| BBS1 | Cre17.g741950 | 6 (12) | 1.00 | 1.64, 1.23 | 5 (4) | 0.95 | 1.94, 1.89 |
| BBS3 | Cre06.g257250 | 8 (14) | 1.00 | 1.63, 1.18 | 5 (5) | 0.96 | 2.01, 2.01 |
| BBS3 | Cre16.g664500 | 1 (3) | 0.88 | 1.94, 2.28 | 11 (14) | 0.97 | 1.85, 1.72 |
| BBS4 | Cre12.g548650 | 3 (5) | 1.00 | 1.31, 1.25 | 5 (4) | 1.04 | 1.96, 1.94 |
| BBS5 | Cre06.g267550 | 1 (3) | 1.09 | 1.39, 0.95 | 1 (1) | 1.00 | 0.68, 0.94 |
| BBS7 | Cre01.g043750 | 6 (16) | 1.01 | 1.68, 1.34 | 10 (11) | 0.98 | 2.06, 1.80 |
| BBS8 | Cre16.g666500 | 2 (7) | 1.02 | 1.52, 2.03 | 2 (2) | 1.01 | 2.17, 2.16 |
| BBS9 | Cre04.g219700 | 2 (8) | 0.92 | 1.50, 1.26 | 3 (2) | NQ | NQ |
| Other | |||||||
| FAP256 | Cre10.g431150 | 10 (23) | 0.98 | 1.29, 1.39 | 14 (17) | 0.95 | 1.49, 1.34 |
| RB47 | Cre01.g039300 | 6 (9) | 0.98 | 1.31, 1.51 | 6 (8) | 1.00 | 2.3, 2.12 |
| tRNA synthetase | Cre02.g143200 | 4 (6) | 1.09 | 1.42, 1.72 | 14 (13) | 1.04 | 1.32, 1.18 |
| HSP90A | Cre09.g386750 | 6 (12) | 1.00 | 1.42, 1.46 | 8 (13) | 1.00 | 1.47, 1.38 |
| ABC4 | Cre04.g222700 | 13 (19) | 1.01 | 1.44, 1.35 | 42 (48) | 1.06 | 1.32, 1.25 |
| GDP-d-mannose phosphorylase | Cre16.g672800 | 3 (3) | 0.94 | 1.61, 1.56 | 2 (4) | 0.99 | 1.63, 1.50 |
| GST | Cre16.g682725 | 4 (10) | 1.02 | 1.71, 1.83 | 6 (7) | 1.03 | 1.47, 1.70 |
| FAP295 | Cre16.g663200 | 41 (138) | 1.03 | 1.79, 1.71 | 44 (61) | 1.02 | 1.89, 1.71 |
| PRX2 | Cre02.g114600 | 17 (56) | 1.00 | 1.95, 2.54 | 33 (47) | 0.98 | 1.57, 1.45 |
| FAP179/GST | Cre17.g742300 | 17 (45) | 1.02 | 2.08, 2.42 | 21 (24) | 0.93 | 3.35, 3.30 |
| ICL1 | Cre06.g282800 | 25 (102) | 1.09 | 2.62, 2.62 | 93 (86) | 0.97 | 2.53, 2.30 |
| Unknown | Cre05.g239500 | 25 (45) | 1.02 | 2.30, 1.68 | 32 (38) | 0.89 | 5.52, 5.19 |
Peptides refers to the number of peptides identified at 95% confidence and the number of peptides used for iTRAQ quantification. Both WT and mutant samples were analyzed in duplicate. WT/WT ratios typically varied <10%. Mutant/WT ratios that were statistically significant (p < 0.05) are shown in bold type. NQ = not quantified.
TABLE 3:
Characteristics of polypeptides altered in d1blic (RNAi-A) flagella.
| Protein | V5.5 genome ID | Amino acid residues (molecular weight, kDa) | Predicted domains | Homo sapiens best hit |
|---|---|---|---|---|
| Decreased | ||||
| RSP17 | Cre07.g340400 | 926 (98) | ND | ND (Volvox, 0.0) |
| WNK1 | Cre17.g700133 | 733 (77) | S/T kinase | AURKA (NP_003591) 2e-39, aa 238–520 |
| FAP24 | Cre02.g081050 | 529 (59) | TroA-d | ND (Volvox, 2e-149) |
| PKD2 | Cre17.g715300 | 1644 (183) | Polycystin cation channel | PKD2L1 (NP_057196) 3e-19, aa 1056–1418 |
| FAP5 | Cre16.g687966 | 1307 (133) | TPR repeats | NHPH3 (NP_694972) 1e-22, aa 1001–1283 |
| FAP208 | Cre11.g482001 | 1101 (114) | ANK repeats | ANK2 (NP_066187) 1e-48, aa 676–1079 |
| Predicted protein | Cre17.g747247 | 1846 (185) | AAA, TPR repeats | KLC1 (NP_005543) 6e-19, aa 1585–1781 |
| FAP26 | Cre04.g215250 | 2462 (partial) | ANK repeats | ND (Volvox, 2e-173) |
| FAP48/IP3R | Cre16.g665450 | 3210 (357) | Ins145_P3, RIH, RYDR_ITP, RYDR_IT | ITPR2 (NP_002214) 9e-30, aa 132–977 |
| Increased | ||||
| FAP256 | Cre10.g431150 | 862 (92) | TPR repeat | CEP104 (NP_055519) 1e-72, aa 7–672 |
| RB47 | Cre01.g039300 | 657 (70) | RRM and PolyA binding | PABPC1 (NP_002559) 0.0, aa 17–657 |
| Alanine tRNA synthetase | Cre02.g143200 | 1164 (122) | Alanyl tRNA synthetase, DHHA1 | Alanine tRNA ligase (NP_001596) 0.0, aa 223–1164) |
| HSP90A | Cre09.g386750 | 705 (81) | ATPase, HSP90 | HSP90α2 (NP_005339) (0.0, aa 4–705) |
| ABC4/EF-3 | Cre04.g222700 | 1053 (115) | ABC transporter, Chromo | ABCF3 (NP_060828) 2e-43, aa 450–788 |
| IPY3 | Cre09.g387875 | 192 (22) | Pyrophosphatase | PPA1 (NP_066952) 2e-11, aa 5–149 |
| GDP-d-mannose phosphorylase | Cre16.g672800 | 360 (40) | Nucleotidyl transferase | GMPPB (NP_068806) 3e-154, aa 1–360 |
| GST | Cre16.g682725 | 217 (24) | GST domains | GSTA1 (NP_665683) 2e-20, aa 4–203 |
| FAP295 | Cre16.g663200 | 707 (80) | PK, cNTP binding | PRKG2 (NP_006250) 1e-99, aa 27–663 |
| PRX2 | Cre02.g114600 | 198 (22) | Peroxiredoxin | PRDX4 (NP_006397) 1e-94, aa 3–198 |
| FAP179 | Cre17.g742300 | 219 (25) | GST domains | GSTP1 (NP_000843) 5e-29, aa 5–216 |
| ICL1 | Cre06.g282800 | 417 (46) | Isocitrate lyase | ND (Volvox, 0.0) |
| Unknown | Cre05.g239500 | 704 (73) | AAA | ND (Volvox, 2e-58) |
The predicted amino acid sequence in the Chlamydomonas genome V5.5 was blasted against the National Center for Biotechnology Information database to identify the most closely related sequence in H. sapiens. The name of the best hit, its accession number, blast score, and region of alignment are shown. If no significant hit was detected (ND), then the blast score for the Volvox orthologue is shown.
A different group of proteins were consistently reduced in RNAi-A flagella (Table 2). As expected, subunits of the retrograde IFT motor (DHC1b, D1bLIC, FAP133/D1bIC2, and FAP163/D1bIC1) were reduced (Table 2), but the decreases in DHC1b and D1bLIC were not as dramatic as those observed in cell extracts (Figure 1A). Thus the remaining dynein 1b complex is still being targeted to the flagella. The three subunits of the kinesin-2 motor, FLA10, FLA8, and KAP, were also reduced in RNAi-A flagella, even though IFT particle subunits were increased (Table 2). These observations suggest that kinesin-2 does not depend on retrograde IFT for exit from the flagella. Nine other proteins were consistently reduced in RNAi-A flagella; these included two large membrane proteins implicated in cell signaling (PKD2 and FAP48/IP3R), two large TPR proteins (FAP5 and Cre17.g747247), two ankyrin repeat proteins (FAP26 and FAP208), FAP24, radial spoke protein 17 (RSP17), and an Aurora A–like S/T kinase annotated as WNK1 (Tables 2 and 3). Western blots confirmed the decrease in PKD2 in RNAi-A flagella (Figure 2E). The potential mechanisms responsible for the changes in the flagellar proteome and their significance for flagellar function are discussed later.
Rescue of flagellar assembly defects in a d1blic-null strain with a D1bLIC-GFP transgene
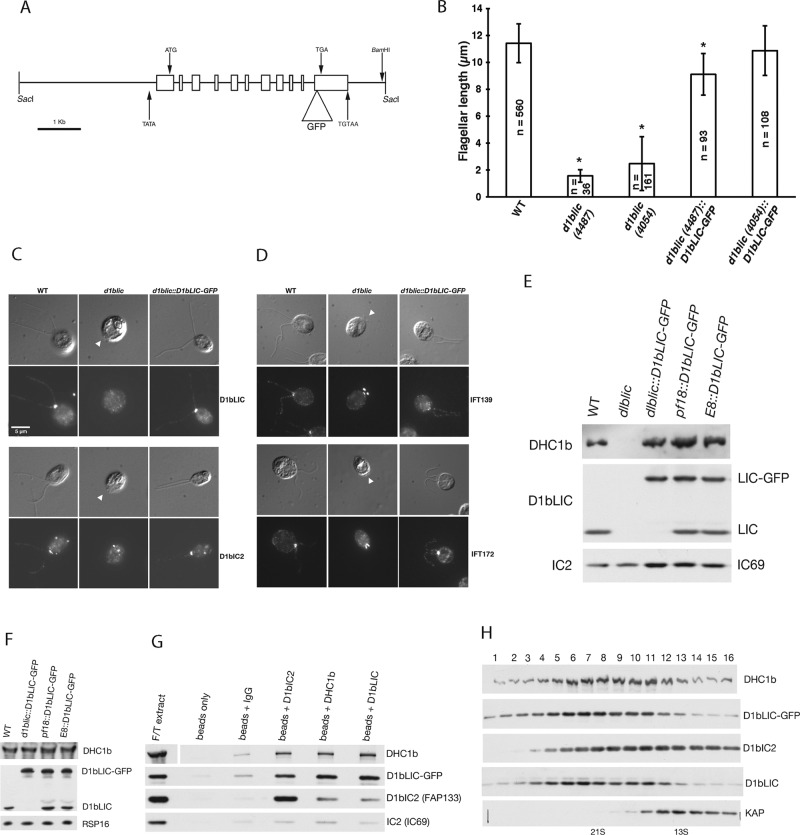
Although D1bLIC was reduced >95% in the RNAi-A strain, the flagellar assembly defects were less severe than that observed for the d1blic-null strain (Figure 1 and Supplemental Figure S2; Hou et al., 2004). To further characterize these differences, we analyzed the phenotype of the d1blic null before and after rescue with a D1bLIC-GFP transgene (Figure 3). A GFP tag was inserted near the C-terminus of a D1bLIC genomic clone (Figure 3A) and introduced into two versions of the null mutant (Supplemental Table S2). Both null strains assembled very short flagella (Figure 3B), with a large number of aflagellate cells (>25%, depending on culture conditions), but a small number of cells occasionally assembled flagella of near-WT length (Supplemental Figure S2). Transformation with D1bLIC-GFP decreased the number of aflagellate cells and increased the average flagellar length to near-WT levels (Figure 3B and Supplemental Figure S2). Staining of fixed cells demonstrated that the recovery of flagellar length was accompanied by increased signals for both D1bLIC and D1bIC2/FAP133 at the basal body region and in the two flagella (Figure 3C). Staining with antibodies to IFT139 and IFT172 showed that IFT particles were concentrated in the short flagellar stumps of the d1blic-null strain (Figure 3D), as previously described (Hou et al., 2004). After rescue with the D1bLIC-GFP transgene, both IFT139 and IFT172 were redistributed to the basal body region in a pattern more similar to that of WT cells (Figure 3D).
FIGURE 3:
A GFP-tagged D1bLIC construct rescues the d1blic mutant and incorporates into the retrograde motor complex. (A) Schematic diagram of the genomic clone encoding D1bLIC-GFP used for transformation. (B) Average length of flagella in WT and mutant cells. d1blic-null cells assembled short flagella, and many cells were aflagellate (Supplemental Figure S2). Transformation with D1bLIC-GFP increased the number of flagellated cells and their average flagellar length (n = cells, *p < 0.005). (C) WT, d1blic, and D1bLIC-GFP rescued cells were fixed and stained with antibodies against D1bLIC and D1bIC2 (FAP133). Top, DIC images; bottom, fluorescence images; arrowheads indicate flagellar stumps in the d1blic mutant. Scale bar, 5 μm. The rescued strains assembled near full-length flagella, and the D1bLIC and D1bIC2 signals at the basal body region and in the two flagella were restored to WT levels. (D) WT, d1blic, and D1bLIC-GFP rescued cells were fixed and stained with antibodies against IFT139 and IFT172. The IFT particle subunits were redistributed from the flagellar stumps in d1blic to a more WT pattern in the rescued cells. (E) A Western blot of cell extracts from WT, d1blic, and D1bLIC-GFP transformed cells probed with antibodies against DHC1b, D1bLIC, and the outer arm dynein subunit IC2 (IC69) as a loading control. Note the shift in size of the D1bLIC band and the increase in the amount of DHC1b in the GFP rescued strain. The E8 and pf18 transformed strains express WT levels of both the endogenous D1bLIC and D1bLIC-GFP. (F) Western blot of isolated flagella from WT and D1bLIC-GFP transformed cells probed with antibodies against DHC1b, D1bLIC, and RSP16 as a loading control. Note that both endogenous and GFP-tagged D1bLIC subunits assembled into flagella at WT levels. (G) Western blot of immunoprecipitates obtained with control antibodies (immunoglobulin G [IgG]) and antibodies against DHC1b, D1bLIC, and D1bIC2 (FAP133). A flagellar extract was prepared from the D1bLIC-GFP rescued strain by freeze-thawing of isolated flagella (lane 1) and then incubated with beads alone or beads plus the indicated antibodies. After several washes, the beads were boiled in sample buffer, loaded on an SDS–PAGE gel, blotted, and probed with antibodies shown on the right. D1bLIC-GFP and DHC1b were coimmunoprecipitated with all antibodies except control IgG. D1bIC2 (FAP133) was more efficiently coimmunoprecipitated with the D1bIC2 antibody than with the DHC1b and D1bLIC antibodies. (H) Western blot of fractions obtained by sucrose density gradient centrifugation of a flagellar extract from a pf28 pf9-2 E8::D1bLIC-GFP strain lacking both outer arm and I1 dyneins. D1bLIC-GFP cosedimented with the endogenous D1bLIC and DHC1b, but D1bIC2 (FAP133) was partially dissociated from the complex.
Assembly of D1bLIC-GFP in the retrograde motor complex
To determine whether D1bLIC-GFP was expressed at levels comparable to those of the endogenous D1bLIC, we transformed D1bLIC-GFP into two other strains (pf18, E8) and compared WT, mutant, and transformed cells on Western blots. D1bLIC-GFP migrated at ∼75 kDa and was expressed in whole cells at levels similar to the endogenous protein in all transformants (Figure 3E). The presence of D1bLIC-GFP was also correlated with an increase in the signal for DHC1b in the rescued strain (Figure 3E, lane 3), consistent with the proposed role of D1bLIC in stabilizing the dynein 1b complex (Hou et al., 2004). To verify that the GFP-tagged D1bLIC could compete with the endogenous D1bLIC for assembly into flagella, we analyzed isolated flagella from WT and transformed cells on Western blots. D1bLIC-GFP was detected in all transformed strains at levels comparable to those for the endogenous D1bLIC (Figure 3F). We then tested whether D1bLIC-GFP was efficiently incorporated into the retrograde motor complex in flagellar extracts. As shown in Figure 3G, D1bLIC-GFP coimmunoprecipitated with both DHC1b and D1bIC2/FAP133 after incubation with antibodies against DHC1b, D1bLIC, or D1bIC2/FAP133 but not after incubation with beads alone or control immunoglobulin G (IgG). Fractionation of flagellar extracts on sucrose density gradients revealed that D1bLIC-GFP and D1bLIC cosedimented at ∼21S with DHC1b and overlapped with the leading edge of the D1bIC2/FAP133 peak (Figure 3H). However, the majority of D1bIC2 sedimented more slowly, between 21S and 13S, consistent with its tendency to dissociate as part of an IC/LC subcomplex (Rompolas et al., 2007; Patel-King et al., 2013). Therefore D1bLIC-GFP incorporates into the retrograde motor complex as efficiently as the endogenous D1bLIC subunit.
Rescue of IFT defects with D1bLIC-GFP
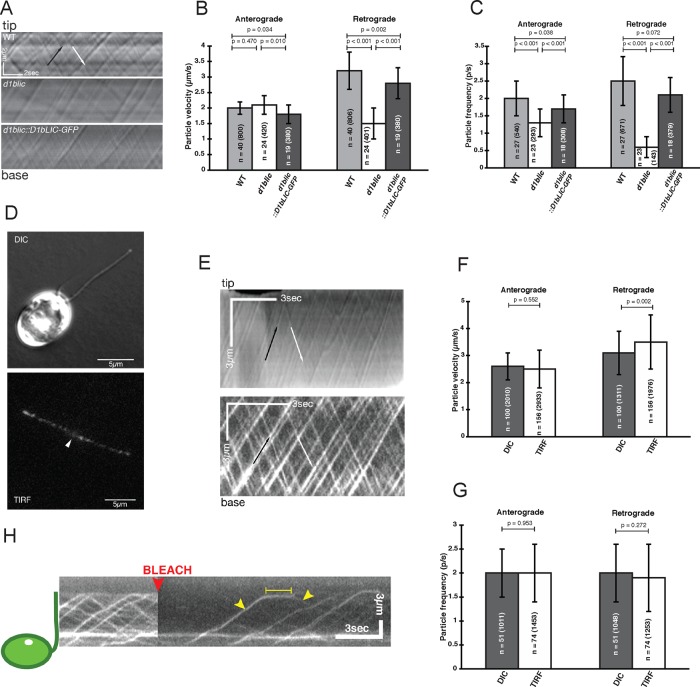
Although previous work demonstrated that disruption of D1bLIC alters flagellar assembly and localization of IFT subunits (Schafer et al., 2003; Hou et al., 2004; Blisnick et al., 2014), fewer studies have quantified the effect of d1blic mutations using in vivo motility assays. We therefore analyzed IFT in wild-type, d1blic, and D1bLIC-GFP cells using DIC microscopy to monitor the IFT particles and total interference reflection fluorescence (TIRF) microscopy to monitor the retrograde motor (Figure 4 and Supplemental Videos S3–S5). Both anterograde and retrograde tracks of IFT particles were visible in all kymographs, but retrograde particles were obviously reduced in d1blic flagella (Figure 4A and Supplemental Video S3). Measurements of particle velocities demonstrated that retrograde velocity was reduced ∼53% in the d1blic null, with no obvious effect on anterograde velocity (Figure 4B). The d1blic knockout had a more significant effect on particle frequency. Retrograde particle frequency was decreased ∼76%, and anterograde particle frequency was decreased ∼35% (Figure 4C). These results show that complete loss of D1bLIC severely reduces but does not eliminate retrograde IFT. It also confirms that the failure to efficiently recycle the IFT particles and/or some other factor(s) influences the frequency of anterograde transport. Transformation with D1bLIC-GFP rescued both anterograde and retrograde IFT velocities and frequencies to near-WT levels (Figure 4, A–C).
FIGURE 4:
Tracking the movement of the retrograde motor with D1bLIC-GFP. (A) Kymographs showing transport of IFT particles observed by DIC microscopy in WT, d1blic, and D1bLIC-GFP rescued flagella. See Supplemental Videos S1 and S3. Quantification of both anterograde and retrograde IFT (B) velocity and (C) frequency demonstrated that the D1bLIC-GFP construct rescued the IFT defects in the d1blic mutant to near wild-type levels (n = number of flagella and particles measured). (D) DIC (top) and TIRF (bottom) images of D1bLIC-GFP rescued cells. The white arrowhead indicates the base of the two flagella in the TIRF image. (E) Kymographs comparing the transport of IFT particles observed by DIC microscopy (top) and the movement of D1bLIC-GFP observed by TIRF microscopy (bottom). Anterograde particles are indicated by the black arrow and retrograde particles by the white arrow. Note the difference in scale bars, as the cells were imaged using different cameras. See Supplemental Videos S4 and S5. (F) Velocities of IFT particles observed by DIC microscopy and D1bLIC-GFP observed by TIRF microscopy (n = number of cells and particles measured). (G) Frequency of IFT particles observed by DIC and D1bLIC-GFP observed by TIRF (n = number of cells and particles measured). (H) Turnaround of D1bLIC-GFP at the flagellar tip observed by TIRF microscopy after photobleaching. A single D1bLIC particle was observed moving in the anterograde direction (first yellow arrowhead), pausing briefly at the flagellar tip (yellow bar), and then dissociating into a smaller particle before moving in the retrograde direction (second yellow arrowhead). See Supplemental Video S6.
To better understand how transport of the IFT particles is coordinated with movement of the retrograde motor, we analyzed parallel samples of D1bLIC-GFP cells using both DIC and TIRF microscopy (Figure 4D). The resulting movies and kymographs showed robust anterograde and retrograde movement of both IFT particles and D1bLIC-GFP (Figure 4E and Supplemental Videos S4–S6). Moreover, the velocity and frequency of transport were nearly identical for the IFT particles and D1bLIC-GFP (Figure 4, F and G). The retrograde motor is therefore tightly coupled to the IFT particles in both directions. We also analyzed the transport of D1bLIC-GFP in the presence of the endogenous D1bLIC subunit using the pf18 and E8 strains (Supplemental Table S2). Measurements of D1bLIC-GFP velocity and frequency were similar in the presence and absence of endogenous D1bLIC (Supplemental Figure S3 and Supplemental Videos S7 and S8). D1bLIC-GFP can therefore reliably report the behavior of the retrograde motor in other mutant backgrounds.
Observations of IFT in many systems have shown that IFT is highly processive and that most IFT particles do not undergo retrograde transport until they reach the flagellar tip (Engel et al., 2009; Buisson et al., 2013). To better understand the parameters that control remodeling and turnaround of IFT particles, we analyzed TIRF images of D1bLIC-GFP before and after photobleaching. Individual flagella were briefly exposed to high light intensities to reduce the D1bLIC-GFP signal. We then monitored the entry of new D1bLIC-GFP–labeled particles into the flagella, their anterograde transport to the tip, and their dwell time at the tip before remodeling and initiating retrograde transport (Figure 4H and Supplemental Video S6). Measurements of 81 particles after photobleaching the GFP signal in 38 flagella indicated that the average dwell time of a DbLIC-GFP particle at the tip was ∼1.7 ± 0.6 s. These values are comparable to the turnaround times previously measured for IFT subunits and axonemal cargoes such as IFT27, IFT52, and DRC4 (Qin et al., 2007; Buisson et al., 2013; Wren et al., 2013).
DISCUSSION
The role of D1bLIC in the stability, targeting, and activity of the retrograde IFT motor
Knockdown or knockout of D1bLIC has a clear, dose-dependent effect on the amount of DHC1b in whole cells (Figure 1A) but a less significant effect on the levels of D1bIC2 (Supplemental Figure S1D). Knockout of DHC1b also reduces the amount of D1bLIC and D1bIC2, but the effect is far less severe (Figure 1A and Supplemental Figure S1D). Thus one major function of D1bLIC is to stabilize the DHC, consistent with other studies (Hou et al., 2004; Blisnick et al., 2014). Yet even when D1bLIC and DHC1b were significantly reduced in the cell body, the retrograde motor complex was still enriched in flagella at ∼30% of WT levels (Figure 2E and Table 2). Therefore D1bLIC is not required for targeting the retrograde motor to the basal body region. Furthermore, retrograde IFT continued in the complete absence of D1bLIC (Figure 4, A–C), demonstrating that D1bLIC is not essential for motor activity. However, reduction or loss of D1bLIC had a significant effect on the frequency of retrograde IFT (Figures 2C and 4C and Supplemental Videos S2 and S3), and all IFT particle subunits were increased in d1blic flagella (Figures 2, D and E, and 3D and Table 2). Reduction or loss of D1bLIC was also associated with a significant decrease in the frequency of anterograde IFT (Figures 2C and 4C).
The phenotypes observed with knockdown of D1bLIC are similar to those described for the temperature-sensitive dhc1b-3 strain (Engel et al., 2012). The RNAi-B strain resembles dhc1b-3 at the permissive temperature with respect to DHC1b stability, retrograde IFT velocity and frequency, and flagellar assembly (Table 1). The RNAi-A strain resembles dhc1b-3 at the restrictive temperature, with decreased DHC1b stability, more significant reductions in retrograde IFT, which ultimately reduce the frequency of anterograde IFT, and decreased flagellar length and assembly (Table 1). Reevaluation of other retrograde IFT mutants (fla2, fla14, fla24) indicated that significant decreases in the frequency of retrograde IFT are usually associated with reductions in the frequency of anterograde IFT and average flagellar length (Supplemental Figure S4, Supplemental Videos S9–S11, and Table 1).
The mechanism by which retrograde motor defects result in a decrease in the frequency of anterograde IFT is not completely understood. Several investigators have proposed that the retrograde IFT is critical for efficient recycling of the IFT particles back to the cell body (e.g., Hou et al., 2004; Blisnick et al., 2014). Alternatively, retrograde motor activity may be critical for the delivery or recycling of other factors that regulate the assembly or entry of IFT particles at the flagellar base. A fluorescence recovery after photobleaching study in trypanosomes showed that retrograde IFT particles are recycled but that there are also two distinct pools of IFT protein at the flagellar base (Buisson et al., 2013). One pool is actively involved in IFT and mixes with returning retrograde particles, whereas the second pool enters the flagellar compartment much more slowly. Additional studies are needed to probe the process of IFT complex assembly at the flagellar base and how this process might be altered in different mutant backgrounds.
The role of D1bLIC in flagellar assembly
One of the most striking results was the degree to which Chlamydomonas cells tolerated reductions in retrograde IFT before these defects affected flagellar assembly. For example, both the RNAi-B strain and dhc1b-3 at the permissive temperature regenerated full-length flagella after pH shock with essentially WT kinetics (Figure 1D; Engel et al., 2012), even though the cellular concentration of DHC1b was reduced ≥50% (Figure 1A). However, when D1bLIC was decreased below 5% (and DHC1b below 25%), both the rate and final extent of flagellar assembly were reduced (Figure 1, A–D). Similar effects were seen with dhc1b-3 and fla24 at the restrictive temperature (Table 1; Engel et al., 2012; Lin et al., 2013). Taken together, these observations demonstrate that the retrograde motor is present in excess with respect to maintenance of flagellar length. Moreover, decreases in retrograde motor activity were not accompanied by increases in flagellar length, as reported in some studies of mammalian cell lines (Li et al., 2011; Palmer et al., 2011; Asante et al., 2014; Taylor et al., 2015). For instance, fibroblasts taken from SRPS patients with compound heterozygote mutations in DYNC2LI1 showed only minor decreases in the percentage of ciliated cells (∼70 vs. 80% in controls) and significant fluctuations in ciliary length relative to control cells, with an increase in the number of hyperelongated cilia (Taylor et al., 2015). Another study using siRNA to knock down DYNC2LI1 reported shorter cilia (Kessler et al., 2015). We occasionally observed full-length flagella in d1blic-null cells, as previously reported (Hou et al., 2004), but most cells were either aflagellate or assembled only short flagella (Supplemental Figure S2). The reasons for these differences in assembly are not readily apparent, but one possible explanation may be differences in growth and/or culture conditions. Flagellar length in Chlamydomonas was measured with vegetatively growing cells, which typically divide two to three times every 24 h, resorbing their flagella before mitosis and regenerating them after cell division (Harris, 2009). The slower kinetics of flagellar assembly in d1blic mutants could increase the fraction of cells with short flagella. In contrast, most studies of primary cilia use cultured cells that have been subjected to serum starvation for several days to inhibit cell division and induce assembly of cilia. Variations in culture conditions and measurement protocols might therefore account for some of the reported differences in ciliary lengths in cultured mammalian cells (Kessler et al., 2015; Taylor et al., 2015; Dummer et al., 2016). In addition, if defects in retrograde motor activity result in delays in ciliary assembly analogous to those observed in Chlamydomonas (Figure 1D; Engel et al., 2012; Schmidts et al., 2015), then dynein mutations may have a more severe effect on ciliary length in developing tissues (Rana et al., 2004; Schmidts et al., 2015).
The role of D1bLIC in flagellar motility and signaling
One of the challenges in measuring flagellar signaling is that many assays are influenced by changes in flagellar length. For example, RNAi-A cells exhibited decreased forward swimming velocity, reduced ability to photoaccumulate, inefficient mating, and decreased ability to adhere to a glass surface for flagellar gliding (Figure 1 and Table 1). However, all of these phenotypes could be the indirect result of reduced flagellar length (Figure 1C). In contrast, the forward swimming velocity, phototaxis behavior, and mating response of the RNAi-B strain were essentially WT, even though retrograde IFT was reduced (Table 1 and Figures 1C and 2, B and C). Of interest, however, the frequency of flagellar gliding by the RNAi-B strain was similar to that of the dhc1b-3 mutant and reduced relative to WT cells in the absence of any major change in flagellar length (Figure 1F; Shih et al., 2013). These results indicate that adhesion-induced flagellar signaling is reduced in RNAi-B cells and further suggest that flagellar gliding may be a more sensitive indicator of defects in retrograde IFT than other assays of flagellar activity in Chlamydomonas. The importance of retrograde IFT for adhesion-induced signaling may explain why dynein-2 mutations in humans are more frequently observed with skeletal ciliopathies than with ciliopathies associated with other tissues (Hou and Witman, 2015). Indeed, primary cilia and IFT proteins play multiple roles in the mechanotransduction signaling pathways that are critical for the differentiation and function of mesenchymal stem cells, chondrocytes, osteoblasts, and osteocytes during skeletal development (reviewed in Schmidts, 2015; Yuan et al., 2015). Thus these cell types might be especially sensitive to defects in retrograde IFT.
The role of D1bLIC in the composition of the flagellar proteome
IFT particle subunits are continuously recycled between the cell body and flagella (Ishikawa and Marshall, 2011; Buisson et al., 2013), and the accumulation of IFT subunits in the flagellar compartment has long been a defining characteristic of retrograde IFT mutants (Cole et al., 1998; Pazour et al., 1998, 1999; Piperno et al., 1998; Signor et al., 1999). However, few studies have analyzed how defects in retrograde IFT affect the total flagellar proteome. We used iTRAQ labeling and mass spectrometry to obtain an unbiased view of the changes in flagellar protein composition in the RNAi-A strain. We considered only those polypeptides whose protein ratios were significantly different from WT (p < 0.05) in two independent experiments with both technical and biological replicates (Table 2). As expected, nearly all of the IFT particle subunits were increased, and most of the retrograde motor subunits (DHC1b, D1bIC1/FAP163, D1bIC2/FAP133, D1bLIC) were decreased. Significant decreases in the LC subunits (LC8, Tctex2b, Tctex1, LC7) were not observed, but these proteins are also subunits of other axonemal dyneins, and thus small changes in these subunits would be difficult to detect.
Most axonemal proteins involved in flagellar motility were not significantly altered by knockdown of D1bLIC, even though several axonemal polypeptides (e.g., DRC4, DRC2, PF16, PF6, ODA16, tubulin) are IFT cargoes (Hou et al., 2007; Ahmed et al., 2008; Lechtreck et al., 2013b; Wren et al., 2013; Craft et al., 2015). However, the transport frequency of axonemal cargoes is relatively low in steady-state flagella (Wren et al., 2013; Craft et al., 2015), and so the residual IFT activity in RNAi-A flagella is likely sufficient to maintain WT levels of axonemal proteins. The sole exception is the radial spoke subunit RSP17, which was significantly reduced in RNAi-A (Table 2). The effect of this change is unclear because RSP17 does not have any well-characterized protein domains, nor does it have obvious orthologues in other species besides Volvox (Table 3).
Only 12 other proteins were significantly reduced in the RNAi-A flagella, and these proteins fell into several distinct classes. Of interest, unlike the IFT particle proteins, all three subunits of the kinesin-2 complex were reduced (Table 2). These results suggest that the anterograde motor may not require retrograde IFT for exit from the flagella. This hypothesis would be consistent with other studies that failed to see an accumulation of kinesin-2 subunits in dhc1b ts and d1blic mutants (Pedersen et al., 2006; Engel et al., 2012) and the difficulty in detecting robust retrograde IFT of KAP-GFP by TIRF microscopy (Engel et al., 2009). Indeed, recent work shows that kinesin-2 may be dissociated from the IFT particles at the flagellar tip by phosphorylation of the FLA8 subunit (Liang et al., 2014), but the mechanism by which phosphorylated kinesin-2 returns to the cell body is unknown.
PKD2 and FAP48/IP3R are two large transmembrane proteins that were reduced in RNAi-A flagella (Table 2). Both have been implicated in calcium signaling, and PKD2 is believed to be important for coupling flagellar adhesion to the increase in flagellar calcium required for completion of the mating response (Huang et al., 2007). Huang et al. (2007) observed that <10% of the flagellar PKD2 moved to the flagellar tip by anterograde IFT, yet PKD2 was increased in half-length fla10 flagella when anterograde IFT was inhibited, leading to the proposal that PKD2 might require IFT for removal from flagella. However, the decrease in PKD2 observed here may suggest instead a role for the retrograde motor in delivery of PKD2 from the cell body to the flagella. Indeed, recent work shows that another protein involved in the Chlamydomonas mating response, the SAG1 agglutinin, requires dynein 1b activity for trafficking from the cytoplasm to the periciliary region and into flagella (Cao et al., 2015). Defects in the trafficking of SAG1 and/or PKD2 may contribute to reduced mating efficiency of d1blic gametes (Table 1).
Most of the other proteins reduced in RNAi-A flagella (Table 2) have been identified as components of the membrane plus matrix fraction (Pazour et al., 2005). Two proteins (FAP5 and Cre17.g747247) contain TPR domains and small regions of limited homology to other proteins in vertebrates (Table 3). Two others (FAP24 and FAP26) have obvious orthologues in Volvox but no homology to any vertebrate proteins. However, FAP24 is enriched in preparations of isolated transition zones (Diener et al., 2015). Both FAP26 and FAP208 contain several ANK repeats—a common protein–protein interaction motif—and FAP208 has several orthologues in other species (Table 3). FAP208 is also one of small number of proteins that were decreased in nphp4 flagella defective in ciliary gating at the transition zone (Awata et al., 2014). The final polypeptide reduced in RNAi-A flagella is annotated in the latest version of the genome as a WNK1 kinase, but closer inspection indicates that this sequence is more closely related to the Aurora A kinases than the WNK (with no lysine) kinases (Table 3). Aurora A kinases have been implicated in ciliary disassembly in many systems, including Chlamydomonas (Cao et al., 2009; reviewed in Ke and Yang, 2014). Additional work is needed to determine whether any of these proteins associate with the retrograde motor during trafficking to the basal body region and/or facilitate the interaction of the retrograde motor with IFT machinery.
Only a small number of proteins besides IFT subunits were consistently elevated in the RNAi-A flagella (Table 2). As expected, most of the BBSome subunits were increased, but the number of peptides detected with high confidence was relatively small, and so not all of the increases were statistically significant (Table 2). The other proteins made up a varied collection (Table 3). For instance, three large, cytoplasmic proteins (70–122 kDa) associated with mRNA translation (alanine tRNA synthetase, EF-3, and RB47) were elevated in RNAi-A flagella. Whether these changes contribute to the reduced kinetics of flagellar assembly (Figure 1D) is unknown, but related proteins were increased in dhc1b-3 (Engel et al., 2012) and also in nphp4 flagella with defects in the transition zone (Awata et al., 2014). Taken together, these observations suggest that defects in retrograde IFT might compromise the barrier function of the transition zone. Several small (<50 kDa) abundant cytoplasmic proteins (ICL1, IPY3, GST, FAP179, GDP-D-mannose phosphorylase, and PRX2) were also reproducibly elevated (Table 2). These results may suggest instead that removal of cytoplasmic proteins by retrograde IFT is compromised in RNAi-A flagella. Indeed, recent studies identified other cytoplasmic proteins that accumulate in flagella in the absence of IFT (Lechtreck et al., 2013a; Lechtreck, 2015; Craft et al., 2015; Harris et al., 2016). However, we observed increases in only a small number of cytoplasmic proteins, and at least three proteins (GST, FAP179, and PRX2) have been linked to metabolic stress responses.
Of interest, PRX2 is one of seven peroxiredoxins in Chlamydomonas previously localized to both the cytoplasm and flagella (Dayer et al., 2008). Peroxiredoxins are antioxidant enzymes that protect cells from metabolic stress and are also involved in non–stress-related signaling (Park et al., 2014; Perkins et al., 2014). The mammalian counterpart of PRX2, PRDX4, was found in the proteome of primary cilia (Ishikawa et al., 2012), and a knockout of PRDX4 has been linked to reduced sperm viability in mice (Perkins et al., 2014; Fujii et al., 2015). Other PRDX isoforms are enriched in human sperm flagella (O’Flaherty and Souza, 2011). In addition, PRDX1 is found in flagella of the microalga Dunaliella, where it becomes elevated during flagellar disassembly (Gong et al., 2014). Overexpression and knockdown of HsPRDX1 were also correlated with loss and reassembly of cilia in esophageal squamous cell carcinoma cells (Gong et al., 2014). These observations suggest that the elevation of PRX2 and other antioxidant proteins in d1blic flagella might be a specific response and not simply cytoplasmic contamination.
The increase in HSP90α (∼80 kDa) might reflect a need for increased chaperone activity due the accumulation of multiple protein complexes in d1blic mutant flagella (Table 2). HSP90 and other chaperones facilitate the assembly of multimeric structures while reducing aggregation in protein-rich environments (Jackson, 2013). HSP90 is associated with tubulin in cilia and flagella of several species (Williams and Nelsen, 1997; Stephens and Lemieux, 1999). It is critical for the maintenance of ciliary beating in postnatal mice (Takaki et al., 2007; Simet et al., 2013), stimulates tubulin polymerization in vitro (Takaki et al., 2007), and protects tubulin against thermal denaturation (Weis et al., 2010). More recently, several PIH (proteins interacting with HSP)-domain proteins were identified as important factors for the preassembly of axonemal dynein complexes (Yamamoto et al., 2010; Dong et al., 2014). HSP90 could therefore stabilize multiple ciliary complexes during assembly and transport. HSP90 has also been found in primary cilia (Gherman et al., 2006), where it interacts with a number of signaling molecules and also plays a role in ciliary maintenance (Prodromou et al., 2012; Brown et al., 2014; Wang et al., 2015).
The last two polypeptides significantly elevated in RNAi-A flagella are FAP256 and FAP295 (Table 2). FAP256/CEP104 is an EB1-interacting protein (92 kDa) associated with the flagellar tip (Satish Tammana et al., 2013) and so its enrichment in short flagella was not unexpected. FAP295 is a cNTP-dependent protein kinase (80 kDa) with a predicted N-myristoylation site that was previously localized along the length of the flagellum (Lechtreck et al., 2009) and more recently as a component of the transition zone (Diener et al., 2015). Additional studies are needed to identify substrates of FAP295 and determine its function in Chlamydomonas flagella.
D1bLIC-GFP is a reliable marker for retrograde motor activity
Transformation of the d1blic null with D1bLIC-GFP rescued all of the defects in IFT, flagellar assembly, and flagellar length (Figures 3B and 4, A–C) and also restored the distribution of IFT particles to their WT pattern (Figure 3D). D1bLIC-GFP assembled into the retrograde motor complex as efficiently as the endogenous D1bLIC subunit in multiple strains (Figure 3, E–H). Comparison of IFT particle movement by DIC microscopy and D1bLIC-GFP movement by TIRF microscopy showed that both the frequency and velocity of transport were essentially identical (Figure 4, E–G, and Supplemental Videos S4 and S5). The retrograde motor was transported as an anterograde cargo from base to tip with very few pauses or reversals (Figure 4E). Once at the tip, the D1bLIC-GFP–containing particles dissociated into smaller particles and then initiated retrograde transport in <2 s (Figure 4H and Supplemental Video S6). If one assumes two copies of the D1bLIC subunit in each retrograde motor and one motor per particle, one might predict that the number of D1bLIC-GFP–labeled particles would be reduced ∼25% in the presence of the endogenous D1bLIC subunit. However, we did not observe any significant decrease in the frequency of either anterograde or retrograde IFT (Supplemental Figure S3 and Supplemental Videos S7 and S8). Further work will be needed to determine the precise number of retrograde motors associated with an individual IFT particle, but the results shown here suggest that D1bLIC-GFP is a reliable marker for retrograde motor behavior, even in the presence of the endogenous D1bLIC subunit.
Summary and future directions
The range of phenotypes displayed by the d1blic strains demonstrates the complex relationship between dynein 1b motor activity, retrograde IFT, flagellar polypeptide composition, flagellar motility, and flagellar assembly. Growing evidence suggests that dynein 1b might be required in the cell body for transport of components to the basal body region and into flagella (Table 2; Cao et al., 2015), but additional work is needed to demonstrate dynein 1b–mediated transport in the cell body. The mechanism by which the retrograde motor becomes assembled onto anterograde IFT particles is also poorly understood. For example, what components are required for attaching dynein 1b to the IFT particles? Several of the proteins reduced in d1blic flagella warrant further investigation as potential candidates (Tables 2 and 3). How is retrograde motor activity suppressed during anterograde transport? We rarely observed D1bLIC-GFP reversal during anterograde IFT (Figure 4). Higher-resolution ultrastructural studies will be required to determine the conformation and arrangement of IFT motors on the IFT particles. Once D1bLIC-GFP reaches the flagellar tip, the larger IFT particles dissociate, and smaller particles begin processive retrograde transport in <2 s (Figure 4). What components are needed for this remodeling, and how is this linked to activation of retrograde transport? Is release of kinesin-2 from the IFT particles sufficient to activate dynein 1b? Simultaneous imaging of both kinesin 2 and dynein 1b under different experimental conditions and in different mutant backgrounds should provide new insights into these questions. In addition, the identification and further characterization of polypeptides located at the flagellar tip may reveal new regulators of this process. Finally, are any of the polypeptides elevated in d1blic flagella (besides the IFT and BBSome subunits) bona fide cargoes of retrograde IFT? Direct observation of tagged candidates will be required to resolve this question and determine their role in flagellar function.
MATERIALS AND METHODS
Cell culture, plasmids, and transformation
Chlamydomonas strains used in this study are listed in Supplemental Table S2. Cells were maintained in constant light on Tris-acetate-phosphate (TAP) medium (Harris, 2009), with the exception of arg7-2, which was supplemented with 0.6 mg/ml arginine-HCl. Two strains with wild-type motility, A54 e18 and arg7-2, were used as hosts for cotransformation with constructs designed to knock down expression of D1bLIC (Supplemental Figure S1). The approach was based on a strategy previously used successfully to knock down expression of chlamyopsin, SF-assemblin, phototropin, and centrin (Fuhrmann et al., 2001; Lechtreck et al., 2002; Huang and Beck, 2003; Koblenz et al., 2003). For the first construct, an 8.5-kb SacI genomic clone (DT04-B1) containing the full-length D1bLIC transcription unit (Perrone et al., 2003) was digested with AscI (in the 11th and final exon of the gene) and BamHI (in the multiple cloning site) to release the 3′ end of the gene. A 2.2-kb cDNA clone (DT05; Perrone et al., 2003) was digested with BamHI and AscI to release exons 1–11 and subcloned into DT04-B1 in the antisense orientation to form the D1bLIC-RNAi plasmid (Supplemental Figure S1A). For the second construct, the D1bLIC-RNAi plasmid was digested with NheI and EcoRI to release a fragment with genomic exons 4–11 in the sense orientation and cDNA exons 1–11 in the antisense orientation. This fragment was subcloned into the plasmid pCB740 after removal of the HSP70B gene (Schroda et al., 2000), between the HSP70A and RBCS2 promoters and the 3′ untranslated region from RBCS2 and upstream from the ARG7 gene to form the pCB740-DLIC-RNAi plasmid (Supplemental Figure S1B).
The LIC RNAi construct was linearized with BamHI and cotransformed into A54 e18 with pSI103, which contains the aphVIII gene (Sizova et al., 2001). Positive transformants were selected by plating on TAP medium plus 10 μg/ml paromomycin. The pCB740-DLIC-RNAi plasmid was digested with PvuI and transformed into arg7-2. Positive transformants were selected by plating on TAP medium lacking arginine. For the initial screens, 250 transformants were picked into 96-well plates containing liquid TAP medium and screened for aberrant flagellar motility or assembly on a dissecting microscope. Thirteen strains with aberrant flagellar motility or assembly were recovered, but none were confirmed as D1bLIC knockdowns by whole-cell Western blots. These strains were discarded as off-target insertional mutants. Subsequent transformants were screened by whole-cell Western blots for knockdown of D1bLIC and then single colony isolated and rescreened at least twice to confirm the D1bLIC phenotype. Ten strains with reduced D1bLIC expression were recovered from 497 transformants and further analyzed for potential defects in flagellar assembly (Supplemental Figure S1C). Two strains were retained for further study, 4a2 and 4e11.
To make the D1bLIC-GFP transgene, a Chlamydomonas codon-optimized GFP tag was amplified from the pCrGFP plasmid (Fuhrmann et al., 1999) using primers with AscI sites (5′-GGCGCGCCCCGGCGCGCCGATGG-3′ and 5′-CCCGCGCCttaCTTGTACAGCTCGTC-3′) and cloned into pGEM T-Easy. The 8.5-kb genomic clone DT04 was digested with AscI, and the GFP tag was subcloned into an AscI site in the last exon, between amino acid residues 401 and 402. DT04-GFP was linearized with BamHI and cotransformed with pSI103 into the d1blic-null strain pf18 and the triple mutant pf9-2 pf28 E8 (Supplemental Table S2). Two versions of the d1blic-null strain are available from the Chlamydomonas Genetics Center (University of Minnesota, Saint Paul, MN). CC-4053 is the original mt+ isolate known as TBD9-1 or T8D9-1 (Hou et al., 2004). It carries an additional, uncharacterized paralyzed flagella mutation (unpublished data). TBD9-1 was backcrossed to yield YH43 (listed as CC-4054); YH43 is mt− and motile (Hou et al., 2004). Transformation mixtures were plated on selective medium, and positive transformants were screened using fluorescence microscopy and Western blotting to identify strains expressing D1bLIC-GFP.
Preparation of antibody against FAP133
Two constructs encoding amino acids 1–140 of FAP133 as fusion proteins were generated by PCR using the primers 5′-GGATCCATGCAAGAGGTCCCGCC-3′ and 5′-GAATTCGGTGGCGTTGAGGTCCTGG-3′, ligation into pGEM, digestion with BamHI and EcoRI, and religation into pET30A (Novagen, EMD Millipore, Billerica, MA) and pGEX-2T (GE Healthcare Life Sciences, Piscataway, NJ). Expression was induced in BL21(DE3) pLysS cells (Promega, Madison, WI), and soluble FAP133-hexahistidine fusion protein was purified with a nickel column (Novagen) and used as an antigen for polyclonal antibody production in rabbits (Spring Valley Laboratories, Sykesville, MD). Soluble FAP133-GST fusion protein was covalently cross-linked to glutathione–Sepharose 4B beads (GE Healthcare Life Sciences) according to manufacturer’s instructions. The FAP133 antibody was affinity purified on the FAP133-GST column as described in Perrone et al. (2003).
Preparation and fractionation of whole-cell and flagellar extracts, immunoprecipitation, SDS–PAGE, and Western blots
Small- and large-scale cultures of cells, isolation and extraction of flagella, and sucrose density centrifugation of flagellar extracts were performed as previously described (Perrone et al., 2003; Bower et al., 2013). Immunoprecipitation of dynein-containing flagellar extracts was done using EZview Red Protein A-Sepharose beads (Sigma Aldrich, St. Louis, MO) and affinity purified antibodies to DHC1b, D1bLIC, FAP133, and GFP as previously described (Perrone et al., 2003). Polypeptides were separated by SDS–PAGE on 5–15% polyacrylamide, 0–2.4 M glycerol gradient gels and blotted to Immobilon P (EMD Millipore). Total protein on the blot was visualized with a reversible Blot FastStain (Chemicon, G-Biosciences, St. Louis, MO). Primary antibodies were diluted in Tropix I-Block (Applied Biosystems, Foster City, CA) at the concentrations indicated in Supplemental Table S3. Alkaline phosphatase–conjugated secondary antibodies and the Tropix chemiluminescent detection system (Applied Biosystems) were used as described in Bower et al. (2013).
To compare the relative amounts of D1bLIC and DHC1b in whole cells of different strains, equivalent numbers of total cells, (1–5) × 106 cells, were loaded per lane. Blots were probed with affinity-purified antibodies for DHC1b and D1bLIC (Perrone et al., 2003) and then reprobed with antibodies against tubulin or IC69 (Sigma-Aldrich) to control for variations in protein loading. Multiple exposures were scanned and analyzed using ImageJ (National Institutes of Health, Bethesda, MD) to compare the relative intensity of different samples (normalized to tubulin or IC69) and ensure that the signals were in the linear range. Each transformant was rescreened at least twice, and two strains, 4a2 and 4e11, were analyzed in six independent experiments. D1bLIC was knocked down below 5% in the 4e11 strain (3.9 ± 1.7%, n = 6) and below 25% in the 4a2 strain (21 ± 11%, n = 6).
Preparation of samples for iTRAQ labeling and mass spectrometry
Flagellar samples were prepared for iTRAQ labeling as previously described (Bower et al., 2013) with minor modifications. Samples of whole flagella were washed briefly with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, centrifuged, and resuspended in dissolution buffer (0.5M triethylammonium bicarbonate, pH 8.5) for determination of protein concentration. Proteins were denatured and reduced, cysteine residues were blocked with methyl-methane thiosulfate, and then samples were digested overnight with trypsin. Aliquots of each sample (50 μg each) were reacted in duplicate with 4-plex iTRAQ labeling reagents (114–117; AB Sciex, Foster City, CA). After labeling, the samples were combined, vacuum dried, and purified with an Oasis MCX 1-cc Vac cartridge according to manufacturer’s instructions (Waters Corporation, Milford, MA) to remove excess trypsin, unreacted iTRAQ reagents, and buffer.
The iTRAQ-labeled sample was next fractionated offline using strong cation exchange (SCX) chromatography or high-pH, C18 reversed-phase chromatography. For SCX, the sample was resuspended in 20% (vol/vol) acetonitrile, 5 mM KH2PO4, pH 2.7, injected into a Magic 2002 HPLC system (Michrom BioResources, Auburn, CA) using a polysulfoethyl A column (PolyLC, Columbia, MD), and eluted with a gradient of 20% (vol/vol) acetonitrile, 5 mM KH2PO4, pH 3.0, and 500 mM KCl as previously described (Lund et al., 2007). For high-pH, C18 reversed-phase chromatography, the sample was resuspended in 10 mM ammonium formate, pH 10, in 98:2 water::acetonitrile, injected into a Magic 2002 HPLC system using a C18 Gemini-NX column (Phenomenex, Torrance, CA), and eluted with a gradient of 10 mM ammonium formate, pH 10, in 10:90 water:acetonitrile, as previously described (Tran et al. 2013). Peptide-containing fractions from the C18 column were divided into two equal-numbered groups, “early” and “late.” The first “early” fraction was concatenated with the first “late” fraction, and so on (Yang et al., 2012).
Fractions from both columns were vacuum dried, resuspended in load solvent (98:2:0.01, water:acetonitrile:formic acid), and loaded in 1- to 1.5-μg aliquots for capillary LC using a C18 column at low pH. The C18 column was mounted in a nanospray source directly in-line with a Velos Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Online capillary LC and tandem mass spectrometry were performed as previously described (Lin-Moshier et al., 2013), with the exception that the higher-energy, collision-induced dissociation activation time was 20 instead of 0.1 ms because iTRAQ-labeled peptides need more energy to dissociate reporter ions.
Database searching and protein identification
The RAW data (ProteoWizard files) from the Velos Orbitrap mass spectrometer were converted to MzXML using MSconvert software (part of ProteoWizard Tools, proteowizard.sourceforge.net/) and to MGF files using TINT mgf converter (toolshed.g2.bx.psu.edu/repos/galaxyp/mgf_formatter). ProteinPilot software version 4.5 (AB Sciex) was used for searching multiple versions of the Chlamydomonas protein FASTA database (the most recent version, V5.5, is available at phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii), and a contaminant database (http://thegpm.org/GPM/index.html, 109 proteins) was appended. Search parameters were as follows: cysteine MMTS; iTRAQ 4plex (Peptide Labeled); trypsin; instrument Orbi MS (1–3 ppm) Orbi MS/MS; biological modifications ID focus; thorough search effort; and False Discovery Rate analysis (with reversed database). The ProteinPilot program calculates a bias adjustment factor based on the protein that represents the median of all proteins identified in that sample, and it automatically sets the ratio of that protein to 1.0. Because the median protein may or may not be the same in each sample, we manually adjusted the bias factor by normalizing each sample on α- and β-tubulin. Tubulin is the most abundant protein in the sample and the most appropriate reference point for comparison with other flagellar proteins.
Measurements of flagellar length, assembly, and motility
To ensure maximal flagellar assembly, strains were grown at room temperature in liquid M medium with aeration for 2–4 d. Samples were removed, fixed with 1% glutaraldehyde, and imaged at 40× using phase contrast microscopy and a halogen light source on a Zeiss Axioskop (Carl Zeiss, Thornwood, NY) and a CoolSnap ES charge-coupled device (CCD) camera (Photometrics, Tucson, AZ). The flagellar lengths were measured using MetaMorph software package 7.6.5.0 (Molecular Devices, Sunnyvale, CA) as described in Mueller et al. (2005). Similar results were observed with strains grown in TAP or M-N/5 medium. For regeneration experiments, flagella were removed by pH shock, and aliquots of cells were taken and fixed at selected time points during regeneration. For resorption experiments, cells were cultured in M medium and induced to shorten their flagella by the addition of 20 mM sodium pyrophosphate, pH 7, as described in Engel et al. (2012). All data are presented as mean ± SD (n = sample number), and samples are compared using the Student’s unpaired t test available on Excel (Microsoft, Redmond, WA).
For assays of cell motility, cells were resuspended in M medium with aeration for 2–4 d or M-N/5 medium overnight. For measurements of forward swimming velocity, cells were imaged over 12–20 s using 20× phase contrast optics and a Rolera-MGi electron-multiplying CCD camera (Q-imaging, Tucson, AZ) with 50-ms exposure (VanderWaal et al., 2011). For assays of photoshock, cell movement was recorded at 20× using a red filter, and the cells were briefly exposed to bright light using a GFP excitation filter on the epi-illuminescence port. The position of each cell before and after light exposure was traced and recorded as no change, paused, changed in forward direction, or reversed. For photoaccumulation assays, cells at a concentration of 2.5 × 106 cells/ml were transferred to 35-mm Petri dishes and dark adapted for 10–15 min. Cells were then exposed to a fluorescent light source located 30 cm away (2 W/m2) for 10 min. Cell densities on both sides of the dish were determined at various time points and also recorded using a Coolpix S620 camera (Nikon, Melville, NY) to determine relative photoaccumulation ability. Each strain was independently assayed at least three times and also tested in several different media (TAP, M-N/5, phototaxis buffer; Wakabayashi et al., 2011). Where applicable, the direction of the movement relative to the light source, positive or negative, was also noted.
To test the ability of flagella to glide on a glass coverslip, cells were resuspended in liquid TAP medium or gliding buffer (Collingridge et al., 2013), aerated overnight, and then incubated for 10–15 min with 15 μM motility inhibitors E (C14 H17 Cl2 N O2) and F (C21 H23 N O2) in 0.5% dimethyl sulfoxide (Engel et al., 2011; Shih et al., 2013). Cells were allowed to settle on glass coverslips and imaged on a Zeiss Axioskop using 40× DIC optics, a halogen light source, and a red filter. Images were recorded for ∼25 s (500 frames × 50-ms exposure) using a CoolSnap ES CCD camera. The total numbers of cells visible, attached to the coverslip, and gliding were counted for each recording.
To test for the ability of cells to mate, cells were induced to form gametes by incubation in M-N/5 overnight and then tested for their ability to form quadriflagellate zygotes after mixing with WT cells of the opposite mating type. Mating mixtures were fixed with 1% glutaraldehyde at various time points, and the number of quadriflagellate cells was counted using phase contrast microscopy. Because some strains were extremely inefficient at forming quadriflagellates, mating mixtures were also plated on Z-plates containing 4% agarose at selected times after mixing. After 24 h in the light, the plates were wrapped in tin foil and kept in the dark for 3–4 d. Nonmated cells were scraped away with a razor blade, and the total number of zygotes adhering to the plates was counted for each strain.
Immunofluorescence light microscopy
Chlamydomonas cells were fixed for immunofluorescence microscopy using the cold methanol procedure of Sanders and Salisbury (1995). Fixed cells were labeled with affinity-purified antibodies against D1bLIC at 1:1000 (Perrone et al., 2003), FAP133/D1bIC2 at 1:200 (this study), and IFT139 and IFT172 at 1:100 (Cole et al., 1998) and the appropriate Alexa Fluor 488–labeled secondary antibodies (Molecular Probes, Eugene, OR). Slides were washed in three changes of phosphate-buffered saline and then mounted in Prolong antifade medium (Molecular Probes). Images were obtained with an X-cite 120 illumination system (EXFO Photonic Solutions, Mississauga, Canada), a Zeiss Axioskop equipped with a 100×/1.3 numerical aperture (NA) Plan Neofluar objective, a 472/30-nm excitation filter and a 520/35-nm emission filter, a CoolSnap ES CCD camera, and MetaMorph software, version 7.6.5.0. Images were cropped and labeled using ImageJ and Photoshop (Adobe, San Jose, CA).
Measurements of IFT
For the initial analysis of IFT using DIC microscopy, strains were immobilized in 0.5–1% low–melting point agarose to limit flagellar motility and imaged using a 100×/1.3 NA Plan Neofluar objective, a 4× TV tube, and a C2400 Newvicon camera (Hamamatsu, Bridgewater, NJ). Supplemental video mages were converted into a digital format, and digital kymographs were created and analyzed using the MetaMorph software package as described in Mueller et al. (2005).
Several variations were used for comparison of GFP-labeled cells using DIC microscopy and TIRF microscopy. For DIC imaging of IFT particles, cells were immobilized in low–melting point agarose and imaged on a Zeiss AxioObserver Z1 microscope using a halogen light source, achromatic-aplanatic 1.4 NA DIC condenser, alpha Plan-Apochromat 100×/1.46 NA oil DIC M27 objective, and 1–1.6× Optovar. IFT was recorded at ∼25 frames/s using the fast acquisition module of the AxioVision 4.8 software package and an AxioCam MRm camera (Carl Zeiss). For TIRF imaging of D1bLIC-GFP, some cells were immobilized with ∼1.85 mM ethylene glycol tetraacetic acid. The GFP signal was detected using a 488-nm laser at 5–7% focused at an angle of ∼61–62° through a 38 HE enhanced GFP filter set with a BP470/40 excitation filter, FT495 dichroic mirror, and BP525/50 emission filter. Images were collected at ∼16 frames/s on a QuantEM 512SC camera (Photometrics). The AxioVision zvi files were converted to 8-bit TIFF stacks and further analyzed using MetaMorph software as described. To analyze IFT after photobleaching, the cells were imaged as before but bleached briefly (100–250 ms) with the 488-nm laser set at 90–99% maximum intensity.
Supplementary Material
Acknowledgments
We thank LeeAnn Higgins, Todd Markowski, and Sue Van Riper of the Center for Mass Spectrometry and Computational Proteomics, University of Minnesota, for assistance with iTRAQ labeling, mass spectrometry, and database management. The Center is supported by multiple grants, including National Science Foundation Major Research Instrumentation Grant 9871237 and National Science Foundation Grant DBI-0215759 as described at cbs.umn.edu/cmsp/about. We also acknowledge the Minnesota Supercomputing Institute at the University of Minnesota for software support (www.msi.umn.edu) and Mark Sanders and the University Imaging Center for assistance with imaging. We give many thanks to Katherine Augspurger and Jason Sakizadeh at the University of Minnesota for assistance with microscope assays and figure preparation. We thank William Dentler (University of Kansas, Lawrenceville, KS) for the T8D9 strain and Matt Laudon and the Chlamydomonas Genetics Center (University of Minnesota, Saint Paul, MN) for other strains used in this study, as described in Supplemental Table S2. Douglas Cole (University of Idaho, Moscow, ID), Steve King (University of Connecticut, Farmington, CT), and Dennis Diener (Yale University, New Haven, CT) generously supplied antibodies, as listed in Supplemental Table S3. Preliminary reports of this work were presented at American Society for Cell Biology meetings in 2006, 2012, and 2013. This work was supported by National Institutes of Health Grant GM-055667 to M.E.P. K.V.M. was supported in part by an American Heart Association Pre-Doctoral Fellowship, a University of Minnesota Graduate School Fellowship, and a University of Minnesota Graduate School Grant to M.E.P.
Abbreviations used:
- D1bIC
dynein 1b–associated IC
- D1bLIC
dynein 1b–associated light intermediate chain
- DHC1b
dynein 1b heavy chain
- DIC
differential interference contrast
- FAP
flagellar-associated polypeptide
- GFP
green fluorescent protein
- IC
intermediate chain
- IFT
intraflagellar transport
- iTRAQ
isobaric tag for relative and absolute quantitation
- LC
light chain
- LIC
light intermediate chain
- PKD
polycystic kidney disease
- RSP
radial spoke protein
- SRPS
short rib polydactyly syndrome
- TAP
Tris-acetate-phosphate
- TIRF
total interference reflection fluorescence
- ts
temperature sensitive
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0191) on June 1, 2016.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante D, Stevenson NL, Stephens DJ. Subunit composition of the human cytoplasmic dynein-2 complex. J Cell Sci. 2014;127:4774–4787. doi: 10.1242/jcs.159038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata J, Takada S, Standley C, Lechtreck KF, Bellve KD, Pazour GJ, Fogarty KE, Witman GB. NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J Cell Sci. 2014;127:4714–4727. doi: 10.1242/jcs.155275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Engel BD, Lorentzen E. Intraflagellar transport complex structure and cargo interactions. Cilia. 2013;2:10. doi: 10.1186/2046-2530-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blisnick T, Buisson J, Absalon S, Marie A, Cayet N, Bastin P. The intraflagellar transport dynein complex of trypanosomes is made of a heterodimer of dynein heavy chains and of light and intermediate chains of distinct functions. Mol Biol Cell. 2014;25:2620–2633. doi: 10.1091/mbc.E14-05-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell. 2013;24:1134–1152. doi: 10.1091/mbc.E12-11-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Santra T, Owens P, Morrison AM, Barry F. Primary cilium-associated genes mediate bone marrow stromal cell response to hypoxia. Stem Cell Res. 2014;13:284–299. doi: 10.1016/j.scr.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Brown JM, Witman GB. Cilia and diseases. BioScience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson J, Chenouard N, Lagache T, Blisnick T, Olivo-Marin JC, Bastin P. Intraflagellar transport proteins cycle between the flagellum and its base. J Cell Sci. 2013;126:327–338. doi: 10.1242/jcs.117069. [DOI] [PubMed] [Google Scholar]
- Cao M, Li G, Pan J. Regulation of cilia assembly, disassembly, and length by protein phosphorylation. Methods Cell Biol. 2009;94:333–346. doi: 10.1016/S0091-679X(08)94017-6. [DOI] [PubMed] [Google Scholar]
- Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;4:e05242. doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge P, Brownlee C, Wheeler GL. Compartmentalized calcium signaling in cilia regulates intraflagellar transport. J Curr Biol. 2013;23:2311–2318. doi: 10.1016/j.cub.2013.09.059. [DOI] [PubMed] [Google Scholar]
- Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol. 2015;208:223–237. doi: 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagoneau N, Goulet M, Genevieve D, Sznajer Y, Martinovic J, Smithson S, Huber C, Baujat G, Flori E, Tecco L, et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet. 2009;84:706–711. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayer R, Fischer BB, Eggen RI, Lemaire SD. The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics. 2008;179:41–57. doi: 10.1534/genetics.107.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella LM, Smith EF, Patel-King RS, Wakabayashi K, King SM. A novel Tctex2-related light chain is required for stability of inner dynein arm I1 and motor function in the Chlamydomonas flagellum. J Biol Chem. 2004;279:21666–21676. doi: 10.1074/jbc.M313540200. [DOI] [PubMed] [Google Scholar]
- Diener DR, Lupetti P, Rosenbaum JL. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr Biol. 2015;25:379–384. doi: 10.1016/j.cub.2014.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Shinohara K, Botilde Y, Nabeshima R, Asai Y, Fukumoto A, Hasegawa T, Matsuo M, Takeda H, Shiratori H, et al. Pih1d3 is required for cytoplasmic preassembly of axonemal dynein in mouse sperm. J Cell Biol. 2014;204:203–213. doi: 10.1083/jcb.201304076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dummer A, Poelma C, DeRuiter MC, Goumans M-JTH, Hierck BP. Measuring the primary cilium length: improved method for unbiased high-throughput analysis. Cilia. 2016;5:7. doi: 10.1186/s13630-016-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ishikawa H, Feldman JL, Wilson CW, Chuang PT, Snedecor J, Williams J, Sun Z, Marshall WF. A cell-based screen for inhibitors of flagella-driven motility in Chlamydomonas reveals a novel modulator of ciliary length and retrograde actin flow. Cytoskeleton. 2011;68:188–203. doi: 10.1002/cm.20504. [DOI] [PubMed] [Google Scholar]
- Engel BD, Ishikawa H, Wemmer KA, Geimer S, Wakabayashi K, Hirono M, Craige B, Pazour GJ, Witman GB, Kamiya R, et al. The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J Cell Biol. 2012;199:151–167. doi: 10.1083/jcb.201206068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Lechtreck KF, Sakai T, Ikebe M, Witman GB, Marshall WF. Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 2009;93:157–177. doi: 10.1016/S0091-679X(08)93009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Stahlberg A, Govorunova E, Rank S, Hegemann P. The abundant retinal protein of the Chlamydomonas eye is not the photoreceptor for phototaxis and photophobic responses. J Cell Sci. 2001;114:3857–3863. doi: 10.1242/jcs.114.21.3857. [DOI] [PubMed] [Google Scholar]
- Fujii J, Ikeda Y, Kurahashi T, Homma T. Physiological and pathological views of peroxiredoxin 4. Free Radic Biol Med. 2015;83:373–379. doi: 10.1016/j.freeradbiomed.2015.01.025. [DOI] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Gong F, Liu H, Li J, Xue L, Zhang M. Peroxiredoxin 1 is involved in disassembly of flagella and cilia. Biochem Biophys Res Commun. 2014;444:420–426. doi: 10.1016/j.bbrc.2014.01.081. [DOI] [PubMed] [Google Scholar]
- Hao L, Efimenko E, Swoboda P, Scholey JM. The retrograde IFT machinery of C. elegans cilia: two IFT dynein complexes. PLoS One. 2011;6:e20995. doi: 10.1371/journal.pone.0020995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use. San Diego, CA: Academic Press; 2009. [Google Scholar]
- Harris JA, Liu Y, Yang P, Kner P, Lechtreck KF. Single-particle imaging reveals intraflagellar transport-independent transport and accumulation of EB1 in Chlamydomonas flagella. Mol Biol Cell. 2016;27:295–307. doi: 10.1091/mbc.E15-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Pazour GJ, Witman GB. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol Biol Cell. 2004;15:4382–4394. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Witman GB. Dynein and intraflagellar transport. Exp Cell Res. 2015;334:26–34. doi: 10.1016/j.yexcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Beck CF. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2003;100:6269–6274. doi: 10.1073/pnas.0931459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C, Wu S, Kim AS, Sigaudy S, Sarukhanov A, Serre V, Baujat G, Le Quan Sang KH, Rimoin DL, Cohn DH, et al. WDR34 mutations that cause short-rib polydactyly syndrome type III/severe asphyxiating thoracic dysplasia reveal a role for the NF-kappaB pathway in cilia. Am J Hum Genet. 2013;93:926–931. doi: 10.1016/j.ajhg.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol. 2001;153:13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Thompson J, Yates JR, 3rd, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SE. Hsp90: structure and function. Top Curr Chem. 2013;328:155–240. doi: 10.1007/128_2012_356. [DOI] [PubMed] [Google Scholar]
- Ke YN, Yang WX. Primary cilium: an elaborate structure that blocks cell division. Gene. 2014;547:175–185. doi: 10.1016/j.gene.2014.06.050. [DOI] [PubMed] [Google Scholar]
- Kessler K, Wunderlich I, Uebe S, Falk NS, Giessl A, Brandstatter JH, Popp B, Klinger P, Ekici AB, Sticht H, et al. DYNC2LI1 mutations broaden the clinical spectrum of dynein-2 defects. Sci Rep. 2015;5:11649. doi: 10.1038/srep11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF. IFT-cargo interactions and protein transport in cilia. Trends Biochem Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, Witman GB. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013a;201:249–261. doi: 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Gould TJ, Witman GB. Flagellar central pair assembly in Chlamydomonas reinhardtii. Cilia. 2013b;2:15. doi: 10.1186/2046-2530-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour G, Ikebe M, Witman GB. HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil Cytoskeleton. 2009;66:469–482. doi: 10.1002/cm.20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Rostmann J, Grunow A. Analysis of Chlamydomonas SF-assemblin by GFP tagging and expression of antisense constructs. J Cell Sci. 2002;115:1511–1522. doi: 10.1242/jcs.115.7.1511. [DOI] [PubMed] [Google Scholar]
- Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, Tomizawa K, Kaitsuka T, Sung CH. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol. 2011;13:402–411. doi: 10.1038/ncb2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YW, Pang YN, Wu Q, Hu ZF, Han X, Xu YS, Deng HT, Pan JM. FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev Cell. 2014;30:585–597. doi: 10.1016/j.devcel.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Lin H, Nauman NP, Albee AJ, Hsu S, Dutcher SK. New mutations in flagellar motors identified by whole genome sequencing in Chlamydomonas. Cilia. 2013;2:14. doi: 10.1186/2046-2530-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS. Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. J Biol Chem. 2013;288:355–367. doi: 10.1074/jbc.M112.405761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund TC, Anderson LB, McCullar V, Higgins L, Yun GH, Grzywacz B, Verneris MR, Miller JS. iTRAQ is a useful method to screen for membrane-bound proteins differentially expressed in human natural killer cell types. J Proteome Res. 2007;6:644–653. doi: 10.1021/pr0603912. [DOI] [PubMed] [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- McInerney-Leo AM, Schmidts M, Cortes CR, Leo PJ, Gener B, Courtney AD, Gardiner B, Harris JA, Lu Y, Marshall M, et al. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am J Hum Genet. 2013;93:515–523. doi: 10.1016/j.ajhg.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill AE, Merriman B, Farrington-Rock C, Camacho N, Sebald ET, Funari VA, Schibler MJ, Firestein MH, Cohn ZA, Priore MA, et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am J Hum Genet. 2009;84:542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J, Perrone CA, Bower R, Cole DG, Porter ME. The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol Biol Cell. 2005;16:1341–1354. doi: 10.1091/mbc.E04-10-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina PJ, Anderson KV. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flaherty C, de Souza AR. Hydrogen peroxide modifies human sperm peroxiredoxins in a dose-dependent manner. Biol Reprod. 2011;84:238–247. doi: 10.1095/biolreprod.110.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Nagaya K, Kawata Y, Asai H, Tsuchida E, Nohara F, Okajima K, Azuma H. Novel compound heterozygous mutations in DYNC2H1 in a patient with severe short-rib polydactyly syndrome type III phenotype. Congenit Anom. 2014;55:155–157. doi: 10.1111/cga.12098. [DOI] [PubMed] [Google Scholar]
- Palmer KJ, MacCarthy-Morrogh L, Smyllie N, Stephens DJ. A role for Tctex-1 (DYNLT1) in controlling primary cilium length. Eur J Cell Biol. 2011;90:865–871. doi: 10.1016/j.ejcb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee S, Lee S, Kang SW. 2-cys peroxiredoxins: emerging hubs determining redox dependency of Mammalian signaling networks. Int J Cell Biol. 2014;2014:715867. doi: 10.1155/2014/715867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King RS, Gilberti RM, Hom EF, King SM. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol Biol Cell. 2013;24:2668–2677. doi: 10.1091/mbc.E13-05-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Geimer S, Rosenbaum JL. Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Curr Biol. 2006;16:450–459. doi: 10.1016/j.cub.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Perkins A, Poole LB, Karplus PA. Tuning of peroxiredoxin catalysis for various physiological roles. Biochemistry. 2014;53:7693–7705. doi: 10.1021/bi5013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Tritschler D, Taulman P, Bower R, Yoder BK, Porter ME. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol Biol Cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen K, Davis EE, Katsanis N. Unique among ciliopathies: primary ciliary dyskinesia, a motile cilia disorder. F1000Prime Rep. 2015;7:36. doi: 10.12703/P7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou NV, Thompson CL, Osborn DP, Cogger KF, Ashworth R, Knight MM, Beales PL, Chapple JP. Heat shock induces rapid resorption of primary cilia. J Cell Sci. 2012;125:4297–4305. doi: 10.1242/jcs.100545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Subrmanian A, Wilkes DE, Pennock DG, Asai DJ. Dynein-2 affects the regulation of ciliary length but it is not required for ciliogenesis in Tetrahymena thermophila. Mol Biol Cell. 2009;20:708–720. doi: 10.1091/mbc.E08-07-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana AA, Barbera JP, Rodriguez TA, Lynch D, Hirst E, Smith JC, Beddington RS. Targeted deletion of the novel cytoplasmic dynein mD2LIC disrupts the embryonic organiser, formation of the body axes and specification of ventral cell fates. Development. 2004;131:4999–5007. doi: 10.1242/dev.01389. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Pedersen LB, Patel-King RS, King SM. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J Cell Sci. 2007;120:3653–3665. doi: 10.1242/jcs.012773. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol. 1995;47:163–169. doi: 10.1016/s0091-679x(08)60805-5. [DOI] [PubMed] [Google Scholar]
- Satish Tammana TV, Tammana D, Diener DR, Rosenbaum J. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J Cell Sci. 2013;126:5018–5029. doi: 10.1242/jcs.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JC, Haycraft CJ, Thomas JH, Yoder BK, Swoboda P. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol Biol Cell. 2003;14:2057–2070. doi: 10.1091/mbc.E02-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M. Clinical genetics and pathobiology of ciliary chondrodysplasias. J Pediatr Genet. 2015;3:46–94. doi: 10.3233/PGE-14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M, Arts HH, Bongers EM, Yap Z, Oud MM, Antony D, Duijkers L, Emes RD, Stalker J, Yntema JB, et al. Exome sequencing identifies DYNC2H1 mutations as a common cause of asphyxiating thoracic dystrophy (Jeune syndrome) without major polydactyly, renal or retinal involvement. J Med Genet. 2013a;50:309–323. doi: 10.1136/jmedgenet-2012-101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M, Hou Y, Cortes CR, Mans DA, Huber C, Boldt K, Patel M, van Reeuwijk J, Plaza JM, van Beersum SE, et al. TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat Commun. 2015;6:7074. doi: 10.1038/ncomms8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M, Vodopiutz J, Christou-Savina S, Cortes CR, McInerney-Leo AM, Emes RD, Arts HH, Tuysuz B, D’Silva J, et al. Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am J Hum Genet. 2013b;93:932–944. doi: 10.1016/j.ajhg.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol. 2013;29:443–469. doi: 10.1146/annurev-cellbio-101512-122335. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Shih SM, Engel BD, Kocabas F, Bilyarn T, Gennerich A, Marshall WF, Yildiz A. Intraflagellar transport drives flagellar surface motility. Elife. 2013;2:e00744. doi: 10.7554/eLife.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simet SM, Pavlik JA, Sisson JH. Proteomic analysis of bovine axonemes exposed to acute alcohol: role of endothelial nitric oxide synthase and heat shock protein 90 in cilia stimulation. Alcohol Clin Exp Res. 2013;37:609–615. doi: 10.1111/acer.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. doi: 10.1016/s0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- Stephens RE, Lemieux NA. Molecular chaperones in cilia and flagella: implications for protein turnover. Cell Motil Cytoskeleton. 1999;44:274–283. doi: 10.1002/(SICI)1097-0169(199912)44:4<274::AID-CM5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Takaki E, Fujimoto M, Nakahari T, Yonemura S, Miyata Y, Hayashida N, Yamamoto K, Vallee RB, Mikuriya T, Sugahara K, et al. Heat shock transcription factor 1 is required for maintenance of ciliary beating in mice. J Biol Chem. 2007;282:37285–37292. doi: 10.1074/jbc.M704562200. [DOI] [PubMed] [Google Scholar]
- Taylor SP, Dantas TJ, Duran I, Wu S, Lachman RS, University of Washington Center for Mendelian Genomics Consortium. Nelson SF, Cohn DH, Vallee RB, Krakow D. Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome. Nat Commun. 2015;6:7092. doi: 10.1038/ncomms8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Dakoji S, Reise KH, Storey KK, Georgieff MK. Fetal iron deficiency alters the proteome of adult rat hippocampal synaptosomes. Am J Physiol Regul Integr Comp Physiol. 2013;305:R1297–R1306. doi: 10.1152/ajpregu.00292.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWaal KE, Yamamoto R, Wakabayashi K, Fox L, Kamiya R, Dutcher SK, Bayly PV, Sale WS, Porter ME. bop5 mutations reveal new roles for the IC138 phosphoprotein in the regulation of flagellar motility and asymmetric waveforms. Mol Biol Cell. 2011;22:2862–2874. doi: 10.1091/mbc.E11-03-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Misawa Y, Mochiji S, Kamiya R. Reduction-oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2011;108:11280–11284. doi: 10.1073/pnas.1100592108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zou X, Wei Z, Wu Y, Li R, Zeng R, Chen Z, Liao K. Hsp90alpha forms a stable complex at the cilium neck for the interaction of signalling molecules in IGF-1 receptor signalling. J Cell Sci. 2015;128:100–108. doi: 10.1242/jcs.155101. [DOI] [PubMed] [Google Scholar]
- Weis F, Moullintraffort L, Heichette C, Chretien D, Garnier C. The 90-kDa heat shock protein Hsp90 protects tubulin against thermal denaturation. J Biol Chem. 2010;285:9525–9534. doi: 10.1074/jbc.M109.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner C, Onnebrink JG, Omran H. Diagnosis and management of primary ciliary dyskinesia. Cilia. 2015;4:2. doi: 10.1186/s13630-014-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- Williams NE, Nelsen EM. HSP70 and HSP90 homologs are associated with tubulin in hetero-oligomeric complexes, cilia and the cortex of Tetrahymena. J Cell Sci. 1997;110:1665–1672. doi: 10.1242/jcs.110.14.1665. [DOI] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF. A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol. 2013;23:2463–2471. doi: 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Hirono M, Kamiya R. Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J Cell Biol. 2010;190:65–71. doi: 10.1083/jcb.201002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Shen Y, Camp DG, 2nd, Smith RD. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev Proteomics. 2012;9:129–134. doi: 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Wirschell M, Davis S. Novel LC8 mutations have disparate effects on the assembly and stability of flagellar complexes. J Biol Chem. 2009;284:31412–31421. doi: 10.1074/jbc.M109.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Sun Z. Expanding horizons: ciliary proteins reach beyond cilia. Annu Rev Genet. 2013;47:353–376. doi: 10.1146/annurev-genet-111212-133243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Serra R, Shuying Y. Function and regulation of primary cilia and intraflagellar transport proteins in the skeleton. Ann NY Acad Sci. 2015;1335:78–99. doi: 10.1111/nyas.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.