GRHL2 suppresses EMT to give a default epithelial phenotype. GRHL2 inhibits this process through the histone acetyltransferase coactivator p300, repressing the partial EMT and preventing induction of MMPs. The results demonstrate novel roles for p300 and GRHL2 in promoting or suppressing EMT in morphogenesis and tumor progression.
Abstract
Developmental morphogenesis and tumor progression require a transient or stable breakdown of epithelial junctional complexes to permit programmed migration, invasion, and anoikis resistance, characteristics endowed by the epithelial–mesenchymal transition (EMT). The epithelial master-regulatory transcription factor Grainyhead-like 2 (GRHL2) suppresses and reverses EMT, causing a mesenchymal–epithelial transition to the default epithelial phenotype. Here we investigated the role of GRHL2 in tubulogenesis of Madin–Darby canine kidney cells, a process requiring transient, partial EMT. GRHL2 was required for cystogenesis, but it suppressed tubulogenesis in response to hepatocyte growth factor. Surprisingly, GRHL2 suppressed this process by inhibiting the histone acetyltransferase coactivator p300, preventing the induction of matrix metalloproteases and other p300-dependent genes required for tubulogenesis. A 13–amino acid region of GRHL2 was necessary for inhibition of p300, suppression of tubulogenesis, and interference with EMT. The results demonstrate that p300 is required for partial or complete EMT occurring in tubulogenesis or tumor progression and that GRHL2 suppresses EMT in both contexts through inhibition of p300.
INTRODUCTION
The protein p300 is a coactivator of transcription that interacts with at least 400 DNA factors to activate transcription from thousands of enhancers/promoters, mostly contingent upon its intrinsic histone acetyltransferase (HAT) activity. In this context, it is viewed as an “integrator” factor that serves as a nexus among multiple signaling pathways and programs of gene expression (Kamei et al., 1996; Vo and Goodman, 2001; Bedford et al., 2010; Yao et al., 2013). Accordingly, p300/CREB-binding protein (CBP)–null mice die at embryonic days 9.5–11.5 with massive deficits of mesenchymal cells and erythroid cells, as well as with defects in neurulation and heart development (Oike et al., 1999; Yao et al., 2013). p300 is essential for the differentiation of muscle, erythroid lineages, osteoblast, oligodendrocytes, and stem cells (Puri et al., 1997; Oike et al., 1999; Blobel, 2000; Polesskaya et al., 2001; Narayanan et al., 2004; Teo and Kahn, 2010; Zhang et al., 2016). A recent study showed that p300 is a marker for superenhancers, positioning it to control cell state transitions (Witte et al., 2015).
Grainyhead-like 2 (GRHL2) is one of the three genes comprising a family of mammalian transcription factors related to Drosophila Grainyhead, the first zygotically encoded transcription factor expressed during the maternal-to-zygotic transition (Harrison et al., 2010). GRHL2 is important for epithelial barrier assembly in, for example, the morphogenesis of kidney collecting ducts, placenta, lung alveoli, and the mammary gland ductal system (through OVOL2, a GRHL2 target gene) (Gao et al., 2013; Watanabe et al., 2014; Aue et al., 2015; Walentin et al., 2015). Grainyhead-like factors are also critical for diverse developmental events that involve the formation of epithelial barriers, including epidermal assembly, neural crest formation, and, in the adult, wound healing (Wilanowski et al., 2002; Gustavsson et al., 2008; Wang and Samakovlis, 2012; Mlacki et al., 2015). GRHL2 activates the expression of multiple genes encoding epithelial cell adhesion molecules (e.g., E-cadherin, desmosomal proteins, tight junction proteins) and, due to its widespread expression, may be generally important for enforcing an epithelial phenotype (Wilanowski et al., 2002; Ting et al., 2003, 2005; Auden et al., 2006; Rifat et al., 2010; Werth et al., 2010; Boglev et al., 2011; Cieply et al., 2012; Senga et al., 2012; Tanimizu and Mitaka, 2013; Aue et al., 2015). Correspondingly, GRHL2 expression plays an important role in suppressing the oncogenic EMT, thereby functioning as a tumor suppressor that enhances anoikis sensitivity (Cieply et al., 2012, 2013; Mlacki et al., 2015); in fact, the gene most tightly correlated with E-cadherin expression is GRHL2 (Kohn et al., 2014).
The opposing roles of GRHL2 in establishing the epithelial state versus p300 in differentiation—including EMT—motivated us to investigate a potential role for GRHL2 in inhibiting p300 function. Kidney tubulogenesis is contingent upon temporal and spatial regulation of epithelial cells, in which GRHL2, which is expressed in the ureteric buds of the developing kidney, plays a critical organizing role (Schmidt-Ott et al., 2005; Aue et al., 2015; Walentin et al., 2015). Madin–Darby canine kidney (MDCK) tubulogenesis in response to hepatocyte growth factor (HGF) models some aspects of kidney collecting-duct tubulogenesis in vivo and requires a transient, partial EMT (Pollack et al., 1998, 2004; O’Brien et al., 2002; Leroy and Mostov, 2007; Hellman et al., 2008; Jung et al., 2012; Zhang et al., 2014). HGF causes MDCK cells to undergo cell scattering in two-dimensional culture and induces tubulogenesis of MDCK cysts that form in a three-dimensional collagen gel (Wang et al., 1990; Pollack et al., 2004). Induction of MMP1 and MMP13 by HGF through AP1/p300 complexes is required for tubulogenesis in the MDCK model (Benbow and Brinckerhoff, 1997; Clark et al., 2008; Hellman et al., 2008; Chacon-Heszele et al., 2014). Targeted knockout studies indicate a role for HGF/Met in kidney development, and, relatedly, HGF/Met signaling is a potent stimulator of Wnt signaling, a crucial signaling pathway in this process, validating the in vivo relevance of this approach (Monga et al., 2002; Bridgewater et al., 2008; Ishibe et al., 2009).
Here we report that GRHL2 promoted cystogenesis but suppressed tubulogenesis. GRHL2 protein interacted functionally with p300, inhibiting its HAT activity and transcriptional activation of target genes, including matrix metalloproteases. A small (13 amino acids [aa]) sequence of GRHL2 was important for inhibition of p300, suppression of tubulogenesis, and reversal of EMT. These results mechanistically position GRHL2, an enforcer of the epithelial default phenotype, as an antagonist of p300, a coactivator of differentiation-specific genes, with important ramifications for developmental and tumor biology.
RESULTS
GRHL2 suppresses cell scattering and tubulogenesis
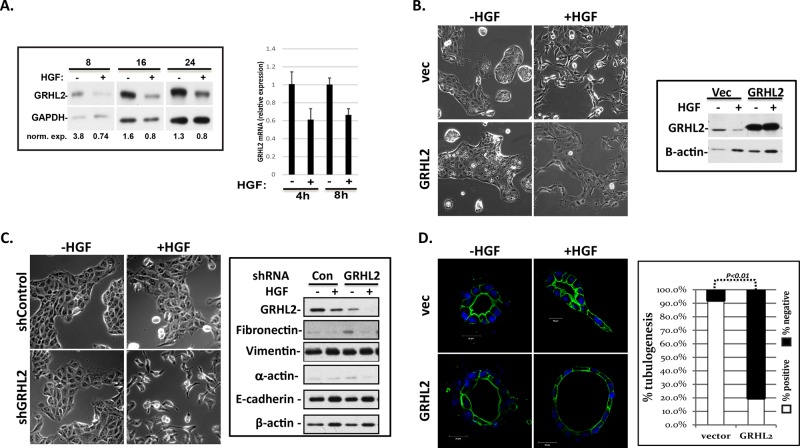
In light of the epithelial programming role of GRHL2, we tested the effect of HGF on GRHL2 levels in MDCK cells. We found that HGF down-regulated the expression of endogenous GRHL2 protein levels at early time points (Figure 1A). When constitutively expressed in MDCK cells, GRHL2 significantly inhibited HGF-induced cell scattering compared with vector control cells (Figure 1B). Conversely, MDCK cells with GRHL2 short hairpin RNA (shRNA) knockdown demonstrated enhanced HGF-induced cell scattering; retention of E-cadherin expression showed that EMT did not occur in response to the knockdown of GRHL2 alone (Figure 1C and Supplemental Figure S1 Video). GRHL2 had only modest effects on the phosphorylation status of two established HGF/Met signaling mediators, Erk and Akt (Supplemental Figure S2).
FIGURE 1:
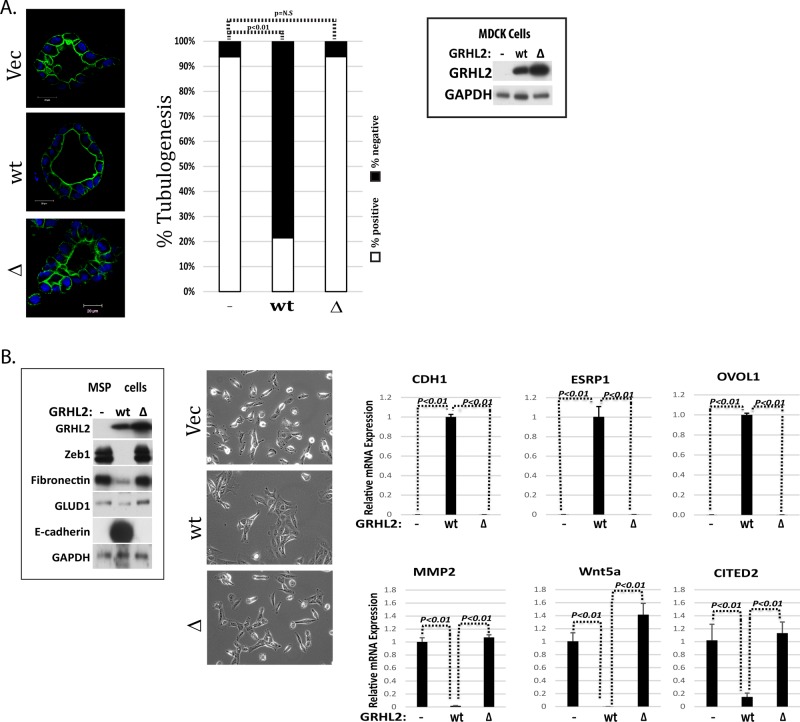
GRHL2 suppresses HGF-induced cell scattering and tubulogenesis and is down-regulated by HGF. (A) HGF induction down-regulates endogenous GRHL2 protein. Western blot and densitometric quantitation of HGF treatment time course in MDCK cells. HGF induction down-regulates endogenous GRHL2 mRNA in MDCK cells; qPCR results expressed as ratios compared with vector control. (B) Constitutive GRHL2 expression in MDCK cells suppresses HGF-induced cell scattering; images represent representative morphologies of cells treated for 42 h. (C) GRHL2 knockdown in enhanced HGF-induced cell scattering (16 h), but MDCK shGRHL2 cells did not undergo an EMT phenotypic change when EMT markers were examined via Western blotting. (D) Constitutive GRHL2 expression in MDCK cells prevents tubulogenesis (blue, nuclei; green, actin). Scale bar, 20 μm. Quantification of percentage of cysts that demonstrated tubulogenesis.
We then examined the effects of GRHL2 on HGF-induced tubulogenesis in the three-dimensional collagen model. GRHL2 expression caused the formation of larger cysts than in vector control cells, consistent with previous reports on hepatic bile duct cells (Figure 1D; Tanimizu and Mitaka, 2013). GRHL2 significantly suppressed tubulogenesis/branching morphogenesis after HGF treatment (Figure 1D); a similar effect of murine Grhl2 was observed in mouse inner medullary collecting-duct cell line (Supplemental Figure S3A), indicating the important role of GRHL2 down-regulation in tubulogenesis. Conversely, the stable knockdown of GRHL2 prevented cystogenesis (Supplemental Figure S3B), presumably by down-regulating epithelial adhesion molecules (Werth et al., 2010; Senga et al., 2012). Transient knockdown of GRHL2 after cyst formation using a doxycycline-induced shRNA vector, however, clearly indicated that the loss of GRHL2 promoted tubulogenesis (Supplemental Figure S4A). There was no effect of the GRHL2 shRNA on cell proliferation (Supplemental Figure S4B). These results indicated that GRHL2 suppressed HGF-activated cell scattering and tubulogenesis.
GRHL2 suppresses the expression of matrix metalloproteases
To identify GRHL2 target genes responsible for the attenuation of cellular responses to HGF/Met signaling, we used RNA-sequencing (RNA-Seq) to compare MDCK cells with constitutive GRHL2 expression versus GRHL2 depletion with either no treatment or HGF induction (24 h). This experimental scheme allowed for two distinct comparisons: genes regulated by HGF in the absence versus presence of GRHL2, and genes regulated by GRHL2 with versus without HGF induction. GRHL2 down-regulated several known matrix metalloprotease (MMP) family members (Table 1; confirmed by quantitative PCR [qPCR] in Figure 2A). In light of the established importance of specific MMPs for tubulogenesis in the MDCK and mIMCD-3 cell culture models (Hotary et al., 2000; Jorda et al., 2005; Hellman et al., 2008; Chacon-Heszele et al., 2014), we verified their importance for branching tubulogenesis in embryonic kidneys cultured ex vivo using the generalized MMP inhibitor batimastat (Supplemental Figure S5). The down-regulation of MMPs by GRHL2 contributed significantly to the tubulogenesis-suppressing effect of constitutive GRHL2 expression, although additional contributions from EMT/mesenchymal-epithelial transition (MET)–related GRHL2 genes are likely.
TABLE 1:
HGF-induced MMP expression in the absence versus the presence of GRHL2.
| Gene regulated by GRHL2 | Fold change (shGRHL2+HGF/GRHL2+HGF) | Significance |
|---|---|---|
| MMP-1 | 31.7 | Interstitial collagenase, necessary for MDCK tubulogenesis, p300-dependent expression |
| MMP-9 | 5.8 | Gelatinase B, p300-dependent cancer invasiveness, up-regulated in EMT |
| MMP-13 | 15.1 | Collagenase 3, necessary for MDCK tubulogenesis, p300-dependent expression |
| MMP-14 | 5.82 | Membrane-type MMP, involved in EMT regulation and invasiveness phenotype |
| MMP-19 | 3.5 | Increases invasion in multiple cancers |
| MMP-24 | 9.6 |
FIGURE 2:
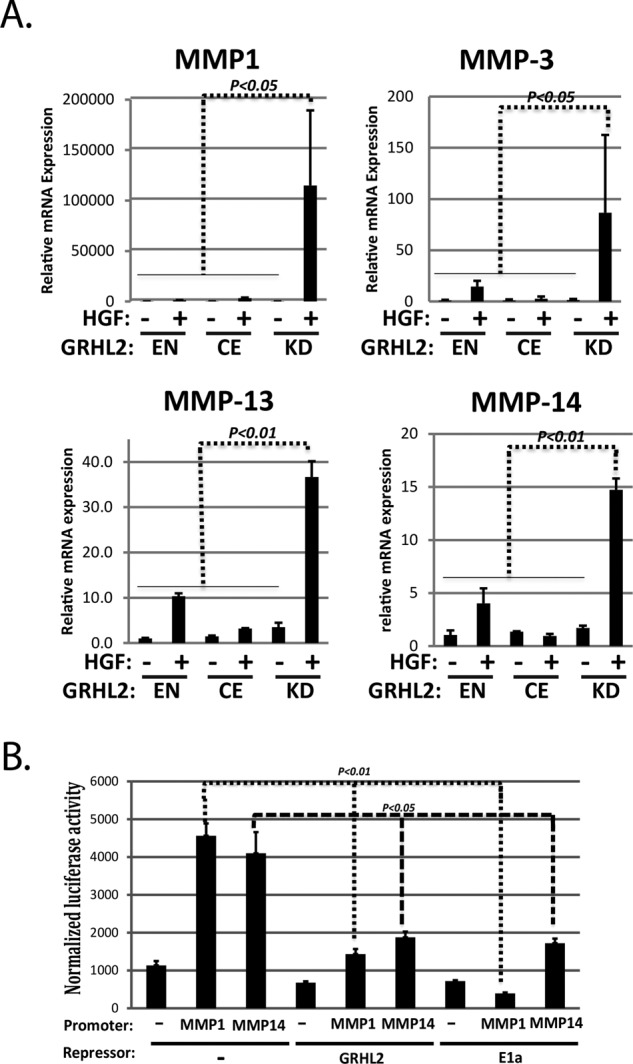
GRHL2 suppresses the induction of MMP genes and MMP promoters by HGF. (A) GRHL2 suppresses the induction of MMP genes (qPCR analysis; EN, endogenous GRHL2; CE, constitutively expressed GRHL2; KD, GRHL2 shRNA knockdown). (B) GRHL2 suppresses the induction of MMP promoters by HGF. HT1080 cells were cotransfected with either MMP1 or MMP14 promoter-luciferase reporter constructs and GRHL2, E1A, or empty expression vectors. Values represent relative luciferase activity normalized to TK-β-galactosidase control.
To investigate the transcriptional mechanisms of MMP gene regulation by GRHL2, we assayed MMP1 and MMP14 promoters as luciferase reporter constructs by cotransfection in HT1080 fibrosarcoma cells, in which the endogenous MMPs are expressed constitutively (Tang and Hemler, 2004; Nonaka et al., 2005). GRHL2 was found to suppress MMP1 and MMP14 promoters using adenovirus E1a protein, a known repressor of MMP genes, as a positive control (Figure 2B). These results demonstrated that GRHL2 repressed MMP1 and MMP14 promoters; however, examination of the promoter sequences used in our reporter constructs did not indicate the presence of GRHL2 DNA–binding consensus sites (Wilanowski et al., 2002; Gao et al., 2013; Aue et al., 2015; Walentin et al., 2015), suggesting a subtler mechanism for repression not involving direct DNA binding.
GRHL2 inhibits p300 function
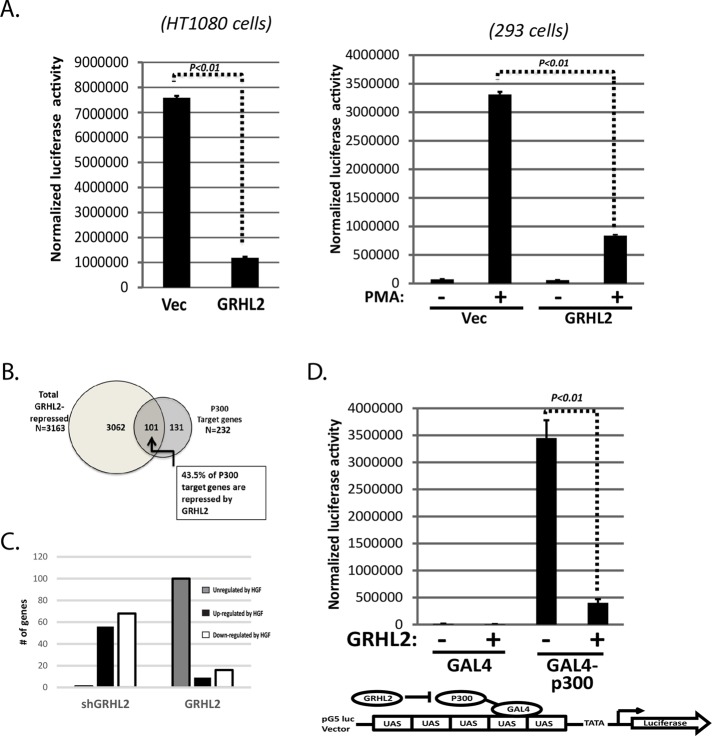
The AP-1 transcription factor family is important in the regulation of most MMP genes (Clark et al., 2008). GRHL2 significantly suppressed AP-1 activity on minimal reporter constructs in HT1080 and 293 cells (Figure 3A). GRHL2 did not appear to affect the expression of FOS or JUN family members after HGF induction (Supplemental Figure S6).
FIGURE 3:
GRHL2 suppresses AP-1 and p300 function. (A) GRHL2 suppresses AP1 function. HT1080 and 293T cells were cotransfected with AP-1 response element-luciferase reporter construct and GRHL2 expression vector or empty vector. PMA was used to induce AP-1 signaling in 293T cells. Values represent relative luciferase activity normalized to TK-β-galactosidase control. (B) Venn diagram comparing GRHL2-repressed genes (Farris et al., 2016) vs. p300 target genes identified via E1a (Ferrari et al., 2014). (C) GRHL2 suppresses the p300 pathway. p300 effector and target genes induced by HGF in MDCK cells with GRHL2 shRNA vs. cells with constitutive GRHL2 determined by Ingenuity Pathway Analysis. Quantitation of the number of unregulated, HGF up-regulated, or HGF down-regulated genes for both cell lines. The p300 interactome diagram on which this is based is shown in Supplemental Figure S7. (D) GRHL2 suppresses p300 function. GAL4-minimal promoter-luciferase activation by a cotransfected GAL4-p300 was assayed in HT1080 cells in the presence or absence of cotransfected GRHL2 expression vector; values represent relative luciferase activity normalized to TK-β-galactosidase control.
In light of the functional similarities between adenovirus E1a and GRHL2 (see Discussion), we compared a published list of genes that E1a down-regulated through p300 interaction (Ferrari et al., 2014) against our list of GRHL2–down-regulated genes; the latter list was derived from the RNA-Seq analysis that we performed on GRHL2-expressing human mesenchymal subpopulation (MSP) cells (Farris et al., 2016) and used because a comparison of canine versus human gene lists proved uninformative for technical reasons. About 43.5% of the genes that E1a repressed through p300 were also repressed by GRHL2 (Figure 3B). Downstream Effector Analysis/Ingenuity Pathway Analysis revealed that the expression of GRHL2 suppressed the regulation of numerous p300-associated genes that were up-regulated or down-regulated by HGF in the absence of GRHL2 expression (Figure 3C and Supplemental Figure S7). These results suggested that GRHL2 could potentially inhibit p300 function.
To investigate this, we assayed the effect of GRHL2 on the activity of a GAL4-DNA binding domain–p300 fusion protein by transient transfection using a GAL4-responsive luciferase promoter, an established method for testing p300 coactivator function without confounding effects from primary DNA-binding activator proteins (Snowden et al., 2000). Adenovirus E1a protein repressed the AP-1 and GAL4/GAL4-p300 reporter activities, but a p300-nonbinding mutant of E1a (Ferrari et al., 2014) failed to do so (Supplemental Figure S8), validating the p300 dependence of transcription in these assays. GRHL2 inhibited the coactivator function of p300 in this assay (Figure 3D). As additional negative controls, GRHL2 did not affect the ability of a p300-independent GAL4-activator fusion protein (VP16-GAL4; Kutluay et al., 2009) to activate the same reporter; another transcription factor unrelated to GRHL2, c-Myc, failed to inhibit GAL4-p300 function in this assay (Supplemental Figure S9). These results indicated that GRHL2 was a potent suppressor of p300 function. The highly related GRHL1 and GRHL3 proteins had similar effects in the reporter assay (Supplemental Figure S10).
GRHL2 inhibits the histone acetyltransferase activity of p300
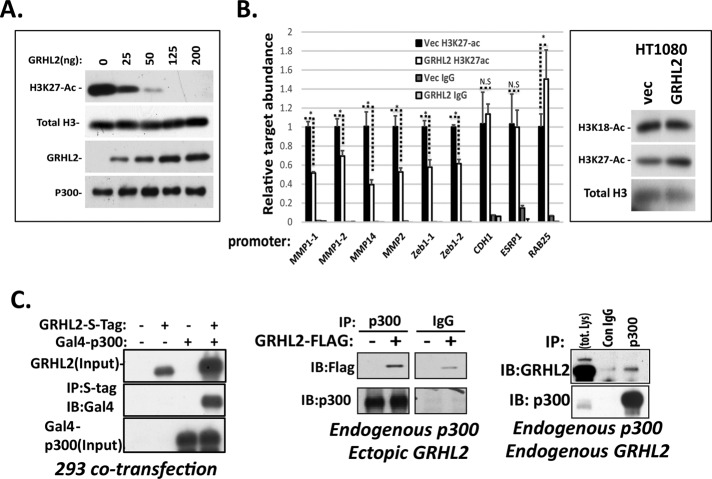
The HAT domain of p300 acetylates numerous substrates, including lysines 18 and 27 in the N-terminal tail of histone 3 (H3; Ogryzko et al., 1996; Kasper et al., 2010; Jin et al., 2011). Using recombinant proteins, we found that GRHL2 inhibited acetylation of H3K27 by p300 in a dose-dependent way in vitro (Figure 4A). We found similar results using the p300 HAT domain alone (aa 1283–1673) rather than full-length p300 (Supplemental Figure S11). Recombinant GRHL1 and GRHL3 proteins also inhibited the HAT activity of p300 (Supplemental Figure S12).
FIGURE 4:
GRHL2 suppresses the HAT activity of p300. (A) In vitro HAT assays with recombinant H3, p300, and GRHL2 proteins. Coomassie stains to assess the quality of recombinant proteins used throughout this study are shown in Supplemental Figure S19. (B) GRHL2 inhibits the acetylation of H3 on GRHL2-repressed but not GRHL2-induced promoters in vivo (ChIP assay). Cross-linked chromatin from HT1080+ vector HT-1080+GRHL2 cells immunoprecipitated with H3K27-Ac or rabbit IgG; the indicated promoters were assayed for H3K27Ac by qPCR using the primers in Materials and Methods. *p < 0.05. GRHL2 does not affect global histone H3K18-Ac or H3K27-Ac (Western blotting of total histones). (C) GRHL2 interacts with p300. Left, cotransfection of indicated expression constructs, followed by coimmunoprecipitation/Western blotting. Middle, coimmunoprecipitation of retrovirally expressed, S-tagged GRHL2 with endogenous p300. Right, coimmunoprecipitation of endogenous GRHL2 and endogenous P300.
To confirm these in vitro effects in cells, we analyzed histone acetylation by chromatin immunoprecipitation (CHIP)–qPCR using the human cell line HT1080 (because Encode reference data reporting histone acetylation are available for the human genome). Consistent with the in vitro results, the acetylation of H3K27 was inhibited in HT1080 cells expressing GRHL2 constitutively, specifically on promoters that GRHL2 repressed, including MMP1, MMP14, MMP2, and ZEB1; acetylation of GRHL2-up-regulated gene promoter–associated H3K27 was either not affected (CDH1, ESRP1) or up-regulated (RAB25; Figure 4B). Unlike the effect of E1a on p300, there was neither an effect of GRHL2 on global H3 acetylation nor global chromatin condensation, as assayed by Western blotting (Figure 4B) or immunofluorescence localization of a LacI-mCherry-GRHL2 protein (Ferrari et al., 2014), respectively (Supplemental Figure S13).
GRHL2 interacted with p300 by the criteria of coimmunoprecipitation of transiently cotransfected genes or of endogenous proteins (Figure 4C). The recombinant proteins interacted only weakly, however (Supplemental Figure S14), suggesting that the interaction is transient or requires posttranslational modifications absent from our recombinant proteins.
GRHL2 inhibits the C-terminal transactivation domain of P300
Multiple domains of p300 can coactivate transcription independently. One of these is the C-terminal domain (aa 1665–2414), which interacts with the p160 family of coactivators (Stiehl et al., 2007). Previous reports suggested that the activity of the C-terminal domain is stimulated by autoacetylation catalyzed by the p300 HAT domain (Stiehl et al., 2007). In light of the HAT-inhibitory effect of GRHL2 described above, we hypothesized that GRHL2 inhibited the C-terminal domain through this mechanism. We confirmed that transcriptional activation by the C-terminal domain, assayed as a GAL4-C-term fusion protein, was stimulated by a cotransfected p300 HAT domain and that the latter domain had no detectable ability to activate transcription independently (Figure 5A). GRHL2 inhibited the enhanced transactivation by the p300 C-terminal domain in the presence of cotransfected p300 HAT (Figure 5B). Embedded within the C-terminal domain is a small subdomain called the IRF-3 binding domain (IBID; aa 2050–2096) that mediates the interaction with the p160 coactivator family, including the family member steroid receptor coactivator-1 (SRC-1; Sheppard et al., 2001). Whereas neither the IBID domain of p300 nor SRC-1 alone activated transcription efficiently, the combination of both activated transcription, as reported previously (Figure 5C; Sheppard et al., 2001). GRHL2 potently inhibited transcription that was driven by the complex of p300-IBID and SRC-1 (Figure 5D). Of interest, mutation of the two lysines (K2086 and K2091) of the IBID domain that are acetylated in vivo abrogated the ability of the p300-IBID/SRC1 complex to activate transcription (Figure 5E). These results indicate that the transcriptional activation by the p300 C-terminus, which is supported by SRC-1 and abrogated by GRHL2, depends on acetylation of at least these (and probably other) lysines by p300 HAT or perhaps other GRHL2-sensitive HATs that remain to be determined.
FIGURE 5:
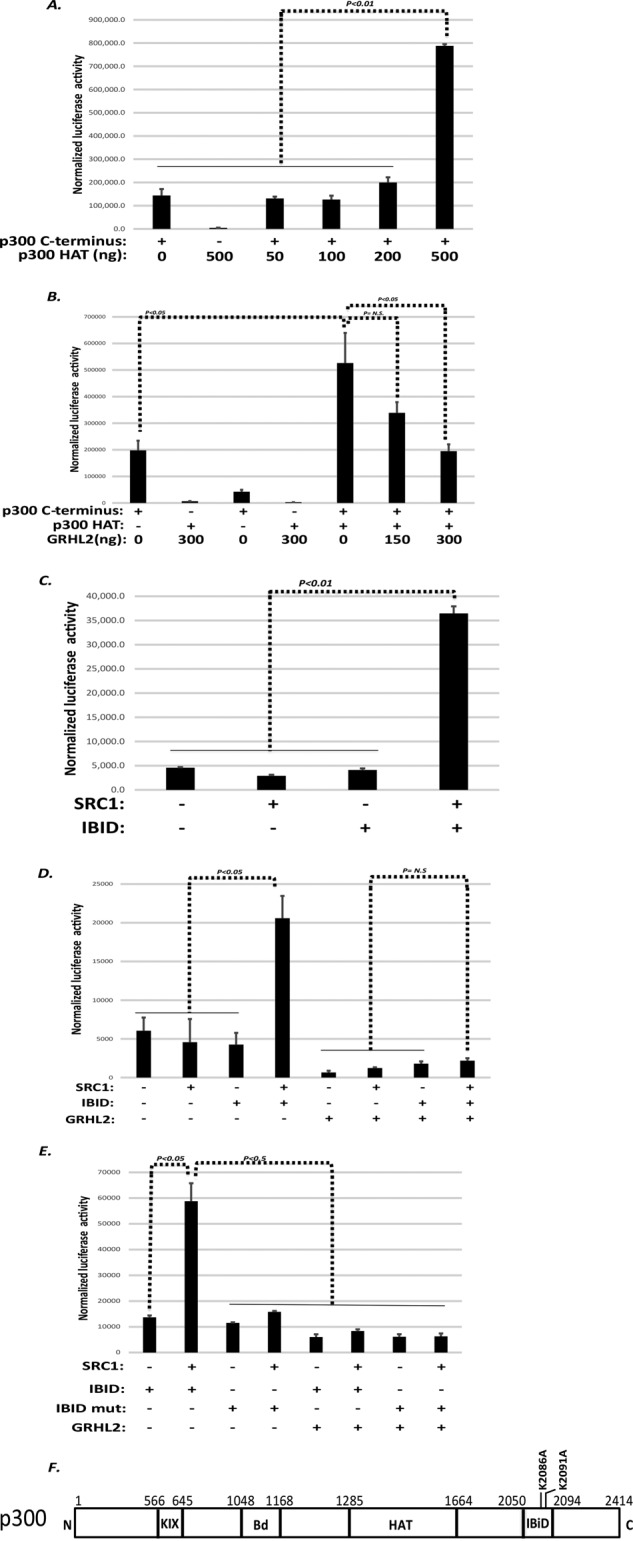
GRHL2 inhibits the C-terminal transactivation domain of p300. (A) The p300 HAT domain stimulates transactivation by the p300 C-terminus (cotransfection in HT1080 cells). (B) GRHL2 inhibits transactivation by the p300 C-terminus. HT1080 cells were cotransfected with GAL4 response element luciferase reporter in the presence of the indicated expression constructs. (C) The IBID domain of p300 and the coactivator SRC-1 synergize to activate transcription. HT1080 cells were cotransfected with GAL4 response element luciferase reporter I in the presence of the indicated expression constructs. (D) GRHL2 inhibits transactivation by the IBID-SRC-1 complex. HT1080 cells were cotransfected with GAL4 response element luciferase reporter in the presence of the indicated expression constructs. (E) Transactivation by the IBID-SRC-1 complex is contingent upon lysines 2086 and 2091. HT1080 cells were cotransfected with GAL4 response element luciferase reporter in the presence of the indicated expression constructs. In A–E, values represent relative luciferase activity normalized to TK-β-galactosidase control. (F) Schematic of p300 domains. Bd, bromodomain; HAT, histone acetyltransferase domain; IBID, IRF-3 binding domain; KIX, CREB-binding domain.
A small region of GRHL2 inhibits p300 and is critical for suppressing tubulogenesis and EMT
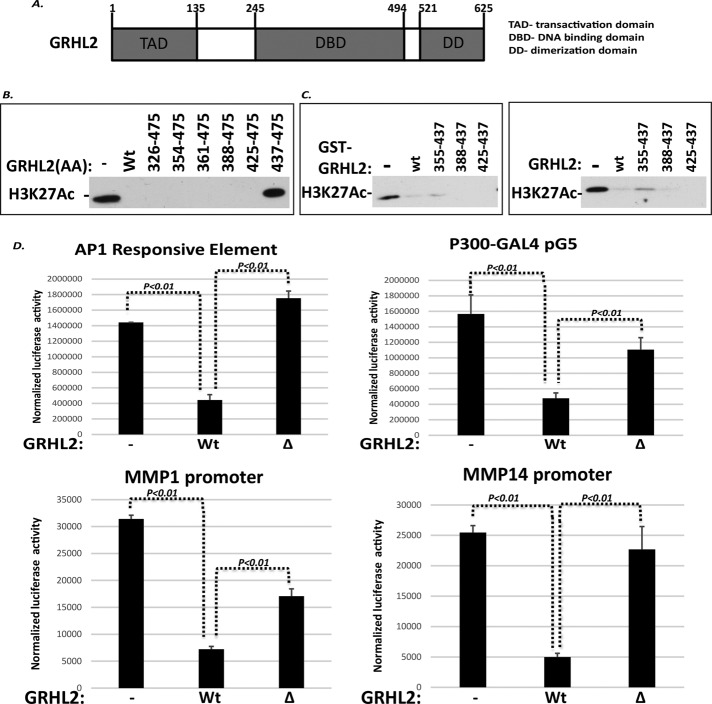
To identify the region of GRHL2 responsible for p300 inhibition, whole domains of GRHL2 were deleted initially (Figure 6A) and assayed for inhibition of HAT activity and transcriptional activation by GAL4-p300. In light of the lack of GRHL2-binding sites in most GRHL2-repressed promoters (unpublished data), the surprising result was that the DNA-binding domain (aa 245–494) was critical for inhibition (Supplemental Figures S15 and S16). Glutathione S-transferase (GST)–GRHL2 protein subfragments derived from the DNA-binding domain were assayed for HAT inhibition (Figure 6B). The region of aa 325–475, and within that, the 13-aa sequence 425–437, inhibited HAT activity potently (Figure 6C), and a synthetic peptide (aa 420–442) was also able to inhibit p300 HAT activity (Supplemental Figure S15C). The aa 425–437 fragment (IRDEERKQNRKKG) is an α-helix conserved among GRHL1, GRHL2, and GRHL3 (Kokoszynska et al., 2008). A GRHL2 Δ425-437 construct (with a synthetic nuclear localization signal) was unable to repress an AP-1-promoter, GAL4-P300 transactivation (assayed on a GAL4-Luc reporter), the MMP1 promoter, or the MMP14 promoter (Figure 6D). We recently reported that GRHL2 repressed the expression of the glutamate dehydrogenase-1 gene (GLUD1), with important consequences for metabolism and anoikis (Farris et al., 2016). Of interest, GRHL2 also repressed this promoter in a manner that required aa 425–437 (Supplemental Figure S17). GRHL2 Δ425-437 was still able to interact, however, with p300 in a cotransfection assay (Supplemental Figure S18).
FIGURE 6:
A small region within the DNA-binding domain of GRHL2, aa 425–437, inhibits p300. (A) Schematic of GRHL2 domains. DBD, DNA-binding domain; DD, dimerization domain; TAD, transactivation domain. (B) HAT assays using the indicated GRHL2 fragments, assayed as GST-fusion proteins. (C) GRHL2 aa 425–437 inhibits p300 HAT activity. The indicated fragments derived from GRHL2 were assayed as GST-fusions (left) or recombinant peptides (right) for inhibition of HAT activity. (D) GRHL2 aa 425–437 are required for inhibition of an AP-1 reporter, GAL4 reporter in conjunction with GAL4-p300, MMP1 reporter, or MMP14 reporter. Values represent relative luciferase activity normalized to TK-β-galactosidase control.
We hypothesized that the inhibition of p300 function could provide a mechanism for some aspects of transcriptional reprogramming by GRHL2. Consistent with the inability of the GRHL2 Δ425-437 to repress the AP1-dependent promoters MMP1 and MMP14 (Figure 6C), this mutant was unable to suppress HGF-induced tubulogenesis in MDCK cells (Figure 7A). Moreover, wild-type GRHL2 reverses EMT in MSP cells, a cancer stem cell–like subpopulation derived from the mammary epithelial cell line HMLE (Cieply et al., 2012, 2013). The GRHL2 Δ425-437 mutant failed to reverse EMT by the criteria of cell morphology as well as EMT marker protein and mRNA expression (Figure 7B). These results indicated that the inhibition of p300 by GRHL2 contributes significantly to transcriptional reprogramming involved in both tubulogenesis and EMT (Figure 8).
FIGURE 7:
The p300-inhibitory domain of GRHL2 is important for the suppression of tubulogenesis and reversion of EMT. (A) MDCK cells expressing GRHL2 wild-type or GRHL2 Δ425-437 were assayed for tubulogenesis (blue, nuclei; green, actin). Scale bar, 20 μm. Middle, quantitation; right, Western blot confirmation of protein levels. (B) MSP cells expressing GRHL2 wild-type or GRHL2 Δ425-437 were assayed for reversion of EMT by Western blotting for EMT markers (left), cell morphology (middle), or qPCR (right).
FIGURE 8:
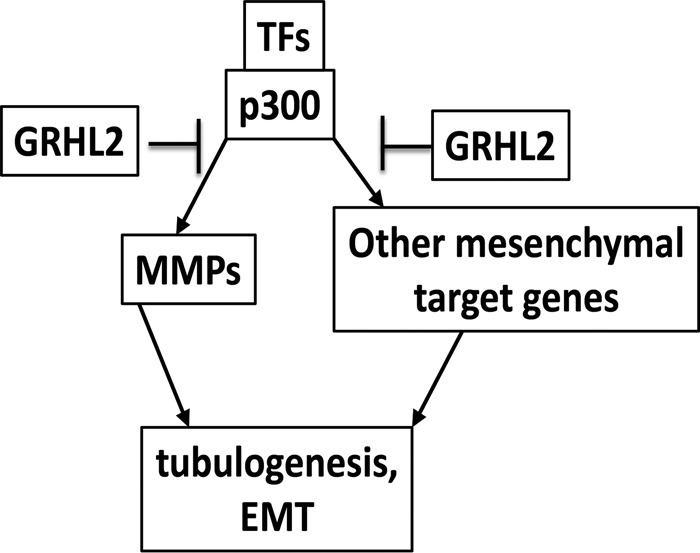
GRHL2 suppresses tubulogenesis and EMT by inhibition of p300.
DISCUSSION
Previously we proposed that the epithelial cell is the default phenotype from both developmental and cancer biological points of view. Under this hypothesis, the epithelial phenotype is adopted automatically by a cell in the absence of genomic/epigenetic alterations or inductive differentiation signaling (Frisch, 1997). Consistent with this idea, E-cadherin–expressing cells are the first to emerge at the late-two-cell stage; these eventually give rise to all differentiated cell types of the adult organism (Hyafil et al., 1980; Vestweber and Kemler, 1985; Fleming et al., 1989; Larue et al., 1994). The adenovirus-5 E1a protein, which interacts with and negatively modulates p300, enforces an epithelial phenotype, suppresses EMT, enhances anoikis, suppresses MMP expression, and exerts a ubiquitously acting tumor suppression effect in human tumor cells (Frisch and Mymryk, 2002).
The default idea has been invoked in recent studies from other laboratories to explain, for example, MET occurring at metastatic sites (Thiery and Sleeman, 2006; Polyak and Weinberg, 2009; Tam and Weinberg, 2013). The default hypothesis could be explained molecularly if critical coactivators such as p300 were required for differentiation-specific but not epithelial-specific gene expression (Frisch, 1997). Accordingly, we reported previously that GRHL2, shown here to antagonize the coactivator p300, inhibits and reverses the oncogenic EMT, suppressing tumor initiation, drug resistance, and anoikis resistance (Cieply et al., 2012, 2013).
The novel observation that GRHL2 antagonizes p300-dependent pathways in the context of tubulogenesis provides a potentially unifying mechanism for both the developmental and oncologic effects of GRHL2. GRHL2 probably regulates tubulogenesis by additional, p300-independent mechanisms. For example, cells that fail to down-regulate GRHL2 appropriately may fail to down-regulate cell adhesion molecules in response to morphogenic factors (Werth et al., 2010; Senga et al., 2012). For example, E-cadherin relocalizes from cell junctions to diffuse membrane pattern in protrusive cell extensions during tubulogenesis; this relocalization might be envisioned to require GRHL2 down-regulation and subsequent loss of junctional complex components in these specific cells (Pollack et al., 1998).
p300 contributes to tubulogenesis and cancer-related EMT in multiple important ways, providing a mechanism for GRHL2, via inhibition of p300, to suppress these processes. With regard to cancer, transcription factors that use p300/CBP and additional coactivators that interact with p300/CBP have been implicated in the oncogenic EMT, tumor drug resistance, and tumor recurrence (Matthews et al., 2007; Qin et al., 2009; Santer et al., 2011; Zhou et al., 2012; Delvecchio et al., 2013; Ringel and Wolberger, 2013; Yang et al., 2013; Xu et al., 2014; Cho et al., 2015). The up-regulation of MMPs associated with the oncogenic EMT requires AP-1, Ets, and other p300-dependent factors (Westermarck and Kähäri, 1999; Sun et al., 2004; Chou et al., 2006; Clark et al., 2008; Lee and Partridge, 2010; Santer et al., 2011).
With regard to tubulogenesis, the p300-dependent Ets family transcription factors Etv4 and Etv5 are key transcriptional regulators of this developmental process (Yang et al., 1998; Jayaraman et al., 1999; Goel and Janknecht, 2003; Foulds et al., 2004; Lu et al., 2009; Oh et al., 2012; Wollenick et al., 2012). In addition, the family of p300-dependent TCF/β-catenin transcription complexes is a central hub for Wnt signaling, crucial for nephron induction (Li et al., 2007; Bridgewater et al., 2008; Miller and McCrea, 2010; Ma et al., 2012).
p300 activates transcription through an array of mechanisms: acetylation of histones, acetylation of transcription factors, binding of the p300 bromodomain to acetylated histones to promote transcriptional activity, and recruitment of histone methyltransferases (e.g., Set1b complex; Ogryzko et al., 1996; Gu and Roeder, 1997; Mizzen and Allis, 1998; Ito et al., 2001; Wolf et al., 2002; Stiehl et al., 2007; Chen et al., 2010; Tang et al., 2013). By inhibiting p300 HAT activity, GRHL2 could enforce an inactive state on p300 in at least two ways. First, p300 HAT activity is substantially increased by autoacetylation of an autoinhibitory loop on the HAT domain, which GRHL2 would suppress, stably inhibiting the enzyme (Thompson et al., 2004). Second, the p300 HAT domain potentiates p300 C-terminal domain (aa 1665–2414) transactivation (Stiehl et al., 2007), which GRHL2 blocked. The minimal domain for p300 C-terminal transactivation is aa 2000–2180, which include the IBID (Lin et al., 2001; Matsuda et al., 2004). The p300 IBID domain cooperates with SRC1, a p160/steroid receptor coactivator family member, to activate transcription (Sheppard et al., 2001). By inhibiting the HAT activity of p300, GRHL2 suppresses coactivation by the p300-IBID complex, which is highly dependent on the two known acetylation sites, K2086 and K2091, for activity. Of interest, SRC1-p300 was shown to induce TWIST expression and EMT in breast cancer (Qin et al., 2009), and GRHL2 is a potent EMT suppressor.
Although >400 DNA-binding proteins interact with p300 and use it as a coactivator, the inhibitory effect of GRHL2 on p300 is nearly unique, shared by only one other family of proteins, the E1a-like inhibitor of differentiation (EID) proteins. EID1 inhibits p300 HAT activity (MacLellan et al., 2000), and EID3 blocks the SRC-1/CBP interaction; the mechanism of EID2 is controversial (Ji et al., 2003; Miyake et al., 2003; Båvner et al., 2005). To our knowledge, the effects of EID proteins on EMT or tubulogenesis have not been investigated.
GRHL2 regulates intracellular metabolism, and this could indirectly affect p300 by altering the levels of the cofactors for p300 HAT, acetyl-CoA, or crotonyl-CoA (Li and Li, 2015; Farris et al., 2016). This may provide an additional, more indirect mechanism for GRHL2 to affect p300 function.
In summary, p300 and related coactivators are crucial for cells to differentiate. Under the default-phenotype hypothesis, this can be extended to critical roles in EMT in contexts such as tubulogenesis and oncogenesis. The inhibition of p300 by GRHL2 provides a mechanism for GRHL2 to enforce the default epithelial phenotype.
MATERIALS AND METHODS
Cell lines
The 293 HEK cells without T antigen (Doug Black, University of California, Los Angeles [UCLA], Los Angeles, CA), MDCK cells (clone M8; Frisch and Francis, 1994), and HT1080 cells (American Type Culture Collection [ATCC], Manassas, VA) were cultured in advanced DMEM (Life Technologies, Carlsbad, CA) with 10% fetal bovine serum (FBS; Hyclone, Chicago, IL) and 1× penicillin-streptomycin-glutamine (PSG) (Invitrogen, Carlsbad, CA). miMCD-3 cells (ATCC) were cultured in advanced DMEM:Ham’s F-12 (Life Technologies) with 10% FBS and 1× PSG. The HMLE and HMLE MSP cells were described previously (Cieply et al., 2013).
Generation of stable cell lines by retroviral transduction and lentiviral transduction
Full-length GRHL2/pMXS-IRES-Puro and retroviral packaging protocol were described previously (Farris et al., 2016). GRHL2 Δ425-437 was subcloned into pMXS-IRES-Puro from GRHL2 Δ425-437-NLS-3xFLAG/pCDNA3.1+. GRHL2 shRNA/pGipZ and scramble control shRNA/pGipz were obtained from Open Biosystems and packaged as previously described (Cieply et al., 2012). The shRNA construct in pTRIPZ was made by subcloning an MluI-XbaI fragment from pGIPZ.
Cell scattering assays
MDCK cells were plated on collagen-coated six-well dishes to give 25% cell confluency on HGF induction. Cells were treated with 60 ng/ml recombinant human HGF (RD Systems, Minneapolis, MN) in DMEM or fresh DMEM for the time periods indicated. The cells were imaged and then harvested for protein or RNA analysis. Images were obtained using a Zeiss Axiovert 200M, AxioCam MRM camera, Phase 20×/0.55 objective, room temperature, and AxioVision, release 4.8 software. Cell scattering time-lapse movies were taken every 15 min over a 24-h period. Time-lapse images were obtained using a Nikon Eclipse TE2000-E with Photometrics CoolSNAP HQ2 Monochrome charge-coupled device with a Phase 40×/0.75 objective using a Bioptechs Delta T4 Culture Dish Heater, Prior ProScan II Encoded Motorized Stage, and Oko Labs Digital Stage Top Incubator (37°C/5% CO2). For each data point, four 40× images were stitched together using 10% overlap with background correction in NIS-Elements AR (Nikon).
Western blotting
Protein lysates were incubated at 95°C for 5 min in 1× SDS lysis buffer (125 mM Tris-HCl, pH 6.8, 0.04% SDS, 0.024% bromophenol blue, 5% β-mercaptoethanol, and 20% glycerol). SDS–PAGE was run in a 4–20% gradient Novex Tris-glycine gel (Invitrogen). Proteins were electrophoretically transferred to a polyvinylidene difluoride membrane (Immobilon, Waltham, MA) in Tris-glycine buffer with 5% methanol. Membranes were blocked for 1 h in 1× Tris-buffered saline (TBS) plus 5% nonfat milk. Primary and secondary antibodies were incubated in 1× TBS plus 5% nonfat milk plus 0.1% Tween-20. Primary antibodies were incubated for 2 h at room temperature or overnight at 4°C at 1:1000 dilution. Primary antibodies used were as follows: GRHL2, rabbit (rb; Sigma-Aldrich, St. Louis, MO); β-actin, mouse (ms; Thermo Pierce); fibronectin, ms (BD Biosciences, Franklin Lakes, NJ); vimentin, ms (Santa Cruz Biotechnology, Dallas, TX); β-actin, ms (Sigma-Aldrich); E-cadherin, ms (BD Biosciences); β-catenin, ms (BD Biosciences); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ms (OriGene, Rockville, MD); acetyl-lysine, rb (Millipore); H3K18AC, rb (Cell Signaling, Danvers, MA); H3K27AC, rb (Cell Signaling); total H3, rb (Cell Signaling); GST, ms (Thermo Pierce, Waltham, MA); p300, rb (sc-584; Santa Cruz Biotechnology); p300, rb (sc-585; Santa Cruz Biotechnology); FLAG, ms (Sigma-Aldrich); and GAL4, rb (Santa Cruz Biotechnology). Goat anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibodies (Bio-Rad) were used at 1:3000 dilution and incubated for 1 h at room temperature RT. Western blot membranes were developed using ECL-West Pico (Pierce) and analyzed using standard chemiluminescence techniques.
MDCK tubulogenesis assay
MDCK tubulogenesis assay was performed as previously described (Elia and Lippincott-Schwartz, 2009). Briefly, collagen matrix was made by mixing 10× MEM (Invitrogen), 100× PSG, 23.5 g/l NaHCO3 (Invitrogen), sterile water, and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Invitrogen) to working concentrations. Vitrogen bovine collagen (5005-b; Advanced Biomatrix, Carlsbad, CA) was added to a final concentration of 2 mg/ml. NaHCO3 was used to bring the solution to pH 7.3 via a pH strip indicator (Elia and Lippincott-Schwartz, 2009). In a 24-well plate, 100 μl of working collagen solution was placed in a 1.0-μm-pore cell culture insert (Falcon, Franklin Lakes, NJ) and allowed to solidify for 3 h in tissue culture incubator. Subconfluent MDCK cells were trypsinized and counted using a Countess Cell counting machine (Invitrogen). Cells were pipetted into the working collagen solution to give a final concentration of 5000 cells/ml. Then 250 μl of cell/collagen solution was pipetted onto the presolidified collagen matrix in the cell culture insert. The cell–collagen matrix was allowed to solidify for 2 h at 37°C and 5% CO2, and then 250 and 500 μl of medium was placed on the cell–collagen matrix cell culture insert and in a 24-well chamber, respectively. Media were changed every 3 d. After 6 d, 60 ng/ml recombinant human HGF was added to indicated wells, and branching morphogenesis was monitored by microscopy. MDCK cyst staining was performed as previously described (Elia and Lippincott-Schwartz, 2009), with the only modification being the substitution of 7 mg/ml fish-skin gelatin for Triton X-100 in permeabilization buffer. Cysts were stained with Alexa Fluor 488–phalloidin (Molecular Probes, Eugene, OR) and Hoescht 33258 (Molecular Probes). Representative 2-d post-HGF images were obtained using a Zeiss 710 Confocal with Airyscan, Apochromat 40×/1.2 W Autocorr M27, room temperature, AxioObserver.Z1 software, and diode 405-nm and argon 488-nm lasers. Images were processed in Photoshop (Adobe). Tubulogenesis was observed by day 2 after HGF addition and quantified on day 5 after HGF addition.
RNA-Seq Assay
MDCK+GRHL2/pMXS and MDCK+shGRHL2/Gipz cells were plated in collagen-coated, 60-mm2 dishes to obtained 25% confluency upon HGF treatment. Cells were treated with either 60 ng/ml HGF in DMEM complete or DMEM complete alone for 24 h; RNA was isolated from all four treatment conditions in triplicate using the RNeasy Plus Kit (Qiagen) and quantified using NanoDrop (Fisher, Waltham, MA). Raw data were aligned to the CanFam3 reference genome using TopHat2 (Kim et al., 2013). For each sample and each gene, the number of reads generated from that sample and mapping to that gene were counted using RSamtools. Genes were as defined in the ensembl database, version 82 (Cunningham et al., 2015). The resulting count table was analyzed using DESeq (Anders and Huber, 2010) with two independent comparisons: samples with HGF compared with those without or with GRHL2 overexpressed, and samples with HGF compared with those without, both expressing GRHL2 shRNA. Genes were considered differentially expressed if they showed a false discovery rate of ≤0.05 and a fold change of ≥1.5 in either direction. Genes regulated by HGF with shRNA against GRHL2 but not regulated by HGF with GRHL2 overexpressed were uploaded to Ingenuity Pathway Analysis (Qiagen), where a Core Analysis was performed using default settings.
Quantitative reverse transcription PCR analysis
MDCK parent, GRHL2/pMXS, and shGRHL2/pGipZ cells were plated on collagen-coated 60-mm2 dishes and treated as indicated. Cells were harvested using RNEasy Plus minikit (Qiagen, Hilden, Germany), analyzed using NanoDrop, and converted to cDNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) with oligo dT primers and 1 μg of RNA. cDNA was analyzed using SYBR Green PCR Master Mix on an Applied Biosystems 7500 Real Time PCR System. Values reported were determined using the ΔΔCT method using canine CBX1 as internal control. Primers were as follows: CBX1-F (AAGTATTGCAAGGCCACCCA), CBX-R (TTTTCCCAATCAGGCCCCAA), GRHL2-F (TTCCTCCCCAGGAGAGATGG), GRHL2-R (GGCCCAACACACGGATAAGA), MMP1-F (TGACTGGAAAGGTCGACACG), MMP1-R (GACCTTGGTGAAGGTCAGGG), MMP3-F (CTGACCCTAGCGCTCTGATG), MMP3-R (GTCCTGAGGGATTTTCGCCA), MMP13-F (GGACTTCCCAGGGATTGGTG), MMP13-R (ATGACACGGACAATCCGCTT), MMP14-F (CCCAGAGGAGGGCATGTTTT), MMP14-R (GACTCCCCACCCTTCCAATG), FOS-F (GGGAGGACCTTATCTGTGCG), FOS-R (ACACACTCCATGCGTTTTGC), FOSL1-F (ACACCCTCCCTGACTCCTTT), FOSL1-R (CTGAAGAGGGCATGGGATTA), FOSB-F (GGAGAAGAGAAGGGTTCGCC), FOSB-R (CACAAACTCCAGACGCTCCT), JUN-F (GACCTTCTACGACGATGCCC), JUN-R (TTCTTGGGGCACAGGAACTG), JUNB-F (GTGAAGACACTCAAGGCCGA), and JUNB-R (AGACTGGGGGAATGTCCGTA).
Luciferase reporter assays
HT1080 or 293 NOT cells were transiently transfected using Lipofectamine 2000 (Invitrogen) at 1 μg of DNA:3 μl of Lipofectamine. After 20 min of incubation in 300 μl of Opti-MEM (Invitrogen), 1.0 μg DNA was transfected into a 12-well plate with DMEM and refed 4 h later with DMEM. After 28–36 h posttransfection, the cells were washed twice with cold phosphate-buffered saline (PBS), lysed in 200 μl of 1× Cell Culture Lysis Buffer (Promega), frozen for at least 1 h at −80°C, thawed on ice, and centrifuged at 13,200 rpm. The supernatants were assayed for luciferase activity (Promega, Madison, WI) and β-galactosidase activity as internal control (2× β-galactosidase assay reagent, 200 mmol/l sodium phosphate, pH 7.3, 2 mmol/l MgCl2, 100 mmol/l 2-mercaptoethanol, and 1.33 mg/ml o-nitrophenylgalactoside). MMP1 promoter (−1200 to +60) and MMP14 promoter (−1114 to −242) luciferase reporter plasmids were generated by subcloning the promoter from genomic DNA using PCR into the pGL3-basic luciferase reporter plasmid. Primer sequences were MMP1prom-F (TTATTACTCGAGCCCTCCCTCTGATGCCTCTGA), MMP1prom-R (TATATAAAGCTTCTCCAATATCCCAGCTAGGAA), MMP14prom-f (TTATTACTCGAGCCCTTTCTCACGGTCAGCTT), and MMP14prom-R (TATATAAAGCTTTCTCGCTCTCTCCTCTGGTC). GRHL2/pcDNA3.1, TK-LacZ, and E1a/pCDNA3.1 plasmids were described previously (Grooteclaes and Frisch, 2000; Cieply et al., 2012). E1A p300 binding mutant was a generous gift from Arnold Berk (UCLA) subcloned into pCDNA3.1+. GRHL2 Δ425-437 was generated by cloning GRHL2 aa 1–424and 438–625 fragments and ligating together with Bglll and then ligating into pCDNA3.1+ using BamHI and Sal1. Primers used were GRHL2-1-424-F (TATATAGGATCCATGTCACAAGAGTCGGACAA), GRHL2-1-424-R (ATATAAAGATCTTTTTCTTTCTGCTCCTTTGT), GRHL2-438-625-F (TAAATTAGATCTAAAGGCCAGGCCTCCCAAAC), and GRHL2-438-625-R (TTATATGTCGACCTAGATTTCCATGAGCGTGA). pGL4.44[luc2P/AP1 RE/Hygro] (Promega) was used to assay for AP-1–responsive promoter activity, and control plasmid was pGL4.27 (Promega). Phorbol 12-myristate 13-acetate (PMA) induction (10 ng/ml; Thermo Fisher) was used for assaying AP-1 activity in transfected 293 cells. Full-length p300-GAL4/VR1012 and EV-GAL4/VR1012 were generous gifts from Neil Perkins (Newcastle University, Newcastle upon Tyne, United Kingdom); they were assayed on the GAL4 responsive reporter pG5-Luc (Promega). GAL4- p300 HAT (aa 1283–1674), GAL4- p300C-terminus (aa 1665–2414), and GAL4- p300 IBID (aa 2050–2096) were generated by subcloning PCR fragments (MluI-NotI) into pBIND (Promega). Primer sequences were p300 C-terminal-F (TATATAACGCGTATCGCTTTGTCTACACCTGCAAT), p300 C-terminal-R (TTTAAAGCGGCCGCCTAGTGTATGTCTAGTGTAC), p300 HAT-F (TATATAACGCGTATAGGAAAGAAAATAAGTTTTCT), p300 HAT-R (TTTAAAGCGGCCGCCTAGTCCTGGCTCTGCGTGTGCAG), p300 IBID-F (TATATAACGCGTATGCCTTACAAAACCTTTTGCG), and p300 IBID-R (TTTAAAGCGGCCGCCTAATTAGAGTTGGCATACTTGG). To generate the p300-IBID mutant (K2086A and K2091A), the p300 IBID lysine nucleotide sequences were mutated to alanine, and flanking Mlu1 and Not1 restriction enzymes were added to the 5′ and 3′ ends, respectively. The sequence was then generated as a gBlock gene fragment (IDT). The p300 mutant sequence was 5′ CACAACCAGG ATTGGGCCAG GTAGGTATCA GCCCACTCGC ACCAGGCACT GTGTCTCAAC AAGCCTTACA AAACCTTTTG CGGACTCTCA GGTCTCCCAG CTCTCCCCTG CAGCAGCAAC AGGTGCTTAG TATCCTTCAC GCCAACCCCC AGCTGTTGGC TGCATTCATC GCGCAGCGGG CTGCCGCGTA TGCCAACTCT AATCCACAAC 3′. The IBID mutant fragment was digested and ligated into pBIND vector. HA-SRC1/pACTAG was a generous gift from Timothy Beischlag (Simon Fraser University, Burnaby, Canada).
Immunoprecipitation
GRHL2-STAP contained full-length human GRHL2 fused to a C-terminal myc–streptavidin binding peptide (SBP)–S-tag sequence derived by ligating a synthetic oligonucleotide containing the S-tag to a PCR product containing the myc-SBP tags from the pCeMM-CTAP vector (Burckstummer et al., 2006). The 293 NOT cells were plated on collagen-coated six-well dishes and transfected using 1 μg of empty vector/pCDNA3.1+ or GRHL2-S-TAP/pCDNA3.1+ and 1 μg of empty vector/VR1012 or p300/VR1012) with 6 μl of Lipofectamine (Invitrogen) in 300 μl of Opti-MEM (Invitrogen). Cells were incubated for 4–6 h with DMEM and then refed with DMEM. At 28 h posttransfection, cells were lysed in 600 μl of cold p300 lysis buffer (25 mM Tris-HCl, pH 8, 75 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.1% NP-40, and protease inhibitor tablet [Roche, Basel, Switzerland]). Sample lysates were passed through a 1-ml syringe with a 27-gauge needle three times and precleared in a 4°C microcentrifuge at 13,200 rpm for 10 min. Total lysate samples were obtained, mixed 1:1 with 2× SDS lysis buffer (250 mM Tris-HCl, pH 6.8, 0.08% SDS, 0.048% bromophenol blue, 10% β-mercaptoethanol, 40% glycerol) and heated for 5 min at 95°C. Lysates were precipitated for 2 h with 40 μl of 50% S protein-agarose (EMD Millipore, Billerica, MA) that had been preequilibrated with p300 lysis buffer containing 10 mg/ml bovine serum albumin (BSA). Immunoprecipitation lysates were washed three times with p300 lysis buffer, diluted with 2× SDS lysis buffer, and heated for 5 min at 95°C. Lysates were processed using previously described Western blotting methods. For endogenous p300 and ectopic GRHL2 coimmunoprecipitation, MSP cells with empty vector/pMXS or GRHL2/pMXS were used in the experiment. For endogenous p300 and endogenous GRHL2 coimmunoprecipitation, parental HMLE cells were used in the experiment. The coimmunoprecipitation method was identical between the different cell lines. For each cell line, two 100-mm dishes were grown to 80% confluency, washed twice with cold PBS, lysed in 1 ml of p300 lysis buffer with protease inhibitors, and passed three times through a 27-gauge needle. After lysates were precleared at 13,200 rpm, 10 μg of p300 antibody (SCBT, SC-584) or rabbit immunoglobulin G (IgG; Jackson, Bar Harbor, ME) was added to lysates and incubated overnight on a 4°C wheel. Preequilibrated Protein A-Sepharose (45 μl; GE Healthcare) was added and incubated for 90 min. Samples were washed three times in p300 lysis buffer, 45 μl of 2× SDS lysis buffer was added, and samples were heated at 95°C for 5 min. Lysates were processed using previously described Western blotting methods.
Histone acetylation assays
Histone acetylation reactions contained indicated amounts of recombinant baculovirus Flag-tagged p300 (37.5 ng; Active Motif, Carlsbad, CA) or baculovirus p300 HAT (1283-1673; Sigma-Aldrich), 250 ng of H3 (Cayman), 100 μM acetyl-CoA (MP Biomedical, Santa Ana, CA), and indicated amounts of PreScission-cleaved GRHL2, GST-GRHL2, or GST alone in 30 μl of p300 acetylation buffer (50 mM Tris-HCl, pH 8.0, 1 mM dithiothreitol [DTT], 0.1 mM EDTA, 10% glycerol, and protease inhibitor). Acetylation reactions were incubated for 15 min at 30°C on heat block. Reactions were diluted with 2× SDS lysis buffer and incubated for 5 min at 95°C. Lysates were processed using previously described Western blotting methods.
Recombinant protein purification
Full-length GRHL2 was subcloned into pGEX-6P-3 vector (GE Healthcare) and transformed into BL21 bacteria (Invitrogen). GRHL2 fragments were subcloned into pGEX-6p-3 using BamH1 and Sal1. A colony was selected and grown in 20 ml of LB+amp overnight at 37°C. The next day, 10 ml of culture was inoculated into 500 ml of LB+amp and incubated for ∼2 h at 37°C with shaking until OD ≈ 0.6 was obtained. Isopropyl-β-d-thiogalactoside was added to final concentration of 0.1 mM and incubated overnight on a shaker at room temperature. The bacteria were spun in a chilled centrifuge for 10 min at 5000 rpm. Bacterial pellet was resuspended in 10 ml of BL21 lysis buffer (1× PBS, 1% Triton-100, 1 mM DTT, 0.4% lysozyme, and protease inhibitor tablet), incubated for 10 min, and sonicated twice for 30 s. The suspension was cleared at 13,000 rpm for 10 min. A 150-μl amount of 50% glutathione-Sepharose beads (GE Healthcare, Little Chalfont, United Kingdom) was added to the supernatant, which was incubated for 2 h with rotation at 4°C. Glutathione-Sepharose beads were washed three times with glutathione washing buffer (1× PBS, 1 mM DTT, and 0.1% Triton-X) and eluted for 30 min with glutathione elution buffer (100 mM Tris, pH 8, 150 mM NaCl, 20 mM glutathione, and protease inhibitors). Alternatively, glutathione-Sepharose beads with GRHL2 protein attached were washed three times in PreScission cleavage buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.0). The GST-GRHL2 on glutathione-Sepharose beads was cleaved by adding 80 μl of PreScission Protease (GE Healthcare) per milliliter of PreScission cleavage buffer at 5°C overnight. The sample was spun at 2000 rpm, and the supernatant with the GST-cleaved GRHL2 was obtained. Proteins were dialyzed against p300 acetylation buffer overnight at 4°C using a 20,000 MWCO Slide-A-Lyzer MINI Dialysis Unit (Thermo Scientific). Protein was analyzed on SDS–PAGE by Coomassie blue staining and quantified by comparison of Coomassie staining to a BSA standard and/or a bicinchoninic acid assay. Synthetic peptides were purchased from ThermoFisher Scientific with N-terminus acetyl group and C-terminus amidation: GRHL2 aa 420–442 (GAERKIRDEERKQNRKKGKGQAS) and scramble (IDEKNKGRERQRK)
Chromatin immunoprecipitation
For ChIP assay, HT1080 cell lines expressing empty vector/pMXS or GRHL2/pMXS cells were used and chromatin generated as described previously, with the following modifications (Kumar et al., 2011): 1) Immunoprecipitation was performed with 3 μg of H3K27-Ac antibody (Cell Signaling) or rabbit IgG (Jackson) to 3.3 × 106 cell equivalents in ChIP RIPA buffer. 2) Protein A-Sepharose was used instead of FLAG-agarose beads. 3) qPCR was performed using 2× SYBR Green Master Mix (10 μl; Applied Biosystems, Foster City, CA), 0.08 μl of 100 mM primer, and 1 μl of ChIP DNA in a 20-μl reaction. Primer sequences were as follows: MMP1-1-F (AACCTCAGAGAACCCCGAAG), MMP1-1-R (TACTAACACTGCGCACCTGA), MMP1-2-F (CAGAGTGGGGCATGAGTAGG), MMP1-2-R (TGTCTTCGGTACTGGTGACC), MMP14-F (AAGTAAGTGAGCTTCCCGGC), MMP14-R (GGAGTTCGCCCCAGTTGTAA), MMP2-F (TCGCCCATCATCAAGTTCCC), MMP2-R (CCCCCAAGCTGTTTACCGAA), ZEB1-1-F (GCGAGGCGTGGGACTGATGG), ZEB1-1-R (AAAGTTGGAGGCTCGGCGGC), ZEB1-2-F (CTGCACGGCGATGACCGCT), ZEB1-2-R (TTCCGCTTGCCAGCAGCCTC), CDH1-F (ACTCCAGGCTAGAGGGTCACC), CHD1-R (CCGCAAGCTCACAGCTTTGCAGTTCC), ESRP1-F (GGGAGCTTGGTCAAGTCAAC), ESRP1-R (TCTTAAATCGGGCCACGCAG), RAB25-F (AGGTCCTGTCCCTTTTTCGC), RAB25-R (TTGGGGGTAAGGGGACTTCT), GAPDH-F (ATGGTTGCCACTGGGGATCT), and GAPDH-R (TGCCAAAGCCTAGGGGAAGA). Results were normalized to GAPDH values.
Ex vivo kidney culture
All experiments involving mice were approved by the West Virginia University Institutional Animal Care and Use Committee. Hoxb7creEGFP transgenic mice and the ex vivo kidney culture method were described previously (Zhao et al., 2004). We removed embryonic kidney at embryonic day 13.5 and cultured the kidneys in 1 μM batimastat (Tocris, Bristol, United Kingdom) or vector in culture medium for 48 h. Images were taken of GFP ureteric buds on an Olympus MVX10 Macro Zoom, ORCA-Flash 4.0 Monochrome camera, objective 2×, zoom 4×, fluorescein isothiocyanate, and using CellSen Imaging Software. Ureteric buds were counted for quantification.
Confocal microscopy of transfected RRE.1 cells
RRE.1 cells, wild-type e1a-NLS-LacI-mCherry, and empty vector NLS-LacI-mCherry were generous gifts from Arnold Berk, and the experiment was performed as previously described (Verschure et al., 2005; Ferrari et al., 2014). GRHL2 was subcloned into empty vector NLS-LacI-mCherry using Sal1. Images were taken on a Zeiss Axioimager Z1 microscope, LSM 510 confocal, 63×/0.75 LDPlan-neofluar, 4′,6-diamidino-2-phenylindole and rhodamine, and Zeiss Zen software. Images were quantified using mean area on ImageJ (National Institutes of Health, Bethesda, MD).
Statistical analysis
Error bars in graphs represent SD. p values were calculated using a Student’s two-tailed t test.
Supplementary Material
Acknowledgments
We thank Neil Perkins (University of Newcastle, Newcastle upon Tyne, United Kingdom), Arnold Berk (UCLA, Los Angeles, CA), and Stephen Jane (Monash University, Melbourne, Australia) for constructs and technical advice, Kathy Brundage for flow cytometry, Karen Martin and Amanda Ammer for imaging expertise, Ryan Percifield for assistance in RNA library construction, Neil Infante for bioinformatics support, Peter Mathers, Sarah McLaughlin, and Emily Ellis for animal management, Paolo Fagone for protein preparation, Carlton Bates and Kenneth Walker for kidney development techniques, and Mary Davis for Ingenuity Pathway Analysis. The work was supported by grants from the Mary Kay Foundation and the National Institute of General Medical Sciences (U54GM104942). J.D. was supported in part by the West Virginia IDeA Network for Biomedical Research Excellence (National Institutes of Health/National Institute of General Medical Sciences Grant P20GM103434). National Institutes of Health Grants GM103488/RR032138, RR020866, OD016165, and GM103434 support the flow cytometry facility. Small-animal imaging and image analysis were performed in the West Virginia University Animal Models and Imaging Facility, which is supported by the Mary Babb Randolph Cancer Center and National Institutes of Health Grants P20 RR016440, P30 GM103488, and S10 RR026378. Imaging experiments and image analysis were performed in the West Virginia University Microscope Imaging Facility, which is supported by the Mary Babb Randolph Cancer Center and National Institutes of Health Grants P20 RR016440, P30 GM103488, and P20 GM103434.
Abbreviations used:
- acetyl-CoA
acetyl-coenzyme A
- AP-1
activator protein 1
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- E1a
adenovirus early region 1A protein
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GLUD1
glutamate dehydrogenase 1
- GRHL1
Grainyhead-like-1
- GRHL2
Grainyhead-like-2
- GRHL3
Grainyhead-like-3
- H3
histone 3
- HAT
histone acetyltransferase domain
- HGF
hepatocyte growth factor
- IBID
IRF-3 binding domain
- MDCK
Madin–Darby canine kidney
- MET
mesenchymal-epithelial transition
- miMCD
mouse inner medullary collecting duct
- MMP
matrix metalloprotease
- MSP
mesenchymal subpopulation
- p300
E1A binding protein p300
- PBS
phosphate-buffered saline
- PMA
phorbol 12-myristate 13-acetate
- PSG
penicillin-streptomycin-glutamine
- RNA-Seq
RNA-sequencing
- SBP
streptavidin binding peptide
- shRNA
short hairpin RNA
- SRC-1
steroid receptor coactivator-1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0249) on June 1, 2016.
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Aue A, Hinze C, Walentin K, Ruffert J, Yurtdas Y, Werth M, Chen W, Rabien A, Kilic E, Schulzke J-D, et al. A Grainyhead-like 2/Ovo-like 2 pathway regulates renal epithelial barrier function and lumen expansion. J Am Soc Nephrol. 2015;26:2704–2715. doi: 10.1681/ASN.2014080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båvner A, Matthews J, Sanyal S, Gustafsson J-Å, Treuter E. EID3 is a novel EID family member and an inhibitor of CBP-dependent co-activation. Nucleic Acids Res. 2005;33:3561–3569. doi: 10.1093/nar/gki667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow U, Brinckerhoff CE. The AP-1 site and MMP gene regulation: what is all the fuss about. Matrix Biol. 1997;15:519–526. doi: 10.1016/s0945-053x(97)90026-3. [DOI] [PubMed] [Google Scholar]
- Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- Boglev Y, Wilanowski T, Caddy J, Parekh V, Auden A, Darido C, Hislop NR, Cangkrama M, Ting SB, Jane SM. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev Biol. 2011;349:512–522. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Bridgewater D, Cox B, Cain J, Lau A, Athaide V, Gill PS, Kuure S, Sainio K, Rosenblum ND. Canonical WNT/β-catenin signaling is required for ureteric branching. Dev Biol. 2008;317:83–94. doi: 10.1016/j.ydbio.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Chacon-Heszele MF, Zuo X, Hellman NE, McKenna S, Choi SY, Huang L, Tobias JW, Park KM, Lipschutz JH. Novel MAPK-dependent and -independent tubulogenes identified via microarray analysis of 3D-cultured Madin-Darby canine kidney cells. Am J Physiol Renal Physiol. 2014;306:F1047–F1058. doi: 10.1152/ajprenal.00589.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ghazawi FM, Li Q. Interplay of bromodomain and histone acetylation in the regulation of p300-dependent genes. Epigenetics. 2010;5:509–515. doi: 10.4161/epi.5.6.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ, Lee CH, Oh SH, Song YS, Kim HS, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun. 2015;6:7821. doi: 10.1038/ncomms8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YT, Wang H, Chen Y, Danielpour D, Yang YC. Cited2 modulates TGF-[beta]-mediated upregulation of MMP9. Oncogene. 2006;25:5547–5560. doi: 10.1038/sj.onc.1209552. [DOI] [PubMed] [Google Scholar]
- Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 2013;73:6299–6309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B, Riley Pt, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 2012;72:2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IM, Swingler TE, Sampieri CL, Edwards DR. The regulation of matrix metalloproteinases and their inhibitors. Int J Biochem Cell Biol. 2008;40:1362–1378. doi: 10.1016/j.biocel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- Elia N, Lippincott-Schwartz J. Culturing MDCK cells in three dimensions for analyzing intracellular dynamics. 2009. Curr Protoc Cell Biol Chapter 4, Unit 4.22. [DOI] [PMC free article] [PubMed]
- Farris JC, Pifer PM, Zheng L, Gottlieb E, Denvir J, Frisch SM. Grainyhead-like 2 reverses the metabolic changes induced by the oncogenic epithelial-mesenchymal transition: effects on anoikis. Mol Cancer Res. 2016;14:528–538. doi: 10.1158/1541-7786.MCR-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK, et al. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe. 2014;16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming T, McConnell J, Johnson MH, Stevenson BR. Development of tight junctions de novo in the mouse early embryo: control of assembly of the tight junction-specific protein, ZO-1. J Cell Biol. 1989;108:1407–1418. doi: 10.1083/jcb.108.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM. The epithelial cell default-phenotype hypothesis and its implications for cancer. Bioessays. 1997;19:705–709. doi: 10.1002/bies.950190811. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Mymryk JS. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH, Reddy TE, Hogan BL. Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc Natl Acad Sci USA. 2013;110:9356–9361. doi: 10.1073/pnas.1307589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Janknecht R. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 2003;23:6243–6254. doi: 10.1128/MCB.23.17.6243-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gustavsson P, Copp AJ, Greene ND. Grainyhead genes and mammalian neural tube closure. Birth Defects Res A Clin Mol Teratol. 2008;82:728–735. doi: 10.1002/bdra.20494. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman NE, Spector J, Robinson J, Zuo X, Saunier S, Antignac C, Tobias JW, Lipschutz JH. Matrix metalloproteinase 13 (MMP13) and tissue inhibitor of matrix metalloproteinase 1 (TIMP1), regulated by the MAPK pathway, are both necessary for Madin-Darby canine kidney tubulogenesis. J Biol Chem. 2008;283:4272–4282. doi: 10.1074/jbc.M708027200. [DOI] [PubMed] [Google Scholar]
- Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyafil F, Morello D, Babinet C, Jacob F. A cell surface glycoprotein involved in the compaction of embryonal carcinoma cells and cleavage stage embryos. Cell. 1980;21:927–934. doi: 10.1016/0092-8674(80)90456-0. [DOI] [PubMed] [Google Scholar]
- Ishibe S, Karihaloo A, Ma H, Zhang J, Marlier A, Mitobe M, Togawa A, Schmitt R, Czyczk J, Kashgarian M, et al. Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development. 2009;136:337–345. doi: 10.1242/dev.024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Lai C-H, Zhao X, Saito Si, Hamilton MH, Appella E, Yao T-P. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, Williams J, Hauser C, Kurkinen M, Dhar R, et al. p300/cAMP-responsive element-binding protein interactions with Ets-1 and Ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem. 1999;274:17342–17352. doi: 10.1074/jbc.274.24.17342. [DOI] [PubMed] [Google Scholar]
- Ji A, Dao D, Chen J, MacLellan WR. EID-2, a novel member of the EID family of p300-binding proteins inhibits transactivation by MyoD. Gene. 2003;318:35–43. doi: 10.1016/j.gene.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, Wang C, Brindle PK, Dent SYR, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda M, Olmeda D, Vinyals A, Valero E, Cubillo E, Llorens A, Cano A, Fabra A. Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J Cell Sci. 2005;118:3371–3385. doi: 10.1242/jcs.02465. [DOI] [PubMed] [Google Scholar]
- Jung YS, Liu X-W, Chirco R, Warner RB, Fridman R, Kim H-RC. TIMP-1 Induces an EMT-like phenotypic conversion in MDCK cells independent of its MMP-inhibitory domain. PLoS One. 2012;7:e38773. doi: 10.1371/journal.pone.0038773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman RA, Rose DW, Glass CK, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn KW, Zeeberg BM, Reinhold WC, Pommier Y. Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype. PLoS One. 2014;9:e99269. doi: 10.1371/journal.pone.0099269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoszynska K, Ostrowski J, Rychlewski L, Wyrwicz LS. The fold recognition of CP2 transcription factors gives new insights into the function and evolution of tumor suppressor protein p53. Cell Cycle. 2008;7:2907–2915. doi: 10.4161/cc.7.18.6680. [DOI] [PubMed] [Google Scholar]
- Kumar S, Park SH, Cieply B, Schupp J, Killiam E, Zhang F, Rimm DL, Frisch SM. A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol Cell Biol. 2011;31:4036–4051. doi: 10.1128/MCB.01342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, DeVos SL, Klomp JE, Triezenberg SJ. Transcriptional coactivators are not required for herpes simplex virus type 1 immediate-early gene expression in vitro. J Virol. 2009;83:3436–3449. doi: 10.1128/JVI.02349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Partridge NC. Parathyroid hormone activation of matrix metalloproteinase-13 transcription requires the histone acetyltransferase activity of p300 and PCAF and p300-dependent acetylation of PCAF. J Biol Chem. 2010;285:38014–38022. doi: 10.1074/jbc.M110.142141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy P, Mostov KE. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Mol Biol Cell. 2007;18:1943–1952. doi: 10.1091/mbc.E06-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 2007;26:2284–2294. doi: 10.1038/sj.emboj.7601667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li W. Epithelial–mesenchymal transition in human cancer: comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Lin CH, Hare BJ, Wagner G, Harrison SC, Maniatis T, Fraenkel E. A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies. Mol Cell. 2001;8:581–590. doi: 10.1016/s1097-2765(01)00333-1. [DOI] [PubMed] [Google Scholar]
- Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, Arber S, Hassell J, MacNeil L, Hoshi M, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Guo M, Shan B, Xia Z. Targeted functional analysis of p300 coactivator in Wnt/β-catenin signaling pathway using phosphoproteomic and biochemical approaches. J Proteomics. 2012;75:2601–2610. doi: 10.1016/j.jprot.2012.03.009. [DOI] [PubMed] [Google Scholar]
- MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A Novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Harries JC, Viskaduraki M, Troke PJF, Kindle KB, Ryan C, Heery DM. A conserved α-helical motif mediates the binding of diverse nuclear proteins to the SRC1 interaction domain of CBP. J Biol Chem. 2004;279:14055–14064. doi: 10.1074/jbc.M310188200. [DOI] [PubMed] [Google Scholar]
- Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Curr Cancer Drug Targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- Miller RK, McCrea PD. Wnt to build a tube: contributions of Wnt signaling to epithelial tubulogenesis. Dev Dyn. 2010;239:77–93. doi: 10.1002/dvdy.22059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Yanagisawa Y, Yuasa Y. A novel EID-1 family member, EID-2, associates with histone deacetylases and inhibits muscle differentiation. J Biol Chem. 2003;278:17060–17065. doi: 10.1074/jbc.M212212200. [DOI] [PubMed] [Google Scholar]
- Mizzen AC, Allis DC. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlacki M, Kikulska A, Krzywinska E, Pawlak M, Wilanowski T. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp Biol Med (Maywood) 2015;240:1396–1401. doi: 10.1177/1535370215588924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monga SPS, Mars WM, Pediaditakis P, Bell A, Mulé K, Bowen WC, Wang X, Zarnegar R, Michalopoulos GK. Hepatocyte growth factor induces Wnt-independent nuclear translocation of β-catenin after Met-β-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064–2071. [PubMed] [Google Scholar]
- Narayanan K, Srinivas R, Peterson MC, Ramachandran A, Hao J, Thimmapaya B, Scherer PE, George A. Transcriptional regulation of dentin matrix protein 1 by JunB and p300 during osteoblast differentiation. J Biol Chem. 2004;279:44294–44302. doi: 10.1074/jbc.M403511200. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Nishibashi K, Itoh Y, Yana I, Seiki M. Competitive disruption of the tumor-promoting function of membrane type 1 matrix metalloproteinase/matrix metalloproteinase-14 in vivo. Mol Cancer Ther. 2005;4:1157–1166. doi: 10.1158/1535-7163.MCT-05-0127. [DOI] [PubMed] [Google Scholar]
- O’Brien LE, Zegers MMP, Mostov KE. Building epithelial architecture: insights from three-dimensional culture models. Nat Rev Mol Cell Biol. 2002;3:531–537. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K-I, Suda T. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood. 1999;93:2771–2779. [PubMed] [Google Scholar]
- Polesskaya A, Naguibneva I, Fritsch L, Duquet A, Ait-Si-Ali S, Robin P, Vervisch A, Pritchard LL, Cole P, Harel-Bellan A. CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J. 2001;20:6816–6825. doi: 10.1093/emboj/20.23.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack AL, Apodaca G, Mostov KE. Hepatocyte growth factor induces MDCK cell morphogenesis without causing loss of tight junction functional integrity. Am J Physiol Cell Physiol. 2004;286:C482-C494. doi: 10.1152/ajpcell.00377.2003. [DOI] [PubMed] [Google Scholar]
- Pollack AL, Runyan RB, Mostov KE. Morphogenetic mechanisms of epithelial tubulogenesis: MDCK cell polarity is transiently rearranged without loss of cell–cell contact during scatter factor/hepatocyte growth factor-induced tubulogenesis. Dev Biol. 1998;204:64–79. doi: 10.1006/dbio.1998.9091. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang X-J, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JYJ, Graessmann A, Nakatani Y, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–3827. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y, Parekh V, Wilanowski T, Hislop NR, Auden A, Ting SB, Cunningham JM, Jane SM. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev Biol. 2010;345:237–245. doi: 10.1016/j.ydbio.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Ringel AE, Wolberger C. A new RING tossed into an old HAT. Structure. 2013;21:1479–1481. doi: 10.1016/j.str.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer FR, Hoschele PP, Oh SJ, Erb HH, Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA, Culig Z. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther. 2011;10:1644–1655. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott KM, Yang J, Chen X, Wang H, Paragas N, Mori K, Li J-Y, Lu B, Costantini F, Schiffer M, et al. Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol. 2005;16:1993–2002. doi: 10.1681/ASN.2004121127. [DOI] [PubMed] [Google Scholar]
- Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell. 2012;23:2845–2855. doi: 10.1091/mbc.E12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard HM, Harries JC, Hussain S, Bevan C, Heery DM. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol Cell Biol. 2001;21:39–50. doi: 10.1128/MCB.21.1.39-50.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden AW, Anderson LA, Webster GA, Perkins ND. A novel transcriptional repression domain mediates p21WAF1/CIP1 induction of p300 transactivation. Mol Cell Biol. 2000;20:2676–2686. doi: 10.1128/mcb.20.8.2676-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiehl DP, Fath DM, Liang D, Jiang Y, Sang N. Histone deacetylase inhibitors synergize p300 autoacetylation that regulates its transactivation activity and complex formation. Cancer Res. 2007;67:2256–2264. doi: 10.1158/0008-5472.CAN-06-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zeng X-R, Wenger L, Firestein GS, Cheung HS. p53 down-regulates matrix metalloproteinase-1 by targeting the communications between AP-1 and the basal transcription complex. J Cell Biochem. 2004;92:258–269. doi: 10.1002/jcb.20044. [DOI] [PubMed] [Google Scholar]
- Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Hemler ME. Caveolin-1 regulates matrix metalloproteinases-1 induction and CD147/EMMPRIN cell surface clustering. J Biol Chem. 2004;279:11112–11118. doi: 10.1074/jbc.M312947200. [DOI] [PubMed] [Google Scholar]
- Tang Z, Chen W-Y, Shimada M, Nguyen, Uyen TT, Kim J, Sun X-J, Sengoku T, McGinty, Robert K, et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Mitaka T. Role of grainyhead-like 2 in the formation of functional tight junctions. Tissue Barriers. 2013;1:e23495. doi: 10.4161/tisb.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo J-L, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: a tale of two coactivators. Adv Drug Deliv Rev. 2010;62:1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003;370:953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D, Kemler R. Identification of a putative cell adhesion domain of uvomorulin. EMBO J. 1985;4:3393–3398. doi: 10.1002/j.1460-2075.1985.tb04095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Walentin K, Hinze C, Werth M, Haase N, Varma S, Morell R, Aue A, Pötschke E, Warburton D, Qiu A, et al. A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development. 2015;142:1125–1136. doi: 10.1242/dev.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AZ, Ojakian GK, Nelson WJ. Steps in the morphogenesis of a polarized epithelium. I. Uncoupling the roles of cell-cell and cell-substratum contact in establishing plasma membrane polarity in multicellular epithelial (MDCK) cysts. J Cell Sci. 1990;95:137–151. doi: 10.1242/jcs.95.1.137. [DOI] [PubMed] [Google Scholar]
- Wang S, Samakovlis C. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr Top Dev Biol. 2012;98:35–63. doi: 10.1016/B978-0-12-386499-4.00002-1. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Villarreal-Ponce A, Sun P, Salmans, Michael L, Fallahi M, Andersen B, Dai X. Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev Cell. 2014;29:59–74. doi: 10.1016/j.devcel.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth M, Walentin K, Aue A, Schönheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M, et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137:3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- Westermarck J, Kähäri V-M. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781–792. [PubMed] [Google Scholar]
- Wilanowski T, Tuckfield A, Cerruti L, O’Connell S, Saint R, Parekh V, Tao J, Cunningham JM, Jane SM. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- Witte S, Bradley A, Enright AJ, Muljo SA. High-density P300 enhancers control cell state transitions. BMC Genomics. 2015;16:1–13. doi: 10.1186/s12864-015-1905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of β-catenin by CREB-binding protein (CBP) J Biol Chem. 2002;277:25562–25567. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- Wollenick K, Hu J, Kristiansen G, Schraml P, Rehrauer H, Berchner-Pfannschmidt U, Fandrey J, Wenger RH, Stiehl DP. Synthetic transactivation screening reveals ETV4 as broad coactivator of hypoxia-inducible factor signaling. Nucleic Acids Res. 2012;40:1928–1943. doi: 10.1093/nar/gkr978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hu B, Qin L, Zhao L, Wang Q, Wang Q, Xu Y, Jiang J. SRC-1 and Twist1 expression positively correlates with a poor prognosis in human breast cancer. Int J Biol Sci. 2014;10:396–403. doi: 10.7150/ijbs.8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Pinello CE, Luo J, Li D, Wang Y, Zhao LY, Jahn SC, Saldanha SA, Chase P, Planck J, Geary KR, et al. Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents. Mol Cancer Ther. 2013;12:610–620. doi: 10.1158/1535-7163.MCT-12-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T-P, Oh SP, Fuchs M, Zhou N-D, Ch’ng L-E, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 2013;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Zhang L, He X, Liu L, Jiang M, Zhao C, Wang H, He D, Zheng T, Zhou X, Hassan A, et al. Hdac3 Interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev Cell. 2016;36:316–330. doi: 10.1016/j.devcel.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan W, Chen X. P63 regulates tubular formation via epithelial-to-mesenchymal transition. Oncogene. 2014;33:1548–1557. doi: 10.1038/onc.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kegg H, Grady S, Truong H-T, Robinson ML, Baum M, Bates CM. Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol. 2004;276:403–415. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Liu Y, Kahn M, Ann DK, Han A, Wang H, Nguyen C, Flodby P, Zhong Q, Krishnaveni MS, et al. Interactions between beta-catenin and transforming growth factor-beta signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP) J Biol Chem. 2012;287:7026–7038. doi: 10.1074/jbc.M111.276311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.