Abstract
TRBP, one of double strand RNA binding proteins (dsRBPs), is an essential cofactor of Dicer in the RNA interference pathway. Previously we reported that TRBP exhibits repetitive diffusion activity on double strand (ds)RNA in an ATP independent manner. In the TRBP-Dicer complex, the diffusion mobility of TRBP facilitates Dicer-mediated RNA cleavage. Such repetitive diffusion of dsRBPs on a nucleic acid at the nanometer scale can be appropriately captured by several single molecule detection techniques. Here, we provide a step-by-step guide to four different single molecule fluorescence assays by which the diffusion activity of dsRBPs on dsRNA can be detected. One color assay, termed protein induced fluorescence enhancement enables detection of unlabeled protein binding and diffusion on a singly labeled RNA. Two-color Fluorescence Resonance Energy Transfer (FRET) in which labeled dsRBPs is applied to labeled RNA, allows for probing the motion of protein along the RNA axis. Three color FRET reports on the diffusion movement of dsRBPs from one to the other end of RNA. The single molecule pull down assay provides an opportunity to collect dsRBPs from mammalian cells and examine the protein-RNA interaction at single molecule platform.
Keywords: double-strand RNA binding protein (dsRBP), TAR RNA binding protein (TRBP), single molecule PIFE (smPIFE), single molecule FRET (smFRET), three color FRET, single molecule pull down (SiMPull), diffusion activity of dsRBP
I. INTRODUCTION
RNA molecules in cells are primarily in the form of single strand (ss) because they are transcribed from the DNA template in this format. The ssRNA, however, often forms secondary structures that encompass segments of double stranded (ds) regions. Some of these secondary structures play a critical role for their biological function. For example, the secondary structure of tRNA and micro RNA (miRNA) are recognized specifically by their host, ribosome and Dicer/TRBP for successful processes of translation and RNA interference, respectively (1,2). Accordingly, a class of proteins named as dsRNA binding proteins (dsRBPs) is responsible for the diverse biological function of dsRNAs by recognizing dsRNAs, which consists of one or more dsRNA binding domains (dsRBDs)(3). Therefore, it is critical to investigate the interaction between dsRNA and dsRBPs, which could unveil the mechanistic role of dsRBPs in regard to their diverse function.
The interaction between dsRNA and dsRBPs has been extensively studied using traditional ensemble methods such as gel electrophoresis or X-ray crystallography (4,5). However, the detection was limited to providing static interaction, but not the dynamic interaction in protein-RNA interaction because of the temporal and spatial averaging inherent to ensemble measurement. Recently, we employed single-molecule approaches to study the interaction between dsRNA and dsRBPs, which unveiled the diffusive motion of dsRBPs on dsRNA in ATP-independent manner (6).
TAR RNA binding protein (TRBP)(7) belongs to dsRBPs and works with Dicer in facilitating pre-miRNA cleavage (8). Our previous work has demonstrated that TRBP diffuses along dsRNA axis repetitively (6), which was probed by single molecule fluorescence in three ways (6). First, singly-labeled (Cy3) TRBP applied to Cy5-labeled dsRNA exhibited fluctuation of Fluorescence Resonance Energy Transfer (FRET) (9), enabling visualization of the repetitive movement of the protein on RNA. This motion is not powered by ATP hydrolysis as it occurs in the absence of ATP. We observed the same FRET fluctuation regardless of the labeling position, indicating that the observed fluctuation is due to diffusion of TRBP, not conformational change of its subdomains. The distance range and kinetics of such movement can be deduced by performing autocorrelation analysis on FRET signal. Using this analysis scheme, we determined that TRBP diffusion is length dependent between 25 and 55 bp dsRNA substrates. Second, we employed protein induced fluorescence enhancement (PIFE) assay (10,11) in which unlabeled TRBP was added to Cy3-labeled dsRNA. Here, we obtained fluctuating Cy3 signal, indicating a repetitive contact between the protein and Cy3 dye positioned at one end of dsRNA. This is consistent with the interpretation that TRBP diffuses on dsRNA repetitively. This demonstrates how single-molecule PIFE (smPIFE) can be a powerful alternative of FRET, especially when protein labeling is difficult. Third, we took advantage of the three-color FRET experiment (12) where two acceptors (Cy5 and Cy7) were positioned at either end of dsRNA substrate and TRBP was labeled with a donor dye, Cy3. This is the most direct way to show that the TRBP protein traverses from one end to the other end of the RNA. The two FRET pairs, Cy3-Cy5 and Cy3-Cy7 were time separated from each other due to the physical separation between the two acceptors (Cy5 and Cy7). Here, we present a detailed protocol on these three single-molecule methods of detecting the dynamics of TRBP-dsRNA interaction.
The single-molecule pull down technique (SiMPull) provides a unique advantage of isolating proteins directly from cell lysate (13). TRBP was overexpressed with a fluorescence protein tag such as EGFP or EYFP in mammalian cells, enabling capture of cellular TRBP on single molecule interface, via interaction between the fluorescence tag and the antibody against the tag. The pull-down efficiency can be determined by anti-TRBP antibody which is commercially available. Since the protein, rather than RNA is immobilized on surface, this represents a reversed geometry from the other three single molecule assays described above. The significant advantage of the SiMPull technique is that the proteins are isolated directly from cell lysate, thus better representing the physiologically relevant species. In addition, this platform is ideal for comparing substrate affinity, since varying concentration of substrates can be added to a fixed concentration of protein bound to surface. We chose to apply six different structured RNA to test the relative affinity toward TRBP, as well as three other dsRBPs, ADAD2, ADAR1 and Staufen1 (14). The SiMPull platform was further used to test whether RNA binding to dsRBPs is static or dynamic in nature. By applying Cy3-labeled RNA to surface tethered TRBP, we observed robust behavior of diffusion, as also seen in the first three single molecule assays described above. It is noteworthy that we observed similar dynamics for both purified and cellular TRBP, suggesting that the diffusion activity likely represents the protein’s behavior in cells.
In this article, we offer a comprehensive review on the single-molecule fluorescence assays including PIFE, 2-color FRET, 3-color FRET, as well as SiMPull assay that may be employed for investigating interaction between dsRBPs and dsRNAs. We envision that combination of these single-molecule techniques will shape a powerful approach to monitor protein-RNA or protein-DNA interaction at high spatial and temporal resolution. This in turn will lead to discovery of novel features of protein-RNA interaction as demonstrated by ATP-independent diffusion dynamics of dsRBPs on dsRNAs.
II. SINGLE MOLECULE IMAGING OF RECONSTITUED DSRBPS
1. SAMPLE PREPARATION
1.1. PROTEIN LABELING
For the three single-molecule fluorescence assays, the labeling of nucleic acids or proteins of interest with a suitable fluorophore is an essential step of sample preparation. Fluorophore labeling of nucleic acids is routinely achieved by amino modification at the target labeling position of the nucleic acid (either 5′-, 3′-, or at an internal position of the nucleic acid), followed by coupling them with amine-reactive fluorophore such as NHS esters. However, fluorophore labeling of proteins is far more challenging. For protein labeling, we can either use the reactive chemical groups in the amino acid residues of the protein of interest; or engineer protein tags or fluorescent proteins in the protein (15). To utilize the former approach, there are two widely-used reactive chemical groups in amino acids, one is primary amine (-NH2) of lysine residues or the N-terminus of the protein; and the other is sulfhydryl group (-SH) of cysteine residues. Because proteins likely possess multiple -NH2 and -SH groups, achieving specificity in labeling of proteins using these chemical groups is difficult. However, this non-specific labeling strategy is sometimes advantageous, particularly when the goal is to observe the overall movement of the entire protein on a substrate, for example rather than conformational dynamics of a specific domain. We will describe here the procedure to achieve non-specific labeling of dsRBPs, with which we observed the movement of dsRBPs on dsRNAs.
The amino group of lysine or the N-terminal amino group has been widely used for protein labeling via acylation reaction with NHS esters. We chose a fluorophore containing NHS ester group for non-specific labeling of dsRBPs, such as Cy3 or Cy5-NHS ester (GE Healthcare). The acylation reaction between amino group and NHS ester is very robust, but since the number of amino groups varies among different proteins, an optimization of the labeling conditions is necessary. Here, we describe the detailed labeling procedure as well as how to optimize experimental conditions for non-specific protein labeling.
Step 1. Make 1 M NaHCO3 buffer by adding 840 mg in 10 ml of milli-Q water. For best results, make it fresh for every labeling reaction. Adjust to pH8.5.
Step 2. Mix 25 ug of dsRBP with 1ul of of Cy3 NHS ester or Alexa 647 NHS ester (2ug/ul) and 3 ul of 1 M NaHCO3, pH 8.5 buffer. Add milli-Q water to make up volume to 30 ul. Incubate the mixture for 30 min at room temperature.
Step 3. Remove excess dye by passing the mix through a Biorad P-6 spin column. If bottom of the P-6 column is stained with excess dye after the first run, then run another P-6 spin column.
Step 4. Calculate the ratio of fluorophore to protein by measuring the absorbance at 280 nm for dsRBPs and the absorption peak wavelength of the fluorophore using a spectrophotometer.
Step 5. The goal is to achieve a ratio as close to 1 as possible. If the ratio is not close to 1, adjust the amount of fluorophore in Step2 and then repeat the following steps.
Step 6. Aliquot the labeled dsRBP and store at −80°C for later use.
1.2. IMMOBILIZATION OF DSRNA
In order to monitor the dynamic interaction between dsRBPs and dsRNAs at single-molecule level, we immobilized dsRNAs onto a poly ethylene glycol (PEG)-coated quartz slide through neutravidin-biotin interaction. PEG-coated surface was prepared to reduce the non-specific binding of dsRBPs and dsRNAs to the imaging surface. Neutravidin (0.05 mg/ml) was incubated for 5 mins with the PEG-coated slide so that it attaches to the 1–2% of biotinlyated PEG on the PEG-coated quartz surface. This was followed by addition of 30–50 pM of biotinlyated dsRNA in T50-BSA buffer (10mM Tris, pH 8, 50mM NaCl, and 0.1mg/ml BSA), finally achieving RNA immobilization via neutravidin-biotin linkage. After 5 min of incubation with the biotinylated dsRNA, the excess RNA was washed away. 10–100 nM of dsRBP was then added to the immobilized dsRNA to monitor the interaction between two.
1.3. OXYGEN SCAVENGER SYSTEM OPTIMIZED FOR RNA IMAGING
Because in single-molecule fluorescence assays of dsRBPs, we observe a single fluorophore on a time scale that is different from ensemble fluorescence assays, it is critical to stabilize the fluorophores so that we are able to observe the fluorescence signal long enough to monitor the interaction between dsRBPs and dsRNAs. Oxygen is known to be an efficient quencher of fluorophores (16), which makes an oxygen scavenger system inevitable for single-molecule fluorescence assays. We employed a glucose oxidase-based oxygen scavenger system (17), that [0.5 w/v % glucose, 100 mg/ml glucose oxidase (Sigma), and 180 U/ml catalase (Calbiochem)], together with 5–10 mM trolox (Sigma) to stabilize the fluorophores during single-molecule data acquisition. It is noteworthy that most commercial catalases are contaminated with RNase that induces degradation of the dsRNA. Therefore, it is important to use minimal amount of catalase for these dsRNA-related experiments. The interaction between dsRBPs and dsRNAs was monitored in 20 mM Tris, pH 7.5, 25 mM NaCl, 1mM DTT, and 0.1 mg/ml BSA in combination with the oxygen scavenger system and trolox (18).
2. SINGLE-MOLECULE FLUORESCENCE ASSAYS
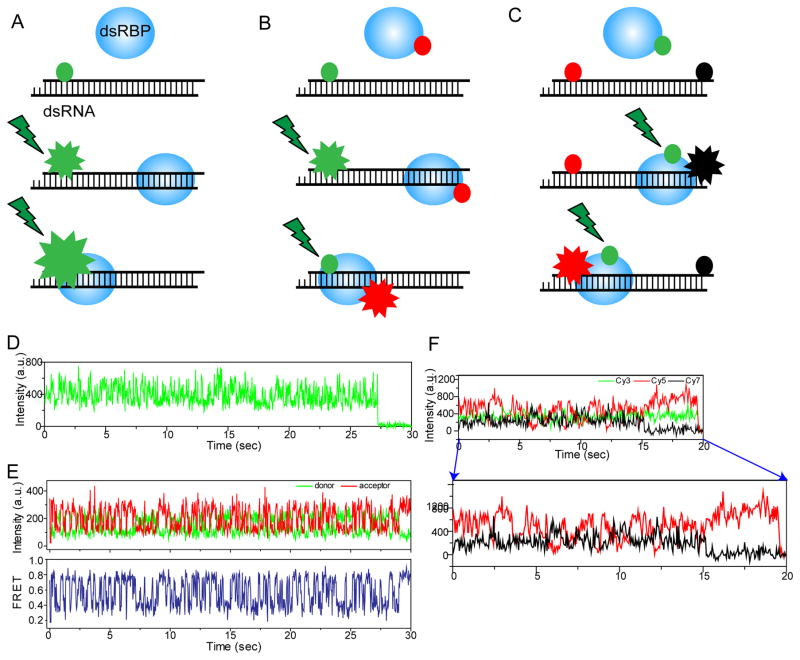
To achieve single-molecule detection sensitivity, we harness the shallow excitation depth (~100 nm) of total internal reflection (TIR) excitation (19), that reduces background signal significantly, thus enabling detection of single fluorophores. We used a custom-built prism-type TIR fluorescence system for studying the interaction between dsRBPs and dsRNAs at a nanometer spatial and sub-second temporal resolution. Here, we describe how we can study the diffusion activity of dsRBPs on dsRNA using 1-color, 2-color and 3-color fluorescence assay (Fig. 1A–C).
Figure 1. Single-molecule fluorescence assays for monitoring the diffusion of dsRBPs on dsRNA.
A–C) Simplistic schemes to show the working principles of 1-color, 2-color and 3-color single-molecule fluorescence assays for studying the diffusion of dsRBPs on dsRNA. D) 1-color fluorescence assay using smPIFE shows the fluctuation of Cy3 intensity induced by the diffusion motion of TRBP on dsRNA. E) 2-color fluorescence assay using smFRET shows the distance change between TRBP and dsRNA due to the diffusion motion of TRBP. F) 3-color fluorescence assay using 3-color smFRET shows the anti-correlated intensity change between two acceptors located at either end of dsRNA due to the diffusion of TRBP on dsRNA.
2.1. 1-COLOR FLUORESCENCE ASSAY
In PIFE experiment, we can monitor RNA-dsRBP interaction without the need to label the protein. This is possible because the brightness of the fluorophore (labeled to the dsRNA) changes according to its proximity to the interacting dsRBP (Fig. 1A) (10,11). Fluorophore labeling is not always possible for proteins and sometimes inhibits or reduces the functional activity of the proteins. Moreover, PIFE senses a shorter distance, 1–3 nm, as compared to the 3–8 nm detected by FRET (10). Thus, the simplicity and convenience of PIFE makes it a powerful and attractive complementary approach to FRET.
The choice of fluorophore however is crucial for PIFE measurement because not all fluorophores change their brightness depending on their proximity to the protein. For instance, Cy3 is one of the most well-known fluorophores showing the PIFE phenomenon, but not Atto 550 or Cy3B despite possessing similar absorption and emission spectral range as Cy3 (10). Hence we labeled the RNA at the position of interest, with Cy3 as a reporter of PIFE. In most cases, we labeled Cy3 at the end of the dsRNA because dsRBPs are known to bind RNA non-specifically. However, if a specific sequence or structure of RNA is critical for the interaction with the protein, we advise to label the position next to it with Cy3, in order to maximize PIFE signal.
A control experiment without dsRBPs should be performed to make sure that no PIFE signal exists for Cy3-labeled RNA alone. Since PIFE depends on the environment of the fluorophore Cy3, it is possible that PIFE could still be observed even without any dsRBPs. It could happen especially when a structured RNA may undergo a conformational change by itself. In this case, we suggest labeling at a different position that is not expected to be affected by conformational change of the RNA. After confirming no fluctuation of Cy3 intensity from Cy3-labeled RNA alone, we added dsRBPs to the Cy3-labeled RNA in order to monitor the RNA-dsRBP interaction. We observed ATP-independent diffusion activity of TRBP using smPIFE as shown in Fig. 1D. For smPIFE data acquisition, we share the same experimental setup as we use for 2-color smFRET which will be described in detail in the following section.
2.2. 2-COLOR FLUORESCENCE ASSAY
The dynamic interaction between dsRNAs and dsRBPs can be efficiently monitored by 2-color smFRET at the nanometer distance sensitivity (9). To study the interaction between dsRBPs and dsRNAs using FRET, we labeled the dsRBP with a donor dye Cy3 and the dsRNA with an acceptor dye, Cy5 or Alexa 647 (Fig. 1B). The distance between the two fluorophores on dsRNA and dsRBP can be measured by calculating FRET efficiency, which is proportional to the reciprocal of sixth power of the distance. We observed a rapid diffusion activity of TRBP by monitoring FRET between Cy3 at the end of dsRNA and Alexa 647 labeled to TRBP non-specifically (Fig. 1E), which is consistent with the result obtained by smPIFE (Fig. 1D).
The smFRET measurement was achieved by a wide-field TIR fluorescence microscopy. Upon excitation by a solid-state 532 nm laser (Spectra physics), Cy3 of dsRNA that is immobilized to a PEG-coated quartz surface emits light and transfers energy to the acceptor dye, Cy5 or Alexa 647 on the dsRBP, according to the distance between two. The fluorescent emission from donor and acceptor dyes were collected through a water-immersion Olympus objective (60 X, NA = 1.2), passed through 555 nm long pass filter for rejection of laser’s Rayleigh scattering. A dichroic mirror (cutoff: 630 nm) separates the fluorescent light from sample into two spectral regions, the one from Cy3 and Cy5/Alexa 647 that were detected by an EMCCD camera (Andor).
2.3. 3- COLOR FLUORESCENCE ASSAY
Adding one more fluorophore in the 2-color FRET geometry described in the previous section will allow us to measure up to three distances at the same time. Ideally, three distances can be monitored simultaneously using three fluorophores which requires at least two laser sources (20). However, we fixed the distance between two acceptor dyes at either end of dsRNA as shown in Fig. 1C to monitor the diffusion of TRBP between two ends of the dsRNA. In this geometry, one laser to excite the donor in TRBP is enough to measure the FRET between one donor and two acceptors (red and black circle in Fig. 1C, Cy5 and Cy7). The anti-correlation change of Cy5 and Cy7 fluorescence intensity was observed as shown in Fig. 1F, supporting that TRBP diffuses along the dsRNA. This assay is a direct way to show that TRBP diffuses along the dsRNA axis.
For 3-color FRET measurement, two dichroic mirrors (cutoff: 630 nm, 725 nm) instead of one as described for 2-color FRET setup, separate the fluorescent light from sample into three different spectral regions, the one from Cy3, Cy5 and Cy7, respectively. The exposure time was 30 msec, and the detected fluorescent signals were analyzed with a custom-written IDL and MATLAB program.
3. DATA ANALYSIS
3.1. FRET HISTOGRAM AND TIME TRACE
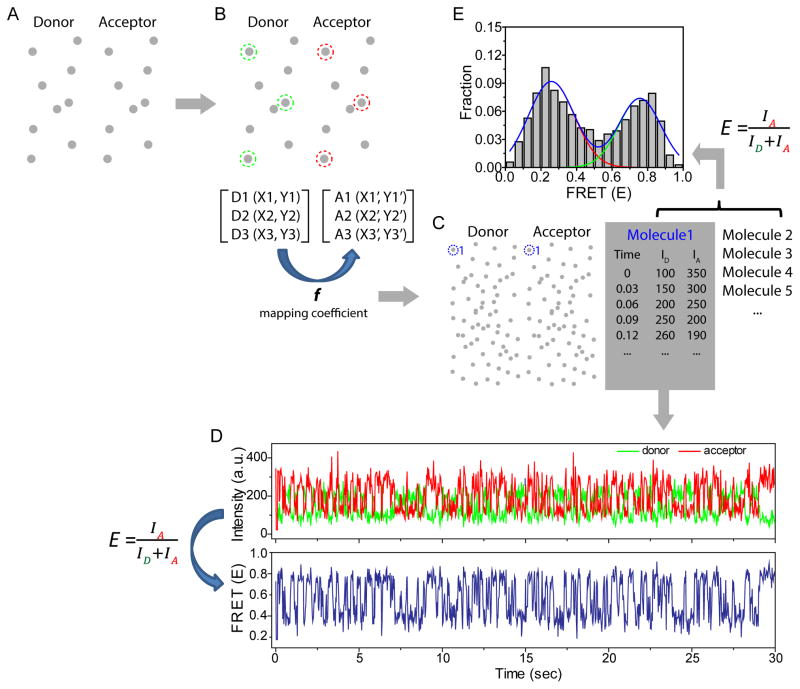
FRET efficiency is calculated by the emission of an acceptor divided by the total emission of a donor and an acceptor, providing the distance information between FRET pair, a donor and an acceptor. With our home-made TIR fluorescence system, we typically observe several hundreds of single molecules per image where the fluorescent emission from a donor and an acceptor were separated into two projections as shown in Fig. 2A. In order to identify the emission spots from the same molecules at both image planes, it is necessary to map between two image planes using a reference sample which produces the emission at both channels. We took an image of crimson latex beads (ThermoFisher) for mapping because their spectral range covers two emission channels, generating the emission spots at both image planes (Fig. 2A). Then, we chose three pairs of emission spots where each pair of the emission comes from the same molecule (Fig. 2B), with which we could calculate the mapping coefficient between two image planes. And this mapping coefficient was used to match the emission spots between two channels which come from the same dsRNA-dsRBP pair when we monitored the diffusion of dsRBP with an acceptor dye on dsRNA with a donor dye (Fig. 2C). For each single molecule, we obtained the information of time, and the intensity of a donor and an acceptor (Fig. 2C), with which we generate the intensity trace and FRET trace over time (Fig. 2D). Also, we can produce a FRET histogram as displayed in Fig. 2E, which contains FRET collected from all molecules that we analyzed. FRET time trace is powerful because it shows the behavior of every single molecule over time, and FRET histogram is also beneficial because it provides the overall trend by showing the FRET distribution of all molecules.
Figure 2. Data analysis procedure for smFRET assay.
A) TIRF image of fluorescent beads of which spectral range covers both donor and acceptor channel. B) Three pairs of fluorescent spots were selected to generate the mapping coefficient correlating two imaging planes. C) The donor fluorescent spot was matched with the acceptor fluorescent spot coming from the same dsRNA-dsRBP complex, and the fluorescent intensity of both channels over time were analyzed for every dsRNA-dsRBP complex. D) Fluorescent intensity time trace and FRET time trace generated using time and fluorescent intensity information per each dsRNA-dsRBP complex obtained in Fig. 2C. E) FRET histogram to show the FRET distribution of hundreds of single molecules.
3.2. AUTOCORRELATION ANALYSIS
To obtain the kinetic information of dynamic motion such as translocation or diffusion, one widely-used analysis is to analyze the dwell-time of a specific state or peak-to-peak for a repetitive event. However, if the change of signal is too quick to define the distinct kinetic steps, the auto-correlation analysis can be a way to go. We analyzed the diffusion kinetics of TRBP on dsRNA using auto-correlation and cross-correlation analysis because the diffusion kinetics is too fast to analyze peak-to-peak dwell time of the repetitive diffusion. The auto-correlation function was analyzed by MATLAB program, using the equation of auto-correlation as below:
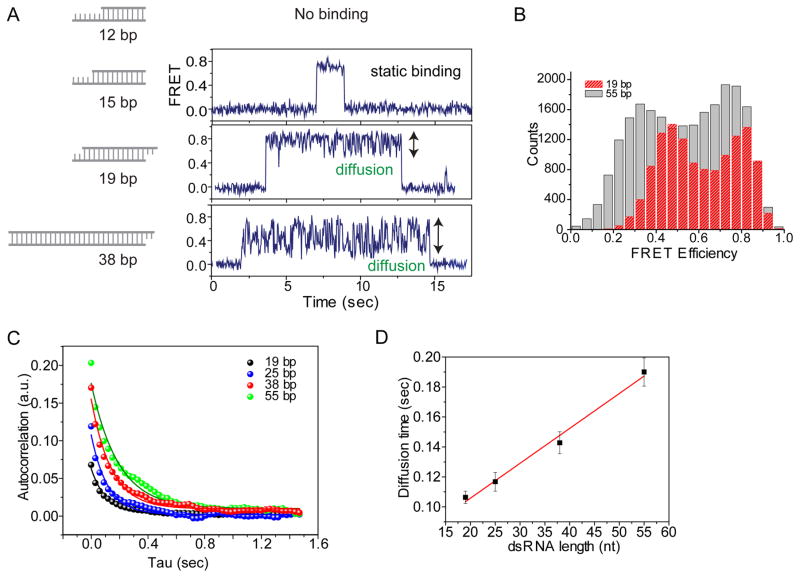
E(t) represents FRET and the FRET auto-correlation, G(τ), gives us an average diffusion rate because FRET changes in our system reflect the diffusion of dsRBPs. We obtained the autocorrelation curve for the diffusion of TRBP on the different length of dsRNA (Fig. 3A–C). More than 30 molecules were analyzed for each auto-correlation curve, and it was fitted with a single exponential decay to obtain an average diffusion rate of the molecules, exhibiting longer diffusion range for a longer dsRNA from the initial value of autocorrelation curve (Fig. 3C). Also, the diffusion time obtained from the half-time of the exponential decay fitting is linearly proportional to the length of dsRNA (Fig. 3D), suggesting that TRBP diffuses along the entire length of dsRNA within the length range of tested dsRNA, 19–55 bp.
Figure 3. Auto-correlation analysis on the diffusion of TRBP depending on the length of dsRNAs.
A, B) The larger FRET fluctuation of TRBP showed the longer diffusion range for longer dsRNA in FRET time trace as well as FRET histogram. C) Auto-correlation analysis on diffusion of TRBP on different length of dsRNA. D) Diffusion time of TRBP calculated from the exponential decay fitting in Fig. 3C linearly correlates with the length of dsRNA.
Similarly, we also tried the cross-correlation analysis to obtain diffusion kinetics from 3-color smFRET measurement. The cross-correlation between the fluorescence intensity of Cy5 and the one of Cy7 was analyzed using the following equation:
III. SINGLE MOLECULE IMAGING OF CELLULAR DSRBPS
The SiMPull assay enables researchers to examine proteins directly from cell lysate, without going through a sophisticated protein purification procedure, hence largely preserving the natural properties of the proteins in cells (13,21). Here, we describe how to investigate the interaction between RNAs and dsRBPs from cell lysates using SiMPull assay. TRBP and other dsRBPs were cloned with a C-terminal EYFP tag and then overexpressed in human A549 lung cancer cells. We pulled down the overexpressed dsRBPs in cell lysates to a single-molecule surface coated with biotinylated anti-GFP antibody and then investigated the dynamic interaction between dsRBPs and various dsRNAs using smPIFE (10,11). TRBP pulled down directly from lysate showed dynamic diffusion activity along dsRNA duplex, which is consistent with purified TRBP. We further tested substrate specificity and dynamic propensity of dsRBPs upon binding to dsRNAs varying their secondary structures.
1. SAMPLE PREPARATION
1.1. CLONING, PLASMID PREPARATION
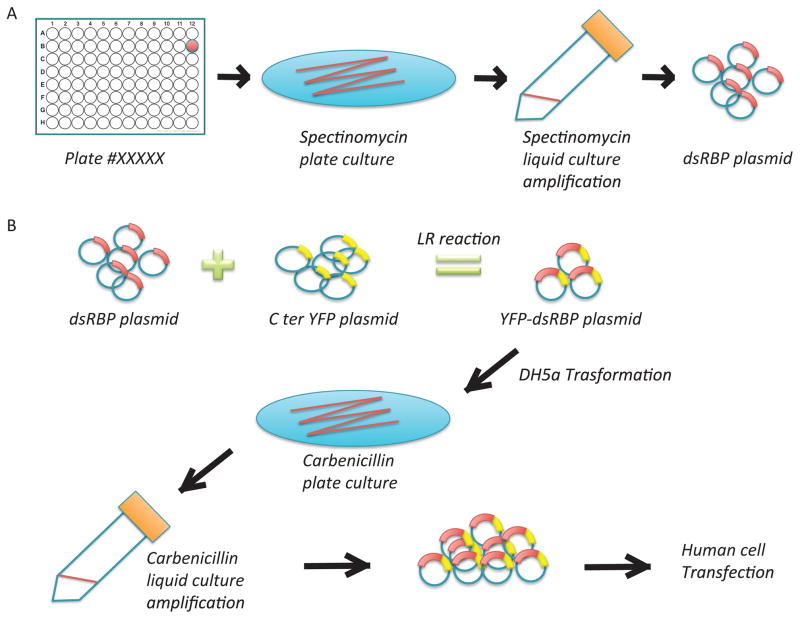
Four different human dsRBPs, ADAD2, ADAR1, TRBP and Staufen1, were cloned from Human Open Reading Frame (hORF) Library (http://horfdb.dfci.harvard.edu/hv5/) (8,22,23). And liquid cultures were built up from single colony, followed by DNA extraction using Mini prep (Fig. 4A). Harvested plasmids of each protein were further modified by adding either N-terminal EGFP or C-terminal EYFP by LR Clonase II enzyme mix (Thermo Fisher Scientific Gateway, Catalog number 11791-100) (Fig 4B).
Figure 4. Sample preparation for SiMPull assay of dsRBPs.
A) Plasmids of dsRBPs were selected from the hORF library, amplified, and purified. B) The selected dsRBP plasmid was conjugated with EYFP or EGFP plasmid by LR reaction, followed by amplification and purification.
1.2. HUMAN CELL TRANSFECTION
We transfected human cells with the engineered dsRBP plasmids using Lipofectamine® 3000 Reagent (Thermo Fisher Scientific) by following their online instruction. C-terminal EYFP-TRBP, C-terminal EYFP-Staufen1 and N-terminal EGFP- ADAR1 were overexpressed in human A549 cells, and C- terminal EYFP-ADAD2 was overexpressed in HEK293 cells for about 24 hours.
1.3. CELL LYSATE PREPARATION
Human cells transfected for 24 hours were lysed using RIPA (Thermo Scientific RIPA Lysis and Extraction Buffer, Catalog number: 89900) buffer for 1 hour in cold room with mild horizontal shaking. For every 8.8 × 106 cells, 1.5ml RIPA buffer was applied. Then cell lysates were collected in 2.0 ml eppendorf tubes and centrifuged at 14,000 rcf for 15min in cold room. We only collected the supernatant for further assay, which could be stored at −80 C for later use. The expression levels of dsRBPs were quantified by, fluorescence measurement based on EYFP or EGFP fluorescent intensity using known concentrations of Cy5 dye as standard.
2. SINGLE MOLECULE ASSAYS USING CELLULAR DSRBPS
In order to study the binding affinity and diffusion activity of dsRBPs from cell lysates, a series of control experiments are required as shown in the Fig. 5A. For the binding affinity test, we quantify the number of dsRNA binding to the dsRBP pulled-down to surface after confirming that the non-specific binding of dsRNA is negligible. Also, it is critical to apply the same concentration of dsRBPs for SiMPull assay to compare the relative binding affinity amongst various dsRBPs. For this reason, we quantified the concentration of dsRBPs as described in detail below. (2.1) We want to ensure that the dsRBPs of our interest are specifically pulled down to single-molecule imaging surface and that dsRNA specifically binds dsRBPs, but not to the bare surface or antibody-coated surface without dsRBPs. Here, we will describe the step-by-step control experiments for the dsRBP binding affinity test as well as the diffusion activity probing by using smPIFE assay.
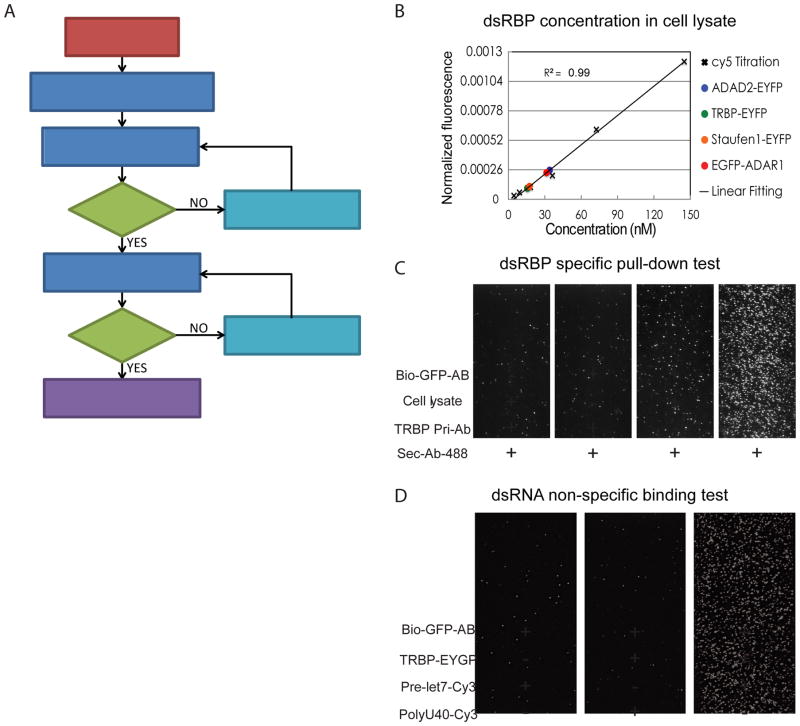
Figure 5. SiMPull assay workflow and the step-by-step control experiments.
A) SiMPull assay workflow for studying the binding affinity and the diffusion activity of dsRBPs to dsRNA. B) Determination of TRBP concentration from lysate based on the calibration curve generated from Cy5 dye of known concentration. C) DsRBP specific pull-down test. TRBP-EYFP in cell lysates was pulled down to the imaging surface using biotinlyated GFP antibody, and the specific pull-down was tested using a pair of TRBP antibody and its interacting secondary antibody. D) RNA non-specific binding test to single-molecule imaging surface. DsRNA (pre-let-7) not ssRNA (polyU40) binds specifically to TRBP pulled-down surface while it show few non-specific binding to the imaging surface coated with biotinylated GFP antibody.
2.1. MEASUREMENT OF PROTEIN CONCENTRATION
It is important to quantify the concentration of dsRBPs in cell lysates for investigating the interaction between dsRBPs and dsRNA using SiMPull assay because the binding affinity is sensitive to the concentration of dsRBPs. To quantify the concentration of dsRBPs in cell lysate, we calibrated it to the known concentration of a commercial Cy5 dye by measuring the fluorescent intensity of EYFP or EGFP of modified dsRBPs and Cy5 dye. It is easier to quantify the concentration of dsRBPs with EYFP or EGFP tag if any EYFP or EGFP sample of known concentration is available. If it is not the case, we can use any commercial organic dye of known concentration instead, but it requires the additional normalization step to compensate for the difference in the extinction coefficient and quantum yield. We generated a standard correlation curve between the concentration and the fluorescent intensity of Cy5 using a serial dilution of Cy5 dye (Fig. 5B), and then calculated the concentration of dsRBPs using this curve after normalizing the fluorescent intensity of EYFP or EGFP to Cy5 intensity considering of the differences in the extinction coefficient and the quantum yield between Cy5 and EYFP or EGFP.
2.2. DSRBP SPECIFIC PULL-DOWN TEST
Since the SiMPull assay utilizes crude cell lysates, it is of utmost importance that we ensure that we have specifically pulled down our dsRBP of interest on the PEG surface, without contamination of other non-specific endogenous proteins from the cell lysate. For pull-down of dsRBPs from cell lysates, the PEG-coated quartz surface was treated with Neutravidin (0.05μg/ml) for 2 minutes and then with biotinylated anti-GFP (RABBIT, 5μg/ml) antibody for 5min in T50 buffer (10 mM Tris pH 8 and 50 mM NaCl). About 400 pM of C-terminal EYFP-TRBP cell lysate was added to the antibody-coated PEG surface and incubated for 5–10 min.
In order to check if we specifically pull down dsRBPs of our interest, for instance, TRBP, we applied anti-TRBP antibody (5μg/ml, Abcam [1D9](ab129325)) and 1nM Alexa Fluor 488 conjugated anti-mouse antibody (Cell Signaling #4408) in T50 buffer and incubated them for 10min. We further tested the specificity of pull-down of TRBP by excluding biotinylated GFP antibody to ensure that the pulled-down proteins are not nonspecifically-bound to the PEG surface (Fig. 5C).
2.3. RNA NON-SPECIFIC BINDING TEST
After checking the specificity of dsRBP pull-down, we applied dsRNAs to study the interaction between dsRBPs and dsRNAs. We have previously quantified relative binding affinity of various dsRBPs to dsRNAs by counting the number of dsRNAs bound to the pulled-down dsRBPs of the same concentration (12). Therefore, it is extremely critical to make sure that RNA binding is specific to the dsRBPs of our interest. To characterize the degree of non-specific RNA binding, we performed several control experiments. We measured the non-specific binding of dsRNA, pre-let-7, by substituting the dsRBP (for instance, TRBP-overexpressed cell lysate) with blank cell lysate where no dsRBP was overexpressed. We added 1nM of dsRNA for the test, incubated for 5min in T50 buffer and then washed away the excess RNA molecules before the measurement. As seen in Fig. 5D, we observed only few RNA binding events in the case of blank lysate, supporting that the dsRNA binding is indeed specific to TRBP. Moreover, we assayed the substrate specificity of TRBP to dsRNA over ssRNA by testing the binding affinity of TRBP to ssRNA, polyU40 labeled with Cy3 dye (Fig. 5D, middle). It showed negligible binding events (< 20) compared with the binding events from dsRNA-TRBP interaction (> 600) (Fig. 5D, right), indicating that dsRNA binds specifically to TRBP at the tested experimental condition.
2.4. BINDING AFFINITY TEST
After testing the non-specific binding of dsRBPs and dsRNAs, we added 1nM of each Cy3-labeled dsRNA substrate to each of the dsRBP pulled-down to imaging surface. This step was followed by incubation for 5min, and washing excess dsRNA molecules away before imaging. The EYFP and EGFP in the modified dsRBPs do not interfere with the detection of Cy3 due to their faster photobleaching (~2–3 sec) as compared to Cy3 (~90–180 sec). Snap shots were taken at multiple locations to calibrate the average amount of dsRNA bound by dsRBPs.
2.5. SMPIFE ASSAY: DYNAMIC VS STATIC INTERACTION
Utilizing the pulled down dsRBPs (via SiMPull), we characterized the dynamic properties of dsRBP-RNA interaction by taking advantage of the smPIFE assay that can detect protein-RNA distance within 3 nm range, without the necessity of labeling the protein (10,11). To study the interaction between pulled down dsRBPs and dsRNA, we added 1nM of Cy3-labeled dsRNA substrates to the the pull-down of dsRBPs, and incubated them for 5mins. To prevent or delay photobleaching, an oxygen scavenging buffer system (0.5% (wt/vol) glucose, 10μg/μl glucose-oxidase (Sigma) and 8.8KU/ml catalase (Calbiochem)) was employed in imaging buffer together with 20 mM Tris pH 7.5, 25 mM NaCl and 2.5mg/ml trolox (Sigma) (21). To capture the dynamic interaction between dsRBPs and dsRNA, we measured the fluorescence intensity of single-fluorophores labeled to dsRNA at exposure times as short as 30 ms for 2–5 min. The time-course of fluorescence intensity fluctuation was analyzed by a custom-deesigned IDL and Matlab program.
3. DATA ANALYSIS
3.1. RELATIVE BINDING AFFINITY
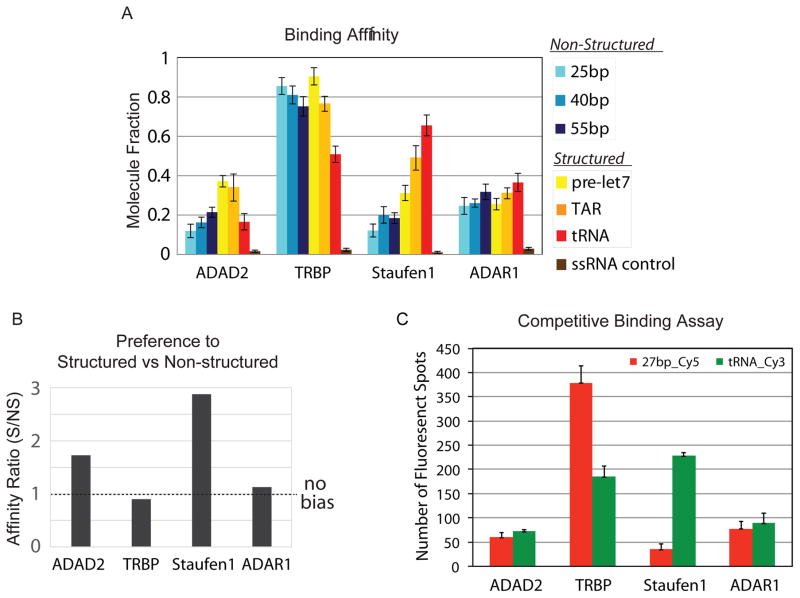
Using SiMPull assay, we can conveniently measure relative dsRNA binding affinity of dsRBPs by counting the number of bound dsRNA molecules to the dsRBP that is pulled down to the single-molecule imaging surface. We tested the relative binding affinity of four different dsRBPs pulled down from cell lysates, to six Cy3-labeled dsRNAs with various secondary structures. For the quantification of the dsRNA binding affinity of dsRBPs, we normalized it by estimating the number of the pulled-down dsRBPs on surface based on the measured concentration (Fig. 5A), and calculated the fraction of protein bound by dsRNA substrate (Fig. 6A). Also, we subtracted the number of non-specific binding of dsRNA measured with blank cell lysate.
Figure 6. Binding affinity of dsRBPs to various dsRNAs.
A) The relative binding affinity of four dsRBPs to six dsRNAs containing various secondary structures. B) The affinity ratio indicates the binding bias of each dsRBP towards structured or non-structured RNA substrate. C) Competitive binding assay to compare binding affinity of each dsRBP to structured (Cy3-tRNA) with non-structured (Cy5-27bp) as a pair.
3.2. PREFERENCE TO STRUCTURED VS. NONSTRUCTURED DSRNA
Based on the relative dsRNA binding affinity of dsRBPs, we noticed that the dsRNA binding affinity of dsRBPs varied depending on the structural features of the dsRNAs. Therefore, we categorized the six dsRNAs we tested into two groups: structured (pre-let7, TAR and tRNA) and non-structured (25, 40 and 55 bp) dsRNA, and compared the average binding affinity between the two groups. Furthermore, we defined a ratio of bound fraction of each dsRBP towards structured over non-structured RNAs, termed as affinity ratio. By this definition, a ratio close to 1 would signify no-biased binding between the two groups, and a ratio greater than 1 would signify preference to structured RNAs. Interestingly, for ADAD2 and Staufen1 the affinity ratio were much greater than 1, suggesting a strong preference for structured dsRNAs, while the affinity ratio for TRBP and ADAR1 were close to 1, suggesting no structural bias (Fig. 6B). Amongst many factors that can contribute to this substrate bias are the number of dsRBDs, linker length in between the dsRBDs, the amino acid composition, and the hydrophobic/hydrophilic environment of dsRBDs in dsRBPs.
3.3. COMPETITIVE BINDING ASSAY
To further characterize the relative binding affinity of dsRBPs towards two different dsRNA substrates, we employed a competitive binding assay. We compared the binding affinity of four dsRBPs (ADAD2, TRBP, Staufen1 and ADAR1) to heavily structured tRNA versus perfectly-matched 27-bp dsRNA. After pull-down of each dsRBP to a single-molecule imaging surface, we added 1nM of Cy3-labeled tRNA (structured) and 1nM of Cy5-labeled 27bp dsRNA (unstructured) simultaneously and quantified the number of Cy3 and Cy5 molecules by dual laser excitation (Fig. 6C). The binding ratio between Cy3-tRNA and Cy5-27bp dsRNA is consistent with that in Fig. 6A, the relative binding affinity independently measured for each dsRNA. This confirms the validity of the affinity testing of dsRBPs performed above.
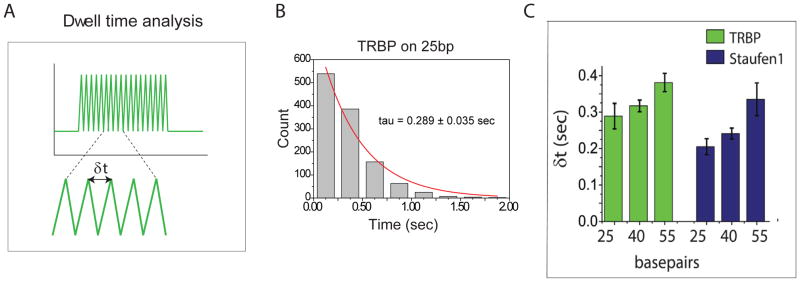
3.4. DWELL TIME ANALYSIS
The dynamic diffusion activity of purified TRBP along dsRNA duplex was reported using smFRET assay (6). We observed consistent dynamic diffusion activity of TRBP from cell lysates on dsRNA by SiMPull and smPIFE assay, and further tested three other dsRBPs from cell lysate to investigate their diffusion activity. In smPIFE assay, the fluorescent intensity of Cy3 on dsRNA would increase and decrease when the protein approaches and moves away, respectively. In other words, dsRNA diffusing on dsRBPs would exhibit fluctuation of Cy3 fluorescence intensity (8,9). By counting the molecules showing the fluctuation of Cy3 fluorescent intensity, we measured if each dsRBP diffuses on dsRNA and if the diffusion propensity differs amongst dsRNA substrates. We found that the diffusion propensity of each dsRBPs varies amongst the dsRBP depending on the type of dsRNA (14). We further characterized the interaction between dsRBPs and dsRNA by performing dwell-time analysis, where we calculated the period of time between two adjacent peaks as shown in Fig. 7A from more than thousands events of 200–300 molecules in average. Fig. 7B shows the result of dwell-time analysis of TRBP on 25-bp dsRNA (Fig. 7B), which provides the average time that it takes for a single dsRBP to move from one end to next and returns to the same position. The diffusion time is correlated linearly with the length of the dsRNA, which indicates that both TRBP and Staufen1 would diffuse along the entire length of dsRNA rather than a fixed distance (Fig. 7C).
Figure 7. Dwell time analysis of dsRBPs.
A) Scheme to show how to analyze dwell time in single molecule trace. B) Dwell time distribution of TRBP on 25-bp dsRNA C) Dwell time analysis of TRBP and Staufen1 interacting with three non-structured duplex RNAs (25, 40, and 55 bp), showing linear dependence of dwell time on the length of RNA duplex.
IV. CONCLUSION
Combination of the various single-molecule fluorescence assays described above constitute a powerful approach to study fast diffusion in a short range as demonstrated by the repetitive diffusion of dsRBPs on dsRNAs because the distance sensitivity of smPIFE and smFRET covers 1–10 nm range. We can also compare the diffusion activity of dsRBPs that are reconstituted in vitro with the one that are pulled-down directly from cells which provides a valuable insight into the potential diffusion behavior of dsRBPs in vivo. These assays may be adapted to study interactions between nucleic acids and proteins, providing the binding affinity as well as the real-time kinetic information at the single-molecule level.
HIGHLIGHTS.
Single-molecule imaging of purified dsRBPs by 1, 2, 3-color fluorescence assay.
Interaction between cellular dsRBPs and dsRNAs studied by single-molecule pull down.
ATP-independent dsRNA diffusion activity exhibited by purified and cellular dsRBPs.
Acknowledgments
We thank Jaya Sarkar for her critical reading and editing of the review article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 3.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita S, Nagata T, Kawazoe M, Takemoto C, Kigawa T, Guntert P, Kobayashi N, Terada T, Shirouzu M, Wakiyama M, et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 2011;20:118–130. doi: 10.1002/pro.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fierro-Monti I, Mathews MB. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- 6.Koh HR, Kidwell MA, Ragunathan K, Doudna JA, Myong S. ATP-independent diffusion of double-stranded RNA binding proteins. Proc Natl Acad Sci U S A. 2013;110:151–156. doi: 10.1073/pnas.1212917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 8.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang H, Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem Soc Rev. 2014;43:1221–1229. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Vukovic L, Koh HR, Schulten K, Myong S. Dynamic profiling of double-stranded RNA binding proteins. Nucleic Acids Res. 2015;43:7566–7576. doi: 10.1093/nar/gkv726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toseland CP. Fluorescent labeling and modification of proteins. J Chem Biol. 2013;6:85–95. doi: 10.1007/s12154-013-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakowicz JR, Weber G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benesch RE, Benesch R. Enzymatic removal of oxygen for polarography and related methods. Science. 1953;118:447–448. doi: 10.1126/science.118.3068.447. [DOI] [PubMed] [Google Scholar]
- 18.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 19.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89:141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Three-color alternating-laser excitation of single molecules: monitoring multiple interactions and distances. Biophys J. 2007;92:303–312. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nature protocols. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrandon D, Elphick L, Nüsslein-Volhard C, St Johnston D. Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, Wang Q, Howell KL, Lee JT, Cho DSC, Murray JM, Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. Journal of Biological Chemistry. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]