Abstract
Magnetic tweezers is a versatile and easy to implement single-molecule technique that has become increasingly prevalent in the study of nucleic acid based molecular motors. Here, we provide a description of the magnetic tweezers instrument and guidelines for measuring and analyzing DNA helicase activity. Along with experimental methods, we describe a robust method of single-molecule trajectory analysis based on the Student’s t-test that accommodates continuous transitions in addition to the discrete transitions assumed in most widely employed analysis routines. To illustrate the single-molecule unwinding assay and the analysis routine, we provide DNA unwinding measurements of Escherichia coli RecQ helicase under a variety of conditions (Na+, ATP, temperature, and DNA substrate geometry). These examples reveal that DNA unwinding measurements under various conditions can aid in elucidating the unwinding mechanism of DNA helicase but also emphasize that environmental effects on DNA helicase activity must be considered in relation to in vivo activity and mechanism.
Keywords: Single-molecule, Helicase, Molecular Motor, Magnetic tweezers, RecQ
1. Introduction
DNA Helicases are molecular motors that unwind DNA duplex or translocate single-stranded DNA by utilizing chemical energy from the hydrolysis of nucleoside triphosphates (NTPs) [1]. Due to their multifaceted and essential roles in DNA metabolism [2], DNA helicases have been extensively investigated experimentally and computationally [1, 3–6]. Fundamental mechanistic questions regarding DNA helicases involve the molecular details of helicase activities; DNA-helicase interactions such as association and dissociation rate constants, unwinding characteristics including unwinding rate, processivity, directionality, physical and kinetic step sizes, and the chemo-mechanical coupling between NTP hydrolysis and unwinding or translocation. Ensemble techniques including gel-based measurements, fluorescent stop-flow measurements [7, 8] and structural studies [2, 4, 9] provide substrate specificity, binding kinetics, molecular interactions and structural conformations of DNA helicases. Single molecule techniques, on the other hand, uniquely measure the dynamics of individual enzymes with high spatial and temporal resolution, permitting a comprehensive picture of how DNA helicases interact with DNA and function as molecular motors [3, 5, 6, 10–12].
Magnetic tweezers (MT) (Fig. 1) is a single-molecule manipulation technique [11, 13–16] that has emerged as an invaluable tool to investigate DNA helicases [11, 17–21]. Despite the relatively low temporal and spatial resolution of magnetic tweezers in comparison with optical tweezers [14], it offers several advantages over other single molecule manipulation techniques [11, 14, 16]. First, the magnetic tweezers approach affords a passive force-clamp since it maintains a constant force over an extended range of probe motion (>10 µm). Second, torque can be readily applied to rotationally constrained DNA via the magnetic bead simply by rotating the magnets. Third, force is applied by a magnetic field rather than a high power laser, which can induce biological damage limiting the useful measurement duration [22, 23]. This aspect also facilitates combining MT with simultaneous single-molecule fluorescent detection. Fourth, the magnetic force is roughly uniform over a large area (~ 100 µm × 100 µm) thereby permitting the simultaneous manipulation and measurement of a large number of molecules [16, 24]. In a typical MT DNA helicase assay a superparamagnetic microsphere attached to an individual DNA molecule functions as a force transducer and a position probe. The three-dimensional position of the microsphere attached to DNA is tracked in real-time via CCD or CMOS camera-based image analysis approaches that can routinely achieve a few nm resolution in the range of 60 – 500 Hz. Two DNA substrate geometries are typically used to measure helicase activity in single-molecule manipulation experiments: A DNA hairpin configuration in which the two free ends of the hairpin are under tension and a DNA duplex containing a gap or a free end on which the helicase can load (Fig. 2a) [3, 21]. In both geometries the helicase unwinding activity transforms double-stranded (ds) DNA to single-stranded (ss) DNA, which results in a change in extension of the probe. For the DNA hairpin geometry each base pair unwound results in an increase in extension of two single-stranded nucleotides. This represents a significant amplification of the unwinding signal, and as a result the hairpin unwinding assay provides the highest resolution measurements of helicase unwinding. For the duplex DNA geometry, unwinding results in the conversion of dsDNA to ssDNA, [3, 11, 15] which can be detected due to the differences in mechanical properties of dsDNA and ssDNA [3]. The resulting change in extension is fraction of a base pair (0.35 nm), resulting in lower signal to noise than the hairpin unwinding assay. These mechanical measurements of helicase unwinding activity have proven efficient for elucidating the mechanistic details of DNA helicases, yet often leave uncertainties concerning DNA-helicase stoichiometry or initial binding. Recently, a hybrid technique [25–28] combining MT with single-molecule fluoresce detection of fluorophore-labeled helicases overcomes this limitation by providing a direct measure of helicase stoichiometry and potentially provides additional measures of DNA-helicase interactions including the binding geometry of the helicase on the DNA substrate, translocation of DNA helicases and the direct coupling of DNA unwinding and helicase domain motion.
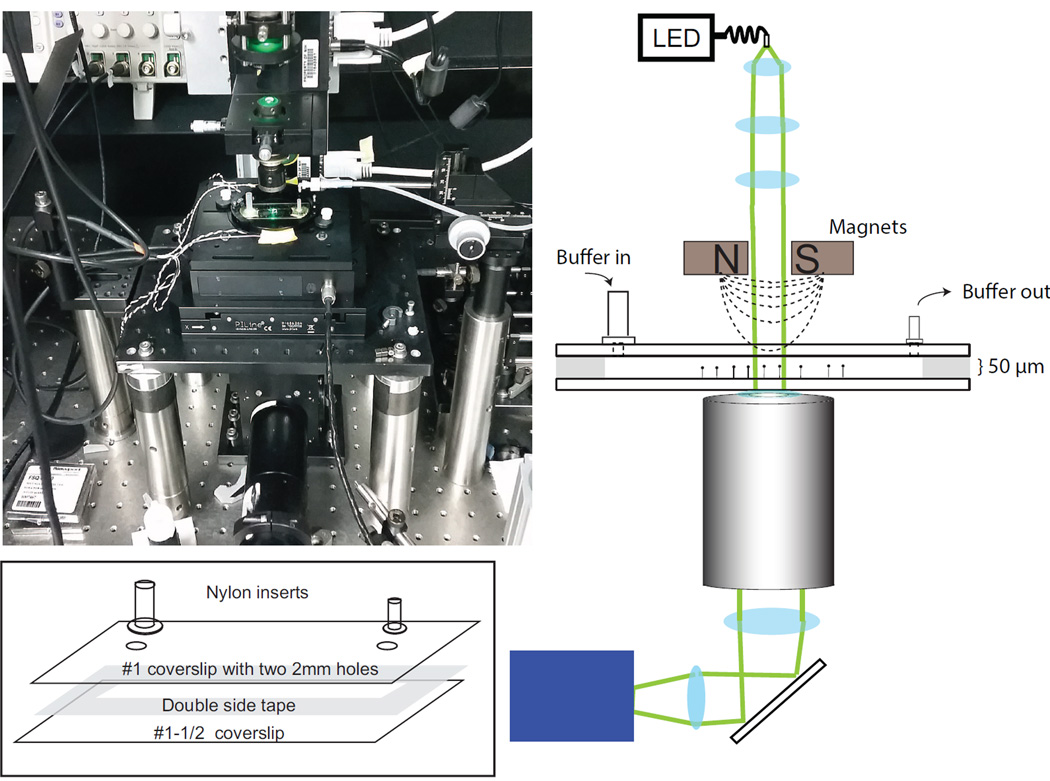
Figure 1.
Picture and schematic (not to scale) of magnetic tweezers. Light from a fiber coupled LED is collimated by three lenses and illuminates the sample cell. Green lines show the illumination path. The light collected by the objective lens is imaged by a CCD. A magnet assembly containing two magnets is located above the sample. The sample chamber has two ports. One serves as a buffer reservoir while the other is connected to a vacuum line to effectuate buffer exchange. The picture shows the magnetic tweezers instrument. Inset: detailed drawing of sample cell components and assembly.
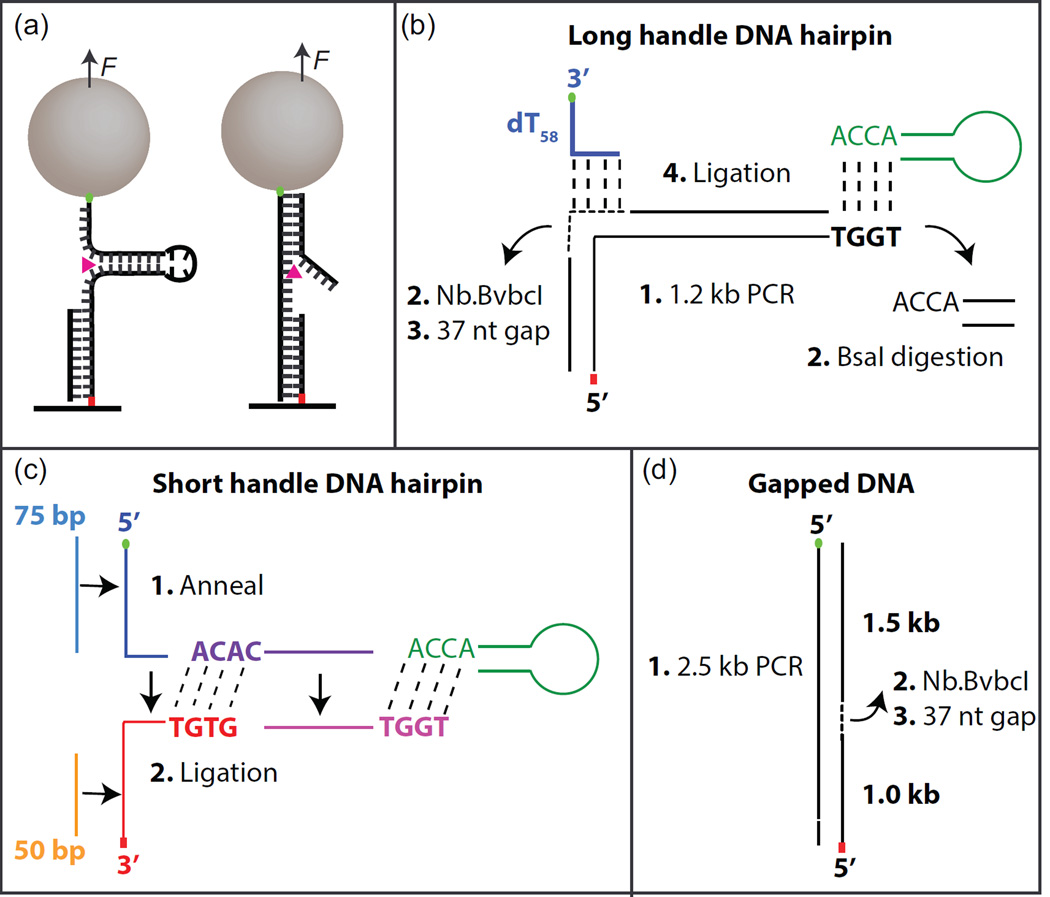
Figure 2.
Hairpin and gapped DNA substrate geometries. (a) Cartoon of hairpin and gapped DNA substrates bound to the surface via anti-digoxigenin and digoxigenin (red dot) and to the streptavidin coated magnetic beads via biotin linkage (green dot). RecQ unwinding DNA duplex is shown as magenta triangle. Assembly of long-handle DNA hairpin (b), short-handle DNA hairpin (c), and Gapped DNA (d). Short dashes indicate gapped region. Long dashes indicate ligation.
Here, we provide guidelines for performing single-molecule DNA unwinding assays using a magnetic tweezers. We illustrate our approach with measurements of Escherichia coli (E. coli) RecQ DNA unwinding. RecQ helicases, conserved from E. coli through humans, play essential roles in DNA recombination and repair [29, 30]. E. coli RecQ is the prototypical RecQ enzyme that contains a well-conserved domain architecture including two RecA domains, Zinc binding and Winged helix domains, and the helicase-and-RNaseD-C-terminal (HRDC) domain [29]. In this study, we include two different RecQ constructs – full length (RecQWT) and an HRDC deletion variant (RecQdH). Starting with a brief description of the instrumentation, we show how the unwinding activity of RecQ is affected by Ionic strength (with RecQdH), temperature (with RecQdH), and DNA substrate geometry (with RecQdH and RecQWT). In addition, we describe an analysis method we developed to obtain fundamental unwinding kinetics of DNA helicase such as unwinding rate, kinetic step-size, and pause duration.
2. Material and methods
2.1 Magnetic tweezers instrumentation
The operational principle of magnetic tweezers and detailed descriptions of the instrumentation have been extensively described elsewhere [15, 31, 32]. The essential feature is that permanent magnets are used to apply force on a micron size super-paramagnetic bead, the three-dimensional position of which is tracked in real time. As shown in the schematic (Fig. 1), the custom-built magnetic tweezers incorporates two permanent magnets (ND52, 4 mm (width) × 3 mm (height) × 2 mm (length), K and J magnets). The position of the magnets, which controls the applied force on the bead, is controlled by a linear motor (M-126.PD1, Physik Instrumente), whereas a rotary motor controls the rotation of the magnets, which rotates the bead (C-150.PD, Physik Instrumente). This assembly is located above the sample cell centered along the axis of illumination and imaging path. The sample cell is mounted on a custom built holder attached to two stacked piezo-driven stages, one for nanometer scale position control in 3 dimensions (PI-527-3CL, Physik Instrumente), the other for large horizontal movements (replacing a conventional manual stage) (PILine M-686.D64, Physik Instrumente). These two stages are mounted on an optical breadboard (MB1218, Thorlabs) that has been modified with a 3” through-hole accommodating the optical imaging path. This magnetic tweezers apparatus is assembled on a vibration isolation table (Micro-g Laboratory table, Technical Manufacturing Corporation). A 95 mm structural rail (XT95-750, Thorlabs) supports the magnet and illumination assemblies. The illumination source is a fiber-coupled 520 nm LED light (Optogenetics LED-520, Prizmatix) that is collimated using three aspheric lenses (C330TMD –A, Thorlabs; 5721 (60×) and 5722 (40×), New Focus). Sample temperature is controlled by a pair of resistive heater elements attached to the bottom of the custom-built sample cell holder. A temperature controller (TC-344C, Warner Instruments) drives the heater elements and maintains a constant temperature at the sample.
Two stacked cage cubes (LC6W, Thorlabs) below the objective (UPLSAPO 100XO, Olympus), are centered on the optical axis. The bottom cube holds a mirror (ME2-P01, Thorlabs) to direct illumination light from the objective to two imaging lenses (AC508-300-A1, Thorlabs; AAP-125.0-25.4-425-675, CVI), which project an image onto a CCD camera (GE-680, Prosilica).
2.2 Protein preparation
2.2.1 Cloning
(His)6 TEV RecQ plasmid
The cDNA coding for E. coli (DH5α) RecQ (provided by J. Keck, University of Wisconsin Madison) was produced by PCR. The N-terminal primer -5’ GGG TTT CAT ATG GAA AAC CTG TAC TTC CAG GGT ATG GCG CAG GCG GAA GTG TTG 3’- (Eurofins MWG Operon) contains an NdeI site and a synthetic coding sequence for a TEV cleavage site, and hybridizes with the N-terminus of RecQ to yield an in-frame TEV-RecQ fusion protein. The C-terminal primer -5’ GCC GGA TCC CTA CTC TTC GTC ATC GCC ATC AAC ATG CGC 3’- hybridizes with the RecQ C-terminus. It contains an Amber codon immediately downstream of the RecQ coding sequence followed by a BamHI site. The NdeI-BamHI digested PCR product was sub-cloned into an NdeI-BamHI linearized pET-15b vector (Novagen - EMD Millipore). The entire coding region of the fusion protein was sequenced (Macrogen USA) with synthetic primers designed specifically for the RecQ construct to ensure the absence of spontaneous mutations.
(His)6 TEV RecQ-ΔHRDC plasmid
The Alanine 522 coding sequence, at position 6964-6966 of the (His)6 TEV RecQ plasmid, was replaced by an Amber codon using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) and two custom primers, -5’ GTA TCG TGG CGC TCA AAC CGA AAT AGA TGC AGA AAT CGT T 3’- and its Reverse Complement Strand, both containing the desired mutation. The coding sequence was verified by sequencing.
2.2.2 Expression of the (His)6 TEV RecQ plasmid
The (His)6 TEV RecQ plasmid was transformed into Rosetta™2 (DE3) pLysS Novagen (EMD Millipore). Cells were grown at 37 °C in 1 L LB medium (MP Biomedicals) containing 100 mg·L−1 of Carbenicillin (EMD Millipore) and 34 mg·L−1 of chloramphenicol (Sigma) for 4 hrs. Expression of the (His)6 TEV RecQ plasmid fusion protein was induced with 1 mM IPTG (EMD Millipore) at 37 °C overnight.
2.2.3 Purification of the (His)6 TEV RecQ protein
The cell pellet was harvested at 6,000rpm for 20 min at 15 °C and stored at −80 °C. The frozen cell pellet (2.6 g) was thawed at room temperature in a stainless steel beaker and re-suspended in 64 mL of resuspension buffer (20 mM Tris·HCl, pH 8.0, 20 mM Imidazole, pH 8.0, 1 mM β-mercapto ethanol, 300 mM NaCl, 10 % v/v glycerol) supplemented with one tablet of EDTA-free complete protease inhibitor (Roche Diagnostics). After three freeze-thaw cycles (dry ice and room temperature), the resuspended cells were lysed with 0.1 mg·mL−1 lysozyme (Sigma) and probe-sonicated on ice for 5 min with a Branson 450 sonicator equipped with a standard disruptor horn at 40 % duty cycle and power level 4. The cells were further disrupted by two passages through an EmulsiFlex-C3 (Avestin). Cell debris was removed by centrifugation at 11,150 × kGmax for 30 min at 20 °C in an 5810R centrifuge (Eppendorf) and the supernatant was further clarified at 185,500 × kGmax for 60 min at 20 °C in an Optima L-100 XP centrifuge (Beckman Coulter).
Immunoaffinity chromatography purification was performed using an ÄKTAprime purification system (GE Healthcare) at room temperature. The high speed supernatant was loaded onto a 1.6 × 5.6 cm Ni-NTA Super-flow (Qiagen) packed XK column (GE Healthcare). After removing unbound proteins by washing with the cell resuspension buffer, the (His)6 TEV RecQ protein was eluted, using a 120 mL linear gradient to 400 mM imidazole, as a shoulder peak with a maximum at 100 mM imidazole. The protein-containing fractions considered pure by SDS-PAGE were pooled. An 30,000 MWCO Amicon® Ultra 15 mL Centrifugal Filter (Millipore) was used for both protein concentration and buffer exchange to the resuspension buffer.
TEV protease cleavage was performed at room temperature for 6 hrs using 0.5 U of pro-TEV (Promega) per µg of (His)6 TEV RecQ. RecQ was purified from the residual uncleaved (His)6 TEV RecQ, the (His)6 TEV tag and the protease by two successive passage through a 5 mL His-Trap™ FF (GE Healthcare) and concentrated using an 30,000 MWCO Amicon® Ultra 15 mL Centrifugal Filter in storage buffer (20 mM Tris·HCl, pH 7.5, 300 mM NaCl, 10 % v/v glycerol). The concentration of the sample was estimated using its A280 (ε = 48820 M−1·cm−1; http://web.expasy.org/protparam/). The expression and protein purification methods of the (His)6 TEV RecQ-ΔHRDC were identical to the ones used for the full length RecQ. The concentration of the sample was estimated using its A280 (ε = 45840 M−1·cm−1; http://web.expasy.org/protparam/). The identities of the all proteins were confirmed by mass spectroscopy analysis (Agilent 6224 ESI-TOF LC-MS).
2.3 DNA substrate preparation
There are potentially many different DNA substrate geometry considerations dependent on the nature of the DNA helicase under study. Here we discuss two common designs, hairpin and gapped DNA, used to measure DNA helicase activity in single molecule manipulation experiments. Our template design accommodates the requirement of a single-stranded DNA region (>20 nucleotides) for initial binding of RecQ DNA helicase and the subsequent 3´ to 5´ translocation of the helicase [33]. In general, the polarity and processivity of the helicase are important considerations when designing single-molecule templates. Duplex handle regions must be long enough to prevent the helicase from unwinding the entire substrate and the polarity of ssDNA attachments to the bead and to the surface must be chosen to prevent the helicase from loading and disrupting either linkage.
2.3.1 DNA hairpin
The DNA hairpin substrate is composed of two DNA handles attached to the ends of a DNA hairpin. We provide two different methods to generate a DNA hairpin substrate, one is based on PCR to create a long DNA handle and the other is based on annealing oligonucleotides to create short DNA handles (fig. 2). In general, the shorter DNA handle affords higher temporal and spatial resolution [34].
DNA hairpin with long handle
174bp DNA hairpin with a ~1.1 kb DNA handle was generated by PCR of pKZ1 using one primer containing a BsaI recognition sequence and the other primer with three digoxigenin modified bases at the 5´end. All oligonucleotides were purchased from Operon. pKZ1 was made by inserting a 44 bp dsDNA segment containing two BbvCI sites separated by 37 nucleotides into pET28b at the BamHI site. The insertion orientation was verified by sequencing. The PCR product was purified using a PCR purification kit (28104, Qiagen). The PCR product was digested for a total of 9 hours with Nt. BbvcI (R0632L, New England Biolabs) and BsaI (R3535L, New England Biolabs) at 37° C to create two nicks at positions 1093 and 1129 and a 5′ overhang at the end of the dsDNA (20 units of each enzyme were added to the sample every 3 hours). After digestion, the 37-nt oligomer was removed by incubating the DNA at 80° C for 20 min and cooling down gradually to 4 °C in the presence of excess complementary 37-nt oligomer, producing a 37-nt single-stranded region (Fig. 2b). The digested products were purified using a PCR purification kit (11732668001, Roche). To complete the 174 bp DNA hairpin, the digestion product was ligated to a 12 bp hairpin DNA containing a 4 dT loop and a 5′ overhang complementary to that of the digestion product, and a 90 bp oligomer consisting of a 33 bp region complementary to the ssDNA region of the digestion product followed by 54 deoxy-thymidines (dT54) labeled on the 3′ end with biotin that works as a binding platform for RecQ as well as providing an attachment to the streptavidin coated magnetic bead.
DNA hairpin with short DNA handle
The DNA hairpin substrate was prepared by annealing and ligation (Fig. 2c). Before annealing, the DNA oligonucleotides were heated for 2 min at 94°C and cooled down quickly on ice. All annealing was done in DNA hybridization buffer (100 mM potassium acetate and 30 mM HEPES, pH 7.5). First, the top and bottom DNA handles and the DNA hairpin main duplex region were separately hybridized by heating the mixed complementary DNA oligonucleotides (75 nt and 95 nt DNA oligomers for the top handle, 50 nt and 99 nt oligomers for the bottom, two 80 nt oligomers for the main duplex region; 10 nmol each) in separate PCR tubes at 94 °C for 5 min followed by gradual cooling to room temperature (−1°C /30s). The top and bottom DNA handles were then annealed via complementary regions from each handle to form a tailed DNA junction by heating the mixture at 80 °C for 5 min and cooling gradually to room temperature (−1°C /30s). Equal molar amounts of this DNA junction molecule and the hairpin main body were mixed with a 10-fold excess of T-loop DNA and ligated with T4 DNA ligase (M1794, Promega) to form the DNA hairpin substrate (Fig. 2c).
2.3.2 Gapped DNA
To prepare the gapped DNA substrate (Fig. 2d), 2.5 kb DNA molecules were generated by PCR of pKZ1 between positions 4550 and 1677 using 5´-biotin- (position 4550) and 5´-digoxigenin- (position 1677) labeled primers. The procedure to generate the gap between positions 1093 and 1129 was similar to that for the hairpin DNA substrate except that Nb. BbvCI (R0631L, New England Biolabs) was used to nick the DNA rather than Nt. BbvCI.
2.4 RecQ helicase unwinding assay
2.4.1 Sample cell preparation
A sample cell was made with two 24mm × 60 mm coverslips (Fig. 1) (top: #1 2975-246; bottom: #1-1/2 2980-240, Corning) held together by double-sided tape (50 µm thick, 8132LE, 3M) (Fig. 1 inset). The double-sided tape was laser-cut to make a 40 mm × 4 mm × 50 µm channel in the sample cell. Two 2 mm diameter through-holes were drilled on the top coverslip using abrasive aluminum oxide beads (50 µm, PD1003-4, ComCo Inc) in a blast cabinet equipped with a fine nozzle (ProCenter, ComCo Inc). All coverslips were cleaned before assembly [35]. Two sizes of nylon shoulder washers (10SC008062; 10SC002037, McMaster Carr) were attached to the holes in the top coverslip creating a buffer reservoir and an outlet port by spreading vacuum grease (Dow Corning) on the bottom surface and applying epoxy (Devcon) around the edge of the washers.
2.4.2 Single molecule assay
DNA substrate (5–10×10−15 moles) was first incubated with ~100 ng of anti-digoxigenin (11333089001, Roche) in 1**X Phosphate buffered saline (pH 7.4) for 15 min at room temperature. This mixture was incubated for either 1 hour at room temperature or overnight at 4°C in a sample cell prepared with non-magnetic 2 micron polystyrene fiducial beads (PS05N, Bangs Laboratories Inc.) melted on the surface [35]. Unbound DNA was washed out with 200 µl of wash buffer (PBS supplemented with 0.04% v/v Tween-20 and 0.3% w/v BSA). 20–30 µl of a 20-fold dilution of 1 µm streptavidin-coated magnetic beads (MyOne, Invitrogen) was then introduced in wash buffer, allowed to tether for 1 hour, and washed with 1 ml of wash buffer. DNA substrates were identified based on their extension under constant force. For DNA hairpin substrates, the extension change due to opening of the DNA hairpin at a force of ~20 pN provided an additional means of identifying suitable molecules. Once a correct DNA substrate was identified, the chamber was washed with 200 µl of RecQ buffer (30 mM Tris pH 8, varying NaCl concentrations (30–250 mM), 5 mM MgCl2, 0.3 % w/v BSA, 0.04 % v/v Tween-20 and 1 mM DTT) and then RecQ buffer containing RecQ at a concentration of 10–50 pM and ATP (0–1.0 mM).
2.4.3 Single molecule measurements of DNA helicase activity
The methods that we employ for three dimensional particle tracking and force (F) calibration have been extensively described [15, 24, 31, 32, 36]. DNA unwinding activity by RecQ was performed at constant force. Higher force increases the spatial and temporal resolution of the measurement, but may also influence the helicase activity. For the DNA hairpin, however, the applied force must be below the critical force at which the hairpin will spontaneously unzip (~15 pN). DNA extension was measured in real-time by tracking the three-dimensional position of the tethered bead while simultaneously tracking a stuck fiducial bead to compensate for drift using a custom-written image analysis program (Labview, National Instruments). The measurements consisting of 3-dimensional tracking data for both beads, and the applied force were stored as either text or binary files. Using the Worm-Like-Chain (WLC) model for dsDNA and ssDNA, the measured DNA extension changes during DNA unwinding by RecQ were converted from nanometers to the number of melted base-pairs to estimate the unwinding activity [21, 37]. For the DNA hairpin, unwinding of one base-pair corresponds to an increase extension of two single-stranded nucleotides. At 8 pN, each bp unwound results in ~0.8 nm increase in the extension of the bead based on the WLC model with an ssDNA contour length (lss) of 0.65 nm/nt and persistence length (Pss) of 1 nm. Due to the short tether length of the hairpin constructs, a force calibration relating the magnet position to force on the magnetic bead could not be performed. Instead, we relied on an average force calibration obtained with the same magnet configuration and magnetic beads with longer (> 10 kb) dsDNA tethers [15]. Relying on an average force calibration results in an ~ 10% uncertainty in the applied force for any given bead, based on the standard deviation of the force calibration distribution [31]. The kinetics of hairpin unwinding by RecQ helicase are largely insensitive to the applied force [21, 31], so the uncertainty in the applied force has little impact on the unwinding behavior of RecQ helicase. However, the change in extension as the hairpin is unzipped depends on the applied force. To eliminate small differences in the relative extension of the unwinding trajectories, they were normalized by the extension difference between the fully closed and fully open hairpin extensions, which is readily extracted from histograms of the raw unwinding trajectories. For gapped DNA, the change in extension results from the conversion of one base-pair of dsDNA to one nucleotide of ssDNA. At 15 pN, the extension of dsDNA is 0.326 nm/bp and that of ssDNA is 0.469 nm/nt resulting in a net increase in extension of ~0.14 nm per bp unwound.
2.4.4 T-test based step-detection analysis to extract helicase kinetics from single-molecule unwinding records
Unwinding trajectories are analyzed to determine kinetic parameters including unwinding and translocation rates, processivity (length of DNA unwound prior to dissociation), the chemical step-size (the number of basepairs unwound per NTP hydrolysis), and the kinetic step-size (the number of basepairs unwound between rate limiting kinetic steps). Helicases, similar to other biological motors, move along their substrate in a step-wise manner. Thus unwinding trajectories are often analyzed with a step detection method. Several step detection methods have been developed to analyze single-molecule trajectories of molecular motors [38–42]. All of these methods assume discrete, essentially instantaneous, step transitions that reflect the fundamental discrete motion of motor proteins moving on polymeric tracks with well-defined unitary steps. However, these approaches do not perform well with trajectories with continuous transitions between dwells for which the instantaneous transitions are either not observed due to the rate or extent of the individual transitions, or for which the underling motion is continuous, such as diffusive motion. We have developed an analysis routine to extract step-sizes and characterize the transitions between dwells for such trajectories. The routine is adapted from a T-test step finding approach that we have extended to handle continuous transitions [38]. The routine requires neither prior knowledge of the step sizes nor a fixed step size [38, 39, 43]. The method is based on the Students t- test, a statistical test that determines the probability that two data sets represent the same underlying distribution [38]. Prior to t-test analysis, the trajectory is smoothed with a 4th order Savitzky-Golay 7 point filter to increase the signal to noise ratio with minimal distortion of the trajectory. By applying a running t-test statistic comparing n prior points to n subsequent points, both instantaneous and continuous transitions in the data can be identified.
For each point i in the extension versus time trajectory the t-statistic (t’) is calculated for two windows (μ1 and μ2) each of n points (Fig. 3):
| (1) |
Where x̄ and s are the mean and the standard deviation of the window μ, respectively. The t’ value is then compared to the critical T value (Tc) for determining if μ1 and μ2 represent different distributions, i.e., if |t’| ≥ Tc, μ1 ≠ μ2. Tc depends on the degrees of freedom, ν, and the significance or probability threshold for rejecting the null hypothesis, α, that is computed numerically by solving the cumulative distribution function, F(x) for x at which F(x) = α/2 (x<0, lower Tc) and F(x) =1-α/2 (x≥0; upper Tc) in the two-tailed test [44]. The expressions for ν and F(x) are as follows:,
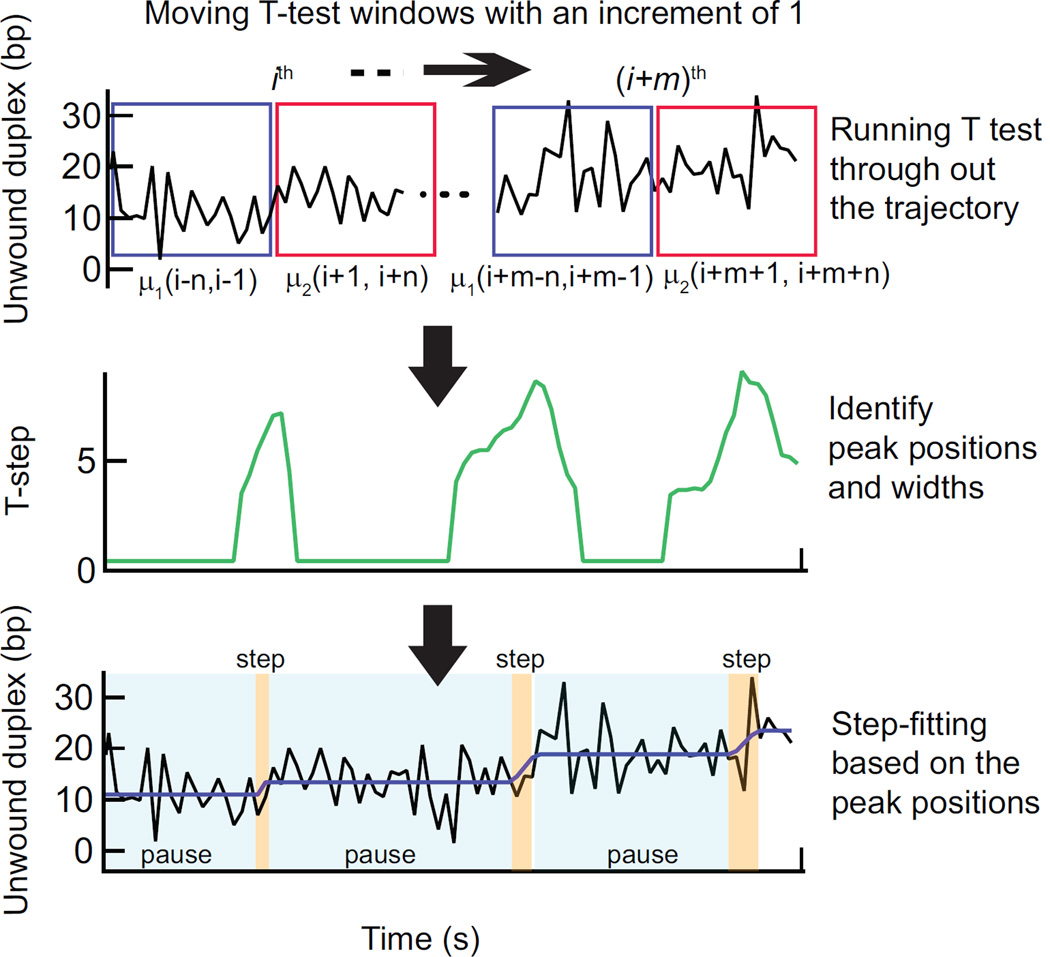
Figure 3.
T-test step detection method. At each point i in the time trajectory a Student’s T-test is performed on two windows of n points prior and subsequent to the ith point (indicated as blue and magenta boxes) (Top). T-test analysis results in a T-step profile (high-pass filtered absolute value of the t-statistic). The peaks correspond to the center of the transitions in the time trajectory and the widths of the peaks are proportional to the duration of the transitions. The peak positions and respective widths are identified (middle). These points are filtered to remove spurious transitions and then used to produce a trial fit of the data (bottom, black line) to a series of dwells (pauses) and runs (steps) of finite duration. Based on this initial fit, the entire data set is then fit with an all-at-once function of the same form. This fit (purple line) provides the durations and amplitudes of pauses and steps (Bottom).
The T-test returns two parameters; a binary test of the null hypothesis, H0 (= 1 if μ1 = μ2 or 0 if μ1 ≠ μ2) and the t-statistic, t’. The product, (1-H0)|t’|, that we define as the T-step profile is a continuous measure of the differences between μ1 and μ2 that has been filtered by the acceptance criteria α. The resulting curve is non-zero only for portions of the trajectory where the probability that the windows are different exceeds α (Fig. 3). Transitions in the extension data are associated with peaks in this T-step profile and the duration of each transition is reflected in the width of the peak (defined as the standard deviation of the Gaussian fit to the t-step profile). The corresponding points in the original trajectory define an idealized trajectory consisting of dwells of constant position between transitions (steps). The step information, i.e., the positions of the steps and the dwells between steps obtained from this analysis, is subsequently filtered to eliminate steps and dwells below the temporal and spatial resolution limits imposed by the noise in the data and the t-test window size, n. For example, steps less than 3 bp and dwells with durations less than 3 points (0.015 sec) are considered as false-positives and thus removed. This filtered set of points serves as the initial guess for an all-at-once fitting function that represents the trajectory as a series of dwells connected by runs of finite duration and velocity (Fig. 4a). The window size, n, is the most important user-defined parameter in the T-test analysis method. In general, n should be sufficiently small to preserve the temporal resolution, since increasing n broadens the T-step values and subsequently decreases the temporal resolution (Fig. S1). However, decreasing n reduces the signal to noise and reduces the statistical probability of identifying steps (Fig. S2). The optimum window size thus represents a trade-off between temporal and spatial resolution. Although the optimum window size depends on the noise level, the step size, and the stepping kinetics, as a rule of thumb, ideally n is large enough to achieve 90–95% rejection of false steps for a given noise amplitude in the signal and small enough so that it is less than the average dwell time. To go beyond these initial guidelines for optimizing the T-test parameters for a given data set, and, equally as importantly, to quantify the unavoidable distortion of the data by the analysis routine, requires analyzing simulated data sets mimicking the experimental data (Fig. 4).
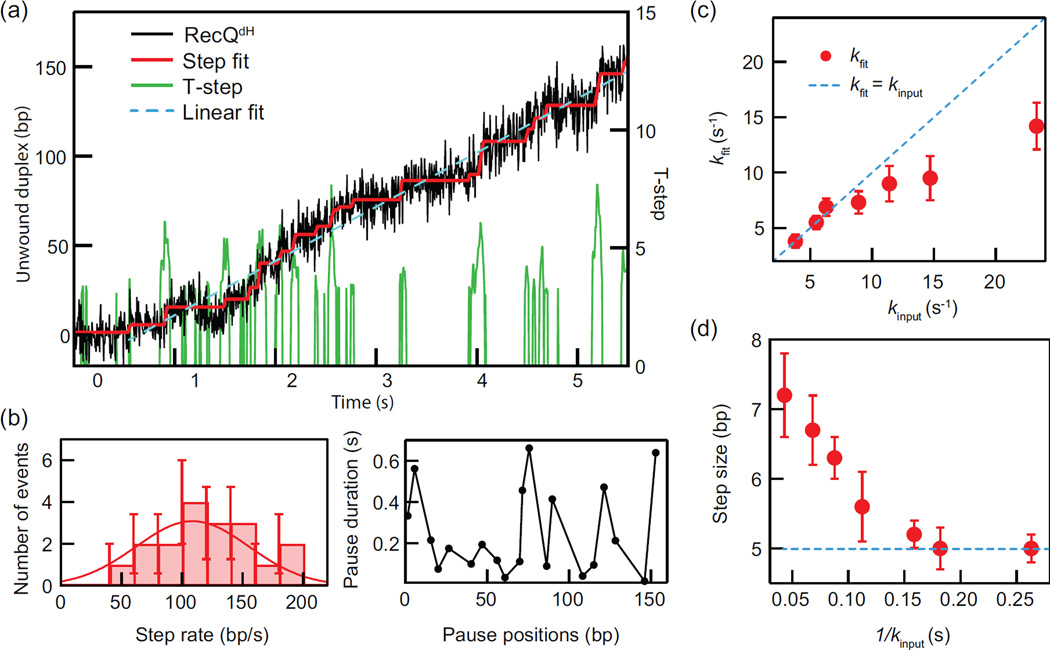
Figure 4.
Example helicase unwinding trajectory (F = 8 pN) and T-test based analysis. (a) The hairpin unwinding trajectory of RecQdH analyzed with the T-test step detection routine yielding T-step profile with n = 20 (0.1 s for 200 Hz frame rate) and α = 0.005. Step fitting of the trajectory was based on the step positions and widths obtained from the peaks in the T-step profile. Pause positions and durations are determined from the step fit as are the “step rates” corresponding to the slopes of the steps between pauses. The mean unwinding rate is obtained from the linear fit to the entire trajectory. (b) The step rates from the step fitting were binned and fitted with a Gaussian to obtain a mean step rate. Average pause durations from the T-test analysis were plotted as a function of pause position. (c) The mean pause escape rates from T-test analysis of simulated data, kfit are plotted as a function of the pause escape rates used in the simulations, kinput. The dashed blue line corresponds to kfit = kinput. (d) Step sizes determined from T-test analysis of simulated data are plotted as a function of the average pause durations used in the simulations, 1/ kinput. The input step-size of 5 bp is indicated as a dashed blue line.
3. Results
3.1 single molecule measurements of DNA unwinding by RecQ helicase
The trajectory of hairpin DNA unwinding by RecQdH (F = 8 pN) shows steps and pauses followed by dissociation from the end of the hairpin substrate (Fig. 4a). T-test analysis using n = 20 and α = 0.005, identifies the steps and pauses from the trajectory and yields the unwinding rates (step-rate in bp s−1), pause durations and pause positions (please see subsection 2.4.4. for details). The mean unwinding rate is obtained by fitting a line from the initiation of unwinding to the maximum unwinding position before either the enzyme pauses or falls off yielding 30.3 ± 0.2 bp/s (Fig.4a). The distribution of step rates is well-fit with a Gaussian distribution to reveal an average step rate of 109 ± 14 bp/s and the average pause duration plotted as a function of pause position can be related to the DNA duplex stability (Fig. 4b). To evaluate the capabilities and limitations of the T-test analysis, we performed stochastic simulations of pauses and steps, varying the pause escape rate (3–23 s−1) while keeping the other parameters fixed (Step size = 5 bp, noise = 5 bp SD). We then obtained the step sizes and pause durations of the simulated trajectories using the same T-test analysis parameters used for the experimental data (n = 20 and α = 0.005). Simulated trajectories contained 35 steps with added Gaussian noise with a standard deviation of 5 bp, close to the experimentally measured value in DNA hairpin unwinding assays. The mean pause escape rates from the fitting, kfit, were slower than the average input pause rates, kinput for kinput > 10 s−1 but the two rates were comparable for kinput < 10 s−1. This temporal limitation is largely associated with the T-test window size, n. In our analysis n was set to 20 points corresponding to a time window of 0.1 s at our camera frame rate (200 frames per second) (Fig. 4c). Similarly, the step-sizes from fitting approach the input step-size, 5 bp, for kinput < 10 s−1 (Fig. 4d). In general, increasing n decreases the standard error of the mean (noise) and subsequently enhances the spatial resolution in finding steps (smaller steps can be determined for a given noise) however it reduces the temporal resolution in determining steps. The stringency of the step finding routine increases as α decreases, we found that an α value below ~0.01 gave reasonable results for our data, but this should be tuned for each data set.
3.2 Unwinding rate measurements under varying Na+ concentration, ATP concentration, and temperature
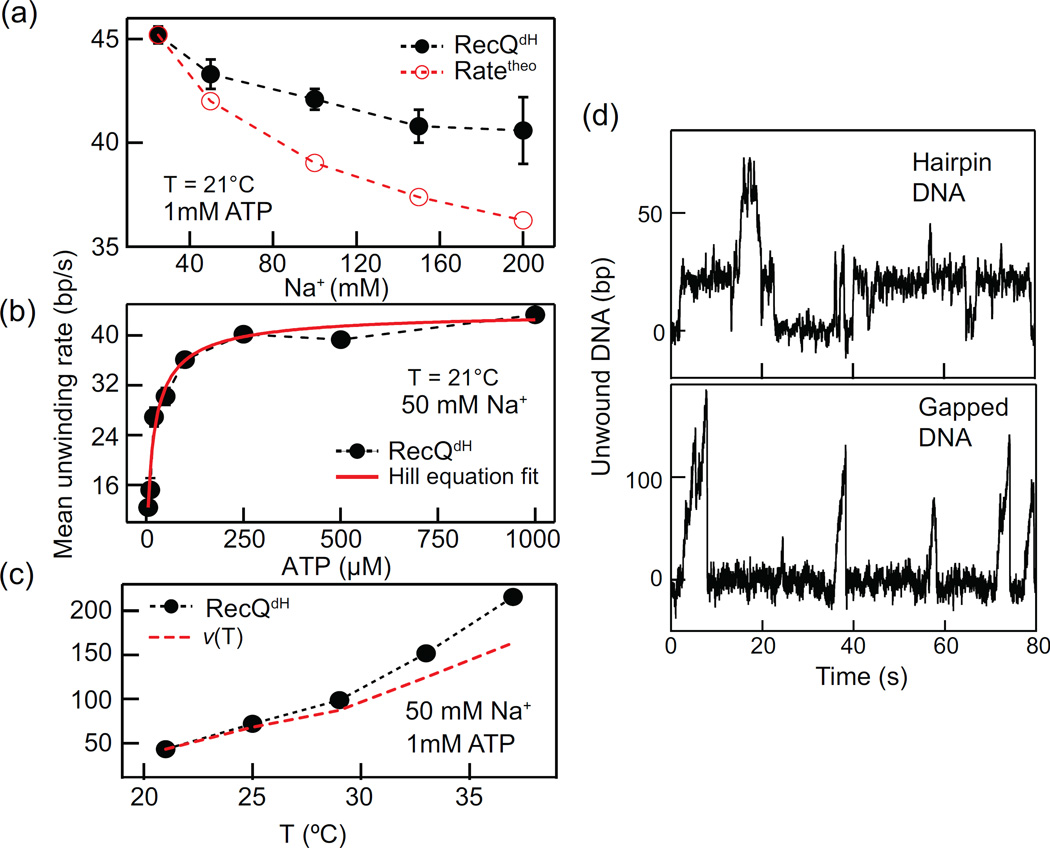
The mean unwinding rate (Fig. 4a) is a convenient way to characterize DNA unwinding activity of DNA helicases as it does not require a step detection analysis. Unlike the step rate, the mean unwinding rate is a convolution of stepping and pausing characteristics, and is an approximation of the ensemble average rate determined in ensemble kinetic assays. Thus, the average of the mean unwinding rates is a good measure of how the helicase unwinding activity varies under different conditions such as ionic strength, ATP concentration, and temperature. Varying these fundamental parameters provides important insights on at least two levels. Mechanistic insights concerning how the enzyme converts the energy of NTP hydrolysis to translocation and DNA unwinding can be obtained by relating changes in enzyme activity to the thermodynamic changes associated with varying the measurement conditions. Although it is impossible to completely recapitulate in vivo conditions in in vitro assays, we can nevertheless assess how enzyme activities may vary in vivo by investigating the effects of physico-chemical parameters. For example, some catalytic activities such as unwinding rate and processivity can be strongly influenced by ionic conditions. To address mechanistic questions and estimate the unwinding activity of RecQ under varying ionic strength, we first investigated how the mean unwinding rate is affected by varying the Na+ concentration using RecQdH (Fig. 5a). The mean unwinding rate decreased with increasing Na+ concentration from 25 mM to 200 mM (Fig. 5c), which may be explained by the increase DNA base-pair energy [45]. However, the measured Na+ concentration dependent rates decreased much less than predicted based on the calculated Na+ concentration-dependent stabilization of the DNA duplex. This is in line with previous findings suggesting that the unwinding activity of RecQ is relatively insensitive to changes in DNA base-pair energy [21].
Figure 5.
RecQ unwinding measurements under different conditions and DNA geometries. (a) Mean unwinding rate plotted as a function of Na+ concentration and compared to the calculated unwinding rate based on models in which the rate is determined solely by the DNA base-pair energy. (b) Mean unwinding rate plotted as a function of ATP concentration and fit with the Hill equation (see main text). (c) The mean unwinding rate plotted as a function of temperature and compared to the predicted rate as a function of temperature: ν(T) = ν(21°C)T/T(21°C). (d) RecQwt shows dramatically different unwinding behavior with two different DNA geometries. RecQwt slowly and inefficiently unwinds hairpin DNA due to long and frequent pauses, whereas it efficiently and rapidly unwinds gapped DNA.
We next measured the ATP concentration-dependent mean unwinding rate for RecQdH (Fig. 5b). ATP is the primary energy source powering the unwinding and translocation activity of most DNA helicases. The ATP dependent rate, ν(ATP) can be analyzed with a Hill equation [46]:
Here vmax is the maximum mean unwinding rate, Km is the ATP concentration at which the rate reaches a half of vmax, and n is the Hill coefficient. The analysis results in vmax = 43.9 ± 0.9 bp/s, Km= 15.7 ± 1.7 µM, and n = 0.8 ± 0.1, indicative of non-cooperative ATP binding. As a last example, we measured the temperature dependent mean unwinding rate of RecQdH. Temperature is one of the most important environmental factors that can affect the catalytic activity of an enzyme. However, temperature dependence of enzymatic rates is infrequently measured in single molecule assays partially due to the difficulty associated with implementing temperature control and mitigating temperature dependent drift. The mean unwinding rate for RecQdH sharply increased with increasing temperature. This change can be explained based on the assumption that the catalytic rate, ν, is proportional to exp(−dG‡/kbT) [47], where dG‡ is the activation energy associated with catalytic activity, kb is the Boltzmann constant, and T is the absolute temperature. Thus, with increasing temperature ν(T) increases as (ν0)T0/T (ν0 is the rate at temperature T0). As shown in Fig. 5c, the measured rates are comparable to the predicted rates from this simple calculation. The residual difference between the measured and predicted rates may be explained by the decreasing DNA base-pair stability due to the temperature increase, consistent with a relatively weak dependence of RecQ unwinding rate on the DNA base-pair stability.
3.3 DNA substrate geometry dependent unwinding
As discussed above, due to the increased signal to noise and facilitated measurements, the DNA hairpin geometry has become the predominant substrate for single-molecule DNA helicase unwinding assays. However, Ribeck and coworkers have shown that DNA substrate geometry can significantly affect helicase unwinding activity in single molecule manipulation measurements [48]. The effect of geometry likely arises from how external force is applied to the DNA substrate as shown for DnaB helicase [48]. In the hairpin geometry both the occluded strand (the strand on which the helicase binds) and the displaced or leaving strand are both under tension, whereas in gapped or “tailed” substrates only the occluded strand or the leaving strand is under tension, respectively. The observed differences in activity result from differences in how the helicase engages with these different substrate geometries and perhaps geometry-dependent differences in how tension is transmitted to the ssDNA-dsDNA junction. We have also observed DNA substrate geometry-dependent unwinding with RecQ helicase. The geometry dependence was largely dependent on the presence of an accessary domain, the HRDC. RecQdH lacking the HRDC was minimally sensitive to substrate geometry, unwinding hairpin and gapped DNA similarly. Wild type RecQ on the other hand unwound gapped DNA efficiently without significant pausing but frequently paused during unwinding of hairpin DNA (Fig. 5d), significantly decreasing the average unwinding rate. In light of these findings, it is apparent that for some helicases the substrate geometry can significantly influence the helicase unwinding activity. These differences can in turn reveal mechanistic details of the unwinding process that could be difficult to ascertain with other methods.
4. Conclusions
We provide procedures for collecting and analyzing single-molecule records of DNA helicase unwinding activity using magnetic tweezers. Although studies of specific helicases may require more specialized procedures relevant to their physiological functions, we focus on procedures that elucidate fundamental mechanistic aspects of DNA helicases such as kinetic step-size, unwinding mechanism, and the effect of environmental conditions that affect the catalytic activity of DNA helicases (ionic strength and temperature). We show details of a T-test based step-detection and trajectory analysis method that can accommodate continuous transitions. This approach is not limited to simply finding steps in the data but has been extended to provide unwinding kinetic parameters including dsDNA unwinding and translocation rates, mean unwinding rate, and pause positions and durations. Two important considerations concerning this analysis routine and data analysis routines more generally are that their performance is ultimately limited by measurement noise and that they necessarily distort the underlying data to varying degrees. We provide guidelines for ameliorating measurement noise by reducing the DNA tether length, for example, which improves both spatial and temporal resolution. Decreasing the unwinding rate is an alternative approach to effectively increase the resolution by increasing the number of data points per dwell resulting in a decrease in noise from increased averaging. This can be achieved by lowering the ATP concentration or alternatively by including non- (or slowly-) hydrolysable ATP analogues such as ATPγS.
We illustrate the limitations of T-test analysis with specific examples demonstrating the gradual loss of faithful representation of the step rate and size as the kinetic rates increase (Fig. 4c and d). These limitations may be difficult to overcome as they are related to the spatial and temporal resolution of the system as well as the noise and the inherent limitations of the analysis routine. However, it is important to recognize, and to the extent possible quantify, the non-ideal performance by analyzing simulated data combined with testing and characterization of the analysis routines. Extending this approach, the underlying motor kinetics could in principle be extracted from running the analysis routine on simulated stepping trajectories and tuning the simulation parameters to minimize differences between the experimental and simulation results from the analysis routine. Put another way, the simulations could be run considering the analysis routine used to analyze the data.
Finally, measuring helicase unwinding activity under a wide range of conditions can provide unanticipated mechanistic details concerning the unwinding process, while also ascertaining the specific activity under conditions of ionic strength, temperature and DNA substrate geometry approximating those found in vivo.
Supplementary Material
Highlights.
Magnetic tweezers afford robust single-molecule measurements of helicase activity
Student’s T-test based trajectory analysis resolves steps of finite duration
Simulations are an important tool for optimizing trajectory analysis routines
DNA substrate geometry can have a dramatic effect on helicase activity
Acknowledgments
We thank Lynda Bradly for comments on the manuscript. This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health (HL001056) and by the Human Frontiers Science Program (RGY0072/2010).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lohman TM, Bjornson KP. Mechanisms of Helicase-Catalyzed DNA Unwinding. Annual Review of Biochemistry. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 2.Fairman-Williams ME, Guenther U-P, Jankowsky E. SF1 and SF2 helicases: family matters. Current Opinion in Structural Biology. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lionnet T, Dawid A, Bigot S, Barre F-X, Saleh OA, Heslot F, Allemand J-F, Bensimon D, Croquette V. DNA mechanics as a tool to probe helicase and translocase activity. Nucleic Acids Research. 2006;34:4232–4244. doi: 10.1093/nar/gkl451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annual Review of Biochemistry. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 5.Sun B, Wang MD. Single-molecule perspectives on helicase mechanisms and functions. Critical Reviews in Biochemistry and Molecular Biology. 2015:1–11. doi: 10.3109/10409238.2015.1102195. [DOI] [PubMed] [Google Scholar]
- 6.Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Quarterly Reviews of Biophysics. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillingham MS, Wigley DB, Webb MR. Direct Measurement of Single-Stranded DNA Translocation by PcrA Helicase Using the Fluorescent Base Analogue 2-Aminopurine†. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 8.Fischer CJ, Lohman TM. ATP-dependent Translocation of Proteins along Single-stranded DNA: Models and Methods of Analysis of Pre-steady State Kinetics. Journal of Molecular Biology. 2004;344:1265–1286. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Bennett RJ, Keck JL. Structure and Function of RecQ DNA Helicases. Critical Reviews in Biochemistry and Molecular Biology. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 10.Khan I, Sommers JA, Brosh RM., Jr Close encounters for the first time: Helicase interactions with DNA damage. DNA Repair. 2015;33:43–59. doi: 10.1016/j.dnarep.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manosas M, Meglio A, Spiering MM, Ding F, Benkovic SJ, Barre F-X, Saleh OA, Allemand JF, Bensimon D, Croquette V. Chapter Thirteen - Magnetic Tweezers for the Study of DNA Tracking Motors. In: Nils GW, editor. Methods in Enzymology, Volume Volume 475. Academic Press; 2010. pp. 297–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg E, Ha T. Single-Molecule FRET Analysis of Helicase Functions. In: Abdelhaleem MM, editor. Helicases. Vol. 587. Humana Press; 2010. pp. 29–43. [DOI] [PubMed] [Google Scholar]
- 13.Dulin D, Berghuis BA, Depken M, Dekker NH. Untangling reaction pathways through modern approaches to high-throughput single-molecule force-spectroscopy experiments. Current Opinion in Structural Biology. 2015;34:116–122. doi: 10.1016/j.sbi.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seol Y, Neuman KC. In: Magnetic Tweezers for Single-Molecule Manipulation. Peterman E J, Wuite G, editors. Vol. 783. Humana press; 2011. pp. 265–293. [DOI] [PubMed] [Google Scholar]
- 16.Vlaminck ID, Dekker C. Recent Advances in Magnetic Tweezers. Annual Review of Biophysics. 2012;41:453–472. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- 17.Gollnick B, Carrasco C, Zuttion F, Gilhooly NS, Dillingham MS, Moreno-Herrero F. Probing DNA Helicase Kinetics with Temperature-Controlled Magnetic Tweezers. Small. 2015;11:1273–1284. doi: 10.1002/smll.201402686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaue D, Kobbe D, Kemmerich F, Kozikowska A, Puchta H, Seidel R. Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat Commun. 2013;4 doi: 10.1038/ncomms3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proceedings of the National Academy of Sciences. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manosas M, Perumal SK, Bianco PR, Ritort F, Benkovic SJ, Croquette V. RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat Commun. 2013;4 doi: 10.1038/ncomms3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Research. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry MP, McCall PM, Qi Z, Chemla YR. Characterization of Photoactivated Singlet Oxygen Damage in Single-Molecule Optical Trap Experiments. Biophysical Journal. 2009;97:2128–2136. doi: 10.1016/j.bpj.2009.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuman KC, Chadd EH, Liou GF, Bergman K, Block SM. Characterization of Photodamage to Escherichia coli in Optical Traps. Biophysical Journal. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeck N, Saleh OA. Multiplexed single-molecule measurements with magnetic tweezers. Review of Scientific Instruments. 2008;79:094301. doi: 10.1063/1.2981687. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q, He Y, Lu HP. Manipulating and probing enzymatic conformational fluctuations and enzyme-substrate interactions by single-molecule FRET-magnetic tweezers microscopy. Physical Chemistry Chemical Physics. 2014;16:13052–13058. doi: 10.1039/c4cp01454e. [DOI] [PubMed] [Google Scholar]
- 26.Kemmerich FE, Swoboda M, Kauert DJ, Grieb MS, Hahn S, Schwarz FW, Seidel R, Schlierf M. Simultaneous Single-Molecule Force and Fluorescence Sampling of DNA Nanostructure Conformations Using Magnetic Tweezers. Nano Letters. 2015 doi: 10.1021/acs.nanolett.5b03956. [DOI] [PubMed] [Google Scholar]
- 27.Lebel P, Basu A, Oberstrass FC, Tretter EM, Bryant Z. Gold rotor bead tracking for high-speed measurements of DNA twist, torque and extension. Nat Meth. 2014;11:456–462. doi: 10.1038/nmeth.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swoboda M, Grieb M, Hahn S, Schlierf M. Measuring Two at the Same Time: Combining Magnetic Tweezers with Single-Molecule FRET. In: Toseland CP, Fili N, editors. Fluorescent Methods for Molecular Motors. Vol. 105. Springer Basel; 2014. pp. 253–276. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA Helicases in DNA Repair. Annual Review of Genetics. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophysical Chemistry. 2010;149:67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Lionnet T, Allemand J-F, Revyakin A, Strick TR, Saleh OA, Bensimon D, Croquette V. Single-Molecule Studies Using Magnetic Traps. Cold Spring Harbor Protocols. 2012;2012 doi: 10.1101/pdb.top067488. pdb.top067488. [DOI] [PubMed] [Google Scholar]
- 32.Lipfert J, Hao X, Dekker NH. Quantitative Modeling and Optimization of Magnetic Tweezers. Biophysical Journal. 2009;96:5040–5049. doi: 10.1016/j.bpj.2009.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kocsis ZS, Sarlós K, Harami GM, Martina M, Kovács M. A Nucleotide-dependent and HRDC Domain-dependent Structural Transition in DNA-bound RecQ Helicase. Journal of Biological Chemistry. 2014;289:5938–5949. doi: 10.1074/jbc.M113.530741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulin D, Cui TJ, Cnossen J, Docter Margreet W, Lipfert J, Dekker Nynke H. High Spatiotemporal-Resolution Magnetic Tweezers: Calibration and Applications for DNA Dynamics. Biophysical Journal. 2015;109:2113–2125. doi: 10.1016/j.bpj.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seol Y, Neuman K. Single-Molecule Measurements of Topoisomerase Activity with Magnetic Tweezers. In: Mashanov GI, Batters C, editors. Single Molecule Enzymology. Vol. 778. Humana Press; 2011. pp. 229–241. [DOI] [PubMed] [Google Scholar]
- 36.te Velthuis AJW, Kerssemakers JWJ, Lipfert J, Dekker NH. Quantitative Guidelines for Force Calibration through Spectral Analysis of Magnetic Tweezers Data. Biophysical Journal. 2010;99:1292–1302. doi: 10.1016/j.bpj.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dessinges M-N, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter BC, Vershinin M, Gross SP. A Comparison of Step-Detection Methods: How Well Can You Do? Biophysical Journal. 2008;94:306–319. doi: 10.1529/biophysj.107.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 40.Kerssemakers JWJ, Laura Munteanu E, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 41.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 42.Syed S, Müllner FE, Selvin PR, Sigworth FJ. Improved Hidden Markov Models for Molecular Motors, Part 2: Extensions and Application to Experimental Data. Biophysical Journal. 99:3696–3703. doi: 10.1016/j.bpj.2010.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seol Y, Zhang H, Pommier Y, Neuman KC. A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proceedings of the National Academy of Sciences. 2012;109:16125–16130. doi: 10.1073/pnas.1206480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zar JH. Biostatistical Analysis. 4th. Englewood Cliffs, New Jersey: Prentice Hall, Englewood Cliffs, New Jersey: 1999. [Google Scholar]
- 45.Huguet JM, Bizarro CV, Forns N, Smith SB, Bustamante C, Ritort F. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proceedings of the National Academy of Sciences. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarlós K, Gyimesi M, Kovács M. RecQ helicase translocates along single-stranded DNA with a moderate processivity and tight mechanochemical coupling. Proceedings of the National Academy of Sciences. 2012;109:9804–9809. doi: 10.1073/pnas.1114468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniel RM, Danson MJ. Temperature and the catalytic activity of enzymes: A fresh understanding. FEBS Letters. 2013;587:2738–2743. doi: 10.1016/j.febslet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 48.Ribeck N, Kaplan DL, Bruck I, Saleh OA. DnaB Helicase Activity Is Modulated by DNA Geometry and Force. Biophysical Journal. 2010;99:2170–2179. doi: 10.1016/j.bpj.2010.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.