Abstract
Purpose
This work proposes the ISMRM Raw Data (ISMRMRD) format as a common MR raw data format, which promotes algorithm and data sharing.
Methods
A file format consisting of a flexible header and tagged frames of k-space data was designed. Application Programming Interfaces were implemented in C/C++, MATLAB, and Python. Converters for Bruker, General Electric, Philips, and Siemens proprietary file formats were implemented in C++. Raw data were collected using MRI scanners from four vendors, converted to ISMRMRD format, and reconstructed using software implemented in three programming languages (C++, MATLAB, Python).
Results
Images were obtained by reconstructing the raw data from all vendors. The source code, raw data, and images comprising this work are shared online, serving as an example of an image reconstruction project following a paradigm of reproducible research.
Conclusion
The proposed raw data format solves a practical problem for the MRI community. It may serve as a foundation for reproducible research and collaborations. The ISMRMRD format is a completely open and community-driven format, and the scientific community is invited (including commercial vendors) to participate either as users or developers.
Keywords: MRI, Image reconstruction, Open source, Raw data format
Introduction
Image reconstruction research has played a pivotal role in driving many advances in magnetic resonance imaging. Examples of paradigm shifting techniques include parallel imaging (1, 2, 3) and, more recently, the introduction of nonlinear reconstruction and compressed sensing (4, 5). Novel reconstruction algorithms build on and improve existing methodology, and most reconstruction articles compare new methods to existing methods in terms of image quality and reconstruction speed.
Reproducible research has drawn a great deal of attention recently, as highlighted for example in a recent special issue of Science (6, 7). The field of computational science in particular has produced several excellent examples of how such research can be carried out, e.g., the wavelab toolbox (8). The ISMRM has begun to take steps to help facilitate reproducible research, e.g., with the MRI unbound website (9). Underlying many of these efforts is a discipline-specific file format specification that allows scientists to exchange data easily. Much of modern astronomy research, for example, relies on telescope data in the FITS standard (10). Other discipline-specific data formats have enabled scientists to escape from the limitations imposed by vendor-specific, proprietary technology, leading to the development of a wide range of post-acquisition data analysis tools, and enabling large-scale collaborations. Medical imaging has the DICOM standard (11), which allows radiology departments to store data from different vendors on a centralized PACS, and computer scientists to implement novel image processing methods in a vendor neutral manner. The subfield of neuroimaging has NIfTI (12), which underlies large-scale projects like the human connectome project (13). The image file formats are useful for disciplines that operate on the reconstructed images, but they do not address the needs of scientists involved in the development of image reconstruction algorithms. This paper proposes an MR-specific raw data format, which is designed to store data from MRI experiments before any reconstruction steps have taken place. Specifically, it is intended to capture the data in acquisition order before it is transformed by any operations such as filtering, interpolation, zero padding, or Fourier transforms.
This ISMRMRD standard was developed by a subcommittee of the ISMRM Sedona 2013 workshop. It is designed to capture the details of the MRI experiment in a way that permits image reconstruction. It is important to note that this goal is fundamentally different from that of proprietary vendor raw data file formats. Generally, proprietary vendor raw file formats are intended to capture protocol parameters for a particular pulse sequence at the time of the scan. The proprietary raw data file formats are intended to store the information needed to reproduce a particular experiment on a specific version of the scanner software and reconstruction framework. The tight coupling between 1) the scanner console user interface, 2) the pulse sequence control parameters, and 3) the image reconstruction control parameters, make the vendor formats depend on a great deal of proprietary, vendor-specific knowledge. The quantity of hidden information makes these formats inconvenient for data sharing. More importantly, the specific implementation details of a particular vendor’s scanner software architecture greatly influences the design of these file formats and the type of information that they contain. In contrast, the proposed standard is designed to facilitate the exchange of the raw k-space data along with the physics parameters of the data acquisition process to provide the information necessary for image reconstruction.
While the ISMRMRD format will continue to evolve as it attracts more users with a more diverse set of applications and needs, the basic format has remained stable for more than two years, and it makes sense to present it to the MRI community. This paper lays out the design and structure of the format and describes several software tools that have been developed to interact with data in the proposed format. A number of vendor-specific tools have been created for converting proprietary data formats to the proposed format. An overview of the current status of these data converters and information needed to locate them is also provided. As a demonstration of how the data format can be used, a phantom has been scanned on four different scanners, the data converted to ISMRMRD and reconstructed with several different open source reconstruction programs written in MATLAB (The MathWorks), Python, and C++ programming languages.
All software, data, figures, scripts to generate the figures, and the text in this manuscript are available as open source.
Methods
Design
The proposed ISMRMRD format combines a flexible header (XML-based) with fixed structures for the data. These are packaged together into a single file. A minimal raw data set is depicted in Fig. 1 and consists of two sections:
XML header. A flexible XML format document that can contain an arbitrary number of fields and accommodate everything from simple values (matrix sizes, etc.) to entire vendor protocols, etc. The purpose of this XML document is to provide parameters that may be meaningful for some experiments but not for others. This XML format is defined by an XML Schema Definition file ( ismrmrd.xsd).
Raw data. This section contains all the acquired data in the experiment organized as a sequence of data items. Each data item corresponds to a single data frame or chunk in an experiment, for example, a single line of data in a Cartesian acquisition or a single interleave in a multi-shot spiral acquisition. Each data item consists of a fixed-size header (C-struct) with encoding numbers, etc., along with the k-space data for all of the acquired channels, and (optionally) the k-space trajectory as depicted in Fig. 2. The raw data structures are defined in a C/C++ header file ( ismrmrd.h).
Figure 1.
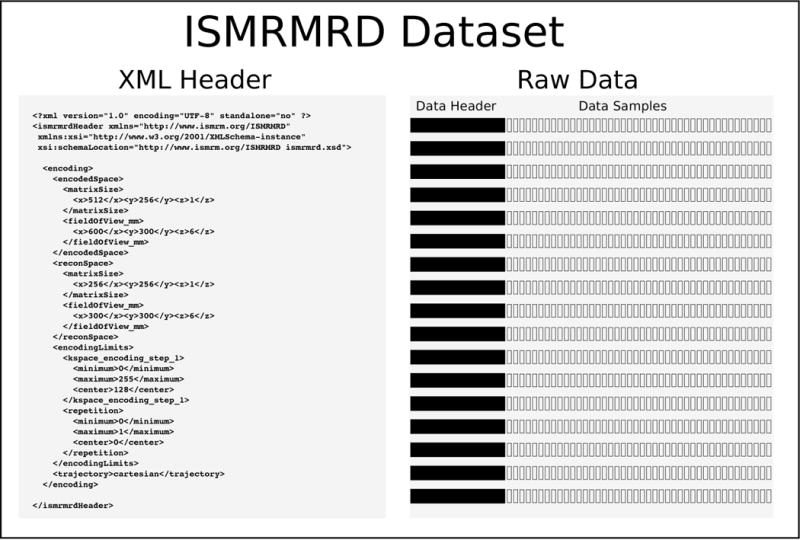
A minimal ISMRMRD dataset consists of a flexible XML header and raw data organized as sequence of data items consisting of fixed-size data headers and the corresponding k-space data for each set of samples or data chunk.
Figure 2.
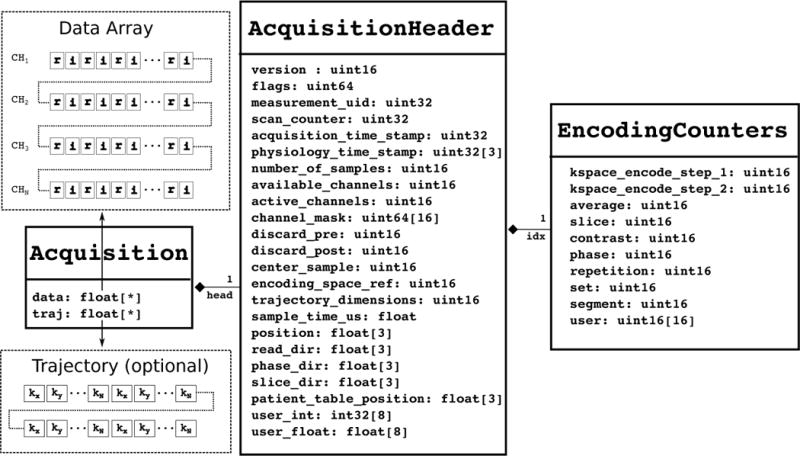
The raw data structure for each data frame or chunk of the acquisition, consisting of a fixed-size header with encoding numbers, location, etc. and the raw k-space data and (optionally) the k-space trajectory sampling locations. r and i indicate the real and imaginary part of the data points respectively.
The proposed format also provides a simple image format for storing the product of reconstructions and a multidimensional array format for storing additional user-specified parameters (e.g., gradient nonlinearity correction maps, etc.).
Encoding Space
One of the key design features of the proposed format is the notion of an “encoding space”, which is a description of the type and limits of the experiment and provides the reconstruction program with the physical size and resolution of the imaging volume and the ranges of the data header labels. An ISMRMRD dataset may contain data from several encoding spaces. Each encoding space is described in the XML header, and each acquisition (data chunk) is tagged with a label for the encoding space from which it was acquired. All encoding spaces have a trajectory type, an “encodedSpace,” a “reconSpace”, “encodingLimits”, and an optional trajectory description, that can contain an arbitrary set of named parameters such as the gradient ramp and flat top times for EPI, or the parameters controlling the shape of the spiral trajectory, etc. A somewhat more complex example of an encoding describing a simple 2D Cartesian acquisition is shown in Fig. 3. In this particular case, the encoding has the following description:
the “encodedSpace” section gives the matrix size (32 × 16 × 1) and field of view in mm (600 × 300 × 10) of the imaging volume encoded by the excitation, i.e., a 10 mm thick slice, 600 mm in x and 300 mm in y, encoded with a nominal matrix size of 32 in × and 16 in y, i.e., at a nominal resolution of 18.75mm.
The “reconSpace” section indicates that the image should be reconstructed on a smaller field of view (300 × 300 × 10), but with half the number of pixels in x.
The combination of the “encodedSpace” and “reconSpace” indicates that the k-space data were acquired on a grid oversampled by a factor of 2 in the x direction.
The “encodingLimits” section indicates the ky center (5) and the minimum and maximum ky values (0 and 11 respectively), i.e., this is a partial Fourier experiment, with a true pixel size that is somewhat larger than the nominal resolution.
Figure 3.
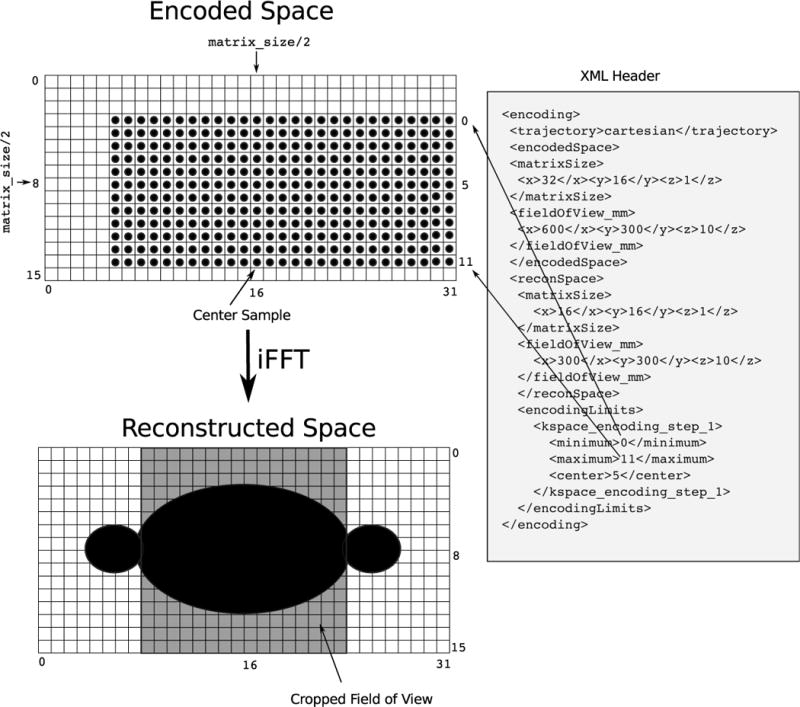
The encoding space of a simple 2D Cartesian acquisition. The XML header describes the image encoding and reconstruction fields of view and matrix sizes and k-space sampling bounding box. The image is acquired at a matrix size of 32 × 16 × 1 and field of view of 600mm × 300mm with a slice thickness of 10mm. It should be reconstructed at a matrix size of 16 × 16 × 1 and field of view of 300mm × 300mm with a slice thickness of 10mm. The k-space data were acquired on a grid oversampled by a factor of 2 in the x direction. The “encodingLimits” section indicates the ky center (5) and the minimum and maximum ky values (0 and 11 respectively), i.e. this is a partial Fourier experiment, with a true pixel size that is somewhat larger than the nominal resolution, min(ky) = −5, max(ky) = +6. The example experiment also employs partial Fourier along the readout dimension, i.e. asymmetric echo.
Encoding counters and flags
Each acquisition data header depicted in Fig. 2, contains a set of encoding counters: kspace_encoding_step_1, slice, etc. For a particular data set, the meaning of the encoding counters depends on the encoding space defined in the data set’s XML header. For the case of cartesian imaging, the mapping between ISMRMRD encoding counters and vendor specific dimensions is shown in Table 1. The encoding counters also include a set of 8 integers and 8 floating point numbers that can be used in an application specific way, for example they could refer to b-value and gradient direction in a diffusion weighted imaging sequence (see below). Or one could use them to label data from a spectroscopy or spectroscopic imaging experiment. Unfortunately, one of the natural counters for spectroscopy ( kspace_encoding step_0) was omitted from version 1 of the acquisition data header. The missing kspace_encoding_step_0 would be specifically useful for 3D chemical shift imaging (CSI) a.k.a. 3D spectroscopic imaging. This oversight will be corrected in version 2.
Table 1.
The relationship between ISMRMRD encoding counters for conventional cartesian imaging and vendor dimension labels.
| ISMRMRD | Bruker | GE | Philips | Siemens |
|---|---|---|---|---|
|
| ||||
| kspace_encode_step_1 | encode step 1 | frame | e1 | line |
| kspace_encode_step_2 | encode step 2 | – | e2 | partition |
| average | – | – | measurement | acquisition |
| slice | slice | slice | location | slice |
| contrast | echo | echo | echo | echo |
| phase | – | – | cardiac phase | phase |
| repetition | repetition | repetition | dynamic scan | repetition |
| set | – | – | row | set |
| segment | – | – | – | segment |
In addition to the encoding counters, each acquisition data header also contains a field for a set of flags (encoded as a 64-bit unsigned integer). These flags can be used to provide hints to the image reconstruction program, for example, an acquisition may be tagged with the flag ISMRMRD_ACQ_FIRST_IN_SLICE. The reader is referred to the header file (ismrmrd.h) for a list of available flags.
File size limitations
The maximum number of acquisitions per data set is ≈4×106 acquisitions because the scan_counter is stored as a 32 bit unsigned integer. This should be sufficient for current scanner technology. Each acquisition can contain 65535 samples because the number_of_samples is stored as a 16 bit unsigned integer. Both of these counters could be increased up to 64 bits, which is the current limit imposed by the the HDF5 file format on the number of elements (acquisitions) in a dataset.
Flexibility vs. structure
One of the main design goals of the format is to strike a balance between flexibility and structure, providing sufficient flexibility to enable the development of novel data acquisition and image reconstruction methods, within a sufficiently well-specified structure that allows for the standardization of accepted methods. It is necessary for the file format to specify a clear set of rules for data organization; this is achieved by the schema for the XML header and the definition of the data structures. For example, for a data set to be valid, it must consist of a minimal ISMRMRD XML header, i.e. one instance of the experimentalConditions, and one encoding space, as well as some number of acquisitions. More complexity is required to describe a spectroscopic imaging experiment, or a diffusion weighted imaging experiment, or an areterial spin labeling experiment, etc., and one would need to combine more information in the XML header and the encoding counters. For example, a data set acquired using a variable density spiral sequence would need to use the trajectory description portion of the encoding space. The XML header for such an experiment is shown in Table 2. The trajectory description provides an identifier and the set of parameters used in the trajectory design, and for this particular case, the encoding counter kspace_encoding_step_1 is used to indicate the interleaf number. At this early stage, it is difficult to foresee the ways in which different practitioners will chose to use the format for their specific applications, and more importantly, the challenge is not technical, but rather sociological. A consensus would need to emerge around the description of a particular type of experiment, and the community would need to agree on the meaning of the various parameters stored in the XML header and in the individual acquisition headers. The file format itself is designed to provide a framework within which these discussions can take place.
Table 2.
XML header encoding space for a variable density spiral sequence, providing an identifier and the set of parameters used to generate the trajectory. The reconstruction program would need to be programmed to use this information.
| <trajectory>spiral</trajectory> |
| <trajectoryDescription> |
| <identifier>HargreavesVDS2000</identifier> |
| <userParameterLong> |
| <name>interleaves</name> |
| <value>32</value> |
| </userParameterLong> |
| <userParameterLong> |
| <name>fov_coefficients</name> |
| <value>1</value> |
| </userParameterLong> |
| <userParameterLong> |
| <name>SamplingTime_ns</name> |
| <value>2600</value> |
| </userParameterLong> |
| <userParameterDouble> |
| <name>MaxGradient_G_per_cm</name> |
| <value>2.400000</value> |
| </userParameterDouble> |
| <userParameterDouble> |
| <name>MaxSlewRate_G_per_cm_per_s</name> |
| <value>14414.413920</value> |
| </userParameterDouble> |
| <userParameterDouble> |
| <name>FOVCoeff_1_cm</name> |
| <value>30.000000</value> |
| </userParameterDouble> |
| <userParameterDouble> |
| <name>krmax per cm</name> |
| <value>3.200000</value> |
| </userParameterDouble> |
| <comment>Using spiral design by Brian Hargreaves |
| (http://mrsrl.stanford.edu/~brian/vdspiral/) |
| </comment> |
| </trajectoryDescription> |
ISMRMRD Library
The ISMRMRD library is a cross-platform implementation based on the domain-independent Hierarchical Data Format (HDF) (http://www.hdfgroup.org/HDF5) for storage and provides C/C++, Python, and MATLAB (Mathworks) interfaces for reading and writing ISM-RMRD files. The project follows an open source development model with a website at (http://ismrmrd.github.io) and a discussion board and code repositories at http://github.com/ismrmrd. The library and associated tools can be compiled in a straightforward way on Linux, Windows, and Apple computers. This section provides a high-level description of the implementation and discuss some of the choices made during the development process. The reader is encouraged to examine and experiment with the source code for more a detailed view.
File container
The HDF format was chosen as the binary file container. The HDF format is very flexible and has been adopted broadly in the scientific community. HDF5 originated with National Center for Supercomputing Applications (NCSA) in the late 1980s and has grown to become the file format used by a wide range of large scale projects in government, private sector, and academic communities, in earth-science, astronomy, nuclear physics, etc. HDF5 has also been adopted by projects in biomedical science, e.g. the Biological Observation Matrix (BIOM) format for “-omics” described at (http://www.biom-format.org). The HDF Group was spun-off from NCSA as a non-profit organization to support the format. The reader is referred to the HDF Group’s website for more information regarding projects using HDF5 (http://www.hdfgroup.org/HDF5/users5.html, and on-going and completed projects involving the HDF Group (http://www.hdfgroup.org/projects). Reading and writing from the HDF format are supported by most programming languages and computational environments. The HDF library has a core C and C++ API, is supported natively in MATLAB (it forms the basis for MATLAB’s own “.mat” file format starting with mat-file version 7.3 (MATLAB R2006b or later), and is supported in Python via the H5Py package (http://www.h5py.org). The HDF Group distributes a viewer (hdfview) and a set of command line utility tools for manipulating and interactive with HDF5 files (https://www.hdfgroup.org/products/hdf5_tools/). The flexibility of the HDF5 format can make its use somewhat complex, for example the format supports multiple creation dates for the file and objects within a file (creation, last access, modification), sometimes causing HDF5 files containing the exact same data to have different checksum values, and making the use of a tool such as the h5diff utility necessary for the accurate comparison of two HDF5 files. Therefore, one of the goals of the implementation of the ISMRMD library is to provide a domain specific layer that hide the details of the HDF5 file format.
XML header data binding
The XML header can be thought of as a text representation of relevant experimental parameters that are needed for meaningful image reconstruction. When writing image reconstruction software, the software developer usually interacts with a binary representation of the XML document (XML Data Binding) to avoid direct, error prone, and tedious interaction with the XML text. The process of generating the binary representation of the XML document (deserialization) and the generation of the XML document from the binary representation (serialization) is handled by dedicated functions. Both the binary representation and the functions for serialization and deserialization can be auto-generated by XML data binding tools, e.g., CodeSynthesis XSD (http://www.codesynthesis.com/products/xsd/) for C++, JAXB for Java (https://jaxb.java.net/), or PyXB (http://pyxb.sourceforge.net/) for Python. The auto-generated data structures and functions are easier to maintain since they can be regenerated when the XML schema ( ismrmrd.xsd) changes, but the syntax they provide can be less convenient. Alternatively, data structures and serialization and deserialization code can be “handcrafted”. This may provide a more convenient interface, but the software maintenance overhead is greater. The proposed data format does not dictate which approach to use when interacting with the XML header, but the provided libraries include data structures and associated functions. Depending on the convenience of available tools these software components have either been “handcrafted” or auto-generated as described below.
Programming language-specific implementations
This work is implemented in three different programming environments or languages to interact with ISMRMRD files. This section contains a few notes regarding the C/C++, MATLAB, and Python APIs that were created:
C library. This code defines fixed data structures ( ismrmrd.h) and the functions used to interact with the HDF5 file.
C++ library. This code is a wrapper around the C library with classes and functions that provide memory management and reduce programming error. It includes a “handcrafted” XML header binding class.
Build system. The C and C++ implementation use the CMake (Kitware https://cmake.org) build configuration system to support multiple operating systems (Linux, Microsoft Windows, Apple OS X).
MATLAB library. This code consists of pure MATLAB data structures and functions (classes) that call the MATLAB interface to HDF5 as well as “handcrafted” Java code for the XML header binding class using the Java provided by MATLAB.
Python library. This code consists of pure Python classes for the data structures, uses NumPy arrays for the data and the k-space trajectories (http://www.numpy.org), and an auto-generated XML header binding class.
The code repositories also contain the source code for the documentation (which use the Doxygen (http://www.doxygen.org) and Sphinx (http://sphinx-doc.org) projects) as well as example utility programs demonstrating how to use the APIs to read and write ISMRMRD files. The utility programs depend on the Boost library (http://boost.org) for command-line user interaction.
Data Conversion Tools
Converters have been developed for several proprietary file formats: Bruker, General Electric, Philips, and Siemens. The Bruker, Philips and Siemens converters are open source with repositories that can also be found at http://github.com/ismrmrd. The GE converter relies on proprietary vendor code and is being distributed via the vendor’s research network. Given the variety in vendor raw data file formats, the architecture of these programs is described in general terms with the aim of giving some guidance or aid to others who are implementing their own conversion tools. The reader is referred to the source code for details, but to give a few examples: the Bruker raw data are stored in a simple directory structure with the acquisition parameters stored in several text files and k-space data stored in a simple binary file as equal length frames of k-space data; there are no data labels associated with each data frame. Siemens raw data are stored in a single integrated file that can contain raw data from multiple scans. For each scan, acquisition parameters are stored in a structured text header, followed by k-space data frames stored in acquisition order, where each frame of k-space data is tagged with data labels containing the frame’s line number, slice number, location, etc. The complexity of the GE and Philips formats fall somewhere between the simple file structure of the Bruker format and the more integrated file format used on the Siemens systems. The GE data does not contain explicit labels for each data frame and more sequence knowledge is needed to convert the data. The Philips raw data format has data labels associated with each data frame and it was possible to design a more generic automated converter.
The converters for the aforementioned vendor formats were written in C++. All four converters use the CMake build system and depend on the ISMRMRD C/C++ library described above as well as Boost for user interaction. For XML file handling, the converters also depend on either 1) libXML2 and libXSLT (http://www.xmlsoft.org) on Linux and Apple, or 2) the Xerces-C XML parser library (http://xerces.apache.org/xerces-c/) on Windows. Acquisition parameters from the vendor-specific data were converted to an intermediate XML file (containing vendor-specific parameter names and values). This XML file was then translated into the vendor-independent ISMRMRD XML header file using a sequence-specific Extensible Stylesheet Language (XSL) style sheet. The frames of k-space data were processed in the order in which they appeared on disk. For formats with tagged data frames (Philips and Siemens), the proprietary binary header tags were parsed and used to populate the fixed-size ISMRMRD data header structures. For formats with untagged data frames (Bruker and GE), sequence and vendor-specific knowledge was used.
It is worth emphasizing that creating the converters required significant work and special knowledge of the vendor formats. However, once code was developed for converting raw data from one pulse sequence on a particular platform, it was straightforward to modify this code to handle other sequences. It is expected that other developers can modify these converters to handle currently unsupported product sequences or their own research sequences. It is also important to note that while some users may work on the implementation of these vendor-specific converters, many users will simply use the converted ISMRMRD files as input for the algorithms and thus be exposed less to these vendor-specific details as a consequence of using the proposed format.
It is also worth emphasizing that the source code for the Siemens, Philips, and Bruker converters has been released publicly since it does not contain propritary files. The parts of the code that interact with the vendor raw data files, parse header and data structures etc, can be reverse engineered in a tedious but straightforward manner. The source code for the GE converter currently contains proprietary code and cannot be shared publicly, it has been released through the GE collaborator network only. The authors have had discussions with GE regarding a new converter that could be shared more freely, but at the time of writing this article, it has not been completed.
Experimental Demonstrations
In a typical MR image reconstruction research project, one often tests and validates an algorithm or a particular implementation of an algorithm. This validation can be done using simulated data and data acquired from phantom or in vivo experiments. In the interest of reproducible research, a scientist could distribute the source code and the raw data sets that were used to generate the figures for the publication and perhaps additional code or data related to the testing or validation of the algorithm.
This section demonstrates how one could use the ISMRMRD format and APIs for the development, implementation, and testing of image reconstruction algorithms suitable for 2D experiments.
Example 1: Cross-vendor and cross-language demonstration
In this example, the k-space data are acquired using a multi-channel receiver array and the data are fully sampled on a Cartesian grid. The workflow for this demonstration project could form a subset of a full development cycle:
An initial implementation or proof of concept of the algorithm is implemented in a rapid prototyping language such as MATLAB or Python.
Synthetic data are generated in simulation and used to test/validate the prototype implementation.
Experimental data are collected and used to test/validate the prototype implementation.
A high performance production version of the algorithm is implemented in a compiled language such as C/C++ or Fortran. In some cases the high performance implementation may require specialized hardware such as a compute cluster or a graphical processing unit (GPU).
The synthetic and experimental data sets are used to test/validate the high performance implementation of the algorithm.
The high-performance implementation of the algorithm is given approval from a local ethics board and put into production in a clinical environment.
The prototype image research example project began with an outline of the steps of the simple 2D image reconstruction algorithm:
Data are read and stored in a buffer (Nkx,Nky,Ncoil).
The Fourier transform is computed in two dimensions (x,y).
The images are cropped if the data were acquired on an oversampled grid in the frequency encode direction.
The prototype implementation of this algorithm was written in Python ( do_recon_python.py) and MATLAB ( do_recon_matlab.m). The high-performance implementation of the algorithm was written in C++ ( ismrmrd_recon_cartesian_2d). All three implementations used the corresponding ISMRMRD APIs to read in an ISMRMRD format file and to save the reconstructed image.
A synthetic data set in ISMRMRD format was generated using a program written in C++ ( ismrmrd_generate_cartesian_shepp_logan). This program simulated a simple experiment collecting data from a single-slice object using an 8-element receive array in the presence of a small amount of noise. The k-space data were generated by multiplying the Shepp-Logan phantom with the receive profile of an 8-rung birdcage coil followed by Fourier transformation and the addition of randomly generated white Gaussian noise with standard deviation 0.05 to the real and imaginary part of each receiver channel.
Data sets were collected from a phantom (kiwi fruit) using scanners from four different vendors:
Bruker 4.7T with a single channel 10cm transmit/receive coil
GE 3T MR750 with a 32-channel head coil
Philips Achieva 3T with a 6-channel knee coil
Siemens 3T Skyra with a 32-channel head coil
Data were acquired using product 2D pulse sequences. The FOV=10 cm, slice thickness=2 mm, and matrix size (Nx=Ny=512) were matched, although no attempt was made to match the pulse sequence type or timings.
Example 2: Non-cartesian trajectories, nominal vs. corrected
A spiral imaging experiment set was used to illustrate the use of the optional k-space trajectory data included in the ISMRMRD format. Data were acquired on a 1.5T Siemens system using an interleaved variable density spiral trajectory (32 spiral interleaves, FOV = 30 cm, slice thickness = 5 cm, matrix size = 192). The parameters describing the nominal trajectory were stored in the encoding section of the ISMRMD XML header. Gradient impulse response function corrected k-space trajectories were predicted using the system-specific gradient impulse response function (14, 15) and stored along with the k-space data in the ISMRMRD data file. A reconstruction program written in MATLAB ( do_spiral_recon_matlab.m) was used to generate images using both the nominal and and corrected spiral trajectories.
Example 3: Accelerated EPI with GRE coil sensitivity calibration and SENSE reconstruction
An echo planar imaging (EPI) experiment was used to illustrate how data from multiple scans can be stored in a single ISMRMRD file. Three data sets were acquired on a 3T Siemens system using two pulse sequences:
a conventional fully-encoded gradient echo sequence
an accelerated (R=2) single-shot EPI sequence with 3 navigators per shot
a noise scan (collected by the EPI sequence)
The three data sets were stored in the same ISMRMRD file. A reconstruction program written in Python (do_epi_recon_python.py) was used to reconstruct images from the EPI time series as follows:
The noise data were used for pre-whitening.
The fully encoded GRE data were reconstructed (2D FFT).
The GRE images were used to estimate SENSE unmixing weights.
Each acquisition (one line in ky) of the data from the EPI sequence was reconstructed in the x-direction using gridding.
For each shot of EPI data, the three navigators were used to estimate EPI phase correction.
After phase correction, the EPI data (R=2) where reconstructed by FFT in the y-direction followed producing aliased images.
The SENSE unmixing coefficients were applied to the aliased images to produce a single un-aliased image.
Results
Experimental Demonstration
The raw data files were transferred off-line and converted into ISMRMRD using the vendor-specific converters described above. It was possible to convert the data from all four vendors. The resulting reconstructions are shown in Fig. 4.
Figure 4.
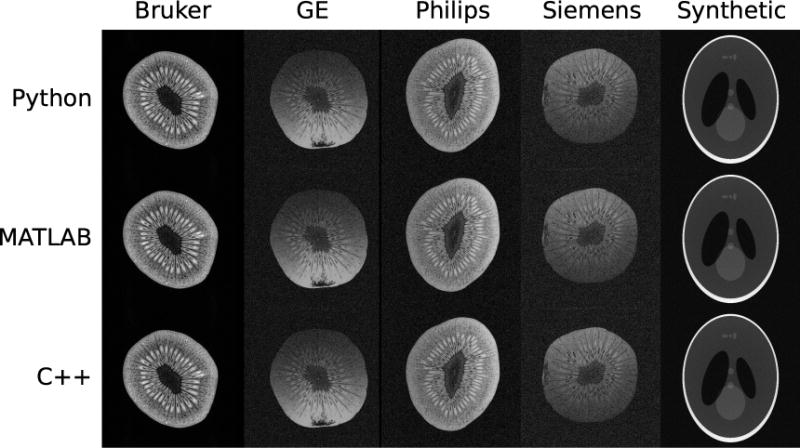
An experimental demonstration where data sets were acquired on scanners from four vendors and converted from the vendor proprietary raw data file formats into ISMRMRD format. A fifth data set was synthesized numerically and stored in ISMRMD format. The five ISMRMRD raw data sets were reconstructed using three image reconstruction programs written in C++, MATLAB, and Python. The resulting images are shown above, from left to right Bruker, General Electric, Philips, Siemens, and the synthetic data set, and from top to bottom C++, MATLAB, and Python reconstruction programs. The differences in SNR and shading between the images are due to the coil geometries: small volume coil vs. knee coil vs. head array.
Figure 5 shows spiral images reconstructed using nominal and corrected variable density spiral trajectories. The corrected trajectories are predicted by the sequence “on the fly” using system-specific calibration data and are stored in the ISMRMRD file along with the raw k-space data. Images reconstructed with knowledge of the corrected trajectories are significantly less distorted and can be produced both in real-time or retrospectively.
Figure 5.
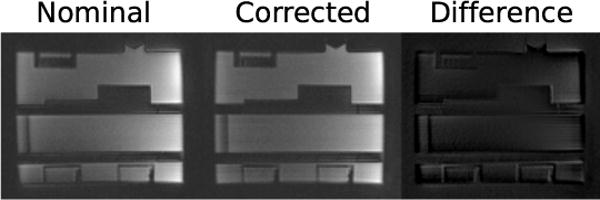
Images reconstructed with nominal variable density spiral trajectories compared to spiral trajectories stored in ISMRMRD format. ISMRMRD trajectories are predicted using contemporary gradient impulse response functions and therefore produce distortion-corrected reconstructed images.
Figure 6 shows images reconstructed from an accelerated EPI sequence. Data from a fully encoded GRE sequence (not shown) were used to estimate the coil sensitivities. Both data sets were stored in a single ISMRMRD file.
Figure 6.

Image reconstructed from an accelerated EPI sequence using SENSE. The coil sensitivities were estimated from a separate GRE sequence (not shown).
Source Code and Raw Data Dissemination
The source code for this project is being distributed in several git repositories hosted on GitHub at the URLs listed bellow. The SHA-1 hashes uniquely identify the particular revision of each of the repositories used for the production of this manuscript. Snapshots of the code repositories can also be found on the E.U funded Zenodo project website hosted at CERN, which provides a digital object identifier (DOI) for each release of each repository:
-
Manuscript LaTeX, figures and MATLAB and Python reconstruction:
https://github.com/ismrmrd/ismrmrd-paper
hash=7f79355e842f2459638921c02b671d7d5ca9c3e2
-
C++ API and C++ synthetic data generation and C++ reconstruction:
https://github.com/ismrmrd/ismrmrd
hash=c485787c98f4c2ad7ad6a2f9f5a0f4dc3927ea94
-
Python API:
https://github.com/ismrmrd/ismrmrd-python
hash=c38cd60c3a69416374c276b7820b0ffd70a6c4e0
-
Bruker converter:
https://github.com/ismrmrd/bruker_to_ismrmrd
hash=b945af3d37bff868d05b23b1dd67d0826f0a993d
-
GE converter (currently private):
Available via the GE collaboration network
-
Siemens converter:
http://github.com/ismrmrd/siemens_to_ismrmrd
hash=a740086b831c5b259aca36a41a3654b04c672b03
-
Philips converter:
https://github.com/ismrmrd/philips_to_ismrmrd
hash=581601fad04dee333ae6ff021d9ad1b39b3044b0
The raw data in vendor proprietary formats and in ISMRMRD format are being distributed via Zenodo (http://dx.doi.org/10.5281/zenodo.33166). Alternatively the raw data can be downloaded using the code/do_get_data.sh script, which is intended for Linux/Apple. Readers are encouraged to download the data and source code.
Discussion and Conclusion
This work presents an initial version of a standard for storing raw MR data and an implementation that supports several common programming languages and operating systems. It has also been demonstrated that data can be converted to ISMRMRD from proprietary vendor formats. Once the data have been converted to a vendor-independent format, generic reconstruction tools can be used to reconstruct data regardless of the scanner origin. The presented data converters are not complete for all proprietary configurations. However, the data converters are either completely available as open source or shared freely in the research communities associated with a specific vendor platform. Following the approach laid out in the source code for the data conversion software it should be a relatively simple task to accommodate other acquisition types.
The methods presented in this paper focused on a 2D Cartesian reconstruction example for simplicity, but more sophisticated reconstruction systems also use the ISMRMRD format. For example, the Gadgetron (16) framework uses the format as its internal data representation. The Graphical Programming Interface (17) has also been used with the ISMRMRD format using a set of externally provided compute nodes (https://github.com/hansenms/gpi_ismrmrd).
Future work will be focused on expanding the header sections in the flexible XML header to support specific applications. Changes to the fixed memory layout (C-struct) header and data may include capabilities to store arbitrary waveform data elements that could be used to capture detailed information about gradient waveforms or physiological telemetry. The format may also be changed to allow more choices for data storage formats and precision. Specifically, data is currently stored as 32-bit floating point values, which causes ISMRMRD files to be larger than the original data from vendors that use fixed point (integer) storage either in 16-bit format or in combination with variable length integer encoding (compression). The proposed format is flexible enough to accommodate different storage forms, possibly in combination with compression, which is easily supported by HDF5, but support for such features is not currently included in the software libraries.
The main aim of the proposed standard is that it will serve as a foundation upon which practitioners can base reproducible research and collaborations. A great deal of work remains and the authors ask for input from the ISMRM community to help shape the direction of the format. Specifically, the project would benefit from volunteers that have a detailed understanding of more specialized research applications in terms of which experimental parameters are needed to produce high quality reconstructions. The authors would also like to encourage groups with existing software packages to adopt this open data format as one of the input formats they support.
It is important to reiterate that the ISMRMRD format is a completely open and community-driven format. As vendors release new software versions, the members of the research community who are users of that platform will need to contribute their expertise to update the conversion tools and to ensure their correctness. As the research community develops new methods, members of the community will need to contribute their expertise to update the format. After informal discussions with ISMRM leadership, it has been decided that a formal governing structure in the form of a committee would not benefit the project at this stage, but a more structured mechanism for prioritization of changes may be needed at a later point in time as the format matures further. The authors and developers invite anybody in the community (including commercial vendors) to participate either as users of the format or as developers of the format or associated tools.
Acknowledgments
Multiple people participated in the discussions that led to this work. The authors acknowledge their contributions (in no particular order): Walter F. Block, Peter Boernert, David O. Brunner, Mark A. Griswold, Brian A. Hargreaves, Craig H. Meyer, Sonia Nielles-Vallespin, Tim Nielsen, Douglas C. Noll, James G. Pipe, Kaveh Vahedipour, and Ghislain Valliant.
This research was supported in part by the Intramural Research Program of the NIH, NHLBI, NIMH, NINDS.
References
- 1.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–62. [PubMed] [Google Scholar]
- 2.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn Reson Med. 1997;38:591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- 3.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47:1202–10. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 4.Donoho DL. Compressed sensing. Information Theory, IEEE Transactions on. 2006;52:1289–1306. [Google Scholar]
- 5.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–95. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 6.Jasny BR, Chin G, Chong L, Vignieri S. Again, and again, and again…. Science. 2011;334:1225–1225. doi: 10.1126/science.334.6060.1225. [DOI] [PubMed] [Google Scholar]
- 7.Peng RD, et al. Reproducible research in computational science. Science (New York, Ny) 2011;334:1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wavelab. WaveLab website. http://statweb.stanford.edu/~wavelab. Accessed August 13, 2015.
- 9.ISMRM MRI Unbound. ISMRM website. http://www.ismrm.org/mri_unbound. Accessed August 13, 2015.
- 10.FITS astronomical image and table format. NASA; website. http://fits.gsfc.nasa.gov/. Accessed August 13, 2015. [Google Scholar]
- 11.DICOM standard. Medical Nema website. http://medical.nema.org. Accessed August 13, 2015.
- 12.NIfTI neuroimaging informatics technology initiative. National Institute of Mental Health; website. http://nifti.nimh.nih.gov. Accessed August 13, 2015. [Google Scholar]
- 13.Human connectome project. Human Connectome website. http://www.humanconnectome.org. Accessed August 13, 2015.
- 14.Vannesjo SJ, Haeberlin M, Kasper L, Pavan M, Wilm BJ, Barmet C, Pruessmann KP. Gradient system characterization by impulse response measurements with a dynamic field camera. Magnetic resonance in medicine. 2013;69:583–593. doi: 10.1002/mrm.24263. [DOI] [PubMed] [Google Scholar]
- 15.CampbellWashburn AE, Xue H, Lederman RJ, Faranesh AZ, Hansen MS. Real-time distortion correction of spiral and echo planar images using the gradient system impulse response function. Magnetic resonance in medicine. 2015 doi: 10.1002/mrm.25788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen MS, Sørensen TS. Gadgetron: an open source framework for medical image reconstruction. Magn Reson Med. 2013;69:1768–76. doi: 10.1002/mrm.24389. [DOI] [PubMed] [Google Scholar]
- 17.Zwart NR, Pipe JG. Graphical programming interface: A development environment for MRI methods. Magn Reson Med. 2014 doi: 10.1002/mrm.25528. [DOI] [PubMed] [Google Scholar]


