SUMMARY
Cellular bodies such as P bodies and PML nuclear bodies (PML NBs) appear to be phase-separated liquids organized by multivalent interactions among proteins and RNA molecules. Although many components of various cellular bodies are known, general principles that define body composition are lacking. We modeled cellular bodies using several engineered multivalent proteins and RNA. In vitro and in cells, these scaffold molecules form phase-separated liquids that concentrate low valency client proteins. Clients partition differently depending on the ratio of scaffolds, with a sharp switch across the phase diagram diagonal. Composition can switch rapidly through changes in scaffold concentration or valency. Natural PML NBs and P bodies show analogous partitioning behavior, suggesting how their compositions could be controlled by levels of PML SUMOylation or cellular mRNA concentration, respectively. The data suggest a conceptual framework for considering the composition and control thereof of cellular bodies assembled through heterotypic multivalent interactions.
Graphical abstract
INTRODUCTION
Eukaryotic cells compartmentalize biological processes to achieve spatial and temporal control over biochemical reactions. Compartmentalization has long been studied in the context of membrane-bound organelles, where mechanisms of biogenesis and transport of molecules into and out of the organelle are well understood. Cells also contain numerous membrane-less organ-elles, collectively referred to as cellular bodies (Mao et al., 2011). These structures, which include P granules, P bodies, nucleoli, and promyelocytic leukemia nuclear bodies (PML NBs), are micron-sized assemblies of proteins and often RNA found in the cytoplasm and nucleoplasm of eukaryotic cells. They appear to be functionally important, as inferred from their conservation among evolutionarily distant species (Handwerger et al., 2005) and their tendency to concentrate functionally related groups of molecules (Mao et al., 2011; Mohamad and Bodén, 2010).
Ultrastructural analysis of cellular bodies suggests that they are porous structures with densities comparable to those of the nucleo- or cytoplasm (Handwerger et al., 2005). Analysis in live cells has revealed that, macroscopically, the bodies persist for hours to days. Yet, they are highly dynamic at the molecular level, turning over their contents within seconds to minutes (Dundr et al., 2004; Weidtkamp-Peters et al., 2008). Recent work has demonstrated that bodies exhibit liquid-like properties (Brangwynne et al., 2009; Brangwynne et al., 2011; Chen et al., 2008; Kroschwald et al., 2015; Patel et al., 2015; Wang et al., 2014). These and other behaviors suggest that cellular bodies are condensed phases that form through liquid-liquid phase separation of the nucleo- or cytoplasm (Hyman et al., 2014).
Cellular bodies are often enriched in multivalent molecules (Li et al., 2012)—proteins that harbor multiple modular domains or stretches of low-complexity amino acid sequence with repeated interaction motifs (Decker et al., 2007; Han et al., 2012; Kato et al., 2012; Reijns et al., 2008) or charged elements (Elbaum-Garfinkle et al., 2015; Nott et al., 2015); RNA species that contain multiple protein-binding elements; or combinations thereof. Interactions between multivalent macromolecules can drive polymerization-driven phase separation (Banjade et al., 2015; Fromm et al., 2014; Li et al., 2012; Mitrea et al., 2016; Nott et al., 2015), resulting in the formation of a condensed, droplet phase suspended in the bulk solution phase. It has been suggested that this fundamental macromolecular behavior may be an important principle governing the organization of cellular bodies (Fromm et al., 2014; Li et al., 2012; Mitrea et al., 2016; Nott et al., 2015). Indeed, expressing engineered multivalent proteins or ectopically tethering high copy numbers of body components (high local valency) in cells is sufficient to form dynamic, membrane-less puncta that resemble bona fide cellular bodies (Chung et al., 2011; Kaiser et al., 2008; Li et al., 2012).
Cellular bodies typically contain tens to hundreds of types of molecules (Buchan and Parker, 2009; Fong et al., 2013). Where characterized in detail, only a small number of these components appear to be essential for the structural integrity of the body (Clemson et al., 2009; Hanazawa et al., 2011; Ishov et al., 1999). We refer here to such molecules as scaffolds. In contrast, the remaining majority of components are dispensable for body assembly and often reside in the bodies only under certain conditions (Dellaire et al., 2006; Grousl et al., 2009). These molecules, which we refer to here as clients, often contain elements that specifically bind to elements in the scaffolds, often via low valency interacting elements of the same class as those in the scaffolds (e.g., Chalupníková et al., 2008; Lin et al., 2006). For example, P bodies assemble in part via scaffolding interactions between RNA binding proteins and RNA but also recruit several RNA binding proteins that are not important for P body assembly (Buchan and Parker, 2009). Within cellular bodies, clients diffuse much more rapidly than scaffolds (Dundr et al., 2004; Weidtkamp-Peters et al., 2008), suggesting that client-scaffold interactions are more transient than the interactions among scaffolds.
Compositional regulation is a general property of many cellular bodies and may be crucial to their function. Cellular body compositions change during the phases of the cell cycle or in response to stresses (Dellaire et al., 2006; Grousl et al., 2009). Despite their importance, the fundamental principles governing cellular body composition have been experimentally difficult to elucidate, owing to the complex nature of both scaffolds and clients and the diversity of species that reside within bodies. However, simplified model systems composed of few types of molecules, each with well-defined interaction elements, can help isolate key molecular parameters and thus have the potential to reveal generalizable concepts.
Here, we describe the biochemical and cellular behavior of three different sets of engineered molecules as simplified but representative multivalent scaffolds and low valency clients, which form model cellular bodies. Clients were differentially recruited into the bodies based on the relative stoichiometries of the scaffolds. Changes in client recruitment occurred sharply and on cellular timescales as the scaffold stoichiometries or valencies changed. Client partitioning also depended on client valency. These findings lead to a simple mass action model that predicts many features of the observed client partitioning behavior and suggests how cellular body compositions could be regulated in cells. Behaviors analogous to those of the model systems were observed in PML NBs in mammalian nuclei and P bodies in yeast cytoplasm. Thus, although natural cellular bodies are complex, their compositions may be governed by simple underlying rules and could be altered based on parameters that are easily tunable through cellular and evolutionary mechanisms.
RESULTS
Scaffold Stoichiometries Dictate Client Recruitment
We began by studying three independent pairs of interacting multivalent scaffolds in vitro. These systems consisted of (1) a protein with ten repeats of human SUMO3 (polySUMO) and a protein with ten repeats of the SUMO Interaction Motif (SIM) from PIASx (polySIM); (2) a protein with four repeats of the second SH3 domain from Nck (polySH3) and a protein containing four repeats of a Proline-Rich Motif (PRM) from Abl1 (polyPRM) (Li et al., 2012); and (3) the PTB protein (contains four RNA recognition motifs [RRMs]) and an RNA with five repeats of the RRM recognition element UCUCU (polyUCUCU) (Li et al., 2012). Each of these pairs phase separated when mixed together, but not when individual components were alone in solution (Li et al., 2012; Figure S1A; and data not shown).
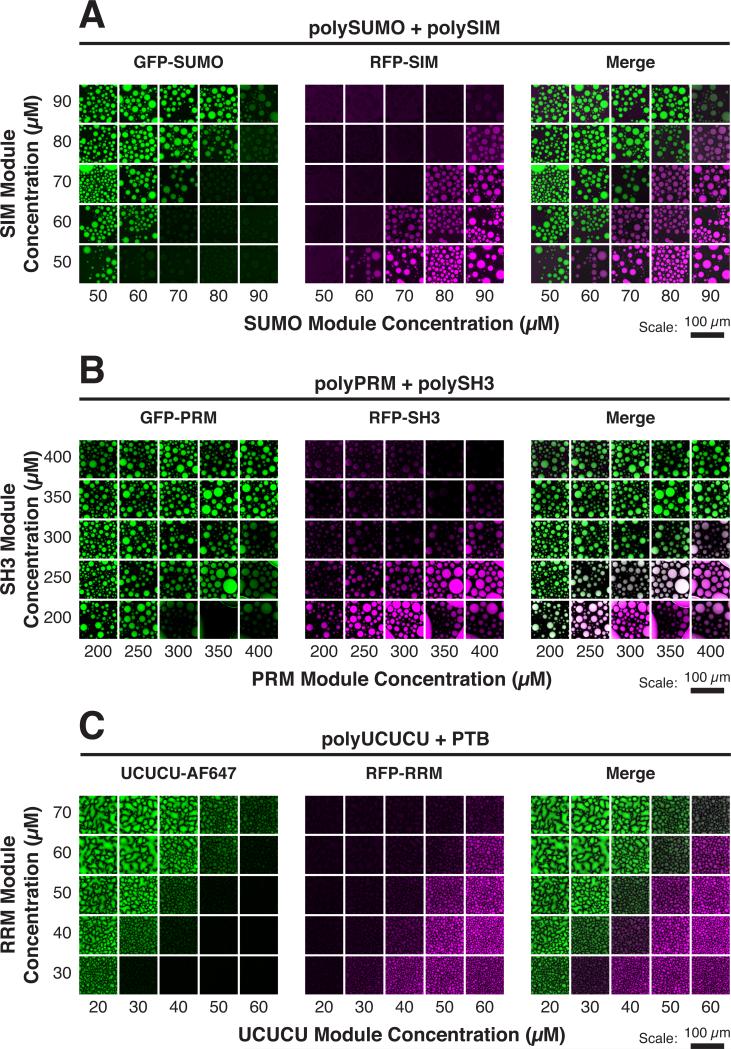
To model client recruitment into the bodies, we engineered a series of fluorescently labeled, monovalent clients (containing a single element that binds the scaffold) and characterized their partitioning into droplets generated by their cognate scaffolds. We mixed (1) GFP-SUMO and RFP-SIM (or GFP-SIM) with polySUMO/polySIM (Figure 1A); (2) GFP-PRM and RFP-SH3 with polySH3/polyPRM (Figure 1B); and (3) GFP-RRM and UCUCU-AlexaFluor647 (AF647) with PTB/polyUCUCU (Figure 1C). Partition coefficients (PCs) for the clients, defined as the ratio of concentrations in the droplet to the bulk phases, ranged from ~1 to 10 across the phase diagram (Figures S1B and S1D). Client recruitment in all three systems was qualitatively similar. Clients partitioned asymmetrically about the diagonal of the phase diagram (the line of equal scaffold stoichiometry) or near to it; each client was enriched only on the side where its cognate scaffold was in stoichiometric excess in the solution. For example, when polySIM was in excess (above the diagonal), GFP-SUMO was enriched in the droplets (PC ~3), but when polySUMO was in excess (below the diagonal), GFP-SUMO concentrated nearly equally in both phases (PC ~1) (Figure S1B). GFP-SIM showed an opposite pattern of enrichment (PC ~3 when polySUMO was in excess; PC ~1 when polySIM was in excess). Recruitment preference transitioned sharply, in switch-like fashion, as the diagonal was crossed. For the poly-SUMO/polySIM system, neither GFP alone nor clients mutated at their binding sites were enriched in droplets on either side of the diagonal (Figures S1B and S1C). Thus, binding to the scaffold proteins is necessary and sufficient for enrichment into the droplets (PC > 1).
Figure 1. Phase Diagram Position Dictates Client Recruitment.
Solutions of multivalent scaffolds plus the indicated clients were imaged for client fluorescence. AF, Alexa fluorophore.
(A) GFP-SUMO (green) and RFP-SIM (magenta) (100 nM each) were mixed with the indicated module concentrations of polySUMO and polySIM. (B) GFP-PRM (green) and RFP-SH3 (magenta) (200 nM each) were mixed with the indicated module concentrations of polyPRM and polySH3. (C) UCUCU-AF647 (green) and RFP-RRM (magenta) (200 nM each) were mixed with the indicated module concentrations of polyUCUCU and PTB.
See also Figure S1.
Together, these data show that, regardless of the molecular system, low valency clients partition asymmetrically into droplets formed by heterotypic scaffold interactions, with a sharp switch in client recruitment preference across the diagonal.
Valency of Client Affects Client Recruitment
Since the clients of a given cellular body can differ in their valencies, we examined how client valency affected partitioning. We fused to GFP 2 or 3 tandem repeats of SUMO or SIM and measured the PC for these clients across the polySUMO/polySIM phase diagram (Figure 2). Like their monovalent counterparts, the di- and trivalent clients partitioned into the droplets predominantly on one side of the phase diagram, transitioning sharply in their PCs across the diagonal. However, both the di- and trivalent clients had larger magnitudes of maximum partitioning than their monovalent counterparts, a feature that increased with valency: max PC was 19 and 37 for GFP-(SUMO)2 and GFP-(SUMO)3, respectively, and 21 and 61 for GFP-(SIM)2 and GFP-(SIM)3, respectively. In all cases, maximum partitioning occurred just past the diagonal, substantially enhancing the sharpness of the switch between client preferences. The increased partitioning was likely due to higher apparent affinity of the di- and tri-valent clients for the scaffold. Indeed, isothermal titration calorimetry (ITC) experiments verified that apparent affinity of the clients to cognate sites increases with increasing valency (Figure S2 and Table S1).
Figure 2. Client Valency Affects Partitioning.
PCs (means of duplicate samples) of the indicated clients (100 nM) into droplets formed by the indicated module concentrations of polySUMO and polySIM. See also Figure S2 and Table S1.
These data demonstrate that, in addition to position on the phase diagram, client valency can strongly influence client partitioning and thus droplet composition.
Mass Action Explains Switch-like Partitioning of Low Valency Clients
We sought to understand the origin of the switch-like nature of client partitioning. Our data suggest that partitioning depends strictly on SUMO-SIM interactions between clients and scaffolds (Figures S1B and S1C). We reasoned that client partitioning should be governed by the relative concentrations of available scaffold binding sites in droplets versus the bulk.
The apparent dissociation constant for polySUMO/polySIM (Kd ≤ 1 nM, based on ITC measurements with (SUMO)5/(SIM)5) is much less than the scaffold concentrations in either phase (Figure 3C), suggesting that most scaffold sites are occupied. Moreover, the apparent client-scaffold dissociation constants (Kd = 70–10,000 nM, estimated from ITC measurements with (SUMO)m + (SIM)m, for m = 1, 2 or 3) (Figure S2 and Table S1) are much weaker than the apparent polySUMO/polySIM affinity. Thus, clients should be poor competitors of scaffold-scaffold interactions. This analysis suggests that, in either phase, only the scaffold that is in stoichiometric excess will have free sites accessible to its cognate client. Conversely, the scaffold that is stoichiometrically deficient will effectively be saturated by scaffold-scaffold interactions and be invisible to its cognate client in either phase.
Figure 3. A Mass Action Model Predicts Client Partitioning Behavior.
(A) Imaging of polySUMO and polySIM (1% labeled with AF488 [green] and AF647 [magenta], respectively) fluorescence.
(B) PCs (means of duplicate samples) of polySUMO (left) and polySIM (right) calculated from imaging (see Experimental Procedures).
(C) Scaffold module concentrations (blue and yellow dots) in the droplet (top) and bulk (bottom) phases at the anti-diagonal data points from panel (B). To model client partitioning, values of concentrations were smoothed and interpolated with a cubic spline to yield continuous curves from discrete data. The continuous, interpolated values were used for subsequent calculations. Error bars represent SEM. Dotted line, phase diagram diagonal.
(D) Blue curve shows the ratio of free SUMO sites in the droplet phase to free SUMO sites in the bulk phase. Yellow curve shows the analogous ratio for free SIM sites.
(E) Mass action model for the partitioning of a low valency client, L, that binds to free scaffold sites R1 and R2 in the droplet and bulk, respectively (see Experimental Procedures).
(F) Predicted PC of clients as a function of affinity for scaffolds. Free site concentrations computed in (E) were used to parameterize the model (C) and predict partitioning of client as a function of their apparent affinity (ranging from 10−2–10−2 μM module) for the scaffolds (see Experimental Procedures).
See also Figures S3 and S4 and Table S2.
The scaffold composition of the droplet and bulk phases varied in a smooth, graded fashion across the phase diagram, with PCs ranging from 30–125 (Figures 3A and 3B). For most of the phase diagram, the PC of the two scaffolds was similar (within an 2-fold range), such that the droplets were essentially concentrated counterparts of the bulk solution (Figure 3C). Thus, at each point in the phase diagram, the scaffold in excess has a higher concentration of free sites in the droplet than in the bulk (Figure 3D) and consequently concentrates its cognate client into the droplets. The scaffold that is stoichiometrically deficient has few free sites in either phase, and its cognate client remains uniformly distributed. Since the stoichio-metric relationship between the two scaffolds switches abruptly in both phases near the diagonal, the capacity of the droplets to recruit one client over the other also switches abruptly.
We modeled client partitioning by mass action (Figure 3E). We allowed clients to equilibrate between two simulated phases while binding to free sites at concentrations computed from our experiments (Figure 3C, Table S1, and Supplemental Information). This simple mass action model suffices to recapitulate the key qualitative features of observed client partitioning (Figure 3F): (1) selective partitioning of clients, restricted to only one side of the diagonal; (2) a sharp change in partitioning as the diagonal is crossed; and (3) the dependence of partitioning on the apparent client-scaffold affinity. As described in the Supplemental Information, the model also predicts less intuitive features of the data, including non-monotonic partitioning, as well as dramatically high partitioning of one client and attenuated partitioning of the other near the diagonal (Figure 3F; Figure 2, trivalent clients; Figures S3 and S4).
Collectively, this analysis suggests that switch-like changes in client partitioning fundamentally arise from the sharp inversion of scaffold excess across the diagonal of the phase diagram.
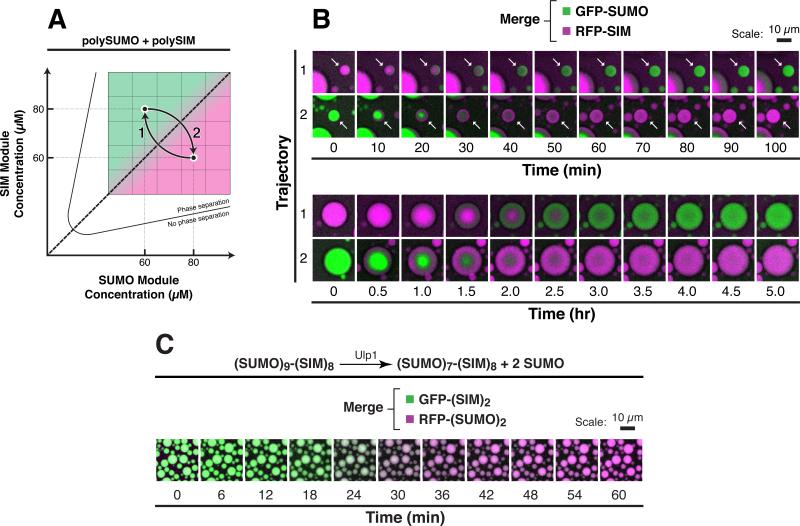
Compositional States Interchange on Cellular Timescales
Our data and analyses suggest how compositional states could be controlled by mass action. We wondered whether transitions between two compositional states were kinetically feasible on cellular timescales. We equilibrated polySUMO/polySIM droplets at a point on the phase diagram where only one of the clients, either GFP-SUMO or RFP-SIM, was preferentially enriched in the droplets (but both were present in solution). We then abruptly changed the concentration of the scaffold components to move the system to a point across the diagonal where the reciprocal recruitment preference was expected (Figure 4A). The droplets remained intact, spherical, and of relatively consistent sizes throughout the experiment. Within ~6 hr, all droplets expelled the initially enriched client in exchange for the other client (Figure 4B). Recruitment of the latter started at the outer edges of the droplets and moved inward, and smaller droplets exchanged clients more rapidly than larger droplets.
Figure 4. Droplets Interchange Composition on Cellular Timescales without Compromising Structural Integrity.
(A) Schematic of experiment. After equilibration of 100 nM GFP-SUMO and 100 nM RFP-SIM with polySUMO and polySIM at module concentrations of 60 μM and 80 μM, respectively, concentrations of the polySUMO and polySIM were abruptly shifted to 80 μM and 60 μM, respectively, for trajectory 1 and vice versa for trajectory 2.
(B) Time lapse imaging of droplets starting immediately after the abrupt change in concentrations of polySUMO and polySIM, showing merged, pseudocolored fluorescence signals from GFP-SUMO (green) and RFP-SIM (magenta). Note that small droplets (white arrows, top) interconvert more quickly than larger droplets (bottom).
(C) 6 μM of a (SUMO)9-(SIM)8 scaffold containing Ulp1 cleavage sites after only the two N-terminal SUMOs was equilibrated with 50 nM of GFP-(SIM)2 (green) and RFP-(SUMO)2 (magenta). Time lapse imaging was started immediately after addition of 10 nM of Ulp1. Pseudocolored images showing merged fluorescent signals from the two clients are shown.
In fluorescence recovery after photobleaching (FRAP) experiments, clients diffused much more rapidly within droplets than did scaffolds (i.e., for droplets ~20 μm in diameter, RFP-SIM and polySUMO had exponential recovery constants, t, of 1.3 min and 38 min, respectively; Figure S5 and Table S3). Thus, scaffold rearrangements likely limit the rate of transitions between compositional states. Scaling recovery times to droplets of 1 μm diameter, as often observed in cells, indicates that compositions should interchange on a timescale of ~6 s in natural systems (Table S3).
We previously demonstrated how covalent modifications of scaffolds could regulate the formation and dissolution of droplet phases (Li et al., 2012). We likewise wondered whether covalent modifications could also regulate droplet compositions. In cells, SUMO modifications are dynamically added by the SUMO ligase cascade and removed by SUMO proteases. We generated a single component, fused (SUMO)9-(SIM)8 scaffold that could be selectively cleaved by Ulp1, the yeast SUMO protease, to produce (SUMO)7-(SIM)8, mimicking natural deSUMOylation (see Experimental Procedures). Such (SUMO)m-(SIM)n (m ≠ n) fusions are essentially fixed on one side of the phase diagram diagonal and have client recruitment preferences analogous to the in trans systems (see Figure 5A). When mixed with GFP-(SIM)2 and RFP-(SUMO)2, (SUMO)9-(SIM)8 droplets recruited the former but not the latter client (Figure 4C). Ulp1 cleavage, which shifted the scaffold to the other side of the phase diagram diagonal, caused the droplets to expel GFP-(SIM)2 and recruit RFP-(SUMO)2. These data suggest that enzymatic modifications of cellular body scaffolds, such as SUMOylation and deSUMOylation, could robustly regulate body composition.
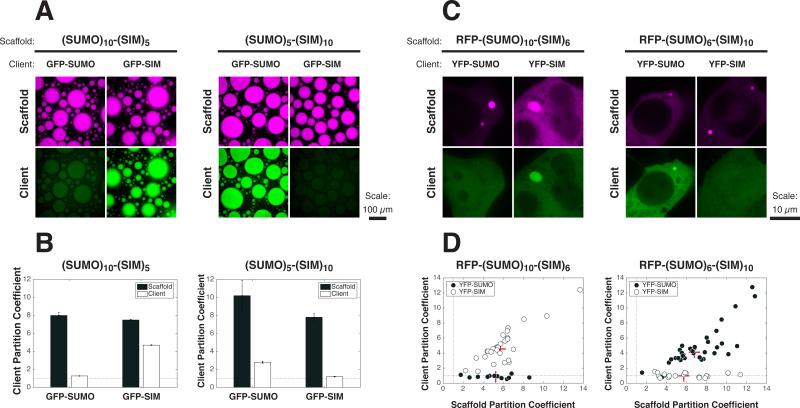
Figure 5. Cellular PolySUMO-PolySIM Puncta Selectively Recruit Low Valency Clients.
(A) 60 nM of GFP-SUMO or GFP-SIM (green) was mixed with 12 μM of (SUMO)10-(SIM)5 (left) or (SUMO)5-(SIM)10 (right) (1% RFP-tagged; magenta), and the resulting droplets were imaged for scaffold and client fluorescence.
(B) PCs for both scaffold (black bars) and clients (white bars) from experiment in (A). Graphs show averages from triplicate experiments. Error bars represent SEM. Dotted line, PC = 1.
(C) Live cell fluorescence images of YFP-SUMO or YFP-SIM (green) co-transfected with RFP-(SUMO)10-(SIM)6 (left) or RFP-(SUMO)6-(SIM)10 (right) (magenta) into HeLa cells.
(D) PCs of scaffolds and client components calculated from cells in the experiment. Each symbol represents the average PC into all puncta (typically 1-3) in a given cell (12-35 cells per sample) when the indicated scaffold was co-transfected with YFP-SUMO (black circles) or YFP-SIM (white circles). Dotted line, PC = 1. Red + sign, median PC.
See also Figure S6.
We conclude that droplets can transition, without compromising structural integrity, between substantially different compositional states on timescales accessible to cells. This can occur with only subtle changes in the concentration or covalent modifications of their polymer scaffolds.
Engineered Cellular Puncta Selectively Concentrate Low Valency Clients
We next asked whether the partitioning behavior observed in vitro could also be observed in cells. For these experiments, we used in cis [(SUMO)m-(SIM)n] scaffolds, which afforded tight experimental control of the relative module concentrations independent of absolute concentrations. Both (SUMO)10-(SIM)5 and (SUMO)5-(SIM)10 phase separated in vitro at micromolar concentrations. (SUMO)10-(SIM)5 droplets enriched GFP-SIM (PC = 4.7), but not GFP-SUMO (PC = 1.3), and (SUMO)5-(SIM)10 showed the reverse (PC = 2.8 for GFP-SUMO; PC = 1.2 for GFP-SIM) (Figures 5A and 5B).
We then individually expressed RFP-(SUMO)10-(SIM)6 or RFP-(SUMO)6-(SIM)10 in HeLa cells, where they each formed spherical, micron-sized puncta in the cytoplasm. In live cells, the puncta occasionally contacted each other and coalesced into larger structures, suggesting that they are phase-separated liquids (data not shown). When co-transfected with individual YFP-tagged clients, RFP-(SUMO)10-(SIM)6 puncta only concentrated YFP-SIM. Reciprocally, RFP-(SUMO)6-(SIM)10 puncta only concentrated YFP-SUMO. In both cases, neither YFP alone nor clients with mutations at their binding sites were enriched in cellular puncta (Figure S6A). We also obtained qualitatively analogous data using co-expression of in trans polySUMO and polySIM scaffolds, along with YFP-tagged clients (Figure S6B). However, experimental uncertainties in the relative concentrations of the scaffold components made it difficult to assign cells confidently to one side the diagonal.
Taken together, our data suggest that mass action-based compositional control can be achieved as robustly in cells as in vitro.
Scaffold Stoichiometries Control Client Recruitment into Natural Cellular Bodies
We sought to determine whether natural cellular bodies could exhibit compositional control analogous to our model systems. Wefocused ontwo naturalcellularbodies, PML NBsin mammalian nuclei and P bodies in the yeast cytoplasm, systems in which the interactions governing client recruitment were well characterized and where their stoichiometries were experimentally perturbable.
PML NBs are micron-sized, membrane-less organelles in mammalian nuclei that are involved in processes including DNA damage repair, apoptosis, and anti-viral responses (Lallemand-Breitenbach and de Thé, 2010). The PML protein appears to be the primary scaffold for these bodies (Ishov et al., 1999). PML can self-assemble via elements within its Tripartite Motif (TRIM) (Antolini et al., 2003; Huang et al., 2014) and also via binding of its conserved SIM element to SUMOs conjugated at up to eight sites in the protein (Nisole et al., 2013; Shen et al., 2006). Though not strictly required for body assembly (Brand et al., 2010; Sahin et al., 2014), SUMO-SIM interactions likely contribute substantially to body architecture, as deletion of the SIM motif or perturbations to PML SUMOylation via mutagenesis, viral infection, or knockdown/overexpression of SUMO ligases/proteases can cause changes in the size, number, morphology, or dynamics of PML NBs (Best et al., 2002; Hattersley et al., 2011; He et al., 2015; Müller and Dejean, 1999; Shen et al., 2006; Weidtkamp-Peters et al., 2008). SUMO-SIM interactions also appear to be critical for the recruitment of many PML NB clients (e.g., Daxx and Sp100) (Lin et al., 2006; Van Damme et al., 2010; Zhong et al., 2000).
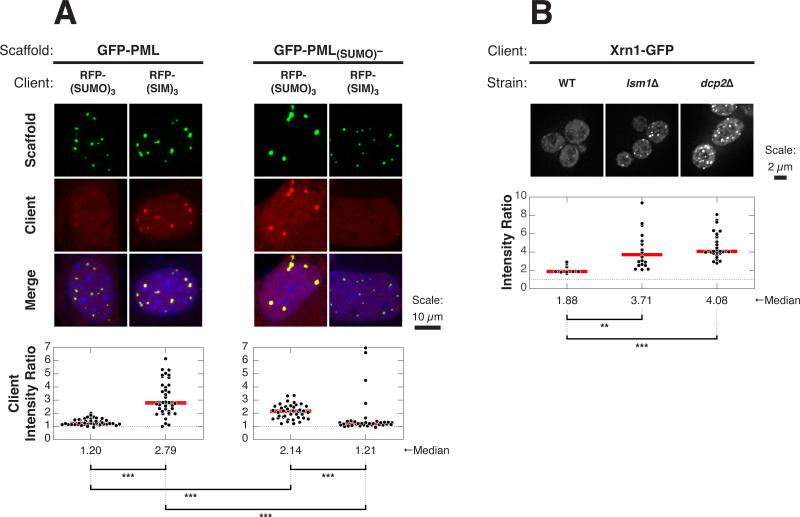
We initially examined partitioning of GFP-tagged SUMO/SIM clients into endogenous PML NBs in U2OS cells (Figure S7A). Immunofluorescence imaging using an antibody against PML revealed numerous PML NBs in nearly all cell nuclei. For each client, we measured the ratio of GFP fluorescence intensity within the PML NBs to that in the bulk nucleoplasm (Intensity Ratio, IR = IntensityPML_NB/Intensitynucleoplasm, see Supplemental Information). GFP-SIM was enriched in these bodies with a median IR of 2.9. In contrast, as previously reported for monovalent SUMO clients (Ayaydin and Dasso, 2004), GFP-SUMO had little enrichment in the PML NBs (median IR = 1.3). Increasing valency increased the enrichment of the preferred client into PML NBs (median IR = 8.1 for GFP-(SIM)3) but had no effect on the impartial client (median IR = 1.4 for GFP-(SUMO)3). Neither GFP alone nor a client with mutated SIM sites were enriched (Figure S7B).
The selective, valency-dependent partitioning into PML NBs is analogous to the behaviors of our polySUMO/polySIM model system on the polySUMO-enriched side of the phase diagram diagonal (i.e., to that of (SUMO)10-(SIM)5, above the diagonal). Our model predicts that PML NBs on the opposite side of the phase diagram diagonal should exhibit inverted partitioning behavior with respect to SUMO versus SIM clients. To create such structures, we expressed either wild-type (WT) GFP-PML or a PML mutant (GFP-PML(SUMO)–) lacking some of the known SUMOylation sites in PML−/− mouse embryonic fibroblasts (MEFs) (Figure 6A). The mutant protein is SUMOylated in cells, but to a lesser degree than the wild-type protein (Figure S7F). Both the WT and mutant scaffolds formed micron-sized puncta in nuclei. Like natural PML NBs, GFP-PML puncta substantially recruited RFP-(SIM)3 (median IR = 2.8), but not RFP-(SUMO)3 (median IR = 1.2). In reciprocal fashion, GFP-PML(SUMO)– puncta efficiently recruited RFP-(SUMO)3 (median IR = 2.1) but recruited RFP-(SIM)3 poorly (median IR = 1.2). Neither scaffold could recruit RFP alone nor clients with mutations at the SUMO-or SIM-binding site (Figures S7C and S7D). Moreover, these results were independent of the SUMO paralog (SUMO1 versus SUMO3) used to construct the client (Figure S7E).
Figure 6. Client Recruitment into Natural Cellular Bodies Is Affected by Scaffold Stoichiometries.
(A) Images of RFP-SUMO or RFP-SIM (red) co-transfected with GFP-PML or GFP-PML(SUMO)– (green) into PML−/− MEFs (top); nuclear staining with Hoecsht 33342 (blue). Plots (bottom) show IRs from individual cells (black dots) and median values (red horizontal lines). Each symbol represents the average IR (see Experimental Procedures) for all puncta in a given cell. 32–44 cells were analyzed per sample, each on average containing 16 or 5 puncta per cell with GFP-PML or GFP-PML(SUMO)–, respectively. Distributions were statistically compared using the Wilcoxon rank sum test followed by the Bonferonni correction for multiple comparisons to determine significance. ***p < 0.001. Dotted line, IR = 1.
(B) Representative images of WT, lsm1Δ, or dcp2Δ yeast strains carrying Xrn1-GFP (green) in their genomes (top). Distributions of Xrn1-GFP IRs (bottom), where each symbol represents IR corresponding to an individual P body. 1–3 P bodies per cell were analyzed from a set of 4–10 cells per sample. Analysis for significance was performed as in (A). **p < 0.01.
See also Figure S7.
These data suggest that decreasing the degree of PML SUMOylation can shift the bodies to a region analogous to the opposite side of the SUMO/SIM diagonal, where recruitment of SUMO-containing clients is favored.
We next explored analogous compositional control in P bodies, protein- and mRNA-rich cellular bodies in the cytoplasm of eukaryotes that promote mRNA decay (Parker, 2012). P bodies assemble through multivalent interactions of RNA binding proteins composed of modular RNA binding domains and self-associating disordered regions and mRNA molecules (Decker and Parker, 2012). mRNAs that have exited translation act as important P body scaffolds (Teixeira et al., 2005). We thus asked whether modulating the levels of mRNA, thereby affecting the relative stoichiometry of an important scaffold component, could affect recruitment of clients into P bodies. We used the lsm1Δ and dcp2Δ yeast strains, which are deficient in mRNA decapping and therefore accumulate deadenylated mRNAs that would otherwise be targets for degradation (Parker, 2012). We then measured the IR of the P body client Xrn1 (Jain and Parker, 2013) fused to GFP (Xrn1-GFP), in the WT, lsm1Δ, or dcp2Δ strains. Xrn1 is predicted to be recruited, at least in part, by interactions with RNA. Compared to its recruitment into P bodies in WT cells (median IR = 1.88), recruitment in the lsm1Δ and dcp2Δ strains increased ~2-fold (median IR = 3.71 and 3.71 and 4.08, respectively) (Figure 6B). This behavior was qualitatively consistent with the behaviors of our engineered clients (Figure 1). The recruitment of the P body scaffold Edc3 (Jain and Parker, 2013) also increased in the two deletion strains concomitant with the increase in deadenylated mRNAs, consistent with the increased partitioning of scaffolds observed in certain regimes of the phase diagram when the concentration of the cognate scaffold increased (Figure 3B). Thus, these data suggest that, like PML NBs, compositional control can be achieved in P bodies by modulation of scaffold stoichiometries.
Collectively, these data indicate that the compositions of both of these natural cellular bodies can be controlled by modulation of stoichiometries of scaffold elements, analogous to the behaviors observed in our model systems. This behavior suggests simple cellular mechanisms to rapidly and dramatically alter the composition, and thus function, of cellular bodies in response to stimuli.
DISCUSSION
Hierarchical Organization of Cellular Bodies
We propose a hierarchical model for the composition of cellular bodies (Figure 7). The model has several key features. First, scaffolds self-associate by multivalent heterotypic interactions and undergo assembly-driven phase separation (Li et al., 2012), forming a condensed phase (Figure 7A)—i.e., the cellular body. Second, clients partition into bodies by interacting with scaffolds, often utilizing the same types of interacting elements as those between scaffolds (Figure 7B). The typically lower valency (and therefore lower apparent affinity) of clients minimizes their competition with the higher affinity scaffold-scaffold interactions. As a result, clients are recruited by binding only to excess, or free, scaffold sites. Thus, their recruitment will be governed by the stoichiometric ratios of the scaffolds (Figure 7C). Third, since the enrichment of excess sites switches sharply across the phase diagram diagonal from one class of sites to the other, bodies can change compositions in a switch-like manner as a function of phase diagram position. Fourth, since scaffold and client valencies can affect position on the phase diagram and the degree of client partitioning, respectively, covalent modifications that change valency can be used to rapidly switch between compositional states (Figure 7D).
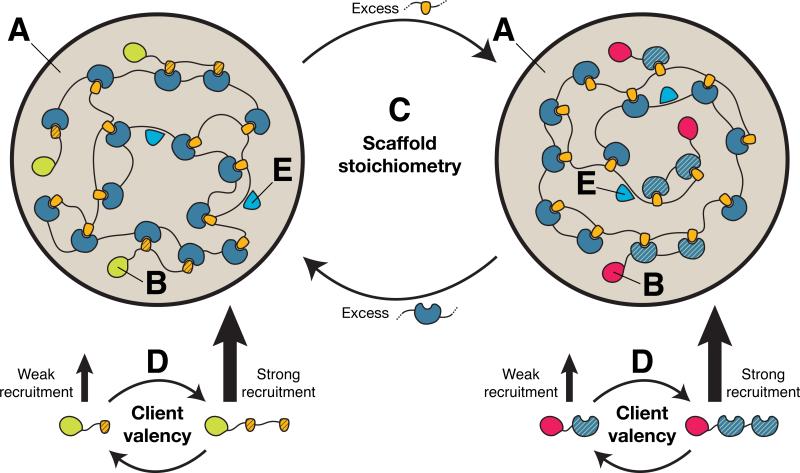
Figure 7. A Model for Compositional Control of Cellular Bodies.
Multivalent scaffold molecules (high valency blue and yellow molecules) assemble and phase separate to form the body (A). Many client molecules (low valency blue and yellow molecules, with additional domains) are enriched in the body through binding to free cognate sites in the scaffold that is in stoichio-metric excess (B). Client modules have a hatched pattern to distinguish them from scaffold modules. Stoichiometric excess of the scaffold modules can be changed either through changes in the scaffold concentrations (C) or through changes in the scaffold valency (not shown). Since stoichio-metric excess of the scaffolds in droplet (A) and bulk (not shown) changes sharply across the phase diagramdiagonal, the nature ofthe clients alsoswitches sharply across the diagonal. Higher valency promotes stronger recruitment of the clients (D). Molecules that bind to other regions of the scaffolds (E, light blue trianguloids) will be recruited independently of the scaffold stoichiometry. Natural bodies are composed of more complicated molecules, with multiple types of interaction elements, but should follow this same logic.
We note that clients that bind regions of the scaffold that are not involved in heterotypic assembly will be recruited but remain relatively insensitive to changes in scaffold stoichiometries (Figure 7E). Moreover, molecules with otherwise appropriate physicochemical properties (e.g., complementary charge) may also partition into droplets due to non-specific interactions (Li et al., 2012). Clients containing multiple types of interaction elements, some mirroring scaffold-scaffold interactions and others not, could show complex behaviors that are essentially superpositions of these individual effects. This reasoning may explain the recruitment of Xrn1-GFP into P bodies without perturbation of mRNA content (i.e., on what may be the non-cognate side of the phase diagram diagonal), as well as the enhanced recruitment when cellular mRNA is increased, as observed in Figure 6B.
Complexities of Natural Cellular Bodies
Although natural cellular bodies are appreciably more complicated than our engineered model systems, their compositions may still be understood with simple extensions to the framework we present here. First, cellular bodies may have multiple scaffolds held together by different types of multivalent interactions. For example, RNA granules likely have multiple scaffolds with contributions from both low-complexity sequence elements, as well as RNA and RNA-binding domains. PML NBs likely assemble by a combination of TRIM and SUMO-SIM interactions. Multiple types of scaffolds and scaffold interactions may cooperate to synergistically promote polymerization and phase separation, as suggested previously (Lin et al., 2015). Moreover, clients may also possess multiple classes of low valency elements that can each interact with scaffolds. Nevertheless, in the absence of cooperativity, one can think of the different interaction motifs independently. For any given class, the corresponding free sites in a scaffold will dictate partitioning of clients that can bind to those sites. Indeed, perturbing one type of interaction motif within PML NBs or P bodies had strong effects on the recruitment of clients that bind to that motif (Figure 6).
Second, for some systems, the distinction between scaffolds and clients may be less stark than in our engineered systems (see also Supplemental Information). As client valency approaches that of the scaffolds, this distinction breaks down, and the client begins to compete with scaffold-scaffold interactions. For such clients, the change in partitioning across the diagonal is likely to be less sharp, as we observe for scaffolds (Figures 3A and 3B). Further investigations are needed to understand the distribution of scaffolds, clients, and such intermediate molecules in the various known natural bodies.
Finally, several cellular bodies contain subcompartments (condensed phases within the primary condensed phase) and thus are not simple droplet/bulk systems (Brangwynne et al., 2011; Feric et al., 2016; Jain et al., 2016; Wang et al., 2014). Each subcompartment can have a unique composition organized by a distinct set of scaffold molecules. A client may bind to free sites in any or all of the body's subcompartments. Partition coefficients between any two sub-compartments or between a sub-compartment and the surrounding bulk will still result from mass action driven by the corresponding free site ratios.
Thus, despite the inherent complexities of natural cellular bodies, they may still be understood through the lens of our simple model.
Biological Mechanisms of Regulating Body Composition
Biological processes could regulate the composition of cellular bodies by acting on either scaffolds or clients on time-scales ranging from physiologic to evolutionary. On the most rapid timescales (seconds to minutes), covalent modifications could change valencies of the scaffold components, shifting the position of the system within the phase diagram. They could also change valencies and affinities of the clients, influencing their degree of partitioning, as suggested here (Figure 2), and previously (Han et al., 2012; Kwon et al., 2013). On slower timescales (hours to days), the scaffold concentrations could change via regulation of expression levels, or their valencies could be changed by alternative splicing. On evolutionary timescales, changes in gene sequences could change the affinity of scaffold components for each other or for clients, shifting composition and function in a more permanent sense.
Some of these processes can be observed in PML NBs. For example, the SUMOylation of PML is substantially decreased during mitosis concomitant with loss of some SIM-containing clients (Dellaire et al., 2006; Everett et al., 1999), and phosphorylation of the SIM in PML increases its affinity for SUMO (Cappadocia et al., 2015). Similarly, phosphorylation of the SIMs of PML NB clients, including Daxx (Chang et al., 2011), increases their interactions with the bodies.
Changes in Body Composition May Dictate Changes in Function
Unlike macromolecular machines, cellular bodies continuously rearrange the bonding interactions and organization of their constituent parts and thus are not stereochemically defined across their lengths. Their functions, therefore, cannot be controlled by allosteric transitions between conformational states, as often occurs with macromolecules. Instead, transitions between compositional states are likely to be key determinants of body function. The differential partitioning of molecules in different regions of the phase diagram implies that it may be most appropriate to consider a given type of cellular body as a distribution of entities (likely defined by a limited number of scaffolds) that lie on a continuum of compositions subject to cellular control. This idea was suggested previously for RNA-based bodies based on the related compositions of P bodies, stress granules, and RNA transport granules (Buchan and Parker, 2009). Similarly, in the case of PML NBs, a variety of structures in different cell types and cell states have been characterized, unified by their enrichment of the PML protein but varying in their composition of other components (Dellaire et al., 2006; Luciani et al., 2006). Our data suggest that this behavior may be generally applicable to many cellular bodies.
Since function is dictated by composition, this reasoning implies that cellular bodies may exhibit a continuum of functions, rather than a limited set of discrete functions as seen for macro-molecular machines in different conformations. Even though cellular body function may be more continuous than discrete, our data suggest that mechanisms could exist, as they do in canonical macromolecular machines, to mediate sharp switches between different functional regimes. Moreover, we and others (Molliex et al., 2015; Patel et al., 2015; Ramaswami et al., 2013; Weber and Brangwynne, 2012) speculate that pathological states of cellular bodies may also lie on the same compositional and functional continuum. As such, manipulation or depletion of certain scaffolds may be a promising approach to mitigate the toxicities associated with these pathological granules. Indeed, in models of ALS, toxicities due to TDP-43 aggregation can be alleviated by removal of the Ataxin-2 scaffold (Elden et al., 2010).
Implications for Cellular Body Function
Precise control of client partitioning could mediate colocalization of reaction partners to accelerate reaction rates and increase reaction specificity. For example, polySUMOylation of cellular substrates was recently demonstrated to activate the ubiquitin E3 ligase RNF4, a process that could be driven or enhanced by such compartmentalization (Rojas-Fernandez et al., 2014). Similarly, metabolic flux could be controlled by colocalization-mediated substrate channeling (Srere, 1987) or the colocalization of a branch point enzyme and downstream molecules in a pathway (Castellana et al., 2014). Compositional control may also help regulate en masse reactions such as SUMOylation of many cellular factors at PML NBs, which, analogous to DNA repair foci (Psakhye and Jentsch, 2012), colocalize not only enzymes of the SUMOylation cascade but also several SUMOylation substrates (Van Damme et al., 2010). Partitioning into a cellular body could also serve to sequester components away from their cellular targets, as has been proposed in the regulation of Daxx (Lallemand-Breitenbach and de Thé, 2010) and the priming of RNA Polymerase II prior to transcription initiation (Kwon et al., 2013). Indeed, strong depletion of clients from bulk solution through dramatically high PC is consistent with behaviors we observe in our mass action model (Figures 3F, S3E, and S3F and Supplemental Information).
Conclusion
We demonstrate how cellular body assembly, when driven by heterotypic polymerization and concomitant phase separation, naturally leads to a simple and predictive model for compositional control of these structures. Our model suggests how bodies could be switched sharply between distinct compositional (and thus functional) states on a range of biological time-scales. Moreover, it suggests that superficially similar cellular bodies composed of a given set of scaffolds may be markedly different in their composition and function, depending on the relative scaffold stoichiometries. Thus, a complete understanding of cellular bodies may require knowing relative scaffold amounts in addition to scaffold identities.
Our studies thus provide a mechanistic framework for studying the biochemical and regulatory function of cellular bodies owing to properties not attributable to any individual molecule but rather to those intrinsic to the macroscopic structure itself.
EXPERIMENTAL PROCEDURES
Genes, RNA, and Plasmids
polyPRM, polySH3, PTB, and the polyUCUCU RNA were described previously (Li et al., 2012). The RNA client UCUCU-AF647 (5′-UCUCUAAAAA-3′; 3′-labeled with AF647), as well as (SUMO)5 and (SIM)5 as synthetic genes, were purchased from Integrated DNA Technologies. Decavalent, fused, and low valency SUMO/SIM constructs were constructed from (SUMO)5 and (SIM)5 by PCR. To prevent conjugation and proteolysis, we mutated the C-terminal di-glycine motif in all SUMO proteins (see Supplemental Information). The RRM client was constructed from the first RRM domain of PTB. The mCherry, mEGFP, mVenus, and mCerulean (referred to as RFP, GFP, YFP, and CFP, respectively) fusion proteins were produced by cloning into corresponding vectors (Clontech). Sequences of molecules used in this study are listed in Table S4.
Protein Expression, Purification, and Labeling
All purified proteins were expressed and purified similarly (see Supplemental Information). Proteins were expressed in E. coli strain BL21 DE3T1R by induction with 1 mM IPTG. Proteins were purified with Ni-NTA Agarose Resin (QIAGEN) or Amylose Resin (NEB), followed by ion exchange (Source 15Q and/or Source 15S [GE Healthcare]) and size exclusion chromatographies using a Superdex 200 or Superdex 75 gel filtration columns (GE Healthcare). Proteins were labeled using maleimide-conjugated Alexa dyes (Life Technologies) following the manufacturer's protocol.
In Vitro Partitioning Assays
Scaffold molecules (1% Alexa-labeled) were mixed with GFP- or RFP-tagged or Alexa-labeled clients in wells of chambered cover glass (GraceBiolabs) or 384-well plates (Sigma) passivated with 30 mg/mL BSA (Sigma). Mixtures were incubated for 2 to 4 hr for SH3/PRM and PTB/ RNA experiments and 20–26 hr for SUMO/SIM experiments and imaged at 20× magnification.
Image Acquisition and Analysis
Yeast cells were imaged using DeltaVision Elite microscope at 100× magnification using a sCMOS camera. In all other experiments, imaging was performed using spinning disk confocal microscopes equipped with EMCCD cameras at 20× or 100× magnification for in vitro or cellular experiments, respectively. Images were analyzed using ImageJ or MATLAB (Mathworks) (see Supplemental Information). Fluorescence intensities were calibrated to concentrations using standard solutions of purified client molecules or corresponding fluorescent proteins, whose concentrations were independently determined. When possible, care was taken to circumvent effects of the PSF in concentration determination (see Supplemental Information).
Mass Action Model for Client Partitioning
Measured concentrations (from imaging) and affinities (from ITC, see Supplemental Information) of polySUMO and polySIM were used to calculate the free sites concentrations in droplet and bulk phases. An equilibrium mass action model was created to describe our systems as two compartments with unequal concentrations of receptors (free scaffold sites) and a permeable ligand (client). The model was numerically solved using MATLAB and predicted PCs were calculated as ratio of the total ligand concentration between the two compartments (see Supplemental Information for details).
Cellular Experiments
For engineered polySUMO/polySIM and PML NB experiments, mammalian cells (HeLa, U2OS, and PML−/− MEFs) were cultured in DMEM supplemented with 10% fetal bovine serum, 1% Penicillin-Streptomycin, and 1% GlutaMAX in 5% CO2 at 37°C. Cells were transfected using Lipofectamine 2000 (Life Technologies) and imaged 18–24 hr after transfection. For P body experiments, WT and mutant yeast strains cells carrying Xrn1-GFP in the genome were grown to log phase at 30°C in yeast minimal media supplemented with a complete set of amino acids and 2% dextrose.
Supplementary Material
Highlights.
Cellular bodies are organized by scaffolds and recruit clients
Clients bind to free sites in the scaffolds, and binding scales with client valency
Relative scaffold stoichiometries control client recruitment in switch-like fashion
Cells can control these parameters and thus regulate cellular body composition
ACKNOWLEDGMENTS
We thank Louie Kerr, Kate Luby-Phelps, and Abhijit Bugde for assistance with imaging; Chad Brautigam and Thomas Scheuermann for assistance with ITC; Mark Kittisopikul for helpful discussions regarding image analysis, numerical fitting, and statistical testing; Pier Paolo Scaglioni for providing PML−/− cells; Rama Ranganathan for critical reading of the manuscript; and members of the Rosen lab for helpful discussions. This work was supported by the Howard Hughes Medical Institute, the HCIA program of HHMI, grants from the NIH (R01-GM56322) and Welch Foundation (I–1544), a Sara and Frank McKnight Graduate Fellowship (to S.F.B.), and the NSF (1000196079; to A.M.R.).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, S.F.B. and M.K.R.; methodology, S.F.B. and M.K.R.; investigation, S.F.B., A.R., W.P., Y.L., S.J.; writing, S.F.B., A.R., R.P., M.K.R.; supervision, R.P. and M.K.R.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.06.010.
REFERENCES
- Antolini F, Lo Bello M, Sette M. Purified promyelocytic leukemia coiled-coil aggregates as a tetramer displaying low α-helical content. Protein Expr. Purif. 2003;29:94–102. doi: 10.1016/s1046-5928(03)00004-4. [DOI] [PubMed] [Google Scholar]
- Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, Rosen MK. Conserved interdomain linker promotes phase separation of the multi-valent adaptor protein Nck. Proc. Natl. Acad. Sci. USA. 2015;112:E6426–E6435. doi: 10.1073/pnas.1508778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, Meluh PB, Pandolfi PP, Zon LI. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell. 2002;10:843–855. doi: 10.1016/s1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- Brand P, Lenser T, Hemmerich P. Assembly dynamics of PML nuclear bodies in living cells. PMC Biophys. 2010;3:3. doi: 10.1186/1757-5036-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Mascle XH, Bourdeau V, Tremblay-Belzile S, Chaker-Margot M, Lussier-Price M, Wada J, Sakaguchi K, Aubry M, Ferbeyre G, Omichinski JG. Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1. Structure. 2015;23:126–138. doi: 10.1016/j.str.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Castellana M, Wilson MZ, Xu Y, Joshi P, Cristea IM, Rabinowitz JD, Gitai Z, Wingreen NS. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 2014;32:1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupníková K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Naik MT, Huang Y-S, Jeng J-C, Liao P-H, Kuo H-Y, Ho C-C, Hsieh Y-L, Lin C-H, Huang N-J, et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell. 2011;42:62–74. doi: 10.1016/j.molcel.2011.02.022. [DOI] [PubMed] [Google Scholar]
- Chen YC, Kappel C, Beaudouin J, Eils R, Spector DL. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol. Biol. Cell. 2008;19:3147–3162. doi: 10.1091/mbc.E08-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Leonhardt H, Rippe K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. 2011;124:3603–3618. doi: 10.1242/jcs.084681. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear non-coding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J. Cell Sci. 2006;119:1034–1042. doi: 10.1242/jcs.02817. [DOI] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J. Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CCH, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 1999;112:4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J. Cell Biol. 2013;203:149–164. doi: 10.1083/jcb.201303145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm SA, Kamenz J, Nöldeke ER, Neu A, Zocher G, Sprangers R. In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew. Chem. Int. Ed. Engl. 2014;53:7354–7359. doi: 10.1002/anie.201402885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frýdlová I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcová I, Nováková L, Janosková D, et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hanazawa M, Yonetani M, Sugimoto A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 2011;192:929–937. doi: 10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell. 2005;16:202–211. doi: 10.1091/mbc.E04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N, Shen L, Jaffray EG, Hay RT. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol. Biol. Cell. 2011;22:78–90. doi: 10.1091/mbc.E10-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Riceberg J, Pulukuri SM, Grossman S, Shinde V, Shah P, Brownell JE, Dick L, Newcomb J, Bence N. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS ONE. 2015;10:e0123882. doi: 10.1371/journal.pone.0123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Naik MT, Chang CF, Fang PJ, Wang YH, Shih HM, Huang TH. The B-box 1 dimer of human promyelocytic leukemia protein. J. Biomol. NMR. 2014;60:275–281. doi: 10.1007/s10858-014-9869-4. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Parker R. The discovery and analysis of P Bodies. Adv. Exp. Med. Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnu-clear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D-Y, Huang Y-S, Jeng J-C, Kuo H-Y, Chang C-C, Chao T-T, Ho C-C, Chen Y-C, Lin T-P, Fang H-I, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and maturation of phase separated liquid droplets by RNA binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ, Megarbane A, Sarda P, Sirma H, Moncla A, et al. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 2006;119:2518–2531. doi: 10.1242/jcs.02965. [DOI] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamad N, Bodén M. The proteins of intra-nuclear bodies: a data-driven analysis of sequence, interaction and expression. BMC Syst. Biol. 2010;4:44. doi: 10.1186/1752-0509-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential Roles of PML Isoforms. Front. Oncol. 2013;3:125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Fernandez A, Plechanovová A, Hattersley N, Jaffray E, Tatham MH, Hay RT. SUMO chain-induced dimerization activates RNF4. Mol. Cell. 2014;53:880–892. doi: 10.1016/j.molcel.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad N, de Thé H, Lallemand-Breiten-bach V. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014;204:931–945. doi: 10.1083/jcb.201305148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin H-K, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere PA. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 2010;6:51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lamb-rus BG, Calidas D, Betzig E, Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.